Abstract
Hydrophobins are highly surface-active proteins that have versatile potential as agents for interface engineering. Due to the large and growing number of unique hydrophobin sequences identified, there is growing potential to engineer variants for particular applications using protein engineering and other approaches. Recent applications and advancements in hydrophobin technologies and production strategies are reviewed. The application space of hydrophobins is large and growing, including hydrophobic drug solubilization and delivery, protein purification tags, tools for protein and cell immobilization, antimicrobial coatings, biosensors, biomineralization templates and emulsifying agents. While there is significant promise for their use in a wide range of applications, developing new production strategies is a key need to improve on low recombinant yields to enable their use in broader applications; further optimization of expression systems and yields remains a challenge in order to use designed hydrophobin in commercial applications.
Introduction
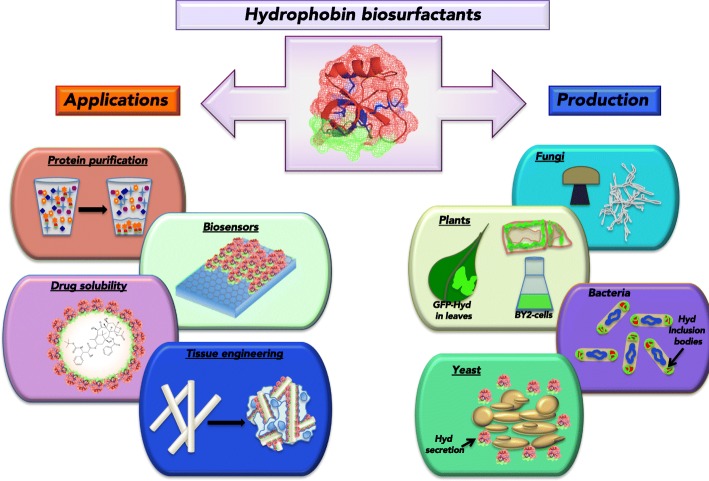
Hydrophobins are a family of small (< 20 kDa), highly surface-active globular proteins that play diverse roles in filamentous fungi growth and development [1–3]; they have been cited as the most surface-active proteins known [3]. Structurally, hydrophobins are characterized by the presence of 8 highly conserved cysteine residues in a specific primary sequence pattern, forming 4 disulfide bonds [4–9]. These 4 disulfide bonds stabilize an amphipathic tertiary structure which imparts surfactant-like activity [5, 6, 10, 11], driving hydrophobin self-assembly into amphipathic layers at hydrophobic-hydrophilic interfaces. Hydrophobins have historically been separated into two groups, class I and class II, based on their hydropathy plots, solubility characteristics, and structures formed during self-assembly [12, 13]. Specifically, class I hydrophobins, like SC3 from Schizophyllum commune, form highly insoluble amyloid-like rodlets at interfaces [2, 8, 14, 15], often proceeding through a conformational change [14–16], that can only be dissolved using strong acids [17, 18]. In contrast, class II hydrophobins, like HFBI or HFBII from Trichoderma reesei, form a highly ordered 2D crystalline monolayer at interfaces [19–21] that can easily be dissolved with detergents, organic solvent solutions, or high pressure [3]. Interestingly, the structural and functional roles of the conserved disulfide bonds differ between the two classes, with disulfides of class I hydrophobin SC3 being necessary to keep the protein soluble and structurally stable, but not affecting the self-assembling ability [10], while class II hydrophobin HFBI disulfides are critical to both protein structure and stability as well as function at interfaces [11]. Recently, intermediate hydrophobin types have been discovered that are between class I and class II either structurally [22, 23] or functionally [24, 25].To this end, Lo et al showed that by producing genetic chimeras of class I hydrophobins EAS and DewA with class II hydrophobin NC2, properties of both classes of hydrophobins could be obtained [21]. The high sequence diversity within the hydrophobin family suggests multiple biological roles of these biosurfactants [18, 26, 27], with complementation studies suggesting, at least for class I hydrophobins, that each was evolved to function at a specific interface [3], which is also demonstrated by the differential expression and localization of different hydrophobins during S. commune development [28]. Recently, Pham et al determined that hydrophobins have a significant level of conformational plasticity, with the nature of the interfacial assemblies being highly dependent on the specific interface the proteins are interacting with [29]. Lienemann et al found that by engineering native surface charges on hydrophobin HFBI, viscoelastic properties of the assembled film at the air-water interface and ability to absorb secondary protein layers were affected [30]. Additionally, Meister et al showed that mutating the surface charges of HFBI does not affect overall protein folding state, but specific charge mutations could be linked to inter-protein interactions at the assembled film, while other mutations were linked to protein orientation at the interface [31]. Meister et al also reported that HFBI adsorbed to the air-water interface reoriented in a pH responsive way due to changes in inter-protein interactions caused by side-chain charge states [32]. Overall, these findings indicate a significant potential to use hydrophobins both directly and in modified forms for many interface-engineering applications, which will be the primary focus of this review. For other areas of active hydrophobin research such as foam stabilization and gushing, the reader is referred to the following reviews [33, 34]. Additionally, the current hydrophobin production modalities and pitfalls will be discussed (Fig. 1).
Fig. 1.
Visual summary of current hydrophobin applications and production systems
Applications
The vast diversity among known hydrophobins, the specificity of particular hydrophobins in their roles in fungal development, and their unique structures and surface activity implicate hydrophobins as advantageous agents in many applications where interfaces need to be altered, bridged, or stabilized.
In biomedical applications, hydrophobins have been particularly useful for hydrophobic drug formulation and delivery. While hydrophobic drugs are often formulated using surfactants such as Tween 80 or Chremophore EL to improve their solubility in an aqueous environment, these surfactants are not innocuous, and have been shown to be immunogenic in immunocompromised patients, such as cancer patients [35]. Aimanianda et al showed that hydrophobins forming the hydrophobic rodlet layer of airborne fungal spores are responsible for the immunological silencing that occurs when a host breathes the spores [36], which suggests that hydrophobins have the opposite effect of industrial surfactants on the immune system, and may act as an immune-suppressive barrier in drug formulations.
Given their high surface activity, hydrophobin-based drug stabilization has been an area of active research [37–42]. Valo et al demonstrated the preparation of class II hydrophobin-coated drug nanoparticles below 200 nm that were stable for at least 5 h in suspension, and for longer times after freeze-drying [37]. They also utilized a hydrophobin fused to green fluorescent protein (GFP) to demonstrate that the particles were indeed decorated with the proteins, and suggested that hydrophobin fusions could be used to further modify the particle surfaces [37]. Hydrophobin HFBI produced as a genetic fusion to cellulose binding domains allowed a cellulose-based nanofibrillar matrix stabilization of hydrophobin stabilized drug particles of around 100 nm, capable of over 10 months storage and enhanced drug dissolution rates [38]. Sarparanta et al. showed that functionalizing thermally hydrocarbonized porous silicon nanoparticles with hydrophobin HFBII altered the biodistribution compared to unfunctionalized particles, as well as altered the protein adsorption profile to the particle surface [39]. Fang et al utilized a commercially available surfactant blend containing class I hydrophobin, H star protein B [43], to solubilize the chemotherapy drug docetaxel [41]. They showed that the formulation was biocompatible and exhibited a high drug loading, high nanoparticle yield, small particles of narrow distribution, and delayed drug release in rats [41]. Moreover, the effective stabilization of model drug oil-in-water emulsions using low concentrations of HFBII with nanofibrillar cellulose suggests an additional advantage of formulation with hydrophobins since less material is needed compared to traditional pharmaceutical surfactant-based emulsion stabilizers [42]. When the class I hydrophobin SC3 was used to solubilize the hydrophobic drugs cyclosporine A and nifedipine, the oral bioavailability was increased by 2- and 6-fold, respectively [44]. Hydrophobins have also been explored, with positive results, as a topical drug formulation agent for nail permeation [45, 46]. Thus, several drug-formulations and administration modalities implicate hydrophobins as effective adjuvants for improved hydrophobic drug solubility, stability, and bioavailability. Furthermore, by using a protein-based biosurfactant capable of manipulation at the genetic level, hydrophobin fusion proteins have also been employed for specific drug targeting. Recently, Reuter et al demonstrated that coating porous silicon nanoproteins with a fusion of T. reesei class II hydrophobins to human transferrin protein resulted in their uptake in cancer cells [47]. Also, the stabilizing effect of the highly conserved disulfide bonds in class II hydrophobins has been exploited as a drug-release mechanism [48]. Maiolo et al used the class II hydrophobin HFBII to organize and stabilize supraparticles of dodecanethiol-protected gold nanoparticles that could be loaded with hydrophobic drug and remain stable in the blood until taken up by tissues, where cytoplasmic glutathione would reduce the disulfides allowing the supraparticles to release the drug load directly in the cytoplasm [48]. This resulted in a two orders of magnitude enhancement of the anticancer drug therapeutic efficiency [48]. Overall, these studies show the feasibility of hydrophobin-based drug formulation and point to a need to continue to understand hydrophobin structure and function as a means to engineer novel hydrophobins for biocompatible coatings that improve both drug bioavailability and targeting.
The self-assembly characteristics of hydrophobins renders them conducive to biosensor applications as well. Corvis et al used class I hydrophobin coating from S. commune to render glassy carbon electrodes catalytic by immobilizing redox enzymes to the hydrophobin layer [49]. Also, Zhao et al utilized class II hydrophobin HFBI as an enzyme immobilization matrix on platinum electrodes to create a selective and efficient glucose biosensor [50]. Later, HFBI was used to alter the surface wettability of a gold surface and immobilize the enzyme choline oxidase [51]. They found that the HFBI layer could withstand pH values from 1 to 13, and was able to behave as an amperometric choline biosensor, further suggesting the potential of hydrophobins in electrochemical biosensing applications [51]. After 7 weeks of storage, the sensor retained > 70% of its initial activity, suggesting the stability of the protein film [51]. More recently, Spadavecchia et al reported that by using gold nanoparticles complexed with a class I hydrophobin Vmh2, which has a natural propensity to bind carbohydrates, a glucose biosensor could be generated using a one-pot synthesis approach [52]. This introduces the idea of using specific hydrophobins with unique and intrinsic biological properties on an application-specific basis. Similarly, a class I hydrophobin-based biosensor for small peptides, specifically yeast pheromones, was reported that enabled an extremely low limit of detection by using combinations of alpha-factor labeled and unlabeled hydrophobins [53]. Recombinant class I EAS hydrophobin was expressed in E. coli with and without the yeast alpha factor, and used to wet a hydrophobic polystyrene surface [53]. The researchers found these biosensors were robust against changes in the sample composition, and due to the high stability of the hydrophobin monolayer, as it was able to withstand hot 2% sodium dodecyl sulfate (SDS) extraction from the polystyrene surface, they could be reused several times without loss of sensitivity [53]. Soikkeli et al designed class II hydrophobin HFBI fusion proteins fused to Protein A or a small peptide ZE produced either in plant or fungal systems to create graphene biosensors that are label free and have femptomolar sensitivities with approximately 1 s readout [54]. The biosensors could be prepared in one-step due to the self-assembling nature of the hydrophobin domain in the fusion proteins, and demonstrated that the receptor modules could be removed and replaced with a different receptor module in situ [54]. Further, they showed that the monolayers survive drying, indicating a reasonable shelf life, and showed that both large and small analytes (immunoglobin and charged peptide) are compatible with this system [54]. In yet another interesting sensor-related application, genetically modified hydrophobin HFBI with an N-terminal cysteine residue were used to selectively nanopattern gold-nanoparticles onto a hydrophobic surface in a pH controlled manner [55]. This allowed production of nanoscale components with a functional electronic interface [55]. The hydrophobin HFBI was also used in a method to exfoliate and functionalize graphene sheets [56].
The surface activity and self-assembly of hydrophobins suggest a broad and growing potential application space. Some additional applications include hydrophobins used as protein purification tags [57–60], protein and cell immobilization [61–65], antimicrobial coatings [66], and biomineralization [67, 68]. Linder et al first demonstrated that class II hydrophobins from T. reesei could be efficiently separated in aqueous two phase systems (ATPS) using nonionic surfactants from crude fungal culture supernatants, and then efficiently back extracted using isobutanol with a partition coefficient over 2500 for HFBI [57]. Joensuu and colleagues later utilized this separation technology to purify Green Fluorescent Protein (GFP)-HFBI fusion expressed in Nicotiana benthamiana leaves, and reported enhanced accumulation of GFP in the leaves due to formation of novel protein bodies, as well as a 91% selective recovery of the GFP-HFBI fusion at concentrations of 10 mg ml− 1 after ATPS separation [58]. Reuter et al explored other class II hydrophobin fusion partners to GFP in the same system and found that efficiency of separation was highly hydrophobin dependent, with only two of the eight new hydrophobins efficiently concentrating GFP to the surfactant phase from plant extracts [59], which suggests specific molecular determinants of separation efficiency. Hydrophobin ATPS separation has also been used to indirectly capture proteins with affinity for the hydrophobin fusion partner. Recently, an HFBI fusion to Protein A, an antibody binding protein, was produced recombinantly in both N. benthamiana leaves and tobacco BY-2 suspension cells, then utilized in a nonionic surfactant ATPS to efficiently bind and purify antibodies in solution [60].
Hydrophobins have also been successfully applied to reversing the wettability of poly(dimethylsiloxane) (PDMS), a material commonly used in microfluidic devices. Wang et al showed that PDMS could effectively be turned from hydrophobic to hydrophilic using a hydrophobin surface layer, which then rendered the surface bioactive so that antigen molecules could be patterned onto the surface layer [61]. Washing the PDMS with water did not remove the stability deposited hydrophobin films from the surface [61]. Hou and colleagues explored the class I hydrophobin HGFI on PDMS wettability, and found that it had higher stability in this application than class II hydrophobin HFBI, able to withstand washes with hot 2% SDS [62]. Furthermore, the versatility of hydrophobin HFBI as a protein-immobilization layer on both hydrophobic and hydrophilic substrates was demonstrated by Qin et al, whereby adsorbed HFBI layers on both PDMS (hydrophobic) and mica (hydrophilic) could immobilize chicken IgG for biosensing applications [63]. They noted that the modified water contact angle due to hydrophobin deposition did not change when the surfaces were stored for several days in air or water, indicating the hydrophobins were stable in this configuration [63]. Similarly, Zhang et al used hydrophobin HFBI to improve hydrophilicity and design bioactive surfaces of electrospun PCL grafts used in tissue engineering [64]. Anti-CD31 antibody could then be immobilized to the PCL surface through the HFBI layer, which promoted the attachment and retention of endothelial cells to the graft [64]. Enhanced cellularization and vascularization of PCL scaffolds was similarly accomplished using a vascular endothelial growth factor fused to class I hydrophobin HGFI produced in the yeast Pichia pastoris [69]. Additionally, Boeuf et al exploited a recombinant class I hydrophobin DewA fused to an integrin binding Arginine-Glycine-Aspartic acid motif (RGD) or laminin domain to enhance adhesion of mesenchymal stem cells, osteoblasts, fibroblasts, and chondrocytes to orthopaedic implant surfaces without affecting the propensity of the bacteria Staphylococcus aureus to adhere [65]. To generate explicitly antibacterial surface coatings, class IIa bacteriocin pediocin PA-1, an antibacterial peptide, was expressed in Saccharomyces cerevisiae fused to the class I hydrophobin HGFI and used to functionalize and greatly improve the bacterial resistance of electrospun PCL grafts [66].
In biomineralization applications, Heinonen et al engineered hydrophobin HFBII modified with a ceramophilic protein sequence to mineralize calcium carbonate [67]. The microparticles produced were uniform and exhibited amphiphilic properties that were demonstrated by preparing pickering emulsions [67]. Melcher et al used a modified class I DewA hydrophobin fusion in a biomimetic approach to enhance hydroxyapatite binding and calcium phosphate nucleation for reconstruction of eroded teeth [68].
In an additional application, Taniguchi et al have used commercially available class I hydrophobin H*protein B [43] in a ligand encapsulation process to phase transfer quantum dots from solvent to aqueous phases. They demonstrated that encapsulating quantum dots allowed for efficient phase transfer while maintaining a significant portion of emission characteristics, and allowing for additional conjugation for biological imaging applications [70]. HFBI fused to an RGD motif was recently employed as a solubilizing agent for a hydrophobic boron-dipyrromethene (BODIPY) dye, with the RGD motif on the encapsulated dye causing effective labeling of tumors in nude mice [71].
Production of hydrophobins
While hydrophobin research has ballooned over the last two decades and application space is growing in terms of impact and diversity, the commercial viability of hydrophobins has been hampered by generally low yields.
Askolin et al were able to overproduce the class II hydrophobin HFBI by homologous expression in T. reesei using a clone with 3 copies of the HFBI gene to a production level of 600 mg L-1 [72]. However, most of the hydrophobin (80%) was bound to the mycelium and required further extraction steps to obtain pure protein [72]. In trying to overproduce the class I hydrophobin SC3 via homologous expression in S. commune using multiple gene copies, Schuurs et al observed gene silencing of the endogenous and introduced SC3 genes at the transcriptional level due to gene methylation [73]. Turning to heterologous production of SC3 in T. reesei yielded approximately the same level of SC3 as the native S. commune [74]. Thus, recombinant production, using either prokaryotic or eukaryotic organisms, has been an attractive choice to try to overproduce both native type or engineered hydrophobins as a means to enhance scalability and avoid pitfalls of using the homologous host. In bacteria, however, hydrophobin production, especially for class I hydrophobin, has typically been on the order of 10 to 100 mg L− 1, but often less [75–78]. In many cases, recombinant hydrophobin production in bacteria proceeds through purification from inclusion bodies, requiring a denaturation/renaturation step to achieve the final product [8, 53, 76, 77, 79]. These denaturation and refolding steps represent added expense for large-scale hydrophobin production using these heterologous systems. On rare occasions for specific hydrophobins these pitfalls were overcome, as for the case of H star A and B proteins where advantageous fusions and expression conditions have yielded industrially feasible amounts of soluble class I hydrophobin from bacteria [43]. More recently, however, eukaryotic heterologous expressions systems have been employed to greatly increase yields of both class I and class II hydrophobins recombinantly with generally better yields than bacteria.
The methylotropic yeast Pichia pastoris has become a popular heterologous host for hydrophobin expression [11, 69, 80–86]. P. pastoris has several advantages for heterologous hydrophobin production. As a fungal host, P. pastoris is expected to share similar chaperone proteins and folding strategies as filamentous fungi. These include glycosylation and proper disulfide bond formation [87, 88], which has been shown to be critical in class II hydrophobin structure and function [11] as well as to class I hydrophobin stability [10]. In addition, recombinant proteins can be secreted into the culture medium of P. pastoris [89], which secretes very low levels of endogenous proteins, under the control of a highly inducible promoter such as the methanol induced AOXI promoter [89, 90]. This means the recombinant proteins are effectively pre-purified by being secreted [89]. In particular, Niu et al have been able to express class II hydrophobin to levels of 120 mg L− 1 in P. pastoris [81], while class I hydrophobins RodA and RodB as well as HGFI were produced to levels of between 200 and 300 mg L− 1 [83, 85]. This represents a substantial improvement to previously reported yields but could be further improved with additional optimization. Of interest is the reported increase in HGFI production in P. pastoris from shake flasks yielding 86 mg L− 1 [82] to fed-batch fermentation yielding 300 mg L− 1 [85]. By optimizing the process parameters using controlled feed rate in the fed-batch fermentation, the recombinant class I hydrophobin HGFI yield was increased over 3-fold between these studies. Of note is the possibility for hydrophobin producing P. pastoris strain optimization at the molecular level [87] to further increase yields at an intrinsic level in conjunction with extrinsically optimized growth conditions, such as growth media pH and composition, temperature, and feed rate. Molecular optimizations might be to increase strain copy number of the target gene, which has been shown to often correlate to higher product expression in P. pastoris [91].
The other emerging heterologous hosts in hydrophobin production are plant based. Transient or stable expression of GFP-HFBI fusion has been carried out in Nicotiana benthamiana leaves [58, 59, 92] and tobacco BY-2 suspension cells [93, 94] to produce high yields of hydrophobins and hydrophobin fusions. In particular, Joensuu et al reported a GFP-HFBI production level of 3.7 mg g− 1 fresh leaf weight [58], comprising approximately 51% of the total soluble protein. Häkkinen et al recently reported the yield of a high-expressing BY-2 clone as 1.1 g L− 1 of GFP-HFBI in suspension, and also reported successful cryopreservation of the cultures, enabling industrial application of this hydrophobin fusion production technology [94]. The interesting development of hydrophobin-induced protein bodies in the leaf cells were credited with the increased accumulation of the recombinant proteins, keeping them in a protected state from proteases in the cytosol [58, 60, 92].
Conclusions and future perspectives
The remarkable surface-activity of hydrophobins has made them attractive candidates in a wide variety of interface-engineering applications to date. While some very specific hydrophobins can be made at industrially feasible levels, there remains an unmet need to produce high levels of both native and engineered forms of hydrophobins before hydrophobin-based technologies can fully realize their commercial potential. Furthermore, a deeper understanding of hydrophobin structure-function relationships would inform novel hydrophobin design for specific applications, which would have tremendous implications in many important fields such as pharmaceuticals, electronics, microfluidics, and food products. To date, biochemical studies have related stability to disulfide bonds [10, 11] and film viscoelastic properties, inter-protein interaction, and pH responsive orientation to surface charge [30–32]. However uncovering other key structure-function relationships in hydrophobins could lead to design from first principles, whereby application specific characteristics could be programmed into the hydrophobin at a genetic level to enable outcomes such as increased binding, enhanced solubilization, switchable surface activity, or specific nanopatterning, although the potential is limitless. To get at this goal, further research into hydrophobin sequence, folding, and the related function needs to be undertaken in order to build a foundation for design.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Science Foundation (award #1452855).
Availability of data and materials
Not applicable.
Abbreviations
- ATP
Aqueous two phase systems
- GFP
Green fluorescent protein
- PDMS
Poly(dimethylsiloxane)
Authors’ contributions
BWB and NDS wrote, read and provided feedback and improvements to the manuscript. NDS prepared figures for the manuscript. Both authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Bryan W. Berger, Email: bryan.berger@virginia.edu
Nathanael D. Sallada, Email: ns6he@virginia.edu
References
- 1.Kershaw MJ, Talbot NJ. Hydrophobins and repellents: proteins with fundamental roles in fungal morphogenesis. Fungal Genet Biol. 1998;23:18–33. doi: 10.1006/fgbi.1997.1022. [DOI] [PubMed] [Google Scholar]
- 2.Wösten HAB, de Vocht ML. Hydrophobins, the fungal coat unravelled. Biochim Biophys Acta (BBA)-Reviews Biomembr. 2000;1469:79–86. doi: 10.1016/S0304-4157(00)00002-2. [DOI] [PubMed] [Google Scholar]
- 3.Wösten HAB. Hydrophobins: multipurpose proteins. Annu Rev Microbiol. 2001;55:625–646. doi: 10.1146/annurev.micro.55.1.625. [DOI] [PubMed] [Google Scholar]
- 4.Kallio, J., Paananen, A., Askolin, S., Nakari-Setälä, T., Parkkinen, T., Penttilä, M., Linder, M., and Rouvinen, J. (2004) Atomic resolution structure of the HFBII Hydrophobin, a self-assembling Amphiphile. [DOI] [PubMed]
- 5.Hakanpää J, Szilvay GR, Kaljunen H, Maksimainen M, Linder M, Rouvinen J. Two crystal structures of Trichoderma reesei hydrophobin HFBI—the structure of a protein amphiphile with and without detergent interaction. Protein Sci. 2006;15:2129–2140. doi: 10.1110/ps.062326706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hakanpää J, Linder M, Popov A, Schmidt A, Rouvinen J. Hydrophobin HFBII in detail: ultrahigh-resolution structure at 0.75 Å. Acta Crystallogr Sect D Biol Crystallogr. 2006;62:356–367. doi: 10.1107/S0907444906000862. [DOI] [PubMed] [Google Scholar]
- 7.Kershaw MJ, Thornton CR, Wakley GE, Talbot NJ. Four conserved intramolecular disulphide linkages are required for secretion and cell wall localization of a hydrophobin during fungal morphogenesis. Mol Microbiol. 2005;56:117–125. doi: 10.1111/j.1365-2958.2005.04547.x. [DOI] [PubMed] [Google Scholar]
- 8.Kwan AHY, Winefield RD, Sunde M, Matthews JM, Haverkamp RG, Templeton MD, Mackay JP. Structural basis for rodlet assembly in fungal hydrophobins. Proc Natl Acad Sci U S A. 2006;103:3621 LP–3623626. doi: 10.1073/pnas.0505704103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ren Q, Kwan AH, Sunde M. Solution structure and interface-driven self-assembly of NC2, a new member of the class II hydrophobin proteins. Proteins Struct Funct Bioinforma. 2014;82:990–1003. doi: 10.1002/prot.24473. [DOI] [PubMed] [Google Scholar]
- 10.de Vocht ML, Reviakine I, Wösten HAB, Brisson A, Wessels JGH, Robillard GT. Structural and functional role of the disulfide bridges in the hydrophobin SC3. J Biol Chem. 2000;275:28428–28432. doi: 10.1074/jbc.M000691200. [DOI] [PubMed] [Google Scholar]
- 11.Sallada ND, Dunn KJ, Berger BW. A structural and functional role for disulfide bonds in a class II Hydrophobin. Biochemistry. 2018;57:645–653. doi: 10.1021/acs.biochem.7b01166. [DOI] [PubMed] [Google Scholar]
- 12.Wessels JGH. Developmental regulation of fungal cell wall formation. Annu Rev Phytopathol. 1994;32:413–437. doi: 10.1146/annurev.py.32.090194.002213. [DOI] [Google Scholar]
- 13.Wessels JGH. Hydrophobins: proteins that change the nature of the fungal surface. In: Poole RK, editor. Advances in Microbial Physiology: Academic Press; 1996. p. 1–45. [DOI] [PubMed]
- 14.Butko P, Buford JP, Goodwin JS, Stroud PA, McCormick CL, Cannon GC. Spectroscopic evidence for amyloid-like interfacial self-assembly of hydrophobin Sc3. Biochem Biophys Res Commun. 2001;280:212–215. doi: 10.1006/bbrc.2000.4098. [DOI] [PubMed] [Google Scholar]
- 15.Mackay JP, Matthews JM, Winefield RD, Mackay LG, Haverkamp RG, Templeton MD. The hydrophobin EAS is largely unstructured in solution and functions by forming amyloid-like structures. Structure. 2001;9:83–91. doi: 10.1016/S0969-2126(00)00559-1. [DOI] [PubMed] [Google Scholar]
- 16.De Vocht ML, Reviakine I, Ulrich W, Bergsma-Schutter W, Wösten HAB, Vogel H, Brisson A, Wessels JGH, Robillard GT. Self-assembly of the hydrophobin SC3 proceeds via two structural intermediates. Protein Sci. 2002;11:1199–1205. doi: 10.1110/ps.4540102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Vries OMH, Fekkes MP, Wösten HAB, Wessels JGH. Insoluble hydrophobin complexes in the walls of Schizophyllum commune and other filamentous fungi. Arch Microbiol. 1993;159:330–335. doi: 10.1007/BF00290915. [DOI] [Google Scholar]
- 18.Wosten HAB, De Vries OMH, Wessels JGH. Interfacial self-assembly of a fungal hydrophobin into a hydrophobic rodlet layer. Plant Cell. 1993;5:1567–1574. doi: 10.1105/tpc.5.11.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szilvay GR, Paananen A, Laurikainen K, Vuorimaa E, Lemmetyinen H, Peltonen J, Linder MB. Self-assembled Hydrophobin protein films at the air−water Interface: structural analysis and molecular engineering. Biochemistry. 2007;46:2345–2354. doi: 10.1021/bi602358h. [DOI] [PubMed] [Google Scholar]
- 20.Yamasaki R, Takatsuji Y, Lienemann M, Asakawa H, Fukuma T, Linder M, Haruyama T. Electrochemical properties of honeycomb-like structured HFBI self-organized membranes on HOPG electrodes. Colloids Surfaces B Biointerfaces. 2014;123:803–808. doi: 10.1016/j.colsurfb.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 21.Lo VC, Ren Q, Pham CLL, Morris VK, Kwan AH, Sunde M. Fungal Hydrophobin proteins produce self-assembling protein films with diverse structure and chemical stability. Nanomaterials. 2014;4:827–843. doi: 10.3390/nano4030827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Littlejohn KA, Hooley P, Cox PW. Bioinformatics predicts diverse aspergillus hydrophobins with novel properties. Food Hydrocoll. 2012;27:503–516. doi: 10.1016/j.foodhyd.2011.08.018. [DOI] [Google Scholar]
- 23.Jensen BG, Andersen MR, Pedersen MH, Frisvad JC, Søndergaard I. Hydrophobins from aspergillus species cannot be clearly divided into two classes. BMC Res Notes. 2010;3:344. doi: 10.1186/1756-0500-3-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seidl-Seiboth V, Gruber S, Sezerman U, Schwecke T, Albayrak A, Neuhof T, von Döhren H, Baker SE, Kubicek CP. Novel hydrophobins from Trichoderma define a new hydrophobin subclass: protein properties, evolution, regulation and processing. J Mol Evol. 2011;72:339–351. doi: 10.1007/s00239-011-9438-3. [DOI] [PubMed] [Google Scholar]
- 25.Gandier J-A, Langelaan DN, Won A, O’Donnell K, Grondin JL, Spencer HL, Wong P, Tillier E, Yip C, Smith SP, Master ER. Characterization of a Basidiomycota hydrophobin reveals the structural basis for a high-similarity class I subdivision. Sci Rep. 2017;7:45863. doi: 10.1038/srep45863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linder MB, Szilvay GR, Nakari-Setälä T, Penttilä ME. Hydrophobins: the protein-amphiphiles of filamentous fungi. FEMS Microbiol Rev. 2005;29:877–896. doi: 10.1016/j.femsre.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 27.Ren Q, Kwan AH, Sunde M. Two forms and two faces, multiple states and multiple uses: properties and applications of the self-assembling fungal hydrophobins. Pept Sci. 2013;100:601–612. doi: 10.1002/bip.22259. [DOI] [PubMed] [Google Scholar]
- 28.Wessels J, De Vries O, Asgeirsdottir SA, Schuren F. Hydrophobin genes involved in formation of aerial hyphae and fruit bodies in Schizophyllum. Plant Cell. 1991;3:793–799. doi: 10.1105/tpc.3.8.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pham CLL, Rodríguez de Francisco B, Valsecchi I, Dazzoni R, Pillé A, Lo V, Ball SR, Cappai R, Wien F, Kwan AH, Guijarro JI, Sunde M. Probing structural changes during self-assembly of surface-active Hydrophobin proteins that form functional amyloids in Fungi. J Mol Biol. 2018;430:3784–3801. doi: 10.1016/j.jmb.2018.07.025. [DOI] [PubMed] [Google Scholar]
- 30.Lienemann M, Grunér MS, Paananen A, Siika-aho M, Linder MB. Charge-based engineering of Hydrophobin HFBI: effect on interfacial assembly and interactions. Biomacromolecules. 2015;16:1283–1292. doi: 10.1021/acs.biomac.5b00073. [DOI] [PubMed] [Google Scholar]
- 31.Meister K, Paananen A, Speet B, Lienemann M, Bakker HJ. Molecular structure of Hydrophobins studied with site-directed mutagenesis and vibrational sum-frequency generation spectroscopy. J Phys Chem B. 2017;121:9398–9402. doi: 10.1021/acs.jpcb.7b08865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meister K, Roeters SJ, Paananen A, Woutersen S, Versluis J, Szilvay GR, Bakker HJ. Observation of pH-induced protein reorientation at the water surface. J Phys Chem Lett. 2017;8:1772–1776. doi: 10.1021/acs.jpclett.7b00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wösten HAB, Scholtmeijer K. Applications of hydrophobins: current state and perspectives. Appl Microbiol Biotechnol. 2015;99:1587–1597. doi: 10.1007/s00253-014-6319-x. [DOI] [PubMed] [Google Scholar]
- 34.Khalesi M, Gebruers K, Derdelinckx G. Recent advances in fungal Hydrophobin towards using in industry. Protein J. 2015;34:243–255. doi: 10.1007/s10930-015-9621-2. [DOI] [PubMed] [Google Scholar]
- 35.Weiszhár Z, Czúcz J, Révész C, Rosivall L, Szebeni J, Rozsnyay Z. Complement activation by polyethoxylated pharmaceutical surfactants: Cremophor-EL, Tween-80 and Tween-20. Eur J Pharm Sci. 2012;45:492–498. doi: 10.1016/j.ejps.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 36.Aimanianda V, Bayry J, Bozza S, Kniemeyer O, Perruccio K, Elluru SR, Clavaud C, Paris S, Brakhage AA, Kaveri SV, Romani L, Latgé J-P. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature. 2009;460:1117. doi: 10.1038/nature08264. [DOI] [PubMed] [Google Scholar]
- 37.Valo HK, Laaksonen PH, Peltonen LJ, Linder MB, Hirvonen JT, Laaksonen TJ. Multifunctional Hydrophobin: toward functional coatings for drug nanoparticles. ACS Nano. 2010;4:1750–1758. doi: 10.1021/nn9017558. [DOI] [PubMed] [Google Scholar]
- 38.Valo H, Kovalainen M, Laaksonen P, Häkkinen M, Auriola S, Peltonen L, Linder M, Järvinen K, Hirvonen J, Laaksonen T. Immobilization of protein-coated drug nanoparticles in nanofibrillar cellulose matrices—enhanced stability and release. J Control Release. 2011;156:390–397. doi: 10.1016/j.jconrel.2011.07.016. [DOI] [PubMed] [Google Scholar]
- 39.Sarparanta M, Bimbo LM, Rytkönen J, Mäkilä E, Laaksonen TJ, Laaksonen P, Nyman M, Salonen J, Linder MB, Hirvonen J, Santos HA, Airaksinen AJ. Intravenous delivery of Hydrophobin-functionalized porous silicon nanoparticles: stability, plasma protein adsorption and biodistribution. Mol Pharm. 2012;9:654–663. doi: 10.1021/mp200611d. [DOI] [PubMed] [Google Scholar]
- 40.Valo H, Arola S, Laaksonen P, Torkkeli M, Peltonen L, Linder MB, Serimaa R, Kuga S, Hirvonen J, Laaksonen T. Drug release from nanoparticles embedded in four different nanofibrillar cellulose aerogels. Eur J Pharm Sci. 2013;50:69–77. doi: 10.1016/j.ejps.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 41.Fang G, Tang B, Liu Z, Gou J, Zhang Y, Xu H, Tang X. Novel hydrophobin-coated docetaxel nanoparticles for intravenous delivery: in vitro characteristics and in vivo performance. Eur J Pharm Sci. 2014;60:1–9. doi: 10.1016/j.ejps.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 42.Paukkonen H, Ukkonen A, Szilvay G, Yliperttula M, Laaksonen T. Hydrophobin-nanofibrillated cellulose stabilized emulsions for encapsulation and release of BCS class II drugs. Eur J Pharm Sci. 2017;100:238–248. doi: 10.1016/j.ejps.2017.01.029. [DOI] [PubMed] [Google Scholar]
- 43.Wohlleben W, Subkowski T, Bollschweiler C, von Vacano B, Liu Y, Schrepp W, Baus U. Recombinantly produced hydrophobins from fungal analogues as highly surface-active performance proteins. Eur Biophys J. 2010;39:457–468. doi: 10.1007/s00249-009-0430-4. [DOI] [PubMed] [Google Scholar]
- 44.Haas Jimoh Akanbi M, Post E, Meter-Arkema A, Rink R, Robillard GT, Wang X, Wösten HAB, Scholtmeijer K. Use of hydrophobins in formulation of water insoluble drugs for oral administration. Colloids Surfaces B Biointerfaces. 2010;75:526–531. doi: 10.1016/j.colsurfb.2009.09.030. [DOI] [PubMed] [Google Scholar]
- 45.Vejnovic I, Simmler L, Betz G. Investigation of different formulations for drug delivery through the nail plate. Int J Pharm. 2010;386:185–194. doi: 10.1016/j.ijpharm.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 46.Vejnovic I, Huonder C, Betz G. Permeation studies of novel terbinafine formulations containing hydrophobins through human nails in vitro. Int J Pharm. 2010;397:67–76. doi: 10.1016/j.ijpharm.2010.06.051. [DOI] [PubMed] [Google Scholar]
- 47.Reuter LJ, Shahbazi M-A, Mäkilä EM, Salonen JJ, Saberianfar R, Menassa R, Santos HA, Joensuu JJ, Ritala A. Coating nanoparticles with plant-produced transferrin–Hydrophobin fusion protein enhances their uptake in Cancer cells. Bioconjug Chem. 2017;28:1639–1648. doi: 10.1021/acs.bioconjchem.7b00075. [DOI] [PubMed] [Google Scholar]
- 48.Maiolo D, Pigliacelli C, Sánchez Moreno P, Violatto MB, Talamini L, Tirotta I, Piccirillo R, Zucchetti M, Morosi L, Frapolli R, Candiani G, Bigini P, Metrangolo P, Baldelli Bombelli F. Bioreducible Hydrophobin-stabilized Supraparticles for selective intracellular release. ACS Nano. 2017;11:9413–9423. doi: 10.1021/acsnano.7b04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corvis Y, Walcarius A, Rink R, Mrabet NT, Rogalska E. Preparing catalytic surfaces for sensing applications by immobilizing enzymes via Hydrophobin layers. Anal Chem. 2005;77:1622–1630. doi: 10.1021/ac048897w. [DOI] [PubMed] [Google Scholar]
- 50.Zhao Z-X, Qiao M-Q, Yin F, Shao B, Wu B-Y, Wang Y-Y, Wang X-S, Qin X, Li S, Yu L, Chen Q. Amperometric glucose biosensor based on self-assembly hydrophobin with high efficiency of enzyme utilization. Biosens Bioelectron. 2007;22:3021–3027. doi: 10.1016/j.bios.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 51.Zhao Z-X, Wang H-C, Qin X, Wang X-S, Qiao M-Q, Anzai J, Chen Q. Self-assembled film of hydrophobins on gold surfaces and its application to electrochemical biosensing. Colloids Surfaces B Biointerfaces. 2009;71:102–106. doi: 10.1016/j.colsurfb.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 52.Spadavecchia JP, De Stefano L, Rea I, Gravagnuolo AM, Giardina P, Methivier C, Casale S, Politi J. One-pot synthesis of a gold nanoparticle–Vmh2 hydrophobin nanobiocomplex for glucose monitoring. Nanotechnology. 2016;27:195701. doi: 10.1088/0957-4484/27/19/195701. [DOI] [PubMed] [Google Scholar]
- 53.Hennig, S., Rödel, G., and Ostermann, K. (2016) Hydrophobin-based surface engineering for sensitive and robust quantification of yeast pheromones. Sensors (Basel). (Saavedra, S. S., Ed.) 16, 602. [DOI] [PMC free article] [PubMed]
- 54.Soikkeli M, Kurppa K, Kainlauri M, Arpiainen S, Paananen A, Gunnarsson D, Joensuu JJ, Laaksonen P, Prunnila M, Linder MB, Ahopelto J. Graphene biosensor programming with genetically engineered fusion protein monolayers. ACS Appl Mater Interfaces. 2016;8:8257–8264. doi: 10.1021/acsami.6b00123. [DOI] [PubMed] [Google Scholar]
- 55.Laaksonen P, Kivioja J, Paananen A, Kainlauri M, Kontturi K, Ahopelto J, Linder MB. Selective Nanopatterning using citrate-stabilized au nanoparticles and Cystein-modified amphiphilic protein. Langmuir. 2009;25:5185–5192. doi: 10.1021/la804001e. [DOI] [PubMed] [Google Scholar]
- 56.Laaksonen P, Kainlauri M, Laaksonen T, Shchepetov A, Jiang H, Ahopelto J, Linder MB. Interfacial engineering by proteins: exfoliation and functionalization of graphene by Hydrophobins. Angew Chemie. 2010;122:5066–5069. doi: 10.1002/ange.201001806. [DOI] [PubMed] [Google Scholar]
- 57.Linder M, Selber K, Nakari-Setälä T, Qiao M, Kula M-R, Penttilä M. The Hydrophobins HFBI and HFBII from Trichoderma reesei showing efficient interactions with nonionic surfactants in aqueous two-phase systems. Biomacromolecules. 2001;2:511–517. doi: 10.1021/bm0001493. [DOI] [PubMed] [Google Scholar]
- 58.Joensuu JJ, Conley AJ, Lienemann M, Brandle JE, Linder MB, Menassa R. Hydrophobin Fusions for High-Level Transient Protein Expression and Purification in <em>Nicotiana benthamiana</em>. Plant Physiol. 2010;152:622 LP–622633. doi: 10.1104/pp.109.149021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reuter L, Ritala A, Linder M, Joensuu J. Novel Hydrophobin fusion tags for plant-produced fusion proteins. PLoS One. 2016;11:e0164032. doi: 10.1371/journal.pone.0164032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kurppa K, Reuter LJ, Ritala A, Linder MB, Joensuu JJ. In-solution antibody harvesting with a plant-produced hydrophobin–protein A fusion. Plant Biotechnol J. 2018;16:404–414. doi: 10.1111/pbi.12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang R, Yang Y-L, Qin M, Wang L-K, Yu L, Shao B, Qiao M-Q, Wang C, Feng X-Z. Biocompatible hydrophilic modifications of poly(dimethylsiloxane) using self-assembled Hydrophobins. Chem Mater. 2007;19:3227–3231. doi: 10.1021/cm070445n. [DOI] [Google Scholar]
- 62.Hou S, Li X, Li X, Feng X-Z, Wang R, Wang C, Yu L, Qiao M-Q. Surface modification using a novel type I hydrophobin HGFI. Anal Bioanal Chem. 2009;394:783–789. doi: 10.1007/s00216-009-2776-y. [DOI] [PubMed] [Google Scholar]
- 63.Qin M, Wang L-K, Feng X-Z, Yang Y-L, Wang R, Wang C, Yu L, Shao B, Qiao M-Q. Bioactive surface modification of Mica and Poly(dimethylsiloxane) with Hydrophobins for protein immobilization. Langmuir. 2007;23:4465–4471. doi: 10.1021/la062744h. [DOI] [PubMed] [Google Scholar]
- 64.Zhang M, Wang Z, Wang Z, Feng S, Xu H, Zhao Q, Wang S, Fang J, Qiao M, Kong D. Immobilization of anti-CD31 antibody on electrospun poly(ɛ-caprolactone) scaffolds through hydrophobins for specific adhesion of endothelial cells. Colloids Surfaces B Biointerfaces. 2011;85:32–39. doi: 10.1016/j.colsurfb.2010.10.042. [DOI] [PubMed] [Google Scholar]
- 65.Boeuf S, Throm T, Gutt B, Strunk T, Hoffmann M, Seebach E, Mühlberg L, Brocher J, Gotterbarm T, Wenzel W, Fischer R, Richter W. Engineering hydrophobin DewA to generate surfaces that enhance adhesion of human but not bacterial cells. Acta Biomater. 2012;8:1037–1047. doi: 10.1016/j.actbio.2011.11.022. [DOI] [PubMed] [Google Scholar]
- 66.Wang X, Mao J, Chen Y, Song D, Gao Z, Zhang X, Bai Y, Saris PEJ, Feng H, Xu H, Qiao M. Design of antibacterial biointerfaces by surface modification of poly (ε-caprolactone) with fusion protein containing hydrophobin and PA-1. Colloids Surfaces B Biointerfaces. 2017;151:255–263. doi: 10.1016/j.colsurfb.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 67.Heinonen H, Laaksonen P, Linder MB, Hentze H-P. Engineered hydrophobin for biomimetic mineralization of functional calcium carbonate microparticles. J Biomater Nanobiotechnol. 2014;5:1. doi: 10.4236/jbnb.2014.51001. [DOI] [Google Scholar]
- 68.Melcher M, Facey SJ, Henkes TM, Subkowski T, Hauer B. Accelerated nucleation of hydroxyapatite using an engineered Hydrophobin fusion protein. Biomacromolecules. 2016;17:1716–1726. doi: 10.1021/acs.biomac.6b00135. [DOI] [PubMed] [Google Scholar]
- 69.Zhao L, Ma S, Pan Y, Zhang Q, Wang K, Song D, Wang X, Feng G, Liu R, Xu H, Zhang J, Qiao M, Kong D. Functional modification of fibrous PCL scaffolds with fusion protein VEGF-HGFI enhanced Cellularization and vascularization. Adv Healthc Mater. 2016;5:2376–2385. doi: 10.1002/adhm.201600226. [DOI] [PubMed] [Google Scholar]
- 70.Taniguchi S, Sandiford L, Cooper M, Rosca EV, Ahmad Khanbeigi R, Fairclough SM, Thanou M, Dailey LA, Wohlleben W, von Vacano BTM, de Rosales R, Dobson PJ, Owen DM, Green M. Hydrophobin-encapsulated quantum dots. ACS Appl Mater Interfaces. 2016;8:4887–4893. doi: 10.1021/acsami.5b11354. [DOI] [PubMed] [Google Scholar]
- 71.Sun F, Yang G, Zhang Q, Xue Z, Gu C, Chen Z, Yan B, Feng Y, Wang Z, Meng S. The self-assembly of monosubstituted BODIPY and HFBI-RGD. RSC Adv. 2018;8:21472–21479. doi: 10.1039/C8RA03687J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Askolin S, Nakari-Setälä T, Tenkanen M. Overproduction, purification, and characterization of the Trichoderma reesei hydrophobin HFBI. Appl Microbiol Biotechnol. 2001;57:124–130. doi: 10.1007/s002530100728. [DOI] [PubMed] [Google Scholar]
- 73.Schuurs TA, Schaeffer EAM, Wessels JGH. Homology-dependent silencing of the Sc3 gene in Schizophyllum commune. Genetics. 1997;147:589–596. doi: 10.1093/genetics/147.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hektor HJ, Scholtmeijer K. Hydrophobins: proteins with potential. Curr Opin Biotechnol. 2005;16:434–439. doi: 10.1016/j.copbio.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 75.Winandy L, Hilpert F, Schlebusch O, Fischer R. Comparative analysis of surface coating properties of five hydrophobins from aspergillus nidulans and Trichoderma reseei. Sci Rep. 2018;8:12033. doi: 10.1038/s41598-018-29749-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kirkland BH, Keyhani NO. Expression and purification of a functionally active class I fungal hydrophobin from the entomopathogenic fungus Beauveria bassiana in E. coli. J Ind Microbiol Biotechnol. 2011;38:327–335. doi: 10.1007/s10295-010-0777-7. [DOI] [PubMed] [Google Scholar]
- 77.Wang Z, Feng S, Huang Y, Qiao M, Zhang B, Xu H. Prokaryotic expression, purification, and polyclonal antibody production of a hydrophobin from Grifola frondosa. Acta Biochim Biophys Sin Shanghai. 2010;42:388–395. doi: 10.1093/abbs/gmq033. [DOI] [PubMed] [Google Scholar]
- 78.Morris VK, Kwan AH, Sunde M. Analysis of the structure and conformational states of DewA gives insight into the assembly of the fungal Hydrophobins. J Mol Biol. 2013;425:244–256. doi: 10.1016/j.jmb.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 79.Rey AA, Hocher A, Kwan AH, Sunde M. Backbone and sidechain 1H, 13C and 15N chemical shift assignments of the hydrophobin MPG1 from the rice blast fungus Magnaporthe oryzae. Biomol NMR Assign. 2013;7:109–112. doi: 10.1007/s12104-012-9394-x. [DOI] [PubMed] [Google Scholar]
- 80.Kottmeier K, Günther TJ, Weber J, Kurtz S, Ostermann K, Rödel G, Bley T. Constitutive expression of hydrophobin HFB1 from Trichoderma reesei in Pichia pastoris and its pre-purification by foam separation during cultivation. Eng Life Sci. 2012;12:162–170. doi: 10.1002/elsc.201100155. [DOI] [Google Scholar]
- 81.Niu B, Wang D, Yang Y, Xu H, Qiao M. Heterologous expression and characterization of the hydrophobin HFBI in Pichia pastoris and evaluation of its contribution to the food industry. Amino Acids. 2012;43:763–771. doi: 10.1007/s00726-011-1126-5. [DOI] [PubMed] [Google Scholar]
- 82.Wang Z, Feng S, Huang Y, Li S, Xu H, Zhang X, Bai Y, Qiao M. Expression and characterization of a Grifola frondosa hydrophobin in Pichia pastoris. Protein Expr Purif. 2010;72:19–25. doi: 10.1016/j.pep.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 83.Pedersen MH, Borodina I, Moresco JL, Svendsen WE, Frisvad JC, Søndergaard I. High-yield production of hydrophobins RodA and RodB from aspergillus fumigatus in Pichia pastoris. Appl Microbiol Biotechnol. 2011;90:1923–1932. doi: 10.1007/s00253-011-3235-1. [DOI] [PubMed] [Google Scholar]
- 84.Niu B, Gong Y, Gao X, Xu H, Qiao M, Li W. The functional role of Cys3–Cys4 loop in hydrophobin HGFI. Amino Acids. 2014;46:2615–2625. doi: 10.1007/s00726-014-1805-0. [DOI] [PubMed] [Google Scholar]
- 85.Song D, Gao Z, Zhao L, Wang X, Xu H, Bai Y, Zhang X, Linder MB, Feng H, Qiao M. High-yield fermentation and a novel heat-precipitation purification method for hydrophobin HGFI from Grifola frondosa in Pichia pastoris. Protein Expr Purif. 2016;128:22–28. doi: 10.1016/j.pep.2016.07.014. [DOI] [PubMed] [Google Scholar]
- 86.Gandier, J.-A., and Master, R. E. (2018) Pichia pastoris is a suitable host for the heterologous expression of predicted class I and class II Hydrophobins for discovery, study, and application in biotechnology. Microorg. . [DOI] [PMC free article] [PubMed]
- 87.Ahmad M, Hirz M, Pichler H, Schwab H. Protein expression in Pichia pastoris: recent achievements and perspectives for heterologous protein production. Appl Microbiol Biotechnol. 2014;98:5301–5317. doi: 10.1007/s00253-014-5732-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cregg, J. M., Tolstorukov, I., Kusari, A., Sunga, J., Madden, K., and Chappell, T. (2009) Chapter 13 expression in the yeast Pichia pastoris, in Methods in Enzymology (Burgess RR, and Deutscher MPB. T.-M. in E., Eds.), pp 169–189. Academic Press. [DOI] [PubMed]
- 89.Cereghino JL, Cregg JM. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol Rev. 2000;24:45–66. doi: 10.1111/j.1574-6976.2000.tb00532.x. [DOI] [PubMed] [Google Scholar]
- 90.Daly R, Hearn MTW. Expression of heterologous proteins in Pichia pastoris: a useful experimental tool in protein engineering and production. J Mol Recognit. 2004;18:119–138. doi: 10.1002/jmr.687. [DOI] [PubMed] [Google Scholar]
- 91.Scorer CA, Clare JJ, McCombie WR, Romanos MA, Sreekrishna K. Rapid selection using G418 of high copy number Transformants of Pichia pastoris for high–level foreign gene expression. Bio/Technology. 1994;12:181. doi: 10.1038/nbt0294-181. [DOI] [PubMed] [Google Scholar]
- 92.Gutiérrez SP, Saberianfar R, Kohalmi SE, Menassa R. Protein body formation in stable transgenic tobacco expressing elastin-like polypeptide and hydrophobin fusion proteins. BMC Biotechnol. 2013;13:40. doi: 10.1186/1472-6750-13-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Reuter LJ, Bailey MJ, Joensuu JJ, Ritala A. Scale-up of hydrophobin-assisted recombinant protein production in tobacco BY-2 suspension cells. Plant Biotechnol J. 2013;12:402–410. doi: 10.1111/pbi.12147. [DOI] [PubMed] [Google Scholar]
- 94.Häkkinen ST, Reuter L, Nuorti N, Joensuu JJ, Rischer H, Ritala A. Tobacco BY-2 media component optimization for a cost-efficient recombinant protein production. Front Plant Sci. 2018;9:45. doi: 10.3389/fpls.2018.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.