Abstract
Objectives
We used an unbiased proteomics approach to identify candidate urine biomarkers (CUBMs) predictive of LN chronicity and pursued their validation in a larger cohort.
Methods
In this cross-sectional pilot study, we selected urine collected at kidney biopsy from 20 children with varying levels of LN damage (discovery cohort) and performed proteomic analysis using isobaric tags for relative and absolute quantification (iTRAQ). We identified differentially excreted proteins based on degree of LN chronicity and sought to distinguish markers exhibiting different relative expression patterns using hierarchically clustered log10-normalized relative abundance data with linked and distinct functions by biological network analyses. For each CUBM, we performed specific ELISAs on urine from a validation cohort (n = 41) and analysis of variance to detect differences between LN chronicity, with LN activity adjustment. We evaluated for CUBM expression in LN biopsies with immunohistochemistry.
Results
iTRAQ detected 112 proteins in urine from the discovery cohort, 51 quantifiable in all replicates. Simple analysis of variance revealed four differentially expressed, chronicity-correlated proteins (P-values < 0.05). Further correlation and network analyses led to selection of seven CUBMs for LN chronicity. In the validation cohort, none of the CUBMs distinguished LN chronicity degree; however, urine SERPINA3 demonstrated a moderate positive correlation with LN histological activity. Immunohistochemistry further demonstrated SERPINA3 staining in proximal tubular epithelial and endothelial cells.
Conclusion
We identified SERPINA3, a known inhibitor of neutrophil cathepsin G and angiotensin II production, as a potential urine biomarker to help quantify LN activity.
Keywords: LN, paediatric rheumatology, biomarker, proteomics, SERPINA3, alpha-1-antichymotrypsin
Rheumatology key messages
SERPINA3 urine levels are elevated with high degrees of histological activity in paediatric lupus nephritis.
Urine protein creatinine ratio does not distinguish the degree of lupus nephritis chronicity on biopsy.
SERPINA3 localizes to proximal tubular epithelial and endothelial cells on lupus nephritis biopsies.
Introduction
Up to 80% of children with lupus experience inflammatory kidney disease with the potential to result in permanent kidney tissue damage, especially if diagnosis is delayed or control of LN cannot be achieved [1]. Traditional LN measures, such as proteinuria, cannot accurately distinguish between irreversible kidney damage and treatment-responsive active inflammation [2]. Indeed, invasive kidney biopsies are still the gold standard for LN diagnosis and classification, as per the International Society of Nephrology/Renal Pathology Society criteria, as well as for measurement of the degree of LN activity and chronicity using the National Institutes of Health (NIH) activity and chronicity indices (NIH-AI and NIH-CI, respectively) [3–5].
Non-invasive biomarkers that can discriminate between LN activity and chronicity are needed. In recent years, advanced urine proteomics techniques have been applied to discover proteins that might serve as novel LN biomarkers [6, 7]. While much progress has been achieved with respect to delineating urine biomarkers (UBMs) that reflect LN activity [8–10], biomarkers for the early identification of LN damage are still lacking. In this setting, urinary proteomics appears especially promising, given its proven track record for delineating biomarkers in various kidney diseases [11] and advances to proteomics techniques in recent years [12].
One of these advanced proteomics techniques is isobaric tags for relative and absolute quantification (iTRAQ), a tandem MS (MS/MS)-based approach for identification and quantification of differentially expressed proteins [13]. Isobaric iTRAQ reagents allow for simultaneous analysis of multiple samples and relative quantification of peptides present. iTRAQ is particularly useful when applied to samples from patients with phenotypically heterogeneous diseases, where the proteome may also vary widely for each individual.
We aimed to discover candidate UBMs (CUBMs) that reflect LN damage using iTRAQ proteomics, and provide initial validation of the usefulness of these CUBMs for reflecting histologically confirmed tissue damage as measured by the NIH-CI on renal biopsy and clinical renal functional loss.
Methods
For this study, we evaluated two cohorts: a discovery cohort, used to perform iTRAQ experiments, and a validation cohort, in which we measured CUBM levels using specific ELISAs.
Patients and clinical data
LN patients with bio-banked urine samples collected within a month of kidney biopsy were recruited from an ongoing paediatric lupus cohort at Cincinnati Children’s Hospital Medical Center. All patients met ACR classification criteria for SLE [14, 15] and had disease onset prior to 18 years of age. The study was approved by the Cincinnati Children’s Institutional Review Board, and all patients and caretakers provided written consent and assent.
Discovery cohort
LN patient samples (n = 20) were selected that contained at least 50 µg protein (1/21 samples were rejected). The discovery cohort test samples (n = 15) were classified as low, moderate or high chronicity (NIH-CI score = 1, 2 or ⩾3, respectively), and compared with a pooled sample consisting of five lower activity and chronicity samples that was used as a common reference for each iTRAQ run.
Validation cohort
A validation cohort (n = 41) was assembled to confirm the usefulness of CUBMs in assessing LN histologic damage. Patients were assigned levels of LN chronicity using the NIH-CI score as follows: ⩽1 or no biopsy performed: no/minimal LN severity; 2: moderate severity; ⩾3: high severity.
In all biopsies, kidney histology was interpreted by one expert nephropathologist (D.W.), who was blinded to clinical and laboratory data. Biopsy interpretation was per the 2003 International Society of Nephrology/Renal Pathology Society classification [3], and assignment of histological activity (NIH-AI, range 0–24; 0 = inactive LN) and chronicity (NIH-CI, range 0–12; 0 = no damage) was done as previously described [4]. We also recorded relevant clinical and laboratory information, including creatinine clearance to estimate the glomerular filtration rate using the modified Schwartz formula [16], proteinuria based on urine protein to creatinine ratio, and disease activity as measured by the SLEDAI (score 0–105; 0 = inactive disease) [17].
Sample preparation, protein separation and digestion
Twenty-five micrograms from each urine sample was concentrated and buffer exchanged against Invitrogen (Thermo Fisher Scientific, Waltham, MA, USA) 0.5× Laemmli buffer using Amicon Ultra 3K filters (UFC500396, MilliporeSigma, Burlington, MA, USA). The final volume was about 40 μL for each sample. Samples were loaded and electrophoresed 1.5 cm into a 1D, 1.5 mm, 4–12% Bis–Tris minigel using MOPS running buffer (Invitrogen NP0001, Thermo Fisher Scientific). The gel region between the well and dye front containing the proteins was excised for trypsin digestion and the resulting peptides extracted following the in-gel iTRAQ protocol (Sciex, Concord, Ontario, Canada).
iTRAQ labelling and nano liquid chromatography coupled electrospray tandem mass spectrometry
The isolated peptides were tagged with the iTRAQ 4-plex reagents using vendor instructions (Sciex, Concord, ON, Canada). Each comparative group contained a control pool sample (114 reporter tag), low, moderate and high chronicity sample (115, 116 and 117 reporter tags, respectively). After iTRAQ labelling, peptides from the four samples were mixed in equal portions. Each iTRAQ labelled protein set (five sets consisting of four samples per set) was run in triplicate.
Nanoflow liquid chromatography coupled electrospray ionization tandem mass spectrometry (nLC-ESI-MS/MS) analysis were performed using 2.5 μg of each 4-plex mixture on a TripleTOF 5600+ (Sciex, Concord, ON, Canada) attached to an Eksigent (Dublin, CA, USA) nLC ultra nanoflow system as described previously [18].
Peptide identification, quantitative profiling and statistics
ProteinPilot software (Sciex, Concord, ON, Canada, version 4.5, revision 1656) was used to identify proteins and determine relative quantification. ProteinPilot Descriptive Statistics Template (ver 3.005pB) was used to process relative quantification data among sample sets and provide statistical probabilities related to confidence of protein identification and provide P-values regarding relative quantification of the four reporter ions for each protein. The ProteinPilot data were then processed through Protein Alignment Template software (Sciex), allowing for comparison of protein changes across designated groups.
Clustering and ranking CUBMs in the discovery cohort
Individual sample relative protein abundance levels and group difference-based P-value data generated from Protein Pilot and Protein Alignment Template were baselined to the median value for the low chronicity samples, subjected to further analysis of variance filtering with a P-value < 0.05 considered significant and hierarchically clustered for heatmap visualization using GeneSpring 14.5 software (Agilent Technologies Inc., Santa Clara, CA, USA) and Morpheus (Broad Institute, Cambridge, MA, USA). Due to large sample variance, we used a conservative high false discovery rate (0.2) to identify CUBMs that could subsequently be validated in a separate cohort using ELISA-based analyses.
To evaluate potential relationships between dysregulated and similarly correlated CUBM and to further prioritize among the CUBM, biological network analysis was carried out using ToppCluster [19] and Cytoscape [20].
Enzyme-linked immunosorbent assays
Urine was collected and stored frozen at −80°C. CUBM levels were measured in urine with commercially available ELISAs: afamin (AFM), retinol binding protein 4 (RBP4) and α1-acid glycoprotein 1 (ORM1) (R&D Systems, Minneapolis, MN, USA), immunoglobulin heavy constant α 1 (IGHA1) (MyBiosource, LLC, San Diego, CA, USA), α1-antichymotrypsin (SERPINA3) and transthyretin (TTR) (Assaypro, St Charles, MO, USA). Transferrin (TF) was measured by immunonephelometry, performed on a Siemens BN2 clinical nephelometer (Siemens, Munich, Germany).
SERPINA3 immunohistochemistry
Immunohistochemistry was performed using 4 µm-thick sections from four paraffin-embedded LN biopsies. Tissue was deparaffinized with xylene and rehydrated in graded ethanol. Slides were placed in 0.1 M sodium citrate buffer, heated and immersed in 0.85% NaCl and 0.5% H2O2. Blocking used one drop of Avidin D in 250 microliters of 1.5% normal horse serum (Avidin/Biotin blocking kit SP-2001, Vector Laboratories, Burlingame, CA, USA). Slides were incubated at 4°C overnight with a primary polyclonal rabbit SERPINA3 antibody at 1: 100 (Thermo Fisher Scientific), treated with a biotinylated anti-rabbit IgG secondary antibody at 1 : 200 for 30 min (Vectastain ABC-HRP Kit, Vector Laboratories) and counterstained with haematoxylin.
Statistical analysis in validation cohort
All CUBM levels assayed by ELISA were found to be right-skewed in their distributions, but their (natural) log-transformed variables were symmetrically distributed and fit the conditions for parametric statistical models. Hence, all analyses were performed using log-transformed CUBM levels, and variation of estimates was measured by standard error or standard deviation. We calculated Spearman’s correlation coefficients to assess relationships between numerical variables. Analysis of variance was performed to detect statistical differences between each of the CUBMs (dependent variables) and LN chronicity groups. Models were repeated after adjusting for renal disease activity (NIH-AI; renal SLEDAI). For SERPINA3, analysis of variance was further performed to detect differences between urine levels in different LN activity groups (no/low, moderate and high, with corresponding NIH-AI of 0–2, 3–9 and ⩾10, respectively).
The Tukey–Kramer test was used for post hoc analyses to determine statistically significant differences of means between LN chronicity and activity groups. In addition, categorical and numerical variables at baseline were summarized using frequency as a percentage and mean (standard deviation), respectively. All statistical analyses were computed using SAS 9.4 (SAS Institute, Cary, NC, USA) package. P-values < 0.05 were considered statistically significant.
Results
Patients
The discovery cohort test samples consisted of 12 patients with Class IV LN and three with Class V LN. Details on the kidney biopsy results of the discovery cohort are summarized in Supplementary Table S1, available at Rheumatology online. Validation cohort participants were commonly female (71%), mostly classified as having proliferative LN (Class III or IV) (67%), and often had both active extrarenal disease as measured by the SLEDAI and active LN (NIH-AI score) based on kidney histology (Table 1). The median (range) NIH-AI and CI were 9 (0–22) and 2 (0–6), respectively.
Table 1.
Clinical and demographic data for validation cohort (n = 41)
| Variable | n (%) | Mean (s.d.) | Median (range) |
|---|---|---|---|
| Demographics | |||
| Females | 29 (71) | ||
| Males | 12 (29) | ||
| Age, years | 16 (3) | 16 (7–26) | |
| Hispanics/non-Hispanics | 5 (12) | ||
| 36 (88) | |||
| White/Black/Asian | 20 (49) | ||
| 20 (49) | |||
| 1 (2) | |||
| Disease activity | |||
| Extrarenal SLEDAI | 3.85 (4.57) | 2 (0–17) | |
| Renal SLEDAI | 6.93 (6.2) | 8 (0–16) | |
| LN | |||
| ISN/RPS Class of LNa | II 3 (8.3%) | ||
| III 7 (19.4%) | |||
| IV 17 (47.2%) | |||
| V 9 (25%) | |||
| NIH Activity Scorea | 8.66 (6.53) | 9 (0–22) | |
| NIH Chronicity Scorea | 2.02 (1.52) | 2 (0–6) | |
| Creatinine clearanceb, mg/ml/1.73 m2 | 109.56 (54.46) | 95.23 (21.1–228.2) | |
| Protein creatinine ratiob | 4.01 (7.06) | 2.25 (0.01–42.5) | |
No biopsy information on five patients given clinical absence of LN, activity and chronicity scores of 0 assigned for purpose of analysis.
Information missing on one patient. ISN/RPS: International Society of Nephrology/Renal Pathology Society; NIH: National Institutes of Health.
Proteomics results
The final iTRAQ dataset was generated from five separate sample sets, each consisting of four samples and run in triplicate. Overall, iTRAQ detected 112 proteins from urine sample sets of the discovery cohort, and 51 proteins were quantifiable in all technical replicates. Statistical analysis of pairwise chronicity group comparisons demonstrated that four proteins differed significantly with P-values < 0.05.
Heatmap analysis
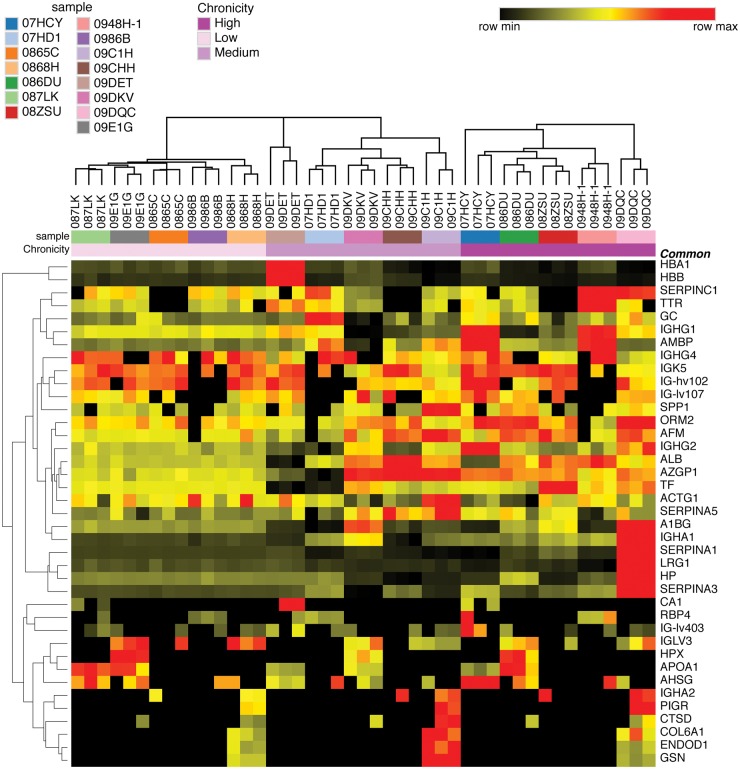
The generated heatmap is shown in Fig. 1 and demonstrates multiple upregulated proteins in the moderate and high chronicity samples in orange/red. Heterogeneity in protein abundance levels is apparent in individual urine samples even at a similar chronicity level. Reproducibility of proteomics results is exhibited by similar protein ratios in replicate sample runs. Similar results were obtained when using either the low chronicity sample within each sample set or control pool as the comparator.
Fig. 1.
Heat map of candidate urine biomarkers associated with LN chronicity in discovery cohort samples
Each column represents average protein expression values from individual samples (low, moderate or high chronicity) relative to the low chronicity sample within the sample set. Low chronicity samples are on the left-hand side of the heat map, followed by moderate chronicity samples in the middle and high chronicity samples on the right. There are three columns for each individual sample, representing replicate runs. Each row is a protein (gene name) that was differentially expressed between samples based on chronicity grouping. Protein expression values are depicted using the colour scale shown with black to red indicating increasing expression.
Identifying CUBMs
Based on cumulative results from iTRAQ data and heatmap analysis, the seven best performing CUBMs and their known biological relevance are summarized in Table 2. The final CUBMs included AFM, IGHA1, SERPINA3, TTR, RBP4, ORM2 and TF.
Table 2.
Top candidate urine biomarkers for LN chronicity from discovery cohort
| Protein | Gene | Function | P-value |
|---|---|---|---|
| Afamin | AFM | Vitamin E binding glycoprotein; upregulated with ovarian cancer and development of metabolic syndrome [21, 22] | 7.6 × 10−2 |
| Immunoglobulin heavy constant α 1 | IGHA1 | Isotype of immunoglobulin A; aberrantly glycosylated forms involved in IgA nephropathy [23] | 2.6 × 10−3 |
| α1-Antichymotrypsin | SERPINA3 | Inhibitor of serine proteinases; acute-phase protein whose concentration can rise up to 5-fold during inflammation [20, 24] | 6.8 × 10−3 |
| Transthyretin | TTR | Carrier protein that transports thyroxine and retinol; misfolding associated with amyloidosis; also known as prealbumin [25] | 8.2 × 10−2 |
| Retinol binding protein 4 | RBP4 | Serves to transport retinol; retinol binding protein 4–retinol complex interacts with transthyretin to prevent loss from kidney; catabolism disturbed in kidney disease [26, 27] | 7.3 × 10−2 |
| α1-Acid glycoprotein 2 | ORM2 | Acute phase reactant and transport protein; probable function in modulation of innate immune response [28] | 8.9 × 10−2 |
| Transferrin | TF | Marker of LN activity and risk factor for renal functional loss [29, 30] | 1.3 × 10−2 |
Network analysis
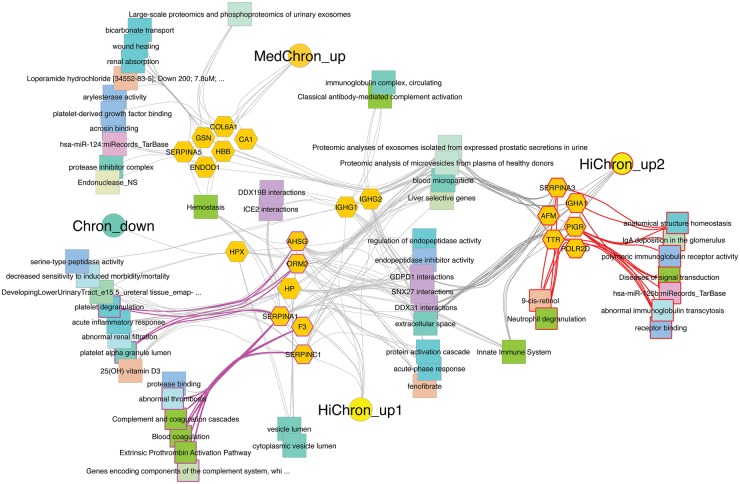
Further network analysis showed involvement of these proteins in pathways of complement activation, coagulation, platelet and neutrophil degranulation, immunoglobulin transcytosis and deposition and renal absorption and filtration (Fig. 2).
Fig. 2.
Network of differentially excreted proteins based on degree of LN chronicity in discovery cohort
Circles represent clusters or designated groupings of genes that are upregulated in both moderate and high chronicity samples from the discovery cohort (HiChron_up2), those upregulated only in high chronicity samples (HiChron_up1), those upregulated only in moderate chronicity samples (MedChron_up) and those downregulated in all moderate and high chronicity samples (Chron_down). Individual genes are represented by orange hexagons and shared molecular pathways among genes by connectivity to coloured squares.
Biomarker assessment in the validation cohort
In the validation cohort, none of the CUBMs were found to differ based on degree of LN chronicity after adjustment for concurrent LN activity (Table 3). Renal SLEDAI and creatinine clearance did differ depending on degree of LN chronicity whereas protein creatinine ratio did not differ. A weak positive association was observed between NIH-CI and four of the seven CUBMs, including SERPINA3 (rs = 0.25, P = 0.13), TF (rs = 0.31, P = 0.047), TTR (rs = 0.22, P = 0.17) and ORM1 (rs = 0.29, P = 0.07).
Table 3.
Candidate urine biomarkers by LN chronicity and adjusted for concurrent LN activity in the validation cohort
| LN biomarker/measure | No/minimal chronicitya (n = 18) (1) | Moderate chronicityb (n = 8) (2) | High chronicityc (n = 15) (3) | P-valued (F-test) | P-valued 1 vs 2 | P-valued 1 vs 3 | P-valued 2 vs 3 |
|---|---|---|---|---|---|---|---|
| Candidate urine biomarkers | |||||||
| Afamin | 4.53 (0.64) | 5.36 (0.94) | 3.67 (0.67) | 0.3182 | 0.4847 | 0.37 | 0.1441 |
| Immunoglobulin heavy constant α 1 | 8.47 (0.46) | 9.07 (0.67) | 8.78 (0.48) | 0.7624 | 0.4729 | 0.6535 | 0.7143 |
| Retinol binding protein 4 | 5.47 (0.48) | 4.69 (0.7) | 5.09 (0.51) | 0.6748 | 0.3846 | 0.6052 | 0.6405 |
| α1-Antichymotrypsin (SERPINA3)e | 7.87 (0.5) | 7.8 (0.91) | 7.26 (0.55) | 0.7096 | 0.9548 | 0.4359 | 0.6077 |
| Transthyretin | 4.83 (0.69) | 4.57 (1.02) | 3.85 (0.74) | 0.6267 | 0.8377 | 0.3518 | 0.5658 |
| Transferrin | 0.82 (0.44) | 1.27 (0.65) | 0.44 (0.47) | 0.5655 | 0.5801 | 0.5689 | 0.2961 |
| α1-Acid glycoprotein 1 | 9.6 (0.64) | 9.7 (0.92) | 9.92 (0.66) | 0.9400 | 0.9371 | 0.7343 | 0.8371 |
| Traditional LN measures | |||||||
| Protein creatinine ratio | 1.58 (1.62) | 5.2 (2.4) | 6.47 (1.83) | 0.1314 | 0.2226 | 0.0531 | 0.679 |
| Creatinine clearance | 125.24 (12) | 122.26 (17.18) | 85.02 (12.37) | 0.0572 | 0.8911 | 0.0285 | 0.0822 |
| Renal SLEDAI | 4.22 (1.37) | 10 (2.06) | 8.53 (1.5) | 0.0364 | 0.025 | 0.041 | 0.5688 |
Data are mean (s.e.).
No kidney biopsy or NIH-CI score ≤1.
Moderate damage NIH-CI score = 2.
High damage NIH-CI score ≥3; CUBM values are expressed as natural log-transformed values.
From Tukey–Kramer test from analysis of variance adjusted for concurrent LN activity as measured by the renal SLEDAI.
For SERPINA3, there were only 36 patient urine samples available for measurement. NIH-CI: National Institutes of Health chronicity index.
Upon evaluation of CUBM association with NIH-AI, SERPINA3 was found to have a moderate positive association with NIH-AI (rs = 0.45, P = 0.005) (Supplementary Fig. S1A, available at Rheumatology online). SERPINA3 levels also significantly increased with higher histological LN activity (no/low, moderate and high LN activity groups with natural log-transformed values (s.e.) of 5.45 (0.89), 7.62 (0.71) and 8.4 (0.57), P = 0.03) (Supplementary Fig. S1B, available at Rheumatology online). TF and ORM1 were also moderately positively associated with NIH-AI (rs = 0.46, P = 0.003 and rs = 0.43, P = 0.005). Multiple CUBMs were associated with one another, including strong positive associations between SERPINA3 and TF/ORM1/AFM (rs = 0.84, 0.88 and 0.76, P < 0.0001), AFM and TF/ORM1 (rs = 0.87 and 0.77, P < 0.0001), moderate positive associations between IGHA1 and TF/AFM (rs = 0.7 and 0.66, P < 0.0001), ORM1 and RBP4 (rs = 0.6, P < 0.0001), and TF and TTR (rs = 0.64, P < 0.0001).
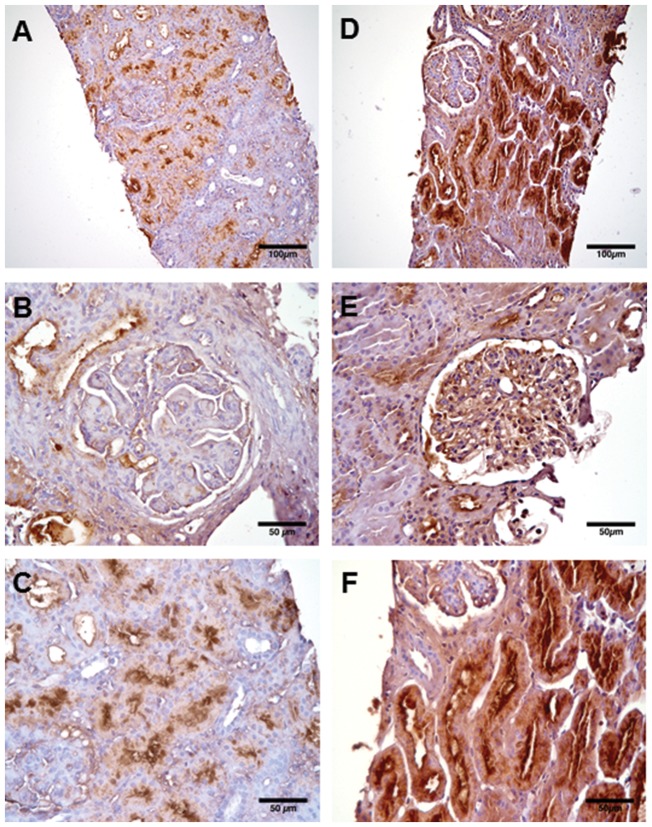
SERPINA3 immunohistochemistry in LN biopsies
SERPINA3 staining localized its expression to endothelial cells and proximal tubular epithelial cells on LN biopsies (Fig. 3). SERPINA3 staining was less consistent in endothelial cells, shown within the glomerulus of Fig. 3E but not Fig. 3B.
Fig. 3.
SERPINA3 immunostaining in LN biopsies
(A–F) Three separate LN biopsies with lower magnification images of larger biopsy region (A and D), and higher magnification images of glomeruli (B and E) and tubules (C and F). SERPINA3 immunostaining demonstrated by brown colour localizes predominantly to proximal tubular epithelial cells (C and F) but also to endothelial cells in (E). Biopsy activity and chronicity information are as follows: (A–C) LN biopsy 1: NIH activity index (NIH-AI) 11/24 and chronicity index (NIH-CI) 0/12; (D and F) LN biopsy 2: NIH-AI 1, NIH-CI 2; (E) LN biopsy 3: NIH-AI 6, NIH-CI 3. Original magnification ×20 for (A and D) and ×40 for (B, C, E and F). SERPINA3: α1-antichymotrypsin.
Discussion
Using advanced proteomics techniques, we identified seven candidate chronicity biomarkers for LN in urine. Although none of the CUBMs validated in an independent cohort with specific ELISAs, we discovered α1-antichymotrypsin (SERPINA3) as a potential urine biomarker to help quantify the degree of LN activity. Immunohistochemistry of LN biopsies further demonstrated staining for SERPINA3 in endothelial and proximal tubular epithelial cells.
SERPINA3 is a member of the serpin superfamily that contains 36 protein-coding genes, of which SERPINA3 and SERPINA1 constitute the two most abundant serpins [31]. The majority of serpins function as specific serine protease inhibitors; however, some serpins serve additional functions in transporting hormones or regulating apoptosis [32, 33]. In particular, SERPINA3 is an extracellular protein that plays a role in regulating inflammation, predominantly through inhibition of neutrophil cathepsin G, and also uniquely binds DNA [34]. In the kidney, SERPINA3 is likely involved in the inhibition of angiotensin converting enzyme, thereby decreasing production of angiotensin II. In fact, elevated serum angiotensin converting enzyme levels have been detected in patients with SERPINA1 deficiency [35]. SERPINA1 or α1-antitrypsin is the better known serpin family member, and its deficiency has been described in association with a spectrum of renal and autoimmune diseases [36].
SERPINA1 and SERPINA3 share similar functions. SERPINA3, like SERPINA1, is an acute phase protein and is known to increase in the circulation by up to 5-fold during periods of inflammation [37]. It is unclear whether increased urinary levels of SERPINA3 found in our study resulted from increased filtration or endogenous renal production in select renal cells. We did measure SERPINA3 serum levels in a sample of our lupus patients (n = 15) and found the natural log-transformed serum SERPINA3 levels to be 1.8 times higher than the natural log-transformed urine SERPINA3 levels, on average. While it certainly appears that filtration of SERPINA3 from plasma is contributing to urine levels, the role of endogenous renal production of SERPINA3 needs to be further investigated. At least in the case of SERPINA1, gene expression in renal cortex and urine protein levels both increased dramatically in a murine model of acute kidney injury with no increase in SERPINA1 plasma levels, suggesting resident kidney production [38]. SERPINA1 production appeared to come principally from proximal tubular epithelial cells, in line with SERPINA3 immunostaining in our study. Furthermore, exogenous administration of SERPINA1 in a murine model of renal ischaemia–reperfusion has been shown to be protective against subsequent inflammation and apoptosis, suggesting a potential therapeutic role for serpins [39].
In a spontaneous murine model of SLE, a peptide fragment of SERPINA1 was reported to improve LN and reduce anti-double stranded DNA antibody levels. No side effects from the SERPINA1 peptide fragment exposures were observed, lending further support to the idea that serpins might be promising therapeutic agents [40]. Additionally, in the same murine lupus model, SERPINA3 mRNA levels were described as elevated in the kidney, spleen and liver tissue when compared with control mice. Serum levels of SERPINA3 were increased with both active renal and extrarenal SLE as compared with healthy controls in this pilot study, although the most significant increase was observed with active renal disease (1.87-fold increase, P < 0.0001) [41].
Further research is needed to better delineate the role of serpins in the setting of inflammation, i.e. determine whether serpins help control inflammation or contribute to increased persistent inflammation or instead play more specific roles. While we understand that serpins certainly function in acute inflammation, less is known regarding a potential role in chronic disease. Interestingly, SERPINA1 has been found to stain diffusely in sclerotic glomeruli and atrophic tubules of patients with chronic diffuse proliferative glomerulonephritis and chronic pyelonephritis [42, 43]. SERPINA3 has also been described as upregulated in the tubulointerstitium of kidneys from cadaveric vs live transplant donors [44], again implicating serpins as indicators of potential damage.
As a renal biomarker, the C-terminal fragment of SERPINA3 has been shown to indicate acute renal allograft rejection. SERPINA3 was identified as a predictor of acute rejection using a proteomics approach, and in a follow-up study, SERPINA3 was shown to be elevated in the urine of patients with acute rejection [45, 46]. A separate study confirmed the potential role of SERPINA3 as a biomarker of acute rejection, demonstrating significantly decreased SERPINA3 plasma levels in those patients with acute rejection [47]. SERPINA3 expression was additionally discovered to be upregulated in the glomeruli and tubulointerstitium of patients with diabetic kidney disease through transcriptome analysis [48].
Based on the current literature in combination with our study findings, it seems plausible that SERPINA3 could have a relevant biological role in LN disease mechanisms. We must first establish whether elevated urine SERPINA3 is reactive to ongoing inflammation or instead functioning more specifically in the LN disease process. In future validation studies, determining the fractional excretion of SERPINA3 may be a useful way of determining the source of SERPINA3 (circulation vs endogenous renal). It will also be important to perform SERPINA3 immunostaining in both healthy kidneys and other renal pathologies to better understand if SERPINA3 could have a unique role in LN. One limitation of this study was the limited sample size due to it being a single-centre, pilot study. Confirmation of our study findings in independent cohorts is needed to elucidate the potential role of SERPINA3 in predicting LN activity. Additionally, none of the patients in our cohorts had either a very high degree of LN chronicity or an NIH-CI of 0 on biopsy, making optimal stratification of our chronicity groupings challenging; however, this was a study of childhood-onset disease, and generally our patients do not present with a high degree of chronicity and often do not get biopsies in the clinical absence of LN. Finally, it was difficult to obtain urine proteomic signatures on healthy controls given lack of proteinuria in patients without underlying disease. For this reason, urine from LN patients with low chronicity was utilized as the comparator and may in fact be better suited as a control group given that we will be using this marker clinically to distinguish specifically between LN patients with and without renal damage. Strengths of our study included use of the criterion standard of NIH-CI and NIH-AI on biopsy to classify our patient groups for analysis, lending support to SERPINA3 as an activity marker, as it can be challenging to differentiate clinical from histological LN activity.
In summary, we demonstrated that urine SERPINA3 holds potential as a novel biomarker of LN activity. Further studies will be important to establish if urine SERPINA3 can be used to direct clinical treatment decisions for LN patients and to delineate the role of SERPINA3 in LN pathogenesis.
Supplementary Material
Acknowledgements
Mass spectrometry data were collected on a system funded through an NIH shared instrumentation grant (S10 RR027015-01; KD Greis-PI). We would like to thank Qing Ma for assisting in performing SERPINA3 immunohistochemistry.
Funding: This work was supported by grants from the National Institutes of Health [P50 DK 096418, U01 AR065098, T32 AR069512, P30 AR070549].
Disclosure statement: The authors have declared no conflicts of interest.
References
- 1. Rianthavorn P, Buddhasri A.. Long-term renal outcomes of childhood-onset global and segmental diffuse proliferative lupus nephritis. Pediatr Nephrol 2015;30:1969–76. [DOI] [PubMed] [Google Scholar]
- 2. Alvarado A, Malvar A, Lococo B. et al. The value of repeat kidney biopsy in quiescent Argentinian lupus nephritis patients. Lupus 2014;23:840–7. [DOI] [PubMed] [Google Scholar]
- 3. Weening JJ, D’Agati VD, Schwartz MM.. The classification of glomerulonephritis in systemic lupus erythematosus revisited. Kidney Int 2004;65:521–30. [DOI] [PubMed] [Google Scholar]
- 4. Austin HA, Muenz LR, Joyce KM. et al. Prognostic factors in lupus nephritis. Contribution of renal histologic data. Am J Med 1983;75:382–91. [DOI] [PubMed] [Google Scholar]
- 5. Austin HA, Muenz LR, Joyce KM, Antonovych TT, Balow JE.. Diffuse proliferative lupus nephritis: identification of specific pathologic features affecting renal outcome. Kidney Int 1984;25:689–95. [DOI] [PubMed] [Google Scholar]
- 6. Alaiya A, Assad L, Alkhafaji D. et al. Proteomic analysis of Class IV lupus nephritis. Nephrol Dial Transplant 2015;30:62–70. [DOI] [PubMed] [Google Scholar]
- 7. Zhang X, Jin M, Wu H. et al. Biomarkers of lupus nephritis determined by serial urine proteomics. Kidney Int 2008;74:799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bennett M, Brunner HI.. Biomarkers and updates on pediatrics lupus nephritis. Rheum Dis Clin North Am 2013;39:833–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li Y, Fang X, Li QZ.. Biomarker profiling for lupus nephritis. Genomics Proteomics Bioinformatics 2013;11:158–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Oates JC, Varghese S, Bland AM. et al. Prediction of urinary protein markers in lupus nephritis. Kidney Int 2005;68:2588–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cutillas P, Burlingame A, Unwin R.. Proteomic strategies and their application in studies of renal function. News Physiol Sci 2004;19:114–9. [DOI] [PubMed] [Google Scholar]
- 12. Suzuki M, Ross GF, Wiers K. et al. Identification of a urinary proteomic signature for lupus nephritis in children. Pediatr Nephrol 2007;22:2047–57. [DOI] [PubMed] [Google Scholar]
- 13. Ross PL, Huang YN, Marchese JN. et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics 2004;3:1154–69. [DOI] [PubMed] [Google Scholar]
- 14. Tan E, Cohen AS, Fries JF. et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 1982;25:1271–7. [DOI] [PubMed] [Google Scholar]
- 15. Hochberg MC. Updating the American college of rheumatology revised criteria for classification of systemic lupus erythematosus. Arthritis Rheumatol 1997;40:1725. [DOI] [PubMed] [Google Scholar]
- 16. Schwartz GJ, Work DF.. Measurement and estimation of GFR in children and adolescents. Clin J Am Soc Nephrol 2009;4:1832–43. [DOI] [PubMed] [Google Scholar]
- 17. Gladman DD, Ibanez D, Urowitz MB.. Systemic lupus erythematosus disease activity index 2000. J Rheumatol 2002;29:288–91. [PubMed] [Google Scholar]
- 18. Bennett MR, Pleasant LT, Haffner C. et al. A novel biomarker panel to identify steroid resistance in childhood idiopathic nephrotic syndrome. Biomark Insights 2017;12:1177271917695832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kaimal V, Bardes EE, Tabar SC, Jegga AG, Aronow BJ.. ToppCluster: a multiple gene list feature analyzer for comparative enrichment clustering and network-based dissection of biological systems. Nucleic Acids Res 2010;38:W96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shannon P, Markiel A, Ozier O. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dieplinger H, Ankerst DP, Burges A. et al. Afamin and apolipoprotein A-IV: novel protein markers for ovarian cancer. Cancer Epidemiol Biomarkers Prev 2009;18:1127–33. [DOI] [PubMed] [Google Scholar]
- 22. Dieplinger H, Dieplinger B.. Afamin—A pleiotropic glycoprotein involved in various disease states. Clin Chim Acta 2015;446:105–10. [DOI] [PubMed] [Google Scholar]
- 23. Maluf D, Mas V.. Molecular pathways involved in loss of kidney graft function with tubular atrophy and interstitial fibrosis. Mol Med 2008;14:5–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jennings P, Crean D, Aschauer L. et al. Interleukin-19 as a translational indicator of renal injury. Arch Toxicol 2015;89:101–6. [DOI] [PubMed] [Google Scholar]
- 25. Banerjee A, Dasgupta S, Mukhopadhyay BP, Sekar K.. The putative role of some conserved water molecules in the structure and function of human transthyretin. Acta Crystallogr Sect D Biol Crystallogr 2015;71:2248–66. [DOI] [PubMed] [Google Scholar]
- 26. Cabré A, Lázaro I, Girona J. et al. Retinol-binding protein 4 as a plasma biomarker of renal dysfunction and cardiovascular disease in type 2 diabetes. J Intern Med 2007;262:496–503. [DOI] [PubMed] [Google Scholar]
- 27. Henze A, Frey SK, Raila J. et al. Alterations of retinol-binding protein 4 species in patients with different stages of chronic kidney disease and their relation to lipid parameters. Biochem Biophys Res Commun 2010;393:79–83. [DOI] [PubMed] [Google Scholar]
- 28. Fattori E, Della Rocca C, Costa P. et al. Development of progressive kidney damage and myeloma kidney in interleukin-6 transgenic mice. Blood 1994;83:2570–9. [PubMed] [Google Scholar]
- 29. Aveles PR, Criminácio CR, Gonçalves S. et al. Association between biomarkers of carbonyl stress with increased systemic inflammatory response in different stages of chronic kidney disease and after renal transplantation. Nephron Clin Pract 2010;116:c294. [DOI] [PubMed] [Google Scholar]
- 30. Abulaban KM, Song H, Zhang X. et al. Predicting decline of kidney function in lupus nephritis using urine biomarkers. Lupus 2016;25:1012–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heit C, Jackson BC, McAndrews M. et al. Update of the human and mouse SERPIN gene superfamily. Hum Genomics 2013;7:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Law RHP, Zhang Q, McGowan S. et al. An overview of the serpin superfamily. Genome Biol 2006;7:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gatto M, Iaccarino L, Ghirardello A. et al. Serpins, immunity and autoimmunity: old molecules, new functions. Clin Rev Allergy Immunol 2013;45:267–80. [DOI] [PubMed] [Google Scholar]
- 34. Baker C, Belbin O, Kalsheker N, Morgan K.. SERPINA3 (aka alpha-1-antichymotrypsin). Front Biosci 2007;12:2821–35. [DOI] [PubMed] [Google Scholar]
- 35. Lieberman J, Sastre A.. Serum angiotensin converting enzyme levels in patients with alpha1-antitrypsin variants. Am J Med 1986;81:821–4. [DOI] [PubMed] [Google Scholar]
- 36. Janciauskiene SM, Bals R, Koczulla R. et al. The discovery of α1-antitrypsin and its role in health and disease. Respir Med 2011;105:1129–39. [DOI] [PubMed] [Google Scholar]
- 37. Kalsheker N, Morley S, Morgan K.. Gene regulation of the serine proteinase inhibitors α1-antitrypsin and α1-antichymotrypsin. Biochem Soc Trans 2002;30:93–8. [DOI] [PubMed] [Google Scholar]
- 38. Zager RA, Johnson ACM, Frostad KB.. Rapid renal alpha-1 antitrypsin gene induction in experimental and clinical acute kidney injury. PLoS One 2014;9:e98380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Daemen MARC, Heemskerk VH, van’t Veer C. et al. Functional protection by acute phase proteins α1-acid glycoprotein and α1-antitrypsin against ischemia/reperfusion injury by preventing apoptosis and inflammation. Circulation 2000;102:1420–6. [DOI] [PubMed] [Google Scholar]
- 40. Shapira E, Proscura E, Brodsky B, Wormser U.. Novel peptides as potential treatment of systemic lupus erythematosus. Lupus 2011;20:463–72. [DOI] [PubMed] [Google Scholar]
- 41. Hutcheson J, Vanarsa K, Min S, Wu TMC. . Dysregulation of clade a serine protease inhibitor expression in murine and human lupus. Arthritis Rheum 2011;63(Suppl):Abstract 564. [Google Scholar]
- 42. Yonezawa S, Sato E, Okamoto K.. Polyanion, immunoprotein and proteinase inhibitor in ischemic glomerular change. Nephron 1981;28:105–11. [DOI] [PubMed] [Google Scholar]
- 43. Yonezawa S, Irisa S, Nakamura T. et al. Deposition of α1-antitrypsin and loss of glycoconjugate carrying Ulex europaeus agglutinin-1 binding sites in glomerular sclerotic process. Nephron 1983;33:38–43. [DOI] [PubMed] [Google Scholar]
- 44. Kainz A, Mitterbauer C, Hauser P. et al. Alterations in gene expression in cadaveric vs. live donor kidneys suggest impaired tubular counterbalance of oxidative stress at implantation. Am J Transplant 2004;4:1595–604. [DOI] [PubMed] [Google Scholar]
- 45. O’Riordan E. Bioinformatic analysis of the urine proteome of acute allograft rejection. J Am Soc Nephrol 2004;15:3240–8. [DOI] [PubMed] [Google Scholar]
- 46. O’Riordan E, Orlova TN, Podust VN. et al. Characterization of urinary peptide biomarkers of acute rejection in renal allografts. Am J Transplant 2007;7:930–40. [DOI] [PubMed] [Google Scholar]
- 47. Ziegler ME, Chen T, LeBlanc JF, Wei X, Gjertson DW.. Apolipoprotein A1 and C-terminal fragment of α-1 antichymotrypsin are candidate plasma biomarkers associated with acute renal allograft rejection. Transplantation 2011;92:388–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Woroniecka KI, Park ASD, Mohtat D. et al. Transcriptome analysis of human diabetic kidney disease. Diabetes 2011;60:2354–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.