Abstract
Intracranial electro-encephalography (icEEG) provides a unique opportunity to record directly from the human brain and is clinically important for planning epilepsy surgery. However, traditional visual analysis of icEEG is often challenging. The typical simultaneous display of multiple electrode channels can prevent an in-depth understanding of the spatial-time course of brain activity. In recent decades, advances in the field of neuroimaging have provided powerful new tools for the analysis and display of signals in the brain. These methods can now be applied to icEEG to map electrical signal information onto a three-dimensional rendering of a patient’s cortex and graphically observe the changes in voltage over time. This approach provides rapid visualization of seizures and normal activity propagating over the brain surface and can also illustrate subtle changes that might be missed by traditional icEEG analysis. In addition, the direct mapping of signal information onto accurate anatomical structures can assist in the precise targeting of sites for epilepsy surgery and help correlate electrical activity with behavior. Bringing icEEG data into a standardized anatomical space will also enable neuroimaging methods of statistical analysis to be applied. As new technologies lead to a dramatic increase in the rate of data acquisition, these novel visualization and analysis techniques will play an important role in processing the valuable information obtained through icEEG.
Keywords: intracranial EEG, cortical power projections, data visualization
Originally introduced in the early 1900s, intracranial electroencephalography (icEEG) has become an important method in neurological diagnosis, particularly in patients with epilepsy. Using electrodes implanted directly on the surface and depths of the brain, this technique can assist physicians in identifying regions of seizure onset and in mapping normal functional areas of the cerebral cortex. However, there are certain challenges faced by clinicians using icEEG as a diagnostic tool, such as the simultaneous visualization of many electrode channels arising from anatomically disparate regions. Epileptologists who use this technique for planning surgical resection are also confronted with localizing a three-dimensional ictal onset region using two-dimensional EEG information. New high-density arrays that combine multiple subdural strips and depth electrodes will continue to increase the number of channels collected during icEEG recordings, making more intuitive and easily interpreted visual representations critical for efficient data processing.
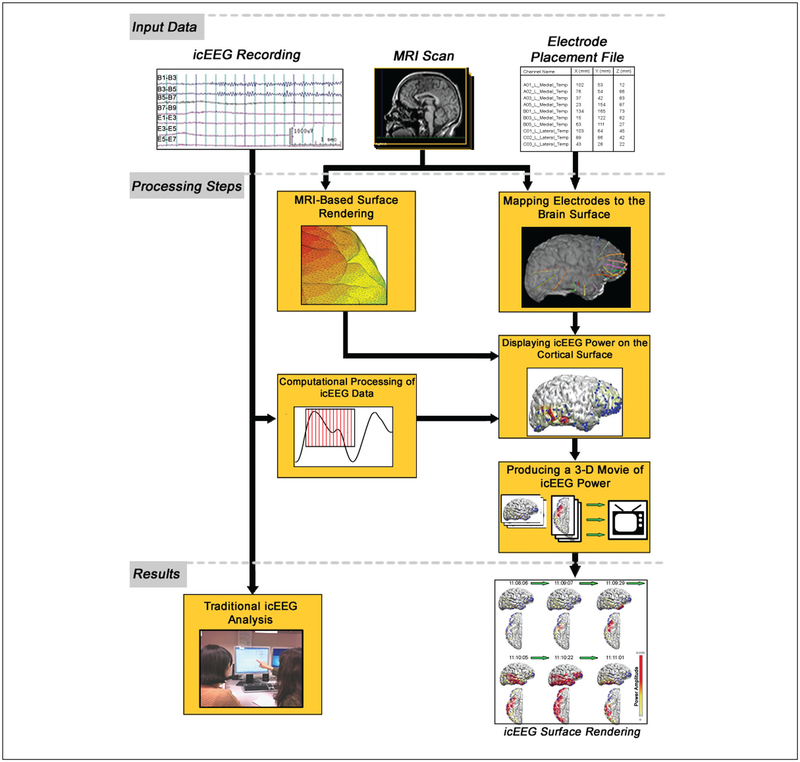
Over the past 20 years, developments in the field of neuroimaging have established widely used methods for making computational models of the human cortex and precisely localizing changes in the brain. By combining these analysis techniques with icEEG information, we can now visualize electrical signals on a cortical surface and illustrate the spatial time course of the collected data. Signal processing and other forms of computational analysis can provide further insight, particularly when displayed in the context of each patient’s unique anatomy. Implementing this approach requires a digital set of icEEG recordings, MRI scans of the patient’s brain, and a map of all electrode locations co-registered with neuroimaging data (Fig. 1). Computational software can assist in integrating these data sets into a three-dimensional, colored projection of icEEG information (Table 1). Data presented in this form can assist both researchers and medical providers in understanding the time course of electrical signals in the brain, using the kind of analysis typically employed in functional neuroimaging to obtain new insights into neural activity during health and disease.
Figure 1.
By combining an intracranial EEG recording, MRI scan, and electrode placement file, an icEEG surface rendering can be generated that displays the spatial time course of collected data (icEEG = intracranial electro-encephalography)
Table 1.
Examples of Software Packages that Can Be Used to Render icEEG Surface Data
| Name | Developer | Comments |
|---|---|---|
| MRI-based surface rendering | ||
| BioImage Suite 3.01—Orthogonal Viewer | Yale University | A segmentation-based method with a relatively short runtime. The resulting cortical surface can be exported as a text file that stores the vertex and face coordinates of the surface. |
| FreeSurfer 5.1.0 | Harvard/MGH | A segmentation-based process for generating high-definition cortical surfaces with a relatively long run-time. Includes several other anatomical analysis tools. |
| Curry 7—Image Analysis Package | Compumedics | A segmentation algorithm is used to generate a triangular mesh surface. |
| Constrained Laplacian Anatomic Segmentation using Proximity (CLASP) | McConnell Brain Imaging Center | An iterative morphing-based technique that minimizes the repulsion-based cost function by deforming a voxel-enclosed surface. |
| Mapping electrodes to the brain surface | ||
| BioImage Suite 3.01—Electrode Editor | Yale University | The electrode editor allows export of a map file with x/y/z coordinates in the MRI space. |
| Statistical Parametric Mapping (SPM) | University College London | A software suite that executes in Matlab. It can also be used for fMRI, SPECT, MEG, and EEG analysis. |
| Flirt 5.5 | University of Oxford | An automated tool for linear image registration between different acquisition methods (i.e., MRI and CT). |
| Curry 7—Image Analysis Package | Compumedics | Electrodes are mapped to the cortical surface through co-registration of MRI and CT images using reference landmarks. |
| Computational processing of icEEG data | ||
| Matlab 7.13 (R2011B) | Mathworks | This software provides a programming environment in which scripts can be written to process electrical data. |
| BCI2000 | Wadsworth Center | A broad collection of signal processing routines and experimental paradigms that are free for research and academic institutions. |
| Displaying icEEG data on the cortical surface | ||
| Matlab 7.13 (R2011B) | Mathworks | The MRI-based mesh surface and postprocessed icEEG data can be combined using the “plot,” “surf,” and “contour” functions in a custom-written script. |
| Curry 7—Image Analysis Package | Compumedics | Data can be rendered on a three-dimensional cortex or shown on a two-dimensional scalp plot. The user retains full control over viewing angle and zoom. |
| Brain Computer Interface 2000 | Wadsworth Center | The SIGFRIED module of BCI2000 can be used to process and display data obtained through icEEG. Data can also be exported and visualized using external software. |
| Producing a 3D color movie of icEEG information | ||
| Matlab 7.13 (R2011B) | Mathworks | After generating projections for each frame of an icEEG recording, code can easily be written to assemble the files into a single *.avi or *.mpg movie. |
| Curry 7—Image Analysis Package | Compumedics | Projections are rendered in real time and allow threedimensional rotation of the cortical surface at any point during playback. |
IcEEG = intracranial electro-encephalography.
As icEEG acquisition tools become increasingly sophisticated, continued development of advanced quantitative techniques is crucial. Providing clinicians with novel and insightful visualization methods could lead to more accurate diagnosis and enhanced treatment of patients with epilepsy and other neurological disorders. Researchers will also benefit from enhanced computational methods that provide unique insight into electrical activity in the brain. This review will explore the basic steps involved in generating icEEG cortical projections, including MRI surface rendering, mapping electrodes to the brain surface, computational data processing, projecting electrical channels onto the cortex, and assembling a three-dimensional movie of icEEG data (Fig. 1; Supplementary Video 1). A brief description of traditional icEEG analysis is also included to provide background and clinical context.
Traditional icEEG Analysis
Intracranial EEG monitoring has long been used as part of presurgical evaluation in patients with medically refractory seizures. Originally developed to precisely localize epileptogenic zones, it is primarily used in cases where noninvasive testing is inconclusive or presurgical mapping of cortical function is required. Different approaches have been studied when interpreting icEEG recordings to improve our abilities at locating seizure onset zones, functional cortical areas, and epileptic networks (Momjian and others 2003).
One such approach is analysis of icEEG power, which has frequently been investigated in both animal and human models. Previous work has examined power bands in the beta, delta, and gamma frequency ranges and attempted to correlate spectral activity to both ictal and interictal periods (Ebersole and Pedley 2002). Recently, much interest has focused on the relationship between high-frequency activity and the region of seizure onset (Gupta and others 2011; Jacobs and others 2010; Schevon and others 2009). It has been suggested that high-frequency oscillations (HFOs) of 250 to 500 Hz are excellent markers for locating the epileptogenic zone, and studies have demonstrated that the removal of HFO-generating tissue increases the likelihood of positive surgical outcomes (Jacobs and others 2010). Until recently, the ability to detect HFOs has been limited in most intracranial acquisition systems, which have a sampling frequency of 0.5 to 2 kHz and use a low pass filter of 100 to 500 Hz (Jirsch and others 2006). Higher bandwidth systems are now becoming increasingly available for icEEG recordings.
To study the dynamic changes seen during icEEG recordings, power spectral analysis is often combined with topographic mapping to illustrate seizure onset and propagation (Akiyama and others 2006; Englot and others 2010). This technique provides important insight in identifying the epileptic network and characterizing regions that should be targeted during surgical resection. Given the limitations of pure visual analysis of icEEG recordings, further development of similar techniques is essential for advancing our understanding of seizure onset, seizure propagation, and epileptic networks. The use of three-dimensional icEEG surface rendering is one such technique that shows promising potential.
MRI-Based Surface Rendering
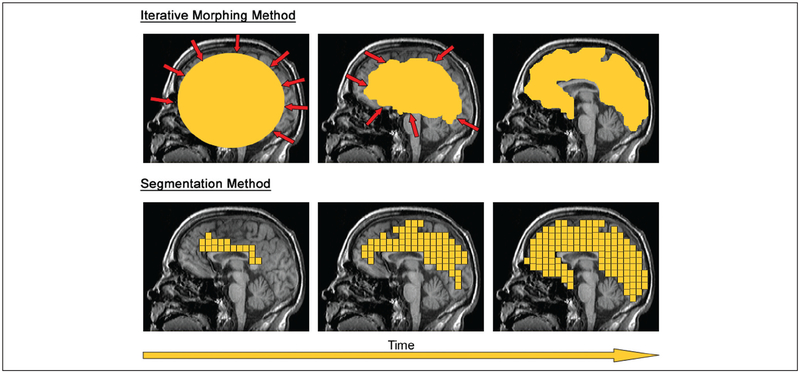
Before EEG information can be projected onto a patient’s brain, two-dimensional MRI scans must be converted into a set of three-dimensional coordinates that accurately describe the cortical surface. A mesh model composed of polygon faces and vertices is typically chosen for this representation because of its efficiency at continuously encoding complex topographies (Carman 1995). Computational methods for performing this transformation have been widely studied since at least the early 1990s, when a wire-frame model was proposed for displaying the sulci and gyri of a cortical sheet (Dale and Sereno 1993). Building on algorithms originally developed for face recognition (Yuille 1991), early methods used iterative deformations of a voxel-constrained sphere to define the interface between white and grey matter in a patient’s brain (Fig. 2, top). Repulsion forces generated by neighboring vertices and areas of MRI contrast (representing the cortical-white matter boundary) determined the final form of the resulting object: a minimal energy state. A mesh grid was tessellated over the sphere-enclosed voxels to create a three-dimensional model of the cortical surface. Since first being described, this algorithm has been progressively refined (Han and others 2001; MacDonald 1998) and is currently implemented in several freely available software packages, such as CLASP (Kim and others 2005).
Figure 2.
Computational cortical models can be generated from MRI data using an iterative morphing (top panels) or segmentation method (bottom panels)
An alternative method for cortical surface rendering is based on segmentation of MRI volumes through tissue classification (Shattuck and Leahy 2001) (Fig. 2, bottom). Segmentation-based surface rendering has been implemented in a number of software platforms, including BioImage Suite, FreeSurfer, and Curry (Table 1). This “bottom-up” approach begins with a small initial region that is incrementally grown through addition of topologically similar voxels. Several improvements to this technique have been proposed since its original introduction, and most variations now employ a “marching cubes” algorithm to arrive at a cortex-constrained surface (Lorensen and Cline 1987). After an accurate model has been constructed from the MRI, tessellation is used to create a polygon mesh. Although “segmentation” is generally considered a more geometrically accurate technique than “iterative morphing,” incorrectly classified voxels (i.e., failure to detect the correct tissue type) can arise from noise and inhomogeneity artifacts (Lee and others 2006).
Developments in the field of cortical surface rendering have largely progressed along either the “iterative morphing” or “segmentation” methods described above (Liu and others 2008). Because the “iterative morphing” method does not use voxel-based classification, noise and artifacts are negligible contributors to the final surface. However, the minimal energy algorithm associated with this technique often fails to accurately represent deep sulci in the brain (Manceaux-Demiau and others 1998). There are trade-offs for each approach that should be considered in the context of the MRI quality and the patient’s cortical topography. Ultimately, the selection of a high-quality rendering algorithm is an important consideration in both clinical work and research.
Mapping Electrodes to the Brain Surface
After generating a computational model of the patient’s brain, the surface must be co-registered with electrode placement information that was recorded during icEEG implantation. Understanding the precise location of each electrode is critical for correct mapping of electrical signals to anatomical regions. A number of imaging techniques have been employed to localize implanted intracranial electrodes, including a postimplant MRI, curvilinear reformatting of the preimplant MRI, digital photography co-registration, and CT/MRI co-registration (LaViolette and others 2011; Tao and others 2009). Currently, CT/MRI co-registration is the most commonly used technique in most epilepsy centers. This method offers the advantages of accurate electrode visualization through CT and high anatomical detail through MRI. Co-registration of CT and MRI occurs through either a reference point–based method (using standard landmarks or external fiducial markers) or an automatic method based on cross-modal image comparison.
Several software packages currently exist for performing the transformation necessary to map electrodes to the brain surface (Table 1). Curry (http://www.neuroscan.com/curry.cfm) is an example of a multimodal imaging software package that employs the reference point–based method. It co-registers the MRI and CT using five landmark points: nasion, preauricular left, preauricular right, inion, and vertex. The subdural electrodes are segmented from postoperative CT images using a thresholding technique and then transformed from CT images onto the MRI (Bai and others 2011; Tao and others 2009). The co-registration error typically ranges from 3 to 10 mm due to the reliability of fiducial markers (Tao and others 2009).
The automatic co-registration technique is implemented in several software packages, including BioImage Suite (http://www.bioimagesuite.org), SPM (http://www.fil.ion.ucl.ac.uk/spm/), and FLIRT (http://www.fmrib.ox.ac.uk/fsl/flirt/index.html). Developed at Yale University, BioImage Suite co-registers postoperative MRI and CT using a six-parameter rigid transformation. A grid-based transformation is then used to co-register the postoperative and preoperative MRI images. This accounts for any distortion of the brain arising from electrode implantation and the craniotomy (Papademetris and others 2009). SPM uses a similar approach that includes a normalized mutual information routine. In the FLIRT algorithm, an efficient search method is applied across many possible registrations to find the optimal transformation. The desired registration will be a global cost minimum with respect to modality overlap. With all these registration methods, a reference intra-operative photo of the electrodes in situ can be useful to correct any residual registration error according to the anatomic shapes of the blood vessels and sulci (LaViolette and others 2011).
Computational Processing of icEEG Data
An accurate cortical mesh model combined with precisely localized electrodes can be used to project icEEG information onto a graphical representation of a patient’s cortex. Although it is possible to display raw broadband amplitude from the recorded signal, the electrical data often undergo computational processing before being displayed on the mesh surface. One common processing method is power spectral analysis, which breaks a channel’s power information into clinically useful, discrete frequency bands. Using this type of analysis, specific bands such as Beta or Delta can be filtered out of the acquired signal and used to localize areas of interest during a seizure or normal cortical activity. Previous studies have found correlations between interictal high-frequency signals and seizure onset regions, suggesting this type of analysis could be useful in a clinical setting (Gupta and others 2011; Jacobs and others 2010; Schevon and others 2009). Signal frequency has also been used as a marker to map normal cortical activity such as language, motor, and other functions (Chang and others 2010; Engell and McCarthy 2010).
Another common form of signal processing is to search for coherence between channels in an icEEG recording. Coherence analysis has widely been used to investigate EEG signals under normal conditions (Bullock and others 1995; Weiss and Mueller 2003); however, recent work has employed this technique to gain insight into abnormal brain function. One recent study used functional connectivity to elucidate the epileptic network in patients undergoing cortical resectioning (Zaveri and others 2009). Connectivity was defined between channels using a frequency-indexed correlation coefficient and was evaluated for all pairs of contacts ipsilateral to the seizure-onset area. The results suggested that significant connectivity exists in the area around the seizure-onset zone and that the correlation is inversely related to distance. Processing for coherence is also commonly used in brain-computer interface research. In one recent study, event-related desynchronization was used to detect patient intent for shoulder abduction and elbow flexion (Zhou and others 2009).
In addition to those mentioned above, many other methods are used to process icEEG signals. BCI2000 is a development platform for signal analysis that includes spatial and temporal filters, linear classifiers, and other signal operators (Schalk and others 2004). Software such as MATLAB (http://www.matlab.com) is a common environment for implementing various other EEG analysis methods, including Teager energy, mutual information, approximate entropy, and time-frequency distributions. In principle, any one of these methods could be similarly rendered on the cortical surface.
Displaying icEEG Data on the Cortical Surface
To visualize icEEG signal information, areas proximal to each electrode can be colored according to the amplitude of the channel’s processed data (Cervenka and others 2011; Englot and others 2010; Voytek and others 2010). A color scale is often established based on the range of values present in the electrical signal data set. The continuous icEEG signal must be broken into discrete time bins that define each brain surface polygon’s value for a particular display frame (e.g., each frame may be chosen to display 0.5 s of icEEG time). The decision of a sampling window size will determine the level of variability and specificity present in an icEEG time course projection (Welch 1967). Although a large frame period will smooth out undesired signal noise, if the window is too large, important epileptic events may also be masked. Selection of an appropriate window size will often depend on the signal-to-noise ratio present in an icEEG data set as well as the sampling rate of the acquisition system. If a power spectral analysis was performed, the sampling window used for the Fourier transform sets a lower limit on the period of time that can be resolved in each display frame.
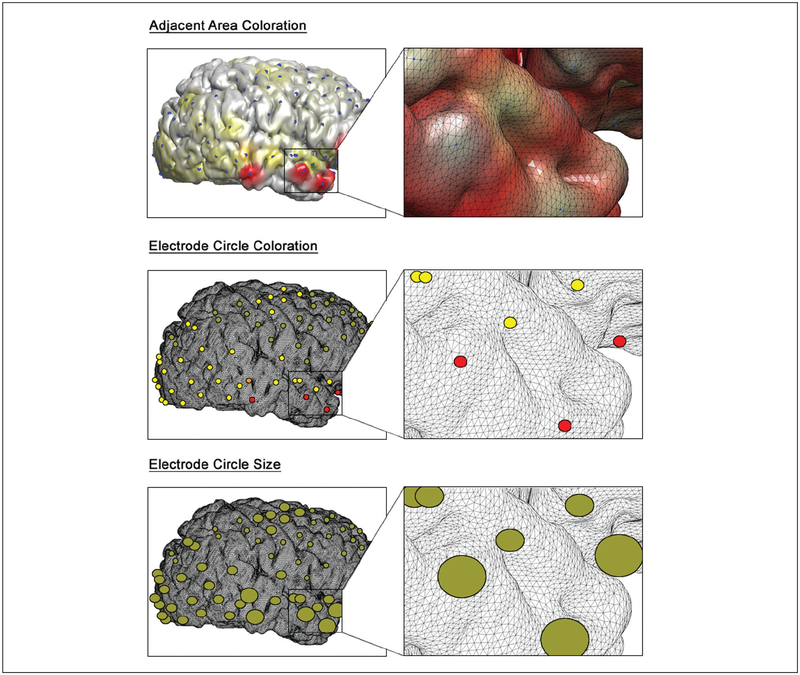
Various approaches have been used when determining the scope and magnitude of coloration for a particular icEEG data set. Although many investigators elect to plot electrical signals on a two-dimensional topographic map (Ebner and others 2011; Gupta and others 2011; Voytek and others 2010), using a three-dimensional template can provide further insight on the anatomical structures involved. Previous studies have used a nearest neighbor method where cortical polygons are colored strongest when near an electrode and fade linearly to zero as distance is increased (Englot and others 2010; Gunduz and others 2012) (Fig. 3, top). The spatial resolution of electrode coverage is one parameter that may be used in determining the rate of decay for polygon opacity. In other cases, small circles are placed at each electrode location and are colored according to the icEEG data (Cervenka and others 2011) (Fig. 3, middle). The coloration scale can be determined by the maximum power intensity or other choice of threshold, which will of course influence interpretation of the results. Some studies have avoided coloration entirely, opting instead to plot scalable circles and size them proportionally to the amplitude of each channel’s signal (Besle and others 2011) (Fig. 3, bottom).
Figure 3.
Common methods for displaying intracranial EEG data on a cortical surface include coloration of areas adjacent to each electrode (top), coloration of electrode-defined circles (middle), and sizing of electrode-defined circles proportional to signal strength (bottom). All three methods show onset of high-frequency beta activity in the right temporal lobe during a seizure
Producing a Three-Dimensional Movie of icEEG Information
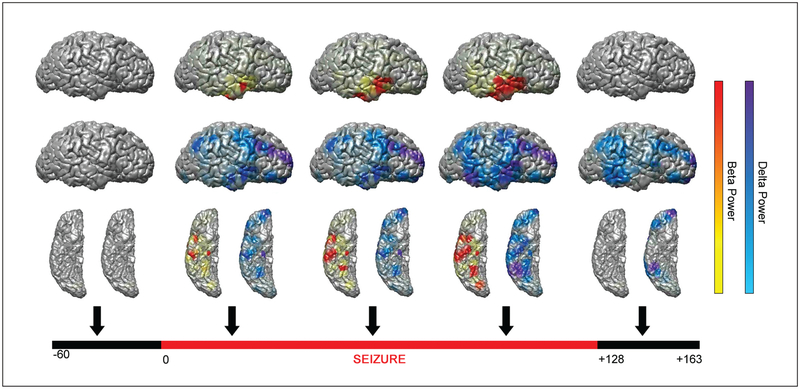
After surface projections of each incremental time period have been rendered, they can be combined to generate a three-dimensional color movie of icEEG signal information. This form of data presentation can be clinically useful for understanding the regions involved in seizure onset and propagation and can also help demonstrate sequential involvement of different cortical areas in normal information processing. Recent studies have used this technique to provide insight on electrical events in the brain and gain an understanding of time-related changes that might not be present in a traditional two-dimensional mapping (Akiyama and others 2006; Brunner and others 2009; Englot and others 2010) (Fig. 4; Supplementary Video 1).
Figure 4.
Time course of single frames from a three-dimensional color movie showing beta frequency (13–25 Hz, warm colors) and delta frequency (0.5–4 Hz, cool colors) power during a seizure arising from the right temporal lobe. Arrows indicate time samples for single frames from a video representing a moving 10 s window at 1 s increments. Signal power changes are normalized relative to 60 s baseline. Lateral (top two rows) and inferior (bottom row) views are shown for the right hemisphere at each time point. For video of the full time course see Supplementary Video 1 online. Reproduced using data originally published in Englot and others (2010), by permission of Oxford University Press
Several important parameters should be considered when generating a movie, such as frame rate, the use of a time-averaged window, and the best view angles of the brain to display. Decisions that affect how data are visualized can have important consequences during interpretation of the results. In cases where noise is a significant component of the icEEG signal, processing techniques such as a continuous sliding window can be used to smooth out changes in the projection display (Englot and others 2010; Harris 1978). Averaging values for each electrode over several data points will not only reduce temporal resolution but also eliminate high-frequency fluctuations that have little neurological meaning. Finding the appropriate scale for a sliding window requires familiarization with the quality and variability of the icEEG signal, which can often be assessed during a baseline, interictal period.
In addition to other parameters that determine how icEEG data will be displayed in a movie, appropriate view angles must be chosen to highlight the anatomical areas important in a study. Because the three-dimensional cortex is viewed on a two-dimensional computer monitor, the view angle will determine what electrodes are visible during review. Lateral, medial, and inferior views are common choices to maximize electrode visibility (Bidet-Caulet and others 2007; Cervenka and others 2011; Englot and others 2010) (see Fig. 4; Supplementary Video 1). Some software packages (such as Curry) allow icEEG movies to be rotated in three-dimensions during playback. This gives the reviewer maximum control over the field of vision during analysis.
Conclusions and Future Directions
Made possible through incremental advances in neuroimaging and computational analysis, projecting icEEG signals onto the cortex is an innovative new technique that has tremendous potential for understanding brain activity in the context of an individual’s unique anatomy. It can provide clinicians and investigators with a rapid and intuitive tool for elucidating the electrical correlates of normal brain function and also lead to a broader understanding of signal propagation during periods of abnormal cortical activity. In settings such as a clinical epilepsy monitoring unit, this method can be a powerful way to identify seizure onset areas and understand the anatomical regions involved in ictal propagation. Research on normal brain function will also benefit from this form of data presentation, which integrates well with other computational methods and offers a highly visual, improved alternative to traditional icEEG data display. By bringing icEEG data into a standard anatomical space, it is also possible to perform group statistical analyses across events or across subjects, using methods similar to those employed in functional neuroimaging. Updating the old saying “a picture is worth a thousand words,” we propose that “a moving picture is worth hundreds of squiggly lines.”
Much potential exists for future developments in icEEG surface renderings, including increased accuracy of cortical reconstruction, improvements in spatial and temporal resolution, and determination of the optimal data processing methods. The adaptability of this method to different blends of data processing is a major benefit of this technique and will allow users to alter their analyses as improvements become available. As with all forms of neuroimaging analysis, the highly compelling visual nature of the results is also a major potential pitfall, and additional work is needed to rigorously establish the optimal thresholds, parameters, and statistical significance levels tht should be used to interpret the displayed data. As improved technology allows the number of electrodes and recording bandwidth to increase in the coming years, innovative methods of icEEG analysis based on neuroimaging will greatly improve our understanding of these valuable sets of human data.
Acknowledgments
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by NIH R01NS055829, R01NS066974, R01MH67528, R01HL059619, P30NS052519, U01NS045911, CTSA UL1 RR0249139, a Donaghue Foundation Investigator Award, and the Betsy and Jonathan Blattmachr Family [HB]; as well as by the Ewha Global Top 5 Grant 2011 of Ewha Womans University and the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology [R01-2011-0015788 to HWL].
Footnotes
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- Akiyama T, Otsubo H, Ochi A, Galicia EZ, Weiss SK, Donner EJ, and others. 2006. Topographic movie of ictal high frequency oscillations on the brain surface using subdural EEG in neo-cortical epilepsy. Epilepsia 47(11):1953–7. [DOI] [PubMed] [Google Scholar]
- Bai X, Towle V, Van Drongelen W, He B. 2011. Cortical potential imaging of somatosensory evoked potentials by means of the boundary element method in pediatric epilepsy patients. Brain Topogr 23:333–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besle J, Schevon CA, Mehta AD, Lakatos P, Goodman RR, McKhann GM, and others. 2011. Tuning of the human neo-cortex to the temporal dynamics of attended events. J Neurosci 31(9):3176–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidet-Caulet A, Fischer C, Besle J, Aguera PE, Giard MH, Bertrand O. 2007. Effects of selective attention on the electrophysiological representation of concurrent sounds in the human auditory cortex. J Neurosci 27(35):9252–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner P, Ritaccio AL, Lynch TM, Emrich JF, Wilson JA, Williams JC, and others. 2009. A practical procedure for real-time functional mapping of eloquent cortex using electrocorticographic signals in humans. Epilepsy Behav 15(3):278–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock T, McClune M, Achimowicz J, Iragui-Madoz V, Duckrow R, Spencer S. 1995. Temporal fluctuations in coherence of brain waves. Proc Natl Acad Sci U S A 92(25):11568–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman G 1995. Computational methods for reconstructing and unfolding the cerebral cortex. Cerebr Cortex 5(6):506–17. [DOI] [PubMed] [Google Scholar]
- Cervenka MC, Boatman-Reich DF, Ward J, Franaszczuk PJ, Crone NE. 2011. Language mapping in multilingual patients: electrocorticography and cortical stimulation during naming. Front Hum Neurosci 5(13):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang EF, Rieger JW, Johnson K, Berger MS, Barbaro NM, Knight RT. 2010. Categorical speech representation in human superior temporal gyrus. Nat Neurosci 13(11):1428–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale AM, Sereno MI. 1993. Improved localization of cortical activity by combining EEG and MEG with MRI cortical surface reconstruction: a linear approach. J Cogn Neurosci 5(2):162–76. [DOI] [PubMed] [Google Scholar]
- Ebersole J, Pedley T. 2002. Current practice of clinical electroencephalography. Philadelphia (PA): Lippincott Williams & Wilkins. [Google Scholar]
- Ebner N, He Y, Fichtenholtz H, McCarthy G, Johnson M. 2011. Electrophysiological correlates of processing faces of younger and older individuals. Soc Cogn Affect Neurosci 6(4):526–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engell A, McCarthy G. 2010. Selective attention modulates face-specific induced gamma oscillations recorded from ventral occipitotemporal cortex. J Neurosci 30(26):8780–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englot DJ, Yang L, Hamid H, Danielson N, Bai X, Marfeo A, and others. 2010. Impaired consciousness in temporal lobe seizures: role of cortical slow activity. Brain 133(12):3764–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunduz A, Brunner P, Daitch A, Leuthardt E, Ritaccio A, Pesaran B, and others. 2012. Decoding covert spatial attention using electrocorticographic (ECoG) signals in humans. NeuroImage 60(4):2285–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta J, Marsh E, Nieh H, Porter B, Litt B. 2011. Discrete gamma oscillations identify the seizure onset zone in some pediatric epilepsy patients. Conf Proc IEEE Eng Med Biol Soc 2011:3095–8. [DOI] [PubMed] [Google Scholar]
- Han X, Xu C, Prince JL. 2001. A topology preserving deformable model using level sets. 2:765–70. [Google Scholar]
- Harris F 1978. On the use of Windows for harmonic analysis with the discrete Fourier transform. Proc IEEE 66(1):51–83. [Google Scholar]
- Jacobs J, Zijlmans M, Zelmann R, Chatillon C, Hall J, Olivier A, and others. 2010. High-frequency electroencephalographic oscillations correlate with outcome of epilepsy surgery. Ann Neurol 67(2):209–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirsch JD, Urrestarazu E, LeVan P, Olivier A, Dubeau F, Gotman J. 2006. High-frequency oscillations during human focal seizures. Brain 129(6):1593–608. [DOI] [PubMed] [Google Scholar]
- Kim JS, Singh V, Lee JK, Lerch J, Ad-Dab’bagh Y, MacDonald D, and others. 2005. Automated 3-D extraction and evaluation of the inner and outer cortical surfaces using a Laplacian map and partial volume effect classification. NeuroImage 27(1):210–21. [DOI] [PubMed] [Google Scholar]
- LaViolette P, Rand S, Ellingson B, Raghavan M, Lew S, Schmainda K, and others. 2011. 3D visualization of subdural electrode shift as measured at craniotomy reopening. Epilepsy Res 94(1–2):102–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, Lee JM, Kim JS, Kim IY, Evans AC, Kim SI. 2006. A novel quantitative cross-validation of different cortical surface reconstruction algorithms using MRI phantom. Neuro-Image 31(2):572–84. [DOI] [PubMed] [Google Scholar]
- Liu T, Nie J, Tarokh A, Guo L, Wong STC. 2008. Reconstruction of central cortical surface from brain MRI images: method and application. NeuroImage 40(3):991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorensen WE, Cline H. 1987. Marching cubes: a high resolution 3D surface construction algorithm. Comput Graphics 21(4):163–9. [Google Scholar]
- MacDonald D 1998. A method for identifying geometrically simple surfaces from three-dimensional images. Montreal, Canada: McGill University. [Google Scholar]
- Manceaux-Demiau A, Bryan RN, Davatzikos C. 1998. A probabilistic ribbon model for shape analysis of the cerebral sulci: application to the central sulcus. J Comput Assist Tomogr 22(6):962–71. [DOI] [PubMed] [Google Scholar]
- Momjian S, Seghier M, Seeck M, Michel C. 2003. Mapping of the neuronal networks of human cortical brain functions. Adv Tech Stand Neurosurg 28:92–142. [DOI] [PubMed] [Google Scholar]
- Papademetris X, DeLorenzo C, Flossmann S, Neff M, Vives K, Spencer D, and others. 2009. From medical image computing to computer-aided intervention: development of a research interface for image-guided navigation. Int J Med Robot 5(2):147–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalk G, McFarland D, Hinterberger T, Birbaumer N, Wolpaw J 2004. BCI2000: a general-purpose brain-computer interface (BCI) system. IEEE Trans Biomed Eng 51(6):1034–43. [DOI] [PubMed] [Google Scholar]
- Schevon C, Trevelyan A, Schroeder C, Goodman R, McKhann G, Emerson R. 2009. Spatial characterization of interictal high frequency oscillations in epileptic neocortex. Brain 132(11):3047–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shattuck DW, Leahy RM. 2001. Automated graph-based analysis and correction of cortical volume topology. IEEE Trans Med Imaging 20(11):1167–77. [DOI] [PubMed] [Google Scholar]
- Tao J, Hawes-Ebersole S, Baldwin M, Shah S, Erickson R, Ebersole J. 2009. The accuracy and reliability of 3D CT/MRI co-registration in planning epilepsy surgery. Clin Neurophysiol 120(4):784–53. [DOI] [PubMed] [Google Scholar]
- Voytek B, Canolty R, Shestyuk A, Crone N, Parvizi J, Knight R. 2010. Shifts in gamma phase-amplitude coupling frequency from theta to alpha over posterior cortex during visual tasks. Front Hum Neurosci 4:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S, Mueller H. 2003. The contribution of EEG coherence to the investigation of language. Brain Lang 85(2):325–43. [DOI] [PubMed] [Google Scholar]
- Welch P 1967. The use of fast Fourier transform for the estimation of power spectra: a method based on time averaging over short, modified periodograms. IEEE Trans Audio Electroacoustics 15(2):70–3. [Google Scholar]
- Yuille AL. 1991. Deformable templates for face recognition. J Cogn Neurosci 3(1):59–70. [DOI] [PubMed] [Google Scholar]
- Zaveri H, Pincus S, Goncharova I, Duckrow R, Spencer D, Spencer S. 2009. Localization-related epilepsy exhibits significant connectivity away from the seizure-onset area. Neuroreport 20(9):891–5. [DOI] [PubMed] [Google Scholar]
- Zhou J, Yao J, Deng J, Dewald J. 2009. EEG-based classification for elbow versus shoulder torque intentions involving stroke subjects. Comput Biol Med 39(5):443–52. [DOI] [PMC free article] [PubMed] [Google Scholar]