Abstract
Background
Although increasing evidence has demonstrated important roles for long non-coding RNAs (lncRNAs) in cancer development, their functions in oral squamous cell carcinoma (OSCC) growth remain largely unknown. Therefore, we aimed to investigate the role of LINC00662 in OSCC.
Methods
The expression of LINC00662 in 61 OSCC tissues and four OSCC cell lines were detected by quantitative reverse-transcription polymerase chain reaction (qRT-PCR). Cell proliferation was detected using Cell Counting Kit-8 (CCK-8) and EdU staining methods. Migration and invasion abilities were analyzed using transwell and wound healing assay. Cell cycle distribution and apoptosis rate were evaluated by flow cytometry. Western blot method was performed to detect protein expression.
Results
We found that the expression of LINC00662 was significantly increased in OSCC tissues, and a higher expression of LINC00662 was detected in larger tumor size, higher stage tumors and with lymph node metastasis. Moreover, overexpression of LINC00662 induced OSCC cell proliferation, increased migration and invasion abilities, and suppressed cell apoptosis. Knockdown of LINC00662 decreased the proliferation, migration, and invasion abilities of OSCC cell, and induced apoptosis. Furthermore, LINC00662 regulated the Wnt/β-catenin pathway.
Conclusion
Our data indicate that LINC00662 may represent a novel indicator of OSCC and may be a potential therapeutic target for diagnosis and therapy.
Keywords: LINC00662, proliferation, migration, Wnt/β-catenin pathway, OSCC
Introduction
Head and neck squamous cell carcinoma (HNSCC) is a common cancer worldwide. Oral squamous cell carcinoma (OSCC) is one of the most lethal cancers of the head and neck.1,2 In spite of significant progress in diagnosis and therapeutic strategies in OSCC including surgery, chemotherapy, and radiation, the general 5-year survival rate of OSCC patients only improved modestly over recent decades and remains less than 20% in patients with advanced conditions.3,4 Although increasing research has been undertaken to understand the basic molecular and cellular activity in OSCC, the precise molecular mechanisms underlying OSCC pathogenesis and identification are still little known. Therefore, to improve the prognosis of patients with OSCC, it is important to develop effective indicators and therapeutic targets.
Long non-coding RNAs (lncRNAs) are a group of RNAs that are over 200 base pairs in length with no protein-coding capacity.5,6 Over the last few decades, a large body of evidence revealed that lncRNAs have contributed to various function, they can act as molecular signals and scaffolds of gene modulation.7 Also, increasing evidence has showed lncRNAs play important roles in cell proliferation, apoptosis, differentiation, and cancer metastasis.8–10 Long intergenic non-protein coding RNA 662 (LINC00662) is located in chromosome 19q11 with 2,085 bp in length.11 Liu et al found that LINC00662 was significantly upregulated in lung squamous carcinoma compared with lung adenocarcinoma.12 Cheng et al suggested that LINC00662 might play a role as a potential tumor suppressor.13 Microarray expression profiling of lncRNAs revealed LINC00662 was increased in nasopharyngeal carcinoma.14 However, it is still unknown whether LINC00662 is involved in OSCC tumorigenesis.
There are few studies in the literature regarding the use of LINC00662 biomarker in human tumors, and it is unknown whether LINC00662 is involved in OSCC tumorigenesis. In the present study, we showed that LINC00662 was aberrantly expressed in human tongue squamous cell carcinoma (TSCC) and that it might play a role as a potential oncogene in promoting proliferation and metastasis of OSCC cells. This is the first time the role of LINC00662 has been evaluated in OSCC. Moreover, systematic analysis revealed that LINC00662 might regulate Wnt and β-catenin expression, indicating that LINC00662 may induce the activation of the Wnt/β-catenin pathway. Our results provide the first evidence in view of the potential role of lncRNA LINC00662 as new biomarker for HNSCC.
Materials and methods
Tissue samples
Sixty-one TSCC samples and adjacent normal mucosal tissues were obtained from patients undergoing surgery at the Department of Thyroid and Neck Surgery, the Second Affiliated Hospital of Nanchang University from October 2014 to March 2017. A detailed description of clinical and tumoral features is shown in Table 1. None of the patients received any radiotherapy and/or chemotherapy before the surgical operation. All tissues were collected and immediately frozen in liquid nitrogen. This study was approved by the Research Ethics Committee of the Second Affiliated Hospital of Nanchang University. Every participant was informed about the aims of specimen collection and gave written informed consent in accordance with the ethical guidelines. This research was conducted in accordance with the Declaration of Helsinki.
Table 1.
Difference in the LINC00662 expression in TSCC patients grouped by clinicopathological characteristics
| Clinicopathological characteristics | Number of patients | Expression of LINC00662a | P-value |
|---|---|---|---|
| Gender | 0.599 | ||
| Male | 28 | 2.450±0.2383 | |
| Female | 33 | 2.272±0.2372 | |
| Age (year) | 0.979 | ||
| <58 | 35 | 2.357±0.2413 | |
| ≥58 | 26 | 2.348±0.2277 | |
| Grade | 0.002 | ||
| I/II | 32 | 1.878±0.1771 | |
| III/IV | 29 | 2.879±0.2637 | |
| Tumor size | 0.007 | ||
| ≤5 cm | 30 | 1.901±0.1693 | |
| >5 cm | 31 | 2.792±0.2480 | |
| Lymph node metastasis | <0.001 | ||
| Yes | 29 | 2.953±0.2573 | |
| No | 32 | 1.810±0.1715 | |
Notes:
The relative expression of LINC00662 was calculated using the 2−ΔΔCq method and was shown as mean ± SD. Bold P-values <0.05 were considered statistically significant.
Cell lines and culture conditions
Human immortal oral epithelial cells (HIOEC) were purchased from BeNa Culture Collection (BNCC340217, Beijing, China). HIOEC cells were cultured in keratinocyte serum-free media. OSCC cell lines CGHNC9, ISG15, SCC9, and SCC25 were obtained from the cell bank of Chinese Academy of Sciences and American Type Culture Collection and routinely cultured in DMEM supplemented with 10% FBS (Thermo Fisher Scientific, Waltham, MA, USA) containing 1% penicillin (100 U/mL)–streptomycin (100 µg/mL) and incubated at 37°C and supplemented with 5% CO2 in a humidified incubator.
Cell transfection
LINC00662 overexpression plasmid pcDNA4.0-LINC00662, LINC00662 interference RNA (siLINC00662) (5′-GCAGGCGTACAACTAACAAdTdT-3′), and control plasmid pcDNA4.0 or control siRNA siCTRL were purchased from GenePharma (Shanghai, China). 1×105 cells were seeded in 24-well plates and incubated for 24 hours, then cells were transfected with plasmids in final concentration with 20 µg/mL or the cells were transfected with siRNA in final concentration with 100 nM using Lipofectamine® 2000 (Thermo Fisher Scientific) in serum-free medium according to the manufacturer’s instructions.
RNA extraction and real-time quantitative PCR analysis
Total RNA was isolated from tissue samples or cultured cells using TRIzol® reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol. For this, 2 µg total RNA in 20 µL reaction mixture was reverse transcribed to complementary DNA (cDNA) using a Reverse Transcription Kit (TaKaRa, Dalian, China). Real-time quantitative PCR analyses were performed with SYBR green real-time Master Mix (TOYOBO, Japan) as described by the manufacturer. The PCR primers used for LINC00662: 5′-ACTAACAAGCTGGGTGCAGA-3′ (forward) and 5′-CCTCCTGGTCTGCGAGAAAT-3′ (reverse); for GAPDH: 5′-TGTTCGTCATGGGTGTGAAC-3′(forward) and 5′-ATGGCATGGACTGTGGTCAT-3′ (reverse). Quantitative reverse-transcription polymerase chain reaction (qRT-PCR) and data collection were performed on Applied Biosystems 7,500 Sequence Detection system (Thermo Fisher Scientific, Waltham, MA, USA). The thermocycling protocol for all experiments was 40 cycles of denaturation at 95°C for 20 seconds, annealing at 58°C for 20 seconds, and extension at 72°C for 20 seconds. The relative expression of LINC00662 was calculated and normalized using the 2−ΔΔCt method relative to GAPDH (internal control).
Western blot
Total proteins were extracted from tissues and cells using RIPA buffer with proteinase inhibitor cocktail. Protein concentration was measured using BCA Protein Assay Kit (Thermo Fisher Scientific). Equal quantities (20 µg) of protein were separated by 10% SDS-PAGE and transferred onto a polyvinylidene fluoride (PVDF) membrane (Merck Millipore, Billerica, MA, USA). The membranes were blocked in 5% skim milk and then incubation with primary antibodies (anti-Wnt3a, ab28472, 1:1,000; anti-β-catenin, ab32572, 1:1,000; anti-GAPDH, ab9485, 1:5,000) for 2 hours at 37°C. Then the membranes were washed with TBST for three times and were incubated with HRP-conjugated secondary antibody (Abcam, Cam-bridge, UK) for 1 hour at room temperature. GAPDH was used as the internal reference. The blots were visualized using an enhanced chemiluminescence kit (Thermo Fisher Scientific).
Immunofluorescence
Cells were fixed with 3% paraformaldehyde and then permeabilized with 0.1% Triton X-100 for 30 min on ice. Anti-β-catenin antibodies (ab32572, 1:100) were added and incubated at 4°C for 12 hours, followed by incubation with TRITC and FITC tagged secondary antibodies for 1 hour at 37°C in a cassette. Subsequently, cells were washed and nuclei were counterstained with DAPI. Cells were visualized and analyzed on an Olympus FluoView 1,000 confocal microscope (Olympus Corporation, Tokyo, Japan).
Cell viability assay
Cell Counting Kit-8 (CCK-8) assay kit (Dojindo, Kumamoto, Japan) was used to determine the cell viability. In this assay, 1×104 treated cells were plated in 96-well plates and cultured for different times. At each time-point, 10 µL of CCK-8 solution was added into the 96-well plate and incubated for 1 hour at room temperature. The absorbance at 450 nm was quantified using a microplate reader (Rayto Life and Analytical Science C. Ltd, Shenzhen, China). Each experiment was performed in triplicate.
EdU incorporation assay
Cell-Light™ EdU Apollo® 567 In Vitro Imaging Kit (Ribobio, Guangzhou, China) was used to perform the EdU incorporation assay. In brief, cells were cultured with 50 µM EdU for 2 hours and then fixed with 4% paraformaldehyde for 20 minutes at 37°C. The cells were then washed with PBS contained 0.5% Triton X-100 in a decolorization shaker three times. Next, cells were stained with 100 µL Apollo 567 stain reaction buffer for 30 minutes in a cassette. Then, the cells were washed and stained with DAPI (5 mg/mL) for 30 minutes at room temperature. Images of the cells were captured using a fluorescence microscope (Olympus), and the EdU-positive cells was calculated using the formula: EdU add-in cells/DAPI-stained cells ×100%.
Flow cytometric analysis
Cells were harvested and washed with ice-cold phosphate-buffered saline (PBS). The PI/RNase staining kits (Multi-sciences, Shanghai, China) and an annexin V-fluorescein isothiocyanate propidium iodide (FITC/PI) apoptosis detection kit (Multisciences, Shanghai, China) were used to detect cell cycle and apoptosis according to the manufacturer’s protocol using a FACScan instrument (BD, Franklin Lakes, NJ, USA), respectively. Three independent experiments were performed.
Hoechst 33342 staining
Hoechst 33342 staining was performed according to the manufacturer’s protocol. Briefly, cells were cultured with Hoechst 33342 nucleic acid stain (Thermo Fisher Scientific) and then examined for nuclear morphology under a fluorescence microscope equipped with a filter at 365 nm.
Transwell invasion assay
Cell invasion ability was measured using transwell Matrigel invasion inserts with an 8 µm pore size (Corning Incorporated, Corning, NY, USA). In this assay, 1×104 cells in serum-free medium were plated in the upper Matrigel-coated chamber, and the lower chamber were filled with 500 µL of culture medium containing 20% FBS. The cells were then allowed to invade for 48 hours at 37°C. The invaded cells were fixed with 100% methanol and stained with 0.5% crystal violet (Sigma-Aldrich Co., St Louis, MO, USA). The permeating cells were counted in at least 10 randomly fields under a phase-contrast microscope (Olympus) at a magnification of ×200. All experiments were repeated three times independently.
Scratch wound healing assay
Cells were plated into 6-well plates and cultured at 37°C in a 5% CO2 cell incubator. Wounds were made by scratching cell monolayers using a 200-µL sterile pipette tip. The cells were continuously cultured in serum-free medium for 24 hours and then photographed. Wound-healing distance was calculated using Image Pro-Plus 6.0 software (Media Cybernetics, Rockville, MD, USA) and the following formula: cell migration distance = (distance at 0 hour − distance at 24 hours)/2.
Statistical analysis
All results were provided as the mean ± SD. Statistical significance between groups was determined using one-way ANOVA or a paired/unpaired Student’s t-test using the SPSS 18.0 (SPSS Inc., Chicago, IL, USA). P-values<0.05 were considered statistically significant. Each experiment was performed at least three times independently.
Results
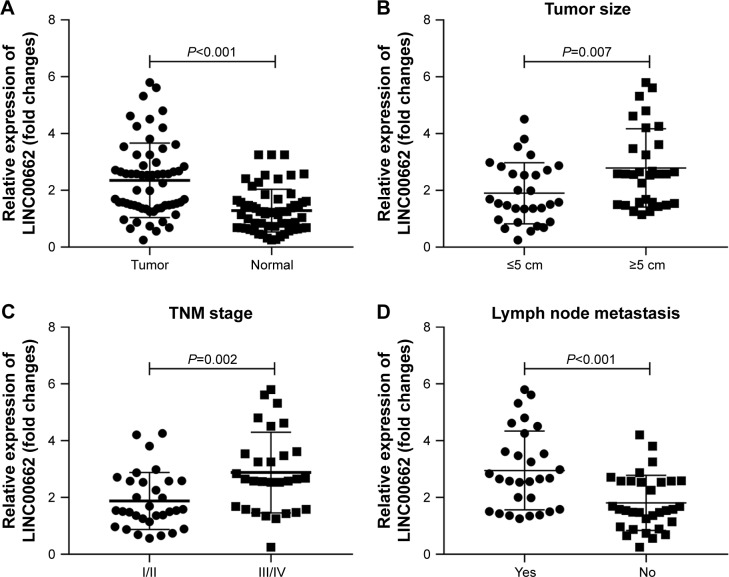
Aberrant upregulation of LINC00662 in TSCC tissues and association with tumor size, TNM stage and lymph node metastasis
The expression levels of lncRNA LINC00662 in TSCC tissues were detected using qRT-PCR. The expression levels of LINC00662 were dramatically higher in TSCC tissues compared with adjacent normal mucosal tissues (Figure 1A). Clinicopathological analysis showed LINC00662 expression was significantly correlated with tumor size, TNM stage, and lymph node metastasis (Figure 1B–D, Table 1). However, LINC00662 expression was not associated with other clinicopathological features such as gender and age (Table 1). Altogether, our results suggest that increase of LINC00662 might be involved in the progression and metastasis of the majority of human TSCC.
Figure 1.
Upregulation of LINC00662 in TSCC tissues and association with tumor size, TNM stage, and lymph node metastasis.
Notes: (A) Relative expression of LINC00662 in 61 pairs of TSCC tissues compared with corresponding adjacent normal tissues. (B) Relative expression of LINC00662 in TSCC patients with different tumor sizes. (C) Relative expression of LINC00662 in TSCC patients with different TNM stages. (D) Relative expression of LINC00662 in TSCC patients with or without lymph node metastasis.
Abbreviation: TSCC, tongue squamous cell carcinoma.
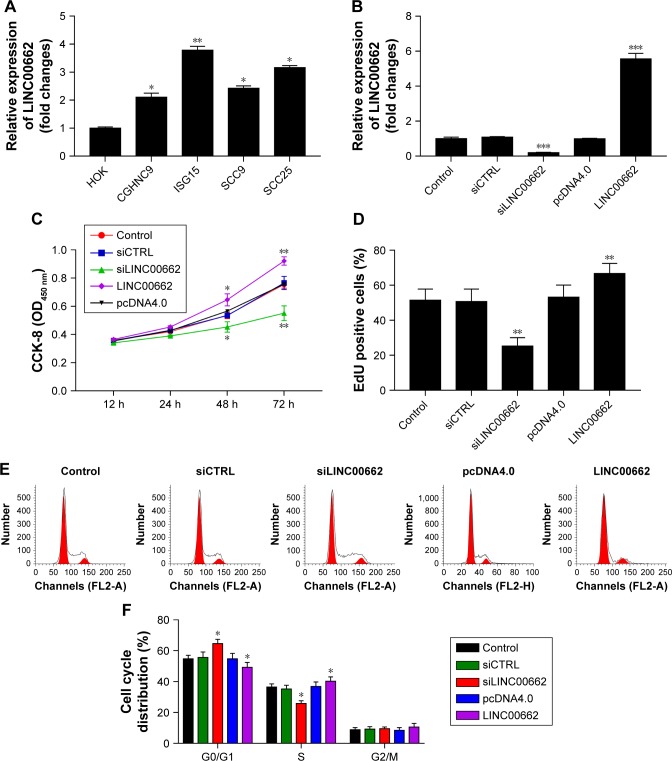
Manipulation of LINC00662 expression regulates cell proliferation in OSCC cell lines
Based on the observations, LINC00662 expression levels in four OSCC cell lines (CGHNC9, ISG15, SCC9, and SCC25) were carried out. Obvious increased LINC00662 levels were identified in OSCC cell lines compared with human normal oral keratinocyte cell line HOK, especially in ISG15 (Figure 2A). Thus, ISG15 cell line was selected as a research representative in the following studies.
Figure 2.
Manipulation of LINC00662 expression regulates cell proliferation in OSCC cells.
Notes: (A) The expression levels of LINC00662 in different OSCC cells. (B) ISG15 cells were transfected with siLINC00662 and LINC00662 overexpression plasmids, and the expression of LINC00662 was measured by qRT-PCR. (C) Cell proliferation ability was detected using CCK-8 assay at 12, 24, 48, and 72 hours. (D) Cell growth viability was measured using EdU staining method at 48 hours. (E, F) Flow cytometry was used to analyze the cell cycle distribution in ISG15 cells. *P<0.05 vs control or NC group. **P<0.01 vs control or NC group. ***P<0.001 vs control or NC group.
Abbreviations: CCK-8, Cell Counting Kit-8; CTRL, control; NC, negative control; OSCC, oral squamous cell carcinoma; qRT-PCR, quantitative reverse-transcription polymerase chain reaction.
Then, we constructed siRNA targeting LINC00662, named siLINC00662 and constructed LINC00662 overexpression plasmid pcDNA4.0-LINC00662, named LINC00662. The knockdown efficiency was approximately 82% in ISG15 cells after transfection with siLINC00662 and a respective 5.7-fold expression increase in the LINC00662-transfected cells compared with the control (Figure 2B). To assess further the potential effects of LINC00662 on cell proliferation, CCK-8 assay was carried out at 12, 24, 48, and 72 hours after siL-INC00662 or LINC00662 transfection (Figure 2C). Compared with the control cells, knockdown of LINC00662 significantly decreased the cell viability and overexpression of LINC00662 promoted ISG15 cell growth. The results of EdU staining were consistent with the CCK-8 results (Figure 2D). In addition, flow cytometry results showed that siRNA-mediated LINC00662 silencing could increase G1 phase and decrease S phase and overexpression of LINC00662 could decrease G1 phase and increase S phase in ISG15 cells (Figure 2E and F). Thus, the results indicated that LINC00662 could control the cell cycle to regulate the proliferation of OSCC cells in vitro.
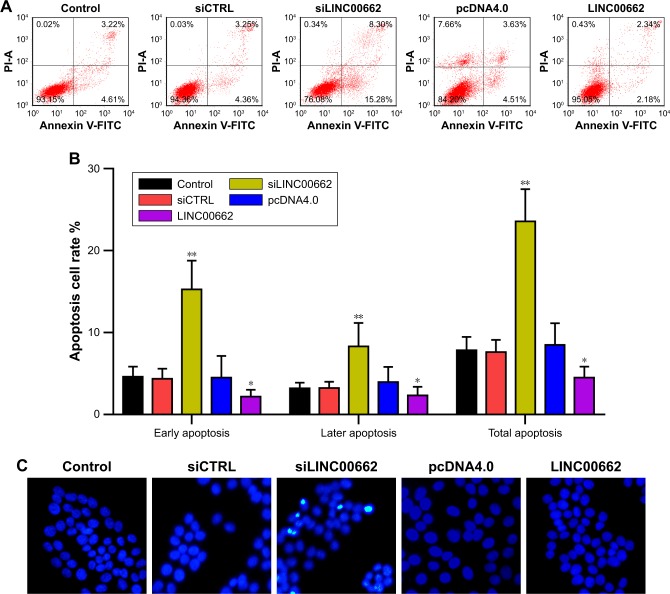
Manipulation of LINC00662 expression regulates cell apoptosis in OSCC cells
To determine whether LINC00662 contribute to cell apoptosis, we performed flow cytometry and Hoechst staining analysis of siLINC00662- and LINC00662-transfected ISG15 cells. The results showed the apoptosis rate was significantly increased in siLINC00662-transfected cells (Figure 3A and B). In addition, the number of cells with condensed and fragmented nuclei indicating the prominent apoptotic cells was significantly different in ISG15 cells with siLINC00662 transfection (Figure 3C). Taken together, our results indicated that knockdown of LINC00662 induced cell apoptosis in vitro.
Figure 3.
Manipulation of LINC00662 expression regulates cell apoptosis in OSCC cells.
Notes: (A, B) Cell apoptosis rate was measured by flow cytometric analysis of annexin V-FITC double staining after transfection of siLINC00662 or LINC00662 for 48 hours. Lower right and upper right in the cell distribution panels represent early and later apoptotic cell proportions, respectively. (C) Hoechst 33,258 staining assay was used to detect cell apoptosis. Magnification ×200. *P<0.05 vs control or NC group. **P<0.01 vs control or NC group.
Abbreviations: CTRL, control; NC, negative control; OSCC, oral squamous cell carcinoma; V-FITC, V-fluorescein isothiocyanate propidium iodide.
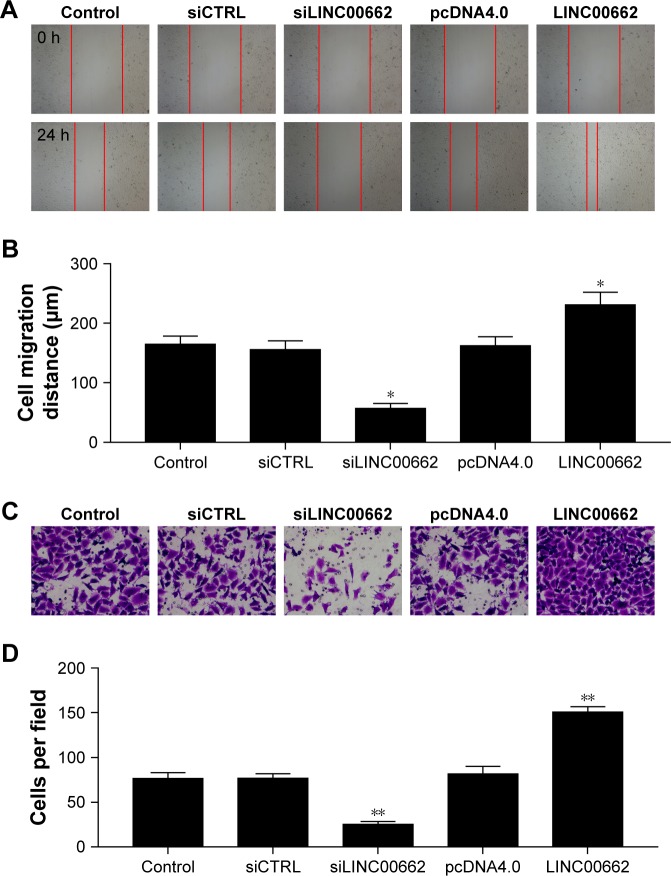
Manipulation of LINC00662 expression regulates OSCC cell migration and invasion
To detect the effect of LINC00662 on cell migration and invasion, the transwell migration assay and scratch wound-healing assay were performed in ISG15 cells. With 24 hours of cultivation, the distance of siLINC00662-transfected ISG15 cells was significantly longer than control group, whereas the width of LINC00662-transfected ISG15 cells was shorter than control group (Figure 4A and B). The transwell invasion assay results showed the invasive ability was suppressed after transfected with siLINC00662 and promoted after transfected with LINC00662 in ISG15 cells (Figure 4C and D).
Figure 4.
Manipulation of LINC00662 expression regulates OSCC cell migration and invasion.
Notes: (A, B) Wound healing assay was used to test the migration ability of OSCC cells after transfection of siLINC00662 or LINC00662. (C, D) Transwell experiment was used to test the invasion ability of OSCC cells after siLINC00662 or LINC00662 transfection. Magnification ×100. **P<0.01 vs control or NC group.
Abbreviations: CTRL, control; NC, negative control; OSCC, oral squamous cell carcinoma.
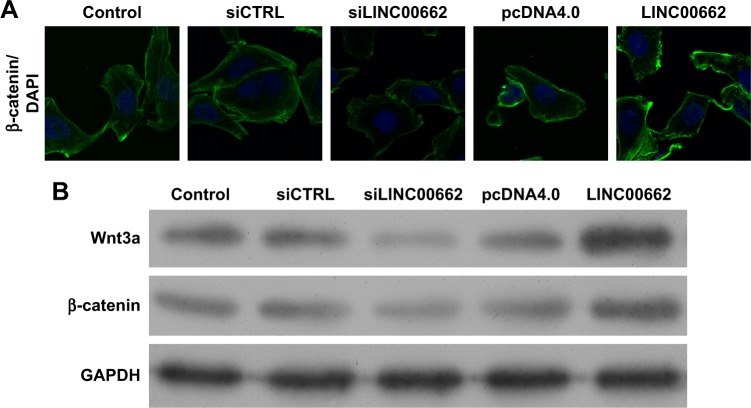
LINC00662 activates the Wnt/β-catenin pathway
As is known, the WNT/β-catenin signaling pathway plays important roles in the modulation of cell proliferation. Here, the results of Western blot and immunofluorescence assay showed knockdown of LINC00662 suppressed the expression of Wnt3a and β-catenin protein and overexpression of LINC00662 induced Wnt3a and β-catenin protein expression (Figure 5). Thus, the upregulation of LINC00662 could activate the Wnt/β-catenin signaling pathway.
Figure 5.
LINC00662 activates the Wnt3a/β-catenin pathway.
Notes: (A) Immunofluorescence assay detected the β-catenin expression. (B) Western blot detected the Wnt3a and β-catenin protein expression. Magnification ×400.
Abbreviations: CTRL, control; NC, negative control.
Discussion
Increasing number of studies strongly indicate the important roles of lncRNAs in the regulation of protein coding genes. They play a crucial role in ontogenetic process and tumor formation at both the transcriptional and posttranscriptional levels.15–17 Emerging researches have reported that large number of lncRNAs are deregulated in human cancers.18–20 These differentially expressed lncRNAs are closely contributed to tumor growth, metastasis, apoptosis or diagnosis, acting as antioncogenes or oncogenes.21–23 In addition, LINC00662 was significantly upregulated in lung squamous carcinoma compared with lung adenocarcinoma,12 and increased in nasopharyngeal carcinoma.14 Thus, more effort should be dedicated to fully expound the potential molecular and cellular mechanisms of LINC00662 action in human tumors.
In the present study, we investigated the expression levels of lncRNA LINC00662 in OSCC tissues and corresponding normal mucosal tissues. The results showed that LINC00662 was increased in OSCC tissues and its expression was observably correlated with tumor size, TNM stage, and lymph node metastasis. However, with losing the volunteers to follow-up, a Kaplan–Meier survival analysis was limited to carry out. We then performed gain- and loss-of-function approaches to measure the function of LINC00662 in OSCC cells. Overexpression of LINC00662 could promote cell proliferation, invasion, and migration and suppress cell apoptosis in OSCC cell lines in vitro, and knockdown of LINC00662 could reduce cell proliferation, migration, and invasion and promote cell apoptosis in OSCC cells in vitro. This is the first report to evaluate the functional significance of LINC00662 in human OSCC, and our data suggest that LINC00662 may act as an oncogene that stimulates malignant progression if OSCC.
In addition, groups of evidence indicate that Wnt/β-catenin signaling pathway may contribute to OSCC development and progression.24,25 Our results elucidated that overexpression of LINC00662 activated Wnt3a and β-catenin protein expression, and knockdown of LINC00662 suppressed Wnt3a and β-catenin protein expression and blocked the activation of the Wnt/β-catenin signaling pathway. Our results revealed a relationship between LINC00662 expression and Wnt/β-catenin pathway activation. Previous studies also found that the Wnt/β-catenin pathway contributed to cancer growth and apoptosis,26,27 which is coincidental to our findings. Therefore, we considered that LINC00662 may be involved in the activation of Wnt/β-catenin signaling pathway. Our results provide a new insight for comprehending the development and progress of OSCC and impart a novel approach for OSCC diagnosis and treatment.
For the limitations, in the present study, we only analyzed the relationship of LINC00662 expression and clinicopathological characteristics, and the survival data needs to be associated with our results. Sixty-one OSCC samples and adjacent normal mucosal tissues were obtained from patients undergoing surgery at the Department of Thyroid and Neck Surgery, the Second Affiliated Hospital of Nanchang University from October 2014 to March 2017. Only eleven OSCC samples were obtained before October 2015. We think the quantity of patients is small for survival data analysis. The overall survival and disease-free survival rates of all 61 patients are tracked at present. This data will be provided later.
In summary, our results showed LINC00662 expression levels were increased in human OSCC tissues and its expression observably correlated with tumor size, TNM stage, and lymph node metastasis. Moreover, knockdown of LINC00662 inhibited OSCC cell proliferation, migration, and invasion and induced cell apoptosis. In addition, knockdown of LINC00662 blocked the activation of Wnt/β-catenin signaling pathway. Our present study suggests that LINC00662 acts as an oncogene by promoting the progression of OSCC and reveals LINC00662-Wnt/β-catenin signaling pathway regulatory network contributes to OSCC. Our data indicate that LINC00662-Wnt/β-catenin signaling pathway may represent a novel indicator of OSCC and may be a potential therapeutic target for diagnosis and therapy.
Acknowledgments
This study was supported by the relationship between human papillomavirus infection and prognosis of oropharyngeal squamous cell carcinoma (No 160044).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Rosebush MS, Rao SK, Samant S. Oral cancer: enduring characteristics and emerging trends. J Tenn Dent Assoc. 2011;91(2):2428–2729. [PubMed] [Google Scholar]
- 3.Fang J, Li X, Ma D, et al. Prognostic significance of tumor infiltrating immune cells in oral squamous cell carcinoma. BMC Cancer. 2017;17(1):375. doi: 10.1186/s12885-017-3317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. 2009;45(4–5):309–316. doi: 10.1016/j.oraloncology.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145(2):178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 7.Eades G, Zhang YS, Ql L, Xia JX, Yao Y, Zhou Q. Long non-coding RNAs in stem cells and cancer. World J Clin Oncol. 2014;5(2):134–141. doi: 10.5306/wjco.v5.i2.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen P, Fang X, Xia B, Zhao Y, Li Q, Wu X. Long noncoding RNA LINC00152 promotes cell proliferation through competitively binding endogenous miR-125b with Mcl-1 by regulating mitochondrial apoptosis pathways in ovarian cancer. Cancer Med. 2018;7(9):4530–4541. doi: 10.1002/cam4.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng W, Deng W, Zhang J, Pei G, Rong Q, Zhu S. Long noncoding RNA ANCR suppresses bone formation of periodontal ligament stem cells via sponging miRNA-758. Biochemical and Biophysical Research Communications. 2018;503(2):815–821. doi: 10.1016/j.bbrc.2018.06.081. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Luo TB, Liu L, Cui ZQ. LncRNA LINC00311 promotes the proliferation and differentiation of osteoclasts in osteoporotic rats through the notch signaling pathway by targeting DLL3. Cell Physiol Biochem. 2018;47(6):2291–2306. doi: 10.1159/000491539. [DOI] [PubMed] [Google Scholar]
- 11.Strausberg RL, Feingold EA, Grouse LH, et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A. 2002;99(26):16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu B, Chen Y, Yang J. LncRNAs are altered in lung squamous cell carcinoma and lung adenocarcinoma. Oncotarget. 2017;8(15):24275–24291. doi: 10.18632/oncotarget.13651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng J, Demeulemeester J, Wedge DC, et al. Pan-cancer analysis of homozygous deletions in primary tumours uncovers rare tumour suppressors. Nat Commun. 2017;8(1):1221. doi: 10.1038/s41467-017-01355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wen X, Tang X, Li Y, et al. Microarray expression profiling of long non-coding RNAs involved in nasopharyngeal carcinoma metastasis. Int J Mol Sci. 2016;17(11):1956. doi: 10.3390/ijms17111956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pauli A, Valen E, Lin MF, et al. Systematic identification of long non-coding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012;22(3):577–591. doi: 10.1101/gr.133009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carlson HL, Quinn JJ, Yang YW, Thornburg CK, Chang HY, Stadler HS. LncRNA-HIT functions as an epigenetic regulator of chondrogenesis through its recruitment of p100/CBP complexes. PLoS Genet. 2015;11(12):e1005680. doi: 10.1371/journal.pgen.1005680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lan Y, Xiao X, He Z, et al. Long noncoding RNA OCC-1 suppresses cell growth through destabilizing HuR protein in colorectal cancer. Nucleic Acids Res. 2018;46(11):5809–5821. doi: 10.1093/nar/gky214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hao S, Yao L, Huang J, et al. Genome-wide analysis identified a number of dysregulated long noncoding RNA (lncRNA) in human pancreatic ductal adenocarcinoma. Technol Cancer Res Treat. 2018;17(4):153303461774842. doi: 10.1177/1533034617748429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paul Y, Thomas S, Patil V, et al. Genetic landscape of long noncoding RNA (lncRNAs) in glioblastoma: identification of complex lncRNA regulatory networks and clinically relevant lncRNAs in glioblastoma. Oncotarget. 2018;9(51):29548–29564. doi: 10.18632/oncotarget.25434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Wang Z, Wang Y, et al. Identification and characterization of lncRNA mediated transcriptional dysregulation dictates lncRNA roles in glioblastoma. Oncotarget. 2016;7(29):45027–45041. doi: 10.18632/oncotarget.7801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang N, Meng X, Mi H, et al. Circulating lncRNA XLOC_009167 serves as a diagnostic biomarker to predict lung cancer. Clinica Chimica Acta. 2018;486:26–33. doi: 10.1016/j.cca.2018.07.026. [DOI] [PubMed] [Google Scholar]
- 22.Guo HM, Yang SH, Zhao SZ, Li L, Yan MT, Fan MC. LncRNA NEAT1 regulates cervical carcinoma proliferation and invasion by targeting AKT/PI3K. Eur Rev Med Pharmacol Sci. 2018;22(13):4090–4097. doi: 10.26355/eurrev_201807_15400. [DOI] [PubMed] [Google Scholar]
- 23.Yin Y, Zhao B, Li D, Yin G. Long non-coding RNA CASC15 promotes melanoma progression by epigenetically regulating PDCD4. Cell Biosci. 2018;8(1):42. doi: 10.1186/s13578-018-0240-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwai S, Yonekawa A, Harada C, et al. Involvement of the Wnt-β-catenin pathway in invasion and migration of oral squamous carcinoma cells. Int J Oncol. 2010;37(5):1095–1103. doi: 10.3892/ijo_00000761. [DOI] [PubMed] [Google Scholar]
- 25.Yang YT, Wang YF, Lai JY, et al. Long non-coding RNA UCA1 contributes to the progression of oral squamous cell carcinoma by regulating the Wnt/β-catenin signaling pathway. Cancer Sci. 2016;107(11):1581–1589. doi: 10.1111/cas.13058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshida T, Sopko NA, Kates M, et al. Three-dimensional organoid culture reveals involvement of Wnt/β-catenin pathway in proliferation of bladder cancer cells. Oncotarget. 2018;9(13):11060–11070. doi: 10.18632/oncotarget.24308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng L, Pan J. The anti-malarial drug artesunate blocks Wnt/β-catenin pathway and inhibits growth, migration and invasion of uveal melanoma cells. Curr Cancer Drug Targets. 2018;18(10):988–998. doi: 10.2174/1568009618666180425142653. [DOI] [PubMed] [Google Scholar]