Abstract
Epithelial-to-mesenchymal transition (EMT) and maturation of collagen fibrils in the tumor microenvironment play a significant role in cancer cell invasion and metastasis. Confinement along fiber-like tracks enhances cell migration. To what extent and in what manner EMT further promotes migration in a microenvironment already conducive to migration is poorly understood. Here, we show that TGFβ-mediated EMT significantly enhances migration on fiber-like micropatterned tracks of collagen, doubling migration speed and tripling persistence relative to untreated mammary epithelial cells. Thus, cell-intrinsic EMT and extrinsic fibrillar tracks have nonredundant effects on motility. To better understand EMT-enhanced fibrillar migration, we investigated the regulation of Golgi positioning, which is involved in front-rear polarization and persistent cell migration. Confinement along fiber-like tracks has been reported to favor posterior Golgi positioning, whereas anterior positioning is observed during 2-day wound healing. Although EMT also regulates cell polarity, little is known about its effect on Golgi positioning. Here, we show that EMT induces a 2:1 rearward bias in Golgi positioning; however, positional bias explains less than 2% of single-cell variability in migration speed and persistence. Meanwhile, EMT significantly stabilizes Golgi positioning. Cells that enhance migration in response to TGFβ maintain Golgi position for 2- to 4-fold longer than nonresponsive counterparts irrespective of whether the Golgi is ahead or behind the nucleus. In fact, 28% of TGFβ-responsive cells exhibit a fully committed Golgi phenotype with the organelle either in the anterior or posterior position for over 90% of the time. Furthermore, single-cell differences in Golgi stability capture up to 18% of variations in migration speed. These results suggest a hypothesis that the Golgi may be part of a core physical scaffold that affects how cell-generated forces are distributed during migration. A stable scaffold would be expected to more consistently and therefore more productively distribute forces over time, leading to efficient migration.
Introduction
Epithelial-to-mesenchymal transition (EMT) and maturation of the fibrillar architecture of the tumor microenvironment (TME) are highly associated with cancer progression (1, 2, 3). Although EMT and fibrillar maturation are each known to enhance cell migration, little is known about how these factors interact and cooperate to promote cell motility and invasion. A better understanding of this cooperation will yield insights into how cell-intrinsic and cell-extrinsic factors conspire to advance cancer progression.
EMT involves major changes in gene expression and cell morphology. During this process, epithelial cells that are ordinarily in contact with and adhered to adjacent cells transform into a mesenchymal phenotype, acquiring an extended uniaxial morphology and enhanced migration properties (4). In cancer, EMT is induced by a number of mechanisms, including the upregulation of transcription factors such as Snail, and by soluble ligands such as TGFβ (5, 6).
Meanwhile, the TME undergoes changes of its own. The matrix is observed to stiffen, and the density and maturity of collagen fibers increases (1, 2). Cancer cells migrate along these fibers in vivo (7, 8). We and others have studied migration along fibers using long, narrow micropatterns as an in vitro model (9, 10, 11, 12, 13, 14, 15). Cells have common features on narrow micropatterns that resemble cells migrating along fibers. This includes an elongated morphology, uniaxial migration, and more effective coordination between leading edge protrusion and trailing edge contraction as well as more efficient retraction of the cell rear.
We recently used fiber-like micropatterns to show that EMT and the fibrillar environment cooperate to regulate migration response to cell-cell interactions (11, 15). When migrating nontransformed mammary epithelial cells encounter each other on fiber-like tracks, they reverse direction and move apart. Inducing EMT with TGFβ treatment changes this migration response to cell-cell contact: instead of reversing direction, cells that have undergone EMT slide past each other. Enhanced sliding enables cells to maintain longer migration paths without changes in direction, a property that is likely to enhance dispersion efficiency, especially in TMEs, where the density of cells and the frequency of cell-cell encounters are high. Notably, cells that advance further into EMT slide on progressively narrower fiber-like tracks, showing that the extent of EMT and the degree of fibrillar maturation cooperate quantitatively to modulate cell-cell interactions during migration.
Here, we investigate to what extent EMT affects migration speed and the persistence of individual cells in a fiber-like microenvironment. Cells confined along fiber-like micropatterns acquire uniaxial morphology and migrate significantly better than cells in two dimensions (13, 16, 17). Meanwhile, EMT in two-dimensional (2D) environments also confers uniaxial morphology and enhances motility (6). Does EMT enhance migration beyond what is already achieved by spatial confinement along fibrils? Or are EMT and fibrillar topology redundant pathways, with little cooperative effect on motility?
To the extent that EMT and fibrillar environment cooperatively regulate individual-cell migration, a potential point of coordination involves the Golgi. Subcellular positioning of the Golgi and the associated centrosome/microtubule-organizing center (MTOC) has been linked to cell polarity and directional migration (18, 19). Anterior positioning of the Golgi was first reported in 2D wound-healing assays (20). From its position at the front of the cell and ahead of the nucleus, the Golgi is thought to mediate the delivery of new membrane and adhesion proteins to the leading edge (21, 22). However, in many cell types and contexts, cell migration occurs without anterior Golgi positioning. No bias in Golgi or MTOC positioning is observed among migrating Rat2 fibroblasts in vitro and neurons in the developing zebrafish cerebellum (23, 24), whereas T cells invading into a collagen gel exhibit posterior positioning of the MTOC (25). Posterior Golgi positioning is also observed in cells migrating on fiber-like micropatterns (12, 13).
Although some of these differences in Golgi positioning can, in part, be attributed to differences among cell systems and microenvironmental context, it raises the possibility that other attributes of the Golgi, not only its subcellular position, are involved in regulating migration. Furthermore, an important consideration is the inherent variability in behaviors at the single-cell level, even for a single-cell system and microenvironmental context. Not every cell in a population moves in the same manner or with the same quantitative migration properties of speed and persistence. Moreover, individual cells may vary in how the Golgi is employed, and these variations could even occur within an individual cell over time. Single-cell analysis of Golgi and cell-migration dynamics are needed to better understand the cell-to-cell variability in Golgi positioning and how these variations are related to the variability in cell-migration properties.
Finally, how EMT regulates Golgi positioning at the single-cell level is unknown. In fiber-like microenvironments, both epithelial-like African green monkey kidney cells and NIH-3T3 fibroblasts have been shown to position the Golgi behind the nucleus when migrating along fiber-like micropatterns and on fibrillar collagen in three-dimensional cell-derived matrix (12, 13). This suggests that cells undergoing EMT would continue to employ posterior Golgi positioning. However, this conclusion is based on epithelial and fibroblast cell types with disparate backgrounds, and the effect of EMT on Golgi positioning remains to be tested directly by taking a cell system, inducing EMT, and analyzing Golgi dynamics during fibrillar migration.
To elucidate the effect of EMT on fibrillar cell migration and to better understand the role of the Golgi in fibrillar migration, we conduct here single-cell, dynamical analysis of Golgi-nuclear position during the fibrillar migration of MCF-10A mammary epithelial cells that have been induced to undergo EMT by TGFβ treatment. Our data show that TGFβ-induced EMT enhances cell motility in a fiber-like context, revealing that these cell-intrinsic and cell-extrinsic factors regulate motility in a nonredundant manner. These results have implications for how EMT and fibrillar maturation work together to promote invasiveness during cancer progression. Moreover, our analysis reveals that the stability of Golgi positioning—independent of whether the Golgi is ahead or behind the nucleus—is associated with EMT-enhanced cell motility. In fact, variations in Golgi positional stability but not a preference for anterior or posterior Golgi position statistically captures variations in single-cell-migration speed. Thus, we identify a novel way, to our knowledge, in which the Golgi is involved in cell migration—through stabilization of its position, more so than the position itself—and propose a structural/physical role for the Golgi in cell migration.
Materials and Methods
Microcontact printing
SU-8 2010 (Microchem, Westbourgh, MA) was spin coated onto 3-inch silicon test wafers (Silicon Sense, Nashua, NH). The coating was exposed to ultraviolet light using a Quintel 4000 mask aligner (Neutronix Quintel, Morgan Hill, CA) through a chrome/soda lime mask with 10-μm-wide line patterns (Front Range PhotoMask, Palmer Lake, CO). Non-cross-linked portions were etched away using SU-8 developer (Microchem), leaving ridges and channels. Polydimethylsiloxane 184 was mixed with cross-linker at an 8:1 ratio (Dow Corning, Midland, MI), poured over the molds, and cured for at least 2 h at 80°C.
Polydimethylsiloxane were peeled off the silicon wafer and cut to form stamps. Before stamping, the stamps were placed ridge side up and plasma treated (Harrick Plasma, Ithaca, NY) for 20 s and quickly covered in a mixture of phosphate-buffered saline (Invitrogen, Carlsbad, CA) and 10 μg/mL rat-tail collagen I (Invitrogen). Stamps were left to incubate at room temperature for 30 min.
Glass-bottomed dishes (Ted Pella, Redding, CA) were flame cleaned before vapor-depositing a monolayer of trimethoxyglycidyl (epoxy) silane (Gelest, Morrisville, PA) overnight.
Stamps were gently rinsed in deionized water, blown dry with air, and immediately placed face down on silane-treated glass dishes. The stamps and surfaces were incubated together at 37°C overnight to allow free amine groups on the collagen to cross-link to the epoxy. After stamp removal, dishes were stored dry until use.
Cell culture and TGFβ treatment
MCF10A cells were infected with retrovirus encoding Golgi (GM130-RFP) and nuclear (histone 2B-GFP) markers and were selected using puromycin and hygromycin, respectively. Cells were cultured by standard methods previously described (26). Briefly, cells were passed at a 1:4 ratio every 3 days after reaching confluence. Cultures were maintained at 37°C and 5% CO2 in standard MCF10A growth media: Dulbecco’s modified Eagle’s medium F12 supplemented with 5% horse serum, 1% penicillin/streptomycin, hydrocortizone, insulin, EGF (Invitrogen), and cholera toxin (Sigma Aldrich, St. Louis, MO).
To induce EMT, MCF10A cells were treated with 20 ng/mL TGFβ (Peprotech, Rocky Hill, NJ) added to growth medium and were cultured for 12 days before experiments. During the 12 days, cells were passaged at the regular frequency using the aforementioned standard methods, with the exception that cells were maintained in 20 ng/mL TGFβ-containing growth medium. This protocol matches exactly that used in our earlier work to quantify morphological and protein-expression changes induced by TGFβ treatment (11).
Imaging and image analysis
In preparation for experiments, micropatterned surfaces were treated with 0.02% Pluronic F-127 (wt/vol) (EMD Biosciences, San Diego, CA) in phosphate-buffered saline for 30 min to prevent cell adhesion to nonstamped areas. Dishes were subsequently washed once with growth media and allowed to sit for 30 min before cell seeding. Just before seeding, the dish was rinsed once more to supply fresh media.
Cells were seeded on micropatterned, glass-bottomed dishes and were allowed to adhere for ∼25 min. Nonadhered cells were removed by aspiration and simultaneous flushing with fresh experimental media (MCF10A growth media ± TGFβ) in a single step to prevent the dish from drying out. Excessive rinsing was avoided to prevent damaging the Pluronic layer. Cells were allowed to fully spread in the incubator (∼30 min) before mounting the dish on the microscope for imaging.
Cell migration was tracked using a Plan-Apochromat 20× objective on an LSM700 confocal microscope equipped with dual photomultipliers, a heated stage, and a chamber with humidity and CO2 control (Zeiss, Oberkochen, Germany). Z-stacks of 8–12 adjacent fields of interest were taken every 2.5 min over a 14-h period. Preparation for track analysis was conducted using post-image processing in ZEN blue (Zeiss) to make 2D projections of the Z-stacks and to stitch together overlapping fields. Cell-migration trajectories consisting of nuclear and Golgi centroid positions were determined in an automated fashion, with manual input as needed, using a customized MATLAB (The MathWorks, Natick, MA) code that takes advantage of the contrast provided by the nuclear and Golgi markers and provides a graphical user interface described previously (27, 28).
Migration properties and Golgi states were subsequently analyzed using MATLAB. For calculating migration speed and persistence, we downsampled the experimentally acquired nuclear positional data. Nucleus position is imaged every 2.5 min. To avoid misappropriating short timescale fluctuations in nuclear position to longer-range migration of the cell body, we downsampled the data by using every fourth time point for the purpose of calculating speed and persistence. Briefly, let xt be the position of the nucleus centroid along the direction of the fiber-like micropattern at downsampled time points τ, where τ = 1, 2, 3… T and the dimensional value of Δτ = 7.5 min. Speed (S) is calculated by
To calculate persistence (P), we subdivided the range of time points [1,T] into persistent blocks bi in which the cell moves in the same direction without stopping or turning around. Thus, the time points of the trajectory of each cell is broken into a sequence of B persistent blocks b1 = [1,τ1], b2 = [τ1 + 1,τ2],…, bB = [τB − 1 + 1,T] with the duration of each block being Δb1 = τ1 − 1, Δb2 = τ2 − τ1, and so on. Persistence (P) is then given by the average duration of these persistent blocks:
For calculating Golgi position relative to the nucleus (GPRN), we use Golgi and nucleus positions measured at the sampling interval of 2.5 min. Here, short-range changes in the relative positions of Golgi and nucleus provide important information regarding Golgi-nucleus orientation relative to the direction of migration. At each time point t = 1, 2, 3…, we take the dot product of the nucleus-to-Golgi vector and the vector of migration, both along the direction the micropattern. If the dot product is positive, the Golgi is ahead of the nucleus and the GPRN state is Ahead (A); if the dot product is negative, the GPRN state is Behind (B). In the event that the cell is not migrating or the Golgi is not identifiable at a particular time point, an Unknown (U) state is assigned. In this manner, the GPRN state Gt for a cell is assigned at every time point t of its trajectory.
Results
Experimental system for automated analysis of Golgi and single-cell-migration dynamics in a fibrillar context
To better understand how EMT affects cell migration in a fibrillar TME, we used narrow 10-μm-wide micropatterns of collagen to model fiber-like spatial constraints and investigated the migration behavior of nontransformed MCF-10A mammary epithelial cells and counterparts induced to undergo EMT. To induce EMT and study its effects on fibrillar migration, MCF-10A cells were treated with 20 ng/mL TGFβ for 12 days. We have shown previously that this dosage and duration of exposure reduces E-cadherin expression, upregulates N-cadherin, disrupts cell-cell adhesions, and induces an extended morphology in MCF-10A cells (11)—all features consistent with progression through EMT (6). MCF-10A cells expressing histone 2B-GFP (H2B-GFP) were used to provide high-contrast images for automated segmentation of cell location and migration trajectories. In addition, to analyze the regulation of GPRN, cells were transduced with an expression construct encoding GM130-RFP (Golgi marker). The migration behaviors of untreated and TGFβ-treated cells expressing these two markers were recorded by time-lapse confocal microscopy, with images acquired at 2.5-min intervals. Automated segmentation and subsequent analysis of migration trajectories were performed in MATLAB.
TGFβ-mediated EMT increases the speed and persistence of cell migration on fiber-like collagen tracks
Migration speed and persistence were calculated from the trajectories of untreated and TGFβ-treated MCF-10A cells. The movement of the cell body was tracked by following the position of the GFP-labeled nucleus over time. The nucleus can undergo short timescale fluctuations in subcellular position even in a stationary cell. To avoid counting short-range fluctuations as migratory displacements of the cell body, we quantified migration speed and persistence using a coarse-grained trajectory in which every fourth experimental time point is sampled, i.e., a time interval of 7.5 min. Thus, migration speed was quantified by averaging instantaneous speeds measured at every 7.5 min interval over the entire migration trajectory of the cell (see Materials and Methods for details). Meanwhile, the persistence of migration was determined by identifying time blocks of consecutive 7.5-min intervals during which the cell moved in the same direction without stopping or turning around. The average duration of these time blocks provided a direct measure of migration persistence.
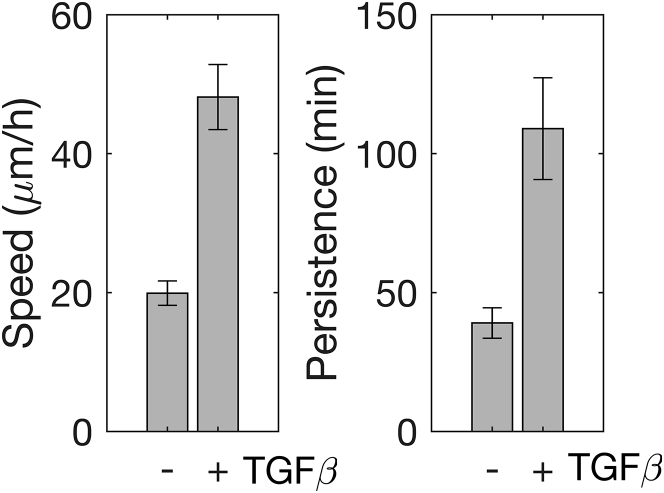
Migration speed was 2.4-fold greater for cells that were treated with TGFβ than those cells left untreated (Fig. 1 ). Untreated cells migrated with a speed of 19.9 μm/h, whereas TGFβ-treated cells moved at a speed of 48.2 μm/h. Meanwhile, TGFβ treatment nearly tripled migration persistence (Fig. 1 ). TGFβ-treated cells migrated with a persistence of 109 min compared to 39.1 min for untreated cells.
Figure 1.
TGFβ treatment enhances speed and persistence of mammary epithelial cells migrating on fiber-like micropatterns. Migration speed (left) and persistence (right) of untreated control (−) and TGFβ-treated (+) cells are shown with error bars indicating mean ± standard error with n = 49 and 61 cells, respectively, across three and six independent trials, respectively. Differences in speed and persistence are statistically significant (p < 0.005, Student’s t-test).
TGFβ-treated cells show a bias in positioning the Golgi behind the nucleus, whereas untreated cells exhibit no bias
The strong enhancement in migration, particularly persistence, suggests that TGFβ-induced EMT confers greater maintenance of front-rear cell polarity. Golgi positioning has been linked to the establishment and maintenance of cell polarity (22, 29, 30). We therefore asked whether TGFβ-induced EMT involves regulation of the GPRN.
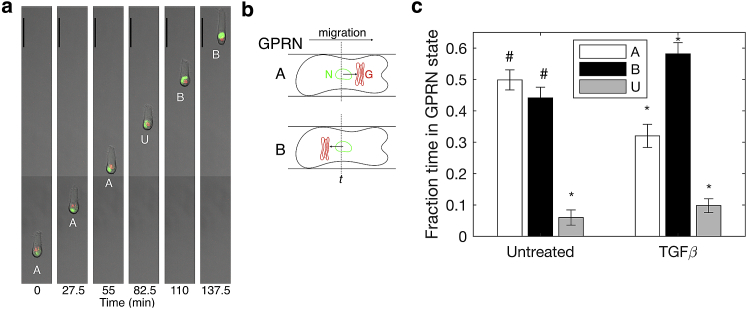
To quantify GPRN, the positions of the Golgi and nucleus centroids were determined from confocal time-lapse images of H2B-GFP and GM130-RFP acquired at 2.5-min intervals (Fig. 2 a; Video S1). At each time point, we related the orientation of the Golgi with respect to the nucleus to the direction in which the cell was migrating (see Materials and Methods for details).
Figure 2.
TGFβ-treated cells exhibit a slight bias in positioning the Golgi behind the nucleus, whereas untreated cells show an equal tendency to position the Golgi ahead or behind the nucleus. (a) A time series of a TGFβ-treated cell migrating from the bottom (t = 0 min) to the top (137.5 min). The nucleus H2B-GFP) is labeled in green and the Golgi (GM130-RFP) in red. The Golgi position relative to the nucleus (GPRN) shifts from the A to the U to the B state. See Video S1. Scale bars, 50 μm. (b) A schematic of scenarios in which GPRN is scored as being in the Ahead or Behind state. The direction of cell migration at time t is determined from the movement of the nucleus (N, green). In relation to migration direction, if the Golgi (G, red) is ahead of the nucleus (top), GPRN at time t is classified in the A state. If the Golgi is behind the nucleus in relation to migration (bottom), GPRN state is B. The Golgi position is unknown (U) when the cell is not observed to move during the time interval, when the Golgi is not observable, or when the centroids of the Golgi and nucleus coincide along the migration axis. (c) The fraction of time untreated and TGFβ-treated cells spend with the Golgi ahead (open bars) or behind (filled) the nucleus or with the Golgi state unknown (gray) was quantified. Error bars are mean ± standard error, with n = 49 cells (untreated) and n = 61 cells (TGFβ treated) spanning three and six independent trials, respectively. Differences in fraction time spent in A, B, and U states are statistically significant within each treatment group (∗p < 0.0001) except that the time spent in A versus B for untreated cells is indistinguishable (#p > 0.35), as determined by ANOVA (analysis of variance) followed by post hoc Tukey’s test.
The cell expresses histone 2B-GFP in its nucleus (green) and GM130-RFP, marking its Golgi (red). Select images from this video are shown in Fig. 2.
We observed and classified three GPRN states at every time point of a cell trajectory (Fig. 2 b). In the ahead state (A), the Golgi centroid was positioned in front of the nuclear centroid along the direction of cell migration. The behind state (B) occurred when the Golgi centroid was behind the nuclear centroid relative to the direction of migration. The unknown state (U) consisted of instances that could not be identified as either A or B state. The U state occurred when the Golgi was undetectable or when the cell did not move, making the direction of migration ambiguous.
With GPRN state recorded at every time point, a migration trajectory is represented by a string of GPRN states, such as BBBAAAUBBAABBBBUBB. This string denotes the GPRN at 2.5-min intervals as the cell migrates from its start point to its final location. In this example, state A occurs in five time intervals, whereas state B occurs in 11 time intervals; thus, the fractions of time spent in states A and B are 0.28 and 0.61, respectively.
To determine whether TGFβ treatment affects Golgi positioning, we quantified the fraction of time that cells spend in the three Golgi states (Fig. 2 c). Among untreated cells, the fraction of time the Golgi is observed ahead or behind the nucleus is statistically indistinguishable. A relatively small fraction of time is spent in the U state (0.06).
In contrast, TGFβ-treated MCF-10A cells exhibit a bias in positioning the Golgi behind the nucleus during cell migration. The fraction of time the Golgi is behind the nucleus (GPRN state B) increases to 0.58, whereas the Golgi leads the nucleus (state A) for a smaller fraction of the time (0.32). The fraction of time in the U state (0.1) is nominally higher for TGFβ-treated cells than for untreated counterparts but remains a minor state relative to GPRN states A and B. These data show that TGFβ treatment induces an apparent bias in Golgi positioning, with cells more likely to position the Golgi behind the nucleus when migrating along fibrillar tracks.
Rearward positioning of the Golgi is specific to the subset of TGFβ-treated cells with enhanced motility
Although the Golgi is, on average, more likely to be behind than in front of the nucleus among TGFβ-treated cells, it is not an absolute requirement: cells move with the Golgi ahead for a non-negligible fraction of time (0.32). Population-level averaging, however, can mask stronger relationships at the single-cell level. It is unclear to what extent individual cells differ in their migration response to TGFβ and to what extent, if any, these differences in migration response are related to the rearward bias in Golgi positioning.
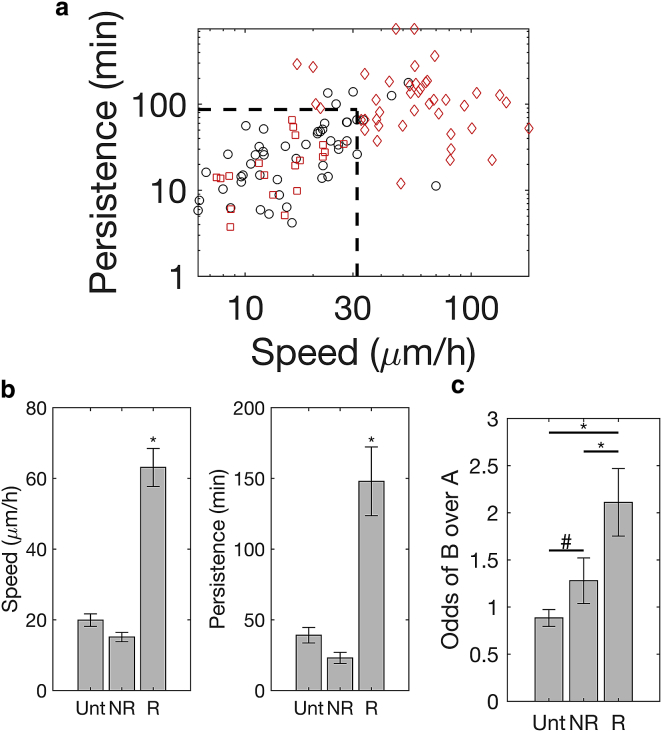
To investigate the relationship between cell migration and Golgi positioning at a single-cell level, we first examined the heterogeneity of cell-migration response to TGFβ treatment. Fig. 3 a shows the speed and persistence of individual cells. Untreated cells clustered into a relatively low-speed and low-persistence quadrant of the migration behavioral space. The cluster of untreated cells provided a reference to which the behavior of individual TGFβ-treated cells could be compared. We demarcated this reference region by determining the 90th percentile values for speed (31.3 μm/h) and persistence (86.7 min) of untreated cells.
Figure 3.
Rearward bias in Golgi position is specific to the subpopulation of TGFβ-treated cells that exhibits enhanced migration. (a) A log-log plot of migration speed and persistence for untreated cells (Unt; black, n = 49) and TGFβ-treated cells (red, n = 61). Dashed vertical and horizontal lines indicate 90th percentile values of speed and persistence, respectively, among untreated cells. TGFβ-treated cells within demarcated region are nonresponsive (NR, red squares, n = 24), whereas those outside are responsive (R) (red diamonds, n = 37). (b) Mean speed (left) and persistence (right) of R cells are significantly higher than that of NR and Unt cells (∗p < 0.0001 by post hoc Tukey’s test). (c) Odds of finding a cell in state B (Golgi behind) are quantified by the ratio of fraction time spent with the Golgi behind to the fraction time spent with Golgi ahead. One-way ANOVA (analysis of variance) followed by post hoc Tukey’s test shows that the odds of state B for R cells are statistically distinguishable from those for NR cells (∗p < 0.05) and Unt cells (∗∗p < 0.0005), whereas odds of B for Unt and NR cells are statistically indistinguishable (#p > 0.8).
In contrast to untreated MCF-10A cells, TGFβ-treated cells spread out over a broader region of the migration behavioral space. Approximately 31% of TGFβ-treated cells occupy the same low-migration quadrant as untreated cells, showing that this subpopulation did not have a motogenic response to TGFβ when compared to untreated controls. The mean speed and persistence of these nonresponsive (NR) cells are 15.1 μm/h and 23.0 min, respectively, compared to 19.9 μm/h and 39.1 min, respectively, for untreated control cells (Fig. 3 b).
In contrast to the NR subgroup, the remaining 69% of TGFβ-treated cells exhibited a quantitatively stronger migration response, with increased migration speed—and in some cases, increased persistence—as compared to untreated cells. The migration speed and persistence of this responsive subpopulation is 63.1 μm/h and 148 min, respectively, ∼4- and 6-fold greater, respectively, than the speed and persistence of NR cells (Fig. 3 b).
We next tested the hypothesis that the subpopulation of cells responsive to TGFβ exhibit a stronger bias for positioning the Golgi in the back during migration than their NR counterparts and untreated cells. We quantified the odds of finding cells in GPRN state B versus A by calculating the ratio of fraction time spent in state B to that spent in state A (ftB/ftA). In untreated cells, GPRN state B occurs slightly less often than A, with the odds of finding cells in state B versus A being 0.9 (Fig. 3 c). The odds of finding NR TGFβ-treated cells in state B is 1.3, a slight bias that is statistically indistinguishable from the no-bias behavior of untreated cells. In contrast, the odds of finding responsive TGFβ cells in GPRN state B is 2.1, a statistically significant increase in the likelihood of finding cells with the Golgi behind the nucleus compared to untreated cells.
These results demonstrate that cells with a strong motogenic response to TGFβ are approximately twofold more likely to position the Golgi in the back when migrating in comparison to counterparts that are untreated or do not have a motile response to TGFβ.
A sizable fraction of TGFβ-responsive cells exhibits an “all-or-none” commitment to Golgi positioning
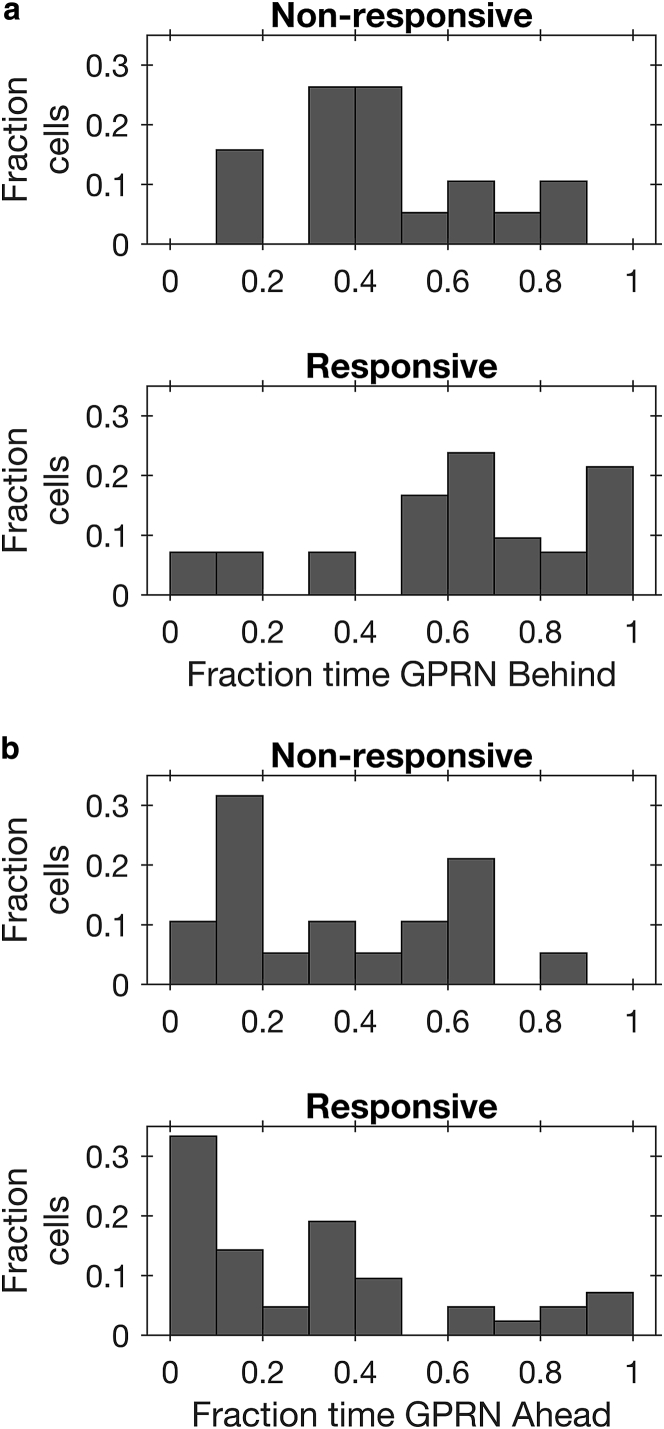
The data show that even among TGFβ-responsive cells, there is a non-negligible probability (0.33) of finding a cell with the Golgi ahead of the nucleus during migration on fiber-like micropatterns. This is consistent with the idea that Golgi positioning in individual TGFβ-responsive cells is plastic, shifting ahead and behind, albeit with twice the amount of time spent with the Golgi behind than ahead of the nucleus. Alternatively, Golgi positioning may not be plastic, and individual TGFβ-responsive cells may spend all their time in either state A or B, with twice as many cells fully committed to state B than A.
To distinguish between these possibilities, we examined the distribution of fraction of time spent in states A and B among individual cells. Among NR, TGFβ-treated cells, the fraction of time spent in state B is distributed around a mean of 0.46 (Fig. 4 a), with no cells occupying the B state for over 90% of their migration trajectory (Fig. 4 a, rightmost bin). In contrast, the distribution of fraction time spent in the B state is shifted to the right among cells that have a motile response to TGFβ, consistent with a higher mean of 0.64. Interestingly, a subset of TGFβ-responsive cells (21%) spend almost all of their time (>90%) in the B state. Thus, unlike NR cells, a subset of cells with enhanced motile response to TGFβ show a full commitment to positioning the Golgi behind the nucleus. The distribution of fraction time spent in state A (Fig. 4 b) shows a similar feature. A fraction of TGFβ-responsive cells (7%) spends over 90% of their time in state A (Fig. 4 b, rightmost bin), whereas none of the NR cells exhibit such full commitment to anterior Golgi positioning.
Figure 4.
A subset of TGFβ-R cells exhibits all-or-none Golgi positioning and fully commits to maintaining the Golgi either always ahead or always behind the nucleus. Histograms of fraction time spent with the Golgi behind (a) or ahead (b) of the nucleus are shown for TGFβ-treated NR (top) and R (bottom) cells. In both (a) and (b), fully committed cells that spend over 90% of their time with the Golgi ahead or behind the nucleus are found only in the TGFβ-R population.
Taken together, this analysis reveals that a considerable 28% of TGFβ-responsive cells fully commit to maintaining a single Golgi position, either ahead or behind the nucleus, while migrating along fiber-like micropatterns. In comparison, none of the NR, TGFβ-treated cells exhibit this fully committed state.
These findings demonstrate that TGFβ-responsive cells have two modes of Golgi positioning during EMT-induced enhanced fibrillar migration: an all-or-none mode in which a cell commits to keeping the Golgi always ahead or behind the nucleus, and a plastic mode in which the cell shifts its Golgi between ahead and behind states. In both modes, the rearward Golgi position is the preferred state. In the all-or-none mode, a cell is threefold more likely to commit to state B than A, as 21% of cells fully commit to the B state versus 7% of cells committing to the A state. In the plastic mode, cells spend more time in B than in state A.
Golgi positioning is stabilized among all TGFβ-responsive cells
Nearly a third of TGFβ-responsive cells (28%) are fully committed to positioning the Golgi either at the front or the rear of the nucleus. Among this subgroup of TGFβ-responsive cells, Golgi positioning is evidently stabilized during their entire migration trajectory. We next asked whether stabilization of Golgi position is limited to this minority, albeit sizeable, subset of TGFβ-responsive cells or whether it is a property more broadly exhibited by all TGFβ-responsive cells. That is, even among TGFβ-responsive cells that do not fully commit Golgi positioning to the A or B state, is the lifetime of one or both states extended?
To test this hypothesis, we quantified in individual TGFβ-treated cells the duration over which a GPRN state is contiguously maintained before it switches to another GPRN state. A GPRN state that persists for long durations before switching to another state has a larger lifetime and greater stability than a state that is occupied for short durations. As an example, for a cell whose GPRN states at 2.5 min intervals are represented by the string BBBAAAUBBAABBBBUBB, we condense the string to B3A3U1B2A2B4U1B2, with each subscript denoting the number of time steps during which the GPRN state is maintained. For each GPRN state, averaging the associated subscript values over the entire migration trajectory provides a direct measurement of its lifetime. For the cell in this example, the average lifetime of GPRN states A, B and U are 1.5, 2.75, and 1 time steps, respectively, or 3.8, 6.9, and 2.5 min, respectively. Using this approach, the stabilities of GPRN states A and B in individual TGFβ-treated cells were quantified.
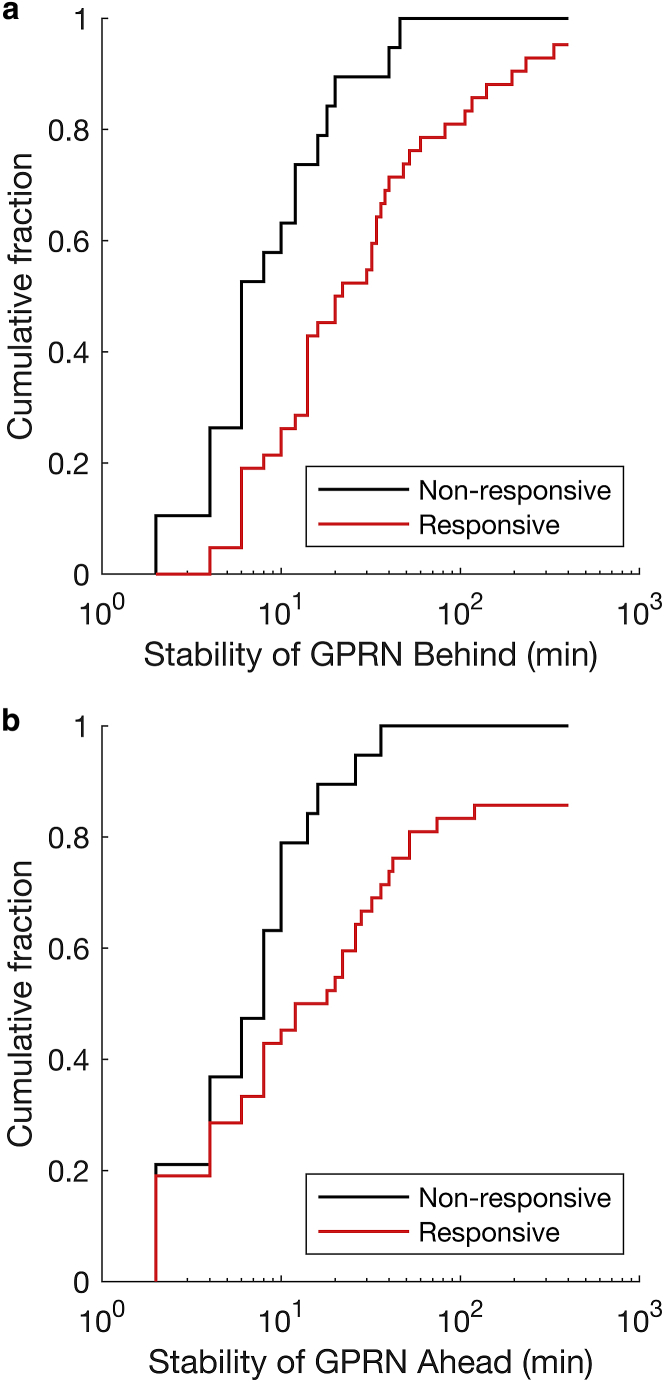
The cumulative distributions of both GPRN states show that Golgi positioning is stabilized among the vast majority of TGFβ-responsive cells (Fig. 5). For the B state, the entire distribution for TGFβ-responsive cells is shifted rightward compared to NR cells, even among cells with the lowest Golgi positional stability. The average lifetime of GPRN state B increases nearly fourfold from 13 min in NR cells to 48 min among TGFβ-responsive cells. A similar rightward shift in the cumulative distribution of the stability of state A is observed. Past the 30th percentile of Golgi positional stability, the lifetime of GPRN state A is increased in individual TGFβ-responsive cells compared to NR counterparts. The average lifetimes of the A state exhibit a twofold difference, with 11 min and 21 min for NR and responsive populations, respectively.
Figure 5.
Golgi position is stabilized among a significant fraction of TGFβ-R cells. Cumulative histograms of the stability of the Golgi behind (a) or ahead (b) of the nucleus for NR (black) and R (red) TGFβ-treated cells are shown.
Taken together, these results show that Golgi positioning is stabilized in all TGFβ-responsive cells, and in a subset (28%) of these cells, the stabilization is so strong that they spend all their time in the A or B state.
Stability of Golgi positioning, irrespective of ahead or behind the nucleus, corresponds to single-cell-migration speed
At the population level, stabilization of GPRN corresponds to enhanced migration speed and persistence of TGFβ-responsive cells relative to NR counterparts. We next asked whether this relationship between GPRN stability and cell-migration behavior applies at the level of individual TGFβ-responsive cells. That is, does the degree of GPRN stabilization correspond to the extent to which TGFβ enhances migration speed and persistence at the single-cell level?
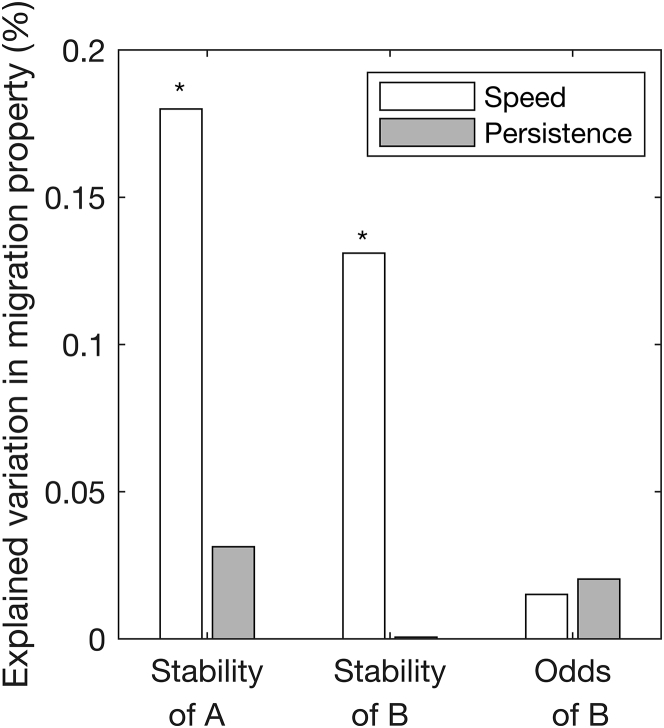
To address this question, we used Pearson’s correlation to analyze to what extent variations in the stability of Golgi state helps to explain variations in migration speed and persistence of individual TGFβ-responsive cells and whether this relationship is stronger for state A versus B. The stabilities of both states A and B exhibit a statistically significant correlation with speed (Fig. 6). Variations in the stability of states A and B capture ∼18 and 13% of the variability in migration speed, respectively. Furthermore, the Spearman correlation indicates a statistically significant monotonic relationship between cell-migration speed and the stability of both Golgi states (data not shown, p < 0.05). In contrast to speed, migration persistence does not show a strong association with Golgi stability in either A or B states.
Figure 6.
The stability of Golgi positioning—but not a bias for one position over another—exhibits a statistically significant relationship to single-cell-migration properties. The square of the Pearson’s correlation coefficient was used to determine the degree to which single-cell variations in the log of stability and odds of positioning the Golgi ahead or behind the nucleus explains variations in the log of migration speed (open) and persistence (filled) among TGFβ-R cells. The Pearson’s correlation coefficients for relating migration speed to stability of the Golgi position are statistically significant (∗p < 0.05).
This analysis demonstrates that stable maintenance of Golgi position either ahead or behind the nucleus is associated with enhanced cell-migration speed at the single-cell level. In contrast, the data show poor correlation between cell-migration properties and the relative abundance or the odds of state B over state A (Fig. 6). Thus, the time that a cell spends in one state over the other is not strongly linked to migration performance.
Discussion
We report here a novel relationship, to our knowledge, between Golgi positioning and cell motility. We show that the stabilization of Golgi positioning with respect to the nucleus—independent of whether the Golgi is ahead or behind the nucleus—corresponds to enhanced migration speed and persistence. TGFβ-mediated EMT stabilizes Golgi positioning by 2- to 4-fold while doubling migration speed and tripling migration persistence along fiber-like micropatterns. Furthermore, single-cell variations in Golgi stability capture nearly 20% of variations in migration speed, whereas the bias in Golgi positioning is not statistically predictive of cell-migration behavior. Because EMT and the development of a fibrillar microenvironment are significant cell-intrinsic and extrinsic drivers of breast cancer progression (3, 31), our findings have implications for understanding and therapeutically targeting cell migration and invasion in cancer.
EMT enhances motility in an already motogoenic fibrillar microenvironment with implications for invasiveness during cancer progression
Here, we show that TGFβ-induced EMT enhances cell migration along fiber-like tracks compared to untreated epithelial cells. That EMT enhances motility is well established in 2D contexts. That EMT also enhances migration in spatially confined fiber-like environments is significant in several ways. First, it shows that cell-intrinsic progression through EMT and fiber maturation in the TME are nonredundant pathways that have the potential to cooperate to enhance motility during cancer progression. Second is the magnitude of the EMT effect: migration speed and persistence increase two- and threefold, respectively, in response to TGFβ-mediated EMT within spatially confined tracks. These effects are on top of the well-documented positive effects that fibrillar topology alone has on cell migration of nontransformed epithelial cells when compared to nonfibrillar 2D and three-dimensional microenvironments (13, 16, 17).
Third, the combined effect of EMT and fibrillar maturation on motility is seen not only at the level of single-cell motility but also at the level of migration response to pairwise cell-cell interactions. We recently showed that when nontransformed epithelial cells encounter another cell along a fiber-like track, they reverse direction (11, 15). Progression through TGFβ-mediated EMT shifts this behavior, enabling cells to slide past each other and continue to move in their original direction. In a tumor environment crowded with cells, gaining the ability to slide allows cells to maintain their migration direction and achieve more effective dispersion, whereas cells that reverse direction at every cell-cell encounter will disperse more slowly.
Thus, the emerging picture is that EMT and fibrillar maturation in the microenvironment cooperate at the levels of individual cells and on cell-to-cell interactions to affect cell migration. A better understanding of this hierarchical cooperation will provide a foundation for predictive multiscale, multicellular models of dispersion and invasion in the TME (32).
A role for stable Golgi positioning in EMT-mediated enhancement of cell motility in fibrillar contexts
How does EMT enhance migration along spatially confined fiber-like tracks? The data indicate a role for the stabilization of GPRN. Cells that respond to TGFβ with enhanced motility also stably maintain GPRN, with the Golgi either ahead (A) or behind (B) the nucleus, for 2- to 4-fold longer duration than cells that are NR to TGFβ. In fact, the stabilization is complete in ∼28% of TGFβ-responsive cells that spend all their time in either the A or B state. Furthermore, among the remaining 72% TGFβ-responsive cells in which Golgi positioning is plastic, shifting between A and B states, the lifetime of each state is much greater than that observed among NR cells.
Untreated nontransformed epithelial cells have relatively unstable GPRN compared to TGFβ-treated cells. Thus, although spatial confinement is sufficient to confer a uniaxial cell morphology and enhance motility compared to 2D environments (13), EMT provides additional cell-intrinsic changes that stabilize Golgi position and promote a 2- and 3-fold increase in migration and persistence, respectively, relative to cells that have not undergone EMT.
In fact, single-cell variations in the stability of Golgi positioning is sufficient to capture nearly 20% of the variability in migration speed among TGFβ-responsive cells. This level of correspondence is remarkable given the many physical and molecular mechanisms involved in a complex behavior such as cell migration. In contrast to positional stability of the Golgi, bias in positioning the Golgi behind the nucleus is statistically insignificant in explaining variation in cell-migration behavior.
In future work, it is reasonable to hypothesize that combining additional molecular and physical properties—e.g., nuclear shape and deformation, properties of focal adhesions, etc.—with the stability of Golgi positioning could yield a multifactorial, predictive model of cell-migration dynamics, akin to systems-level models of apoptosis composed of MAP kinase predictors (33). The inclusion of physical properties, such as Golgi stabilization, as predictors is consistent with the physiochemical nature of the cell-migration process.
The role of the Golgi in migration is independent of its position relative to the nucleus
Our data indicate that the role of the Golgi in EMT-mediated enhanced fibrillar migration is independent of whether it is ahead or behind the nucleus. Whether a cell spends more time in state B than in state A has less than 2% association with enhanced migration. In contrast, the stabilities of both the A and B states are more strongly associated with enhanced migration speed. Furthermore, treatment with TGFβ enhances cell migration and stabilizes both A and B states, indicating that overall positional stability is more important than stabilizing one state preferentially. Thus, although TGFβ-responsive cells exhibit a population-averaged 2:1 bias in the prevalence of GPRN state B versus A, single-cell analysis shows that stability of Golgi positioning, more so than the actual position of the Golgi, is quantitatively related to cell-migration behavior.
The most developed paradigm of Golgi positional bias in migration is based on 2D wound healing models in which the Golgi, together with the MTOC/centrosome, is positioned ahead of the nucleus from which it traffics proteins to the leading edge, resupplying molecular components, such as integrins, necessary for adhesion and traction (20, 21, 22). The Golgi has also been shown to nucleate its own set of microtubules that are preferentially oriented toward the leading edge, potentially providing avenues for polarized trafficking (34, 35).
However, there are many exceptions to anterior Golgi positioning. Even during wound healing in 2D, centrosome positioning—and by inference, Golgi position—is cell-type specific, with anterior positioning in Chinese hamster ovarian cells and posterior positioning in rat-kangaroo epithelial kidney cells (36). Meanwhile, chicken embryo fibroblasts exhibit anterior positioning of the centrosome on 2D glass but exhibit no bias when migrating in collagen gels and along micron-scale grooves (37). Indeed, among individual Rat2 fibroblasts, no correlation is observed between Golgi orientation and migration speed on 2D surfaces (24).
Furthermore, rearward bias in Golgi positioning is observed in cells migrating on narrow micropatterns. African green monkey kidney cells (Bsc1) position the Golgi posteriorly 70% of the time during migration along 6–14 μm micropatterns (12). Another group observed rearward bias in 3T3 fibroblasts, albeit less pronounced (13). Meanwhile, we report here no bias in untreated mammary epithelial cells and a 60% rearward bias in TGFβ-treated cells migrating along 10-μm fiber-like micropatterns.
Considering these observations altogether, it is evident that cells are capable of migrating with anterior, posterior, and even no bias in Golgi position in a context- and cell-type-dependent manner. Furthermore, in some situations, such as the aforementioned studies in fiber-like contexts, the positional bias is quantitative and not qualitatively exclusive, suggesting that anterior versus posterior positioning of the Golgi is not an overriding factor in motility in these systems. Meanwhile, our findings point to a significant role for the stability of Golgi position in fibrillar migration, and in future studies, it will be important to test the relationship between Golgi positional stability and motility across a wide range of cell types and contexts.
A working hypothesis for a structural/physical role for the Golgi in cell motility
How is stable positioning of the Golgi—independent of whether it is ahead or behind the nucleus—advantageous to migration along fiber-like tracks? We propose a structural and physical role for the Golgi in cell motility. The Golgi is part of a multicomponent intracellular scaffold. It physically interacts and associates with microtubules, the MTOC, and the nucleus (19). The Golgi is also associated with the actin cytoskeleton (38, 39), and particularly relevant in the context of EMT, it interacts with the vimentin intermediate filament network (40). We hypothesize that the Golgi, as a component of this intracellular scaffold, bears load from cell-generated forces and affects the transmission of these forces across the cell. If the Golgi position undergoes frequent changes, its effect on the spatial distribution of forces at one instant in time could be counteracted by contributing to a different, conflicting force distribution later in time. To the extent that the Golgi position can be maintained, its role in distributing forces is consistent over time, and migration is made more efficient.
Additionally, a canonical arrangement of the Golgi next to the MTOC next to the nucleus is well known (19). Every change in Golgi position could disrupt this three-member structure, and the rate at which the Golgi-MTOC-nucleus arrangement is reacquired may limit motility. Increased Golgi positional stability would minimize this disruption.
The extent to which the Golgi regulates the assembly and maintenance of an intracellular “core” scaffold mediating migration is unclear. Most likely, the role of the Golgi must be understood in the context of an integrative, systems-level cross talk among organelles and cytoskeletal components. Whether stable Golgi positioning regulates or is an indicator of a stable migration-promoting core scaffold will help determine whether it could be targeted as part of a therapeutic strategy or be utilized in an imaging-based drug screening platform, respectively. In either case, the reported findings and implications underscore a need to more deeply understand the regulatory mechanisms and downstream consequences of stable Golgi positioning during cell migration.
Conclusions
EMT and maturation of collagen fibrils in the microenvironment are associated with cancer progression. To the extent that these factors can act as nonredundant promoters of motility, their simultaneous occurrence during cancer progression has the potential to accelerate motility and invasion. In this context, we find that EMT significantly enhances migration in a spatially confined, one-dimensional microenvironment that is already known to be more conducive to motility than an unconstrained 2D context. Moreover, single-cell analysis shows that EMT-mediated stabilization of Golgi position and not a preference for anterior or posterior positioning is associated with enhancements in migration speed and persistence. These results suggest a model in which the Golgi is part of a core physical scaffold whose stability provides a reliable platform for coordinating and transducing forces across the cell, thereby enabling more efficient migration.
Author Contributions
R.J.N. and A.R.A. conceived and designed the experiments. R.J.N. performed the experiments. R.J.N. and M.L.L. conducted the image analysis. R.J.N. and A.R.A. analyzed results and wrote the manuscript. All authors edited the manuscript.
Acknowledgments
We thank Nick Ngai for making and sharing the MCF-10A cell line expressing nuclear and Golgi markers.
This work was funded by National Institutes of Health grant R01CA138899 to A.R.A., National Institutes of Health grant R01CA098830, and the Era of Hope Scholar Award from the Department of Defense Breast Cancer Research Program to S.K.M.
Editor: Stanislav Shvartsman.
Footnotes
One video is available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)31145-7.
References
- 1.Provenzano P.P., Eliceiri K.W., Keely P.J. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 2006;4:38. doi: 10.1186/1741-7015-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conklin M.W., Eickhoff J.C., Keely P.J. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am. J. Pathol. 2011;178:1221–1232. doi: 10.1016/j.ajpath.2010.11.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nieto M.A., Huang R.Y., Thiery J.P. EMT: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 4.Kalluri R., Weinberg R.A. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Massagué J. TGFbeta in cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamouille S., Xu J., Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang W., Wyckoff J.B., Condeelis J.S. Single cell behavior in metastatic primary mammary tumors correlated with gene expression patterns revealed by molecular profiling. Cancer Res. 2002;62:6278–6288. [PubMed] [Google Scholar]
- 8.Sidani M., Wyckoff J., Condeelis J. Probing the microenvironment of mammary tumors using multiphoton microscopy. J. Mammary Gland Biol. Neoplasia. 2006;11:151–163. doi: 10.1007/s10911-006-9021-5. [DOI] [PubMed] [Google Scholar]
- 9.Maiuri P., Rupprecht J.F., Voituriez R. Actin flows mediate a universal coupling between cell speed and cell persistence. Cell. 2015;161:374–386. doi: 10.1016/j.cell.2015.01.056. [DOI] [PubMed] [Google Scholar]
- 10.Sharma V.P., Beaty B.T., Eddy R.J. Reconstitution of in vivo macrophage-tumor cell pairing and streaming motility on one-dimensional micro-patterned substrates. Intravital. 2012;1:77–85. doi: 10.4161/intv.22054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milano D.F., Natividad R.J., Asthagiri A.R. Positive quantitative relationship between EMT and contact-initiated sliding on fiber-like tracks. Biophys. J. 2016;111:1569–1574. doi: 10.1016/j.bpj.2016.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pouthas F., Girard P., Reynaud E.G. In migrating cells, the Golgi complex and the position of the centrosome depend on geometrical constraints of the substratum. J. Cell Sci. 2008;121:2406–2414. doi: 10.1242/jcs.026849. [DOI] [PubMed] [Google Scholar]
- 13.Doyle A.D., Wang F.W., Yamada K.M. One-dimensional topography underlies three-dimensional fibrillar cell migration. J. Cell Biol. 2009;184:481–490. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scarpa E., Roycroft A., Mayor R. A novel method to study contact inhibition of locomotion using micropatterned substrates. Biol. Open. 2013;2:901–906. doi: 10.1242/bio.20135504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milano D.F., Ngai N.A., Asthagiri A.R. Regulators of metastasis modulate the migratory response to cell contact under spatial confinement. Biophys. J. 2016;110:1886–1895. doi: 10.1016/j.bpj.2016.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kushiro K., Asthagiri A.R. Modular design of micropattern geometry achieves combinatorial enhancements in cell motility. Langmuir. 2012;28:4357–4362. doi: 10.1021/la204872c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li S., Bhatia S., Chien S. Effects of morphological patterning on endothelial cell migration. Biorheology. 2001;38:101–108. [PubMed] [Google Scholar]
- 18.Vinogradova T., Miller P.M., Kaverina I. Microtubule network asymmetry in motile cells: role of Golgi-derived array. Cell Cycle. 2009;8:2168–2174. doi: 10.4161/cc.8.14.9074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yadav S., Linstedt A.D. Golgi positioning. Cold Spring Harb. Perspect. Biol. 2011;3:a005322. doi: 10.1101/cshperspect.a005322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kupfer A., Louvard D., Singer S.J. Polarization of the Golgi apparatus and the microtubule-organizing center in cultured fibroblasts at the edge of an experimental wound. Proc. Natl. Acad. Sci. USA. 1982;79:2603–2607. doi: 10.1073/pnas.79.8.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bergmann J.E., Kupfer A., Singer S.J. Membrane insertion at the leading edge of motile fibroblasts. Proc. Natl. Acad. Sci. USA. 1983;80:1367–1371. doi: 10.1073/pnas.80.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yadav S., Puri S., Linstedt A.D. A primary role for Golgi positioning in directed secretion, cell polarity, and wound healing. Mol. Biol. Cell. 2009;20:1728–1736. doi: 10.1091/mbc.E08-10-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Distel M., Hocking J.C., Köster R.W. The centrosome neither persistently leads migration nor determines the site of axonogenesis in migrating neurons in vivo. J. Cell Biol. 2010;191:875–890. doi: 10.1083/jcb.201004154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uetrecht A.C., Bear J.E. Golgi polarity does not correlate with speed or persistence of freely migrating fibroblasts. Eur. J. Cell Biol. 2009;88:711–717. doi: 10.1016/j.ejcb.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ratner S., Sherrod W.S., Lichlyter D. Microtubule retraction into the uropod and its role in T cell polarization and motility. J. Immunol. 1997;159:1063–1067. [PubMed] [Google Scholar]
- 26.Kim J.H., Asthagiri A.R. Matrix stiffening sensitizes epithelial cells to EGF and enables the loss of contact inhibition of proliferation. J. Cell Sci. 2011;124:1280–1287. doi: 10.1242/jcs.078394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lalli M.L., Asthagiri A.R. Collective migration exhibits greater sensitivity but slower dynamics of alignment to applied electric fields. Cell. Mol. Bioeng. 2015;8:247–257. doi: 10.1007/s12195-015-0383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lalli M.L., Wojeski B., Asthagiri A.R. Label-free automated cell tracking: analysis of the role of E-cadherin expression in collective electrotaxis. Cell. Mol. Bioeng. 2017;10:89–101. doi: 10.1007/s12195-016-0471-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li C.C., Kuo J.C., Vaughan M. Effects of brefeldin A-inhibited guanine nucleotide-exchange (BIG) 1 and KANK1 proteins on cell polarity and directed migration during wound healing. Proc. Natl. Acad. Sci. USA. 2011;108:19228–19233. doi: 10.1073/pnas.1117011108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bershadsky A.D., Futerman A.H. Disruption of the Golgi apparatus by brefeldin A blocks cell polarization and inhibits directed cell migration. Proc. Natl. Acad. Sci. USA. 1994;91:5686–5689. doi: 10.1073/pnas.91.12.5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Insua-Rodríguez J., Oskarsson T. The extracellular matrix in breast cancer. Adv. Drug Deliv. Rev. 2016;97:41–55. doi: 10.1016/j.addr.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 32.Szabó A., Merks R.M. Cellular potts modeling of tumor growth, tumor invasion, and tumor evolution. Front. Oncol. 2013;3:87. doi: 10.3389/fonc.2013.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janes K.A., Albeck J.G., Yaffe M.B. A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science. 2005;310:1646–1653. doi: 10.1126/science.1116598. [DOI] [PubMed] [Google Scholar]
- 34.Miller P.M., Folkmann A.W., Kaverina I. Golgi-derived CLASP-dependent microtubules control Golgi organization and polarized trafficking in motile cells. Nat. Cell Biol. 2009;11:1069–1080. doi: 10.1038/ncb1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Efimov A., Kharitonov A., Kaverina I. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev. Cell. 2007;12:917–930. doi: 10.1016/j.devcel.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yvon A.M., Walker J.W., Wadsworth P. Centrosome reorientation in wound-edge cells is cell type specific. Mol. Biol. Cell. 2002;13:1871–1880. doi: 10.1091/mbc.01-11-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schütze K., Maniotis A., Schliwa M. The position of the microtubule-organizing center in directionally migrating fibroblasts depends on the nature of the substratum. Proc. Natl. Acad. Sci. USA. 1991;88:8367–8371. doi: 10.1073/pnas.88.19.8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Egea G., Lázaro-Diéguez F., Vilella M. Actin dynamics at the Golgi complex in mammalian cells. Curr. Opin. Cell Biol. 2006;18:168–178. doi: 10.1016/j.ceb.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 39.Guet D., Mandal K., Manneville J.B. Mechanical role of actin dynamics in the rheology of the Golgi complex and in Golgi-associated trafficking events. Curr. Biol. 2014;24:1700–1711. doi: 10.1016/j.cub.2014.06.048. [DOI] [PubMed] [Google Scholar]
- 40.Gao Y., Sztul E. A novel interaction of the Golgi complex with the vimentin intermediate filament cytoskeleton. J. Cell Biol. 2001;152:877–894. doi: 10.1083/jcb.152.5.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The cell expresses histone 2B-GFP in its nucleus (green) and GM130-RFP, marking its Golgi (red). Select images from this video are shown in Fig. 2.