ABSTRACT
Rab proteins regulate vesicular transport in eukaryotic cells and establish connections to various cellular structures and processes by interacting with so-called effector molecules. Several of these effectors are known to not only bind a single Rab protein, but to be able to bind multiple different Rabs simultaneously. In this review we will give a short overview of effectors in general and (putative) functions of the aforementioned multivalent Rab:effector interactions.
KEYWORDS: bMERB, EHBPs, Gcc185, Micals, multivalency, MyoV, Rab effectors, Rabenosyn-5
Introduction
The superfamily of Ras-related Small GTPases regulates a variety of different physiological functions in cells, switching certain pathways on or off in a spatiotemporally controlled manner.1 One major subfamily within this superfamily are Rab proteins (Ras-related in brain)2 that control vesicular trafficking in eukaryotic cells, including all steps involved from budding of vesicles from a donor membrane, transport along cytoskeletal tracks, and finally tethering and fusion with a target membrane.3,4
Two interconnected cycles are tightly regulated to allow Rab proteins to fulfill their function: First, Rab proteins are C-terminally modified at one or usually two cysteine residues with geranylgeranyl- moieties, allowing the Rabs to reversibly bind to intracellular membranes.5 In addition, a solubilizing factor, the GDP dissociation inhibitor (GDI) can bind prenylated Rab proteins to allow shuttling of the Rab protein through the hydrophilic cytosol.6 Second, Rab proteins (like all members of the superfamily) can bind either GDP (guanosine-5’-diphosphate; “off-state”) or GTP (guanosine-5’-triphosphate; “on-state”). The cycling between these two states is regulated by additional factors, guanine nucleotide exchange factors (GEFs) that catalyze the GDP-GTP exchange reaction and GTPase activating proteins (GAPs) that accelerate the hydrolysis of bound GTP, thus switching the Rabs on or off, respectively.7 Structurally, switching between the active and inactive states is characterized by conformational changes of two important regions within the small GTPases, the switch I and switch II regions, which adopt a mostly disordered state in the GDP-bound form due to loss of interactions with the γ-phosphate group, and an ordered and structurally defined conformation in the active state.8
The interconnectivity and mutual dependence of the two cycles of reversible membrane attachment and nucleotide exchange arises from the fact that GDI only binds Rabs in their inactive GDP-bound state with high affinity, but not in their GTP-bound active state.9 This leads to a general tendency for Rab proteins to be attached to membranes in their active state, but to be bound to GDI and solubilized in the cytosol in the inactive state.
Finally, in addition to the regulatory factors mentioned above, Rab proteins interact with proteins collectively referred to as “effector” proteins. These proteins are characterized by high specificity toward the conformationally defined active state of Rabs, and not surprisingly, were found to interact with the switch regions of Rab proteins in all structurally characterized Rab:effector complexes.10,11 In this review, we present an overview of (structurally characterized) Rab:effector complexes with a focus on the binding stoichiometry and multivalent effectors that can bind more than one Rab molecule simultaneously, as well as putative functions thereof.
Rab effectors vary greatly in size and structure
Several structures of Rab proteins and their effectors have been solved in the past (see overview in Fig. 1 and Table 1) and it has been shown that structures of different effectors, despite being mostly α-helical, especially within their Rab-binding interfaces, vary greatly both in size and overall domain architecture.11 Besides containing (predicted) coiled-coils, Rab effectors usually also contain a variety of additional functional domains, such as domains interacting and modifying cytoskeletal components (e.g. MyoVb and Micals), membrane binding and remodeling motifs (e.g., OCRL, Slac2-a, Rabenosyn-5, EEA1, Fip2) or other Rab/GTPase interaction and regulatory motifs and domains (e.g., GEF and GAP domains in AnkRD27, OCRL and Rab6IP1), and this modular architecture allows them to establish a connection between active GTP-bound Rabs and physiological downstream effects (Fig. 1). Most effectors characterized so far bind to the Rab proteins via one or two (anti-parallel) helices (e.g., Slac2-a, Slp2-a, Rabenosyn-5, Rabphilin-3a, Rabaptin-5, RILP, etc.), however Fig. 1 also shows several exceptions to this ‘rule’. AnkRD27 (VARP) binds Rab32 via 4 parallel helices of its ankyrin repeats,12 MyoVb interacts with Rab11a via 3 helices and a loop,13 EEA1 binds Rab5 via a short β-hairpin and an α-helix14 and OCRL binds to Rab8 via an Immunoglobulin-like β-strand domain and an α-helix.15
Figure 1.
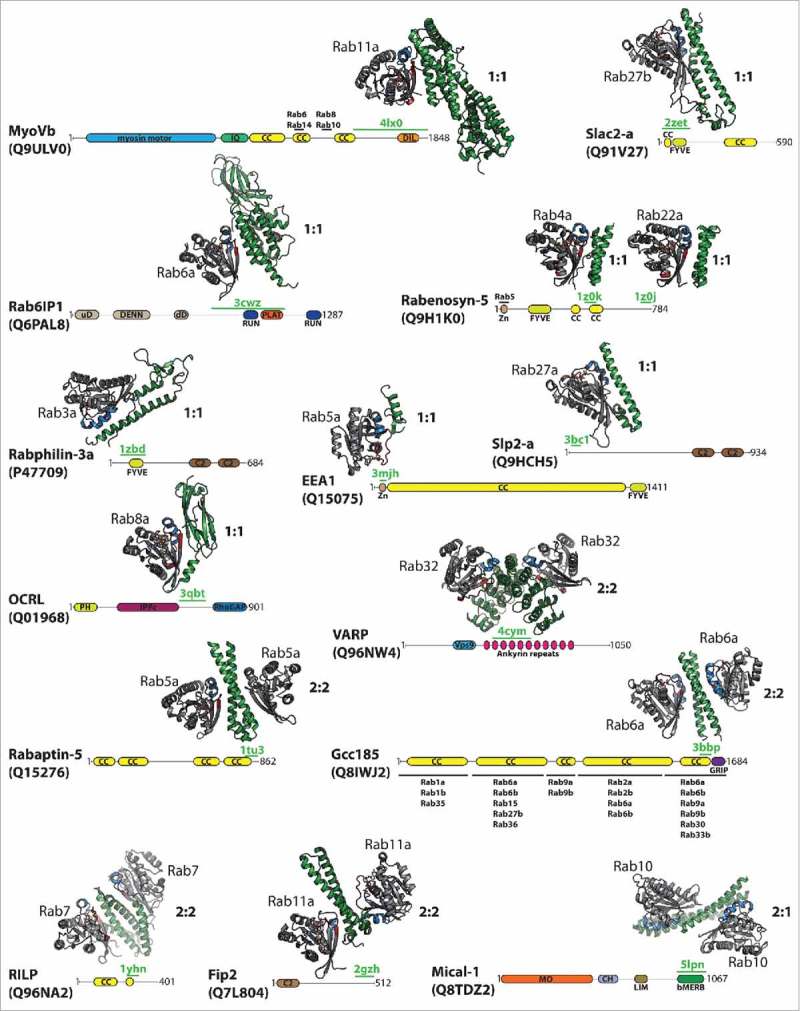
Structurally characterized Rab:effector complexes. The structures of several Rab:effector complexes are indicated together with a scheme showing the overall domain architecture of the effector proteins (according to the SMART and uniprot databases42,43). The structurally resolved regions involved in Rab-binding are highlighted by a green line including the corresponding pdb code, additional Rab-binding sites/regions are indicated by a black line including the Rab proteins known to bind at the corresponding site (the additional Rab binding sites in MyoVb are inferred from studies on MyoVa37). Structures of the Rab:effector complexes are shown in cartoon style (Rab: gray with switch I in red and switch II in blue; effector: green). Besides the Rab-binding regions, Rab effectors usually contain several additional domains, as indicated (bMERB – bivalent Mical/EHBP Rab-binding domain; C2 – C2 Ca2+-binding domain; CC – (predicted) coiled-coil regions; CH – calponin-homology domain; DENN – Rab GEF domain (uD – upstream DENN, dD – downstream DENN); DIL – Dilute domain; FYVE – FYVE Zinc finger domain; GRIP - golgin-97, RanBP2α, Imh1p and p230/golgin-245 (Golgi targeting) domain; IPPc - Inositol polyphosphate phosphatase; IQ – IQ Calmodulin-binding motif; LIM – Zinc-binding domain (Lin-11, Isl-1, Mec-3); MO – Monooxygenase domain; PLAT – Polycystin-1, Lipoxygenase, α-Toxin domain; RUN – RUN domain; Vps9 – Vps9 GEF domain; Zn – Zinc finger C2H2 domain).
Table 1.
Structurally characterized Rab:effector complexes.
| Rab: effector complex | stoichiometry | pdb id | KD | references |
|---|---|---|---|---|
| Rab3a: Rabphilin-3a/Exophilin1 | 1:1 | 1zbd | — | 44 |
| Rab4: Rabenosyn-5440–503 | 1:1 | 1z0k | 6.2 µM | 19 |
| Rab5a: EEA1 | 1:1 | 3mjh | 2.4 µM | 14 |
| Rab6: Rab6IP1 | 1:1 | 3cwz | — | 45 |
| Rab8a: OCRL1 | 1:1 | 3qbt | 0.9 µM | 15 |
| Rab11a: MyoVb | 1:1 | 4lx0 | 254 nM | 13 |
| Rab22: Rabenosyn-5728–784 | 1:1 | 1z0j | 9.7 µM | 19 |
| Rab27a: Slp2-a/Exophilin4 | 1:1 | 3bc1 | 13.4 nM | 46,47 |
| Rab27a: Slac2-a/Melanophilin | 1:1 | 2zet | 112 nM | 47,48 |
| Rab5a: Rababtin-5 | 2:2 | 1tu3 | — | 23 |
| Rab6: Gcc185 | 2:2 | 3bbp | 2.3 µM | 22 |
| Rab7: RILP | 2:2 | 1yhn | — | 24 |
| Rab11a: Fip2 | 2:2 | 2 gzh | 40 nM | 25,49 |
| Rab32: VARP/AnkRD27 | 2:2 | 4cym | 2.5 µM | 12 |
| Rab10:Mical-1 | 2:1 | 5lpn | — | 18 |
The specificity of effector proteins for the active form of Rab proteins can be explained by the fact that they interact with the conformationally defined switch and interswitch regions of GTP-bound Rabs. Several interactions between amino acids located in these regions and the effector are highly conserved between different Rab:effector complexes. The interface includes several hydrophobic residues (among which is a conserved aromatic triad consisting of Trp, Phe and Phe/Tyr within the interswitch and switch II regions) that are also shared by Arf GTPases.7,16 The specificity toward certain Rabs or Rab families on the other hand is obtained by interactions of the effectors with stretches of amino acids known as RabSF motifs 1–4 or CDRs (complementarity-determining regions) that differ strongly between different Rab families.16
Several reviews11,16,17 give a more extensive overview of Rab:effector interactions and the reader is referred to these for further information.
Multivalency in Rab effectors and its functions
Whereas some Rab effectors contain only a single Rab-binding site, several others are known to contain two or even more (different) binding sites (Table 2) or to dimerize and thus allow binding of two Rabs to similar or identical binding sites within the dimeric effectors (structurally characterized Rab:effector complexes are shown in Fig. 1 including representatives of all cases mentioned above). With respect to the stoichiometry of Rab:effector interactions, the recently published bMERB domains are the first known examples of Rab effectors that provide two separate binding sites within a single domain, and due to their high similarity, the sites have been suggested to have evolved by gene duplication.18 Another example of Rab effectors that presumably originated from gene duplication are the two separate Rab-binding sites in Rabenosyn-5 (pdb ids 1z0k and 1z0j) that share a very similar relative orientation toward their bound Rab molecules (Fig. 1).19,20 Similar to the case in bMERB domains, the sequences have diverged and two Rab binding sites have been obtained in Rabenosyn-5 with similar overall structure, but different Rab specificity.20 However, in contrast to bMERB domains, the Rab binding sites are separated by a linker in Rabenosyn-5 and thus form two separate Rab binding domains (in addition to the two structurally characterized Rab binding sites, Rabenosyn-5 contains a third Rab5 binding site at the N-terminal Zn2+ finger domain21).
Table 2.
Multivalent Rab effectors.
| Effector | Rabs (No. of sites) | reference |
|---|---|---|
| bMERB domains | Rab1, Rab8, Rab10, Rab13, Rab15, Rab35 (2 sites within 1 domain) | 18 |
| dGcc88 | Rab6, Rab19, Rab30 (2 sites) | 29 |
| dGolgin-97 | Rab6, Rab19, Rab30 (3 sites) | 29 |
| Gcc185 | Rab1a, Rab1b, Rab35, Rab6a, Rab6b, Rab9a, Rab9b, Rab15, Rab27b, Rab30, Rab33b, Rab35, Rab36 (5–6 sites) | 22,26 |
| MyosinVa | Rab3, Rab6, Rab8, Rab10, Rab11, Rab14, Rab39b (3 sites) | 37 |
| Rab6IP1 | Rab6 and Rab11 | 33 |
| Rabip4’ | Rab4 and Rab5 (2 sites) | 32 |
| Rabenosyn-5 | Rab4 and Rab5 (2 sites) | 31 |
The observation that many effectors form dimers and thus establish two separate (but identical) binding sites (e.g., Rabaptin-5, Gcc185, RILP, Fip2, AnkRD27)12,22-25 and the presence of (non-identical) Rab-binding sites in multiple separate domains (e.g., Gcc185, Rabenosyn-5)19,26 or in one single domain (e.g., Micals, EHBPs)18 argues for specific functions of this multivalency. One possible function of this multivalency might be that it contributes to a stabilization of active Rabs in complex with their effectors at membranes by simply multiplying the number of membrane-anchored isoprenoids within the complex.27 In this case, formation of effector dimers with two identical binding sites is sufficient, but several other functions can be envisaged that would benefit from or would only be possible if two non-identical Rab binding sites were present. Such multivalent effectors could, for example, assist in the formation of Rab microdomains, small membrane areas with high local concentrations of a certain Rab protein (Fig. 2).3,28 The idea behind this is that activated Rabs could bind to a multivalent effector protein, which subsequently binds and recruits a second (and depending on the number of available Rab-binding sites a third or fourth) Rab molecule. In a dynamic equilibrium and depending on the association and dissociation kinetics of the Rab:effector complexes, a certain number of Rab proteins would again dissociate and be available to bind another effector molecule, and this would finally lead to an effective clustering of Rabs and effectors at a certain site on the membrane (Fig. 2B). The availability of two different sites would facilitate this, since one high affinity site could be available for the initial recruitment and stabilization of the effector at a certain membrane domain whereas a second lower affinity site would allow for the necessary dynamics of recruitment and release.
Figure 2.
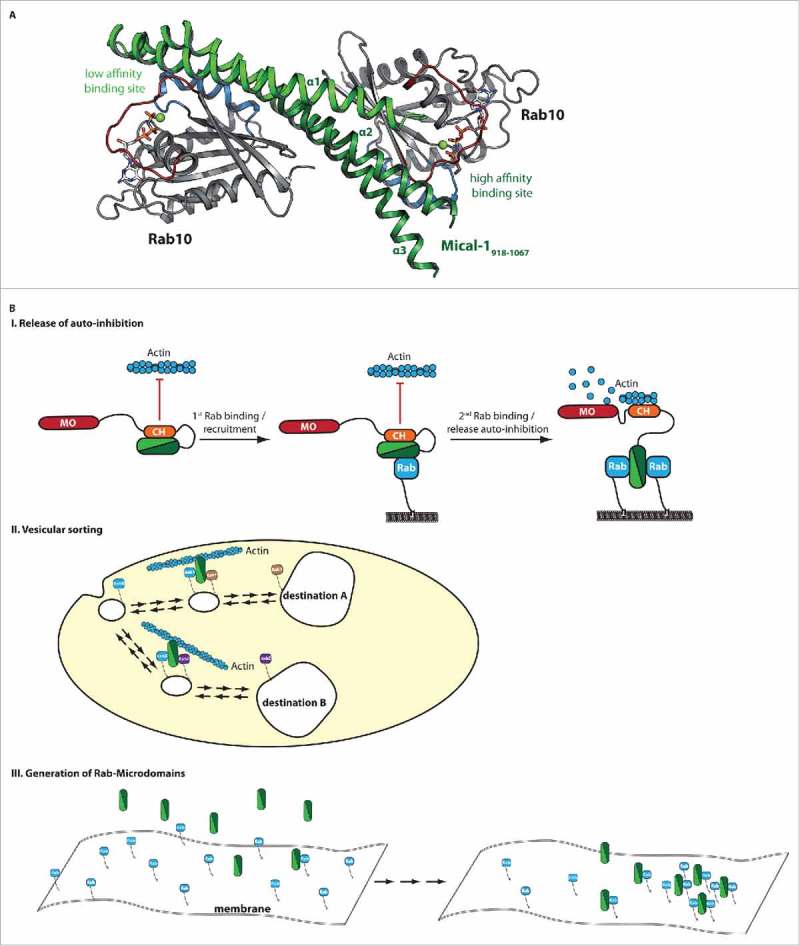
Structure and putative functions of bMERB domains. (A) The X-ray crystallographic structure of Mical-1 in complex with Rab10 revealed two separate Rab binding sites (additional biochemical characterization showed that the binding sites have different affinities to Rab proteins and the corresponding low and high affinity binding sites at the N- and C-terminus, respectively, are indicated).18 (B) Putative functions of bMERB domains are illustrated. These include the release of auto-inhibition by competitive replacement of the auto-inhibitory interaction due to Rab-binding (I.), sorting of vesicular cargo via different specificities of the two separate Rab-binding sites toward certain Rab proteins and establishment of a connection between different Rab compartments (II.) and finally the formation of Rab microdomains on intracellular membranes (III.).
Of course, the presence of non-identical binding sites not only allows binding of two similar Rabs, but also allows evolution of different specificities and binding of multiple different Rabs to the different binding sites to establish connections between these. Several Golgi-localized proteins such as Gcc185 (which contains 5–6 separate Rab-binding sites with varying specificities, see Fig. 1)26 for example have been suggested to function in the stabilization of Golgi stacks via simultaneous interaction with several different Golgi-localized Rab proteins and/or to reach into the cytosol to act as capturing tentacles of Rabs and associated vesicles.26,29,30 In some multivalent effectors, binding to multiple different Rabs simultaneously and/or subsequently might also function to establish connections between trafficking steps regulated by different Rabs, as previously suggested for Rabenosyn-5 and Rabip4’ (both linking Rab4 and Rab531,32), Rab6IP1 (linking Rab6 and Rab1133) and bMERB domains (connecting Rab8 family members, Arf6 and Rab3518,34). In many of these cases, the Rab-binding repertoire, consisting of mostly endosomal Rab proteins, further suggests a possible role in sorting of endocytosed cargo and transport to different destinations within the cell (Fig. 2B). A similar sorting function can also be envisaged for exocytic trafficking.35,36 It should also be noted that multivalency in some effectors is not exclusive to Rab proteins, since some have been shown to also bind members of other small GTPase families (e.g., Gcc185 and Mical-L1 interact with members of the Arl and Arf GTPases, respectively)21,34 Furthermore, previous observations that binding of one small GTPase at one site of an effector can either positively or negatively influence the affinity of another GTPase at a second site in Gcc185 are very interesting, since they would allow for an additional level of allosteric control in regulated Rab and/or general GTPase cascades.21,22
In addition to providing platforms for the coordinated recruitment of different Rab proteins, multiple binding sites in certain effector proteins could also simply ensure binding to multiple different Rabs due to a conserved function of the effector within different trafficking pathways. In this regard, MyoVa, a molecular motor for vesicular transport, has been shown to contain 3 different Rab-binding sites with varying specificity. Although the exact function of the multiple binding sites is not understood,37 this might generally allow binding and transport of vesicles from different origins within the cell, with different Rab proteins attached to their surface.
Yet another putative function of the two separate binding sites specific to bMERB domains comes from the observation that Micals form an intramolecular auto-inhibitory interaction between their CH/LIM domains and the bMERB domain (Fig. 2B),35,38-40 suggesting that while one Rab-binding site might have a function for recruitment of the effector protein to a certain membrane, the presumably high concentration of active Rabs at this membrane could subsequently lead to displacement of the auto-inhibitory interaction via competitive binding (Fig. 2B). This model assumes an overlapping binding interface of one of the Rab sites and the auto-inhibitory intra-molecular interaction. The release of auto-inhibition would subsequently allow Mical proteins to bind to actin and depolymerize actin via their monooxygenase domain (Fig. 2B).41
Conclusions
In many publications on Rabs and their effectors it has been shown that multivalency is a common theme in Rab:effector interactions that can be established either by dimerization of the effector proteins, implying binding of two identical Rab molecules, or the presence of separate Rab binding sites within the effectors. In many cases, binding of separate Rabs presumably functions to establish connections between Rab domains and coordinate subsequent vesicular trafficking steps and sorting of vesicular cargo. However, besides this function several other roles of multivalency can be envisaged, as discussed above. bMERB domains might additionally represent a special case due to the propensity of proteins harboring this domain to form an auto-inhibitory interaction. Further research on the Rab specificity and other proteins or domains interacting with the bMERB domain will presumably shed light on their specific role in vesicular trafficking as well as actin interaction and regulation.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev 2013; 93(1):269-309; PMID:23303910; http://dx.doi.org/ 10.1152/physrev.00003.2012 [DOI] [PubMed] [Google Scholar]
- [2].Touchot N, Chardin P, Tavitian A. Four additional members of the ras gene superfamily isolated by an oligonucleotide strategy: molecular cloning of YPT-related cDNAs from a rat brain library. Proc Natl Acad Sci U S A 1987; 84(23):8210-4; PMID:3317403; http://dx.doi.org/ 10.1073/pnas.84.23.8210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol 2001; 2(2):107-17; PMID:11252952; http://dx.doi.org/ 10.1038/35052055 [DOI] [PubMed] [Google Scholar]
- [4].Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev 2011; 91(1):119-49; PMID:21248164; http://dx.doi.org/ 10.1152/physrev.00059.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhang FL, Casey PJ. Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem 1996; 65:241-69; PMID:8811180; http://dx.doi.org/ 10.1146/annurev.bi.65.070196.001325 [DOI] [PubMed] [Google Scholar]
- [6].Pfeffer S, Aivazian D. Targeting Rab GTPases to distinct membrane compartments. Nat Rev Mol Cell Biol 2004; 5(11):886-96; PMID:15520808; http://dx.doi.org/ 10.1038/nrm1500 [DOI] [PubMed] [Google Scholar]
- [7].Itzen A, Goody RS. GTPases involved in vesicular trafficking: structures and mechanisms. Semin Cell Dev Biol 2011; 22(1):48-56; PMID:20951823; http://dx.doi.org/ 10.1016/j.semcdb.2010.10.003 [DOI] [PubMed] [Google Scholar]
- [8].Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science 2001; 294(5545):1299-304; PMID:11701921; http://dx.doi.org/ 10.1126/science.1062023 [DOI] [PubMed] [Google Scholar]
- [9].Wu YW, Tan KT, Waldmann H, Goody RS, Alexandrov K. Interaction analysis of prenylated Rab GTPase with Rab escort protein and GDP dissociation inhibitor explains the need for both regulators. Proc Natl Acad Sci U S A 2007; 104(30):12294-9; PMID:17640890; http://dx.doi.org/ 10.1073/pnas.0701817104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Khan AR. Oligomerization of rab/effector complexes in the regulation of vesicle trafficking. Prog Mol Biol Transl Sci 2013; 117:579-614; PMID:23663983; http://dx.doi.org/ 10.1016/B978-0-12-386931-9.00021-0 [DOI] [PubMed] [Google Scholar]
- [11].Oesterlin LK, Pylypenko O, Goud B. Effectors of Rab GTPases: Rab binding specificity and their role in coordination of Rab function and localization, in Ras Superfamily Small G Proteins: Biology and Mechanisms 2, Wittinghofer A, Editor 2014, Springer International Publishing; p. 39-66. [Google Scholar]
- [12].Hesketh GG, Pérez-Dorado I, Jackson LP, Wartosch L, Schäfer IB, Gray SR, McCoy AJ, Zeldin OB, Garman EF, Harbour ME, et al.. VARP is recruited on to endosomes by direct interaction with Retromer, where together they function in export to the cell surface. Dev Cell 2014; 29(5):591-606; PMID:24856514; http://dx.doi.org/ 10.1016/j.devcel.2014.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pylypenko O, Attanda W, Gauquelin C, Lahmani M, Coulibaly D, Baron B, Hoos S, Titus MA, England P, Houdusse AM. Structural basis of myosin V Rab GTPase-dependent cargo recognition. Proc Natl Acad Sci U S A 2013; 110(51):20443-8; PMID:24248336; http://dx.doi.org/ 10.1073/pnas.1314329110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Mishra A, Eathiraj S, Corvera S, Lambright DG. Structural basis for Rab GTPase recognition and endosome tethering by the C2H2 zinc finger of Early Endosomal Autoantigen 1 (EEA1). Proc Natl Acad Sci U S A 2010; 107(24):10866-71; PMID:20534488; http://dx.doi.org/ 10.1073/pnas.1000843107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hou X, Hagemann N, Schoebel S, Blankenfeldt W, Goody RS, Erdmann KS, Itzen A. A structural basis for Lowe syndrome caused by mutations in the Rab-binding domain of OCRL1. EMBO J 2011; 30(8):1659-70; PMID:21378754; http://dx.doi.org/ 10.1038/emboj.2011.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Khan AR, Menetrey J. Structural biology of Arf and Rab GTPases' effector recruitment and specificity. Structure 2013; 21(8):1284-97; PMID:23931141; http://dx.doi.org/ 10.1016/j.str.2013.06.016 [DOI] [PubMed] [Google Scholar]
- [17].Mott HR, Owen D. Structures of Ras superfamily effector complexes: What have we learnt in two decades? Crit Rev Biochem Mol Biol 2015; 50(2):85-133; PMID:25830673; http://dx.doi.org/ 10.3109/10409238.2014.999191 [DOI] [PubMed] [Google Scholar]
- [18].Rai A, Oprisko A, Campos J, Fu Y, Friese T, Itzen A, Goody RS, Gazdag EM, Müller MP. bMERB domains are bivalent Rab8 family effectors evolved by gene duplication. Elife 2016; 5; http://dx.doi.org/ 10.7554/eLife.18675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Eathiraj S, Pan X, Ritacco C, Lambright DG. Structural basis of family-wide Rab GTPase recognition by rabenosyn-5. Nature 2005; 436(7049):415-9; PMID:16034420; http://dx.doi.org/ 10.1038/nature03798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Morrison HA, Dionne H, Rusten TE, Brech A, Fisher WW, Pfeiffer BD, Celniker SE, Stenmark H, Bilder D. Regulation of early endosomal entry by the Drosophila tumor suppressors Rabenosyn and Vps45. Mol Biol Cell 2008; 19(10):4167-76; PMID:18685079; http://dx.doi.org/ 10.1091/mbc.E08-07-0716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lee MTG, Mishra A, Lambright DG. Structural mechanisms for regulation of membrane traffic by Rab GTPases. Traffic 2009; 10(10):1377-1389; PMID:19522756; http://dx.doi.org/ 10.1111/j.1600-0854.2009.00942.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Burguete AS, Fenn TD, Brunger AT, Pfeffer SR. Rab and arl GTPase family members cooperate in the localization of the golgin GCC185. Cell 2008; 132(2):286-298; PMID:18243103; http://dx.doi.org/ 10.1016/j.cell.2007.11.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhu G, Zhai P, Liu J, Terzyan S, Li G, Zhang XC. Structural basis of Rab5-Rabaptin5 interaction in endocytosis. Nat Struct Mol Biol 2004; 11(10):975-83; PMID:15378032; http://dx.doi.org/ 10.1038/nsmb832 [DOI] [PubMed] [Google Scholar]
- [24].Wu M, Wang T, Loh E, Hong W, Song H. Structural basis for recruitment of RILP by small GTPase Rab7. EMBO J 2005; 24(8):1491-501; PMID:15933719; http://dx.doi.org/ 10.1038/sj.emboj.7600643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Jagoe WN, Lindsay AJ, Read RJ, McCoy AJ, McCaffrey MW, Khan AR. Crystal structure of rab11 in complex with rab11 family interacting protein 2. Structure 2006; 14(8):1273-83; PMID:16905101; http://dx.doi.org/ 10.1016/j.str.2006.06.010 [DOI] [PubMed] [Google Scholar]
- [26].Hayes GL, Brown FC, Haas AK, Nottingham RM, Barr FA, Pfeffer SR. Multiple Rab GTPase binding sites in GCC185 suggest a model for vesicle tethering at the trans-Golgi. Mol Biol Cell 2009; 20(1):209-17; PMID:18946081; http://dx.doi.org/ 10.1091/mbc.E08-07-0740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Goody RS, Wu Y, Itzen A. Prenylation of RabGTPases, their delivery to membranes, and Rab recycling, in Ras Superfamily Small G Proteins: Biology and Mechanisms 2: Transport, Wittinghofer A, Editor 2014, Springer International Publishing: Cham: p. 3-16. [Google Scholar]
- [28].Nottingham RM, Pfeffer SR. Defining the boundaries: Rab GEFs and GAPs. Proc Natl Acad Sci U S A 2009; 106(34):14185-6; PMID:19706500; http://dx.doi.org/ 10.1073/pnas.0907725106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sinka R, Gillingham AK, Kondylis V, Munro S. Golgi coiled-coil proteins contain multiple binding sites for Rab family G proteins. J Cell Biol 2008; 183(4):607-15; PMID:19001129; http://dx.doi.org/ 10.1083/jcb.200808018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Goud B, Gleeson PA. TGN golgins, Rabs and cytoskeleton: regulating the Golgi trafficking highways. Trends Cell Biol 2010; 20(6):329-36; PMID:20227882; http://dx.doi.org/ 10.1016/j.tcb.2010.02.006 [DOI] [PubMed] [Google Scholar]
- [31].de Renzis S, Sonnichsen B, Zerial M. Divalent Rab effectors regulate the sub-compartmental organization and sorting of early endosomes. Nat Cell Biol 2002; 4(2):124-33; PMID:11788822; http://dx.doi.org/ 10.1038/ncb744 [DOI] [PubMed] [Google Scholar]
- [32].Fouraux MA, Deneka M, Ivan V, van der Heijden A, Raymackers J, van Suylekom D, van Venrooij WJ, van der Sluijs P, Pruijn GJ. Rabip4′ is an effector of rab5 and rab4 and regulates transport through early endosomes. Mol Biol Cell 2004; 15(2):611-24; PMID:14617813; http://dx.doi.org/ 10.1091/mbc.E03-05-0343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Miserey-Lenkei S, Waharte F, Boulet A, Cuif MH, Tenza D, El Marjou A, Raposo G, Salamero J, Héliot L, Goud B, et al.. Rab6-interacting protein 1 links Rab6 and Rab11 function. Traffic 2007; 8(10):1385-403; PMID:17725553; http://dx.doi.org/ 10.1111/j.1600-0854.2007.00612.x [DOI] [PubMed] [Google Scholar]
- [34].Rahajeng J, Giridharan SS, Cai B, Naslavsky N, Caplan S. MICAL-L1 is a tubular endosomal membrane hub that connects Rab35 and Arf6 with Rab8a. Traffic 2012; 13(1):82-93; PMID:21951725; http://dx.doi.org/ 10.1111/j.1600-0854.2011.01294.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sun Y, Jaldin-Fincati J, Liu Z, Bilan PJ, Klip A. A complex of Rab13 with MICAL-L2 and alpha-actinin-4 is essential for insulin-dependent GLUT4 exocytosis. Mol Biol Cell 2016; 27(1):75-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Grigoriev I, Yu KL, Martinez-Sanchez E, Serra-Marques A, Smal I, Meijering E, Demmers J, Peränen J, Pasterkamp RJ, van der Sluijs P, et al.. Rab6, Rab8, and MICAL3 cooperate in controlling docking and fusion of exocytotic carriers. Curr Biol 2011; 21(11):967-74; PMID:21596566; http://dx.doi.org/ 10.1016/j.cub.2011.04.030 [DOI] [PubMed] [Google Scholar]
- [37].Lindsay AJ, Jollivet F, Horgan CP, Khan AR, Raposo G, McCaffrey MW, Goud B. Identification and characterization of multiple novel Rab-myosin Va interactions. Mol Biol Cell 2013; 24(21):3420-34; PMID:24006491; http://dx.doi.org/ 10.1091/mbc.E13-05-0236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Schmidt EF, Shim SO, Strittmatter SM. Release of MICAL autoinhibition by semaphorin-plexin signaling promotes interaction with collapsin response mediator protein. J Neurosci 2008; 28(9):2287-97; PMID:18305261; http://dx.doi.org/ 10.1523/JNEUROSCI.5646-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sakane A, Honda K, Sasaki T. Rab13 regulates neurite outgrowth in PC12 cells through its effector protein, JRAB/MICAL-L2. Mol Cell Biol 2010; 30(4):1077-87; PMID:20008558; http://dx.doi.org/ 10.1128/MCB.01067-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Vitali T, Maffioli E, Tedeschi G, Vanoni MA. Properties and catalytic activities of MICAL1, the flavoenzyme involved in cytoskeleton dynamics, and modulation by its CH, LIM and C-terminal domains. Arch Biochem Biophys 2016; 593:24-37; PMID:26845023; http://dx.doi.org/ 10.1016/j.abb.2016.01.016 [DOI] [PubMed] [Google Scholar]
- [41].Hung RJ, Pak CW, Terman JR. Direct redox regulation of F-actin assembly and disassembly by Mical. Science 2011; 334(6063):1710-3; PMID:22116028; http://dx.doi.org/ 10.1126/science.1211956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A 1998; 95(11):5857-64; PMID:9600884; http://dx.doi.org/ 10.1073/pnas.95.11.5857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].UniProt C. UniProt: a hub for protein information. Nucleic Acids Res 2015; 43(Database issue):D204-12; PMID:25348405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ostermeier C, Brunger AT. Structural basis of Rab effector specificity: crystal structure of the small G protein Rab3A complexed with the effector domain of rabphilin-3A. Cell 1999; 96(3):363-74; PMID:10025402; http://dx.doi.org/ 10.1016/S0092-8674(00)80549-8 [DOI] [PubMed] [Google Scholar]
- [45].Recacha R, Boulet A, Jollivet F, Monier S, Houdusse A, Goud B, Khan AR. Structural basis for recruitment of Rab6-interacting protein 1 to Golgi via a RUN domain. Structure 2009; 17(1):21-30; PMID:19141279; http://dx.doi.org/ 10.1016/j.str.2008.10.014 [DOI] [PubMed] [Google Scholar]
- [46].Chavas LM, Ihara K, Kawasaki M, Torii S, Uejima T, Kato R, Izumi T, Wakatsuki S. Elucidation of Rab27 recruitment by its effectors: structure of Rab27a bound to Exophilin4/Slp2-a. Structure 2008; 16(10):1468-77; PMID:18940603; http://dx.doi.org/ 10.1016/j.str.2008.07.015 [DOI] [PubMed] [Google Scholar]
- [47].Fukuda M. Distinct Rab27A binding affinities of Slp2-a and Slac2-a/melanophilin: Hierarchy of Rab27A effectors. Biochem Biophys Res Commun 2006; 343(2):666-74; PMID:16554019; http://dx.doi.org/ 10.1016/j.bbrc.2006.03.001 [DOI] [PubMed] [Google Scholar]
- [48].Kukimoto-Niino M, Sakamoto A, Kanno E, Hanawa-Suetsugu K, Terada T, Shirouzu M, Fukuda M, Yokoyama S. Structural basis for the exclusive specificity of Slac2-a/melanophilin for the Rab27 GTPases. Structure 2008; 16(10):1478-90; PMID:18940604; http://dx.doi.org/ 10.1016/j.str.2008.07.014 [DOI] [PubMed] [Google Scholar]
- [49].Junutula JR, Schonteich E, Wilson GM, Peden AA, Scheller RH, Prekeris R. Molecular characterization of Rab11 interactions with members of the family of Rab11-interacting proteins. J Biol Chem 2004; 279(32):33430-7; PMID:15173169; http://dx.doi.org/ 10.1074/jbc.M404633200 [DOI] [PubMed] [Google Scholar]


