Abstract
The plant pathogen Agrobacterium tumefaciens infects plants and introduces the transferred-DNA (T-DNA) region of the Ti-plasmid into nuclear DNA of host plants to induce the formation of tumors (crown galls). The T-DNA region carries iaaM and iaaH genes for synthesis of the plant hormone auxin, indole-3-acetic acid (IAA). It has been demonstrated that the iaaM gene encodes a tryptophan 2-monooxygenase which catalyzes the conversion of tryptophan to indole-3-acetamide (IAM), and the iaaH gene encodes an amidase for subsequent conversion of IAM to IAA. In this article, we demonstrate that A. tumefaciens enhances the production of both IAA and phenylacetic acid (PAA), another auxin which does not show polar transport characteristics, in the formation of crown galls. Using liquid chromatography-tandem mass spectroscopy, we found that the endogenous levels of phenylacetamide (PAM) and PAA metabolites, as well as IAM and IAA metabolites, are remarkably increased in crown galls formed on the stem of tomato plants, implying that two distinct auxins are simultaneously synthesized via the IaaM-IaaH pathway. Moreover, we found that the induction of the iaaM gene dramatically elevated the levels of PAM, PAA and its metabolites, along with IAM, IAA and its metabolites, in Arabidopsis and barley. From these results, we conclude that A. tumefaciens enhances biosynthesis of two distinct auxins in the formation of crown galls.
Keywords: Agrobacterium tumefaciens, Auxin, Crown gall, Indole-3-acetic acid, Phenylacetic acid
Introduction
Auxin is the first discovered plant hormone that plays crucial roles in various aspects of plant growth and development including embryogenesis and tropism (Zhao 2010). The most studied naturally occurring auxin is indole-3-acetic acid (IAA). Plants regulate cell expansion and differentiation in an IAA concentration-dependent manner. In plants, IAA is mainly produced from tryptophan (Trp) through indole-3-pyruvate (IPA) in a two-step reaction using the TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS (TAA) family and the YUCCA (YUC) flavin-containing monooxygenase family of enzymes (Stepanova et�al. 2008, Tao et�al. 2008, Mashiguchi et�al. 2011, Won et�al. 2011) (Fig. 1). The TAA and YUC genes contribute to the spatiotemporal regulation of IAA concentrations in plant growth and development (Kasahara 2016). Plants evolutionally acquired various metabolic enzymes for inactivation of IAA, including IAA-amino acid conjugate synthetase (GH3), DIOXYGENASE FOR AUXIN OXIDATION (DAO), UDP-glucosyltransferase UGT84B1, and IAA METHYLTRANSFERASE 1 (IAMT1) (Korasick et�al. 2013, Zhao et�al. 2013). Among them, GH3 genes are known as a typical family of early auxin responsive genes and play important roles for rapid inactivation of IAA in various plant tissues. IAA displays polar transport characteristics and rapidly changes its distribution pattern. In plants, membrane-localized PIN efflux carrier proteins, AUXIN1/LIKE-AUX1 (AUX1/LAX) influx transporters and ATP-binding cassette subfamily B (ABCB) transporters regulate cellular concentration of IAA (Friml 2010). Among these membrane-localized proteins, multiple PIN efflux carrier proteins are thought to play a primary role in polar auxin transport to form auxin concentration gradients. In the nucleus, IAA is perceived by auxin receptors TRANSPORT INHIBITOR RESPONSE 1/AUXIN SIGNALING F-BOX (TIR1/AFB) and AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) proteins and promotes the ubiquitination of Aux/IAAs by the SKP1 (S-PHASE KINASE-ASSOCIATED PROTEIN 1)-CULLIN-F-BOX (SCF) E3-ligase complex (Salehin et�al. 2015). Degradation of polyubiquitinated Aux/IAAs by the 26S–proteasome pathway activates auxin response factors (ARFs) and induces the expression of various auxin-responsive genes.
Phenylacetic acid (PAA) has been recognized as another type of auxin that widely occurs in plants at various concentration ranges (Wightman and Lighty 1982, Korasick et�al. 2013, Sugawara et�al. 2015). In general, plants accumulate much higher levels of PAA than IAA. Similar to IAA, PAA forms TIR1/AFB-Aux/IAA auxin co-receptor complexes and induces various auxin responsive genes in Arabidopsis (Shimizu-Mitao and Kakimoto 2014, Sugawara et�al. 2015). PAA displays less auxin activity than IAA in most plant systems, especially in the Avena test which requires polar auxin transport (Haagen Smit and Went 1935, Muir et�al. 1967), except for the lateral root formation test in pea (Schneider et�al. 1985). Previous study suggests a possibility that TAA and YUC genes may also contribute to PAA synthesis in Arabidopsis, but the PAA biosynthesis pathway is still elusive (Dai et�al. 2013, Sugawara et�al. 2015, Cook et�al. 2016) (Fig.�1). PAA is also metabolized to its amino acid conjugates by the GH3 family in Arabidopsis (Sugawara et�al. 2015) (Fig.�1). An important difference between IAA and PAA is that IAA is actively transported in a polar manner, but PAA is not. Unlike IAA, 14C-labeled PAA does not display polar movement in various plant tissues (Prochazka and Borkovec 1984, Suttle and Mansager 1986, Morris and Johnson 1987). Moreover, IAA forms concentration gradients in maize coleoptiles in response to gravitropic stimulation, whereas PAA does not show these characteristics (Sugawara et�al. 2015). All of these facts suggest that PAA likely plays important roles as an auxin in plants; however its physiological significance, including in plant-microbe interactions, is still largely unknown.
Fig. 1.
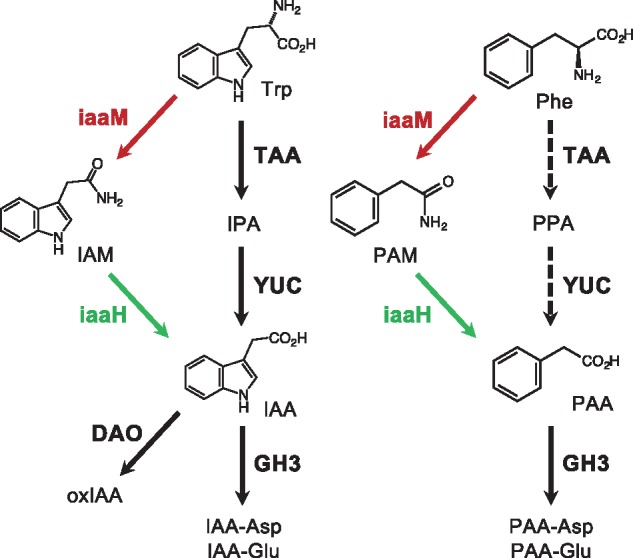
Auxin biosynthesis and inactivation pathways in plants and A. tumefaciens-induced crown galls. The IaaM-IaaH pathway is shown by red and green arrows. Auxin biosynthesis and inactivation enzymes identified in Arabidopsis are shown in bold. Dashed arrow indicates possible PAA biosynthesis pathway in Arabidopsis.
Agrobacterium tumefaciens is a well-studied soil bacterium that causes crown gall disease. By introducing the transferred-DNA (T-DNA) region of the Ti-plasmid into nuclear DNA of its host plants, A. tumefaciens modulates plant cell division and induces the formation of tumors. The T-DNA region carries two genes, iaaM and iaaH, for biosynthesis of auxin, as well as cytokinin biosynthesis genes (Patten and Glick 1995). The iaaM gene encodes a flavin-containing monooxygenase, TRYPTOPHAN 2-MONOOXYGENASE (TMO/IaaM), which catalyzes the conversion of Trp to indole-3-acetamide (IAM) (Fig.�1). The iaaH gene encodes an amidase, INDOLE-3-ACETAMIDE HYDROLASE (IaaH), which converts IAM to IAA (Fig.�1). The iaaM and iaaH genes were originally found on the pIAA1 plasmid in the plant pathogen Pseudomonas savastanoi, which causes a disease known as olive or oleander knot, and later homologous genes were found on the Ti-plasmid of A. tumefaciens (Comai and Kosuge 1980, Yamada et�al. 1985). Previous study demonstrated that P. savastanoi IaaM proteins expressed in E. coli catalyze the conversion of Trp to IAM in vitro (Emanuele et�al. 1995). These facts suggest that A. tumefaciens can modify the endogenous levels of IAA to induce the formation of crown galls by sidestepping the main IAA biosynthesis pathway of plants. Intriguingly, P. savastanoi IaaM proteins expressed in E. coli also catalyze the conversion of phenylalanine (Phe) to phenylacetamide (PAM) in vitro (Emanuele et�al. 1995). In addition, Kemper et�al. (1985) suggested that IAM and PAM are likely to be the most important substrates for IaaH by enzyme activity tests. These facts imply that A. tumefaciens may produce both IAA and PAA via the IaaM-IaaH pathway to induce the formation of crown galls.
In this study, we investigated auxin biosynthesis in the formation of crown galls by A. tumefaciens. We found that biosynthesis of both IAA and PAA increases in crown galls and in transgenic Arabidopsis and barley expressing the A. tumefaciens iaaM gene. The IaaM-IaaH pathway is a predominant route for biosynthesis of these two auxin in tumors as treatment with inhibitors specific for plant auxin biosynthesis enzymes do not affect tumor formation. Our results suggest that A. tumefaciens likely utilizes two different types of auxins in the formation of crown galls.
Results
The endogenous levels of PAA metabolites increase in plant tumors
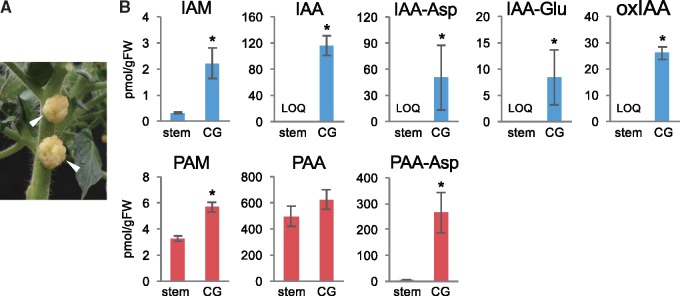
A previous enzyme activity study suggested that Phe is also a good substrate for P. savastanoi TMO/IaaM in vitro (Emanuele et�al. 1995). Since A. tumefaciens iaaM and iaaH genes are homologous to the corresponding iaaM and iaaH genes in P. savastanoi, we hypothesized that A. tumefaciens may also modulate the endogenous levels of PAA using the IaaM-IaaH pathway to induce the formation of crown galls. To address this, we took an auxin metabolite analysis-based approach using a liquid chromatography-tandem mass spectroscopy (LC-MS/MS). We generated tumors on the stems of tomato plants by inoculating cells of wild-type A. tumefaciens (Fig.�2A) and analyzed endogenous amounts of PAA and its metabolites, as well as IAA and its metabolites, in tumors. Based on the results from previous studies (Kemper et�al. 1985, Emanuele et�al. 1995), we predicted PAM as a possible precursor in the IaaM-dependent PAA biosynthesis pathway (Fig.�1). To analyze PAM, we synthesized 15N-labeled PAM as an internal standard and used it for quantification by LC-MS/MS. Two PAA-amino acid conjugates, PAA-aspartate (PAA-Asp) and PAA-glutamate (PAA-Glu), were also analyzed using corresponding 13C-labeled internal standards, because plants can metabolize overproduced auxins to their amino acid conjugates for homeostatic regulation (Sugawara et�al. 2015). For auxin metabolite analysis, we used 3-week-old crown galls formed on the stems of tomato plants. As shown in Fig.�2B, we found that the endogenous levels of IAM, IAA and its primary amino acid conjugates, IAA-aspartate (IAA-Asp) and IAA-glutamate (IAA-Glu), and a major IAA metabolite, 2-oxindole-3-acetic acid (oxIAA), were significantly increased in tumors compared with those in the stem tissues of uninoculated plants. Increase of IAM, IAA and its metabolites clearly supports a previously proposed model that introduction of the A. tumafaciens IaaM-IaaH pathway enhances IAA biosynthesis in the formation of crown galls. LC-MS/MS analysis of PAA metabolites demonstrated that the stem of uninoculated plants accumulates PAM, PAA and PAA-Asp, and the levels of PAA were much higher than those of IAA. Although the increase of PAA levels was slight in tumors, endogenous amounts of PAM and PAA-Asp were substantially increased, similar to IAM and IAA-Asp (Fig.�2B). The levels of PAA-Glu were under our limit of quantification (LOQ). This evidence strongly suggests that the bacterial IaaM-IaaH pathway also enhances PAA biosynthesis.
Fig. 2.
Auxin metabolite concentrations in crown galls. (A) Crown galls formed on the stem of tomato plants (indicated by white arrows). (B) Auxin metabolite concentrations in the stem of noninoculated tomato plants (stem) and crown galls (CG). Values are mean � SD (n = 3). *Differences between the stems and crown galls are statistically significant at P < 0.05 (Student’s t-test).
Expression of iaaM gene enhances PAA production in Arabidopsis
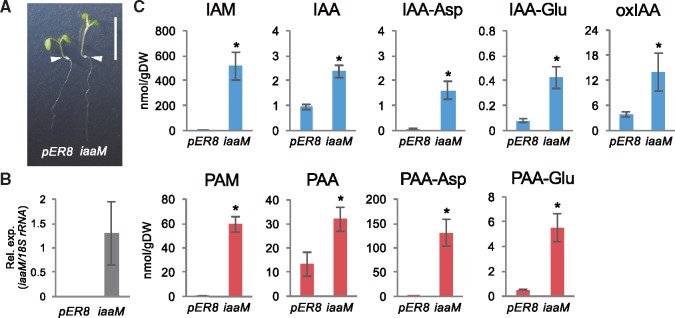
Previous studies show that expression of the iaaM gene results in increases of IAA concentrations and high-auxin phenotypes such as epinastic cotyledons and elongated hypocotyls and stems in Arabidopsis and other plants (Klee et�al. 1987, Sitbon et�al. 1991, Romano et�al. 1995). These observations indicate that not only does IaaM catalyze a rate-limiting step in the bacterial IaaM-IaaH-dependent IAA biosynthesis, but also suggest that certain amidases which can catalyze the subsequent conversion of IAM to IAA likely occur in various plants. To provide evidence that IaaM also catalyzes a rate-limiting step of PAA synthesis in vivo, we next studied the impact of iaaM gene expression on auxin metabolite concentrations in Arabidopsis as a dicot model system. We generated transgenic Arabidopsis harboring the iaaM gene in estradiol-inducible expression system using a pER8 vector (pER8: iaaM). Expression of iaaM gene in pER8: iaaM transgenic seedlings causes high-auxin phenotypes including elongated hypocotyls and epinastic cotyledons (Fig.�3A, B), similar to what has been previously reported for transgenic Arabidopsis expressing iaaM gene under control of the cauliflower mosaic virus-derived 35S promoter (Romano et�al. 1995). For auxin metabolite analysis, seedlings of vector control (pER8) or pER8: iaaM were treated with estradiol for 2 days in MS liquid media. A substantial level of iaaM transcripts was detected in estradiol treated plants based on quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis (Fig.�3B). LC-MS/MS analysis of auxin metabolites indicates that the induction of iaaM expression dramatically increased the endogenous levels of IAM, IAA, IAA-Asp, IAA-Glu and oxIAA by 945-, 2.5-, 20-, 5- and 3.6-fold, respectively, compared with those in pER8 plants (Fig.�3C). These results are consistent with the notion that IaaM catalyzes a rate-limiting step in the IaaM-IaaH-dependent IAA biosynthesis. Similar to IAA, we found that the induction of iaaM expression also elevated the levels of PAM, PAA, PAA-Asp and PAA-Glu by 83-, 2.4-, 274- and 10-fold, respectively (Fig.�3C). These metabolite profiles indicate that IaaM remarkably enhances the synthesis of PAM in Arabidopsis and mediates a rate-limiting step in PAA synthesis.
Fig. 3.
Auxin metabolite concentrations in transgenic Arabidopsis plants expressing the A. tumefaciens iaaM. (A) Phenotypes of pER8 and pER8: iaaM (iaaM) plants treated with β-estradiol (2 �M) for 5 days. Bars indicate 1 cm. White arrows indicate the root-shoot junctions in each plant. (B) Expression levels of iaaM gene of plants shown in (A). Values are mean � SD (n = 4). (C) Endogenous concentrations of auxin metabolites in plants, 5 days after treatment with β-estradiol. Values are mean � SD (n = 3). *Differences between pER8 and iaaM plants are statistically significant at P < 0.05 (Student’s t-test).
Intriguingly, the levels of IAM were higher by 2–3 orders of magnitude than those of IAA and IAA-Asp in iaaM-induced plants, whereas the levels of PAA and PAA-Asp were similar or higher than those of PAM. These results suggest that Arabidopsis has amidases that can more efficiently metabolize PAM than IAM in vivo. In addition, although PAA-Asp and PAA-Glu levels increased 274- and 10-fold, respectively, the levels of PAA were elevated only 2.4-fold in pER8-iaaM compared with pER8. This indicates that GH3 genes also play a role in determining the endogenous levels of PAA concentrations.
We note that transgenic tomato plants harboring the iaaM gene in estradiol-inducible expression system were also generated using a pMDC7 vector. However, iaaM gene expression levels were not increased by estradiol treatment probably due to a weak promotor activity of this vector in tomato plant system (Valdivia et�al. 2013).
IaaM can promote PAA biosynthesis in barley
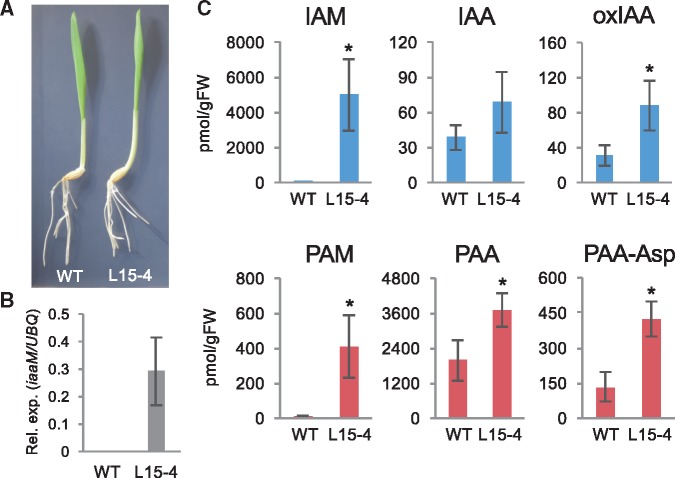
We further studied the effect of iaaM gene expression on IAA and PAA production in monocot plants using barley as a model system. We used barley in the experiments because of the availability of complete genome sequence, transformation, and easy cultivation in growth chambers (Tingay et�al. 1997, Sakata et�al. 2010, Mascher et�al. 2017). Transgenic barley harboring iaaM gene in an estradiol-inducible expression system were generated using a pERV1-hyg vector (Valdivia et�al. 2013). Using a qRT-PCR analysis method, we identified more than 10 independent transgenic lines of barley in which iaaM expression was induced significantly by estradiol treatment. For auxin metabolite analysis, we used mature fifth leaves (five-leaf stage) of wild-type barley plants (WT) and transgenic barley plants expressing iaaM (L15-4) treated with 10 �M estradiol for 2 days. No significant high-auxin phenotypes were observed in seedlings or fifth leaves of L15-4 plants by expression of iaaM gene under our growth conditions (Fig.�4A, B). However, we found that the induction of iaaM expression markedly increased the endogenous levels of IAM (2,014-fold) and oxIAA in L15-4 plants compared with those in WT plants (Fig.�4C). For PAA metabolites, we found that the induction of iaaM expression in L15-4 plants results in the elevation of PAM (49-fold), PAA (1.9-fold) and PAA-Asp (3.2-fold) levels compared with WT plants (Fig.�4C). The levels of IAA-Asp, IAA-Glu and PAA-Glu were under our limit of quantification. These results further support the idea that the A. tumefaciens iaaM gene can also enhance the synthesis of both IAA and PAA in monocot plants. We also found that the levels of IAM were two orders of magnitude higher than those of IAA in iaaM-induced barley leaf tissues, whereas the levels of PAA were higher than those of PAM. Moreover, PAA levels increased 54-fold more than IAA levels in L15-4 plants. These results suggest that amidases in barley more efficiently metabolize PAM than IAM, as with the case of Arabidopsis.
Fig. 4.
Auxin metabolite concentrations in transgenic barley expressing the A. tumefaciens iaaM. (A) Phenotypes of control (WT) and pERV1: iaaM (L15-4) plants treated with β-estradiol (10 �M) for 8 days. Bar indicates 1 cm. (B) Expression levels of iaaM gene. Values are mean � SD (n = 4). (C) Endogenous concentrations of auxin metabolites in WT and L15-4 transgenic plants. Values are mean � SD (n = 3). *Differences between WT and L15-4 plants are statistically significant at P < 0.05 (Student’s t-test).
Auxin receptor inhibitor can suppress the formation of crown galls
We provided evidence suggesting that the IaaM-IaaH pathway is a predominant route for synthesis of both IAA and PAA in crown galls. We predicted that application of specific inhibitors for auxin receptors, but not for IAA biosynthesis enzymes of plants, would suppress the formation of crown galls, if the IaaM-IaaH pathway is a predominant route for IAA biosynthesis in tumors. Recently, various specific inhibitors for the auxin-mediated pathway such as L-kynurenine (Kyn) for TAA1 (He et�al. 2011), yucasin DF (YDF) for YUC (Tsugafune et�al. 2017) and auxinole (AXL) for auxin receptor TIR1/AFB proteins (Hayashi et�al. 2012) were developed. We observed that Kyn, YDF and AXL suppressed normal seedling development in Arabidopsis. However, only AXL could suppress high-auxin phenotypes of Arabidopsis seedlings caused by expression of the iaaM gene (Supplementary Fig. S1). This result supports that Kyn and YDF specifically inhibit the IPA pathway but not the IaaM-dependent auxin biosynthesis in plants. To test our hypothesis, we investigated the effect of these inhibitors on formation of crown galls (Supplementary Fig. S2). We induced the formation of tumors on the stem of tomato plants, surgically removed tumor tissues, applied solutions containing effective concentrations of AXL (100 �M), Kyn (200 �M), YDF (200 �M), or DMSO (mock) to cut end surfaces of stem tissues, and grew the test plants for 7 days. As shown in Fig.�5, tumors were formed on mock-treated tissues and they were substantially suppressed to 55% by treatment with AXL, suggesting that plant auxin receptor inhibitors can effectively block the auxin-mediated formation of tumors. In contrast, tumors are formed on Kyn- or YDF-treated tissues at levels similar to that observed in mock-treated tissues (Fig.�5), supporting our idea that auxins are synthesized predominantly from the IaaM-IaaH pathway in crown galls. Efficient suppression of tumor formation by AXL suggests a possibility that specific inhibitors of IaaM activity would be applicable for local treatment of crown gall disease.
Fig. 5.
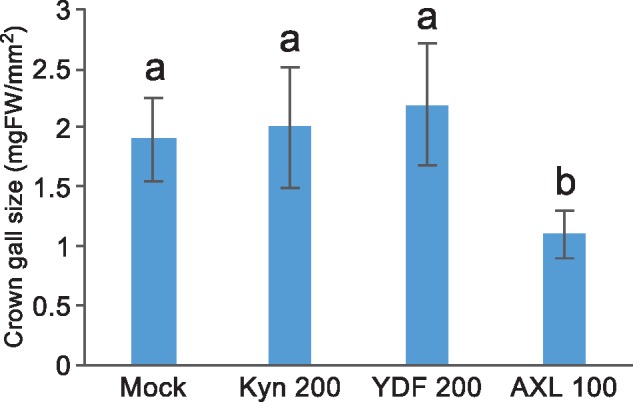
Effect of auxin synthesis or receptor inhibitors on formation of crown galls. Tumor size was measured after treatment with inhibitors for 7 days. Values are mean � SD (n = 20). *Differences between mock and AXL-treated plants are statistically significant at P < 0.05 (Steel-Dwass’s test).
Discussion
PAA biosynthesis pathway in microbes
IAA is mainly synthesized from Trp through the IPA pathway in plants. It has been suggested that up to 80% of bacteria isolated from the rhizosphere can synthesize IAA (Patten and Glick 1995). Among several proposed IAA biosynthesis pathways in bacteria, the bacterial IaaM-IaaH and IPA pathways have been extensively studied. Plant pathogens such as A. tumefaciens and P. savastanoi synthesize IAA from Trp in tumors through the bacterial IaaM-IaaH pathway as previously described. The bacterial IPA pathway is similar but distinct from the IPA pathway in plants. In plants, IAA is synthesized from Trp via IPA by TAA and YUC family enzymes (Mashiguchi et�al. 2011). In the bacterial IPA pathway, it has been suggested that Trp is converted to IPA in the first step by tryptophan transferase, however, IPA is further metabolized to indole-3-acetaldehyde (IAAld) and then IAAld is oxidized to IAA (Patten and Glick 1995). So far, it has been demonstrated that INDOLE-3-PYRUVATE DECARBOXYLASE (IPDC) encoded by the ipdC gene catalyzes the conversion of IPA to IAAld in a plant growth-promoting bacteria Azospirillum brasilense (Costacurta et�al. 1994). Recently, McClerklin et�al. (2018) identified that INDOLE-3-ACETALDEHYDE DEHYDROGENASE (ALD) encoded by AldA and AldB genes converts IAAld to IAA in Pseudomonas syringae strain DC3000. These facts suggest that various microbes can produce IAA using the bacterial IaaM-IaaH or IPA pathways by sidestepping the main IAA biosynthesis pathway of host plants.
Similar to IAA, PAA also occurs widely in plants, however the biosynthesis pathway for PAA is still under investigation (Sugawara et�al. 2015). In bacteria, Somers et�al. (2005) previously demonstrated that A. brasilense likely produces PAA from Phe via phenylpyruvate (PPA) and phenylacetaldehyde (PAAld). They also suggested that IPDC for IAA synthesis most likely contributes to PAA biosynthesis in A. brasilense. We here show that in crown galls PAA is predominantly synthesized through the IaaM-IaaH pathway (Fig.�1). In this pathway, IaaM catalyzes a rate-limiting step since the endogenous levels of PAM, PAA and its amino acid conjugates remarkably increased upon induction of iaaM expression in Arabidopsis and barley. This evidence suggests that bacteria likely produce PAA through the IaaM-IaaH and PPA pathways.
Metabolic regulation of two auxins in crown galls
In this article, we showed a significant difference in auxin metabolism between crown galls and transgenic plants expressing iaaM gene. In tumor cells, the endogenous levels of IAM were increased, but to a much lower degree than those of IAA and its metabolites (Fig.�2). A similar metabolic profile was observed for PAA. These facts indicate that both IAM and PAM were efficiently converted to IAA and PAA, respectively, by IaaH in crown galls. In contrast, expression of only the iaaM gene in transgenic plants resulted in a remarkable accumulation of IAM compared to IAA and its metabolites in Arabidopsis and barley, whereas the levels of PAM were similar or less than those of PAA and its metabolites (Figs.�3 and 4). Although substrate specificity of A. tumefaciens IaaM has not been investigated, our results suggest that plant amidases in Arabidopsis and barley likely possess higher affinity for PAM rather than IAM. Consistent with our results, recent biochemical study showed that rice, Sorghum bicolor, Medicago truncatula, and Populus trichocarpa have homologous genes of Arabidopsis amidase (AMI1) and each recombinant enzyme displayed much higher affinity for PAM rather than IAM in vitro (S�nchez-Parra et�al. 2014). Considering the fact that IAM concentration was controlled to much lower levels than IAA and its metabolites in tumor cells, we assume that A. tumefaciens IaaH likely possesses higher affinity for IAM than PAM and efficiently hydrolyzes IAM to IAA in plants.
Our auxin metabolite analysis indicates that not only IaaM and IaaH, but also GH3 and DAO contribute to determine the levels and ratio of two auxin concentrations in crown galls. Staswick et�al. (2005) previously demonstrated that six GH3 members of Arabidopsis can produce IAA-amino acid conjugates with several amino acids in vitro. Similarly, GH3 members efficiently produced PAA-amino acid conjugates with several amino acids. Sugawara et�al. (2015) showed that induction of GH3 gene expression enhances the inactivation of IAA and PAA to their amino acid conjugates in Arabidopsis. In addition, defect of DAO gene increased IAA levels and caused high-auxin phenotypes in rice (Zhao et�al. 2013). Our metabolite-based evidences suggest that GH3 and DAO enzymes actively metabolize overproduced auxins to regulate cellular auxin levels, but IaaM and IaaH produce more auxins and form tumors. Given that the endogenous levels of PAA were much higher than those of IAA in crown galls, iaaM-induced plants, and control plants we analyzed, auxin metabolic genes such as GH3 and DAO likely play important roles to modulate the levels and ratio of IAA and PAA concentrations in both normal plants and crown galls.
Physiological roles of PAA in plants
Although IAA has a pivotal role as a signal molecule in cell to cell communication, physiological roles of PAA are still unknown in plants. We demonstrate that A. tumefaciens increases not only IAA but also PAA in the formation of crown galls. This result suggests that PAA likely also plays an important role as an auxin in plant growth regulation and plant-microbe interaction.
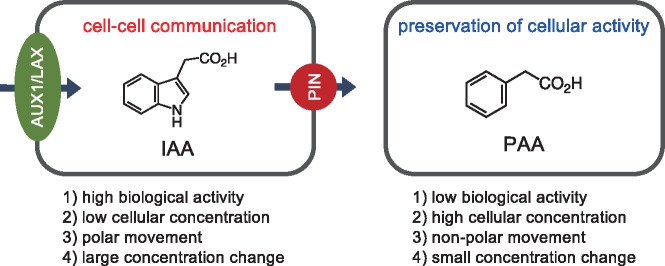
Previous studies indicate that there are four major differences between IAA and PAA in plants; (1) strength of biological activity, (2) endogenous concentration, (3) transport characteristics, and (4) cellular concentration change (Haagen Smit and Went 1935, Muir et�al. 1967, Wightman and Lighty 1982, Prochazka and Borkovec 1984, Suttle and Mansager 1986, Morris and Johnson 1987, Sugawara et�al. 2015). PAA generally shows much lower biological activity as an auxin than IAA, but it occurs at much higher concentrations than IAA in numerous plant species. Moreover, IAA moves between plant cells in a polar manner, but PAA does not have such a property. Plants rapidly and dramatically change cellular concentrations of IAA by polar auxin transport during plant growth and development such as embryogenesis, leaf development, lateral root formation, and gravitropism. In contrast, PAA moves relatively slowly between plant cells in a diffusive manner. Based on the distinct properties, we propose that PAA may play a role to maintain the steady-state levels of auxin activity for preservation of cellular activity (Fig.�6). To test our hypothesis, it is important to clarify the PAA biosynthesis pathway and analyze the phenotypes of PAA-deficient mutants.
Fig. 6.
A proposed physiological role of PAA in plants. Arrows indicate the direction of IAA movement mediated by PIN and AUX1/LAX proteins in plant cells.
It is also unclear why A. tumefaciens increases the production of two distinct auxins in crown galls. Previous studies demonstrated that IAA and PAA have different affinities for various TIR1/AFB and AUX/IAA co-receptors and may generate different outputs from auxin perception (Shimizu-Mitao and Kakimoto 2014, Sugawara et�al. 2015). Thus, A. tumefaciens may regulate the ratio of PAA and IAA concentrations to modulate cell division for vigorous formation of tumors in host plants.
Suppression of crown gall formation by auxin inhibitors
It is well known that A. tumefaciens modulates biosynthesis of two plant hormones, auxin and cytokinin, to induce the formation of crown galls. A. tumefaciens has the Tmr gene for cytokinin biosynthesis on its T-DNA region and Tmr proteins promote the production of trans-zeatin riboside 5′-monophosphate, one type of major cytokinin derivatives, in the plastids of host plants (Sakakibara et�al. 2005). Defects in auxin or cytokinin biosynthesis genes in A. tumefaciens result in the formation of abnormal shoot-like or root-like tumor tissues, respectively, on plants. Thus, combined application of inhibitors of bacterial auxin and cytokinin biosynthesis enzymes may be efficient for chemical treatment of crown galls. We demonstrated here that the auxin receptor inhibitor AXL suppresses generation of tumors on the stems of tomato plants, but auxin biosynthesis inhibitors for TAA1 and YUC did not display this effect. These facts indicate that the bacterial IaaM-IaaH pathway is a predominant route for IAA and PAA biosynthesis in crown galls and specific inhibitors of this pathway are possibly useful for chemical treatment of crown gall disease. However, based on our results, IaaH would not be a good target for the development of inhibitors. When iaaM gene expression was induced in transgenic Arabidopsis and barley, we observed a remarkable increase of IAA and PAA production in these plant systems. These facts suggest that plants possess amidases which can convert IAM or PAM to auxins independently of IaaH. Therefore, inhibitors of IaaH may not effectively block production of IAA and PAA. Alternatively, IaaM is likely a worthwhile target for chemical treatment of crown gall disease. IaaM is a flavin-containing monooxygenase and crystal structure of P. savastanoi IaaM protein has been clarified. A chemical screening-based approach would possibly give a good candidate of inhibitors, since potent and persistent inhibitors of YUC flavin-containing monooxygenase has been successfully developed by this approach (Tsugafune et�al. 2017).
Materials and Methods
Plant materials and growth conditions
Tomato (Solanum lycopersicum) cv. Micro-Tom was obtained from the National BioResources Project (NBRP) at the University of Tsukuba (http://tomatoma.nbrp.jp). Tomato plants were germinated and grown on soil in a photoperiod of 16 h light at 25�C. For analysis of auxin metabolites in crown galls, tomato plants were germinated and grown on soil for 4 weeks. Cells of wild-type A. tumefaciens strain (MAFF302307) were inoculated onto wounded stem tissues of tomato plants as previously reported (Sakakibara et�al. 2005). After incubation for another 4 weeks, crown galls formed on stem tissues were collected, frozen by liquid N2, and kept at −80�C until use. For treatment of auxin inhibitors, 4-week-old crown galls were removed from stem tissues using a razor blade, and 10 �L of 5% DMSO/H2O solution containing AXL (100 �M), Kyn (200 �M), YDF (200 �M) or mock solution (5% DMSO/H2O) were applied, respectively, to the cut end surface. After incubation for 7 days, regenerated tumors were collected and measured.
Transgenic Arabidopsis pER8: iaaM and pER8 (vector control) were used for experiments. A cDNA of iaaM gene was amplified using gene-specific primer pairs listed in Supplementary Table S1 and the YUC1pro: iaaM plasmid as template. The PVR fragment was cloned into the pER8 vector using the XhoI site by Gibson assembly system (New England Biolabs). The pER8: iaaM plasmid was transformed into Arabidopsis plants by floral dipping. Arabidopsis seeds were stratified at 4�C for 2 days in the dark. Seedlings were grown vertically on Murashige and Skoog (MS) agar medium with 1% sucrose under continuous light conditions at 22�C. For β-estradiol treatment, seeds were germinated and grown on MS agar medium containing β-estradiol (2 �M) for 4 days. For qRT-PCR and auxin metabolite analysis, seedlings were grown vertically on MS agar medium for 10 days, transferred to MS agar media and grown for 2 days, then grown for another 2 days in the presence of β-estradiol (2 �M final concentration). After β-estradiol treatment, plants were collected, freeze-dried, and kept at −80�C until use.
For preparation of a pERV1-hyg: iaaM construct, a cDNA of iaaM was amplified using gene-specific primer pairs listed in Supplementary Table S1 and introduced into the pERV1-hyg vector using Gateway system (Invitrogen). Agrobacterium-mediated transformation was performed for producing transgenic barley (Hordeum vulgare) plants by basically following the previously described method (Hisano and Sato 2016). Briefly, immature embryos of barley cv. ‘Golden Promise’ were infected with A. tumefaciens strain AGL1 carrying pERV1-hyg: iaaM. To check transgenes in regenerated hygromycin-registant plants (T0 generation), PCR was performed as previously described (Hisano and Sato 2016) using specific primer pairs for the hygromycin phosphotransferase (HPH) and iaaM genes (iaaM-F3; 5′-aatggtggatctgacaatgg-3′ and iaaM-R3; 5′-gtccactatcggaacattgg-3′). T1 plants were used for each analysis. Barley plants were germinated and grown on soil in a photoperiod of 16 h light at 15�C. For β-estradiol treatment, fifth leaves of barley plants were sprayed with β-estradiol (10 �M in 0.2% DMSO/H2O) and then grown for another 2 days. After β-estradiol treatment, plants were collected, freeze-dried, and kept at −80�C until use. Alternatively, sterilized barley seeds were aseptically germinated and grown on 1� MS agar plates containing β-estradiol (10 �M final concentration) for 8 days in a photoperiod of 16 h light at 20�C.
qRT-PCR analysis
The qRT-PCR analysis was performed on a 7500 Real-time PCR system (Applied Biosystems) using a THUNDERBIRD SYBR qPCR mix (Toyobo) and specific primers listed in Supplementary Table S1 as previously described (Sugawara et�al. 2015). 18S rRNA expression was used as an internal standard in Arabidopsis.
Chemicals and general experimental conditions
All chemicals were purchased from Sigma-Aldrich unless otherwise stated. [phenyl-13C6]IAA was purchased from Cambridge Isotope Laboratories. [phenyl-13C6]PAA, [phenyl-13C6]IAM, [phenyl-13C6]oxIAA, [13C4, 15N]IAA-Asp, [13C5, 15N]IAA-Glu, [13C4, 15N]PAA-Asp and [13C5, 15N]PAA-Glu were previously synthesized (Sugawara et al. 2009, 2015, Mashiguchi et�al. 2011, Tanaka et�al. 2014).
15N-2-Phenylacetamide was synthesized to according to the previously published methods (Reuther et�al. 2012). Briefly, 15N-enriched ammonium chloride (150 mg, 2.75 mmol, 15N ≥ 99%) was dissolved in water (9 mL) and then diethyl ether (15 mL) was added. Phenylacetyl chloride (2130 mg, 13.8 mmol) was directly added dropwise to the ether layer and aqueous KOH solution (KOH, 930 mg, 16.6 mmol in 5 mL water) was directly added to the water layer at 0 C�. The reaction mixture was gently stirred for 15 min on ice bath and then vigorously stirred for 40 min at room temperature. The resultant mixture was extracted with diethyl ether and then the solvent layer washed with brine. The solvent layer was concentrated in vacuo and then purified by silica gel column chromatography (hexane:EtOAc = 4:1). 15N-2-Phenylacetamide was obtained as crystal (323 mg, yield 86%). The spectroscopic data were identical with the reported data (Reuther et�al. 2012).
Measurement of auxin metabolites
Frozen plant materials (ca 30 mg) were homogenized with zirconia beads (3 mm) in 0.3 mL of 80% acetonitrile/1% acetic acid/H2O containing [phenyl-13C6]IAA, [phenyl-13C6]PAA, [phenyl-13C6]IAM, 15N-PAM, [phenyl-13C6]oxIAA, [13C4, 15N]IAA-Asp, [13C5, 15N]IAA-Glu, [13C4, 15N]PAA-Asp and [13C5, 15N]PAA-Glu using a Tissue Lyser (Qiagen) for 3 min. Extracts were centrifuged at 13,000 � g for 3 min under 4�C, and the supernatant was collected. Extraction was repeated twice without internal standards. Extracts were combined, 1% acetic acid/H2O (1 ml) was added, and the total volume was reduced to <1 ml by evaporation using a Speed Vac (Thermo Fisher Scientific). For plant materials with high amounts of IAM and PAM (crown galls, pER8: iaaM and L15-4), extraction was performed without [phenyl-13C6]IAM and 15N-PAM, and after the total volume was reduced to <1 ml by evaporation, [phenyl-13C6]IAM and 15N-PAM were added to aliquots of extracts (1/100). Concentrated extracts were loaded onto an Oasis HLB column (1 ml, Waters), and then washed with 1% acetic acid/H2O (1 ml). IAA, PAA and their metabolites were eluted with 80% acetonitrile/1% acetic acid/H2O (2 ml). After addition of 1% acetic acid/H2O (1 ml) to the eluted fraction, the volume was reduced to <1 ml by evaporation of acetonitrile using a Speed Vac. The eluted fractions were loaded onto an Oasis WAX column (1 ml). After washing with 1% acetic acid/H2O (1 ml), IAM and PAM were eluted with 30% acetonitrile/H2O (2 ml). After subsequent washing with 80% acetonitrile/H2O (2 ml), IAA, PAA and oxIAA were eluted with 80% acetonitrile/1% acetic acid/H2O (2 ml). IAA-Asp, IAA-Glu, PAA-Asp and PAA-Glu were eluted from the column by 80% acetonitrile/0.8% formic acid/H2O (2 ml). Each fraction was evaporated to dryness using a Speed Vac. Then, fractions were re-dissolved in 1% acetic acid/H2O (30 �l) and injected into an Agilent 6420 Triple Quad system (Agilent) with a ZORBAX Eclipse XDB-C18 column (1.8 mm, 2.1 � 50 mm). The HPLC separation and MS/MS analysis conditions were shown in Supplementary Table S2.
Funding
This study was supported by the Japan Science and Technology Agency (JST) [PRESTO], Japan Society for the Promotion of Science (JSPS) KAKENHI [JP18H02457] to H.K., and National Institute of Health (NIH) grant [R01GM114660] to Y.Z.
Supplementary Material
Acknowledgments
We thank Dr. Barbara Kunkel and Dr. Sam Cook for critical reading of the manuscript, Dr. Javier Sampedro for generously sharing the pERV1-hyg vector, Dr. Atsushi Higashitani and Dr. Ken Komatsu for helpful suggestions, Dr. Hitoshi Sakakibara for generously sharing a cDNA of iaaM gene, and Ms. Aya Ide for excellent technical assistance. This study was supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) as part of the Joint Research Program implemented at the Institute of Plant Science and Resources, Okayama University in Japan. Barley and tomato seeds were provided by the National Bioresource Project of the MEXT in Japan.
Disclosures
The authors have no conflicts of interest to declare.
Glossary
Abbreviations
- IAA
indole-3-acetic acid
- IAA-Asp
IAA-aspartate
- IAA-Glu
IAA-glutamate
- gDW
gram dry weight
- gFW
gram fresh weight
- LC-ESI-MS/MS
liquid chromatography-electrospray ionization-tandem mass spectrometry
- PAA
phenylacetic acid
- PAM
phenylacetamide
- PAA-Asp
PAA-aspartate
- PAA-Glu
PAA-glutamate
References
- Comai L., Kosuge T. (1980) Involvement of plasmid deoxyribonucleic acid in indoleacetic acid synthesis in Pseudomonas savastanoi. J. Bacteriol. 143: 950–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook S.D., Nichols D.S., Smith J., Chourey P.S., McAdam E.L., Quittenden L., et al. (2016) Auxin biosynthesis: are the indole-3-acetic acid and phenylacetic acid biosynthesis pathways mirror images? Plant Physiol. 171: 1230–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costacurta A., Keijers V., Vanderleyden J. (1994) Molecular cloning and sequence analysis of an Azospirillum brasilense indole-3-pyruvate decarboxylase gene. Mol. Gen. Genet. 243: 463–472. [DOI] [PubMed] [Google Scholar]
- Dai X., Mashiguchi K., Chen Q., Kasahara H., Kamiya Y., Ojha S. (2013) The biochemical mechanism of auxin biosynthesis by an Arabidopsis YUCCA flavin-containing monooxygenase. J. Biol. Chem. 288: 1448–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele J., Heasley C., Fitzpatrick P. (1995) Purification and characterization of the flavoprotein tryptophan 2-monooxygenase expressed at high levels in Escherichia coli. Arch. Biochem. Biophys. 316: 241–248. [DOI] [PubMed] [Google Scholar]
- Friml J. (2010) Subcellular trafficking of PIN auxin efflux carriers in auxin transport. Eur. J. Cell Biol. 89: 231–235. [DOI] [PubMed] [Google Scholar]
- Haagen Smit A., Went F. (1935) A physiological analysis of the growth substance. Proc. R. Acad. Amsterdam 38: 852–857. [Google Scholar]
- Hayashi K., Neve J., Hirose M., Kuboki A., Shimada Y., Kepinski S., et al. (2012) Rational design of an auxin antagonist of the SCF(TIR1) auxin receptor complex. ACS Chem. Biol. 7: 590–598. [DOI] [PubMed] [Google Scholar]
- He W., Brumos J., Li H., Ji Y., Ke M., Gong X. (2011) A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell 23: 3944–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisano H., Sato K. (2016) Genomic regions responsible for amenability to Agrobacterium-mediated transformation in barley. Sci. Rep. 6: 37505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara H. (2016) Current aspects of auxin biosynthesis in plants. Biosci. Biotechnol. Biochem. 80: 34–42. [DOI] [PubMed] [Google Scholar]
- Kemper E., Wafenschmidt S., Weiler E.W., Rausch T., Schr�der J. (1985) T-DNA-encoded auxin formation in crown-gall cells. Planta 163: 257–262. [DOI] [PubMed] [Google Scholar]
- Klee H., Horsch R., Hinchee M., Hein M., Hoffmann N.L. (1987) The effects of overproduction of two Agrobacterium tumefaciens T-DNA auxin biosynthetic gene products in transgenic petunia plants. Genes Dev. 1: 86–96. [Google Scholar]
- Korasick D.A., Enders T.A., Strader L.C. (2013) Auxin biosynthesis and storage forms. J. Exp. Bot. 64: 2541–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher M., Gundlach H., Himmelbach A., Beier S., Twardziok S.O., Wicker T., et al. (2017) A chromosome conformation capture ordered sequence of the barley genome. Nature 544: 427–433. [DOI] [PubMed] [Google Scholar]
- Mashiguchi K., Tanaka K., Sakai T., Sugawara S., Kawaide H., Natsume M., et al. (2011) The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 18512–18517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClerklin S.A., Lee S.G., Harper C.P., Nwumeh R., Jez J.M., Kunkel B.N. (2018) Indole-3-acetaldehyde dehydrogenase-dependent auxin synthesis contributes to virulence of Pseudomonas syringae strain DC3000. PLoS Pathog. 14: e1006811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris D., Johnson C. (1987) Regulation of auxin transport in pea (Pisum sativum L.) by phenylacetic acid: inhibition of polar auxin transport in intact plants and stem segments. Planta 172: 408–416. [DOI] [PubMed] [Google Scholar]
- Muir R., Fujita T., Hansch C. (1967) Structure–activity relationship in the auxin activity of mono-substituted phenylacetic acids. Plant Physiol. 42: 1519–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten C., Glick B. (1995) Bacterial biosynthesis of indole-3-acetic acid. Can. J. Microbiol. 42: 207–220. [DOI] [PubMed] [Google Scholar]
- Prochazka S., Borkovec V. (1984) Transport and regulative properties of phenylacetic acid. Biol. Plant. 26: 358–363. [Google Scholar]
- Reuther J., DeSousa J., Novak B. (2012) Direct probing of regioregularity for polycarbodiimide systems via 15N NMR analysis. Macromolecules 45: 7719–7728. [Google Scholar]
- Romano C.P., Robson P.R.H., Smith H., Estelle M., Klee H. (1995) Transgene-mediated auxin overproduction in Arabidopsis: hypocotyl elongation phenotype and interactions with the hy6-1 hypocotyl elongation and axr1 auxin-resistant mutants. Plant Mol. Biol. 27: 1071–1083. [DOI] [PubMed] [Google Scholar]
- Sakakibara H., Kasahara H., Ueda N., Kojima M., Takei K., Hishiyama S., et al. (2005) Agrobacterium tumefaciens increases cytokinin production in plastids by modifying the biosynthetic pathway in the host plant. Proc. Natl. Acad. Sci. USA 102: 9972–9977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata T., Oshino T., Miura S., Tomabechi M., Tsunaga Y., Higashitani N., et al. (2010) Auxins reverse plant male sterility caused by high temperatures. Proc. Natl. Acad. Sci. USA 107: 8569–8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehin M., Bagchi R., Estelle M. (2015) SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. Plant Cell 27: 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- S�nchez-Parra B., Frerigmann H., Alonso M., Loba V., Jost R., Hentrich M., et al. (2014) Characterization of four bifunctional plant IAM/PAM-amidohydrolases capable of contributing to auxin biosynthesis. Plants (Basel) 3: 324–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider E., Kazakoff C., Wightman F. (1985) Gas chromatography–mass spectrometry evidence for several endogenous auxins in pea seedling organs. Planta 165: 232–241. [DOI] [PubMed] [Google Scholar]
- Shimizu-Mitao Y., Kakimoto T. (2014) Auxin sensitivities of all Arabidopsis Aux/IAAs for degradation in the presence of every TIR1/AFB. Plant Cell Physiol. 55: 1450–1459. [DOI] [PubMed] [Google Scholar]
- Sitbon F., Sundberg B., Olsson O., Sandberg G. (1991) Free and conjugated indoleacetic acid (IAA) contents in transgenic tobacco plants expressing the iaaM and iaaH IAA biosynthesis genes from Agrobacterium tumefaciens. Plant Physiol. 95: 480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers E., Ptacek D., Gysegom P., Srinivasan M., Vanderleyden J. (2005) Azospirillum brasilense produces the auxin-like phenylacetic acid by using the key enzyme for indole-3-acetic acid biosynthesis. Appl. Environ. Microbiol. 71: 1803–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P.E., Serban B., Rowe M., Tiryaki I., Maldonado M.T., Maldonado M.C., et al. (2005) Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17: 616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova A.N., Robertson-Hoyt J., Yun J., Benavente L.M., Xie D.Y., Dolezal K., et al. (2008) TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191. [DOI] [PubMed] [Google Scholar]
- Sugawara S., Hishiyama S., Jikumaru Y., Hanada A., Nishimura T., Koshiba T., et al. (2009) Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 5430–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara S., Mashiguchi K., Tanaka K., Hishiyama S., Sakai T., Hanada K., et al. (2015) Distinct characteristics of indole-3-acetic acid and phenylacetic acid, two common auxins in plants. Plant Cell Physiol. 56: 1641–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle J., Mansager E. (1986) The physiological significance of phenylacetic acid in abscising cotton cotyledons. Plant Physiol. 81: 434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Hayashi K., Natsume M., Kamiya Y., Sakakibara H., Kawaide H., et al. (2014) UGT74D1 catalyzes the glucosylation of 2-oxindole-3-acetic acid in the auxin metabolic pathway in Arabidopsis. Plant Cell Physiol. 55: 218–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Ferrer J.L., Ljung K., Pojer F., Hong F., Long J.A., et al. (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tingay S., McElroy D., Kalla R., Fieg S., Wang M., Thornton S., et al. (1997) Agrobacterium tumefaciens-mediated barley transformation. Plant J. 11: 1369–1376. [Google Scholar]
- Tsugafune S., Mashiguchi K., Fukui K., Takebayashi Y., Nishimura T., Sakai T., et al. (2017) Yucasin DF, a potent and persistent inhibitor of auxin biosynthesis in plants. Sci. Rep. 7: 13992.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia E.R., Herrera M.T., Gianzo C., Fidalgo J., Revilla G., Zarra I., et al. (2013) Regulation of secondary wall synthesis and cell death by NAC transcription factors in the monocot Brachypodium distachyon. J. Exp. Bot. 64: 1333–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman F., Lighty D. (1982) Identification of phenylacetic acid as a natural auxin in the shoots of higher plants. Physiol. Plant. 55: 17–24. [Google Scholar]
- Won C., Shen X., Mashiguchi K., Zheng Z., Dai X., Cheng Y., et al. (2011) Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 18518–18523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Palm C., Brooks B., Kosuge T. (1985) Nucleotide sequences of the Pseudomonas savastanoi indoleacetic acid genes show homology with Agrobacterium tumefaciens T-DNA. Proc. Natl. Acad. Sci. USA 82: 6522–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. (2010) Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 61: 49–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Zhang Y., Liu X., Zhang X., Liu S., Yu X. (2013) A role for a dioxygenase in auxin metabolism and reproductive development in rice. Dev. Cell 27: 113–122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.