Abstract
Background:
Genome-wide association studies have been extensively applied in identifying SNP associated with major psychiatric disorders. However, the SNPs identified by the prevailing univariate approach only explain a small percentage of the genetic variance of traits, and the extensive data have shown the major psychiatric disorders have common biological mechanisms and the overlapping pathophysiological pathways.
Methods:
We applied the genetic pleiotropy-informed metaCCA method on summary statistics data from the Psychiatric Genomics Consortium Cross-Disorder Group to examine the overlapping genetic relations between the five major psychiatric disorders. Furthermore, to refine all genes, we performed gene-based association analyses for the five disorders respectively using VEGAS2. Gene enrichment analysis was applied to explore the potential functional significance of the identified genes.
Results:
After metaCCA analysis, 1147 SNPs reached the Bonferroni corrected threshold (p < 1.06 × 10−6) in the univariate SNP-multivariate phenotype analysis, and 246 genes with a significance threshold (p < 3.85 × 10−6) were identified as potentially pleiotropic genes in the multivariate SNP-multivariate phenotype analysis. By screening the results of gene-based p-values, we identified 37 putative pleiotropic genes which achieved significance threshold in metaCCA analyses and were also associated with at least one disorder in the VEGAS2 analyses.
Limitations:
Alternative approaches and experimental studies may be applied to check whether novel genes could still be identified/substantiated with these methods.
Conclusions:
The metaCCA method identified novel variants associated with psychiatric disorders by effectively incorporating information from different GWAS datasets. Our analyses may provide insights for some common therapeutic approaches of these five major psychiatric disorders based on the pleiotropic genes and common mechanisms identified.
Keywords: Psychiatric disorder, Multivariate statistical analysis, Genome-wide association study, Pleiotropic
1. Introduction
Psychiatric disorders represent a mix of common, chronic, and complex conditions affecting populations with highly prevalence (Gaebel et al., 2015; Jeste et al., 2015; Merikangas et al., 2010). The main clinical disorders include schizophrenia (SCZ), bipolar disorder (BP), major depression (MD), autism spectrum disorders (ASD) and attention deficit-hyperactivity disorder (ADHD), which are characterized by perceptive and cognitive impairments resulting in abnormalities of behavior, volition, and emotion. According to statistics from the American National Institute of Mental Health, the life-time prevalence of MD and ADHD in adolescents are up to 16% and 9%, respectively (Merikangas et al., 2010; Ripke et al., 2013). Family and twin studies have shown these five common psychiatric disorders each show high rates of heritability: 90% for ASD (Lee et al., 2013a), 60–85% for BP, ADHD and SCZ (Hunt et al., 2016; Nothen et al., 2010; Visscher et al., 2012), and 40% for MD (Ripke et al., 2013; Sullivan et al., 2013; Visscher et al., 2012). With most individual patients experiencing multiple episodes throughout their life and increased morbidity, excess mortality and substantial health costs, psychiatric disorders represent major public health problems (Contreras et al., 2018; Ski et al., 2016).
Pleiotropy describes the genetic effect of a single nucleotide polymorphisms (SNP) or gene on two or more phenotypic traits and its outcome is genetic correlation. Largely, this concept concerns across-trait architecture (Stearns, 2011). Previous different types of studies have repeatedly indicated that the genetic pleiotropy exists in these five correlated psychiatric disorders. For example, CACNA1C is a susceptibility gene for bipolar disorder, schizophrenia, and major depressive disorder (Bigos et al., 2010; Consortum, 2013; Ripke et al., 2011; Thimm et al., 2011). SNP within two different genes on chromosomes 3p21 and 10q24 are associated with diagnostic boundaries among several psychiatric disorders (Consortum, 2013; McMahon et al., 2010; Sklar et al., 2011). The estimated genetic correlation calculated using common SNPs was high between SCZ and BP (0.68 ± 0.04 s.e.), moderate between SCZ and MD (0.43 ± 0.06 s.e.), BP and MD (0.47 ± 0.06 s.e.), and ADHD and MD (0.32 ± 0.07 s.e.), and lower between SCZ and ASD (0.16 ± 0.06 s.e.) (Lee et al., 2013a). It is therefore important to identify pleiotropic genes that acting through common biological mechanisms and assess overlapping pathophysiological relationships of these five disorders using effective analytical approaches.
Genome-wide association studies (GWAS) is a most powerful, systematic, and standard univariate approach to investigate and identify potentially causal or risk-conferring genetic variants for common and complex diseases in the individual level measurement (Kettunen et al., 2012; Tang and Ferreira, 2012). Although GWAS, especially those with large sample size and meta-analysis of multiple studies, have successfully identified many loci/genes associated with psychiatric disorders, however, they only explain about 30% of observed heritability for psychiatric disorders (Cross-Disorder Group of the Psychiatric Genomics Consortum, 2013; Mistry et al., 2018). Indeed, multivariate analysis may have more statistical power to detect the unexplained heritability and could lead to more extensive findings due to correlations among different phenotypes, considered by combining the independent GWASs from associated traits or diseases (Inouye et al., 2012; Marttinen et al., 2013). Existing studies of genetic risk factors for psychiatric disorders have used bivariate analysis, and multivariate analysis based on multiple correlated psychiatric disorders are rare (Evangelou and Ioannidis, 2013). Therefore, a multivariate analysis to identify pleiotropic genes, especially using the publicly available summary statistics of GWAS, is worth pursuing.
Cichonska et al. (2016) recently developed a multivariate meta-analysis based on summary statistics from GWAS using a canonical correlation analysis (metaCCA) method allowing multivariate representation of both genotypic and phenotypic variables. This new approach’s aim is to increase statistical power to identify novel genetic associations, and the core principle uses the method of canonical correlation analysis (CCA) to identify linear relationships between two sets of variables: multiple SNPs against multiple traits based on already published univariate summary statistics from GWAS by meta-analysis (Cichonska et al., 2016; Tang and Ferreira, 2012). Cichonska et al. (2016) have successfully applied this method to 9 lipid measures related from studies of three Finnish cohorts. They showed metaCCA highly improved the statistical power by considering the correlations among multiple SNPS and multiple phenotypes.
In this study, we applied the genetic pleiotropy-informed metaCCA method on summary statistics data from Psychiatric Genomics Consortium (PGC) Cross-Disorder Group to examine the overlapping genetic relation between the five psychiatric disorders. By using this method, we could identify more common variants that are genetic risk factors for one or more common psychiatric disorders and resulting potentially shared genetic influences should provide novel insights for specifying clinical presentations and generating new models for prevention and, ultimately, treatment.
2. Methods
2.1. GWAS datasets
The large scale GWAS datasets containing the association summary statistics for the 5 correlated disorders in this present study were downloaded from Psychiatric Genomics Consortium(PGC, website: http://www.med.unc.edu/pgc/). GWAS results for ADHD were based on a meta-analysis of 8,094,094 genotyped or imputed SNPs from 19,099 cases and 34,194 control individuals performed by the PGC and the Lundbeck Foundation Initiative for Integrative Psychiatric Research (iPSYCH) released in June 2017 (Demontis et al., 2017). Genotypes for the ASD GWAS were also a meta-analysis including data from 6,440,259 common variants in 6,197 cases and 7,377 controls from fourteen independent cohorts (Autism Spectrum Disorders Working Group of The Psychiatric Genomics Consortium, 2017). The SCZ is the second PGC schizophrenia mega-analysis, and included 46 non-overlapping case-control samples of European ancestry (32,375 cases and 42,186 controls) plus deCODE European ancestry genetics (1,513 cases and 66,236 controls) (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). The MD included 9,227 cases and 7,383 controls were tested based on the Stage 2 polygenic risk profile analyses for 1,232,793 common variants (Ripke et al., 2013). Data of 10,410 BP cases and 10,700 controls are the association analysis between BP and SCZ using polygenic risk scores (Ruderfer et al., 2014). All the samples in the GWAS datasets came from populations of European ancestry. Further detail descriptions of the sample ascertainment and stringent quality control procedures can be found in the corresponding consortium publications (Autism Spectrum Disorders Working Group of The Psychiatric Genomics Consortium, 2017; Demontis et al., 2017; Ripke et al., 2013; Ruderfer et al., 2014; Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). We avoided reduplicating control individuals when selecting these datasets. The data contain summary statistics, only including p-values, regression coefficients and standard error after meta-analysis. Finally, 1,011,503 overlapping SNPs of the 5 disorders were selected on which we performed the multivariate analysis. The study details are shown in Table 1.
Table 1.
Details of the samples (all European ancestry).
| Disorders | Number of individuals | Number of SNPs | MAF | INFO | Imputation reference panel | |
|---|---|---|---|---|---|---|
| Cases | Controls | |||||
| ADHD | 19,099 | 34,194 | 8,094,094 | ≥ 0.01 | ≥0.8 | 1000 Genomes Project Phase 3 |
| MD | 9,227 | 7,383 | 1,232,793 | 0.02~0.98 | ≥0.9 | HapMap Phase3 CEU + TSI data |
| BP | 10,410 | 10,700 | 1,252,022 | ≥ 0.01 | ≥0.60 | HapMap Phase3 CEU + TSI data and BEAGLE |
| ASD | 6,197 | 7,377 | 6,440,259 | ≥0.05 | ≥0.60 | HapMap Phase3 CEU |
| SCZ | 33,888 | 108,422 | 15,358,497 | ≥ 0.01 | ≥ 0.6 | 1000 Genomes Project |
2.2. Data preparation
Before the implementation of the metaCCA method, several steps were undertaken. First, we combined the summary statistics for the 1,011,503 common SNPs included in the studies of all five disorders and completed the gene annotation for the five GWASs according to the 1000 Genome datasets using PLINK1.9. The reference data, which contained 26,291 genes, were downloaded from the website: https://www.cog-genomics.org/static/bin/plink/glist-hg19. Second, a linkage disequilibrium (LD) based SNP pruning method was used to remove SNPs with large pairwise correlations. The SNP pruning method was proceeded by a window of 50 SNPs where LD was calculated between each pair of SNPs. The minor allele frequency (MAF) is also considered for the SNP pruning, and SNP with smaller MAF for pairs with r2 > 0.2 were removed. Following this initial removal of SNPs in high LD, each sliding window of 5 SNPs forward and the process repeated until there were no pairs of SNPs with high LD (Andreassen et al., 2015). All datasets were pruned using the HapMap 3 CEU genotypes as a reference panel. After gene annotation and SNP pruning, there remained 47,217 SNPs located in 12,989 gene regions on which we performed the metaCCA analysis. The regression coefficient beta was normalized before conducting the metaCCA analysis because the individual-level data set genotype and phenotype matrices were not standardized. Standardization was achieved afterwards by:
| (1) |
where SEgp is the standard error of βgp, as given by the original GWAS result, g is the number of genotypic, p is the number of phenotypic variables, and n is the sample number of each disorder.
2.3. MetaCCA analysis
To identify the potential pleiotropic genes, we computed metaCCA for all five psychiatric diseases. MetaCCA is an extension of the method of CCA, which required a cross-covariance matrix between all genotypic and phenotypic variables (∑XY), a genotypic correlation structure between SNPs (∑^XX), and a phenotypic correlation structure between traits (∑^YY) (Cichonska et al., 2016).
In metaCCA, ∑XY is constructed as the normalized regression coef-ficient βgp, and ∑^XX is calculated using a reference database representing the study population, such as the 1000 Genomes database, or other genotypic data available on the target population. There will be better results if ∑^XX were estimated from the target population or the same ethnicity instead of interracial populations (Cichonska et al., 2016). In our study, ∑^XX was estimated using the reference SNP dataset of the HapMap 3 CEU. The phenotypic correlation structure ∑^YY is computed based on ∑XY. Each entry of ∑^YY corresponds to a Pearson correlation coefficient between the vector of β estimates from p phenotypic variables across g genetic variants. It has been demonstrated that the bigger the number of genotypic variables g, the more accurate the quality of the estimate. Thus, ∑^YY were calculated from summary statistics for all available genetic variants (1,011,503 overlapping SNPs), even if only a part of were used for further analysis.
After calculation, the full covariance matrix (∑), consisting of three covariance matrices, can be obtained. We should determine whether the full covariance matrix is positive semidefinite (PSD). If it is not PSD, an iterative procedure is used to shrink the full covariance matrix until ∑ becomes PSD. In the next analysis, the PSD of the full covariance matrix is plugged into the CCA framework to get the final genotypephenotype association result (Cichonska et al., 2016). The correlation between genotype and phenotype is called the canonical correlation r (Seoane et al., 2014).
In this study, two types of multivariate analysis were considered. First, univariate SNP-multivariate phenotype association analysis was tested at the SNP level. Manhattan plots presented all SNPs within an LD block in relation to their chromosomal locations. To identify any potential pleiotropic gene, we did multivariate SNP-multivariate phenotype association analysis at the gene level. The result was the canonical correlation of a gene with five disorders. The most significant SNP within each gene region and its p-value were also obtained through these two types of multivariate analysis. We checked the summary statistics of GWAS for a set of 47,217 pre-selected SNPs and found that the mean standardized β’s is close to zero and the median p-value for those β’s close to 0.5 in all the five datasets, which means the sample of SNPs behaves like a random sample of the whole genome, then Bonferroni corrected p-value < 0.05 was used as the threshold for nominal significance. If the p-value of the canonical correlation r of any SNP was smaller than 1.06 × 10−6 (= 0.05/47,217), it was deemed significantly associated with the five disorders. Because the β’s of genes could not be obtained and computed from the summary statistics of GWAS, a conservative corrected method-Bonferroni corrected threshold-is used in the gene level. Similarly, genes with a canonical correlation p-value smaller than 3.85 × 10−6 (= 0.05/12,989) were significantly associated with the five disorders.
2.4. Gene-based analyses
To refine the identified genes by MetaCCA, we performed gene-based association analyses, using the VEGAS2 (Versatile Gene-based Association Study–2) method (performed at: https://vegas2.qimrberghofer.edu.au/) (Mishra and Macgregor, 2015). This method calculates the correlation analysis of multiple SNPs in a gene region with one phenotype using GWAS summary statistics, which has previously shown higher sensitivity and lower false positive rates compared to other gene-based approaches (Autism Spectrum Disorders Working Group of The Psychiatric Genomics Consortium, 2017; Wojcik et al., 2015). All SNPs in each gene was analysed using the 1000 Genomes European reference genotypes. We obtained the gene-based p-value of each gene for each disorder and selected the pleiotropic genes that were associated with at least one disorder using the threshold adjusted p-value < 0.05(= 1E-06).
2.5. Functional annotation and gene enrichment analysis
An useful way to understand polygenic associations is to determine whether the implicated genetic variants occur in genes that comprise a biological pathway or not (Lencz et al., 2013). To evaluate the potential biological function of all putative pleiotropic genes, we conducted the GO enrichment analysis using Enrichr (http://amp.pharm.mssm.edu/ Enrichr/). All significant genes re-identified by VEGAS2 in our study were annotated and enriched based on three main categories: biological processes, cellular component and molecular functions. An adjusted p-value < 0.05 in the enrichment analysis became the threshold for nominal significance.
3. Results
To identify common variants shared among different psychiatric disorders, we undertook a two-step analysis strategy. First, by using the metaCCA method, we inspected the potential pleiotropic SNPs and genes among all five phenotypes. Next, by adopting the VEGAS2 method, we checked those potential common variants for their specific associations with individual disorders.
3.1. Potential pleiotropic SNPs and genes by metaCCA analysis
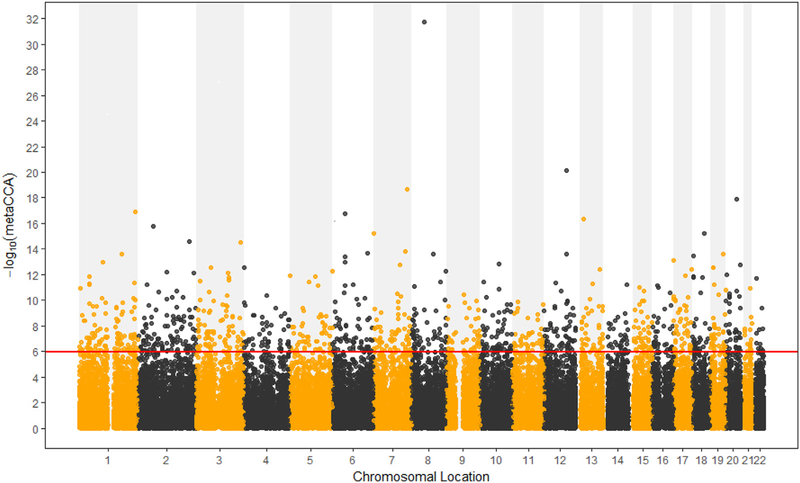
After gene annotation and SNP pruning, there were 47,217 SNPs located in 12,989 gene regions available for the metaCCA analysis. The size of SNP representation of the genes ranged from 1 to 147 SNPs; the average number of SNPs in each gene was 4. For the univariate SNP-multivariate phenotype analysis, 1147 SNPs reached the Bonferroni corrected threshold (p < 1.06 × 10−6), and the canonical correlation r between each SNP and phenotype ranged from 0.0269 to 0.0566. The results are presented by the Manhattan plot in Fig 1. If the −log10 (metaCCA) value of a certain SNP was greater than 5.98, this SNP was flagged as a potential pleiotropic SNP for these five correlated disorders. For the multivariate SNP-multivariate phenotype analysis, 246 genes with a significance threshold of p-value < 3.85 × 10−6 were identified as the potential pleiotropic genes for the five disorders. The canonical correlation r between genotype and phenotype ranged from 0.0217 to 0.2143.
Fig. 1.
Manhattan plot of −log10 (metaCCA) values for univariate SNP-five disorders analysis. The red line marks the −log10 (metaCCA) value of 5.98 corresponds to p < 1.06 × 10−6. If the −log10 (metaCCA) value of a certain SNP was greater than 5.98, this SNP was identified as a pleiotropic SNP for the five correlated disorders.
3.2. Refining the pleiotropic genes by gene-based analyses
After the metaCCA analysis, we refined the list of 246 pleotropic genes associated with more than one disorder to identify their association with specific traits using the gene-based p-value calculation using the VEGAS-2 algorithm. For BP, 5 genes achieved a significance threshold of the adjusted p-value < 0.05. For SCZ, 340 genes associated with SCZ were identified. For MD, ADHD, and ASD, no significant genes identified.
By screening the results of gene-based analysis p-values, we identified 37 putative pleiotropic yielding significance in the metaCCA analyses and were associated with at least one trait in the VEGAS2 analyses. All of these possible 37 pleiotropic genes were identified as the associated genes with SCZ, and 1 of these 37 genes (CACNA1C) was identified as being associated with BP in the original GWAS study. The findings of the metaCCA and VEGAS2 analyses are summarized in Table 2. The most significant SNP within 37 putative pleiotropic genes region that reached the significance threshold in the metaCCA study are illustrated in S1 Table, and the top SNP that reached genome-wide significance in the original GWAS study are listed in S2 Table.
Table 2.
The 37 pleiotropic genes identified by the metaCCA and VEGAS2 analysis.
| Locus | Gene | MetaCCA adjusted p-value | VEGAS adjusted p-value | ||||
|---|---|---|---|---|---|---|---|
| ADHD | MD | BP | ASD | SCZ | |||
| 1 | SRPK2 | 0 | 0.37 | 0.40 | 0.05 | 0.06 | 1.0E-06 |
| 2 | DPYD | 8.5E-292 | 0.58 | 0.19 | 0.75 | 0.35 | 1.0E-06 |
| 3 | MAD1L1 | 4.0E-120 | 0.66 | 0.92 | 2.1E-04 | 0.03 | 1.0E-06 |
| 4 | F0XP1 | 3.1E-107 | 0.37 | 0.02 | 0.46 | 0.01 | 1.0E-06 |
| 5 | CACNA1C | 1.0E-65 | 0.37 | 0.20 | 1.0E-06 | 0.41 | 1.0E-06 |
| 6 | RGS7 | 2.7E-74 | 0.05 | 0.53 | 0.30 | 0.50 | 1.0E-06 |
| 7 | IMMP2L | 9.5E-63 | 0.35 | 0.08 | 0.21 | 0.81 | 1.0E-06 |
| 8 | SLC8A3 | 9.6E-33 | 0.14 | 0.38 | 0.96 | 0.06 | 1.0E-06 |
| 9 | CEMIP | 1.4E-39 | 0.60 | 0.15 | 1.5E-03 | 0.14 | 1.0E-06 |
| 10 | CACNB2 | 4.0E-47 | 0.07 | 0.02 | 6.1E-03 | 0.46 | 1.0E-06 |
| 11 | SCAPER | 5.2E-28 | 0.31 | 0.54 | 0.11 | 0.33 | 1.0E-06 |
| 12 | ANKRD44 | 4.6E-24 | 0.52 | 0.44 | 0.03 | 0.51 | 1.0E-06 |
| 13 | ISC20 | 1.5E-17 | 1.9E-03 | 0.36 | 0.01 | 0.06 | 1.0E-06 |
| 14 | TMEM120B | 4.8E-13 | 0.12 | 0.21 | 0.44 | 0.15 | 1.0E-06 |
| 15 | ZSWIM6 | 1.2E-15 | 0.04 | 0.24 | 0.04 | 0.05 | 1.0E-06 |
| 16 | TRANK1 | 1.1E-12 | 0.62 | 0.56 | 0.01 | 0.08 | 1.0E-06 |
| 17 | ALPI | 4.2E-11 | 0.43 | 0.03 | 0.03 | 0.13 | 1.0E-06 |
| 18 | EFHD1 | 2.2E-11 | 0.56 | 0.02 | 0.01 | 0.03 | 1.0E-06 |
| 19 | RCOR1 | 3.9E-08 | 0.06 | 0.14 | 0.03 | 0.77 | 1.0E-06 |
| 20 | OPN3 | 1.8E-07 | 0.28 | 0.68 | 0.02 | 0.75 | 1.0E-06 |
| 21 | ITIH4 | 8.8E-10 | 0.02 | 0.06 | 1.4E-04 | 0.10 | 1.0E-06 |
| 22 | LINC01004 | 1.3E-09 | 0.65 | 0.81 | 0.01 | 0.10 | 1.0E-06 |
| 23 | BTN2A1 | 8.1E-11 | 0.85 | 0.34 | 0.01 | 0.91 | 1.0E-06 |
| 24 | GPX6 | 3.1E-09 | 0.19 | 0.10 | 0.04 | 0.39 | 1.0E-06 |
| 25 | ABHD17C | 2.0E-07 | 0.43 | 0.21 | 1.9E-03 | 0.12 | 1.0E-06 |
| 26 | ETF1 | 5.4E-09 | 0.09 | 0.06 | 2.3E-03 | 0.20 | 1.0E-06 |
| 27 | ARFGEF2 | 3.1E-07 | 0.30 | 0.31 | 0.02 | 0.84 | 1.0E-06 |
| 28 | CREB3L1 | 3.2E-07 | 0.15 | 0.76 | 0.47 | 6.3E-04 | 1.0E-06 |
| 29 | KMO | 2.5E-09 | 0.43 | 0.83 | 0.04 | 0.55 | 1.0E-06 |
| 30 | SNAP91 | 1.4E-07 | 0.33 | 0.35 | 2.8E-03 | 1.00 | 1.0E-06 |
| 31 | HIST1H3D | 1.2E-07 | 0.35 | 0.25 | 0.42 | 0.24 | 1.0E-06 |
| 32 | C3orf49 | 1.7E-06 | 0.54 | 0.15 | 0.06 | 0.73 | 1.0E-06 |
| 33 | C2 | 1.6E-07 | 0.83 | 0.11 | 0.01 | 0.63 | 1.0E-06 |
| 34 | ZSCAN23 | 2.3E-06 | 0.36 | 0.08 | 0.03 | 0.23 | 1.0E-06 |
| 35 | ZNF391 | 3.0E-06 | 0.49 | 0.02 | 0.03 | 0.70 | 1.0E-06 |
| 36 | STAB1 | 3.5E-06 | 0.12 | 0.01 | 4.5E-03 | 0.07 | 1.0E-06 |
| 37 | HFE | 2.4E-06 | 0.43 | 0.11 | 0.07 | 0.49 | 1.0E-06 |
Specifically, 13 of these 37 putative pleiotropic genes (DPYD, MAD1L1, FOXP1, CACNA1C, IMMP2L, CACNAB2, TRANK1, ITIH4, KMO, SNAP91, C2, STAB1, HFE) have been previously reported to be associated with more than one of these five disorders. Of these 13 genes, CACNA1C and CACNAB2 were repeatedly replicated in published studies, an association was identified between DPYD and BP, SCZ, MD, and ASD, and TRANK1 previously reported to be associated with BP, SCZ and ADHD. Of the 24 detected novel putative pleiotropic genes, 3 genes (RGS7, ZSWIM6, EFHD1) were reported to be associated with SCZ by the contributing GWAS study, but ETF1 was reported to be associated with BP. Other significant genes, especially for the remaining top 5 significant genes (SRPK2, CEMIP, SLC8A3, SCAPER, ANKRD44), might represent novel pleiotropic candidate genes for these five disorders. The detailed features of 37 significant pleiotropic genes are shown in Table 3.
Table 3.
The features of 37 significant pleiotropic genes.
| Locus | Gene | Chr | Gene type | r-value | Adjusted p-value | Number of SNPs |
|---|---|---|---|---|---|---|
| 1 | SRPK2 | 7 | Novel | 0.21 | 0 | 5 |
| 2 | DPYD | 1 | Confirmed(Noor et al., 2010; Witt et al., 2017) | 0.15 | 8.5E-292 | 26 |
| 3 | MAD1L1 | 7 | Confirmed(Ruderfer et al., 2014) | 0.10 | 4.0E-120 | 10 |
| 4 | F0XP1 | 3 | Confirmed(Autism Spectrum Disorders Working Group of The Psychiatric and Consortium, 2017; | 0.09 | 3.1E-107 | 25 |
| Nurnberger et al., 2014) | ||||||
| 5 | CACNA1C | 12 | Confirmed(Consortum, 2013; Dedic et al., 2017; Gasso et al., 2016) | 0.08 | 1.0E-65 | 31 |
| 6 | RGS7 | 1 | Novel*(Jayaraman et al., 2009) | 0.08 | 2.7E-74 | 31 |
| 7 | IMMP2L | 7 | Confirmed(Casey et al., 2012; Elia et al., 2010; Goes et al., 2015) | 0.07 | 9.5E-63 | 16 |
| 8 | SLC8A3 | 14 | Novel | 0.06 | 9.6E-33 | 8 |
| 9 | CEMIP | 15 | Novel | 0.06 | 1.4E-39 | 18 |
| 10 | CACNAB2 | 12 | Confirmed(Soldatov, 2015) | 0.06 | 4.0E-47 | 21 |
| 11 | SCAPER | 15 | Novel | 0.06 | 5.2E-28 | 3 |
| 12 | ANKRD44 | 2 | Novel | 0.05 | 4.6E-24 | 10 |
| 13 | ISG20 | 15 | Novel | 0.04 | 1.5E-17 | 3 |
| 14 | TMEM120B | 12 | Novel | 0.04 | 4.8E-13 | 5 |
| 15 | ZSWIM6 | 5 | Novel*(Twigg et al., 2016) | 0.04 | 1.2E-15 | 3 |
| 16 | TRANK1 | 3 | Confirmed(Ruderfer et al., 2014; Schimmelmann et al., 2013) | 0.04 | 1.1E-12 | 4 |
| 17 | ALPI | 2 | Novel | 0.04 | 4.2E-11 | 2 |
| 18 | EFHD1 | 2 | Novel*(Alkelai et al., 2011) | 0.04 | 2.2E-11 | 2 |
| 19 | RCOR1 | 14 | Novel | 0.03 | 3.8E-08 | 4 |
| 20 | OPN3 | 1 | Novel | 0.03 | 1.8E-07 | 2 |
| 21 | TTIH4 | 3 | Confirmed(Cross-Disorder Group of the Psychiatric Genomics Consortum, 2013) | 0.03 | 8.8E-10 | 1 |
| 22 | UNC01004 | 7 | Novel | 0.03 | 1.3E-09 | 1 |
| 23 | BTN2A1 | 6 | Novel | 0.03 | 8.1E-11 | 2 |
| 24 | GPX6 | 6 | Novel | 0.03 | 3.1E-09 | 1 |
| 25 | ABHD17C | 15 | Novel | 0.03 | 2.0E-07 | 2 |
| 26 | ETF1 | 5 | Novel*(Kao et al., 2016) | 0.03 | 5.4E-09 | 1 |
| 27 | ARFGEF2 | 20 | Novel | 0.03 | 3.1E-07 | 2 |
| 28 | CREB3L1 | 11 | Novel | 0.03 | 3.2E-07 | 2 |
| 29 | KMO | 1 | Confirmed(Borsini et al., 2017; Wurfel et al., 2017) | 0.03 | 2.5E-09 | 6 |
| 30 | SNAP91 | 6 | Confirmed(Goes et al., 2012) | 0.03 | 1.4E-07 | 5 |
| 31 | MST1H3D | 6 | Novel | 0.03 | 1.2E-07 | 1 |
| 32 | C3orf49 | 3 | Novel | 0.03 | 1.7E-06 | 2 |
| 33 | C2 | 6 | Confirmed(Cortabitarte et al., 2017; Khoury et al., 2017) | 0.03 | 1.6E-07 | 1 |
| 34 | ZSCAN23 | 6 | Novel | 0.03 | 2.3E-06 | 1 |
| 35 | ZNF391 | 6 | Novel | 0.03 | 3.0E-06 | 1 |
| 36 | STAB1 | 3 | Confirmed(Lee et al., 2013b; Witt et al., 2014) | 0.03 | 3.5E-06 | 1 |
| 37 | HFE | 6 | Confirmed(Buretic-Tomljanovic et al., 2012; Guerini et al., 2009; Nigg et al., 2016) | 0.03 | 2.4E-06 | 4 |
Note:
Confirmed: This gene was previously reported to be associated with more than one psychiatric disorder.
Novel
: This gene had been reported to be associated with only one psychiatric disorder.
Novel: This gene had never been reported to be associated with any psychiatric disorder.
3.3. Functional term enrichment analysis
When pleiotropic genes annotated by the variants associated with psychiatric disorders were used as the gene sets for the GO term enrichment analysis, several functional terms were identified as being enriched in psychiatric disorders. For the GO cellular component, the top five significant GO terms were L-type voltage-gated calcium channel complex, apical recycling endosome, basolateral recycling endosome, recycling endosome, and peri-centrosomal recycling endosome. For the GO molecular function, the top five significant GO terms were flavin adenine dinucleotide binding, FADH2 binding, voltage-gated calcium channel activity involved in regulation of cytosolic calcium levels, intermediate voltage-gated calcium channel activity, and voltage-gated calcium channel activity involved in cardiac muscle cell action potential. This GO term enrichment analysis furnished supporting evidence for our results from a functional aspect and may contribute to the illumination of etiology of psychiatric disorders. Detailed information is shown in Table 4.
Table 4.
Top five significant GO term enrichment of the pleiotropic genes.
| Term(GO_Cellular_Component) | p-value | Adjusted p-value | Genes |
| L-type voltage-gated calcium channel complex(GO:1990454) | 7.0E-05 | 1.0E-02 | CACNB2;CACNA1C |
| apical recycling endosome(GO:0090653) | 9.2E-03 | 4.8E-02 | AKFGEF2;HFE |
| basolateral recycling endosome(GO:0090654) | 9.2E-03 | 4.8E-02 | AKFGEF2;HFE |
| recycling endosome(GO:0055037) | 9.2E-03 | 4.8E-02 | AKFGEF2;HFE |
| peri-centrosomal recycling endosome(GO:0098832) | 9.2E-03 | 4.8E-02 | ARFGEF2;HFE |
| Term(GO_Molecular_Function) | p-value | Adjusted p-value | Genes |
| flavin adenine dinucleotide binding(GO:0050660) | 7.5E-04 | 4.8E-02 | DPYD;KMO |
| FADH2 binding(GO:0071950) | 7.5E-04 | 4.8E-02 | DPYD;KMO |
| voltage-gated calcium channel activity involved in regulation of cytosolic calcium levels(GO:0099511) | 1.4E-03 | 4.8E-02 | CACNB2;CACNA1C |
| intermediate voltage-gated calcium channel activity(GO:1990028) | 1.4E-03 | 4.8E-02 | CACNB2;CACNA1C |
| voltage-gated calcium channel activity involved in cardiac muscle cell action potential (GO:0086007) | 1.4E-03 | 4.8E-02 | CACNB2;CACNA1C |
4. Discussion
In the present study, five independent GWAS meta-analyses with available summary statistics were combined to explore the common genetic variants for five major psychiatric disorders using a novel analytical approach—metaCCA. After verification using gene-based analyses, we successfully identified a total of 37 putative pleiotropic genes and performed the functional term enrichment analysis based on these results. In particularly, 13 confirmed genes were identified as pleiotropic in previous different types of studies and were validated in the present study. Twenty-four significant genes might represent novel pleiotropic candidate genes for at least two of the five psychiatric traits, which include 3 genes previously reported associated with SCZ, 1 gene previously reported as associated with BP and 20 novel findings. The improved detection not only yielded the potential shared genetic components but also provide better understanding for further exploring potential common biological pathogenesis of these major psychiatric disorders.
Among the 13 confirmed pleiotropic genes, some were shown to play an important role on the pathomechanism of psychiatric disorders. For example, CACNA1C and CACNAB2 are expressed in the brain and encode L-type voltage-gated calcium-channel subunits, as well as being among the most commonly reported genes in the scientific literature. Gain-of-function mutations in CACNA1C cause Timothy syndrome, a developmental disorder in which includes ASD can be a component phenotype (Cross-Disorder Group of the Psychiatric Genomics Consortum, 2013). Consistent with their suggested pleiotropic role, neuroimaging studies have documented the effects of CACNA1C gene on a range of brain expressions implicating a molecular and neural system mechanism, including circuitry involved in emotion processing, executive function, attention, and memory (Bigos et al., 2010; Thimm et al., 2011). CACNB2 encodes an auxiliary voltage-gated calcium-channel subunit that interacts with L-type calcium-channel subunits to promote their trafficking to the plasma membrane, increase their function, and regulate their modulation by other signaling proteins and molecules (Smoller and Finn, 2003). GO terms enrichment analysis results also suggest voltage-gated calcium signaling and calcium-channel activity could be an important biological process in psychiatric disorders. Another important and confirmed gene is MAD1L1, which has been associated with plasma Ndel1 enzyme activity (Gadelha et al., 2016). Ndel1 is a DISC1-interacting oligopeptidase. It influences neuronal migration and neurite outgrowth that may contribute to psychiatric physiopathology. Several recent GWASs and pathway/GO terms enrichment analyses have found variants in MAD1L1 were associated with risk of SCZ and BP (Budde et al., 2017; Chang et al., 2015; Ikeda et al., 2017).
Interestingly, 4 (RGS7, ZSWIM6, EFHD1, ETF1) of the 24 novel pleiotropic candidate genes had been validated associated with some kind of psychiatric disorders. RGS7 is a member of the RGS (regulator of G protein signaling) proteins, which regulate vision, postnatal development, working memory and the action of psychostimulants or morphine in vertebrates (Jayaraman et al., 2009). On the other hand, previous molecular, linkage and association studies in five independently ascertained samples have implicated another RGS member, RGS4, in the etiology of both SCZ and BP (Cordeiro et al., 2004; Ding and Hegde, 2009). ZSWIM6 is a protein of unknown function associated with SCZ and acromelic frontonasal dysostosis with intellectual disability by independent GWASs (Twigg et al., 2016). Fortunately, a new experimental study has reported on the generation of ZSWIM6 knockout mice and has provided a detailed anatomical and behavioral characterization of the resulting phenotype, showing that ZSWIM6 is indispensable to normal brain function and supports the notion that ZSWIM6 might act as an important contributor to the pathogenesis of SCZ and other neurodevelopmental disorders (Tischfield et al., 2017; Twigg et al., 2016). Similar to CACNA1C and CACNAB2, EFHD1 functions as a novel mitochondrial Ca2+ sensor underlying Ca2+-dependent activation of mitoflashes, and was identified as associated with risk to SCZ in a family-based GWAS strategy in an enlarged, ethnically homogeneous, Arab-Israeli family sample (Alkelai et al., 2011; Hou et al., 2016). The ETF1 gene, located in the 5q31, plays important roles in regulating the activity in the translational termination process and transcriptional regulation, and has been reported as a novel susceptible gene for pure BP-II in the latest research literature (Kao et al., 2016; Larkin et al., 2017).
Compared with genetic findings in previous GWASs of psychiatric disorders, there are 20 novel genes not previously reported. However, active SRPK2 is increased in the cortex of APP/PS1 mice and the pathological structures of human Alzheimer’s disease (AD) brain (Hong et al., 2012; Wang et al., 2017). Neuropsychiatric disorders involve various pathological mechanisms, and AD is one of the most severe and most common progressive neurodegenerative diseases of aging. BP and AD have common mechanisms such as aberrant neurogenesis and neurotoxicity (Correa-Velloso et al., 2018; Knezevic and Mizrahi, 2018). Therefore, SRPK2 may play some important role in the development and therapeutics of other psychiatric disorders. ARFGEF2 gene was recently recognized in children with movement disorder, severe developmental delay and microcephaly including psychiatric disorders (Ellis et al., 2015; Yilmaz et al., 2016). From a biochemistry view, the GO terms have also shown that ARFGEF2 had functional enrichment including apical recycling endosome, basolateral recycling endosome, recycling endosome, and peri-centrosomal recycling endosome. Apparently, detailed pathomechanisms of the 20 novel genes identified here are unclear, and further experimental studies will need to be conducted to confirm our novel findings.
Many genes and pathways may have pleiotropic effects on more than one disease, which is a common phenomenon in psychiatric disorders (Ripke et al., 2013). Systematically and comprehensively searching for the pleiotropic genes and their effects is essential and necessary. Compared to the conventional standard single phenotype GWAS, the advantages of this study are as follows. First, statistical power is increased through the metaCCA method by leveraging five large GWAS summary statistics, which provided an increase in effective sample size to detect potential pleiotropic genes. Second, simultaneously analyzing multiple related traits can lead to richer findings compared univariate analyses, since a few of the complex associations become detectable only when multiple variants are tested jointly with multiple phenotypes. Because of these advantages, metaCCA and other multivariate analyses based on GWAS summary statistics are an emerging and powerful tool for detection of the pleiotropic genes of multiple correlated traits. However, this study could not relate to the information about the direction of effects of pleiotropic genes on risk to these psychiatric disorders because of a lack of detailed original individual measures. Alternative approaches and experimental studies may be applied to check whether novel genes could still be identified/substantiated with these methods in order to confirm novel findings in the further study.
In summary, we have performed the first systematic multivariate analysis of genome-wide data using metaCCA, identified 13 confirmed pleiotropic genes in the previous studies and highlighted 24 significant genes that may be the novel pleiotropic candidate genes for at least two of the five psychiatric traits. Furthermore, we also showed potential functional significance of the pleiotropic genes is relevant and illustrate how our results may provide with novel insights into the shared genetic factors in development of psychiatric disorders.
Supplementary Material
Acknowledgments
The authors are grateful to members of the Center for Bioinformatics and Genomics, School of Public Health and Tropical Medicine, Tulane University, for their advice and suggestions on the project.
Funding
This study was funded by grants from the National Institutes of Health (https://www.nih.gov/) to HWD (AR069055, U19 AG055373, R01 MH104680, R01AR059781 and P20GM109036), Key Technologies R & D Program of Henan Province in China (http://www.hnkjt.gov.cn/) to YLY (152102310263, 152102410007), and Edward G. Schlieder Endowment to HWD.
Footnotes
Conflict of interest
All authors declared no financial/personal interest.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jad.2018.07.046.
References
- Alkelai A, Lupoli S, Greenbaum L, Giegling I, Kohn Y, Sarner-Kanyas K, Ben-Asher E, Lancet D, Rujescu D, Macciardi F, Lerer B, 2011. Identification of new schizophrenia susceptibility loci in an ethnically homogeneous, family-based, Arab-Israeli sample. FASEB J. 25, 4011–4023. [DOI] [PubMed] [Google Scholar]
- Andreassen OA, Thompson WK, Schork AJ, Ripke S, Mattingsdal M, Kelsoe JR, Kendler KS, O’Donovan MC, Rujescu D, Werge T, Sklar P, Roddey JC, Chen C-H, McEvoy L, Desikan RS, Djurovic S, Dale AM, Psychiatric Genomics C, Bipolar Disorder and Schizophrenia Working, G., 2015. Correction: improved detection of common variants associated with schizophrenia and bipolar disorder using pleiotropy-informed conditional false discovery rate. PLos Genet. 11 e1005544–e1005544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autism Spectrum Disorders Working Group of The Psychiatric Genomics Consortium, 2017. Meta-analysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol. Autism 8, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigos KL, Mattay VS, Callicott JH, Straub RE, Vakkalanka R, Kolachana B, Hyde TM, Lipska BK, Kleinman JE, Weinberger DR, 2010. Genetic variation in CACNA1C affects brain circuitries related to mental illness. Arch. Gen. Psychiatry 67, 939–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsini A, Alboni S, Horowitz MA, Tojo LM, Cannazza G, Su K-P, Pariante CM, Zunszain PA, 2017. Rescue of IL-1 beta-induced reduction of human neurogenesis by omega-3 fatty acids and antidepressants. Brain Behav. Immun 65, 230–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde M, Forstner AJ, Adorjan K, Schaupp SK, Noethen MM, Schulze TG, 2017. Genetics of bipolar disorder. Nervenarzt 88, 755–759. [DOI] [PubMed] [Google Scholar]
- Buretic-Tomljanovic A, Vranekovic J, Rubesa G, Jonovska S, Tomljanovic D, Sendula-Jengic V, Kapovic M, Ristic S, 2012. HFE mutations and transferrin C1/C2 polymorphism among Croatian patients with schizophrenia and schizoaffective disorder. Mol. Biol. Rep 39, 2253–2258. [DOI] [PubMed] [Google Scholar]
- Casey JP, Magalhaes T, Conroy JM, Regan R, Shah N, Anney R, Shields DC, Abrahams BS, Almeida J, Bacchelli E, Bailey AJ, Baird G, Battaglia A, Berney T, Bolshakova N, Bolton PF, Bourgeron T, Brennan S, Cali P, Correia C, Corsello C, Coutanche M, Dawson G, de Jonge M, Delorme R, Duketis E, Duque F, Estes A, Farrar P, Fernandez BA, Folstein SE, Foley S, Fombonne E, Freitag CM, Gilbert J, Gillberg C, Glessner JT, Green J, Guter SJ, Hakonarson H, Holt R, Hughes G, Hus V, Igliozzi R, Kim C, Klauck SM, Kolevzon A, Lamb JA, Leboyer M, Le Couteur A, Leventhal BL, Lord C, Lund SC, Maestrini E, Mantoulan C, Marshall CR, McConachie H, McDougle CJ, McGrath J, McMahon WM, Merikangas A, Miller J, Minopoli F, Mirza GK, Munson J, Nelson SF, Nygren G, Oliveira G, Pagnamenta AT, Papanikolaou K, Parr JR, Parrini B, Pickles A, Pinto D, Piven J, Posey DJ, Poustka A, Poustka F, Ragoussis J, Roge B, Rutter ML, Sequeira AF, Soorya L, Sousa I, Sykes N, Stoppioni V, Tancredi R, Tauber M, Thompson AP, Thomson S, Tsiantis J, Van Engeland H, Vincent JB, Volkmar F, Vorstman JAS, Wallace S, Wang K, Wassink TH, White K, Wing K, Wittemeyer K, Yaspan BL, Zwaigenbaum L, Betancur C, Buxbaum JD, Cantor RM, Cook EH, Coon H, Cuccaro ML, Geschwind DH, Haines JL, Hallmayer J, Monaco AP, Nurnberger JI Jr., Pericak-Vance MA, Schellenberg GD, Scherer SW, Sutcliffe JS, Szatmari P, Vieland VJ, Wijsman EM, Green A, Gill M, Gallagher L, Vicente A, Ennis S, 2012. A novel approach of homozygous haplotype sharing identifies candidate genes in autism spectrum disorder. Hum. Genet 131, 565–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Fang K, Zhang K, Wang J, 2015. Network-based analysis of schizophrenia genome-wide association data to detect the joint functional association signals. PLoS One 10, e0133404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichonska A, Rousu J, Marttinen P, Kangas AJ, Soininen P, Lehtimaki T, Raitakari OT, Jarvelin M-R, Salomaa V, Ala-Korpela M, Ripatti S, Pirinen M, 2016. metaCCA: summary statistics-based multivariate meta-analysis of genome-wide association studies using canonical correlation analysis. Bioinformatics 32, 1981–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 381, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras J, Hare E, Chavarria-Soley G, Raventos H, 2018. Genome-wide QTL analysis for anxiety trait in bipolar disorder type I. J. Affect. Disord 234, 105–108. [DOI] [PubMed] [Google Scholar]
- Cordeiro Q, Talkowski ME, Chowdari KW, Wood J, Nimgaonkar V, Vallada H, 2004. Association and linkage analysis of RGS4 polymorphisms with schizophrenia and bipolar disorder in Brazil. Am. J. Med. Genet. Part B-Neuropsychiatr. Genet 130B 126–126. [DOI] [PubMed] [Google Scholar]
- Correa-Velloso JC, Goncalves MC, Naaldijk Y, Oliveira-Giacomelli A, Pillat MM, Ulrich H, 2018. Pathophysiology in the comorbidity of bipolar disorder and Alzheimer’s disease: pharmacological and stem cell approaches. Progr. Neuro-Psychopharmacol. Biol. Psychiatry 80, 34–53. [DOI] [PubMed] [Google Scholar]
- Cortabitarte A.d.S., Degenhardt F, Strohmaier J, Lang M, Weiss B, Roeth R, Giegling I, Heilmann-Heimbach S, Hofmann A Rujescu D, Fischer C, Rietschel M, Noethen MM, Rappold GA, Berkel S, 2017. Investigation of SHANK3 in schizophrenia. Am. J. Med. Genet. Part B Neuropsychiatr. Genet 174, 390–398. [DOI] [PubMed] [Google Scholar]
- Dedic N, Pohlmann ML, Richter JS, Mehta D, Czamara D, Metzger MW, Dine J, Bedenk BT, Hartmann J, Wagner KV, Jurik A, Almli LM, Lori A, Moosmang S, Hofmann F, Wotjak CT, Rammes G, Eder M, Chen A, Ressler KJ, Wurst W, Schmidt MV, Binder EB, Deussing JM, 2017. Cross-disorder risk gene CACNA1C differentially modulates susceptibility to psychiatric disorders during development and adulthood. Mol. Psychiatry [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E, Belliveau R, Bybjerg-Grauholm J, Baekved-Hansen M, Cerrato F, Chambert K, Churchhouse C, Dumont A, Eriksson N, Gandal M, Goldstein J, Grove J, Hansen C, Hauberg M, Hollegaard M, Howrigan DP, Huang H, Maller J, Martin AR, Moran J, Pallesen J, Palmer DS, Pedersen CB, Pedersen MG, Poterba T, Poulsen JB, Ripke S, Robinson EBSFK, Stevens C, Turley P, Won H, ADHD Working Group of the, Psychiatric Genomics Consortium (PGC), E.L.G.E.E., Consortium, Andreassen a.R.T., Burton, C. OA, Boomsma D, Cormand BDS, Franke B, Gelernter J, Geschwind D, Hakonarson H, Haavik J, Kranzler HKJ, Langley K, Lesch K-P, Middeldorp C, Reif A, Rohde LA, Roussos P, Schachar R, Sklar P, Sonuga-Barke E, Sullivan PF, Thapar A, Tung J, Waldman I, Nordentoft M, Hougaard DM, Werge T, Mors O, Mortensen PB, Daly MJ, Faraone SV,B,AD, Neale BM, 2017. Discovery of the First Genome-Wide Significant Risk Loci for ADHD. [Google Scholar]
- Ding L, Hegde AN, 2009. Expression of RGS4 splice variants in dorsolateral prefrontal cortex of schizophrenic and bipolar disorder patients. Biol. Psychiatry 65, 541–545. [DOI] [PubMed] [Google Scholar]
- Elia J, Gai X, Xie HM, Perin JC, Geiger E, Glessner JT, D’Arcy M, deBerardinis R, Frackelton E, Kim C, Lantieri F, Muganga BM, Wang L, Takeda T, Rappaport EF, Grant SFA, Berrettini W, Devoto M, Shaikh TH, Hakonarson H, White PS, 2010. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol. Psychiatry 15, 637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis KL, Zhou Y, Beshansky JR, Ainehsazan E, Selker HP, Cupples LA, Huggins GS, Peter I, 2015. Genetic modifiers of response to glucose-insulin-potassium (GIK) infusion in acute coronary syndromes and associations with clinical outcomes in the IMMEDIATE trial. Pharmacogenomics J. 15, 488–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelou E, Ioannidis JPA, 2013. Meta-analysis methods for genome-wide association studies and beyond. Nat. Rev. Genet 14, 379–389. [DOI] [PubMed] [Google Scholar]
- Gadelha A, Coleman J, Breen G, Mazzoti DR, Yonamine CM, Pellegrino R, Ota VK, Belangero SI, Glessner J, Sleiman P, Hakonarson H, Hayashi MAF, Bressan RA, 2016. Genome-wide investigation of schizophrenia associated plasma Ndel1 enzyme activity. Schizophr. Res 172, 60–67. [DOI] [PubMed] [Google Scholar]
- Gaebel W, Grossimlinghaus I, Heun R, Janssen B, Johnson B, Kurimay T, Montellano P, Muijen M, Munk-Jorgensen P, Roessler W, Ruggeri M, Thornicroft G, Zielasek J, 2015. European Psychiatric Association (EPA) guidance on quality assurance in mental healthcare. Eur. Psychiatry 30, 360–387. [DOI] [PubMed] [Google Scholar]
- Gasso P, Sanchez-Gistau V, Mas S, Sugranyes G, Rodriguez N, Boloc D, de la Serna E, Romero S, Moreno D, Moreno C, Diaz-Caneja CM, Lafuente A, Castro-Fornieles J, 2016. Association of CACNA1C and SYNE1 in offspring of patients with psychiatric disorders. Psychiatry Res. 245, 427–435. [DOI] [PubMed] [Google Scholar]
- Goes FS, Hamshere ML, Seifuddin F, Pirooznia M, Belmonte-Mahon P, Breuer R, Schulze T, Noethen M, Cichon S, Rietschel M, Holmans P, Zandi PP, Craddock N, Potash JB, and Bipolar Genome Study Bi, G.S., 2012. Genome-wide association of mood-incongruent psychotic bipolar disorder. Transl. Psychiatry 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goes FS, McGrath J, Avramopoulos D, Wolyniec P, Pirooznia M, Ruczinski I, Nestadt G, Kenny EE, Vacic V, Peters I, Lencz T, Darvasi A, Mulle JG, Warren ST, Pulver AE, 2015. Genome-wide association study of schizophrenia in Ashkenazi Jews. Am. J. Med. Genet. Part B Neuropsychiatr. Genet 168, 649–659. [DOI] [PubMed] [Google Scholar]
- Guerini FR, Bolognesi E, Manca S, Sotgiu S, Zanzottera M, Agliardi C, Usai S, Clerici M, 2009. Family-based transmission analysis of HLA genetic markers in Sardinian children with autistic spectrum disorders. Hum. Immunol 70, 184–190. [DOI] [PubMed] [Google Scholar]
- Hong Y, Chan CB, Kwon I-S, Li X, Song M, Lee H-P, Liu X, Sompol P, Jin P, Lee H. g., Yu SP, Ye K, 2012. SRPK2 phosphorylates tau and mediates the cognitive defects in Alzheimer’s disease. J. Neurosci 32, 17262–17272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou T, Jian C, Xu J, Huang AY, Xi J, Hu K, Wei L, Cheng H, Wang X, 2016. Identification of EFHD1 as a novel Ca2+ sensor for mitoflash activation. Cell Calcium 59, 262–270. [DOI] [PubMed] [Google Scholar]
- Hunt GE, Malhi GS, Cleary M, Lai HMX, Sitharthan T, 2016. Prevalence of comorbid bipolar and substance use disorders in clinical settings, 1990–2015: systematic review and meta-analysis. J. Affect. Disord 206, 331–349. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Takahashi A, Kamatani Y, Okahisa Y, Kunugi H, Mori N, Sasaki T, Ohmori T, Okamoto Y, Kawasaki H, Shimodera S, Kato T, Yoneda H, Yoshimura R, Iyo M, Matsuda K, Akiyama M, Ashikawa K, Kashiwase K, Tokunaga K, Kondo K, Saito T, Shimasaki A, Kawase K, Kitajima T, Matsuo K, Itokawa M, Someya T, Inada T, Hashimoto R, Inoue T, Akiyama K, Tanii H, Arai H, Kanba S, Ozaki N, Kusumi I, Yoshikawa T, Kubo M, Iwata N, 2017. A genome-wide association study identifies two novel susceptibility loci and trans population polygenicity associated with bipolar disorder. Mol. Psychiatry [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye M, Ripatti S, Kettunen J, Lyytikainen L-P, Oksala N, Laurila P-P, Kangas AJ, Soininen P, Savolainen MJ, Viikari J, Kahonen M, Perola M, Salomaa V, Raitakari O, Lehtimaki T, Taskinen M-R, Jaervelin M-R, Ala-Korpela M, Palotie A, de Bakker PIW, 2012. Novel loci for metabolic networks and multi-tissue expression studies reveal genes for atherosclerosis. PLos Genet. 8, e1002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman M, Zhou H, Jia L, Cain MD, Blumer KJ, 2009. R9AP and R7BP: traffic cops for the RGS7 family in phototransduction and neuronal GPCR signaling. Trends Pharmacol. Sci 30, 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeste DV, Palmer BW, Rettew DC, Boardman S, 2015. Positive psychiatry: its time has come. J. Clin. Psychiatry 76, 675–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao C-F, Chen H-W, Chen H-C, Yang J-H, Huang M-C, Chiu Y-H, Lin S-K, Lee Y-C, Liu C-M, Chuang L-C, Chen C-H, Wu J-Y, Lu R-B, Kuo P-H, 2016. Identification of Susceptible loci and enriched pathways for bipolar ii disorder using genome-wide association studies. Int. J. Neuropsychopharmacol 19, pyw064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettunen J, Tukiainen T, Sarin A-P, Ortega-Alonso A, Tikkanen E, Lyytikainen L-P, Kangas AJ, Soininen P, Wuertz P, Silander K, Dick DM, Rose RJ, Savolainen MJ, Viikari J, Kahonen M, Lehtimaki T, Pietilainen KH, Inouye M, McCarthy MI, Jula A, Eriksson J, Raitakari OT, Salomaa V, Kaprio J, Jarvelin M-R, Peltonen L, Perola M, Freimer NB, Ala-Korpela M, Palotie A, Ripatti S, 2012. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat. Genet 44, 269–U265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury JM, Neves M.d.C.L.d., Roque MAV, Queiroz D.A.d.B., de Freitas Correa, de Fatima AA, Moreira, F.A. A, Garcia FD, 2017. Is there a role for cannabidiol in psychiatry? World J. Biol. Psychiatry 1–16. [DOI] [PubMed] [Google Scholar]
- Knezevic D, Mizrahi R, 2018. Molecular imaging of neuroinflammation in Alzheimer’s disease and mild cognitive impairment. Progr. Neuro-Psychopharmacol. Biol. Psychiatry 80, 123–131. [DOI] [PubMed] [Google Scholar]
- Larkin D, Kirtchuk G, Yamaguchi M, Martin CR, 2017. A proposal for the inclusion of ‘obesity dysmorphia’ in the Diagnostic and statistical manual of mental disorders. Aust. N. Z. J. Psychiatry 4867417722641–4867417722641. [DOI] [PubMed] [Google Scholar]
- Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, Perlis RH, Mowry BJ, Thapar A, Goddard ME, Witte JS, Absher D, Agartz I, Akil H, Amin F, Andreassen OA, Anjorin A, Anney R, Anttila V, Arking DE, Asherson P, Azevedo MH, Backlund L, Badner JA, Bailey AJ, Banaschewski T, Barchas JD, Barnes MR, Barrett TB, Bass N, Battaglia A, Bauer M, Bayes M, Bellivier F, Bergen SE, Berrettini W, Betancur C, Bettecken T, Biederman J, Binder EB, Black DW, Blackwood DHR, Bloss CS, Boehnke M, Boomsma DI, Breen G, Breuer R, Bruggeman R, Cormican P, Buccola NG, Buitelaar JK, Bunney WE, Buxbaum JD, Byerley WF, Byrne EM, Caesar S, Cahn W, Cantor RM, Casas M, Chakravarti A, Chambert K, Choudhury K, Cichon S, Cloninger CR, Collier DA, Cook EH, Coon H, Cormand B, Corvin A, Coryell WH, Craig DW, Craig IW, Crosbie J, Cuccaro ML, Curtis D, Czamara D, Datta S, Dawson G, Day R, De Geus EJ, Degenhardt F, Djurovic S, Donohoe GJ, Doyle AE, Duan J, Dudbridge F, Duketis E, Ebstein RP, Edenberg HJ, Elia J, Ennis S, Etain B, Fanous A, Farmer AE, Ferrier IN, Flickinger M, Fombonne E, Foroud T, Frank J, Franke B, Fraser C, Freedman R, Freimer NB, Freitag CM, Friedl M, Frisen L, Gallagher L, Gejman PV, Georgieva L, Gershon ES, Geschwind DH, Giegling I, Gill M, Gordon SD, Gordon-Smith K, Green EK, Greenwood TA, Grice DE, Gross M, Grozeva D, Guan W, Gurling H, De Haan L, Haines JL, Hakonarson H, Hallmayer J, Hamilton SP, Hamshere ML, Hansen TF, Hartmann AM, Hautzinger M, Heath AC, Henders AK, Herms S, Hickie IB, Hipolito M, Hoefels S, Holmans PA, Holsboer F, Hoogendijk WJ, Hottenga J-J, Hultman CM, Hus V, Ingason A, Ising M, Jamain S, Jones EG, Jones I, Jones L, Tzeng J-Y, Kaehler AK, Kahn RS, Kandaswamy R, Keller MC, Kennedy JL, Kenny E, Kent L, Kim Y, Kirov GK, Klauck SM, Klei L, Knowles JA, Kohli MA, Koller DL, Konte B, Korszun A, Krabbendam L, Krasucki R, Kuntsi J, Kwan P, Landen M, Langstrom N, Lathrop M, Lawrence J, Lawson WB, Leboyer M, Ledbetter DH, Lee PH, Lencz T, Lesch K-P, Levinson DF, Lewis CM, Li J, Lichtenstein P, Lieberman JA, Lin D-Y, Linszen DH, Liu C, Lohoff FW, Loo SK, Lord C, Lowe JK, Lucae S, MacIntyre DJ, Madden PAF, Maestrini E, Magnusson PKE, Mahon PB, Maier W, Malhotra AK, Mane SM, Martin CL, Martin NG, Mattheisen M, Matthews K, Mattingsdal M, McCarroll SA, McGhee KA, McGough JJ, McGrath PJ, McGuffin P, McInnis MG, McIntosh A, McKinney R, McLean AW, McMahon FJ, McMahon WM, McQuillin A, Medeiros H, Medland SE, Meier S, Melle I, Meng F, Meyer J, Middeldorp CM, Middleton L, Milanova V, Miranda A, Monaco AP, Montgomery GW, Moran JL, Moreno-De-Luca D, Morken G, Morris DW, Morrow EM, Moskvina V, Muglia P, Muehleisen TW, Muir WJ, Mueller-Myhsok B, Murtha M, Myers RM, Myin-Germeys I, Neale MC, Nelson SF, Nievergelt CM, Nikolov I, Nimgaonkar V, Nolen WA, Noethen MM, Nurnberger JI, Nwulia EA, Nyholt DR, O’Dushlaine C, Oades RD, Olincy A, Oliveira G, Olsen L, Ophoff RA, Osby U, Owen MJ, Palotie A, Parr JR, Paterson AD, Pato CN, Pato MT, Penninx BW, Pergadia ML, Pericak-Vance MA, Pickard BS, Pimm J, Piven J, Posthuma D, Potash JB, Poustka F, Propping P, Puri V, Quested DJ, Quinn EM, Antoni Ramos-Quiroga J, Rasmussen HB, Raychaudhuri S, Rehnstroem K, Reif A, Ribases M, Rice JP, Rietschel M, Roeder K, Roeyers H, Rossin L, Rothenberger A, Rouleau G, Ruderfer D, Rujescu D, Sanders AR, Sanders SJ, Santangelo SL, Sergeant JA, Schachar R, Schalling M, Schatzberg AF, Scheftner WA, Schellenberg GD, Scherer SW, Schork NJ, Schulze TG, Schumacher J, Schwarz M, Scolnick E, Scott LJ, Shi J, Shilling PD, Shyn SI, Silverman JM, Slager SL, Smalley SL, Smit JH, Smith EN, Sonuga-Barke EJS, St Clair D, State M, Steffens M, Steinhausen H-C, Strauss JS, Strohmaier J, Stroup TS, Sutcliffe JS, Szatmari P, Szelinger S, Thirumalai S, Thompson RC, Todorov AA, Tozzi F, Treutlein J, Uhr M, van den Oord EJCG, Van Grootheest G, Van Os J, Vicente AM, Vieland VJ, Vincent JB, Visscher PM, Walsh CA, Wassink TH, Watson SJ, Weissman MM, Werge T, Wienker TF, Wijsman EM, Willemsen G, Williams N, Willsey AJ, Witt SH, Xu W, Young AH, Yu TW, Zammit S, Zandi PP, Zhang P, Zitman FG, Zoellner S, Devlin B, Kelsoe JR, Sklar P, Daly MJ, O’Donovan MC, Craddock N, Sullivan PF, Smoller JW, Kendler KS, Wray NR, Cross-Disorder Grp Psychiat G, Int Inflammatory Bowel Dis, G., 2013a. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat. Genet 45 984–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Kim J-H, Song GG, 2013b. Pathway analysis of a genome-wide association study in schizophrenia. Gene 525, 107–115. [DOI] [PubMed] [Google Scholar]
- Lencz T, Guha S, Liu C, Rosenfeld J, Mukherjee S, DeRosse P, John M, Cheng L, Zhang C, Badner JA, Ikeda M, Iwata N, Cichon S, Rietschel M, Noethen MM, Cheng ATA, Hodgkinson C, Yuan Q, Kane JM, Lee AT, Pisante A, Gregersen PK, Pe’er I, Malhotra AK, Goldman D, Darvasi A, 2013. Genome-wide association study implicates NDST3 in schizophrenia and bipolar disorder. Nat. Commun 4, 2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marttinen P, Gillberg J, Havulinna A, Corander J, Kaski S, 2013. Genome-wide association studies with high-dimensional phenotypes. Stat. Appl. Genet. Mol. Biol 12, 413–431. [DOI] [PubMed] [Google Scholar]
- McMahon FJ, Akula N, Schulze TG, Muglia P, Tozzi F, Detera-Wadleigh SD, Steele CJM, Breuer R, Strohmaier J, Wendland JR, Mattheisen M, Muehleisen TW, Maier W, Noethen MM, Cichon S, Farmer A, Vincent JB, Holsboer F, Preisig M, Rietschel M, Bipolar Disorder Genome Study Bi, G.S., 2010. Meta-analysis of genome-wide association data identifies a risk locus for major mood disorders on 3p21.1. Nat. Genet 42, 128–U152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merikangas KR, He JP, Burstein M, Swanson SA, Avenevoli S, Cui LH, Benjet C, Georgiades K, Swendsen J, 2010. Lifetime prevalence of mental disorders in U.S. adolescents: results from the National Comorbidity Survey Replication-Adolescent Supplement (NCS-A). J. Am. Acad. Child Adolesc. Psychiatry 49, 980–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A, Macgregor S, 2015. VEGAS2: software for more flexible gene-based testing. Twin Res. Hum. Genet 18, 86–91. [DOI] [PubMed] [Google Scholar]
- Mistry S, Harrison JR, Smith DJ, Escott-Price V, Zammit S, 2018. The use of polygenic risk scores to identify phenotypes associated with genetic risk of bipolar disorder and depression: a systematic review. J. Affect. Disord 234, 148–155. [DOI] [PubMed] [Google Scholar]
- Nigg JT, Elmore AL, Natarajan N, Friderici KH, Nikolas MA, 2016. Variation in an iron metabolism gene moderates the association between blood lead levels and attention-deficit/hyperactivity disorder in children. Psychol. Sci 27, 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor A, Whibley A, Marshall CR, Gianakopoulos PJ, Piton A, Carson AR, Orlic-Milacic M, Lionel AC, Sato D, Pinto D, Drmic I, Noakes C, Senman L, Zhang X, Mo R, Gauthier J, Crosbie J, Pagnamenta AT, Munson J, Estes AM, Fiebig A, Franke A, Schreiber S, Stewart AFR, Roberts R, McPherson R, Guter SJ, Cook EH Jr., Dawson G, Schellenberg GD, Battaglia A, Maestrini E, Jeng L, Hutchison T, Rajcan-Separovic E, Chudley AE, Lewis SME, Liu X, Holden JJ, Fernandez B, Zwaigenbaum L, Bryson SE, Roberts W, Szatmari P, Gallagher L, Stratton MR, Gecz J, Brady AF, Schwartz CE, Schachar RJ, Monaco AP, Rouleau GA, Hui C. c., Raymond FL, Scherer SW, Vincent JB, Autism Genome Project C, 2010. Disruption at the PTCHD1 Locus on Xp22.11 in Autism spectrum disorder and intellectual disability. Sci. Transl. Med 2 49ra68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothen MM, Nieratschker V, Cichon S, Rietschel M, 2010. New findings in the genetics of major psychoses. Dialogues Clin. Neurosci 12, 85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurnberger JI Jr., Koller DL, Jung J, Edenberg HJ, Foroud T, Guella I, Vawter MP, Kelsoe JR, Psychiat Genomics C, 2014. Identification of pathways for bipolar disorder a meta-analysis. Jama Psychiatry 71, 657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, Lin D-Y, Duan J, Ophoff RA, Andreassen OA, Scolnick E, Cichon S, Clair DS, Corvin A, Gurling H, Werge T, Rujescu D, Blackwood DHR, Pato CN, Malhotra AK, Purcell S, Dudbridge F, Neale BM, Rossin L, Visscher PM, Posthuma D, Ruderfer DM, Fanous A, Stefansson H, Steinberg S, Mowry BJ, Golimbet V, De Hert M, Jonsson EG, Bitter I, Pietilainen OPH, Collier DA, Tosato S, Agartz I, Albus M, Alexander M, Amdur RL, Amin F, Bass N, Bergen SE, Black DW, Borglum AD, Brown MA, Bruggeman R, Buccola NG, Byerley WF, Cahn W, Cantor RM, Carr VJ, Catts SV, Choudhury K, Cloninger CR, Cormican P, Craddock N, Danoy PA, Datta S, De Haan L, Demontis D, Dikeos D, Djurovic S, Donnelly P, Donohoe G, Duong L, Dwyer S, Fink-Jensen A, Freedman R, Freimer NB, Friedl M, Georgieva L, Giegling I, Gill M, Glenthoj B, Godard S, Hamshere M, Hansen M, Hansen T, Hartmann AM, Henskens FA, Hougaard DM, Hultman CM, Ingason A, Jablensky AV, Jakobsen KD, Jay M, Juergens G, Kahn R, Keller MC, Kenis G, Kenny E, Kim Y, Kirov GK, Konnerth H, Konte B, Krabbendam L, Krasucki R, Lasseter VK, Laurent C, Lawrence J, Lencz T, Lerer FB, Liang K-Y, Lichtenstein P, Lieberman JA, Linszen DH, Lonnqvist J, Loughland CM, Maclean AW, Maher BS, Maier W, Mallet J, Malloy P, Mattheisen M, Mattingsdal M, McGhee KA, McGrath JJ, McIntosh A, McLean DE, McQuillin A, Melle I, Michie PT, Milanova V, Morris DW, Mors O, Mortensen PB, Moskvina V, Muglia P, Myin-Germeys I, Nertney DA, Nestadt G, Nielsen J, Nikolov I, Nordentoft M, Norton N, Noethen MM, O’Dushlaine CT, Olincy A, Olsen L, O’Neill FA, Orntoft TF, Owen MJ, Pantelis C, Papadimitriou G, Pato MT, Peltonen L, Petursson H, Pickard B, Pimm J, Pulver AE, Puri V, Quested D, Quinn EM, Rasmussen HB, Rethelyi JM, Ribble R, Rietschel M, Riley BP, Ruggeri M, Schall U, Schulze TG, Schwab SG, Scott RJ, Shi J, Sigurdsson E, Silverman JM, Spencer CCA, Stefansson K, Strange A, Strengman E, Stroup TS, Suvisaari J, Terenius L, Thirumalai S, Thygesen JH, Timm S, Toncheva D, van den Oord E, van Os J, van Winkel R, Veldink J, Walsh D, Wang AG, Wiersma D, Wildenauer DB, Williams HJ, Williams NM, Wormley B, Zammit S, Sullivan PF, O’Donovan MC, Daly MJ, Gejman PV, Schizophrenia Psychiat G-W, 2011. Genome-wide association study identifies five new schizophrenia loci. Nat. Genet 43, 969–U977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM, Breen G, Byrne EM, Blackwood DHR, Boomsma DI, Cichon S, Heath AC, Holsboer F, Lucae S, Madden PAF, Martin NG, McGuffin P, Muglia P, Noethen MM, Penninx BP, Pergadia ML, Potash JB, Rietschel M, Lin D, Muller-Myhsok B, Shi J, Steinberg S, Grabe HJ, Lichtenstein P, Magnusson P, Perlis RH, Preisig M, Smoller JW, Stefansson K, Uher R, Kutalik Z, Tansey KE, Teumer A, Viktorin A, Barnes MR, Bettecken T, Binder EB, Breuer R, Castro VM, Churchill SE, Coryell WH, Craddock N, Craig IW, Czamara D, De Geus EJ, Degenhardt F, Farmer AE, Fava M, Frank J, Gainer VS, Gallagher PJ, Gordon SD, Goryachev S, Gross M, Guipponi M, Henders AK, Herms S, Hickie IB, Hoefels S, Hoogendijk W, Hottenga JJ, Iosifescu DV, Ising M, Jones I, Jones L, Jung-Ying T, Knowles JA, Kohane IS, Kohli MA, Korszun A, Landen M, Lawson WB, Lewis G, Macintyre D, Maier W, Mattheisen M, McGrath PJ, McIntosh A, McLean A, Middeldorp CM, Middleton L, Montgomery GM, Murphy SN, Nauck M, Nolen WA, Nyholt DR, O’Donovan M, Oskarsson H, Pedersen N, Scheftner WA, Schulz A, Schulze TG, Shyn SI, Sigurdsson E, Slager SL, Smit JH, Stefansson H, Steffens M, Thorgeirsson T, Tozzi F, Treutlein J, Uhr M, van den Oord EJCG, Van Grootheest G, Volzke H, Weilburg JB, Willemsen G, Zitman FG, Neale B, Daly M, Levinson DF, Sullivan PF, Major Depressive Disorder Working Group of the Psychiatric, G., Consortium, 2013. A mega-analysis of genome-wide association studies for major depressive disorder. Mol. Psychiatry 18, 497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruderfer DM, Fanous AH, Ripke S, McQuillin A, Amdur RL, Gejman PV, O’Donovan MC, Andreassen OA, Djurovic S, Hultman CM, Kelsoe JR, Jamain S, Landen M, Leboyer M, Nimgaonkar V, Nurnberger J, Smoller JW, Craddock N, Corvin A, Sullivan PF, Holmans P, Sklar P, Kendler KS, Schizophrenia Working Grp P, Bipolar Disorder Working Grp, P., Cross-Disorder Working Grp, P., 2014. Polygenic dissection of diagnosis and clinical dimensions of bipolar disorder and schizophrenia. Mol. Psychiatry 19, 1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmelmann BG, Hinney A, Scherag A, Puetter C, Pechlivanis S, Cichon S, Joeckel KH, Schreiber S, Wichmann HE, Albayrak O, Dauvermann M, Konrad K, Wilhelm C, Herpertz-Dahlmann B, Lehmkuhl G, Sinzig J, Renner TJ, Romanos M, Warnke A, Lesch KP, Reif A, Hebebrand J, 2013. Bipolar disorder risk alleles in children with ADHD. J. Neural Transm. 120, 1611–1617. [DOI] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane JA, Campbell C, Day INM, Casas JP, Gaunt TR, 2014. Canonical correlation analysis for gene-based pleiotropy discovery. PLoS Comput. Biol 10, e1003876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ski CF, Jelinek M, Jackson AC, Murphy BM, Thompson DR, 2016. Psychosocial interventions for patients with coronary heart disease and depression: a systematic review and meta-analysis. Eur. J. Cardiovasc. Nurs 15, 305–316. [DOI] [PubMed] [Google Scholar]
- Sklar P, Ripke S, Scott LJ, Andreassen OA, Cichon S, Craddock N, Edenberg HJ, Nurnberger JI Jr., Rietschel M, Blackwood D, Corvin A, Flickinger M, Guan W, Mattingsdal M, McQuillin A, Kwan P, Wienker TF, Daly M, Dudbridge F, Holmans PA, Lin D, Burmeister M, Greenwood TA, Hamshere ML, Muglia P, Smith EN, Zandi PP, Nievergelt CM, McKinney R, Shilling PD, Schork NJ, Bloss CS, Foroud T, Koller DL, Gershon ES, Liu C, Badner JA, Scheftner WA, Lawson WB, Nwulia EA, Hipolito M, Coryell W, Rice J, Byerley W, McMahon FJ, Schulze TG, Berrettini W, Lohoff FW, Potash JB, Mahon PB, McInnis MG, Zoellner S, Zhang P, Craig DW, Szelinger S, Barrett TB, Breuer R, Meier S, Strohmaier J, Witt SH, Tozzi F, Farmer A, McGuffin P, Strauss J, Xu W, Kennedy JL, Vincent JB, Matthews K, Day R, Ferreira MA, O’Dushlaine C, Perlis R, Raychaudhuri S, Ruderfer D, Hyoun PL, Smoller JW, Li J, Absher D, Thompson RC, Meng FG, Schatzberg AF, Bunney WE, Barchas JD, Jones EG, Watson SJ, Myers RM, Akil H, Boehnke M, Chambert K, Moran J, Scolnick E, Djurovic S, Melle I, Morken G, Gill M, Morris D, Quinn E, Muehleisen TW, Degenhardt FA, Mattheisen M, Schumacher J, Maier W, Steffens M, Propping P, Noethen MM, Anjorin A, Bass N, Gurling H, Kandaswamy R, Lawrence J, McGhee K, McIntosh A, McLean AW, Muir WJ, Pickard BS, Breen G, St Clair D, Caesar S, Gordon-Smith K, Jones L, Fraser C, Green EK, Grozeva D, Jones IR, Kirov G, Moskvina V, Nikolov I, O’Donovan MC, Owen MJ, Collier DA, Elkin A, Williamson R, Young AH, Ferrier IN, Stefansson K, Stefansson H, Porgeirsson P, Steinberg S, Gustafsson O, Bergen SE, Nimgaonkar V, Hultman C, Landen M, Lichtenstein P, Sullivan P, Schalling M, Osby U, Backlund L, Frisen L, Langstrom N, Jamain S, Leboyer M, Etain B, Bellivier F, Petursson H, Sigurosson E, Mueller-Mysok B, Lucae S, Schwarz M, Schofield PR, Martin N, Montgomery GW, Lathrop M, Oskarsson H, Bauer M, Wright A, Mitchell PB, Hautzinger M, Reif A, Kelsoe JR, Purcell SM, Psychiat GCBD, 2011. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet 43 977–U162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoller JW, Finn CT, 2003. Family, twin, and adoption studies of bipolar disorder. Am. J. Med. Genet Part C 123C, 48–58. [DOI] [PubMed] [Google Scholar]
- Soldatov NM, 2015. CACNB2: an emerging pharmacological target for hypertension, heart failure, arrhythmia and mental disorders. Current Mol. Pharmacol 8, 32–42. [DOI] [PubMed] [Google Scholar]
- Stearns FW, 2011. One hundred years of pleiotropy: a retrospective. 186, pg 767, 2010. Genetics 187 355–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Daly MJ, Ripke S, Lewis CM, Lin D-Y, Wray NR, Neale B, Levinson DF, Breen G, Byrne EM, Rietschel M, Hoogendijk W, Hamilton SP, Weissman MM, Breuer R, Cichon S, Degenhardt F, Frank J, Gross M, Herms S, Hoefels S, Maier W, Mattheisen M, Noeethen MM, Schulze TG, Steffens M, Treutlein J, Boomsma DI, De Geus EJ, Hottenga JJ, Jung-Ying T, Middeldorp CM, Nolen WA, Penninx BP, Smit JH, van Grootheest G, Willemsen G, Zitman FG, Coryell WH, Knowles JA, Lawson WB, Potash JB, Scheftner WA, Shi J, Holsboer F, Muglia P, Tozzi F, Blackwood DHR, MacIntyre DJ, McIntosh A, McLean A, van den Oord EJCG, Lucae S, Binder E, Mueller-Myhsok B, Czamara D, Kohli MA, Ising M, Uhr M, Bettecken T, Barnes MR, Craig IW, Farmer AE, McGuffin P, Byrne E, Gordon SD, Heath AC, Henders AK, Hickie IB, Madden PAF, Martin NG, Montgomery GM, Nyholt DR, Pergadia ML, McGrath PJ, Shyn SI, Slager SL, Oskarsson H, Sigurdsson E, Stefansson H, Stefansson K, Steinberg S, Thorgeirsson T, Guipponi M, Lewis G, O’Donovan M, Tansey KE, Uher R, Castro VM, Churchill SE, Fava M, Gainer VS, Gallagher PJ, Goryachev S, Iosifescu DV, Kohane IS, Murphy SN, Perlis RH, Smoller JW, Weilburg JB, Kutalik Z, Preisig M, Grabe HJ, Nauck M, Schulz A, Teumer A, Voelzke H, Landen M, Lichtenstein P, Magnusson P, Pedersen N, Viktorin A, Psychiat GC, 2013. A mega-analysis of genome-wide association studies for major depressive disorder. Mol. Psychiatry 18, 497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang CS, Ferreira MAR, 2012. A gene-based test of association using canonical correlation analysis. Bioinformatics 28, 845–850. [DOI] [PubMed] [Google Scholar]
- Thimm M, Kircher T, Kellermann T, Markov V, Krach S, Jansen A, Zerres K, Eggermann T, Stoecker T, Shah NJ, Noethen MM, Rietschel M, Witt SH, Mathiak K, Krug A, 2011. Effects of a CACNA1C genotype on attention networks in healthy individuals. Psychol. Med 41, 1551–1561. [DOI] [PubMed] [Google Scholar]
- Tischfield DJ, Saraswat DK, Furash A, Fowler SC, Fuccillo MV, Anderson SA, 2017. Loss of the neurodevelopmental gene Zswim6 alters striatal morphology and motor regulation. Neurobiol. Dis 103, 174–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg SRF, Ousager LB, Miller KA, Zhou Y, Elalaoui SC, Sefiani A, Bak GS, Hove H, Hansen LK, Fagerberg CR, Tajir M, Wilkie AOM, 2016. Acromelic frontonasal dysostosis and ZSWIM6 mutation: phenotypic spectrum and mosaicism. Clin. Genet 90, 270–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Brown MA, McCarthy MI, Yang J, 2012. Five years of GWAS discovery. Am. J. Hum. Genet 90, 7–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-H, Liu P, Liu X, Manfredsson FP, Sandoval IM, Yu SP, Wang J-Z, Ye K, 2017. Delta-secretase phosphorylation by srpk2 enhances its enzymatic activity, provoking pathogenesis in Alzheimer’s disease. Mol. Cell 67 812–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt SH, Juraeva D, Sticht C, Strohmaier J, Meier S, Treutlein J, Dukal H, Frank J, Lang M, Deuschle M, Schulze TG, Degenhardt F, Mattheisen M, Brors B, Cichon S, Noethen MM, Witt CC, Rietschel M, 2014. Investigation of manic and euthymic episodes identifies state- and trait-specific gene expression and STAB1 as a new candidate gene for bipolar disorder. Transl. Psychiatry 4, e426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt SH, Streit F, Jungkunz M, Frank J, Awasthi S, Reinbold CS, Treutlein J, Degenhardt F, Forstner AJ, Heilmann-Heimbach S, Dietl L, Schwarze CE, Schendel D, Strohmaier J, Abdellaoui A, Adolfsson R, Air TM, Akil H, Alda M, Alliey-Rodriguez N, Andreassen OA, Babadjanova G, Bass NJ, Bauer M, Baune BT, Bellivier F, Bergen S, Bethell A, Biernacka JM, Blackwood DHR, Boks MP, Boomsma DI, Borglum AD, Borrmann-Hassenbach M, Brennan P, Budde M, Buttenschon HN, Byrne EM, Cervantes P, Clarke TK, Craddock N, Cruceanu C, Curtis D, Czerski PM, Dannlowski U, Davis T, de Geus EJC, Di Florio A, Djurovic S, Domenici E, Edenberg HJ, Etain B, Fischer SB, Forty L, Fraser C, Frye MA, Fullerton JM, Gade K, Gershon ES, Giegling I, Gordon SD, Gordon-Smith K, Grabe HJ, Green EK, Greenwood TA, Grigoroiu-Serbanescu M, Guzman-Parra J, Hall LS, Hamshere M, Hauser J, Hautzinger M, Heilbronner U, Herms S, Hitturlingappa S, Hoffmann P, Holmans P, Hottenga JJ, Jamain S, Jones I, Jones LA, Jureus A, Kahn RS, Kammerer-Ciernioch J, Kirov G, Kittel-Schneider S, Kloiber S, Knott SV, Kogevinas M, Landen M, Leber M, Leboyer M, Li QS, Lissowska J, Lucae S, Martin NG, Mayoral-Cleries F, McElroy SL, McIntosh AM, McKay JD, McQuillin A, Medland SE, Middeldorp CM, Milaneschi Y, Mitchell PB, Montgomery GW, Morken G, Mors O, Muehleisen TW, Mueller-Myhsok B, Myers RM, Nievergelt CM, Nurnberger JI, O’Donovan MC, Loohuis LMO, Ophoff R, Oruc L, Owen MJ, Paciga SA, Penninx BWJH, Perry A, Pfennig A, Potash JB, Preisig M, Reif A, Rivas F, Rouleau GA, Schofield PR, Schulze TG, Schwarz M, Scott L, Sinnamon GCB, Stahl EA, Strauss J, Turecki G, Van der Auwera S, Vedder H, Vincent JB, Willemsen G, Witt CC, Wray NR, Xi HS, Tadic A, Dahmen N, Schott BH, Cichon S, Noethen MM, Ripke S, Mobascher A, Rujescu D, Lieb K, Roepke S, Schmahl C, Bohus M, Rietschel M, Psychiat Genomics C, 2017. Genome-wide association study of borderline personality disorder reveals genetic overlap with bipolar disorder, major depression and schizophrenia. Transl. Psychiatry 7, e1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik GL, Kao WHL, Duggal P, 2015. Relative performance of gene- and pathway-level methods as secondary analyses for genome-wide association studies. BMC Genet. 16, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurfel BE, Drevets WC, Bliss SA, McMillin JR, Suzuki H, Ford BN, Morris HM, Teague TK, Dantzer R, Savitz JB, 2017. Serum kynurenic acid is reduced in affective psychosis. Transl. Psychiatry 7, e1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz S, Gokben S, Serdaroglu G, Eraslan C, Mancini GMS, Tekin H, Tekgul H, 2016. The expanding phenotypic spectrum of ARFGEF2 gene mutation: cardiomyopathy and movement disorder. Brain Dev. 38, 124–127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.