ABSTRACT
NUT carcinoma (NC) is an aggressive squamous tumor characterized by NUT gene rearrangement, and the most common fusion form is BRD4-NUT. However, NC diagnosis is difficult for its rareness and often being confused with a variety of poorly differentiated tumors. A 21-year-old Chinese woman was referred to our hospital for cough and intermittent fever. Chest computed tomography (CT) imaging revealed a left lobe hilar mass. Fiberoptic bronchoscopy results showed that tumor cells were poorly differentiated. In combination with immunohistochemistry staining, she was misdiagnosed with Ewing’s sarcoma/primitive neuroectodermal tumor. Next-generation sequencing (NGS) revealing BRD4-NUT fusion, and NUT immunohistochemistry confirmed the diagnosis of NC. Subsequently, left pneumonectomy and lymph node dissection were performed, and the patient received pemetrexed and lobaplatin treatment. NGS technology played an important role in NC diagnosis in this case, and it may have clinical use for rare cancer diagnosis and guidance of potential targeted therapies.
Keywords: NUT carcinoma, BRD4-NUT, next-generation sequencing
Background
NUT carcinoma (NC), an aggressive subtype of squamous cell carcinoma, is defined by an acquired rearrangement involving the NUT (NUT midline carcinoma family member 1) gene on chromosome 15. In most cases, NUTM1 is fused to BRD4 (bromodomain-containing protein 4) on chromosome 19.1 NUT carcinoma can arise in different organs, including lung, mediastinum, bladder, pancreas, kidney and salivary gland,1 which can affect any age or gender group.2 Ewing’s sarcoma (ES)/primitive neuroectodermal tumor (PNET) is a malignant prototypical primitive small round cell tumor that arises from lung, bone and other sites in children or adolescents,3,4 which can be indistinguishable from poorly differentiated NC.5
Here, we present a case that was previously misdiagnosed as ES/PNET but was later confirmed as NC through next-generation sequencing (NGS).
Case report
A 21-year-old Chinese woman presented with cough and intermittent fever. The patient had no hemoptysis, chest pain, shortness of breath, hoarseness, headache or bone pain. Chest computed tomography (CT) imaging revealed a left lobe hilar mass measuring 5.7 cm x 4.5 cm x 5.4 cm, enlarged hilar lymph nodes, mediastinal vessels invasion and inflammatory obliteration in the left lung lobe (Figure 1(a)). Bone scan and magnetic resonance imaging (MRI) for brain showed no abnormality. Laboratory tests for blood showed elevated AFP (137.26 ng/mL; normal range 0.89–8.78 ng/mL) and β-HCG of 1.20 mIU/ml (negative). She underwent fiberoptic bronchoscopy, and pathologic examination showed poorly differentiated malignant tumor composing of cords and nests of small-sized cells with round conspicuous nucleus and translucent cytoplasm (Figure 2(a)) and demonstrated the presence of bronchus and blood vessel involvement, neutrophil infiltration and alveolar cleft-like defect (data not shown). Morphologically, squamous differentiation was absent by hematoxylin-eosin staining. A battery of immunohistochemistry markers was performed for tumor cells, but only CD99 was positive (Figure 2(b)). Despite the young age, malignant ES/PNET was considered. Differential diagnosis included adenocarcinoma (TTF-1, napsin A, CK7 and pancytokeratin negative), squamous carcinoma (P40 and CK5/6 negative), neuroendocrine carcinoma (NSE, chromogranin A and CD56 negative), embryonal rhabdomyosarcoma (desmin, MyoD1, myogenin and synaptophysin negative). There were surgery indications, so left pneumonectomy by video-assisted thoracic surgery (VATS) and lymph node dissection were performed (Figure 1(b)). Immunohistochemistry staining of the resection specimen demonstrated that tumor cells were positive for CD99, synaptophysin and FLI1, and negative for pancytokeratin, CK18, and P40 (data not shown). Fluorescence in situ hybridization (FISH) for resection specimen was negative for EWSR1 (Ewing sarcoma breakpoint region 1) rearrangement. A few days after surgery, the level of AFP dramatically decreased to 14.05 ng/ml (normal range 0.89–8.78 ng/ml).
Figure 1.
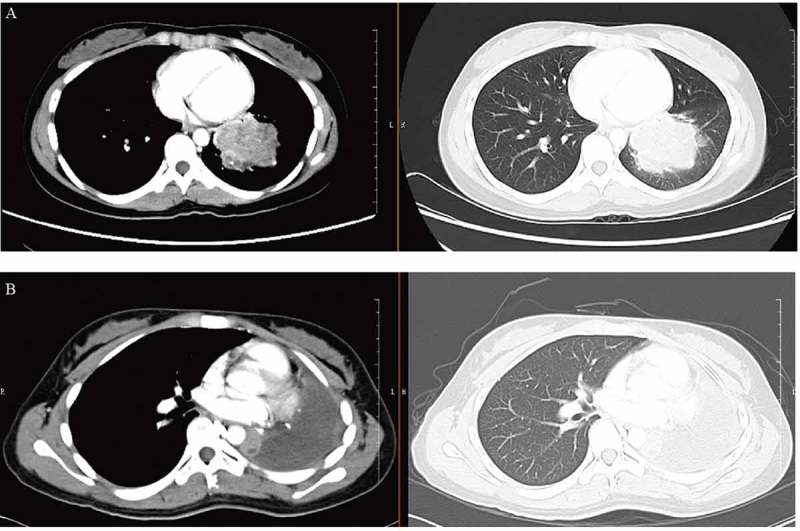
Disease status at diagnosis (a) and after left pneumonectomy and lymph node dissection (b) by CT scan (axial). CT, computed tomography.
Figure 2.
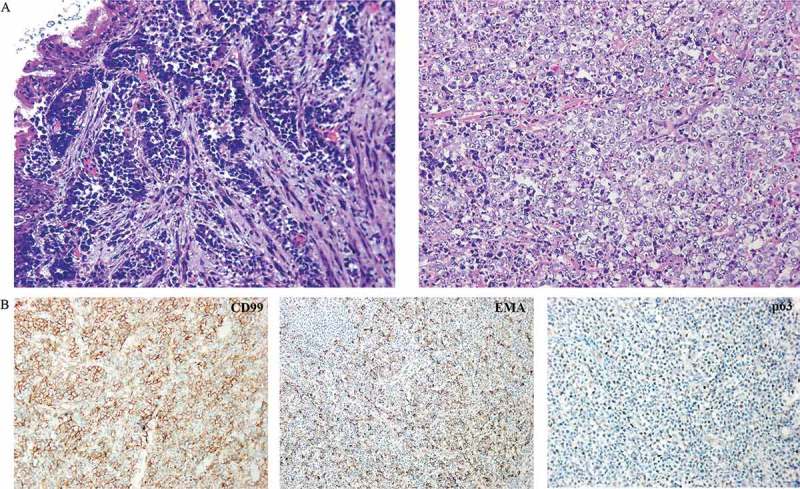
Clinicopathologic characteristics of this case. (a) Pathology image of fiberoptic bronchoscopy deposits by hematoxylin and eosin stain showing round undifferentiated tumor cells. (b) Immunohistochemistry staining of tumor cells from a resected specimen that are positive for CD99, EMA and p63.
The resected specimen was assayed using adaptor-ligation and hybrid capture NGS (Yuansu; Origimed Inc, Shanghai, China) for all coding exons from 450 cancer-related genes plus introns from 35 genes frequently rearranged in cancer. At least 50 ng of DNA was extracted from formalin-fixed paraffin-embedded (FFPE) tumor tissue from unstained slides, and peripheral blood was used as a control. The mean exon coverage depth of sequencing was >1000× and >200× for tissue and blood controls, respectively. Molecular profiling revealed a BRD4 exon11-NUT1 exon2 fusion (Figure 3(a,b)). The tumor mutational burden (TMB) was 0.8 muts/Mb. Microsatellite status was stable in this specimen. Immunohistochemistry staining of previous fiberoptic bronchoscopy deposits demonstrated positive staining for p63 and EMA (Figure 2(b)). According to the NGS result, NUT antibody (C52B1 clone, Cell Signaling Technology, Danvers, MA) was used to confirm the diagnosis of NC (Figure 3(c)). It is worth mentioning that immunohistochemistry staining for NUT was performed by Guangdong General Hospital because this test is not available at our hospital.
Figure 3.
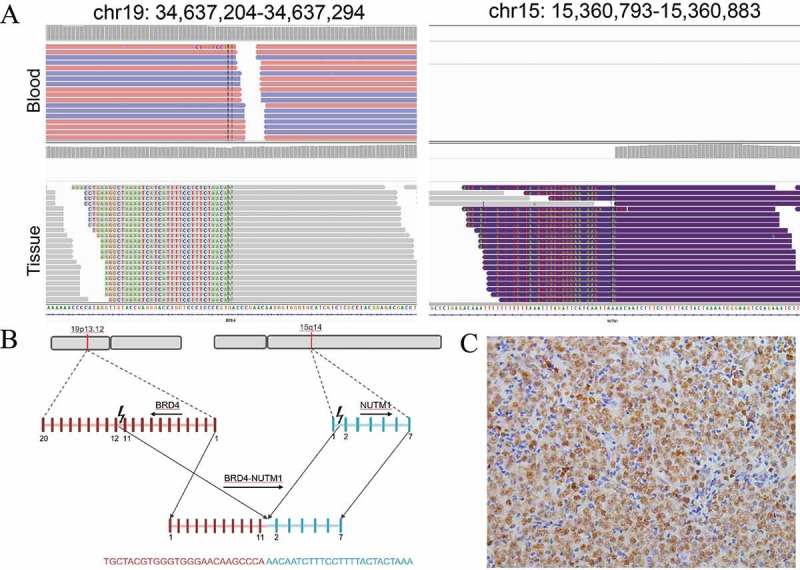
Detail presentation of BRD4-NUT fusion. (a) Screenshot of Integrative Genomics Viewer demonstrating uniquely mapped paired-end reads in the intronic regions of BRD4 (Intron 11) on chromosome 19 and NUT (Intron 1) on chromosome 15. For no hybrid capture probes were designed for NUT gene, tissue sample showed aberrant arrangement reads. (b) Schematic representation of the translocation responsible for the generation of the BRD4-NUT fusion gene. (c) Immunohistochemistry staining of tumor cells from resected specimen were positive for anti-NUT antibody.
Considering the poor prognosis of NC, postoperative adjuvant chemotherapy was performed. The patient received docetaxel plus carboplatin, but she was significantly allergic to docetaxel. Thus, chemotherapy was changed to pemetrexed 700 mg and lobaplatin 40 mg on day 1. After three weeks, the patient came back to the hospital for dull ache in the chest and upper back and intermittent abdominal pain. Laboratory tests for blood showed that CEA, CA125, CA15-3, CA19-9, SCCAg and AFP were negative, while TK1 was elevated (4.91 pM/L; normal range 0–2.0 pM/L). Brain MRI showed no obvious abnormalities. Chest CT revealed decreased absorption of pleural fluid, left mediastinal shift, pleural thickening and soft tissue density on the left lung region (Figure 4). We suggested ultrasound-guided fine needle aspiration of pleural fluid, but the patient and her family refused all testing and treatment and left the hospital. The patient died within five months after the onset of symptoms.
Figure 4.
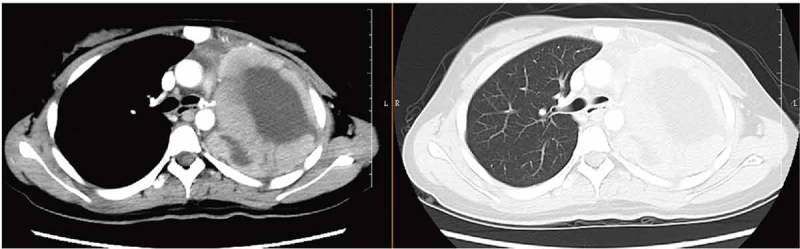
Disease status after three weeks of chemotherapy by CT scan (axial). CT, computed tomography.
Here, we have summarized the clinicopathologic characteristics of NC cases of lung origin with BRD4-NUT fusion in previous studies along with our present study (Table 1).
Table 1.
Clinicopathologic characteristics of NUT carcinomas of lung origin.
| Case | Age | Sex | Initial diagnosis | Squamous differentiated | AFP | Immunohistochemistry staining |
RT-PCR | NC confirmation | Survival time (months) | Reference | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | ||||||||||
| 1 | 16 | M | Squamous cell carcinoma | positive | NA | pancytokeratin | PLAP, CD34 | NA | BRD4-NUT (FISH) | 35 | French et al., 20046 |
| 2 | 14 | M | Unspecified sarcoma or undifferentiated carcinoma | NA | NA | CK1/5/10/14, CK5, CK8, vimentin, TTF-1, p63, CD99 | CK7, CK19, CK20, EMA, CD5, CD34, neurofilament, synaptophysin, chromogranin A, desmin, muscle actin, SMA, WT1 | EWSR1-FLI1, EWSR1-ERG, EWSR1-WT1 | anti-NUT antibody (IHC) and BRD4-NUT (RT-PCR) | 12 | Tanaka et al., 20127 |
| 3 | 7 | F | squamous cell carcinoma | positive | NA | CK1/5/10/14, CK5, CK7, CK19, EMA, p63 | CK20,vimentin, TTF-1, CD5, CD34, CD99, neurofilament, synaptophysin, chromogranin A, desmin, muscle actin, SMA, WT1 | NA | anti-NUT antibody (IHC) and BRD4-NUT (RT-PCR) | 4 | Tanaka et al., 20127 |
| 4 | 36 | M | nonseminomatous primary mediastinal germ cell tumor | NA | NA | NA | pancytokeratin, CK7, OSCAR, CAM 5.2, EMA, CD45, S100, CD117, TTF-1,PLAP, Oct-4, CD31, CD34, FLI1 | NA | anti-NUT antibody (IHC) and BRD4-NUT (FISH) | 2 | Parikh et al., 20138 |
| 5 | 12 | F | NC | positive | NA | pancytokeratin, CAM5.2, p63, CD56 | chromogranin A, synaptophysin, neurofilament, S-100 | NA | BRD4-NUT (FISH) | 21 | Ueki et al., 20149 |
| 6 | 68 | F | Lymphoma | NA | NA | p63, MNF-116 | TTF-1, synaptophysin, chromogranin, CD56 | NA | anti-NUT antibody (IHC) and BRD4-NUT (FISH) | 1 | Sholl et al., 201510 |
| 7 | 21 | F | NC | NA | NA | p63, pancytokeratin, Cam5.2 | NA | NA | anti-NUT antibody (IHC) and BRD4-NUT (FISH) | 4 | Sholl et al., 201510 |
| 8 | 23 | M | Thymic carcinoma | NA | NA | p63, pancytokeratin | TTF-1 | NA | anti-NUT antibody (IHC) and BRD4-NUT (FISH) | 2 | Sholl et al., 201510 |
| 9 | 63 | F | Poorly differentiated NSCLC | NA | NA | p63, TTF-1, pancytokeratin, CK5/6, CK7, CK20 | synaptophysin, chromogranin | NA | variant NUT rearrangement (FISH) and anti-NUT antibody (IHC) | 2 | Sholl et al., 201510 |
| 10 | 37 | M | Germ cell tumor | NA | NA | p63, TTF-1, Cam5.2, pancytokeratin, CD56 | synaptophysin, chromogranin | NA | anti-NUT antibody (IHC) and BRD4-NUT (FISH) | 1 | Sholl et al., 201510 |
| 11 | 63 | M | NC | NA | NA | p63, CK7, CK20, CK5/6 | TTF-1, synaptophysin, chromogranin, CD56 | NA | anti-NUT antibody (IHC) and BRD4-NUT (FISH) | 1 | Sholl et al., 201510 |
| 12 | 26 | M | Adenosquamous carcinoma | NA | NA | p63, Cam5.2, pancytokeratin, CK5/6 | TTF-1, synaptophysin | NA | BRD4-NUT (FISH) and anti-NUT antibody (IHC) | 2 | Sholl et al., 201510 |
| 13 | 30 | M | Poorly differentiated carcinoma | NA | NA | p63, Cam5.2, pancytokeratin, synaptophysin | TTF-1, chromogranin | NA | BRD4-NUT (FISH) and anti-NUT antibody (IHC) | 5 | Sholl et al., 201510 |
| 14 | 29 | F | NSCLC | NA | NA | p63, TTF-1 | NA | NA | BRD4-NUT (FISH) and anti-NUT antibody (IHC) | 3 | Sholl et al., 201510 |
| 15 | 34 | M | NC | negative | NA | p63, Ki-67 | CAM5.2, CK7, CK20, CK5/6, lymphoid, myeloid, chromogranin, synaptophysin, CD56, S-100 | NA | anti-NUT antibody (IHC) | 1 | Policarpio-Nicolas et al., 201511 |
| 16 | 36 | F | NC | positive | NA | EMA, p63, pancytokeratin, CAM5.2, CD138, vimentin | NA | NA | anti-NUT antibody (IHC) and BRD4-NUT (FISH) | 10 | Kuroda et al., 201512 |
| 17 | 23 | F | non-Hodgkin’s lymphoma | negative | NA | synaptophysin, polytypic keratin, OSCAR keratin, CD99, EMA | LCA, CD20, BCL-2, CAM5.2, desmin, CD3, CD30, BCL-6, CK7, myogenin, CD5, CD34, MUM-1, p16, FLI1, CD10, CD79a, Ber-Ep-4, p63, OCT3/4, CD15, BCL-1, pancytokeratin, TTF-1, S100 | NA | anti-NUT antibody (IHC) and BRD4-NUT (FISH) | 2 | Shatavi et al., 201613 |
| 18 | 15 | F | ES/PNET | NA | NA | pancytokeratin, CD99 | NA | NA | BRD4-NUT (FISH) | 17 | Lemelle et al.,201714 |
| 19 | 16 | F | Undifferentiated carcinoma |
NA | NA | KL1 cytokeratins | CD99 | NA | t(15;19) (q13;p13)(cytogenetics) | 3 | Lemelle et al.,201714 |
| 20 | 39 | M | non-small cell lung cancer | positive | NA | p63 | TTF-1, napsin A, synaptophysin | NA | NGS and anti-NUT antibody (IHC) | 4 | Baras et al.,201815 |
| 21 | 21 | F | ES/PNET | negative | elevated | CD99, EMA, p63, FLI1, synaptophysin | TTF-1, napsin A, CK7, p40, CK5/6, NSE, CgA, CD56, pancytokeratin, desmin, MyoD1, myogenin | NA | NGS and anti-NUT antibody (IHC) | 5 | present study |
IHC: Immunohistochemistry staining; NC: NUT carcinoma
Discussion
NC is a rare aggressive squamous malignancy and is often difficult to diagnosis due to its rarity and non-specific histology 1. NC is histologically described as poorly differentiated, which is indistinguishable from other poorly differentiated cell carcinomas such as ES/PNET, especially in the absence of squamous differentiation. ES/PNET is positive for CD99, a cell surface glycoprotein, and has been reported to be positive in all 52 cases.16 EWSR1 was reportedly fused with FLI1 in only 85% of cases3; thus, ES/PNET was initially diagnosed, although EWSR1 rearrangement was negative in our case. Additionally, D’Ambrosio et al. presented an NC case in which elevated AFP was observed.17 However, not all previous NC cases presented with AFP.18
NC is characterized by a NUT rearrangement, which can be detected by immunohistochemistry, karyotype, FISH, RT-PCR or NGS. Immunohistochemistry staining for NUT is effective for NUT diagnosis. However, in this case, we did not determine NC in the initial differential diagnosis. Because, an immunohistochemistry antibody for NUT is not available in our hospital or other hospitals in our province. FISH has been adopted to detect NUT gene rearrangement in NC using dual-color NUT split-apart probes, but is not widely available or commercialized.8,19,20 BRD4-NUT fusion can also be detected using RT-PCR.14 BRD4-NUT fusion, the most common rearrangement in NC cases, has attracted interest for the development of small molecules that target BRD4,21 a member of the BET (bromodomain and extra terminal domain) family that contains two bromodomains.1 A novel oral BET inhibitor, OTX015/MK-8628, is currently in clinical development for targeting BRD2/3/4/T, and two NC patients have responded rapidly to this inhibitor.22 NGS can be applied for detecting the NUT rearrangement,23–26 especially for the BRD4-NUT fusion, which is crucial for guiding targeted therapy. Thus, NGS can be used not only to detect multiple NUT rearrangements for NC diagnosis but also to guide targeted therapy of NC.
For now, there is no effective treatment for NC. The rarity of NC renders multiple treatment paradigms used in the clinic, such as those for sarcoma and non-small lung cancer.27 Bauer et al. reported improved survival for gross total resection in NC patients. However, the NC patients still had poor prognosis, and the median overall survival for patients was 6.7 months.28 In addition to the BET inhibitor mentioned above, the histone deacetylase inhibitor (HDACi) CUDC-907 showed effectiveness in NC xenograft tumors.29 A child with NC showed an objective response to the FDA-approved HDAC inhibitor vorinostat after five weeks of therapy.30
In summary, NC is an extremely aggressive carcinoma that can be diagnosed by NGS. It is necessary to consider NC when poor differentiation is observed by pathological examination. NGS technology can be an effective tool for rare disease diagnosis and further targeted therapy. Poor prognosis is the main characteristic of NC. Surgery and radiotherapy are still the main treatment options for NC. However, some novel targeted therapies such as BETi and HDACi have been developed.
Funding Statement
This work was supported by funding from the Natural Science Foundation of Guangxi, Grant No. 2011GXNSFA018248.
Abbreviations
- NC:
NUT carcinoma
- NUT:
NUT carcinoma, family member 1, former known as NUT
- CT:
computed tomography
- ES/PNET:
Ewing’s sarcoma/primitive neuroectodermal tumor
- BRD4:
bromodomain-containing protein 4
- TTF-1:
thyroid transcription factor 1
- FLI1:
a member of the ETS transcription factor family
- CK7:
cytokeratin 7
- CK5/6:
cytokeratin 5/6
- MyoD1:
myogenic differentiation 1
- CK18:
cytokeratin 18
- AFP:
alpha-fetoprotein
- NGS:
next-generation sequencing
- FISH:
Fluorescence in situ hybridization
- RT-PCR:
reverse transcription polymerase chain reaction.
- EWSR1:
Ewing sarcoma breakpoint region 1
- FFPE:
formalin-fixed, paraffin-embedded
- TMB:
tumor mutational burden
- BET:
bromodomain and extra terminal domain
- CEA:
carcinoembryonic antigen
- CA125:
cancer antigen 125
- CA15-3:
cancer antigen 15-3
- CA19-9:
cancer antigen 19-9
- SCCAg:
human squamous cell carcinoma related antigen
- TK1:
thymidine kinase 1
- MRI:
magnetic resonance imaging.
Ethics approval and consent to participate
The patient provided written informed consent and gave permission for the use of biopsies and publication of case details.
This study was approved by the Medical Ethics Committee of the Affiliated Tumor Hospital of Guangxi Medical University.
Consent for publication
Written informed consent was obtained from the patient for the publication of this case report.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We owe thanks to the patient and her family. We also thank Guangdong General Hospital for performing immunohistochemistry staining for NUT.
Authors’ contributions
Naiquan Mao, Zhiling Liao, Junwei Wu, Kai Liang, Shoufeng Wang, Shaomian Qin, Hanqing Lin and Xiaowei Dong were involved in diagnostic flow and patient follow-up. Hanqing Lin and Ying Dou contributed to the interpretation of published data and were involved in drafting of the manuscript. Zhiling Liao and Xiaowei Dong are licensed pathologists. All the authors read and gave their final approval of the version to be published.
References
- 1.French C. NUT midline carcinoma. Nature Rev Cancer. 2014;14:149–150. [DOI] [PubMed] [Google Scholar]
- 2.French CA. Pathogenesis of NUT midline carcinoma. Annu Rev Pathol. 2012;7:247–265. doi: 10.1146/annurev-pathol-011811-132438. [DOI] [PubMed] [Google Scholar]
- 3.Balamuth NJ, Womer RB. Ewing’s sarcoma. Lancet Oncol. 2010;11:184–192. doi: 10.1016/S1470-2045(09)70286-4. [DOI] [PubMed] [Google Scholar]
- 4.Lessnick SL, Ladanyi M. Molecular pathogenesis of Ewing sarcoma: new therapeutic and transcriptional targets. Annu Rev Pathol. 2012;7:145–159. doi: 10.1146/annurev-pathol-011110-130237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bishop JA, French CA, Ali SZ. Cytopathologic features of NUT midline carcinoma: A series of 26 specimens from 13 patients. Cancer Cytopathol. 2016;124:901–908. doi: 10.1002/cncy.21761. [DOI] [PubMed] [Google Scholar]
- 6.French CA, Kutok JL, Faquin WC, Toretsky JA, Antonescu CR, Griffin CA, Nose V, Vargas SO, Moschovi M, Tzortzatou-Stathopoulou F, et al. Midline carcinoma of children and young adults with NUT rearrangement. J Clin Oncol. 2004;22:4135–4139. doi: 10.1200/JCO.2004.02.107. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka M, Kato K, Gomi K, Yoshida M, Niwa T, Aida N, Kigasawa H, Ohama Y, Tanaka Y. NUT midline carcinoma: report of 2 cases suggestive of pulmonary origin. Am J Surg Pathol. 2012;36(3):381–388. doi: 10.1097/PAS.0b013e31824230a8. [DOI] [PubMed] [Google Scholar]
- 8.Parikh SA, French CA, Costello BA, Marks RS, Dronca RS, Nerby CL, Roden AC, Peddareddigari VG, Hilton J, Shapiro GI, et al. NUT midline carcinoma: an aggressive intrathoracic neoplasm. J Thorac Oncol. 2013;8:1335–1338. doi: 10.1097/JTO.0b013e3182a00f41. [DOI] [PubMed] [Google Scholar]
- 9.Ueki H, Maeda N, Sekimizu M, Yamashita Y, Moritani S Horibe K. A case of NUT midline carcinoma with complete response to gemcitabine following cisplatin and docetaxel. J Pediatr Hematol Oncol. 2014;36(8):e476-80. doi: 10.1097/MPH.0000000000000082. [DOI] [PubMed] [Google Scholar]
- 10.Sholl LM, Nishino M, Pokharel S, Mino-Kenudson M, French CA, Janne PA, Lathan C. Primary pulmonary NUT midline carcinoma: clinical, radiographic, and pathologic characterizations. J Thorac Oncol. 2015;10:951–959. doi: 10.1097/JTO.0000000000000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Policarpio-Nicolas ML, de Leon EM, Jagirdar J. Cytologic findings of NUT midline carcinoma in the hilum of the lung. Diagn Cytopathol. 2015;43:739–742. doi: 10.1002/dc.23291. [DOI] [PubMed] [Google Scholar]
- 12.Kuroda S, Suzuki S, Kurita A, Muraki M, Aoshima Y, Tanioka F, Sugimura H. Cytological features of a variant NUT midline carcinoma of the lung harboring the NSD3-NUT fusion gene: a case report and literature review. Case Rep Pathol. 2015;2015:572951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shatavi S, Fawole A, Haberichter K, Jaiyesimi I, French C. Nuclear protein in testis (NUT) midline carcinoma with a novel three-way translocation (4;15;19)(q13;q14;p13.1). Pathology. 2016;48:620–623. doi: 10.1016/j.pathol.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 14.Lemelle L, Pierron G, Freneaux P, Huybrechts S, Spiegel A, Plantaz D, Julieron M, Dumoucel S, Italiano A, Millot F, et al. NUT carcinoma in children and adults: A multicenter retrospective study. Pediatr Blood Cancer. 2017;64(12). doi: 10.1002/pbc.26693. [DOI] [PubMed] [Google Scholar]
- 15.Baras AS, Naidoo J, Hann CL, Illei PB, Reninger CW 3rd, Lauring J. Rediagnosis of lung cancer as NUT midline carcinoma based on clues from tumor genomic profiling. J Natl Compr Canc Netw. 2018;16:467–472. doi: 10.6004/jnccn.2017.7203. [DOI] [PubMed] [Google Scholar]
- 16.Folpe AL, Goldblum JR, Rubin BP, Shehata BM, Liu W, Dei Tos AP, Weiss SW. Morphologic and immunophenotypic diversity in Ewing family tumors: a study of 66 genetically confirmed cases. Am J Surg Pathol. 2005;29:1025–1033. [PubMed] [Google Scholar]
- 17.D’Ambrosio L, Palesandro E, Moretti M, Pelosi G, Fabbri A, Carnevale Schianca F, Aglietta M, Grignani G. Alpha-fetoprotein elevation in NUT midline carcinoma: a case report. BMC Cancer. 2017;17:266. doi: 10.1186/s12885-017-3262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamura H, Tsuta K, Tsuda H, Katsuya Y, Naka G, Iizuka T, Igari T. NUT midline carcinoma of the mediastinum showing two types of poorly differentiated tumor cells: a case report and a literature review. Pathol Res Pract. 2015;211:92–98. doi: 10.1016/j.prp.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 19.French CA. Demystified molecular pathology of NUT midline carcinomas. J Clin Pathol. 2010;63:492–496. doi: 10.1136/jcp.2007.052902. [DOI] [PubMed] [Google Scholar]
- 20.Haack H, Johnson LA, Fry CJ, Crosby K, Polakiewicz RD, Stelow EB, Hong S-M, Schwartz BE, Cameron MJ, Rubin MA, et al. Diagnosis of NUT midline carcinoma using a NUT-specific monoclonal antibody. Am J Surg Pathol. 2009;33:984–991. doi: 10.1097/PAS.0b013e318198d666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stathis A, Zucca E, Bekradda M, Gomez-Roca C, Delord JP, de La Motte Rouge T, et al. 2016. Clinical response of carcinomas harboring the brd4-nut oncoprotein to the targeted bromodomain inhibitor otx015/mk-8628. Cancer Discov. 6:492––500.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaefer IM, Dal Cin P, Fletcher CDM, Hanna GJ, Ca F. CIC-NUTM1 fusion: a case which expands the spectrum of NUT-rearranged epithelioid malignancies. Genes Chromosomes Cancer. 2018. doi: 10.1002/gcc.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dickson BC, Sung Y-S, Rosenblum MK, Reuter VE, Harb M, Wunder JS, Swanson D, Antonescu CR. NUTM1 gene fusions characterize a subset of undifferentiated soft tissue and visceral tumors. Am J Surg Pathol. 2018;42:636–645. doi: 10.1097/PAS.0000000000001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watson S, Perrin V, Guillemot D, Reynaud S, Coindre J-M, Karanian M, Guinebretière J-M, Freneaux P, Le Loarer F, Bouvet M, et al. Transcriptomic definition of molecular subgroups of small round cell sarcomas. J Pathol. 2018;245:29–40. doi: 10.1002/path.5053. [DOI] [PubMed] [Google Scholar]
- 26.Thompson-Wicking K, Francis RW, Stirnweiss A, Ferrari E, Welch MD, Baker E, Murch AR, Gout AM, Carter KW, Charles AK, et al. Novel BRD4-NUT fusion isoforms increase the pathogenic complexity in NUT midline carcinoma. Oncogene. 2013;32:4664–4674. doi: 10.1038/onc.2012.487. [DOI] [PubMed] [Google Scholar]
- 27.Bair RJ, Chick JF, Chauhan NR, French C, Madan R. Demystifying NUT midline carcinoma: radiologic and pathologic correlations of an aggressive malignancy. AJR Am J Roentgenol. 2014;203:W391–9. doi: 10.2214/AJR.13.12401. [DOI] [PubMed] [Google Scholar]
- 28.Bauer DE, Mitchell CM, Strait KM, Lathan CS, Stelow EB, Lüer SC, Muhammed S, Evans AG, Sholl LM, Rosai J, et al. Clinicopathologic features and long-term outcomes of NUT midline carcinoma. Clin Cancer Res. 2012;18:5773–5779. doi: 10.1158/1078-0432.CCR-12-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun K, Atoyan R, Borek MA, Dellarocca S, Samson MES, Ma AW, Xu G-X, Patterson T, Tuck DP, Viner JL, et al. Dual HDAC and PI3K inhibitor CUDC-907 downregulates MYC and suppresses growth of MYC-dependent cancers. Mol Cancer Ther. 2017;16:285–299. doi: 10.1158/1535-7163.MCT-16-0390. [DOI] [PubMed] [Google Scholar]
- 30.Schwartz BE, Hofer MD, Lemieux ME, Bauer DE, Cameron MJ, West NH, Agoston ES, Reynoird N, Khochbin S, Ince TA, et al. Differentiation of NUT midline carcinoma by epigenomic reprogramming. Cancer Res. 2011;71:2686–2696. doi: 10.1158/0008-5472.CAN-10-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.


