ABSTRACT
Breast cancer threatened the health of millions of people around the world. Here we explored the influence of TCDD on the expression of circRNA_BARD1 (circ_0001098) in breast cancer and studied the potential molecular mechanism of circRNA_BARD1. The data from GSE76608 was applied to analyze differentially expressed circRNAs and mRNAs. The expressions of circRNA_BARD1, BARD1, miR-3942-3p, miR-4760-3p and apoptosis-related protein p53 were detected by qRT-PCR or western blot. Circinteractome, TargetScan, CIRCNET and dual luciferase reporter assay were employed to uncover the target relationship between circRNA_BARD1/BARD1 and miR-3942-3p/miR-4760-3p. Flow cytometric analysis was used to reveal cell cycle and cell apoptosis. Immunofluorescence was applied to determinate γ-H2AX level. Xenograft assay and in vivo 3-D imaging was implemented to further verify the conclusions in vitro. CircRNA_BARD1 (circ_0001098) was up-regulated in breast cancer with the treatment of TCDD and the up-regulation of circRNA_BARD1 could restrain cell proliferation, block cell cycle and promote cell apoptosis. Moreover, the target relationship between circRNA_BARD1/BARD1 and miR-3942-3p was confirmed. In addition, miR-3942-3p overexpression promoted the disease progression and BARD1 up-regulation inhibited the disease progression in the breast cancer. Similarly, circRNA_BARD1 overexpression induced by TCDD suppressed the growth and metastasis of tumor in vivo. In conclusion, TCDD induced circ_0001098 overexpression and then suppressed breast cancer tumorigenesis via miR-3942-3p/BARD1 axis. The finding of TCDD-circRNA-miRNA-mRNA axis might bring a new perspective for cure strategy of breast cancer.
KEYWORDS: Breast cancer, CircRNA_BARD1 (circ_0001098), TCDD, MiR-3942-3p, BARD1
Introduction
According to the data of International Agency for Research on Cancer, breast cancer has been the second common cancer in 2012, only following lung cancer [http://gco.iarc.fr/]. Besides, breast cancer has been the most common cancer that causes high mortality in female all around the world [1]. Therefore, it is urgent to make the internal mechanisms of breast cancer progression and metastasis clear.
2, 3, 7, 8-tetrachlorodibenzo-p-dioxin (TCDD) has been identified as a persistent environmental pollutant and a well-known human carcinogen [2–4]. Until now, quantities of studies have been implemented to expose the function and the mechanisms of TCCD in therapy of breast cancer. Signaling pathways caused by TCDD are various. It has been reported that TCDD can bind to the aryl hydrocarbon receptor (AHR) [2,3,5], estrogen receptor alpha (ERα) [6,7] and progesterone receptor (PR) [4]. Chen et al. revealed that PR was involved in TCDD-stimulated breast cancer cell proliferation for the first time [4]. The findings of Yoshioka et al. suggested that the principal pathways of TCDD-induced anti-proliferation in breast cancer were not AhR dependent [8].
Non-coding RNAs have been uncovered that they are related to the progression of cancer [3]. Circular RNAs (circRNAs) are non-coding RNA molecules with covalently closed continuous loops, not possessing polarity or a polyadenylated tail [8,9]. They have been discovered in multiple cell lines and across variety of species, relatively conserved and stable in cytoplasm [10]. According to the previous studies, circRNAs play an important role in the regulation of cancer, such as gastric cancer, breast cancer and so on [8,10]. However, the specific mechanisms remain largely unknown.
MicroRNAs (miRNAs), non-coding RNAs with 19–25 nucleotides, can control the expression of many mRNAs via targeting the 3´ untranslated region (3´UTR) of matched mRNAs [1,8,11]. It has been made clear that circRNAs can act as miRNAs sponges through binding to cancer-related miRNAs [8,10]. For example, Ma et al. disclosed that the latent regulation of circRNA-000284 on cell proliferation and invasion of cervical cancer via sponging miR-506 [10]. Wang et al. found that circRNA-000911/miR-449a pathway played a regulatory role in breast carcinogenesis [8].
In our study, has_circ_0001098 (circRNA-BARD1) was detected to be highly expressed with TCDD treatment in breast cancer through the analysis of gene chip GSE76608. Overexpression of circRNA-BARD1 could inhibit cell proliferation, block cell cycle and enhance apoptosis. Further, it was revealed the roles of TCDD/circRNA-BARD1/miR-3942-3p/BARD1 axis in tumor progression.
Methods
GEO data
CircRNA and mRNA filtration were carried out on Gene Expression Omnibus (GEO) database (GEO accession: GSE76608). The differential expression of circRNA was screened out among 3 naive breast cancer cells and 4 TCDD treated breast cancer cells through prediction tools of circRNA finder and find circ. Similarly, the differential expression of mRNA was also compared among 3 naive breast cancer tissues and 4 TCDD treated breast cancer tissues.
Clinical tissue samples
Breast cancer tissues (15 paired tissues with or without TCDD treatment) with full materials and distinct diagnosis in this study were acquired from Shengjing Hospital of China Medical University. Liquid nitrogen was used to freeze the specimens, which were not with chemotherapy or radiotherapy history. All patients enrolled this study were provided informed consent and the experiments associated with specimens were supported by the Ethics Committee of Shengjing Hospital of China Medical University.
Cell and cell culture
Breast cancer cells MCF-7 were got from BeNa Culture Collection (Beijing, China). Mixture of DMEM (Dulbecco’s modified Eagle’s medium, Thermo Fisher Scientific, Waltham, USA) supplemented with 10% FBS, 100 units/mL penicillin and 100 ug/mL streptomycin (Sigma-Aldrich, St. Louis, MO, USA) were used to culture the cells and were stored in a humidified incubator with 5% CO2 at 37°C. At 70% confluency, cells were cultured with either dimethyl sulfoxide (DMSO) or 10 nM TCDD (Sigma-Aldrich, St. Louis, MO, USA) diluted in DMSO for 24 hours. The concentration of TCDD has been proved to reproducibly cause differential gene expression in variety of cells. And then the cells were collected under the condition of 0.25% trypsin plus EDTA and counted by trypan blue exclusion. Cell lysates and supernatants were stored at −80°C for application.
Cell transfection
The 3rd–5th generations of the cells were inoculated in 60 mm cell culture bottles at the density of 1 × l05 cell/bottle and stored in 5% CO2 at 37°C. Cells in miRNA transfection group were transfected with miR-3942-3p mimics or negative control (Genepharma, Shanghai, China). Recombinants pcDNA3.1-BARD1 and si-circ_BARD1 were purchased from Invitrogen and were transfected into cells to upregulate BARD1 expression or downregulate circRNA_BARD1 expression. Transfection was carried out with the Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). 48 hours later, transfection efficiency was detected by qRT-PCR.
Quantitative real-time PCR analysis (qRT-PCR)
TRIzol reagent (Invitrogen) was used to extract total RNA in cells or tissues and the procedures were all implemented based on the manufacturer’s instruction. Reverse transcription was performed according to the protocol of TransScript One-Step RT-PCR SuperMix Kit (Transgen, Beijing, China). The quantification of mRNA and miRNA were calculated by the 2−∆∆Ct method.
Cell proliferation assay
Cells were inoculated in 96-well plates at the density of 1 × 105 cell/well and cultured to 80% confluence with 5% CO2 at 37°C and then incubated for 48 hours. The CCK-8 kit (Dojindo Molecular Technologies, Kumamoto, Japan) was employed to measure the cell proliferation according to the manufacturer’s introductions. CCK-8 solution was added into each well, and cells were incubated for 2 hours. The optical density (OD) value was detected at 450 nm using a microplate reader.
Cell cycle distribution analysis
The MCF-7 cells were divided into several different groups and separately cultured with DMSO, TCDD, si-circ_BARD1, TCDD plus si-circ_BARD1, miR-3942-3p mimics, pcDNA3.1-BARD1, TCDD plus miR-3942-3p mimics, TCDD plus pcDNA3.1-BARD1 or TCDD plus miR-3942-3p mimics plus pcDNA3.1-BARD1 for 48 hours. The concentration of TCDD was 10 nmol/L. Then, cells were washed twice by PBS before and after fixed with iced 70% ethanol overnight at 4°C. Next, the cells were treated with RNase A at 37°C and stained with PI in the dark for 30 min. Eventually, the DNA content was detected with a flow cytometer (Accuri C6, BD Biosciences, USA) and the data were analyzed by Flow Jo 7.6 (Tree star, Ashland, OR, USA).
Cell apoptosis assay
Cell apoptosis was detected by Annexin V-FITC apoptosis detection kit (Transgen, Beijing, China). MCF-7 cells were treated with different reagents (TCDD, si-circ_ BARD1, TCDD plus si-circ_ BARD1, miR-3942-3p mimics, pcDNA3.1-BARD1, TCDD plus miR-3942-3p mimics, TCDD plus pcDNA3.1-BARD1 and TCDD plus miR-3942-3p mimics plus pcDNA3.1-BARD1) for 48 hours. The concentration of TCDD was 10 nmol/L. All the procedures were conducted according to the protocol of the apoptosis kit (http://www.transgen.com.cn/attached/down/FA101-01_2016090508.pdf). Apoptosis was measured with flow cytometer (Accuri C6).
Western blot
The total protein in cells was extracted with RIPA lysis buffer containing protease inhibitors. BCA Kit (Sigma-Aldrich, St. Louis, MO, USA) were used to measure protein concentration. 10% SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis) was used to separate protein and PVDF polyvinylidene fluoride membranes were used to transfer the protein. Blocked with 5% BSA (Bull Serum Albumin) at room temperature for 1 h, the membranes were incubated with corresponding primary antibodies: anti-BARD1 (1: 1000, Abcam, Cambridge, MA, US), anti-p53 (1: 1000, Abcam) and anti-β-actin (1: 2000, Abcam) at 4°C overnight. Then the membranes were washed and combined with the secondary antibody-HRP (1: 4000, Abcam). Signals were detected with the enhanced chemiluminescence (ECL) and the bands were visualized.
Dual luciferase report assay
Cells were seeded into a 12 pore plates and were incubated to approximately 70% confluency, and then transfection was conducted with Promega’s (Madison, WI, USA) pGL3 luciferase plasmid (1 μg) and pGL4.73 [hRluc/SV40] (500 ng) using Lipo8000TM (Beyotime, Shanghai, China). After transfection for 48 hours, cells were incubated with specific treatment for 24 hours. And then after lysed with passive lysis buffer, the lysates were centrifuged. The analysis for luciferase activity was performed using the FLUOstar OPTIMA luminometer (BMG Labtechnologies, Ortenberg, Germany) based on the Promega’s Dual-luciferase® Reporter Assay System protocol.
Immunofluorescence (IF)
Cells in different groups were fixed by 4% paraformaldehyde for 10 to 15 min at room temperature (RT), and were then incubated using the blocking buffer (containing 0.8× PBS, 0.5% Triton-X 100, 50 mM NaCl, 3% BSA) for 1 hours, followed by incubation with the primary antibody to γ-H2AX (ab2893, Abcam, Cambridge, MA, USA) in blocking buffer overnight at 4°C. Cells were then washed three times for 10 min in 0.8× PBS, 50 mM NaCl, and 1.5% BSA at RT. Incubation with donkey polyclonal anti-rabbit Alexa Fluor 555 (A0453; Beyotime, Shanghai, China) antibodies was conducted in the dark for 1 hours at 37°C in the blocking buffer. All antibody incubations in a moist chamber were performed. Cells were then washed three times for 10 min in 0.8× PBS, 0.5% Triton-X 100, and 50 mM NaCl. Cells were then rinsed in PBS, counterstained using 4´, 6-diamidino-2-phenylindole (DAPI), mounted in VECTASHIELD, and stored in the dark at 4°C.
Animal experiment
CircRNA_BARD1 short hairpin RNAs (sh-circ_BARD1) were obtained (Genepharma) and bedded into the hU6-MCS-CMV-Puromycin lentiviral vector to establish the stable circ_BARD1-knockdown MCF-7 cells. The mice subcutaneously injected with specific treated MCF-7 cells were divided into four groups: empty vector control group, co-treatment group of TCDD and empty vector control group, sh-circ_BARD1 stable knock-down group and co-treatment group of TCDD and sh-circ_BARD1 stable knock-down. The concentration of TCDD was 10 nmol/L. After inoculation for approximate 1 week, the size of the tumors of each group was measured each week using 3-D imaging in vivo. At the 5th week of experiment, the mice were executed, dissected, and the entire tumor was removed and weighed. The volume of the tumors was evaluated according to the formula: volume = length × width2/2. The miR-3942-3p expression in tumor tissue was detected by qRT-PCR. The BARD1 expression in tumor tissue was detected using qRT-PCR and western blot. Experiments in vivo were supported by Animal Ethic and Care Committee of Shengjing Hospital of China Medical University.
In vivo 3-D imaging
Mice were anesthetized using Nembutal (50 mg/kg body weight, IP), followed by in vivo 3-D fluorescence tomography imaging. Meanwhile, the hair of breast was removed with Nair lotion (Ovation Pharmaceuticals Inc, Deerfield, Illinois) to allow light transmission. And then, the mice were positioned into the imaging chamber of the FMT 2500 LX system (PerkinElmer, Waltham, MA, USA). The entire image acquisition sequence was performed for 3 to 5 minute each mouse. The 3-D distribution of light source and spatial quantification of the fluorophore were reconstructed by a built-in reconstruction algorithm.
Statistical analysis
Statistical analysis was implemented with GraphPad Prism 6 software (GraphPad Software Inc, CA, USA). All data were presented as mean ± standard deviation (SD). Student’s t-test and variance analysis (ANOVA) were used to make the comparisons between two groups or among multi-groups, respectively. P value less than 0.05 was considered statistically significant.
Results
Circ_0001098 (circRNA_BARD1) was highly expressed with the treatment of TCDD in breast cancer
According to the differential expression analysis of circRNAs based on GSE76608, circ_0001098 (circRNA_BARD1), the abnormally expressed circRNA simultaneously screened out by prediction tools of circRNA finder and find circ, was highly expressed in breast cancer with TCDD treatment (Figure 1(a–c)). The conclusion was confirmed that the expression of circRNA_BARD1 was higher in breast cancer cells (MCF-7) with TCDD (10 nmol/L) treatment for 24 hours, compared with control group (Figure 1(d), P < 0.05).
Figure 1.
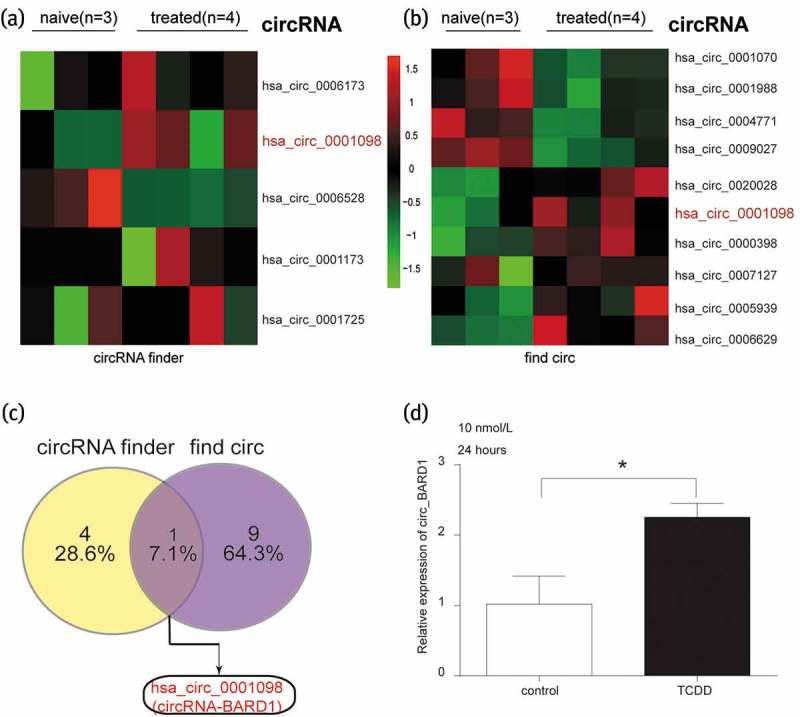
Circ_0001098 was highly expressed with the treatment of TCDD.
(a–b) Differentially expressed circRNAs with TCDD treatment were predicted by circRNA finder and find circ, respectively, based on data from GSE76608. Of which, circ_0001098 (circRNA_BARD1), was remarkably highly expressed in breast cancer. The number of tissue in naive and treated group was respectively 3 and 4. (c) Highly expressed circRNA_BARD1 was both predicted by circRNA finder and find circ. (d) The expression of circRNA_BARD1 in MCF-7 cell was considerably up-regulated after treated with 10nmol/L TCDD for 24 hours. *P < 0.05 compared with the control group.
CircRNA_BARD1 inhibited cell proliferation, block cell cycle and promote cell apoptosis
As shown in Figure 2(a), it was confirmed again that circRNA_BARD1 was up-regulated by TCDD treatment. When knocking down circRNA_BARD1, the expression of circRNA_BARD1 was successfully downregulated. However, the circRNA_BARD1 expression was not upregulated by the TCDD treatment after knocking-down circRNA_BARD1. As shown in Figure 2(b–d), when adding TCDD in MCF-7 cells, OD value was less but the occupation of G0/G1 in cell cycle and apoptosis rate was more compared with control group. However, the level of above parameters displayed a contrary trend after knocking down circRNA_BARD1. In conclusion, cell proliferation could be restrained, cell cycle could be arrested in G0/G1 phase, and cell apoptosis could be facilitated with the circRNA_BARD1 upregulation induced by TCDD treatment.
Figure 2.
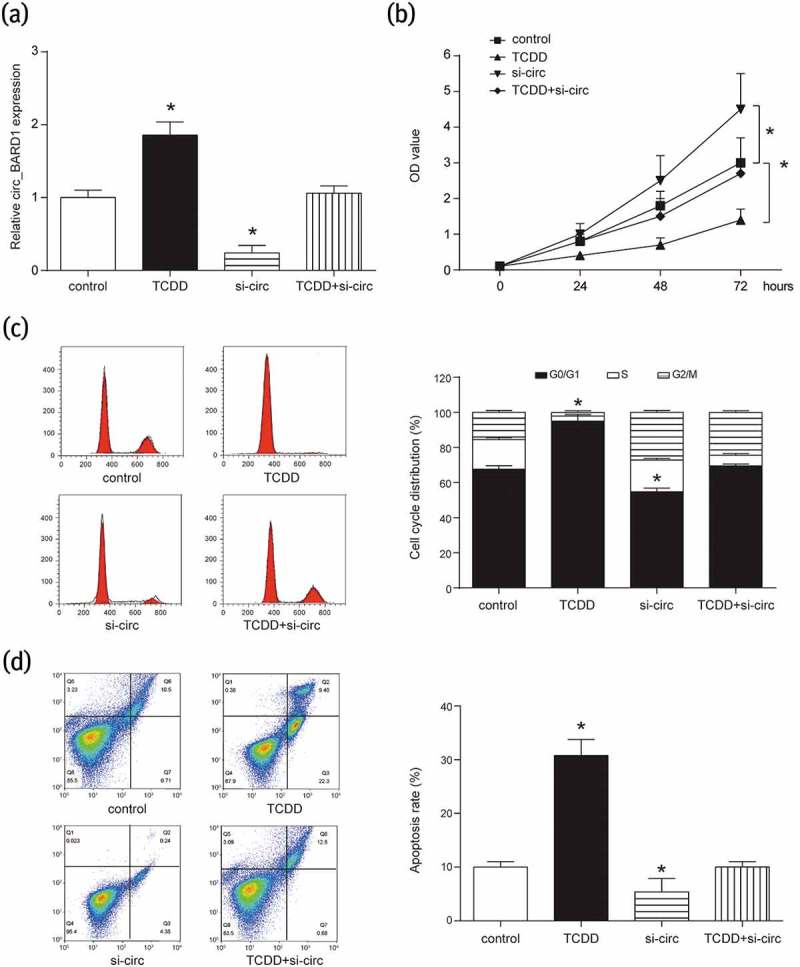
Overexpression of circRNA_BARD1 could inhibit cell proliferation and promote cell apoptosis.
(a) CircRNA_BARD1 was increased with TCDD treatment and decreased after knocking down circRNA_BARD1. *P < 0.05 compared with control group. (b) Cell proliferation was inhibited by the up-regulation of circRNA_BARD1 induced by TCDD and promoted by the down-regulation of circRNA_BARD1.*P < 0.05 compared with control group. (c) In cell cycle analysis, TCDD made G0/G1 longer and the down-regulation of circRNA_BARD1 shortened G0/G1. *P < 0.05 compared with control group. (d) Cell apoptosis was enhanced by TCDD and inhibited by the down-regulation of circRNA_BARD1. *P < 0.05 compared with control group.
BARD1 targeted miR-3942-3p and was regulated by circRNA_BARD1 induced by TCDD
BARD1 was significantly up-regulated after breast cancer tissues were treated with TCDD based on the analysis of GSE76608 (Figure 3(a)). Western blot assay revealed that the expression of BARD1 in MCF-7 cells was indeed obviously promoted with the addition of TCDD (Figure 3(b), P < 0.05). Meanwhile, Circinteractome, TargetScan and CIRCNET indicated a target relationship between circRNA_BARD1/BARD1 and miR-4760-3p, also between circRNA_BARD1/BARD1 and miR-3942-3p (Figure 3(c–d)).
Figure 3.
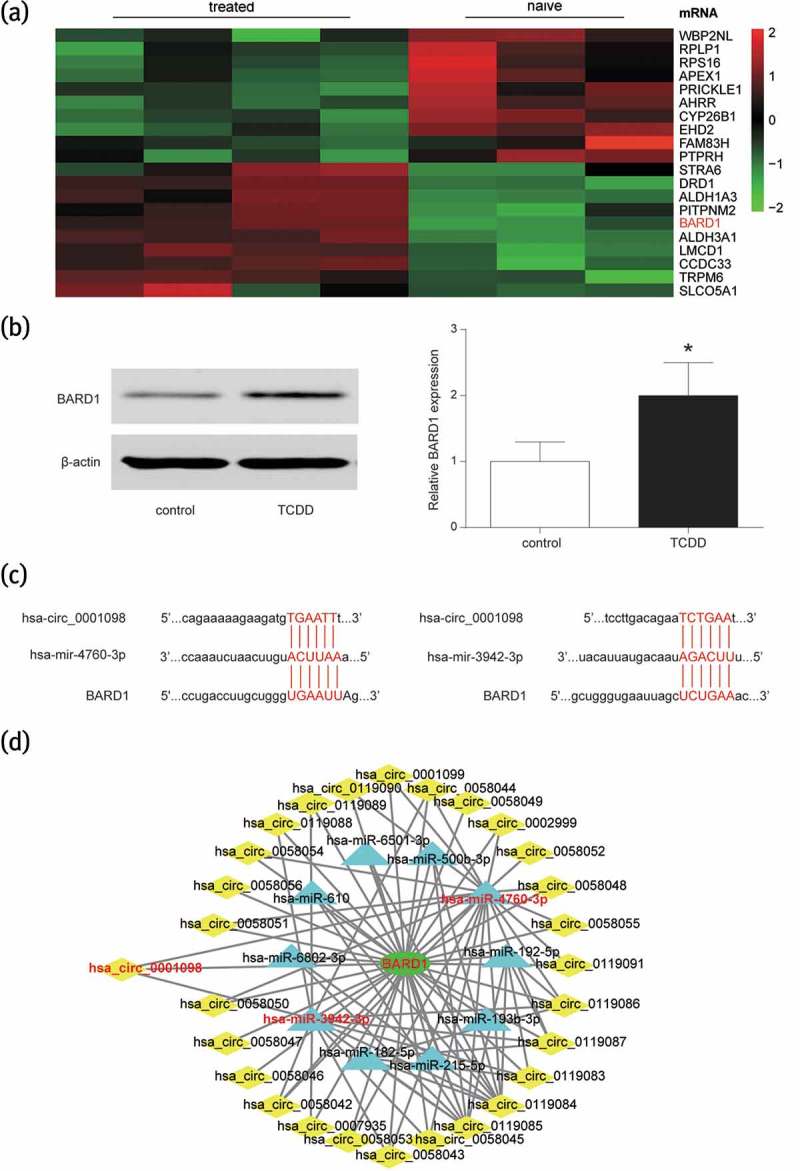
BARD1 was overexpressed in the breast cancer tissues with TCDD treatment and targeted at miR-4760-3p and miR-3942-3p.
(a) BARD1 was revealed to be overexpressed with TCDD treatment according to the analysis of gene chip GSE76608. The number of tissue in treated and naive group was respectively 4 and 3. (b) After treated with TCDD, the expression of BARD1 was promoted in MCF-7 cell. *P < 0.05 compared with control group. (c) Circinteractome and TargetScan indicated that miR-4760-3p and miR-3942-3p could target at circ_0001098 and BARD1. (d) CIRCNET forecasted that miR-4760-3p and miR-3942-3p could target at circ_0001098 and BARD1.
In order to further verify the above mentioned target relationship, we first investigated the expression of miR-3942-3p and miR-4760-3p expressions in breast cancer tissue with or without TCDD teatment. The results showed that miR-3942-3p was down-regulated and miR-4760-3p was up-regulated under TCDD treatment (Figure 4(a)). As mentioned earlier, the addition of TCDD could enhance the expression of circRNA_BARD1. Theoretically, the miRNA targeted by circRNA_BARD1 should be down-regulated by TCDD treatment. Therefore, miR-3942-3p down-regulated induced by TCDD treatment met the anticipation for further study. Moreover, circRNA_BARD1, miR-3942-3p and BARD1 expressions in 15 paired tissue samples were measured using qRT-PCR. A negative correlation was found between circRNA_BARD1 and miR-3942-3p (Figure 4(c), P < 0.05), miR-3942-3p and BARD1 (Figure 4(e), P < 0.05), and a positive correlation between circRNA_BARD1 and BARD1 (Figure 4(d), P < 0.05). Meanwhile, the target bindings between circRNA_BARD1/BARD1 and miR-3942-3p were verified by the dual luciferase report assay (Figure 4(f)). In addition, the correlation was proved by the experiments in Figure 4(g–h). MiR-3942-3p was down-regulated with TCDD treatment, up-regulated after down-regulating circRNA_BARD1, and was not obviously changed with TCDD treatment and circRNA_BARD1 down-regulation, which meant that circRNA_BARD1 played a reverse regulation on miR-3942-3p. Similarly, the conclusion could be drawn in Figure 4(h) that circRNA_BARD1 played a positive regulation on BARD1 and indirectly suggested that miR-3942-3p played a negative regulation on BARD1.
Figure 4.
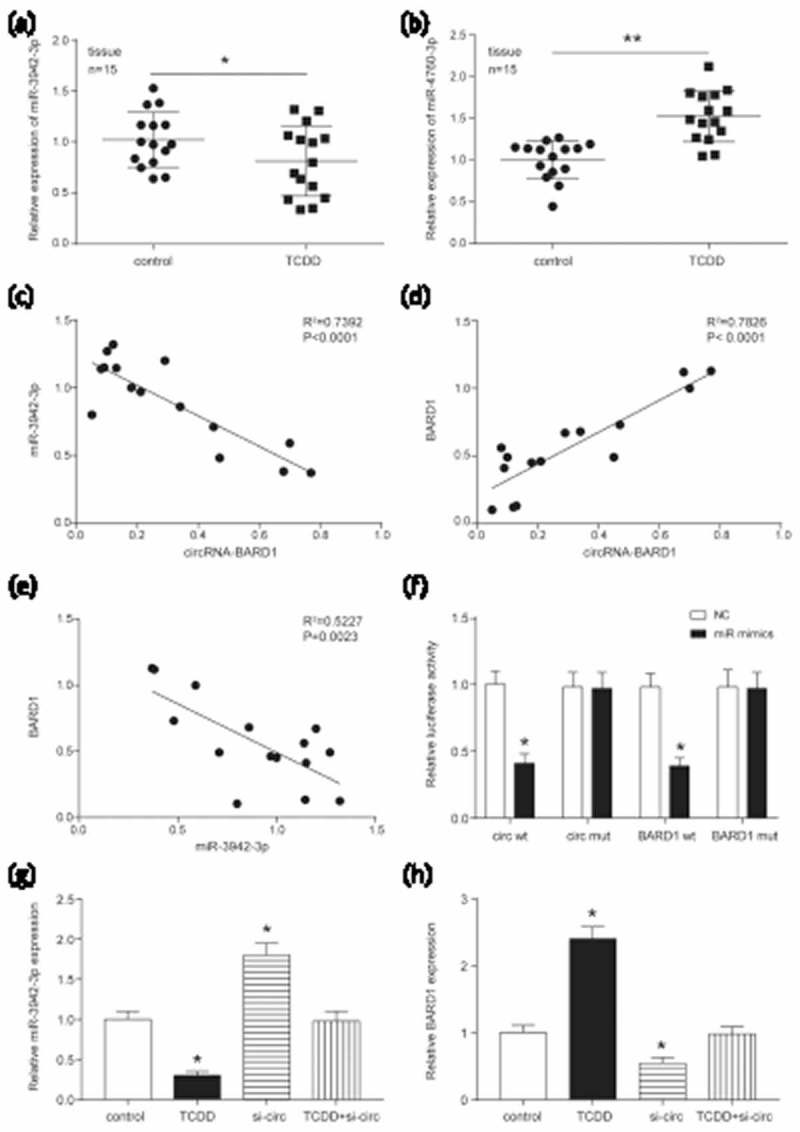
CircRNA_BARD1/BARD1 targeted at miR-3942-3p.
(a–b) The expression of miR-3942-3p and miR-4760-3p was shown in breast cancer tissues. The number of tissues in control and TCDD group was separately 15. Data were represented as the mean ± SEM. *P < 0.05, **P < 0.01 compared with control group. (c and e) A negative correlation was found between miR-3942-3p and circRNA-BARD1 (R2 = 0.7392, P < 0.0001), BARD1 and miR-3942-3p (R2 = 0.5227, P = 0.0023). (d) A negative correlation was explored between circRNA-BARD1 and BARD1 (R2 = 0.7826, P < 0.0001). (f) The dual luciferase report assay confirmed the target relationship between circRNA_BARD1/BARD1 and miR-3942-3p. (g–h) The miR-3942-3p expression was decreased by TCDD and increased by down-regulating circRNA_BARD1. *P < 0.05 compared with control group. (h) The BARD1 expression was promoted by TCDD and inhibited by knocking down circRNA_BARD1. *P < 0.05 compared with control group.
TCDD inhibited cell proliferation and promoted apoptosis through regulating miR-3942-3p and BARD1
As indicated earlier, TCDD induced circRNA_BARD1 up-regulation, circRNA_BARD1 and BARD1 targeted by miR-3942-3p. Therefore, the effects of miR-3942-3p, BARD1 and TCDD on cell phenotype were explored. Firstly, western blot assay discovered that the expression of BARD1 was restrained by the miR-3942-3p up-regulation (group of miR-mimics), promoted by the BARD1 up-regulation (group of BARD1), TCDD treatment plus BARD1 up-regulation (group of TCDD+BARD1) and TCDD treatment plus the miR-3942-3p up-regulation plus BARD1 up-regulation (group of TCDD+miR-mimics+BARD1). However, TCDD treatment and miR-3942-3p up-regulation (group of TCDD+miR-mimics) did not significantly change BARD1 expression (Figure 5(a)). These findings indirectly verified the target relationship between circRNA_BARD1/BARD1 and miR-3942-3p based on TCDD induced the up-regulation of circRNA_BARD1.
Figure 5.
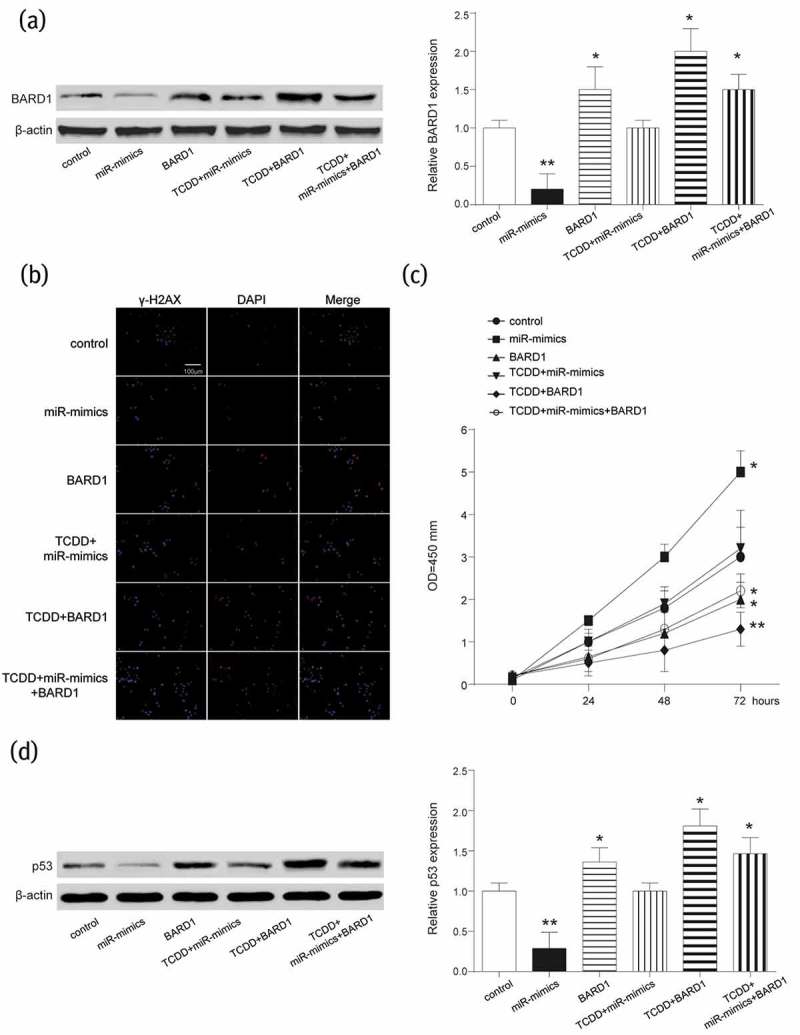
BARD1 was inhibited by miR-3942-3p and its up-regulation inhibited DNA lesion and promoted cell proliferation.
(a) BARD1 was significantly down-regulated by the up-regulation of miR-3942-3p and up-regulated by up-regulating BARD1 or TCDD treatment. *P < 0.05, **P < 0.01 compared with control group. (b and d) γ-H2AX and p53 were the proteins related to repair of DNA damage and cell apoptosis, respectively. They were restrained by miR-3942-3p mimics but promoted by BARD1 up-regulation or TCDD treatment. *P < 0.05, **P < 0.01 compared with control group. (d) Cell proliferation was enhanced by miR-3942-3p mimics, and suppressed by BARD1 up-regulation or TCDD treatment. *P < 0.05, **P < 0.01 compared with control group.
The γ-H2AX was used to evaluate the lesions of DNA double-strand breaks (DSBs) and detected by IF. Compared with control group, the number of γ-H2AX foci was more in groups of BARD1, TCDD+BARD1 and TCDD+miR-mimics+BARD1 and was less in miR-mimics group (Figure 5(b)), which suggesting TCDD, circRNA_BARD1 and BARD1 promoted tumor cell DNA lesion, but miR-3942-3p inhibited that. CCK-8 assay showed that in MCF-7 cells, miR-3942-3p mimics could enhance the viability, but TCDD and BARD1 had the adverse effect on its viability (Figure 5(c)). P53, as a tumor suppressor, was correspondingly up-regulated with the upregulation of BARD1 and TCDD treatment, and was down-regulated with miR-3942-3p overexpression (Figure 5(d)). Additionally, the influences of TCDD, BARD1, and miR-3942-3p on cell cycle and cell apoptosis were revealed. MiR-3942-3p mimics could cut down G0/G1 phrase and enhance S phrase in cell cycle, while the effectivity of TCDD and BARD1 on cell cycle was opposite to that of miR-3942-3p (Figure 6(a)). Furthermore, the cell apoptosis was enhanced by TCDD and BARD1, but weakened by miR-3942-3p (Figure 6(b)).
Figure 6.
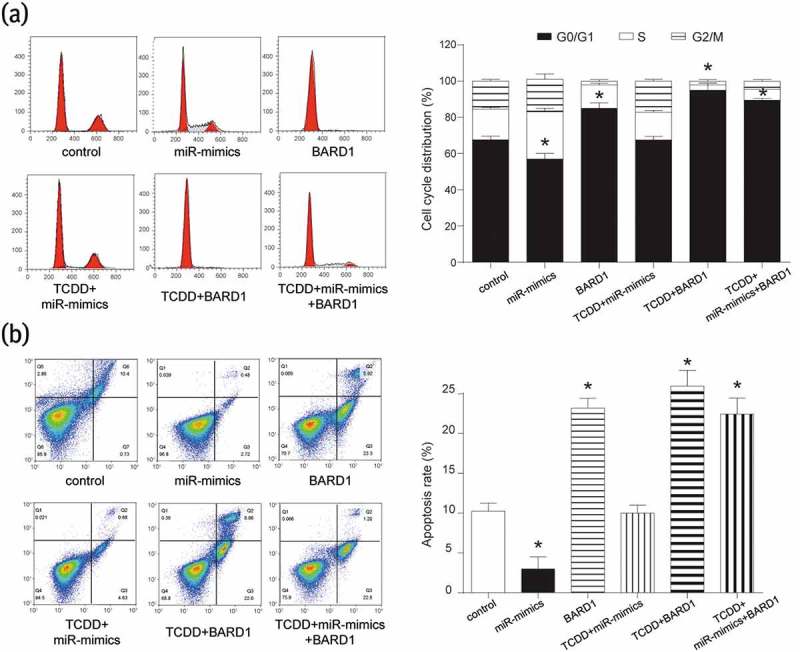
BARD1 was inhibited by miR-3942-3p and its up-regulation arrested cell cycle in G0/G1 phrase and promoted cell apoptosis.
(a) In the cell cycle analysis, miR-3942-3p mimics promoted cell cycle into S phrase and BARD1 up-regulation or TCDD treatment arrested cell cycle in G0/G1 phrase. *P < 0.05 compared with control group. (b) Cell apoptosis was restrained by miR-3942-3p mimics, and enhanced by BARD1 up-regulation or TCDD treatment. *P < 0.05 compared with control group.
All above disclosed that BARD1 had the same function to circRNA_BARD1 on DNA lesion, cell proliferation, cell cycle and cell apoptosis while miR-3942 played an opposite role. In brief, TCDD induced circRNA_BARD1 overexpression to inhibit cell proliferation and promote apoptosis through regulating miR-3942-3p and BARD1.
CircRNA_Bard1 overexpression induced by TCDD suppressed tumor growth and metastasis in vivo
To deeply explore the role of TCDD and circRNA_BARD1 in tumor growth, in vivo experiment was conducted. As displayed in Figure 7(a–b), tumor volume of mice in the TCDD group was obviously smaller than that in control group. However, when knocking down circRNA_BARD1, the volume was obviously larger, moreover, metastasis was observed. The change trend of tumor weight was the same to that of tumor volume in specific group (Figure 7(c), P < 0.05). These results suggested that TCDD induced circRNA_BARD1 up-regulation to inhibit tumor growth and metastasis.
Figure 7.
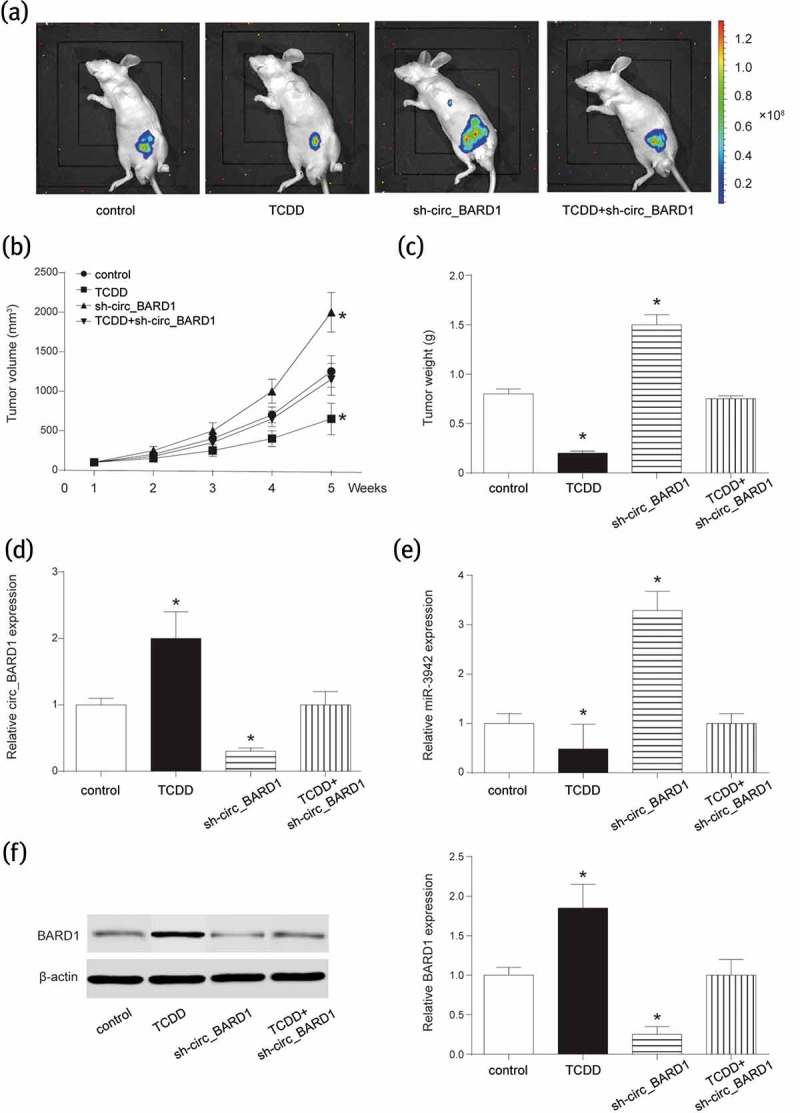
TCDD contributed to circRNA_BARD1 overexpression and restrained the growth of tumor in vivo.
(a) In vivo 3-D imaging revealed the tumor growth in vivo after xenograft for 5 weeks. (b) Tumor volume of mice in TCDD group was significantly smaller but larger in sh-circ_BARD1 group than that in control group. *P < 0.05 compared with the control group. Tumor metastasis was observed in sh-circ_BARD1 group. (c) Tumor weight was decreased in TCDD group and increased in sh-circ_BARD1 group. *P < 0.05 compared with the control group. (d–f) TCDD increased circRNA_BARD1 and BARD1 and decreased miR-3942. Sh-circ_BARD1 inhibited the expression of circRNA_BARD1 and BARD1, and promoted the expression of miR-3942-3p. *P < 0.05 compared with the control group.
In addition, expressions of miR-3942-3p and BARD1 were detected. As shown in Figure 7(d–f), miR-3942-3p was suppressed by TCDD and promoted by circRNA_BARD1 down-regulation. However, BARD1 was enhanced by TCDD and inhibited by circRNA_BARD1 down-regulation. These results of in vivo experiment indicated that CircRNA_BARD1 overexpression induced by TCDD could suppress tumor growth and metastasis via regulating miR-3942-3p and BARD1.
Discussion
In the current study, we concentrated on the role of a new gene regulator circRNA-BARD1 in breast cancer, which maintained unclear. CircRNA-BARD1 was found to be up-regulated in breast cancer with TCDD treatment. Besides, circRNA-BARD1 up-regulation inhibited cell proliferation and enhanced cell apoptosis through regulating miR-3942-3p and BARD1.
Recently, attention is focused on the circular RNAs. Since Sanger et al. proposed the concept of circular RNA, properties of circular RNA have been discovered partly [8]. Several circular RNAs have been believed to be mediated in the regulation of breast cancer. In breast cancer cells, hsa_circ_006054, hsa_circ_100219, hsa_circ_0001982 and hsa_circ_406697 were highly expressed, while hsa_circ_0011946 was down-regulated [12,13]. It was revealed that circRNA_Foxo3 could promote p53 via inhibiting MDM2 and then give rise to cell apoptosis [14,15]. Moreover, Wang et al. reported that circ_000911 could work with miR-449a to cause the inhibition of cell proliferation, migration, and invasion, meanwhile, upregulated notch1 in breast cancer [8]. In addition, Tang et al. discovered that circ_0001982 induced the carcinogenesis of breast cancer cell based on the decrease of miR-143 [13]. Our study showed that circRNA_BARD1 could decrease cell viability, increase cell apoptosis in breast cancer cell.
TCDD is an environmental toxicant and plays a valid role in signaling pathways and protein kinase activity [4]. There are few studies about the effect of TCDD on circular RNAs. As we knew, TCDD functions in vivo through AHR. After triggering AHR, a series of molecules was involved to enhance or suppress the tumor extension. Recently, researchers have found that it acts as a tumor suppressor in the breast cancer. Zhang et al. found that TCDD combined with AHR and then up-regulated miR-335, which was a suppressor of SOX4, thus decreased the metastasis of breast cancer [16]. The study of Hanieh et al. suggested that TCDD activated miR-212/132 complex via combining with AHR to further suppress SOX4, followed by the suppression of breast cancer metastasis [17]. However, there were also several reports indicated that TCDD might enhance the metastasis of breast cancer. For instance, Al-Dhfyan et al. reported that the activation of AHR/Cyp1A1 could increase the metastasis ability of breast cancer when upregulating β-catenin and AKT [18]. In our study, TCDD could inhibit the advance of breast cancer. Therefore, the mechanism of TCDD in therapy for breast cancer was still controversial.
In addtion, studies about the influence of TCDD on microRNA expression are numerous. Matthew et al. performed a research associated with the effects of short-term exposure to TCDD on the expression of microRNA in zebrafish embryos [19]. In that study, the expression of miR-451, 23a, 23b, 24 and 27e were abnormal. The study of Yoshioka et al. in the mouse showed that TCDD could inhibit the expressions of miR-101a and miR-122 [20]. Hanieh explored miR-212/132 cluster was promoted in breast cancer cells MDA-MB-231 and T47D with TCDD treatment [21]. In our study, TCDD induced the up-regulation of circRNA-BARD1 and BARD1 and the down-regulation of miR-3942-3p in breast cancer tissues and cells.
BARD1 was one of the molecules that participated in the DNA double strand break (DSB) repair. When H2AX was phosphorylated into γ-H2AX, it would be recruited, thus the DSB repair would be initiated. Nevertheless, BARD1 had distinct functions in different condition. Stewart et al. reported that BARD1 and BRCA1 complex could inhibit Cyp1A1 and Cyp3A4 [22]. As we mentioned before, the activation of AHR/Cyp1A1 could enhance the breast cancer metastasis [18]. Furthermore, Liu et al. found that FOXK2 would stabilize ERα with the combination of BRCA1 and BARD1, so that it could decrease breast cancer growth [23]. BARD1 was also suggested as a mediator between proapoptotic stress and p53 dependent apoptosis [24]. There were studies showed that BARD1 had many splice variants, which might have different roles in different subtype of breast cancer [25]. BARD1 in our study was discovered to have a positive relationship with circRNA_BARD1 and a negative correlation with miR-3942-3p. Furthermore, BARD1 functioned as a tumor suppressor in this study. We suspected that endogenous competition mechanism existed between circRNA_BARD1 and BARD1 with target binding to miR-3942-3p and the detailed regulation network among circRNA_BARD1, miR-3942-3p and BARD1 was shown in Figure 8.
Figure 8.
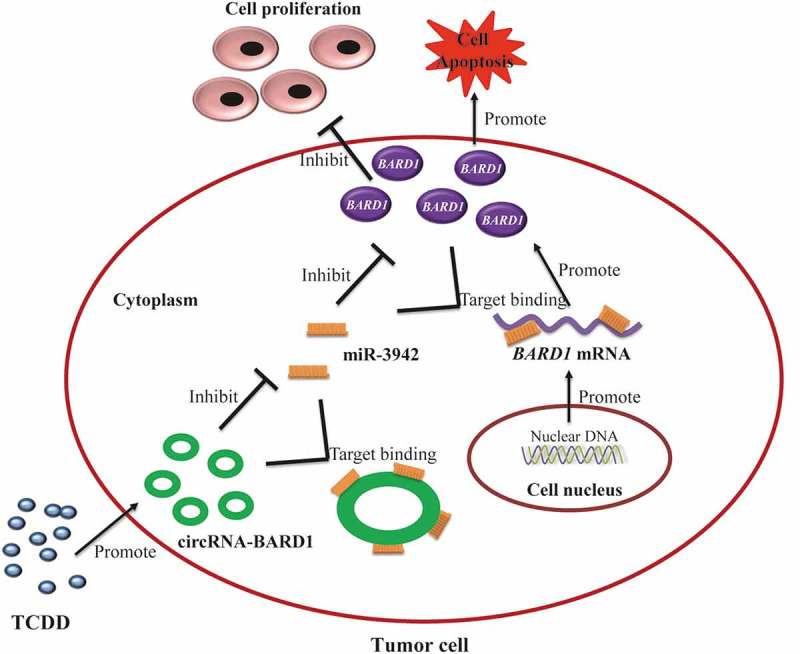
The regulation network among TCDD, circRNA_BARD1, miR-3942-3p and BARD1 demonstrated the potential endogenous competition mechanism.
This study demonstrated the function of TCDD/cricRNA_BARD1/miR-3942-3p/BARD1 axis in breast cancer development for the first time. However, the internal mechanisms that TCDD up-regulated circRNA_BARD1 remain to be studied. Furthermore, the downstream signaling of BARD1 could be further explored. In conclusion, circRNA-BARD1 was remarkably up-regulated in breast cancer with TCDD treatment and inhibited carcinogenesis via regulating miR-3942-3p and BARD1. TCDD/cricRNA_BARD1/miR-3942-3p/BARD1 axis could be a novel regulator in the therapy of breast cancer.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].van Schooneveld E, Wildiers H, Vergote I, et al. Dysregulation of microRNAs in breast cancer and their potential role as prognostic and predictive biomarkers in patient management. BCR. 2015;17:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Prokopec SD, Houlahan KE, Sun RX, et al. Compendium of TCDD-mediated transcriptomic response datasets in mammalian model systems. BMC Genomics. 2017;18:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Yoshioka H, Hiromori Y, Aoki A, et al. Possible aryl hydrocarbon receptor-independent pathway of 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced antiproliferative response in human breast cancer cells. Toxicol Lett. 2012;211:257–265. [DOI] [PubMed] [Google Scholar]
- [4].Chen YJ, Hung CM, Kay N, et al. Progesterone receptor is involved in 2,3,7,8-tetrachlorodibenzo-p-dioxin-stimulated breast cancer cells proliferation. Cancer Lett. 2012;319:223–231. [DOI] [PubMed] [Google Scholar]
- [5].Bock KW. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD)-mediated deregulation of myeloid and sebaceous gland stem/progenitor cell homeostasis. Arch Toxicol. 2017;91:2295–2301. [DOI] [PubMed] [Google Scholar]
- [6].Furrer D, Lemieux J, Cote MA, et al. Evaluation of human epidermal growth factor receptor 2 (HER2) single nucleotide polymorphisms (SNPs) in normal and breast tumor tissues and their link with breast cancer prognostic factors. Breast. 2016;30:191–196. [DOI] [PubMed] [Google Scholar]
- [7].Marquez-Bravo LG, Gierthy JF.. Differential expression of estrogen receptor alpha (ERalpha) protein in MCF-7 breast cancer cells chronically exposed to TCDD. J Cell Biochem. 2008;103:636–647. [DOI] [PubMed] [Google Scholar]
- [8].Wang H, Xiao Y, Wu L, et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-000911/miR-449a pathway in breast carcinogenesis. Int J Oncol. 2018;52:743–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chen B, Huang S. Circular RNA: an emerging non-coding RNA as a regulator and biomarker in cancer. Cancer Lett. 2018;418:41–50. [DOI] [PubMed] [Google Scholar]
- [10].Ma HB, Yao YN, Yu JJ, et al. Extensive profiling of circular RNAs and the potential regulatory role of circRNA-000284 in cell proliferation and invasion of cervical cancer via sponging miR-506. Am J Transl Res. 2018;10:592–604. [PMC free article] [PubMed] [Google Scholar]
- [11].Mulrane L, McGee SF, Gallagher WM, et al. miRNA dysregulation in breast cancer. Cancer Res. 2013;73:6554–6562. [DOI] [PubMed] [Google Scholar]
- [12].Zhou J, Zhang WW, Peng F, et al. Downregulation of hsa_circ_0011946 suppresses the migration and invasion of the breast cancer cell line MCF-7 by targeting RFC3. Cancer Manag Res. 2018;10:535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tang YY, Zhao P, Zou TN, et al. Circular RNA hsa_circ_0001982 promotes breast cancer cell carcinogenesis through decreasing miR-143. DNA Cell Biol. 2017;36:901–908. [DOI] [PubMed] [Google Scholar]
- [14].Zhang HD, Jiang LH, Sun DW, et al. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25:1–7. [DOI] [PubMed] [Google Scholar]
- [15].Du WW, Fang L, Yang W, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhang S, Kim K, Jin UH, et al. Aryl hydrocarbon receptor agonists induce microRNA-335 expression and inhibit lung metastasis of estrogen receptor negative breast cancer cells. Mol Cancer Ther. 2012;11:108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bond Edmond M, Aletraris L, Roman PM. Rural substance use treatment centers in the United States: an assessment of treatment quality by location. Am J Drug Alcohol Abuse. 2015;41:449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Al-Dhfyan A, Alhoshani A, Korashy HM. Aryl hydrocarbon receptor/cytochrome P450 1A1 pathway mediates breast cancer stem cells expansion through PTEN inhibition and beta-Catenin and Akt activation. Mol Cancer. 2017;16:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hanieh H. Aryl hydrocarbon receptor-microRNA-212/132 axis in human breast cancer suppresses metastasis by targeting SOX4. Mol Cancer. 2015;14:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Jenny MJ, Aluru N, Hahn ME. Effects of short-term exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin on microRNA expression in zebrafish embryos. Toxicol Appl Pharmacol. 2012;264:262–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yoshioka W, Higashiyama W, Tohyama C. Involvement of microRNAs in dioxin-induced liver damage in the mouse. Toxicol Sci. 2011;122:457–465. [DOI] [PubMed] [Google Scholar]
- [22].Stewart MD, Zelin E, Dhall A, et al. BARD1 is necessary for ubiquitylation of nucleosomal histone H2A and for transcriptional regulation of estrogen metabolism genes. Proc Natl Acad Sci U S A. 2018;115:1316–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Liu Y, Ao X, Jia Z, et al. FOXK2 transcription factor suppresses ERalpha-positive breast cancer cell growth through down-regulating the stability of ERalpha via mechanism involving BRCA1/BARD1. Sci Rep. 2015;5:8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Irminger-Finger I, Leung WC, Li J, et al. Identification of BARD1 as mediator between proapoptotic stress and p53-dependent apoptosis. Mol Cell. 2001;8:1255–1266. [DOI] [PubMed] [Google Scholar]
- [25].Marzec KA, Martino-Echarri E, Irminger-Finger I, et al. BARD1 splice variants display mislocalization in breast cancer cells and can alter the apoptotic response to cisplatin. Cancer Lett. 2016;381:149–155. [DOI] [PubMed] [Google Scholar]


