ABSTRACT
Natural killer (NK) cells are innate effector lymphocytes widely involved in cancer immunosurveillance. In this study, we described three circulating NK cell subsets in patients with non-small cell lung cancer (NSCLC). Compared to healthy donors (HD), lower rate of the cytotoxic CD56dim CD16+ NK cells was found in NSCLC patients (76.1% vs 82.4%, P = 0.0041). In contrast, the rate of CD56bright NK cells was similar between patients and HD. We showed in NSCLC patients a higher rate of a NK cell subset with CD56dim CD16− phenotype (16.7% vs 9.9% P = 0.0001). The degranulation property and cytokines production were mainly drive by CD56dim CD16− NK cell subset in patients. Analysis of natural cytotoxicity receptors (NCRs) expression identified four distinct clusters of patients with distinct NK cell subset profiles as compared to one major cluster in HD. Notably the cluster characterized by a low circulating level of NKp46+ NK cell subsets was absent in HD. We showed that the rate of circulating NKp46+ CD56dim CD16+ NK cells influenced the patients’ survival. Indeed, the median overall survival in patients exhibiting high versus low level of this NK cell subset was 16 and 27 months respectively (P = 0.02). Finally, we demonstrated that blocking NKp46 receptor in vitro was able to restore spontaneous tumor specific T cell responses in NSCLC patients. In conclusion, this study showed a distinct distribution and phenotype of circulating NK cell subsets in NSCLC. It also supports the regulatory role of NKp46+ NK cell subset in NSCLC patients.
KEYWORDS: NK cells, NSCLC ; NKp46, CD16, prognosis
Introduction
Natural killer (NK) cells, the most important effectors of innate lymphoid cells (ILCs), play a fundamental role in tumor immunosurveillance.1–3 NK cells are subdivided into several subsets based on CD56 and CD16 relative expression, with different antitumor functions.4 CD56dim CD16+ NK cells are largely predominant in blood and have high cytotoxic properties mediated by a strong production of granzymes and perforin.5 This function is regulated by a balance between a set of inhibitory and activating receptors such as CD16 which is critical to mediate antibody dependent cell mediated cytotoxicity (ADCC). The recognition of tumor cells by CD56dim CD16+ NK cells involves other main activating receptors such as NKG2D and natural cytotoxicity receptors (NCRs) NKp30 and NKp46, which bind their respective ligands expressed on tumor cells.6–8 CD56bright CD16− NK cells (CD56bright NK) which are poorly cytotoxic and have a regulatory function,9,10 contribute also in tumor immune control by secreting large amounts of IFN-γ.11 More recently NK cell subsets such as CD56bright CD16+ NK cells and CD56dim CD16− NK cells have been described but they are poorly found in healthy individuals and their functions are largely unknown.10,12,13
Tumor-infiltrating NK cells are associated with a better prognosis in several tumors14,15 including in lung cancer, suggesting their implication in tumor control.16 Thus, NK cells were characterized within the tumors and their phenotype and function were mainly investigated in patients’ blood due to an easier access. A downregulation of NKG2D, NKp30 and NKp46 expression on NK cells was found in tumor microenvironment as well as in blood of patients with breast cancer, colorectal cancer, gastric and pancreatic cancer and melanoma.17–23 This NK cells altered phenotype is correlated with a defective functionality highlighted by a reduction of both perforin level19 and IFN-γ production.20,24 It has also been reported in melanoma and colorectal cancer that the decrease of NKp46+ NK cells in advanced disease is associated with a decreased survival.18,24 In this study, we described in NSCLC patients three main circulating NK cell subsets such as CD56dim CD16+, CD56dim CD16− and CD56bright, NK cells. Subsequently, we evaluated their cytotoxic potential, analyzed the NCRs expression and also addressed the issue of their clinical influence.
Results
Distribution of peripheral NK cell subsets in NSCLC patients
NK cell subsets were measured in 176 NSCLC patients free of any treatment and 41 HD. Patients’ main clinical characteristics at the time of sampling are indicated in Supplementary Table S1. For the flow cytometry analysis, total NK cells were gated among the lymphocyte population on CD3− CD56+ cells. No differences were observed between the rate of total circulating NK cells from NSCLC patients and HD (15.1x vs 15.5% respectively) (Figure 1A). NK cells were further divided according to the expression of CD56 and CD16 markers. We found that both HD and NSCLC patients exhibited three distinct NK cell subsets: CD56dim CD16+, CD56dim CD16− and CD56bright NK cells (Figure 1B). Like in HD the majority of circulating NK cells in patients were of CD56dim CD16+ phenotype (76.1%). But, there was a substantial decrease of the percentage of CD56dim CD16+ NK cells in NSCLC patients as compared to HD (76.1 vs 82.4% respectively, P = 0.0041) (Figure 1B-C). The CD56dim CD16− and CD56bright NK cell subpopulations represented 16.7% and 5.6% of total NK cells respectively in NSCLC patients (Figure 1B).While we found a significant higher rate of CD56dim CD16− NK cells in NSCLC patients (16.7 vs 9.9%, P = 0.0001) (Figure 1D), no difference was observed between the frequency of CD56bright NK cells from patients and HD (Figure 1E). We confirmed that the three NK cell subsets found in the blood of NSCLC patients differ from NCRs+ ILC3 by their lack of CD127 expression (Supplementary Fig. S1). These different subsets were homogeneously distributed according to patients’ main clinical characteristics (Figure 1F and Supplementary Table S1). Thus, our results indicate that NSCLC patients exhibit high level of CD56dim CD16− NK cells and low rate of CD56dim CD16+ NK cells in blood.
Figure 1.

Distribution of NK cell subsets in the blood of NSCLC patients.
Representative flow cytometry gating strategy and proportion of circulating (A) total CD3− CD56+ NK cells among PBMCs in HD (n = 41) and NSCLC patients (n = 176); and (C) CD56dim CD16+, (D) CD56dim CD16− and (E) CD56bright CD16− NK cell subsets among total NK cells in HD (n = 41) and NSCLC patients (n = 176). (B) Frequency of each NK cell subsets in blood of HD (n = 41) and NSCLC patients (n = 176) represented by stacked bars. (F) Proportion of total NK cells among PBMCs and proportions of CD56dim CD16+, CD56dim CD16− and CD56bright CD16− NK cell subsets among total NK cells in NSCLC patients according to the disease stage. Statistical analysis were performed using the Mann-Whitney test (two groups) or the Kruskal-Wallis test (more than two groups) (ns = not significant; ** P < 0.01; *** P < 0.001).
Cytotoxic and regulatory functions of NK cell subsets in NSCLC patients
Next, we asked for the cytotoxic property of each NK cell subsets. To this end we evaluated their degranulation capacity when exposed to NK-sensitive K562 tumor cells. As for HD, we showed that the percentage of total CD107a+ NK cells from patients significantly increased in presence of K562 target cells (P < 0.0001 and P = 0.003 respectively) (Figure 2A). Conversely, the rate of total Granzyme B+ NK cells similarly decreased after K562 exposition both in patients and HD (P = 0.004 and P = 0.015 respectively) (Figure 2A). However, at the steady state, NSCLC patients exhibited higher circulating rate of Granzyme B+ NK cells than HD (P = 0.003). Next, we evaluated K562 cell death after culture with PBMCs from patients and HD. We found similar rates of annexin V+ 7-AAD− (early apootosis) and annexin V+ 7-AAD+ (late apoptosis) K562 cells, between patients and HD (Figure 2B).
Figure 2.

Blood NK cell cytotoxicity in NSCLC patients.
PBMCs from patients and HD were cultured with K562 cells for 6 hours at a ratio E:T of 1:1. (A) Degranulation of total NK cells among PBMCs was assessed by flow cytometry: percentages of total NK cells expressing CD107a (right panel) and Granzyme B (left panel) in HD (n = 10) and NSCLC patients (n = 10), Mann-Whitney test (ns = not significant; ** P < 0.01). Spontaneous (open circles) and K562-activated (solid circles) values are indicated, paired t-test (* P < 0.05; ** P < 0.01; **** P < 0.0001). (B) Percentages of K562 cell death represented by rates of annexin V+ 7-AAD+ and annexin V+ 7-AAD− K562 cells, after 6 hours of interactions with PBMCs from HD (n = 10) and NSCLC patients (n = 10), Mann-Whitney test (ns = not significant). Columns, mean of rates; bars, SEM. On the left panels, (C) degranulation and (D) Granzyme B production by NK cell subsets in response to K562 stimulation of a representative patient are showed. The percentages indicate the rate of CD107a+ and Granzyme B+ NK cells after gating on CD56dim CD16+, CD56dim CD16− or CD56bright cells. On the right panels, are summarized rates of (C) CD107a+ and (D) Granzyme B+ among CD56dim CD16+, CD56dim CD16− and CD56bright NK cell subsets of either both HD (grey n = 10) and patients (black n = 10), Mann-Whitney test (ns = not significant; * P < 0.05; *** P < 0.001; **** P < 0.0001), or only patients (black n = 10), Friedman test (ns = not significant; * P < 0.05; *** P < 0.001; **** P < 0.0001). Red bars represent the mean of each group.
When we focused on each NK cell subset, the degranulation activity characterized by CD107a expression was mainly driven by CD56dim CD16− NK cells as compared to CD56dim CD16+ commonly described as high cytotoxic subpopulation (23.4 vs 9.4% respectively, P = 0.041) (Figure 2C). However, the ability to produce Granzyme B against K562 was preferentially relied on CD56dim CD16+ NK cell subset in NSCLC patients (Figure 2D). As expected CD56bright NK cells, a subset with regulatory properties,10 showed a lower level of Granzyme B compared to CD56dim CD16+ NK (19.3 vs 54.8% respectively, P = 0.0002), that was associated with a poor degranulation property against K562 cells (Figure 2C-D).
We next investigated the property of NK cell subsets to produce effector cytokines in response to K562 cells. Overall, circulating total NK cells both from NSCLC patients and HD were able to significantly produce IFN-γ and TNF-α in response to K562 cells (Figure 3A). We found that IFN-γ was mainly produced by CD56dim CD16− NK cells as compared to CD56bright NK cells (14.2 vs 6.3% respectively, P = 0.048) (Figure 3B). Similar observation was made about TNF-α production (5.5 vs 1.1% respectively, P = 0.015) (Figure 3C). As expected, CD56dim CD16+ NK cell showed a low production of IFN-γ and TNF-α following K562 cells activation (Figure 3B-C). Of note, no obvious difference was observed between patients and HD about the capacity of NK cell subsets to produce IFN-γ and TNF-α (Figure3A-C). Moreover, we analyzed the capacity of NK cell subsets to secrete effector cytokines in response to different conditions of activation using cytokines involved in NK cell homeostasis and function.5 We showed that the two main CD56dim CD16− and CD56bright NK cell subsets particularly secreted IFN-γ and TNF-α following stimulation with IL-2, IL-12 or IL-21 (Figure 3D). Collectively our results show that NK cells and especially CD56dim CD16− NK cells display both regulatory and degranulation functions in NSCLC patients.
Figure 3.
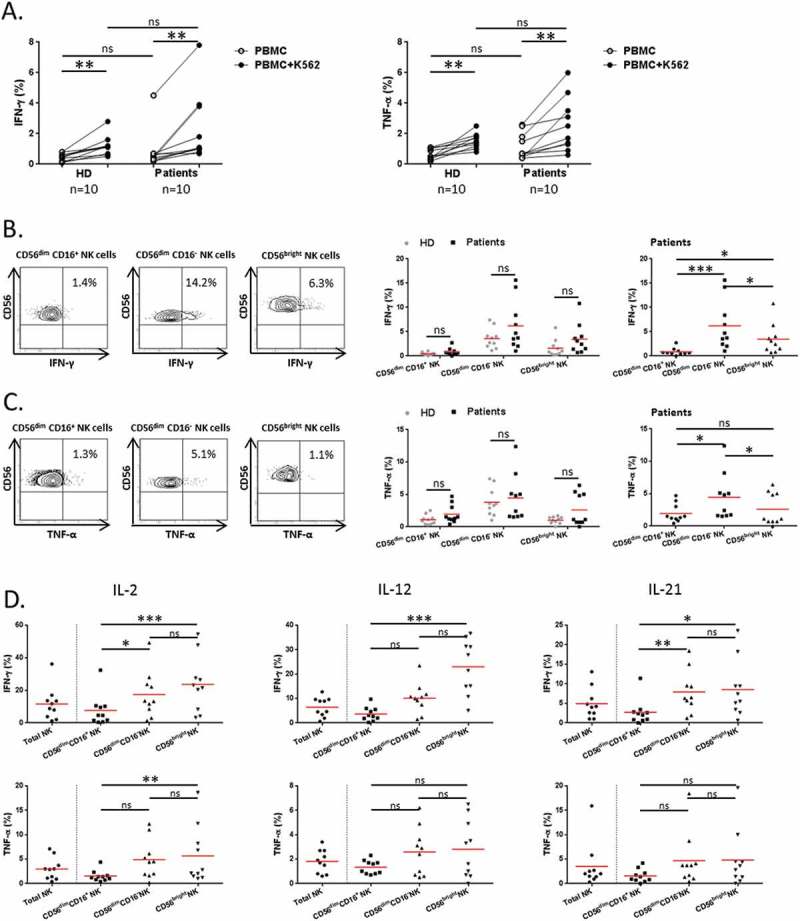
Regulatory function of NK cells in NSCLC patients.
PBMCs from patients and HD were activated with K562 cells for 6 hours at a ratio E:T of 1:1. (A) Cytokines production of total NK cells among PBMCs was assessed by flow cytometry: percentages of total NK cells producing IFN-γ (left panel) and TNF-α (right panel) in HD (n = 10) and NSCLC patients (n = 10), Mann-Whitney test (ns = not significant). Spontaneous (open circles) and K562-activated (solid circles) values are indicated, paired t-test (** P < 0.01). On the left panels, (B) IFN-γ and (C) TNF-α production by NK cell subsets in response to K562 stimulation of a representative patient are showed. The percentages indicate rates of IFN-γ+ and TNF-α+ NK cells after gating on CD56dim CD16+, CD56dim CD16− or CD56bright cells. On the right panels, are summarized rates of (B) IFN-γ+ and (C) TNF-α+ among CD56dim CD16+, CD56dim CD16− and CD56bright NK cell subsets of either both HD (grey n = 10) and patients (black n = 10), Mann-Whitney test (ns = not significant), or only patients (black n = 10), Friedman test (ns = not significant; * P < 0.05; *** P < 0.001) Red bars represent the mean of each group. (D) PBMCs from patients were activated with IL-2, (1000U/mL), IL-12 (10ng/mL) or IL-21 (50ng/mL) for 20 hours. Proportions of IFN-γ+ and TNF-α+ among total, CD56dim CD16+, CD56dim CD16− and CD56bright NK cell subsets in NSCLC patients (n = 10), Friedman test (ns = not significant; * P < 0.05; ** P < 0.01; *** P < 0.001). Red bars represent the mean of each group.
Distinct profile of ncr-expressing NK cell subsets in NSCLC patients
To further characterize these circulating NK cell subsets, we analyzed the cell-surface expression of NKG2D and NCRs (NKp30, NKp44, NKp46) activating receptors. About NKG2D, only the rate of NKG2D+ CD56bright NK cells was found increased in NSCLC patients as compared to HD (72.5 vs 61.4% respectively, P = 0.0001) (Figure 4A). No obvious difference was observed between patients and HD about NKp30+ and NKp44+ NK cell subsets (Figure 4B-C). In contrast, the percentage of each NKp46+ NK cell subset was significantly lower in NSCLC patients than in HD (Figure 4D). Notably, we distinguished two groups of patients according to the low (< 37%) or high rate (> 37%) of circulating NKp46+ NK cells (referred as NKp46+ NKlow or NKp46+ NKhigh cells respectively) (Supplementary Fig. S2A). Of note, NSCLC stages did not seem to influence the rate of circulating NKp46+ NK cell subsets (Supplementary Fig. S2B). As expected, we observed a strong positive correlation between NKp46 on NK cells and the other activating receptors among NSCLC patients (Supplementary Table S2).
Figure 4.
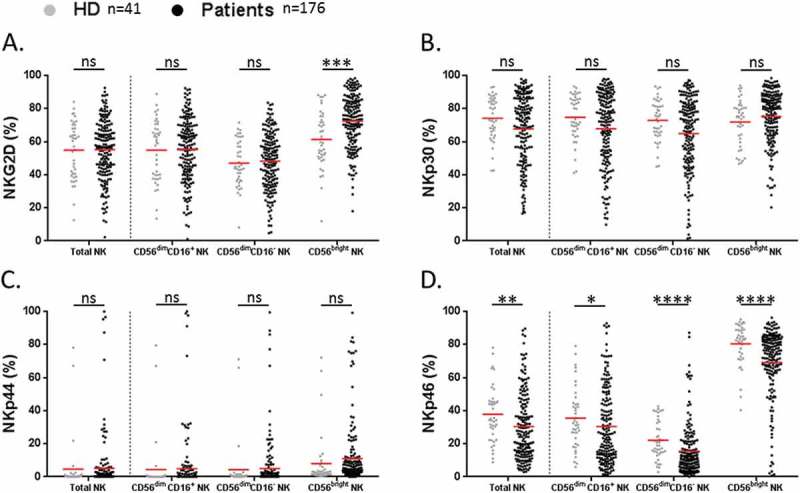
Analysis of activating receptors on NK cell subsets in NSCLC patients.
Proportions of (A) NKG2D+, (B) NKp30+, (C) NKp44+ and (D) NKp46+ among total, CD56dim CD16+, CD56dim CD16− and CD56bright NK cell subsets in HD (grey n = 41) and NSCLC patients (black n = 176). Red bars represent the mean of each group. Statistical analysis were performed using the Mann-Whitney test (ns = not significant; * P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001).
Unsupervised hierarchical clusterings (heatmap) according to the NCRs-expressing NK cell subsets identified four clusters of NSCLC patients (Figure 5A). The cluster 1 (CL1) gathered patients with globally high level of NCRs on all NK cell subsets. The cluster 2 (CL2) was characterized by a lower rate of NKp44+ NK cells compared to CL1. Patients from cluster 3 (CL3) have both a reduced level of NKp44+ and NKp46+ NK cells. Finally, the last cluster (CL4) with heterogeneous expression of NCR included only 5 patients, thus it was not included for further analysis. In contrast, the heatmap showed one major cluster in HD corresponding to CL2 found in patients (Figure 5B). Remarkably, the cluster 3 from NSCLC patients characterized by a low/moderated level of circulating NKp46+ NK cells, was totally absent in HD. The cluster distribution was similar for patients’ main clinical characteristics (data not shown). We further analyzed the expression of co-inhibitory receptors such as TIM3 and PD-1 known to be expressed by NK cells.25–28 TIM3 was significantly higher in NK cells from NSCLC patients than in HD (19.7 vs 5.9% respectively, P < 0.0001) and was preferentially expressed by CD56dim CD16+ NK cell subset (Supplementary Fig. S2C). Similarly, the percentage of PD-1+ NK cells was significantly increased in NSCLC patients as compared to HD (1.6 vs 0.4% respectively, P = 0.008) and also for the whole of subsets (Supplementary Fig. S2C).
Figure 5.

Hierarchical clustering of NK cell activating receptors.
Hierarchical clustering of (A) NSCLC patients (n = 176) and (B) HD (n = 41) was realized according to NKp44, NKp30, NKG2D and NKp46 expression on NK cell subsets. The percentage values obtained for the Figure 3 were used and the clustering was performed using Morpheus Software.
These results show distinct levels of NCRs+ NK cells in NSCLC patients and especially a subgroup that exhibiting a low rate of circulating NKp46+ NK cells.
The level of circulating nkp46+ NK cells influences the overall survival
We investigated the prognostic value of circulating NK cell subsets on four years-overall survival (OS) of patients. There was no association of the level (low or high according to the mean percentage) of total circulating NK cells or the three described NK cell subsets with the OS (Supplementary Fig. S3A-D). Next we wondered whether the level NCRs+ NK cell subsets influenced the outcome. To this end we defined two groups of patients according to the low or high circulating level of each NCRs+ NK cell subset. No association of the level of circulating NKp44+ or NKp30+ NK cell subsets with survival was observed (data not shown).
However, we showed that the level of NKp46+ NK cells influenced the OS. Indeed the total NKp46+ NKlow is significantly associated with a better OS (median OS, 24 vs 16 months, P = 0.04) (Figure 6A). Notably, we found that this association with OS was significantly driven by the NKp46+ CD56dim CD16+ subset (median OS, 27 vs 16 months, P = 0.02) (Figure 6B-D). Indeed, no association between the level of NKp46+ CD56bright or NKp46+ CD56dim CD16− NK cells and the OS was found (Figure 6C-D). Thus, the level of circulating NKp46+ CD56dim CD16+ NK cells predicts distinct outcome in NSCLC.
Figure 6.
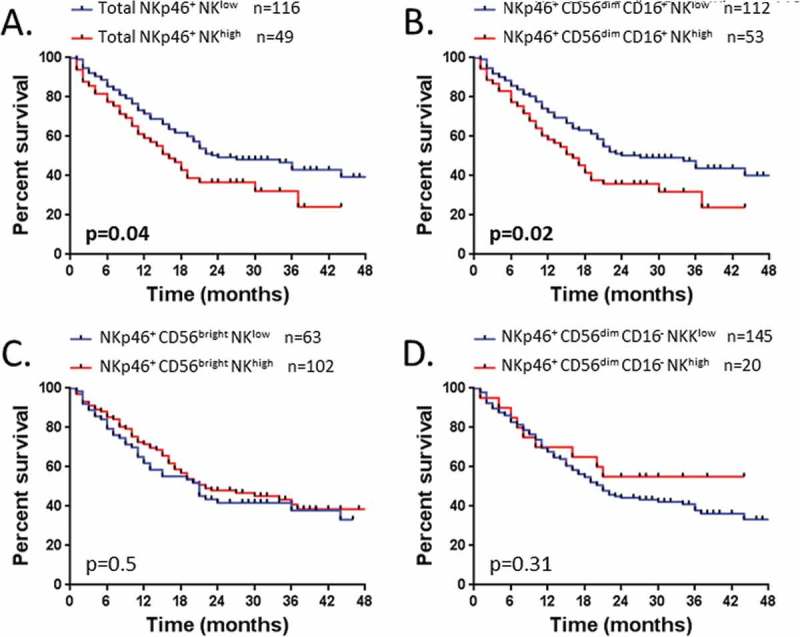
Patients’ survival according to the rate of NK cell subsets expressing NKp46.
Survival of patients is depicted as Kaplan-Meier curves. Patients (n = 165) were divided into two groups low and high as detailed in result section, graphically defined by a cut off value of 37% for (A) NKp46+ NK cells and (B) NKp46+ CD56dim CD16+ NK cells; and defined by the percentage mean value of: (C) NKp46+ CD56bright NK cells (mean = 59%) and (D) NKp46+ CD56dim CD16− NK cells (mean = 29%). Statistical analysis was performed using the Log-Rank test.
Blockade of nkp46 pathway restored tumor-specific th1 response
A recent study reported the inhibitory effect of a subset of tumor-infiltrating NKp46+ CD3− CD56+ cells against effector T cells in the tumor microenvironment of ovarian cancer patients.29 So, we assessed whether the circulating NKp46+ NK cell subset could exert similar effect on tumor-specific T cell responses detected in blood. To this end, PBMCs from NSCLC patients were cultured with telomerase (TERT)-derived peptides in presence or not of anti-NKp46 monoclonal blocking antibody previously described.29,30 The presence of tumor-specific T cell response was measured by IFN-γ ELISpot assay (Figure 7A). As shown in patients in Figure 7B, the addition of anti-NKp46 in the culture was able to restore or increase the antitumor Th1 response in vitro (P = 0.052), suggesting that NKp46+ NK cells could exert an inhibitory effect in a contact depend-manner. In line with this, we showed a positive correlation between the concentration of TGF-β and the level of NKp46+ NK cells. Conversely, the level of this NK cell subset was inversely correlated with the IL-10 concentration (Supplementary Fig. S4A-B). We also found a low level of inflammatory cytokines such as TNF-α and IL-21 associated with high circulating rate of NKp46+ CD56dim CD16+ NK cells in the serum of NSCLC patients (Figure 7C).
Figure 7.
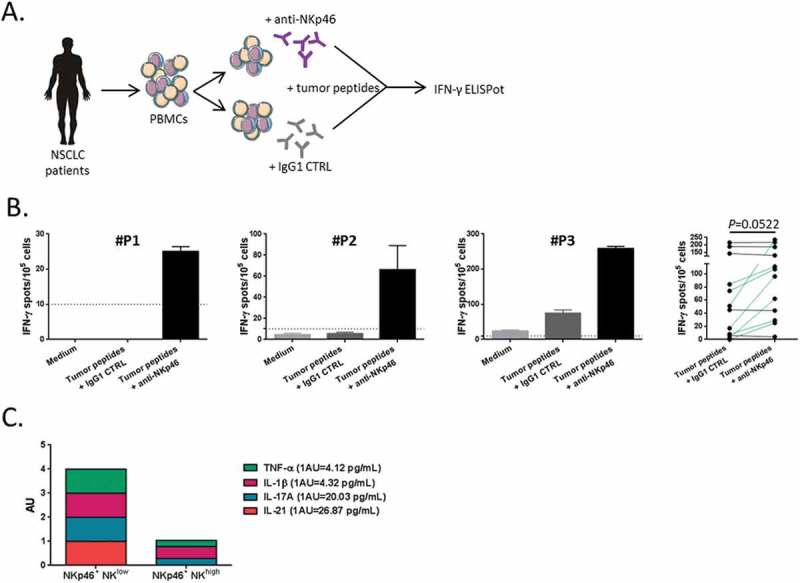
Blockade of NKp46 restored tumor-specific T cell response.
(A) Blood lymphocytes from NSCLC patients were stimulated with tumor-derived peptides in presence or not of blocking antibodies (mAb) against NKp46. (B) Three representative examples and the whole of number of IFN-γ-producing antitumor Th1 cells measured by ELISpot assay with IgG1 isotype control or anti-NKp46 mAb are showed in NSCLC patients (n = 11), paired t-test. Columns, mean of spots from triplicate wells; bars, SEM. (D) Cumulative representation of normalized (arbitrary unit) cytokines concentrations (TNF-α, IL-1β, IL-17A and IL-21) in serum of patients with low or high rate of NKp46+ CD56dim CD16+ NK cells.
Collectively, our results demonstrate that low level of peripheral NKp46+ CD56dim CD16+ NK cells is associated with better survival in lung cancer and also highlights a potential regulatory property of this NK cell subset.
Discussion
Tumor-induced modifications of NK cell phenotype and function is a well-known mechanism contributing to the tumor escape.31 Here we described three circulating NK cell subsets in NSCLC patients such as CD56dim CD16+, CD56dim CD16− and CD56bright NK cells. While the cytotoxic and regulatory functions of NK cells, mainly driven by CD56dim CD16− NK cell subset, were similar between patients and HD, we found differences about NK cell phenotype. Indeed, the rate of all of these NK cell subsets expressing NKp46 is significantly lower in NSCLC patients as compared to HD. Notably, we showed in NSCLC patients that a low rate of NKp46+ CD56dim CD16+ NK cells is significantly associated with a better OS.
The analysis of NK cells based on CD56 and CD16 expression highlighted three main NK cell subsets in both NSCLC patients and HD. Although similar rate of the regulatory CD56bright NK cell subset12 was found between patients and HD, lower circulating level of the cytotoxic CD56dim CD16+ NK subsets10 was observed in NSCLC patients. We also observed in peripheral blood of NSCLC patients a higher rate of a NK cell subset with CD56dim CD16− phenotype as compared to HD. This subset differs from NCR+ ILC33 by its lack of CD127 expression. According with our results, the rate of NK cells expressing CD16 which has been reported in NSCLC patients,32,33 such as 76.9%34 or 80%35 is similar to those of CD56dim CD16+ NK cells in our study (76.1%). Moreover, Bruno et al. described around 17% of CD16− NK cells in the blood of NSCLC patients which is very close to the value of the rate of CD56dim CD16− NK cells in our cohort (16.7%).35 All of these studies support our results that NSCLC patients display a high rate of circulating CD56dim CD16− NK cells. Nevertheless, although recent studies identified these CD56dim CD16− NK cells as a distinct subset their ontogeny remains unclear.10,12,13 Indeed, several studies described a quick shedding of CD16 from NK cell surface upon activation due to metalloproteases.36,37 Future investigations are required to assess whether CD56dim CD16− NK cells represent a specific differentiation of circulating NK cells or derived from another mechanisms.38–40
While we found a similar NK cells-cytotoxic and regulatory functions in both patients and HD, our results also demonstrated that these CD56dim CD16− NK cells exert a superior degranulation capacity against K562 cells and produce more amounts of IFN-γ and TNF-α than the two others circulating NK cell subsets found in NSCLC. Although CD56dim NK cells are well known for their cytotoxic activity, CD56dim CD16+ NK cells were rather described as responsible for ADCC and CD56dim CD16− NK cells for natural cytotoxicity.41,42 The high capacity of response of CD56dim CD16− NK cells to K562 cells may suggest a potential involvement of this subset in cancer immunosurveillance.
Analysis of NCRs expression on NK cells revealed several groups of patients and HD. Two clusters CL1 and CL2 were shared by both HD and NSCLC patients with a main cluster represented by CL2. Another third cluster CL3 was only found in lung cancer patients. This cluster showed a lower expression of NCRs and particularly the activating receptor NKp46 on NK cells. Grouped analysis of NCRs and NKG2D expression on NK cells confirmed the heatmap results by revealing significant lower rates of NKp46+ NK cell subsets in NSCLC patients as compared to HD. Nonetheless, the disease stage didn’t have impact on the rate of activating receptor expression on NK cell subsets.
Although NKp46 is the main activating receptor involved in NK cell functions,43,44 the downregulation of this receptor observed in our study has also been reported in many other cancers.17–19,24,45,46 This suggests an involvement of tumor-related factors in the phenotypic alteration of NK cells. Indeed, some immune suppressive cytokines or soluble factors such as TGF-β or IDO secreted by tumor cells or stromal cells have been shown to decrease the expression of NCRs or NKG2D on NK cells.47,48 Moreover, tumor cells are able to release a soluble form of MIC ligands which keeps away NK cells and downregulates NKG2D expression.49,50 Thus possible NKp46 ligands or immunosuppressive factors released from tumor cells and could also promote a downregulation of NKp46 on NK cells. On this view, we investigated the correlation between the rate of NKp46+ NK cells and the concentration of several cytokines in the serum of patients. We found a positive correlation between the concentration of TGF-β and the level of NKp46+ NK cells while IL-10 is inversely correlated.
While other circulating NK cells expressing NCRs or NKG2D don’t have any prognostic role, we notably found an inverse correlation between patients’ survival and the rate of NKp46 CD56dim CD16+ NK cells. Thus, we showed that patients with NKp46+ NKlow cells had a better survival than those with NKp46+ NKhigh cells. This observation is unexpected because previous studies reported conflicting results in melanoma and colorectal cancer where a low rate of NKp46+ NK cells in blood was associated with poor prognosis.18,24 Several parameters could explain this discrepancy. A very low number of patients have been evaluated in these studies.18,24 Furthermore, in the study by Fregni et al., the association between the level of NKp46+ NK cells and the survival had only found in metastatic melanoma patients.24 Moreover, the cut-off value of NKp46+ NK cells rate used to define the groups of patients is around 80% in melanoma and colorectal cancer versus 37% in our study.18,24
One hypothesis that could explain the worse prognosis of patients with high rate of NKp46+ NK cells likely involved the crosstalk between NKp46+ NK cells and adaptive T cell immunity. Indeed, it has been reported that NK cells are able to regulate T cell immunity by soluble factors or by direct cell contact.29,51 For example, the IFN-γ production by NK cells is well known to promotes the priming of Th1-polarized immunity.52 A recent study reported that a subset of tumor-infiltrating NKp46+ CD3− CD56+ cells prevent the expansion of antitumor T cells by mechanism involving NKp46.29 In line with this study, we also showed that the blockade of Nkp46 receptor in vitro effectively reinvigorates the IFN-γ production by TERT-specific CD4 Th1 cells. We previously demonstrated that the presence of circulating anti-TERT Th1 cells in cancer patients is associated with better survival.53–55 Thus we believed that the worse prognostic value found in population with NKp46+ NKhigh cells could be due to the ability of these NK cells to suppress anti-tumor Th1 immunity. Although our observation also suggests that NKp46+ NK cells could exert its negative control on antitumor T cells in a contact depend-manner, we demonstrated a positive correlation between the rate of NKp46+ NK cells and the level of TGF-β in the serum of patients, which is a cytokine known to be of poor prognosis in NSCLC.56 In contrast to immune profile shown in patients with NKp46+ NKhigh cells, we showed a trend of inflammatory signature profile in patients with NKp46+ NKlow cells. Another hypothesis that could explain the worse prognosis of patients with high rate of NKp46+ NK cells is the expression of co-inhibitory receptors such as PD-1 or TIM-3. Indeed, a previous study in lung cancer reported that a high expression of TIM-3 on NK cells was associated with poor survival of patients.57 However in our study, no obvious difference of TIM3 and PD-1 levels was found regardless NKp46 expression (data not shown). Thus, we speculated that NKp46+ NK cells could drive an immunosuppressive environment which prevents adaptive T cell immunity. However, future investigations are needed to address the mechanisms by which NKp46 acts on antitumor T cell responses.
In conclusion, our study describes the presence of distinct circulating NK cell subsets and highlights an alteration of NKp46 pathway in NSCLC patients. This study also supports a regulatory role of NKp46+ NK cells in lung cancer.
Materials and methods
Patients and healthy donors
Non-Small Cell Lung Cancer (NSCLC) patients were from TeloCap01 cohort, a prospective multicenter immunomonitoring study conducted in the European Hospital Georges Pompidou (Paris) and the University Hospital of Besançon between July 2010 and January 2014 (N°EUDRACT: 2009-A00642-55). Patient blood samples were collected before any treatment including surgery. Information about patients’ survival was collected at one and two-years after the inclusion. All patients had given their written consent and the protocol was approved by local and national ethic committees. For healthy volunteer donors, peripheral blood mononuclear cells (PBMCs) were collected from anonymous donors at the Etablissement Français du Sang (EFS, Besançon, France) as apheresis kit preparation after the signature of the informed consent and following the EFS guidelines. Patients’ and donors’ blood samples were isolated and frozen until use.
Flow cytometry analysis
PBMCs were stained for 20 minutes at 4°C with conjugated antibodies: anti-CD3-V500 (Biolegend, 613406), anti-CD16-APC H7 (BD Biosciences, 554529), anti-CD56-eFluor 710 (eBioscience, 46–0567-42), anti-NKp30-PE (BD Biosciences, 558407), anti-NKp44-APC (R&D System, IC0041P), anti-NKp46-V450 (BD Biosciences, 562099), anti-NKG2D-PeCy7 (BD Biosciences, 557924), anti-PD-1-Pecy7 (Biolegend, 367414) and anti-TIM3-APC (Biolegend, 345012). Cells were analyzed with BD Facs Canto II flow cytometer (BD Biosciences) and data were analyzed with BD Facs Diva software.
Cell culture and cd107a degranulation assay
PBMC were cultured in RPMI 1640 medium (Gibco Invitrogen) supplemented with 10% human serum and 1% Penicillin-Streptomycin. K562 cells derived from human leukemia cell line were cultured in RPMI 1640 medium (Gibco Invitrogen) supplemented with 10% fetal calf serum and 1% Penicillin-Streptomycin.
For CD107a degranulation assay, PBMCs were cultured with K562 cells at 1:1 effector/target (E/T) ratio, for 6 hours at 37°C with golgi stop (BD Biosciences, 554724) and anti-CD107a-PE (BD Biosciences, 555801) or isotype control (BD Biosciences, 555749). Cells were then stained for 20 minutes at 4°C with anti-CD3-PB (BD Biosciences, 558117), anti-CD56-eFluor 710 (eBioscience, 46–0567-42) and anti-CD16-FITC (BD Biosciences, 555406).
Cytokines secretion assay
For measurement of intracellular IFN-γ, TNF-α and Granzyme B, PBMCs were cultured with K562 cells at 1:1 E/T ratio for 6 hours or were stimulated with 1000U/mL IL-2 (Peprotech, 200–02), 10ng/mL IL-12 (Miltenyi Biotec, 130–096-704) or 50ng/mL IL-21 (Shenandoah Biotechnology Inc, 100–90) for 20 hours, at 37°C. Golgi plug (BD Biosciences, 555029) was added to cultures during the last 6 hours of stimulation. PBMCs were then stained for 20 minutes at 4°C with anti-CD3-PB (BD Biosciences, 558117), anti-CD56-PerCP-Cy5.5 (BD Biosciences, 560842) and anti-CD16-FITC (BD Biosciences, 555406). After cell surface staining, cells were fixed and permeabilized using Cytofix/CytoPerm kit (BD Biosciences, 554714) and stained with anti-IFN-γ-APC (BD Biosciences, 554702), anti-TNF-α-PE (Biolegend, 502909) and anti-Granzyme B-BV510 (BD Biosciences, 563388).
Cytotoxicity assay
K562 cells were labeled with CFSE (CellTrace CFSE Cell proliferation Kit, Molecular Probe) at 5µM in PBS 1X (Gibco Invitrogen) for 15 min under agitation. Then 2 volumes of fetal calf serum were added and cells were again incubated under agitation for 10 min. Finally, cells were washed twice with PBS 1X before culture. K562 cells were cultured with PBMCs at 1:1 E/T ratio, for 6 hours at 37°C. Cells were then stained for 15 minutes at room temperature with anti-Annexin V-APC (BD Biosciences, 550475) and anti-7-AAD (BD Biosciences, 559925) in 1X Annexin V binding buffer (BD Biosciences, 556454). Dead cells were analyzed after gating on CFSE positive K562 cells.
Synthetic peptides
We previously reported four HLA-DR and four HLA-DP4-restricted telomerase (TERT)-derived peptides that can be used for the monitoring of anti-TERT Th1 responses.53,58 These peptides were purchased from CTL (Cellular Technology Ltd, Germany).
Assessment of tumor antigen-specific t-cell responses
Tumor-specific Th1 response was assessed using a short-term in vitro stimulation with TERT-derived peptides as described previously.53,58 Briefly, PBMCs from patients or HD were cultured in 24-well plates with TERT-derived peptides (5µg/mL) in presence of anti-NKp46 mAb (5µg/mL, BD Biosciences, 557847) or IgG1 isotype control (5µg/mL, BD Biosciences, 554721). At day 10, the TERT-specific T cell specificity was investigated by IFN-γ ELISpot assay. Cells were seeded in triplicates in anti-human IFN-γ monoclonal antibody pre-coated ELISpot plates with TERT-derived peptides (5µg/mL) or with medium in X-VIVO 15 (control) for 18 hours at 37°C. The IFN-γ spots were revealed following the manufacturer’s instructions (Diaclone). Spot-forming cells were counted using the CTL Immunospot system. The number of specific T-cells expressed as spot forming cells per 105 cells was calculated after subtracting negative control values (background).
Cytokine assay by CBA
Concentrations of IL-1β, IL-10, IL-17A, IL-21, TNF-α and TGF-β in undiluted sera samples of patients were measured using Cytometric Bead Array (CBA) Flex Set kits according to the manufacturer’s instructions (BD Biosciences, 558279, 558274, 560383, 560358, 560112, 560429 respectively). Samples were acquired using BD Facs Canto II flow cytometer (BD Biosciences) and data were analyzed with FCAP Array Software (BD Biosciences).
Statistical analysis
Data were analyzed using GraphPad Prism 6.0 software. Descriptive analyses are expressed as mean or median. For two-group comparisons, the nonparametric Mann-Whitney U-test was used. For multiple-group comparisons, the nonparametric unpaired Kruskal-Wallis test or paired Friedman test were used. Proportions were compared using the X2-test. Correlations were performed using the nonparametric Spearman test. Hierarchical cluster analysis and dendrograms were performed using the online Morpheus software and robust Z-score normalization (https://software.broadinstitute.org/morpheus/). Overall survival (OS) was calculated from the date of study enrollment to the date of death from any cause. Patients known to be alive were censored at the time of their last follow-up assessment. The survival curves were plotted according to the Kaplan-Meier method and compared using the Log-Rank test. P values less than 0,05 were considered significant (* P < 0,05; ** P < 0,01; *** P < 0,001; **** P < 0.0001).
Funding Statement
This work was supported by the Conseil Régional de Franche-Comté;Ligue Contre le Cancer;Assistance Publique des Hôpitaux de Paris;
Acknowledgments
We thank all patients who contributed to this study. We thank all of the medical doctors, nurses from thoracic oncologic department of university hospital of Besançon and European Georges Pompidou hospital in Paris, for their contributions. The authors also thank the Biomonitoring plateform of CIC-1431 for their technical support.
Supplemental data
Supplemental data for this article can be accessed here.
Disclosure of interest
The authors report no conflict of interest.
References
- 1.Waldhauer I, Steinle A.. NK cells and cancer immunosurveillance. Oncogene. 2008;27(45):5932–5943. doi: 10.1038/onc.2008.267. [DOI] [PubMed] [Google Scholar]
- 2.Guillerey C, Mj S. NK Cells and Cancer Immunoediting. Curr Top Microbiol Immunol. 2016;395:115–145. doi: 10.1007/82_2015_446. [DOI] [PubMed] [Google Scholar]
- 3.Del Zotto G, Marcenaro E, Vacca P, Sivori S, Pende D, Della Chiesa M, Moretta F, Ingegnere T, Mingari MC, Moretta A, et al. Markers and function of human NK cells in normal and pathological conditions. Cytometry B Clin Cytom. 2017;92(2):100–114. doi: 10.1002/cyto.b.21508. [DOI] [PubMed] [Google Scholar]
- 4.Nagler A, Lanier LL, Cwirla S, Phillips JH. Comparative studies of human FcRIII-positive and negative natural killer cells. J Immunol. 1989. 143(10):3183–3191. [PubMed] [Google Scholar]
- 5.Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends Immunol. 2001;22(11):633–640. doi: 10.1016/S1471-4906(01)02060-9. [DOI] [PubMed] [Google Scholar]
- 6.Brandt CS, Baratin M, Yi EC, Kennedy J, Gao Z, Fox B, Haldeman B, Ostrander CD, Kaifu T, Chabannon C, et al. The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. J Exp Med. 2009;206(7):1495–1503. doi: 10.1084/jem.20090681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hecht M-L, Rosental B, Horlacher T, Hershkovitz O, De Paz JL, Noti C, Schauer S, Porgador A, Seeberger PH. Natural cytotoxicity receptors NKp30, NKp44 and NKp46 bind to different heparan sulfate/heparin sequences. J Proteome Res. 2009;8(2):712–720. doi: 10.1021/pr800747c. [DOI] [PubMed] [Google Scholar]
- 8.El-Gazzar A, Groh V, Spies T. Immunobiology and conflicting roles of the human NKG2D lymphocyte receptor and its ligands in cancer. J Immunol. 2013;191(4):1509–1515. doi: 10.4049/jimmunol.1301071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper MA, Fehniger TA, Turner SC, Chen KS, Ghaheri BA, Ghayur T, Carson WE, Caligiuri MA. Human natural killer cells: a unique innate immunoregulatory role for the CD56(bright) subset. Blood. 2001. 97(10):3146–3151. [DOI] [PubMed] [Google Scholar]
- 10.Poli A, Michel T, Thérésine M, Andrès E, Hentges F, Zimmer J. CD56bright natural killer (NK) cells: an important NK cell subset. Immunology. 2009;126(4):458. doi: 10.1111/j.1365-2567.2008.03027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9(5):503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 12.Michel T, Poli A, Cuapio A, Briquemont B, Iserentant G, Ollert M, Human ZJ. CD56bright NK Cells: an Update. J Immunol. 2016;196(7):2923–2931. doi: 10.4049/jimmunol.1502570. [DOI] [PubMed] [Google Scholar]
- 13.Penczek A, Sienczyk M, Wirtz CR, Burster T. Cell surface cathepsin G activity differs between human natural killer cell subsets. Immunol Lett. 2016;179:80–84. doi: 10.1016/j.imlet.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Coca S, Perez-Piqueras J, Martinez D, Colmenarejo A, Saez MA, Vallejo C, Martos JA, Moreno M. The prognostic significance of intratumoral natural killer cells in patients with colorectal carcinoma. Cancer. 1997. 79(12):2320–2328. [DOI] [PubMed] [Google Scholar]
- 15.Ishigami S, Natsugoe S, Tokuda K, Nakajo A, Che X, Iwashige H, Aridome K, Hokita S, Aikou T. Prognostic value of intratumoral natural killer cells in gastric carcinoma. Cancer. 2000. 88(3):577–583. [PubMed] [Google Scholar]
- 16.Villegas FR, Coca S, Villarrubia VG, Jiménez R, Chillón MJ, Jareño J, Zuil M, Callol L. Prognostic significance of tumor infiltrating natural killer cells subset CD57 in patients with squamous cell lung cancer. Lung Cancer Amst Neth. 2002. 35(1):23–28. [DOI] [PubMed] [Google Scholar]
- 17.Nieto-Velázquez NG, Torres-Ramos YD, Muñoz-Sánchez JL, Espinosa-Godoy L, Gómez-Cortés S, Moreno J, Moreno-Eutimio MA. Altered Expression of Natural Cytotoxicity Receptors and NKG2D on Peripheral Blood NK Cell Subsets in Breast Cancer Patients. Transl Oncol. 2016;9(5):384–391. doi: 10.1016/j.tranon.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rocca YS, Roberti MP, Juliá EP, Pampena MB, Bruno L, Rivero S, Huertas E, Sánchez Loria F, Pairola A, Caignard A, et al. Phenotypic and Functional Dysregulated Blood NK Cells in Colorectal Cancer Patients Can Be Activated by Cetuximab Plus IL-2 or IL-15. Front Immunol. 2016;7. doi: 10.3389/fimmu.2016.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng Y-P, Zhu Y, Zhang -J-J, Xu Z-K, Qian Z-Y, Dai -C-C, Jiang K-R, Wu J-L, Gao W-T, Li Q, et al. Comprehensive analysis of the percentage of surface receptors and cytotoxic granules positive natural killer cells in patients with pancreatic cancer, gastric cancer, and colorectal cancer. J Transl Med. 2013;11:262. doi: 10.1186/1479-5876-11-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mamessier E, Sylvain A, Bertucci F, Castellano R, Finetti P, Houvenaeghel G, Charaffe-Jaufret E, Birnbaum D, Moretta A, Olive D. Human breast tumor cells induce self-tolerance mechanisms to avoid NKG2D-mediated and DNAM-mediated NK cell recognition. Cancer Res. 2011;71(21):6621–6632. doi: 10.1158/0008-5472.CAN-11-0792. [DOI] [PubMed] [Google Scholar]
- 21.Konjević G, Mirjacić Martinović K, Vuletić A, Jović V, Jurisić V, Babović N, Spuzić I. Low expression of CD161 and NKG2D activating NK receptor is associated with impaired NK cell cytotoxicity in metastatic melanoma patients. Clin Exp Metastasis. 2007;24(1):1–11. doi: 10.1007/s10585-006-9043-9. [DOI] [PubMed] [Google Scholar]
- 22.Mamessier E, Sylvain A, Thibult M-L, Houvenaeghel G, Jacquemier J, Castellano R, Gonçalves A, André P, Romagné F, Thibault G, et al. Human breast cancer cells enhance self tolerance by promoting evasion from NK cell antitumor immunity. J Clin Invest. 2011;121(9):3609–3622. doi: 10.1172/JCI45816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen Y, Shen Y, Lu C, Lu C, Tian W, Tian W, Wang L, Wang L, Cui B, Cui B, et al. Possible association of decreased NKG2D expression levels and suppression of the activity of natural killer cells in patients with colorectal cancer. Int J Oncol. 2012;40(4):1285–1290. doi: 10.3892/ijo.2011.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fregni G, Messaoudene M, Fourmentraux-Neves E, Mazouz-Dorval S, Chanal J, Maubec E, Marinho E, Scheer-Senyarich I, Cremer I, Avril M-F, et al. Phenotypic and functional characteristics of blood natural killer cells from melanoma patients at different clinical stages. PloS One. 2013;8(10):e76928. doi: 10.1371/journal.pone.0076928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beldi-Ferchiou A, Lambert M, Dogniaux S, Vély F, Vivier E, Olive D, Dupuy S, Levasseur F, Zucman D, Lebbé C, et al. PD-1 mediates functional exhaustion of activated NK cells in patients with Kaposi sarcoma. Oncotarget. 2016;7(45):72961–72977. doi: 10.18632/oncotarget.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Cheng Y, Xu Y, Wang Z, Du X, Li C, Peng J, Gao L, Liang X, Ma C. Increased expression of programmed cell death protein 1 on NK cells inhibits NK-cell-mediated anti-tumor function and indicates poor prognosis in digestive cancers. Oncogene. 2017;36(44):6143–6153. doi: 10.1038/onc.2017.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Sun J, Gao W, Song B, Shao Q, Zhao L, Zhang Y, Wang Q, Zhang Y, Qu X. Preoperative Tim‑3 expression on peripheral NK cells is correlated with pathologic TNM staging in colorectal cancer. Mol Med Rep. 2017;15(6):3810–3818. doi: 10.3892/mmr.2017.6482. [DOI] [PubMed] [Google Scholar]
- 28.Ip S, Da, Gallois A, Jimenez-Baranda S, Khan S, Ac A, Vk K, Osman I, Bhardwaj N. Reversal of NK-Cell Exhaustion in Advanced Melanoma by Tim-3 Blockade. Cancer Immunol Res. 2014. February 11. doi: 10.1158/2326-6066.CIR-13-0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crome SQ, Nguyen LT, Lopez-Verges S, Yang SYC, Martin B, Yam JY, Johnson DJ, Nie J, Pniak M, Yen PH, et al. A distinct innate lymphoid cell population regulates tumor-associated T cells. Nat Med. 2017;23(3):368–375. doi: 10.1038/nm.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du N, Zhou J, Lin X, Zhang Y, Yang X, Wang Y, Shu Y. Differential activation of NK cells by influenza A pseudotype H5N1 and 1918 and 2009 pandemic H1N1 viruses. J Virol. 2010;84(15):7822–7831. doi: 10.1128/JVI.00069-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zitvogel L, Tesniere A, Kroemer G. Cancer despite immunosurveillance: immunoselection and immunosubversion. Nat Rev Immunol. 2006;6(10):715–727. doi: 10.1038/nri1936. [DOI] [PubMed] [Google Scholar]
- 32.Platonova S, Cherfils-Vicini J, Damotte D, Crozet L, Vieillard V, Validire P, André P, M-C D-N, Alifano M, J-F R, et al. Profound coordinated alterations of intratumoral NK cell phenotype and function in lung carcinoma. Cancer Res. 2011;71(16):5412–5422. doi: 10.1158/0008-5472.CAN-10-4179. [DOI] [PubMed] [Google Scholar]
- 33.Le Maux Chansac B, Moretta A, Vergnon I, Opolon P, Lécluse Y, Grunenwald D, Kubin M, Soria J-C, Chouaib S, Mami-Chouaib F. NK cells infiltrating a MHC class I-deficient lung adenocarcinoma display impaired cytotoxic activity toward autologous tumor cells associated with altered NK cell-triggering receptors. J Immunol. 2005. 175(9):5790–5798. [DOI] [PubMed] [Google Scholar]
- 34.Carrega P, Morandi B, Costa R, Frumento G, Forte G, Altavilla G, Ratto GB, Mingari MC, Moretta L, Ferlazzo G. Natural killer cells infiltrating human nonsmall-cell lung cancer are enriched in CD56 bright CD16(-) cells and display an impaired capability to kill tumor cells. Cancer. 2008;112(4):863–875. doi: 10.1002/cncr.23239. [DOI] [PubMed] [Google Scholar]
- 35.Bruno A, Focaccetti C, Pagani A, Imperatori AS, Spagnoletti M, Rotolo N, Cantelmo AR, Franzi F, Capella C, Ferlazzo G, et al. The proangiogenic phenotype of natural killer cells in patients with non-small cell lung cancer. Neoplasia N. Y. N.. 2013;15(2):133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grzywacz B, Kataria N, Verneris MR. CD56(dim)CD16(+) NK cells downregulate CD16 following target cell induced activation of matrix metalloproteinases. Leukemia. 2007;21(2):356–359. doi: 10.1038/sj.leu.2404499. [DOI] [PubMed] [Google Scholar]
- 37.Romee R, Foley B, Lenvik T, Wang Y, Zhang B, Ankarlo D, Luo X, Cooley S, Verneris M, Walcheck B, et al. NK cell CD16 surface expression and function is regulated by a disintegrin and metalloprotease-17 (ADAM17). Blood. 2013;121(18):3599–3608. doi: 10.1182/blood-2012-04-425397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nielsen CM, White MJ, Goodier MR, Riley EM. Functional Significance of CD57 Expression on Human NK Cells and Relevance to Disease. Front Immunol. 2013;4:422. doi: 10.3389/fimmu.2013.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krzywinska E, Cornillon A, Allende-Vega N, Vo D-N, Rene C, Lu Z-Y, Pasero C, Olive D, Fegueux N, Ceballos P, et al. CD45 Isoform Profile Identifies Natural Killer (NK) Subsets with Differential Activity. PloS One. 2016;11(4):e0150434. doi: 10.1371/journal.pone.0150434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Celis-Gutierrez J, Boyron M, Walzer T, Pandolfi PP, Jonjić S, Olive D, Dalod M, Vivier E, Nunès JA. Dok1 and Dok2 proteins regulate natural killer cell development and function. Embo J. 2014;33(17):1928–1940. doi: 10.15252/embj.201387404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Penack O, Gentilini C, Fischer L, Asemissen AM, Scheibenbogen C, Thiel E, Uharek L. CD56dimCD16neg cells are responsible for natural cytotoxicity against tumor targets. Leukemia. 2005;19(5):835–840. doi: 10.1038/sj.leu.2403704. [DOI] [PubMed] [Google Scholar]
- 42.Takahashi E, Kuranaga N, Satoh K, Habu Y, Shinomiya N, Asano T, Seki S, Hayakawa M. Induction of CD16+ CD56bright NK Cells with Antitumour Cytotoxicity not only from CD16− CD56bright NK Cells but also from CD16− CD56dim NK Cells. Scand J Immunol. 2007;65(2):126–138. doi: 10.1111/j.1365-3083.2006.01883.x. [DOI] [PubMed] [Google Scholar]
- 43.Mandelboim O, Porgador A. NKp46. Int J Biochem Cell Biol. 2001;33(12):1147–1150. doi: 10.1016/S1357-2725(01)00078-4. [DOI] [PubMed] [Google Scholar]
- 44.Glasner A, Levi A, Enk J, Isaacson B, Viukov S, Orlanski S, Scope A, Neuman T, Enk CD, Hanna JH, et al. NKp46 Receptor-Mediated Interferon-γ Production by Natural Killer Cells Increases Fibronectin 1 to Alter Tumor Architecture and Control Metastasis. Immunity. 2018;48(2):396–398. doi: 10.1016/j.immuni.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fend L, Rusakiewicz S, Adam J, Bastien B, Caignard A, Messaoudene M, Iribarren C, Cremer I, Marabelle A, Borg C, et al. Prognostic impact of the expression of NCR1 and NCR3 NK cell receptors and PD-L1 on advanced non-small cell lung cancer. Oncoimmunology. 2017;6(1):e1163456. doi: 10.1080/2162402X.2016.1163456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fregni G, Perier A, Pittari G, Jacobelli S, Sastre X, Gervois N, Allard M, Bercovici N, Avril MF, Caignard A. Unique functional status of natural killer cells in metastatic stage IV melanoma patients and its modulation by chemotherapy. Clin Cancer Res. 2011;17(9):2628–2637. doi: 10.1158/1078-0432.CCR-10-2084. [DOI] [PubMed] [Google Scholar]
- 47.Castriconi R, Cantoni C, Della Chiesa M, Vitale M, Marcenaro E, Conte R, Biassoni R, Bottino C, Moretta L, Moretta A. Transforming growth factor beta 1 inhibits expression of NKp30 and NKG2D receptors: consequences for the NK-mediated killing of dendritic cells. Proc Natl Acad Sci. 2003;100(7):4120–4125. doi: 10.1073/pnas.0730640100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pietra G, Manzini C, Rivara S, Vitale M, Cantoni C, Petretto A, Balsamo M, Conte R, Benelli R, Minghelli S, et al. Melanoma Cells Inhibit Natural Killer Cell Function by Modulating the Expression of Activating Receptors and Cytolytic Activity. Cancer Res. 2012;72(6):1407–1415. doi: 10.1158/0008-5472.CAN-11-2544. [DOI] [PubMed] [Google Scholar]
- 49.Groh V, Wu J, Yee C, Spies T. Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature. 2002;419(6908):734–738. doi: 10.1038/nature01112. [DOI] [PubMed] [Google Scholar]
- 50.Salih HR, Rammensee H-G SA. Cutting edge: down-regulation of MICA on human tumors by proteolytic shedding. J Immunol. 2002. 169(8):4098–4102. [DOI] [PubMed] [Google Scholar]
- 51.Loyon R, Picard E, Mauvais O, Queiroz L, Mougey V, Pallandre J-R, Galaine J, Mercier-Letondal P, Kellerman G, Chaput N, et al. IL-21-Induced MHC Class II+ NK Cells Promote the Expansion of Human Uncommitted CD4+ Central Memory T Cells in a Macrophage Migration Inhibitory Factor-Dependent Manner. J Immunol. 2016;197(1):85–96. doi: 10.4049/jimmunol.1501147. [DOI] [PubMed] [Google Scholar]
- 52.Martín-Fontecha A, Thomsen LL, Brett S, Gerard C, Lipp M, Lanzavecchia A, Sallusto F. Induced recruitment of NK cells to lymph nodes provides IFN-gamma for T(H)1 priming. Nat Immunol. 2004;5(12):1260–1265. doi: 10.1038/ni1138. [DOI] [PubMed] [Google Scholar]
- 53.Godet Y, Fabre E, Dosset M, Lamuraglia M, Levionnois E, Ravel P, Benhamouda N, Cazes A, Le Pimpec-Barthes F, Gaugler B, et al. Analysis of spontaneous tumor-specific CD4 T-cell immunity in lung cancer using promiscuous HLA-DR telomerase-derived epitopes: potential synergistic effect with chemotherapy response. Clin Cancer Res. 2012;18(10):2943–2953. doi: 10.1158/1078-0432.CCR-11-3185. [DOI] [PubMed] [Google Scholar]
- 54.Beziaud L, Mansi L, Ravel P, Marie-Joseph EL, Laheurte C, Rangan L, Bonnefoy F, Pallandre J-R, Boullerot L, Gamonet C, et al. Rapalogs Efficacy Relies on the Modulation of Antitumor T-cell Immunity. Cancer Res. 2016;76(14):4100–4112. doi: 10.1158/0008-5472.CAN-15-2452. [DOI] [PubMed] [Google Scholar]
- 55.Kim S, François E, André T, Samalin E, Jary M, Hajbi FE, Baba-Hamed N, Pernot S, Kaminsky M-C, Bouché O, et al. Docetaxel, cisplatin, and fluorouracil chemotherapy for metastatic or unresectable locally recurrent anal squamous cell carcinoma (Epitopes-HPV02): a multicentre, single-arm, phase 2 study. Lancet Oncol. 2018;19(8):1094–1106. doi: 10.1016/S1470-2045(18)30321-8. [DOI] [PubMed] [Google Scholar]
- 56.Wang S, Han X, Hu X, Wang X, Zhao L, Tang L, Feng Y, Wu D, Sun Y, Shi Y. Clinical significance of pretreatment plasma biomarkers in advanced non-small cell lung cancer patients. Clin Chim Acta Int J Clin Chem. 2014;430:63–70. doi: 10.1016/j.cca.2013.12.026. [DOI] [PubMed] [Google Scholar]
- 57.Xu L, Huang Y, Tan L, Yu W, Chen D, Lu C, He J, Wu G, Liu X, Zhang Y. Increased Tim-3 expression in peripheral NK cells predicts a poorer prognosis and Tim-3 blockade improves NK cell-mediated cytotoxicity in human lung adenocarcinoma. Int Immunopharmacol. 2015;29(2):635–641. doi: 10.1016/j.intimp.2015.09.017. [DOI] [PubMed] [Google Scholar]
- 58.Laheurte C, Galaine J, Beziaud L, Dosset M, Kerzerho J, Jacquemard C, Gaugler B, Ferrand C, Dormoy A, Aubin F, et al. Immunoprevalence and magnitude of HLA-DP4 versus HLA-DR-restricted spontaneous CD4(+) Th1 responses against telomerase in cancer patients. Oncoimmunology. 2016;5(5):e1137416. doi: 10.1080/2162402X.2015.1137416. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


