Abstract
Most anaplastic lymphoma kinase (ALK)-rearranged non-small cell lung tumours initially respond to small molecule ALK inhibitors but drug resistance often develops1–4. After tumours develop resistance to highly potent 2nd generation ALK inhibitors, approximately half harbor ALK resistance mutations, while the other half have other mechanisms of resistance. The latter often have activation of at least one of several different tyrosine kinases driving resistance5. Such tumours are not expected to respond to the 3rd generation ALK inhibitor, lorlatinib, which is able to overcome all clinically identified ALK resistance mutations5,6 and further therapeutic options are limited5. Herein, we deployed an shRNA screen of 1000 genes in multiple ALK inhibitor resistant patient derived cells (PDC) to discover sensitizers to ALK inhibition. This approach identified SHP2, a non-receptor protein tyrosine phosphatase, as a common targetable resistance node in multiple PDCs. SHP2 provides a parallel survival input downstream of multiple tyrosine kinases that promote resistance to ALK inhibitors. The recently discovered small molecule SHP2 inhibitor, SHP099, in combination with the ALK TKI (tyrosine kinase inhibitor), ceritinib, halted the growth of resistant PDCs by preventing compensatory RAS and ERK1/2 reactivation. These findings suggest that combined ALK and SHP2 inhibition may be a promising therapeutic strategy for resistant cancers driven by several different ALK-independent resistance mechanisms.
Multiple molecular mechanisms can lead to acquired resistance to ALK inhibitors. Secondary ALK kinase domain mutations are observed in 20% and 50% of patients after 1st (crizotinib) and 2nd (ceritinib, alectinib) generation ALK inhibitors, respectively5,7–9. Other resistance mechanisms include ALK gene amplification8,10,11 and activation of alternate kinases (EGFR10,12, KIT10, SRC13 and IGF1R14). These tyrosine kinases cause resistance by maintaining activation of downstream ERK and/or PI3K-AKT signaling despite ALK inhibition10,12–14.
The highly potent and selective 3rd generation ALK inhibitor lorlatinib, which is currently in clinical trials, is active against all known secondary ALK kinase domain mutations that confer resistance to 1st and 2nd generations ALK inhibitors in preclinical models5,6. However, lorlatinib fails to suppress growth of resistant cells with alternate kinase bypass signaling pathways5, and new therapeutic options are needed for these patients. The heterogeneity of resistance mechanisms makes clinical development of new candidate therapeutic strategies challenging, and this is compounded by the fact that many are not readily identified through genomic analyses13.
We sought to identify a treatment approach that could more broadly address acquired resistance in cases without ALK secondary mutations. First, we performed a pooled short hairpin (shRNA) dropout screen on a panel of seven ALK-rearranged non-small cell lung cancer (NSCLC) cell lines generated directly from patients who developed resistance to crizotinib or ceritinib in clinic (Supplementary Table 1). Cells were infected with a lentiviral shRNA library (termed tDRIVE) targeting ~1000 cancer related genes (20 hairpins per gene, see Supplementary Table 2 for complete gene list), and treated with the ALK inhibitor ceritinib or vehicle for 14 days (Fig. 1a). To identify shRNAs that were selectively depleted after ceritinib treatment, we quantified the abundance of barcoded shRNA vectors by next-generation sequencing and calculated the relative fold depletion after ceritinib treatment compared to vehicle treatment. We identified members of known resistance pathways13 and additional putative resistance drivers including kinases (EGFR, FGFR1, ERBB2, SRC, MAPK1, RAF1), phosphatases (PTPN11), the adaptor protein FRS2, and several transcription factors such as MYC (Fig. 1b). The full list of identified hits is shown in Supplementary Table 3. All cell lines used in the shRNA screen underwent targeted NGS of 1000 known cancer genes (Supplementary Table 4)5,13. Notably, no activating genetic alterations were identified in the genes scored in the shRNA screen or in their pathways, underlying the challenge of identifying therapeutic alternatives for resistant tumors solely based on DNA sequencing5,13.
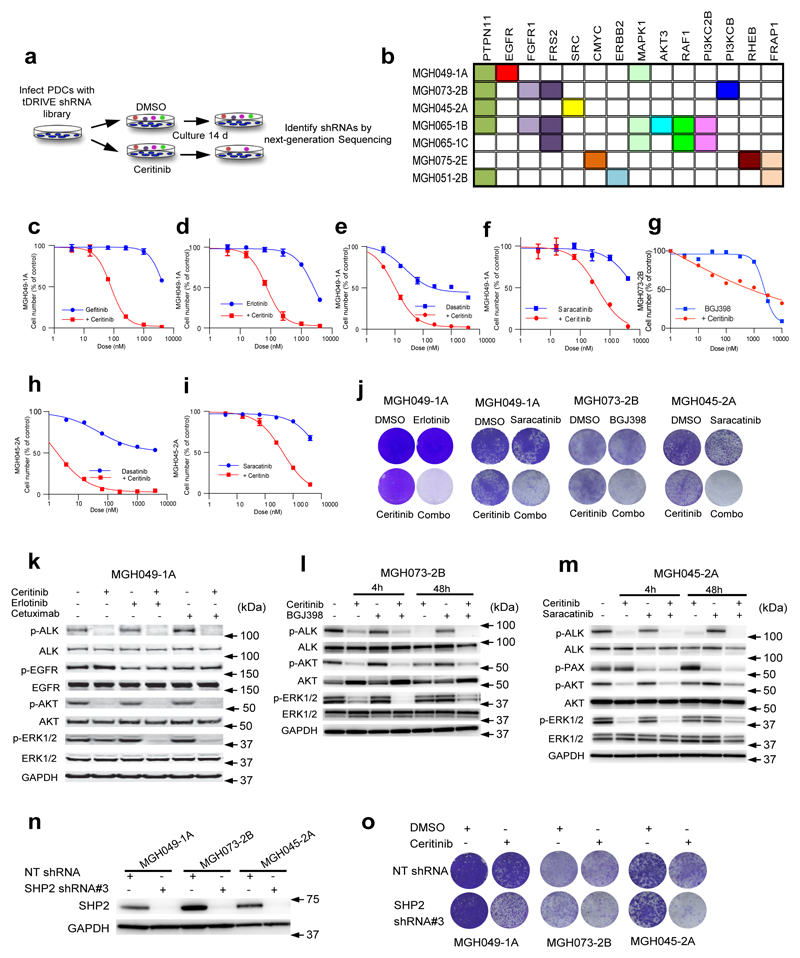
Figure 1. Complementary shRNA screens and high-throughput drug combination screens identify therapeutic approaches to overcome resistance in ALK-rearranged PDCs.
(a) Schema depicting the tDRIVE pooled shRNA screen to identify sensitizers to ceritinib. (b) Top gene candidates driving resistance to ceritinib identified by the pooled shRNA screen (please see raw data in Supplementary Table 3). Each gene is color-coded as indicated. (c-f) EGFR or SRC inhibition restore sensitivity to ALK inhibition in MGH049-1A cells. Primary drug screen data depicting the shift in the dose response to gefitinb, erlotinib, dasatinib or saracatinib in the presence of ceritinb (500 nM). (g) FGFR inhibition restores sensitivity to ALK inhibition in MGH073-2B cells. Primary drug screen data depicting the shift in the dose response to BGJ398 in the presence of ceritinb (500 nM). (h,i) SRC inhibition restores sensitivity to ALK inhibition in MGH045-2A cells. Primary drug screen data depicting the shift in the dose response to dasatinib or saracatinib in the presence of ceritinib (500 nM). For figures c-i error bars are mean ± s.e.m. of n=3 thechnical replicates. (j) Long-term proliferation assay of MGH049-1A, MGH073-2B, and MGH045-2A that were grown in the presence of the indicated drugs (all used at 500nM) for 14 days followed by crystal violet staining. The image is representative of at least two independent experiments. (k) MGH049-1A cells were treated with ceritinib (500 nM), erlotinib (1 μM), or cetuximab (100 nM) or the indicated combinations for 4h. Lysates were analyzed by immunoblot to detect the indicated proteins. The image is representative of at least two independent experiments. (l,m) MGH073-2B and MGH045-2A cells were treated for 4h and 48h with the indicated drugs at 500 nM and lysates were analyzed by immunobloting using antibodies against the indicated proteins. The image is representative of at least two independent experiments. (n) Representative western blot analysis for SHP2 expression in MGH049-1A, MGH073-2B and MGH045-2A cells that were transduced with lentivirus encoding a SHP2-specific or non-targeting (NT) shRNA. SHP2 shRNA#3 is selected from pooled shRNA library. GAPDH was used as a loading control. Please see full scans of all blots in Supplementary figure 11-1. (o) Effect of SHP2 depletion on sensitivity to ceritinib in PDCs is shown in a colony formation assay. Cells were treated with indicated drugs for 14 days and stained by crystal violet. The image is representative of two independent experiments.
Second, in a complementary approach, we utilized combination drug screening strategy that is effective for identifying bypass resistance mechanisms13. We treated resistant patient-derived cell lines with a panel of 112 targeted agents alone or in combination with ceritinib. By comparing the ability of these drugs to suppress cell viability in the presence or absence of ceritinib13, we identified EGFR (in MGH049-1A), FGFR (in MGH073-2B) and SRC (in MGH045-2A and MGH049-1A) as key resistance mechanisms (Fig. 1c-i). These results were confirmed in long-term viability assays: erlotinib (EGFR inhibitor) and saracatinib (SRC inhibitor) restored sensitivity to ALK inhibition in MGH049-1A, BGJ398 (FGFR inhibitor) in MGH073-2B, and saracatinib in MGH045-2A (Fig. 1j). Overall, there was very good agreement between the results of the drug and shRNA screens. Expectedly however, not all drug screen hits were seen in the shRNA screen as multiple paralogues can often be targeted by drugs but not through gene specific shRNA strategies. Thus, although SRC family small molecule inhibitors were found to sensitize MGH049-1A to ceritinib (Fig. 1e,f,j)13, SRC or other related kinases were not shRNA screen hits. In support of the high sensitivity and specificity of the shRNA screens, none of the 4 cell lines with no FGFR or related pathway hits (FRS2) were sensitive to the ceritinib plus BGJ398 combination (Supplementary Fig. 1a).
Analysis of downstream signaling revealed that ERK reactivation rapidly occurred after ceritinib treatment in multiple ALK-positive models, and that effective combinations suppressed this reactivation. In MGH049-1A cells, ceritinib effectively suppressed ERK1/2 phosphorylation one hour after drug treatment, however, ERK1/2 reactivation was observed after four hours (Supplementary Fig. 1b). Co-treatment with either erlotinib or cetuximab blocked reactivation of ERK1/2 (Fig. 1k). In the MGH073-2B and MGH045-2A cell lines, ceritinib treatment led to acute suppression of phospho-ERK1/2, but ERK1/2 reactivation was seen by 48 hours (Fig. 1l-m). Addition of the appropriate second kinase inhibitor (corresponding to the respective drug screen and shRNA screen hits) overcame this rebound reactivation and led to durable ERK1/2 suppression (Fig. 1l-m). Together, these results confirm that targeting bypass signaling pathways may be a useful strategy against these resistant cancers. However, these results also demonstrate that distinct resistant cancers use different tyrosine kinases to drive resistance, underscoring the challenge of developing a single therapeutic approach that would be effective against a majority of resistant cancers.
In addition to the different kinases driving resistance in each cell line, the shRNA screen revealed that depletion of SHP2 (PTPN11) re-sensitized multiple PDCs to ceritinib (Fig. 1b). SHP2, an SH2 domain containing tyrosine phosphatase, mediates GTP loading of RAS downstream of multiple tyrosine kinases including EGFR, FGFR and SRC15–23. Thus, we hypothesized that SHP2 might serve as a common signaling node in cells with different resistance drivers leading to reactivation of downstream RAS/MAPK pathway signaling. To test whether ALK TKI resistant cells have up-regulation of SHP2 compared to ALK TKI sensitive cells, we compared SHP2 expression level in both sensitive and resistant models and did not observe significant differences (Supplementary Fig. 2a). We further expressed SHP2 in sensitive cells and showed that SHP2 expression also did not confer resistance to ceritinib in these cells (Supplementary Fig. 2b-c). These data suggest that SHP2 expression levels do not predict sensitivity to ALK inhibition. To validate the initial screen results, we used two independent shRNAs to knockdown SHP2 in MGH049-1A (EGFR bypass), MGH073-2B and MGH065-1B (FGFR bypass), and MGH045-2A (SRC bypass) (Fig. 1n, Supplementary Fig. 2d). SHP2 depletion increased sensitivity to ceritinib in all models (Fig. 1o, Supplementary Fig. 2e). To test whether pharmacological inhibition of SHP2 might re-sensitize resistant cell lines to ceritinib (the original combination drug screen did not include a SHP2 inhibitor), we utilized SHP099, a highly potent and selective SHP2 allosteric inhibitor24,25. As a single agent, SHP099 had minimal effect on cell proliferation (consistent with the shRNA results), however the combination of SHP099 and ceritinib led to marked inhibition of colony formation in all PDCs examined (Fig. 2a, Supplementary Fig. 3a). Moreover, in MGH049-1A and MGH045-2A, SHP099 enhanced the apoptotic activity of ceritinib. In MGH073-2B and MGH065-1B, combination of SHP099 and ceritinib did not induced apoptosis and rather blocked cell proliferation (Supplementary Fig. 3b).
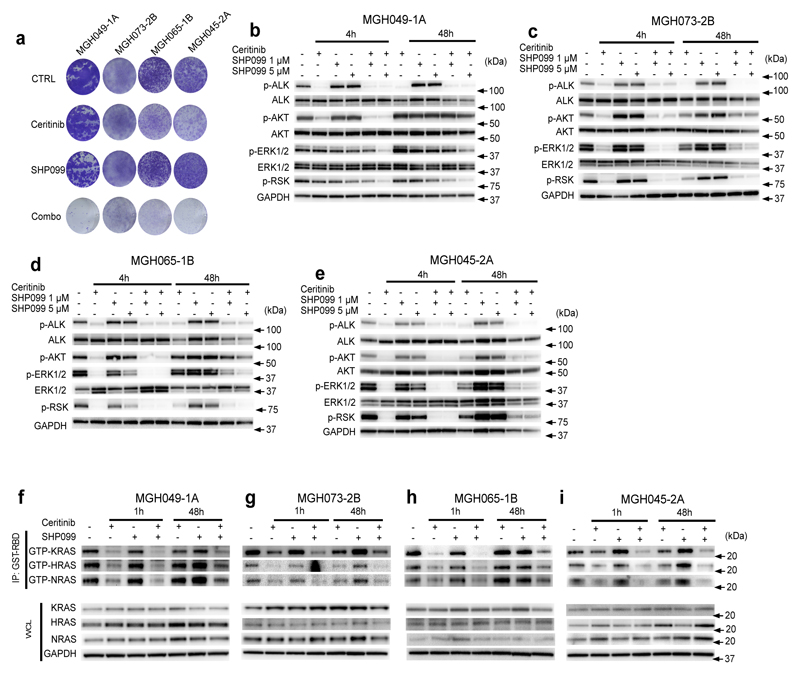
Figure 2. The SHP2 inhibitor, SHP099, attenuates ceritinib-induced ERK reactivation and increases ceritinib efficacy in vitro.
(a) Long-term proliferation assay of MGH049-1A, MGH073-2B, MGH065-1B and MGH045-2A cells treated with ceritinib (500 nM), SHP099 (5 μM) or a combination of those for 14 days. Cells were stained using crystal violet. The image is representative of three independent experiments. (b-e) PDCs were treated with 500 nM ceritinib, 1 or 5 μM of SHP099, or a combination thereof for the indicated time points. Lysates were analyzed by immunoblot to detect the indicated proteins. GAPDH was assessed as a loading control. The image is representative of three independent experiments. Please see full scans of all blots in Supplementary figure 11-2. (f-i) PDCs were treated with ceritinib (500 nM), SHP099 (5 μM), or their combination for 1hr and 48hr. Whole cell lysates (WCL) or GST-RBD-immunoprecipitated (IP) lysates were analyzed by immunoblotting using specific antibodies against indicated proteins. The image is representative of at least two independent experiments. Please see full scans of all blots in Supplementary figure 11-3.
We further tested the effect of SHP2 inhibition in ALK TKI sensitive cells, H3122 and MGH026-1. Similar to what we observed in ALK TKI resistant cells, SHP099 alone did not have anti-proliferative activity in these ceritinib sensitive cells. Furthermore, the addition of SHP099 did not substantially sensitize these cells to ceritinib (Supplementary Fig. 4a-b). Analysis of downstream signaling in H3122 cells revealed that ceritinib alone effectively suppressed ERK1/2 and AKT phosphorylation and addition of SHP099 to ceritinib did not further downregulate pathway. Furthermore, SHP099 alone did not affect ERK1/2 or AKT activation status (Supplementary Fig. 4c).
Consistent with previous observations that SHP2 mediates RAS/MAPK pathway activation19,25,26, co-treatment of resistant cell lines with SHP099 and ceritinib resulted in durable suppression of phosphorylation of ERK1/2 and the ERK substrate p90 RSK (Fig. 2b-e). Consistent with the viability data, SHP099 alone (without ALK inhibition) did not suppress ERK activity at 48h. However, SHP2 inhibition blocked ERK1/2 reactivation following continued suppression of ALK by ceritinib. Consistent with the hypothesis that SHP099 blocks re-activation of ERK driven by RTKs, a short late time exposure (last 4h of 48h) with SHP099 was able to suppress ERK phosphorylation observed in the presence of continuous ALK inhibition (Supplementary Fig. 5a-d). We next examined the ERK1/2-dependent gene expression of DUSP6 (a phosphatase and negative regulator of ERK), ETV5, and SPRY2 as an additional readout of flux through the ERK signaling pathway, as this allows for a more sensitive measurement of ERK signaling status than p-ERK western blotting27. Compared to ceritinib alone, the combination treatment led to greater suppression of ERK-dependent transcripts (Supplementary Fig. 6). To further test the specificity of action of SHP099 on tyrosine kinase driven bypass resistance mechanisms, we utilized a PDC (MGH034-2A) harboring a MEK activating mutation (MAP2K1 K57N) as the mechanism of resistance13. In this model ERK activation should be maintained in the presence of ceritinib without the need for upstream RAS reactivation and thus should be impervious to the addition of SHP099. Consistent with this hypothesis, phospho-ERK and phospho-RSK were maintained in MGH034-2A cells after treatment with the certinib plus SHP099 combination (Supplementary Fig. 7a).
There were some other notable findings in the screens. FGFR1 was a shRNA screen hit in MGH065-1B. However, in the MGH065-1C cells (generated from the MGH065-1B cultured in croztinib), FRS2, an adaptor downstream of FGFR, was a hit but FGFR1 was not. Notably, the FGFR inhibitor BGJ398 that acts on FGFR1/2/3 sensitized this model to ceritinib, suggesting that other FGF receptors could mediate resistance in addition to FGFR1 in this model (Supplementary Fig. 8a-c). Consistent with other cases of FGFR driven resistance, SHP099 could sensitize MGH065-1C cells to ceritinib and suppress ERK reactivation (Supplementary Fig. 8d-e). It is not clear why the shRNA screen failed to detect PTPN11 as a hit in this cell line. MGH075-2E was not sensitized by PTPN11 in the shRNA screen, consistent with the lack of RTK hits and was the only model with a MYC hit. We confirmed that SHP099 did not affect viability or inhibit ERK activity rebound at 48h in this model (Supplementary Fig. 7b-c). Interestingly, previous work had shown that in some triple negative breast cancer models, suppression of ERK yielded to MYC degradation, RTK upregulation and adaptive resistance28. However, MYC downregulation did not appear to support ERK reactivation in our context (Supplementary Fig 7d). Taken together, our results demonstrate that targeted SHP2 inhibition can overcome resistance in multiple contexts of tyrosine kinase mediated bypass signaling leading to survival in the presence of ALK inhibition, most likely through the suppression of ERK signaling.
EML4-ALK was recently shown to promote cell survival in NSCLC by activation of MAPK pathway signaling via all three RAS isoforms (KRAS, HRAS, NRAS)29. To better understand the relationship between RAS isoforms, ALK and SHP2 in the resistant models, we performed GTP-RAS pull down assays using the RAS binding domain of RAF. Consistent with a previous report29, ALK inhibition led to initial loss of GTP loading of all three RAS isoforms in all models (Fig. 2f-i). Neither short nor long exposure to single-agent SHP099 alone decreased RAS activity in any of the PDCs. In contrast, short-term treatment with the combination of ceritinib and SHP099 further decreased the level of GTP-RAS in all models compared to ALK inhibition alone. After 48 hours of single-agent ceritinib treatment, all three RAS isoforms were reactivated to varying degrees in MGH049-1A, MGH073-2B, MGH065-1B and MGH045-2A cells (consistent with the reactivation of ERK signaling observed with single-agent ceritnib). Notably, addition of SHP099 invariably led to greater suppression of all RAS isoforms in all PDCs (Fig. 2f-i). It is clear that SHP2 can modulate ERK1/2 activity downstream of several tyrosine kinases19. Interestingly, short time treatment with SHP099 did substantially impact ERK activity in the PDCs. In addition, SHP099 did not suppress ERK activity in ceritinib sensitive models (Supplementary Fig. 4c). Taken together, our results suggest that SHP2 is not a key modulator of RAS and ERK activity downstream of EML4-ALK, but rather mediates RAS and ERK reactivation downstream of a diverse set of tyrosine kinases activated upon EML4-ALK inhibition.
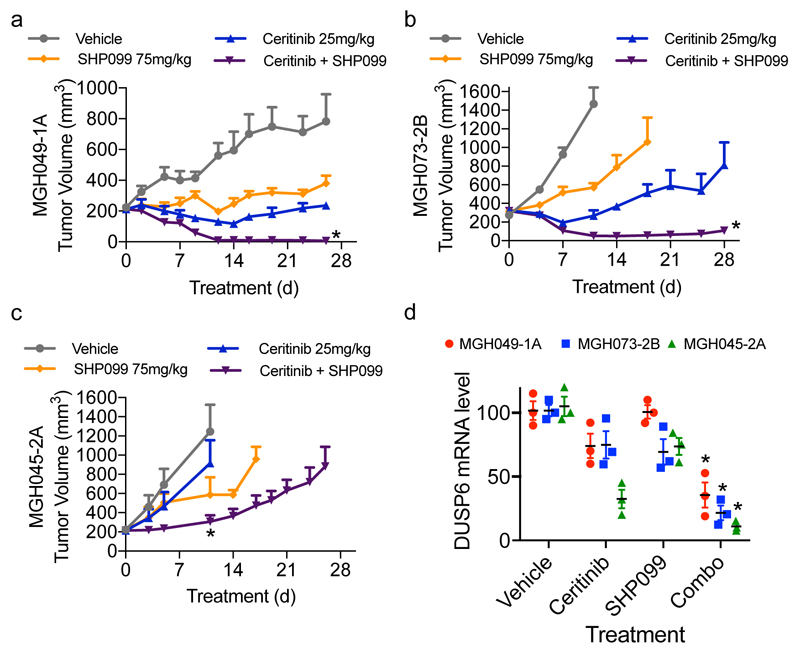
To evaluate the in vivo efficacy of combined SHP2 and ALK inhibitors, we treated mice bearing PDC subcutaneous xenografts with SHP099, ceritinib or their combination. Treatment of MGH049-1A and MGH073-2B xenograft tumors with ceritinib resulted in modest and short-lived responses while MGH045-2A xenografts were completely resistant (Fig. 3a-c). The combination of SHP099 and ceritinib resulted in profound tumor regression in MGH049-1A and MGH073-2B and modestly reduced the tumor growth in MGH045-2A (Fig. 3a-c), and coincided with significantly decreased DUSP6 mRNA levels (Fig. 3d). Similar results were observed when combining ceritinib with the EGFR antibody cetuximab or the FGFR inhibitor BGJ398, targeting the specific bypass mechanisms in MGH049-1A and MGH073-2B cells, respectively (Supplementary Fig. 9a-b). We also evaluated tumor regrowth after withdrawal of the combination of SHP099 and ceritinib. Tumors that had regressed did re-grow after treatment cessation although growth was slow at least during the 22 days of follow up in the MGH-073-B model. We tested this in another model (MGH049-1A) and found that tumors also grew back upon drug withdrawal but remained sensitive to drug retreatment (Supplementary Fig. 9 c-d). As reported25, single-agent SHP099 was well tolerated with minor or no body weight loss over the course of treatment. The SHP099 plus ceritinib exhibited minor toxicity following the initiation of the treatment with no more than 7% body weight loss. However, over the course of treatment toxicity decreased (Supplementary Fig. 10a-c). The same pattern of toxicity was observed in BGJ398 plus ceritinib treated mice (Supplementary Fig. 10b). Thus, SHP2 inhibition suppresses reactivation of ERK1/2 downstream of different tyrosine kinases leading to tumor regression in vivo, suggesting that this combination approach might be broadly useful against acquired resistance to ALK inhibitors.
Figure 3. Combined inhibition of ALK and SHP2 enhances response and overcomes resistance in drug resistant ALK-rearranged PDCs in vivo.
(a) The combination of ceritinib and SHP099 more effectively treats MGH049-1A subcutaneous xenografts than either drug alone. Mice were treated with vehicle, ceritinib (25 mg/kg, daily), SHP099 (75 mg/kg, daily) or the combination of both drugs. Changes in tumor volume were measured twice weekly during the course of treatment. Error bars are mean ± s.e.m. *p<0.05 vs vehicle, SHP099 by one way ANOVA followed by Tukey test. n=5 mice per group. (b) MGH073-2B subcutaneous xenografts grown in mice were treated as indicated: vehicle (n=5 mice), ceritinib 25 mg/kg daily (n=5 mice), SHP099 75 mg/kg daily (n=5 mice), or the combination of both (n=8 mice). Error bars are mean ± s.e.m. *p<0.05 vs control by one way ANOVA followed by Dunnett’s test. (c) MGH045-2A xenografts were treated as in (b); n=4 mice per group. *p<0.05 vs vehicle by one way ANOVA followed by Tukey test. (d) Mice bearing MGH049-1A,MGH073-2B or MGH045-2A subcutaneous tumors were administered with vehicle, ceritinib (25 mg/kg), SHP099 (75 mg/kg) or the combination of both drugs for 3 days. n=3 mice per condition were used. Tumors were collected 3h after the last treatment. DUSP6 transcription was assessed by quantitative RT-PCR. Error bars are mean ± s.e.m. *p<0.01 vs vehicle or ceritinib or SHP099 by two-tailed Student’s t-test.
In conclusion, our study suggests that inhibition of SHP2 may abrogate RAS and ERK1/2 reactivation following treatment with an ALK inhibitor in NSCLC. Combined ALK and SHP2 inhibition may provide a broad reaching therapeutic strategy for overcoming or preventing heterogeneous ALK-independent mechanisms of acquired drug resistance in patients with ALK-rearranged positive NSCLC.
Online Methods
Cell Culture and compounds
Patient-derived cell lines were established as previously described1. MGH034-2A, MGH049-1A, MGH026-1A, and MGH075-2E patient-derived cell lines were previously described5,13. MGH073-2B was developed from crizotinib-resistant pleural effusion. MGH065-1B was developed from crizotinib-resistant lymph node biopsy. MGH065-1C was developed from MGH065-1B in media containing 300 nM crizotinib. MGH051-2B was established from ceritinib-resistant liver biopsy. MGH045-2A was developed from crizotinib-resistant pleural effusion. All patients signed informed consent to participate in a Dana-Farber-Harvard Cancer Center Institutional Review Board-approved protocol giving permission for research to be performed on their samples. Cell lines were sequenced to confirm the presence of ALK rearrangements identified by clinical testing of biopsy specimens from the same patients and tested negative for mycoplasma contamination. Additional authentication was performed for MGH049-1A, MGH045-2A, MGH051-2B, MGH034-2A, and MGH065-1B by SNP fingerprinting. Cells were grown either in DMEM or in RPMI-1640 (Corning) supplemented with 10% FBS and 1X Antibiotic-Antimycotic. Ceritinib, BGJ398 and SHP099 were synthesized in the Global Discovery Chemistry Department at NIBR (Novartis). Saracatinib, erlotinib and gefitinib were purchased from Selleckchem. Each compound was dissolved in DMSO for cell culture experiments. Cetuximab (eribitux) was obtained from Imclone LLC, Eli Lilly subsidiary.
Drug screen
Combination drug screen consisting of 112 agents was performed as previously described1. Cells were treated with vehicle or varying concentrations of drugs to be screened in the absence or presence of 300 nM ceritinib. After incubation for 5 days in drug, cell counts were determined using CellTiter-Glo (Promega) as per manufacturer’s instructions. GraphPad Prism version 5.0 was used to graphically display data by a nonlinear regression model utilizing a four-parameter analytic method.
tDRIVE shRNA screen and next generation sequencing
The synthetic lethal pooled shRNA screen was conducted using a custom-made shRNA library (tDRIVE) containing 20,000 hairpins that target 1000 known cancer genes with 20 shRNAs per gene. Cells were transduced with tDRIVE lentivirus at MOI~0.3 using spin infection protocol (1hr at 2100 rpm), selected with puromycin and expanded to 60 million cells. These cells were then divided into three equivalent pools of 20 million cells (1000 cells per shRNA): Day 0, DMSO vehicle and ceritinib treated groups. Day 0 cells were harvested at the time of treatment start and snap-frozen using liquid nitrogen. The other 2 pools were treated with DMSO vehicle or 500 nM ceritinib for 14 days and then next generation sequencing was performed to determine hairpin distribution. For each individual cell line, genes for further functional studies were selected based on the fold depletion of abundance of shRNAs targeting these genes in ceritinib-treated samples compared with that in DMSO-treated samples. P values based on the RSA statistic for ceritinib-treated cell lines relative to DMSO-treated cell lines were calculated by pooling the counts of barcodes in each cell line. In addition, a z-score was calculated using the mean and standard deviation for the fold change in counts of the shRNAs in the library. A z-score for each gene was calculated as the mean of the z-scores of the shRNAs targeting the gene.
Infection and transfection
Custom packaged lentiviral particles carrying sh_hPTPN11 #165861 (shRNA#3, Hairpin sequence:ACCGGCGGTTTGATTCTTTGATAGATGTTAATATTCATAGCATCTGTCAAAGAATCAAACCGTTTT), sh_hPTPN11 #165860 (shRNA#4, Hairpin sequence: ACCGGGCAATGATGGCAAGTTTAAAGGTTAATATTCATAGCCTTTAGACTTGCCGTCATTGCTTTT), or shNT (non-targeting control in pRSI16-U6-sh-ubic-TagRFP-2A-Puro) were obtained from Cellecta. Cells were seeded into six-well plates at density of 3 X 105 cells/well. 24 hours later, cells were infected with indicated lentiviral particles. Cells were selected with puromycin 48 hours after viral transduction before using for experiments.
SMARTpool ON-TARGET plus MYC siRNA was obtained from Dharmacon. Cells were seeded into six-well plates at density of 2 X 105 cells/well. 24 hours later, cells were transfected with indicated siRNA according to manufacturer instructions. Cells were used for experiments 48 hours post transfection. Production of doxycycline (dox)-inducible wild-type streptavidin-binding peptide tagged (SBP) SHP2 viral particles performed as described25. Cells were seeded into six-well plates at density of 3 X 105 cells/well. 24 hours later, cells were infected with SBP-SHP2 wild-type viral particles. Cells were selected with G418 48 hours after viral transduction before using for experiments.
Antibodies and immunoblotting
Cells were seeded in 6-well plates and treated with indicated agents for specified timepoints. Lysates were prepared as previously described, and equal volumes of total cell lysate were processed for immunoblotting10. Antibodies against phospho-ALK Y1282/1283 (9687), ALK (3633), phospho-AKT S473 (4060), AKT (4691), Phospho-ERK T202/Y204 (9101), ERK (9102), Phospho-Paxillin Y118 (2541), SHP2 (Cell Signaling 3752), and EGFR (4267), were obtained from Cell Signaling Technology and used at 1:1000 dilution. Antibodies against Phospho-EGFR Tyr1068 (AB5644) and Phospho-RSK 359/S363 (AB32413) were purchased from Abcam and used at 1:1000 dilution. GAPDH (MAB374) antibody was purchased from Millipore and used at 1:5000 dilution. K-RAS (F234), H-RAS (F235) and N-RAS (F155) antibodies were purchased from Santa Cruz and used at 1:500 dilution. All secondary antibodies (anti-mouse IgG HRP-linked (7076S) and anti-rabbit IgG HRP-linked (7074S)) were purchased from Cell Signaling and used at 1:50000 dilution.
RAS activation assay
Cells were seeded in 10 cm dishes and treated with indicated agents for specified timepoints. The RAS GST-RBD activation assay was performed using RAS activation assay kit (Cytoskeleton, Catalog No. BK008) according to manufacturer instructions.
Crystal violet assay
Cells were seeded at a density of 5,000 - 10,000/well in 12 well plates and were drugged the following day. Media and drug were replaced every 72 hours for 12 days. Cells were fixed with glutaraldehyde for 10 minutes, washed 2x with H2O and stained with 0.1% crystal violet (Sigma) for 30 minutes. Cells were then washed 3x with H2O and plates were dried overnight.
Quantitative PCR
Cells were seeded 24 h before give a confluency of 50%. Cells were treated with drugs for 4h and 48h and RNA was extracted using the RNeasy Kit (Qiagen). Likewise, RNA was extracted from tumor samples for pharmacodynamic analyses. cDNA was prepared from 500 ng total RNA with the First Strand Synthesis Kit (Invitrogen) using oligo-dT primers. Quantitative PCR was performed on a Light Cycler 480 (Roche) system using FastStart SYBR green master mix (Roche). mRNA expression relative to actin mRNA levels was calculated using the delta-delta threshold cycle (ΔΔCT) method. Primers used: SPRY2 F 5’-TTGCACATCGCAGAAAGAAG-3’, R 5’-GGTCACTCCAGCAGGCTTAG-3’; ETV5 F 5’-CCTACATGAGAGGGGGTTATTTC-3’, R 5’-CGTCAAAGTATAATCGGGGATCT-3’ DUSP6 F 5’-CGACTGGAACGAGAATACGG-3’, R 5’- TTGGAACTTACTGAAGCCACCT-3’; HPRT F 5’-TCAGGCAGTATAATCCAAAGATGGT-3’, R 5’- AGTCTGGCTTATATCCAACACTTCG-3’; SDHA F 5’-TGGGAACAAGAGGGCATCTG-3’, F 5’- CCACCACTGCATCAAATTGATG-3’; TBP F 5’-CACGAACCACGGCACTGATT-3’, R 5’-TTTTCTTGCTGCCAGTCTGGAC-3’.
Xenograft studies
All animal studies were conducted in accordance with the guidelines as published in the Guide for the Care and Use of Laboratory Animals and Novartis International Animal Care and Use Committee (IACUC) regulations and guidelines and by the Institutional Animal Care and Use Committee (IACUC) of Massachusetts General Hospital. Female Nu/Nu mice aged 6 to 8 weeks were obtained from Charles River laboratories Inc, Wilmington, MA. Mice were maintained in laminar flow units in sterile filter-top cages with Alpha-Dri bedding. The MGH045-2A and MGH073-2B cells were harvested during exponential growth. Each mouse was inoculated subcutaneously in the upper right flank with 5 X 106 cells suspended either in 0.2 mL cold PBS (MGH045-2A) or in 20% BD Matrigel Basement Membrane Matrix in PBS (MGH073-2B). The development of MGH049-1A xenograft tumors comprised 2 steps. In the first step, 1 X 107 MGH049-1A cells harvested during exponential growth were suspended in a 1:1 mixture of cold PBS and Matrigel in a total volume of 0.2 mL, and injected subcutaneously into the upper right flank of mice. Tumors established in this step were collected and fragmented. Tumor fragments were then implanted into the upper right flank of mice. Tumor volumes and mice weights were monitored twice per week. Tumor size was measured by caliper twice weekly and weights were determined at the same time. Volume was calculated as follows: (length*(width^2)*(0.51). When tumor volume reached approximately 200 mm3, mice were randomized and orally administered with vehicle, 25 mg/kg ceritinib (0.5% Methyl Cellulose and 0.5% Tween 80), 75 mg/kg SHP099 (0.5% Methyl Cellulose and 0.5% Tween 80), 30 mg/kg BGJ398 (50% Acetic Acid/Acetate Buffer (pH 4.68) and 50% PEG300), 25 mg/kg ceritinib plus 75 mg/kg SHP099 daily, or 25 mg/kg ceritinib plus 30 mg/kg BGJ398 daily respectively. Cetuximab 20 mg/kg was used ip twice a week as monotherapy or in combination with 25 mg/kg ceritinib. For pharmacodynamic analyses, tumor-bearing mice were administered with drugs or vehicle for 3 days. Tumor tissue was excised 3h after last treatment and snap-frozen in liquid nitrogen for DUSP6 level measurement by quantitative PCR. Ceritinib, SHP099, and BGJ398 were provided by Novartis. Cetuximab (eribitux) was obtained from Imclone LLC, Eli Lilly subsidiary. When combinations were administered 30 minutes elapsed between drug doses.
Next-Generation Sequencing:
Performed as previously described13. Briefly, RNA bait-based hybridization capture was performed to capture 1000 known cancer genes (RightOn Cancer Sequencing Kit, developed in collaboration with Elim BioPharma). Next-Generation sequencing data for MGH075-2E, MGH051-2B, MGH034-2A and MGH049-1A was previously reported5,13.
Statistical analyses
All analyses for invivo experiments were performed with SigmaPlot version 13.0. Invivo data sets were analyzed for statistical significance using either one way ANOVA followed by Tukey test or one way ANOVA followed by Dunnett’s test. GraphPad Prism version 6.0 was used to analyse invitro data. Invitro data sets were analyzed for statistical significance using an unpaired two-tailed Student’s t-test.
Supplementary Material
Acknowledgments
We thank D. Rakiec for help with next-generation sequencing and C. Liu for help with invivo experiments. This study was supported by grants from Novartis Institutes for BioMedical Research, AACR-AstraZeneca Fellowship in Lung Cancer Research (17-40-12-DARD to L.D.), National Cancer Institute (R01CA164273 to A.T.S.), a grant from the Wellcome Trust (102696 to C.H.B) and the National Foundation for Cancer Research (to ATS), by Be a Piece of the Solution, and by LungStrong.
Footnotes
Data Availability
The authors declare that the main data supporting the findings of this study are available within the article and its Supplementary Information files. A Life Sciences Reporting Summary is available.
Author contributions:
L.D., F.L., A.T.S., A.N.H., C.H.B. and J.A.E. designed the study and analyzed the data. L.D., C.H.B. and J.A.E. wrote the paper. G.C., F.L., J.A.E and C.H.B supervised the studies. L.D., F.L., M.S., P.F., S.Y., K.Y., Y.C., Y.C., E.L. performed cell line and biochemical studies. L.D., D.T.M., J.L., H.Q.W. performed tumor xenograft studies. M.P., R.H.D., and D.L. developed or maintained patient-derived cell lines. G.K., F.L., and D.R., performed shRNA screen analysis. M.S.L. and A.L. performed next generation sequencing analysis. J.F.G, I.D.J, L.F., E.L., S.M., H.H., M.M., M.L., J.W., K.H., G.C. were involved with study design. All authors discussed the results and commented on the manuscript.
Competing Financial Interests Statement:
H.Q.W., P.F., G.K., K.Y., J.L., Y.C., Y.C., E.L., D.R., S.M., H.H., M.M., M.L., J.W., K.H., G.C., F.L., J.A.E. are employees of Novartis. J.F.G. has served as a compensated consultant or received honoraria from Bristol-Myers Squibb, Genentech, Ariad, Loxo, Incyte, Novartis, Merck, and Clovis. A.T.S. has served as a compensated consultant or received honoraria from Pfizer, Novartis, Genentech, Roche, Ignyta, Blueprint Medicine, Daiichi-Sankyo, Ariad, Chugai, Taiho Pharmaceuticals, and EMD Serono. C.H.B. has received research funding from Novartis and Amgen. A.N.H. has received research funding from Novartis, Relay Therapeutics and Amgen. The remaining authors have no financial interests to declare.
References
- 1.Solomon BJ, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med. 2014;371:2167–2177. doi: 10.1056/NEJMoa1408440. [DOI] [PubMed] [Google Scholar]
- 2.Shaw AT, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368:2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 3.Shaw AT, et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med. 2014;370:1189–1197. doi: 10.1056/NEJMoa1311107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Camidge DR, Doebele RC. Treating ALK-positive lung cancer--early successes and future challenges. Nat Rev Clin Oncol. 2012;9:268–277. doi: 10.1038/nrclinonc.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gainor JF, et al. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov. 2016;6:1118–1133. doi: 10.1158/2159-8290.CD-16-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zou HY, et al. PF-06463922, an ALK/ROS1 Inhibitor, Overcomes Resistance to First and Second Generation ALK Inhibitors in Preclinical Models. Cancer Cell. 2015;28:70–81. doi: 10.1016/j.ccell.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi YL, et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med. 2010;363:1734–1739. doi: 10.1056/NEJMoa1007478. [DOI] [PubMed] [Google Scholar]
- 8.Doebele RC, et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin Cancer Res. 2012;18:1472–1482. doi: 10.1158/1078-0432.CCR-11-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katayama R, et al. Therapeutic strategies to overcome crizotinib resistance in non-small cell lung cancers harboring the fusion oncogene EML4-ALK. Proc Natl Acad Sci U S A. 2011;108:7535–7540. doi: 10.1073/pnas.1019559108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katayama R, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med. 2012;4:120ra117. doi: 10.1126/scitranslmed.3003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim S, et al. Heterogeneity of genetic changes associated with acquired crizotinib resistance in ALK-rearranged lung cancer. J Thorac Oncol. 2013;8:415–422. doi: 10.1097/JTO.0b013e318283dcc0. [DOI] [PubMed] [Google Scholar]
- 12.Sasaki T, et al. A novel ALK secondary mutation and EGFR signaling cause resistance to ALK kinase inhibitors. Cancer Res. 2011;71:6051–6060. doi: 10.1158/0008-5472.CAN-11-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crystal AS, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science. 2014;346:1480–1486. doi: 10.1126/science.1254721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lovly CM, et al. Rationale for co-targeting IGF-1R and ALK in ALK fusion-positive lung cancer. Nat Med. 2014;20:1027–1034. doi: 10.1038/nm.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett AM, Hausdorff SF, O'Reilly AM, Freeman RM, Neel BG. Multiple requirements for SHPTP2 in epidermal growth factor-mediated cell cycle progression. Mol Cell Biol. 1996;16:1189–1202. doi: 10.1128/mcb.16.3.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kouhara H, et al. A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell. 1997;89:693–702. doi: 10.1016/s0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- 17.Shi ZQ, Yu DH, Park M, Marshall M, Feng GS. Molecular mechanism for the Shp-2 tyrosine phosphatase function in promoting growth factor stimulation of Erk activity. Mol Cell Biol. 2000;20:1526–1536. doi: 10.1128/mcb.20.5.1526-1536.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang SQ, et al. Shp2 regulates SRC family kinase activity and Ras/Erk activation by controlling Csk recruitment. Mol Cell. 2004;13:341–355. doi: 10.1016/s1097-2765(04)00050-4. [DOI] [PubMed] [Google Scholar]
- 19.Neel BG, Gu H, Pao L. The 'Shp'ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem Sci. 2003;28:284–293. doi: 10.1016/S0968-0004(03)00091-4. [DOI] [PubMed] [Google Scholar]
- 20.Bennett AM, Tang TL, Sugimoto S, Walsh CT, Neel BG. Protein-tyrosine-phosphatase SHPTP2 couples platelet-derived growth factor receptor beta to Ras. Proc Natl Acad Sci U S A. 1994;91:7335–7339. doi: 10.1073/pnas.91.15.7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng GS, Hui CC, Pawson T. SH2-containing phosphotyrosine phosphatase as a target of protein-tyrosine kinases. Science. 1993;259:1607–1611. doi: 10.1126/science.8096088. [DOI] [PubMed] [Google Scholar]
- 22.Noguchi T, Matozaki T, Horita K, Fujioka Y, Kasuga M. Role of SH-PTP2, a protein-tyrosine phosphatase with Src homology 2 domains, in insulin-stimulated Ras activation. Mol Cell Biol. 1994;14:6674–6682. doi: 10.1128/mcb.14.10.6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Vactor D, O'Reilly AM, Neel BG. Genetic analysis of protein tyrosine phosphatases. Curr Opin Genet Dev. 1998;8:112–126. doi: 10.1016/s0959-437x(98)80070-1. [DOI] [PubMed] [Google Scholar]
- 24.Garcia Fortanet J, et al. Allosteric Inhibition of SHP2: Identification of a Potent, Selective, and Orally Efficacious Phosphatase Inhibitor. J Med Chem. 2016;59:7773–7782. doi: 10.1021/acs.jmedchem.6b00680. [DOI] [PubMed] [Google Scholar]
- 25.Chen YN, et al. Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature. 2016;535:148–152. doi: 10.1038/nature18621. [DOI] [PubMed] [Google Scholar]
- 26.Cunnick JM, et al. Regulation of the mitogen-activated protein kinase signaling pathway by SHP2. J Biol Chem. 2002;277:9498–9504. doi: 10.1074/jbc.M110547200. [DOI] [PubMed] [Google Scholar]
- 27.Pratilas CA, et al. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci U S A. 2009;106:4519–4524. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duncan JS, et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 2012;149:307–321. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hrustanovic G, et al. RAS-MAPK dependence underlies a rational polytherapy strategy in EML4-ALK-positive lung cancer. Nat Med. 2015;21:1038–1047. doi: 10.1038/nm.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.