Summary
KRAS can bind numerous effector proteins, which activate different downstream signaling events. The best known are RAF, PI-3’ kinase and RalGDS families, but many additional direct and indirect effectors have been reported. We have assessed how these effectors contribute to several major phenotypes in a quantitative way, using an arrayed combinatorial siRNA screen in which we knocked down 41 KRAS effectors nodes in 92 cell lines. We show that every cell line has a unique combination of effector dependencies, but in spite of this heterogeneity, we were able to identify two major subtypes of KRAS mutant cancers of the lung, pancreas and large intestine, which reflect different KRAS effector engagement and opportunities for therapeutic intervention.
Keywords: KRAS, RSK, RNAi screen, paralogs, redundancy
Introduction
Oncogenic Ras proteins promote multiple phenotypes, including uncontrolled proliferation, loss of contact inhibition, increased motility, altered metabolism and loss of genome integrity. This range of phenotypes may relate to the number of different effector pathways that Ras activates. The best validated of these is the Raf/MAPK pathway. Ras proteins can also activate PI3K pathways directly, though this seems to vary between tissue types. Ras proteins can also bind and activate RalGDS, but this pathway is less well understood. In addition to these three major pathways, active Ras proteins have been reported to interact directly with PKC, TIAM1, RASSF and indirectly with other effectors.
Analysis of the contributions that these pathways make towards cancer phenotypes using genetic approaches or RNA interference has been complicated by redundancies within each effector pathway. For example, there are three RAF isoforms, ARAF, BRAF and RAF1, and three class IA PI3K isoforms, PIK3CA, PIK3CB and PIK3CD, which all share some degree of functional redundancy. Compensatory activation by redundant paralogs explains why screens for single-gene dependencies in Ras mutant cells have not identified classical Ras effectors. Furthermore, each pathway may have several relevant phenotypes, such as effects on cell cycle progression, survival and metabolic stress. Screens that read out only one phenotype, such as viability, might miss other important gene functions. In addition to these technical complications, it is important to recognize differences in signaling between different tumor types. For example, in lung adenocarcinoma, the RTK/RAS/MAPK pathway can be activated by mutations in RTKs, activation of SOS, loss of NF1, mutation in KRAS, or activation of BRAF, with similar clinical outcomes. In pancreatic cancer, only mutation in KRAS appears capable of driving malignancy. Similarly, in lung adenocarcinoma and pancreatic cancer, KRAS mutations are likely initiating events, whereas in colorectal cancer they are definitely not, but rather exist in the context of un-regulated beta-catenin signaling.
To address these issues, we analyzed the importance of each canonical effector pathway, plus 37 other potential effectors, in 92 cell lines derived from lung, pancreas and colorectal tissues, measuring 5 different parameters on a single cell basis (size, proliferation, apoptosis, ROS and viability). We used combinations of highly potent and well characterized siRNAs that enabled us to knock down all the paralogs within an effector node at once. For example, we were able to knock down ARAF, BRAF and RAF1, together, in the “RAF node”. We took advantage of the variable nature of siRNA knockdown to generate dose-response curves for each node, measuring effects on 5 parameters in single cells.
This analysis revealed the expected dependence on tissue of origin, with pancreatic cells being more dependent on Ras signaling than lung adenocarcinoma. Unexpectedly, we found that most KRAS mutant tumor cell lines fell into two major classes: those that depend strongly on KRAS itself, and the Raf/MAPK pathway, and those that had lost dependence on KRAS, but were strongly dependent on RSK p90 S6 kinase. These cells were less dependent on PI3K genes themselves, but rather on PDK1, loss of LKB1 and other aspects of downstream PI3K signaling. We also identified a small number of cells that depended strongly on signaling through RalGDS. Each of these subsets had remarkably different properties, in terms of energy metabolism, EMT status and cell-cell interactions.
Results
siREN screen
Small-interfering RNAs (siRNAs) were designed using RNAi Sensor assay technology against RAS pathway genes (Fellmann et al., 2011; Pelossof et al., 2017; Yuan et al., 2014) (Table S1, Figure S1A-B). The selectivity and potency of these siRNAs allow for simultaneous knockdown of multiple functional paralogs (i.e. ARAF, BRAF and RAF1). This allows for near-complete ablation of an effector node by eliminating compensatory activity by redundant paralogs.
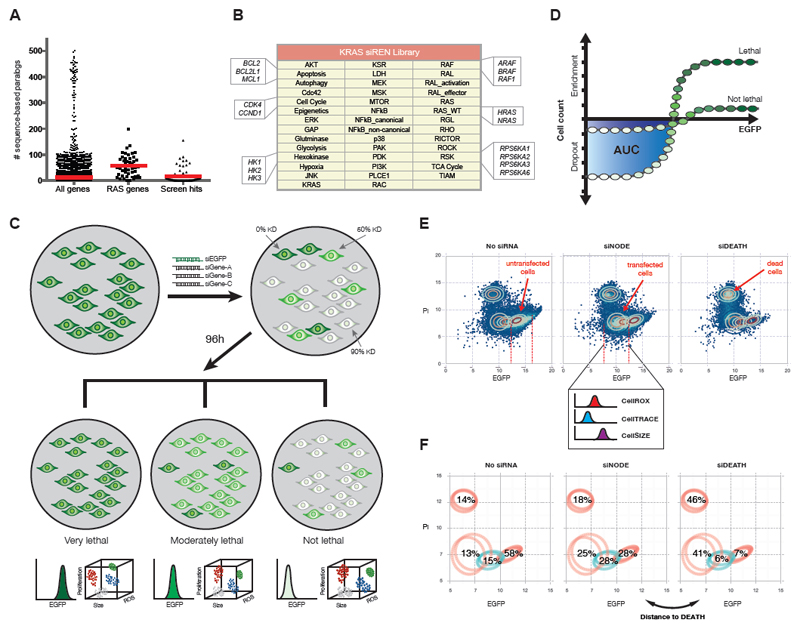
Complete paralog knockdown is particularly important for perturbation of the RAS pathway. Core RAS pathway genes are highly conserved and have a higher than average number of sequence-based paralogs (Figure 1A). These redundant paralogs can buffer against genotoxic damage to one member, however they are prohibitive when interrogating the RAS pathway using single-gene knockdown synthetic lethal screens. As expected, hits from such screens are biased for genes with no paralogs and may underestimate the value of targeting multiple paralogs, which is technically feasible and often routine with small molecules(Downward, 2015). We thus generated a multiplexed siRNA library targeting 41 complete RAS effector “nodes”, represented by 84 genes (Figure 1B, Table S2). Twenty-five nodes knock down 2-4 functional paralogs. Five nodes knock down multiple members of a protein complex, and 5 nodes knock down multiple steps within a pathway as additional approaches to achieve complete node ablation. Only six nodes knock down single genes.
Figure 1.
The siREN assay measures node dependencies across cell lines. (A) The number of sequence-based paralogs for all genes, core RAS pathway genes and validated hits from RNAi synthetic lethal screens was compiled from GenesLikeMe (GeneCardsSuite). Means are indicated by red lines. Zinc-finger genes and olfactory receptors were excluded from the analysis, given their extraordinary number of sequence-based paralogs (~500). (B) Forty-one key effector nodes are targeted in the KRAS siREN library, with most nodes represented by multiple single genes. For example, the KRAS node targets only KRAS, the RAS node targets H-, K-, and NRAS and the RAS_WT node targets H- and NRAS. (C) The siREN assay utilizes EGFP as a marker of node knockdown and measures drop-out/enrichment of cells with all levels of knockdown. Multi-color flow cytometry further captures phenotypic readouts such as proliferation rate, cell size and ROS accumulation upon node knockdown. (D) Cell viability is measured by counting the remaining cells with low EGFP 96h after siRNA transfection. Integration of the area under the curve (AUC) in the range of low EGFP cells represents the effect on cell viability. (E) Cells are analyzed by flow cytometry and assigned to one of four distinct Gaussian distributions, which represent untransfected, transfected, and dead cells. Effects on ROS (CellROX), proliferation (CellTRACE) and cell size (CellSIZE) are measured only the population of transfected cells. (F) Cell death (Distance to DEATH) is measured by the similarity in Gaussian profiles between a sample transfected with a positive control (siDEATH) and samples transfected with a siNODE.
We developed a flow cytometry-based assay, siREN (siRNA Effector Node), to measure the effects of node knockdown in 92 cell lines, including 64 KRAS mutant lines from multiple lineages (Table S2). For all lines, we transduced EGFP expression vectors using lentiviral infection and sorted for uniform EGFP-expressing populations (Supplemental Methods). In an arrayed format, we transfected these cells with siRNAs against each node, plus siRNA against EGFP (Figure 1C). siRNA transfection resulted in heterogeneous knockdown across cells due to differential endocytic uptake of lipid-RNA complexes. This resulted in a population of ~30,000 single cells, each cell with a different level of node knockdown. The relative degree of node knockdown in each cell was measured by EGFP positivity, given that EGFP knockdown closely matched the extent of knockdown of all intended targets (Figure S1C).
We monitored cell viability 96h-post transfection by counting the number of cells that drop-out or are enriched across all EGFP-expressing populations (Figure S1D, Supplemental Methods). Essential nodes are characterized by extreme drop-out of EGFPlow populations with a concomitant enrichment of EGFPhigh populations (Figure 1C). The selective depletion of cells with high knockdown (EGFPlow) generated a dose-response curve, which allowed us to integrate an area under the curve (AUC) to quantify the effect of node knockdown on viability (Figure 1D, S1E). Importantly, unless a node was lethal in which case no EGFPlow cells survive, a viable EGFPlow population represents healthy cells with high node knockdown. We thus could attribute resistance to node knockdown directly to independence from the gene(s) and not to incomplete gene knockdown. We further measured other cellular parameters (reactive oxygen (CellROX), proliferation (CellTRACE), growth (CellSIZE) and cell death (Distance to DEATH)) in the EGFPlow population using multi-color flow cytometry to elucidate the phenotypic effects of node knockdown (Figure 1E-F, Supplemental Methods).
Analysis of single cells allowed us to achieve levels of sensitivity well beyond traditional assays. Single cell analysis allowed for very sensitive viability measurements, equivalent to a 75-point dose-response curve, which maximized dynamic range and minimized false negatives (Figure S1F). Single cell analysis of cell death (Distance to DEATH) measured the accumulation of PI-positive cells as well as the depletion of live cell populations, which captured the penetrance of the cell death effect in the whole population (Supplemental Methods). Furthermore, by analyzing single cells, we extracted phenotypic data only from cells with target knockdown (EGFPlow), thereby circumventing effect dilution from whole population averages (Supplemental Figure 1G). Lastly, single cell analysis allowed us to pinpoint the degree of target knockdown necessary to achieve loss of viability, which may have implications on druggability (Figure S1H). The hexokinases, for example, require only low knockdown to achieve loss of viability, while other nodes require high knockdown and could be indicative of high protein reserves or long half-lives. Interestingly, we also show that the same amount of target knockdown was required to elicit a viability effect, regardless of the degree of dependency (Figure S1I).
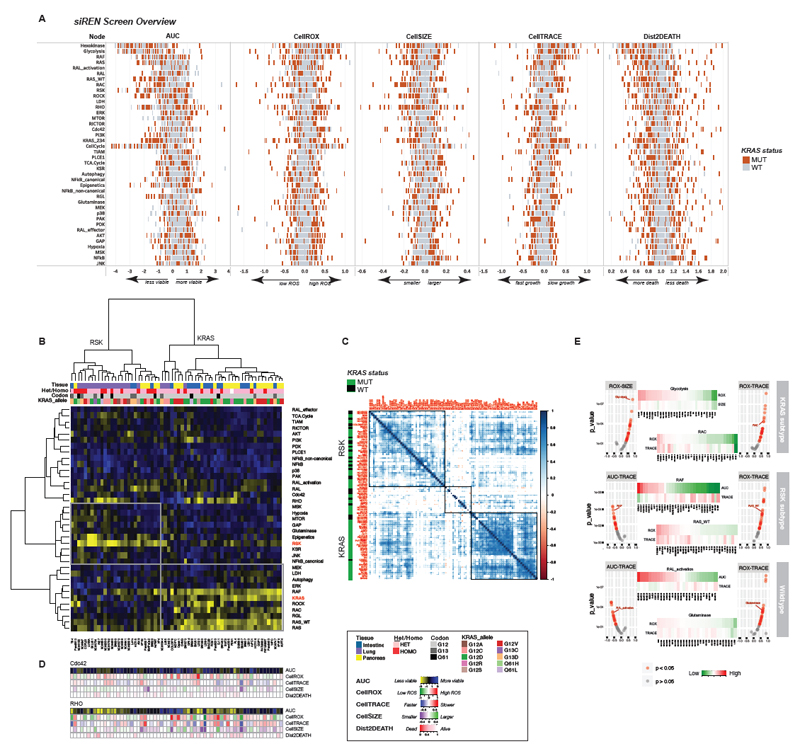
An overview of the siREN screen results is presented in Figure 2A, showing the phenotypic effects of node knockdown across all 92 cell lines. Strikingly, KRAS mutant lines as a whole (red bars) did not exhibit a pattern of dependency particularly distinct from KRAS wildtype lines (gray bars), save the expected sensitivity to RAS isoform knockdown. This demonstrates the heterogeneity of KRAS cancers and helps to explain the inability to identify an efficacious treatment for KRAS mutant patients as a uniform class.
Figure 2.
KRAS mutant lines consist of two major subtypes. (A) The results of the siREN screen are summarized, showing 5 phenotypic effects of node knockdown (viability, ROS, growth, proliferation and cell death) across 92 cell lines. Each cell line is depicted as a tick mark, with KRAS mutant lines shown in red. All lines were run in technical duplicates (B) Unsupervised hierarchical clustering of KRAS mutant cell lines by AUC reveal two distinct subtypes. Prominent dependencies in each subtype include KRAS and RSK. Tissue lineage (but not zygosity, KRAS codon or KRAS allele) is correlated with subtype membership. (C) Cell line – cell line correlations (Pearson, p < 0.05) based on AUC show that RSK-type lines correlate more strongly with KRAS wildtype lines than mutant lines from the KRAS subtype. (D) The phenotypic effects of Cdc42 or RHO knockdown are featured. Cell lines are ordered as depicted in (B). (E) Correlations between phenotypic readouts in the KRAS subtype, RSK subtype and KRAS wildtype lines are shown for select nodes. Volcano plots show Pearson correlation scores and p-values and heat maps show the raw phenotypic output across lines for the indicated node.
Also notable is the sizable effect of complete node knockdown across all 5 measured parameters (Figure 2A, Figure S2A). This is in contrast to pooled CRISPR screens that knock down one gene at a time, where smaller effects are observed due to gene redundancy (Figure S2B). In the siREN assay, only 5 nodes (PDK, RAL_effector, NFkB_non-canonical, PLCE, and PAK) failed to appreciably affect viability (AUCmedian > 0.5; MAD < 0.4) in any line. Conversely, only 4 nodes (Cell Cycle, Glycolysis, Hexokinase, Apoptosis) elicited large effects across > 80% of lines (Figure S2C). These data indicate that complete node knockdown using multiple siRNAs maximizes effect size without causing wide-spread pan-lethality. Across KRAS mutant lines, substantial heterogeneity in node dependency was observed, which segregated largely by lineage (Figure 2B). KRAS mutant lung lines clustered away from pancreas and large intestine, indicating that tissue of origin is a strong predictor of node dependency. Most notably, KRAS dependency across these 64 KRAS mutant lines varied widely, with greater than one-third of lines exhibiting KRAS-independence, i.e. full viability of the EGFPlow population with maximal KRAS knockdown. These results, obtained with a quantitative and highly sensitivity assay, are consistent with previous findings of KRAS-independence among KRAS mutant cell lines(Singh et al., 2009). Furthermore, we show that KRAS knockdown in dependent lines corresponds to a striking loss of proliferation, but rarely translates into appreciable cell death (Figure S2A, Dist2DEATH). The complete siREN dataset is available in Table S3.
Differential effector engagement by KRAS mutant subtypes
In vitro, oncogenic RAS can bind RAF, p110α and RalGDS via the switch I region of RAS and related RAS binding domains (RBD) of the effectors. While the affinity and kinetics of binding vary, there is little known about the cellular context that dictates effector activation. Here, we find that some KRAS mutant cell lines are indeed dependent on RAF, presumably through direct binding. A second major group depends on components of the PI3K pathway, though not on PI-3 kinases themselves. This group engages the RSK p90 S6 kinases to drive RSK-MTOR signaling. A third minor group depends strongly on RalGDS, presumably, like RAF, through direct binding.
Figure 2B shows the differential dependence (AUC) on 37 effector nodes across 64 KRAS mutant lines. Unsupervised hierarchical clustering shows two distinct subtypes. The KRAS-subtype is dependent on KRAS itself, as well as H- and NRAS. Among the effectors, this subtype is very dependent on RAF (and to a lesser extent, MEK and ERK) as well as RAC, RGL and autophagy. The RSK-subtype is strikingly resistant to KRAS (and RAF/MEK/ERK) knockdown and is instead dependent on numerous indirect RAS effectors such as RSK, glutaminase, MTOR, and KSR, among others. While resistant to loss of KRAS, this subtype retains dependence on the wildtype RAS isoforms, suggesting that non-canonical RAS effector activation may be driven in part by H- and NRAS. Surprisingly, ERK, a potent activator of RSK in many cell types, does not appear to be linked to RSK activation in this subtype (Chen et al., 1992).
The co-dependencies across the KRAS and RSK subtypes are surprisingly non-overlapping, with the exception of RAL and RHO (Figure 2B). This striking heterogeneity in effector engagement may help to explain our failure as yet to identify a single therapeutic target for all KRAS mutant cancers. However, it also serves to validate numerous studies identifying context-specific vulnerabilities in KRAS mutant cancer. RAC1 and autophagy, for example, have been reported as requirements in some KRAS mutant tumors, and here we validate those findings in the KRAS-subtype (Guo et al., 2011; Kissil et al., 2007). Other studies have shown that some KRAS mutant tumors rely on oxidative phosphorylation or canonical NFkB signaling, and here we validate those findings in the RSK-subtype (Barbie et al., 2009; Meylan et al., 2009; Viale et al., 2014). Diversity in effector engagement has thus been well documented in the literature. In fact, correlation of node dependencies (AUC) showed that RSK-type lines more closely resemble KRAS wild-type lines than their mutant counterparts in the KRAS-subtype (Figure 2C). This warns against the simple comparison of KRAS mutant versus KRAS wild-type lines to uncover the complex dependencies of KRAS mutant cell lines.
Effector node knockdown also caused differential phenotypic responses among cell lines, revealing interesting cell biology (Figure 2C, Figure S2A). Nodes can have little effect on viability, yet cause prominent phenotypic effects, as in the case of Cdc42 loss on reactive oxygen levels (Figure 2D, top). Targeting Cdc42 could thus serve to increase ROS, a specific liability in KRAS mutant cancers, without narrowing therapeutic index in combination with a more toxic drug. Knockdown of RHO represents how a node can affect viability in both subtypes, but with different biological outcomes (Figure 2D, bottom). In the KRAS subtype, loss of viability upon RHO knockdown is explained by dramatic changes in ROS, proliferation and cell size. In contrast, the loss of viability upon RHO knockdown in RSK-type lines was likely due to cellular processes not measured in this assay.
Measuring phenotypic parameters thus provides additional cell biological insight into node function. Rarely do all cell lines respond similarly, as in the case of knocking out the CellCycle node, which results in decreased proliferation and increased ROS and cell size in all lines (Figure S2A). More typically, phenotypic responses among subtypes were unique. Elevated ROS levels particularly affect the KRAS subtype and frequently correlate with decreased cell size and slowed proliferation, as seen in response to Glycolysis or RAC node knockdown (Figure 2E). The RSK subtype and wildtype lines, on the other hand, most frequently exhibited slowed proliferation in response to loss of viability or ROS (Figure 2E). Overall, the phenotypic response to node knockdown was very similar between RSK-type and wildtype lines, while KRAS-type lines uniquely exhibited large effects on ROS and cell size (Figure S2D). As ROS and cell size are intimately linked to growth and metabolism, this suggests that some KRAS mutant lines are more metabolically volatile than wild-type lines.
Properties of KRAS- and RSK-type cell lines
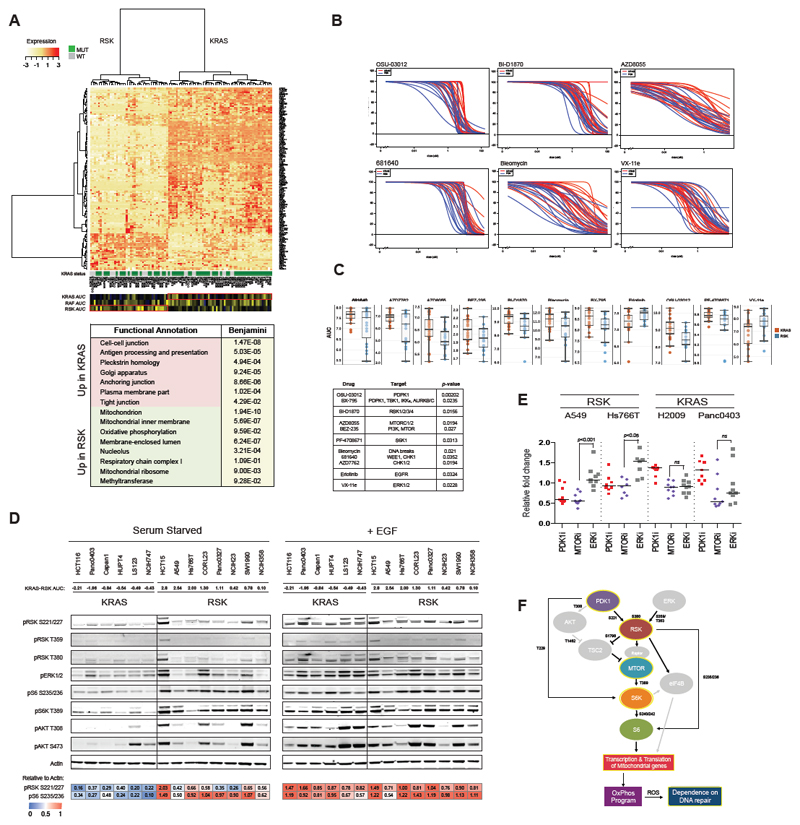
To better elucidate the biological properties resulting from differential effector activation, we assigned cell lines to the KRAS and RSK subtype based on dependency on KRAS, RAF and RSK and analyzed the differential gene expression between these two groups (moderated t-test with the R package, limma). We found 1150 differentially expressed genes (Figure 3A, Table S4). Functional annotation (Huang da et al., 2009) of genes upregulated in the KRAS-subtype reveal genes involved in cell-cell junctions and membrane integrity, confirming that KRAS-type lines are more epithelial than the RSK-type lines, as shown by others (Singh et al., 2009). Notably, CDH1 and EPCAM, regulators of epithelial fate, are upregulated in KRAS lines (2.62- and 3.92-fold, respectively), while ZEB1 and VIM, regulators of mesenchymal fate, are upregulated in RSK lines (1.95- and 3.81-fold, respectively).
Figure 3.
Screen reveals subtype-specific sensitivities to small molecules. (A) Differential gene expression between KRAS- and RSK-type lines reveal 1150 differentially expressed genes (FDR < 0.25). Genes with greater than 2-fold change in expression between subtypes are shown. (B) Forty KRAS mutant lines (21 KRAS-type lines and 19 RSK-type lines) were screened for sensitivity to 11 compounds. All lines were run in biological duplicates. AUCs are shown in (C). (D) Basal and EGF-stimulated signaling in 14 KRAS mutant lines were assessed by western blot. One measure of KRAS-“ness” and RSK-“ness” is the difference in KRAS and RSK AUC, where negative values are more KRAS-like. Blots were quantified with ImageJ. (E) Relative fold change in expression of mitochondrial genes in KRAS- and RSK-type lines was assessed in the presence of PDK1, MTOR and ERK inhibitors. (F) Model of signaling in RSK-type lines features activation of the MTOR pathway to drive oxidative phosphorylation. Processes outlined in yellow were experimentally derived.
Overwhelmingly, the genes upregulated in the RSK-subtype are involved in oxidative phosphorylation and mitochondrial ribosome maintenance. In particular, numerous members of complexes I and V of the electron transport chain and diverse regulators of oxidative stress are upregulated. As both subtypes are sensitive to knockdown of the glycolysis and hexokinase nodes (Figure S2C), this suggests that RSK-type lines fulfill additional metabolic demands through aerobic respiration. Differential gene expression thus revealed that epithelial/mesenchymal morphology and glycolytic/oxidative metabolism are two major divergent biological properties of the KRAS or RSK subtypes. This gene expression signature allowed us to re-classify several “border-line” cell lines more firmly into the KRAS- or RSK-subtype in subsequent analyses.
To identify a mechanism by which RSK could drive aerobic metabolism, we conducted a small molecule screen on 40 KRAS mutant lines (21 KRAS-type and 19 RSK-type) to find drugs differentially effective in either subtype (Table S5). We showed that RSK-type lines are more sensitive than KRAS-type lines to inhibitors of PDK1 (3-phosphoinositide-dependent kinase 1, aka, PDPK1), RSK, MTOR, S6K1 and DNA repair enzymes, while KRAS-type lines were more sensitive to EGFR and ERK inhibition (Figure 3B,C). By western blot, we show that RSK-type lines have high basal RSK-MTOR-S6K activity (Figure 3D). High levels of phosphorylated RSK at S221 and AKT at T308, both PDK1 sites, suggest that RSK activation can be driven by PDK1, perhaps more predominantly than by the KRAS-ERK axis, further consistent with the KRAS/RAF/MEK/ERK-independence of this subtype. This was surprising, as RSK proteins are generally considered to be major targets of ERK1/2.
As MTOR has been shown to drive the transcription and translation of mitochondrial genes involved in respiration, we showed that either PDK1 or MTOR inhibition, but not ERK inhibition, represses the transcription of mitochondrial genes (found upregulated in Figure 3A) in RSK-type lines but not KRAS-type lines (Figure 3E) (Cunningham et al., 2007; Morita et al., 2013). A byproduct of oxidative phosphorylation is reactive oxygen species, which can induce DNA damage and dependency on DNA repair enyzmes. RSK-type lines have slightly higher basal levels of γH2AX, which is amplified by the addition of a CHK1 inhibitor and normalized by PDK1/MTOR/oxphos inhibition but not ERK inhibition (Figure S2E). We thus speculate that accumulation of ROS through respiration may explain the enhanced sensitivity of the RSK subtype to inhibitors of DNA repair.
By integrating the siREN screen, gene expression and small molecule screen, we demonstrate that KRAS-type lines depend on canonical RAS effectors and downstream pathways, including RAF and RAC, and upregulate genes involved in the maintenance of the epithelial phenotype. The relative sensitivity of the KRAS subtype to erlotinib and the wildtype RAS isoforms, HRAS and NRAS, supports the notion that KRAS-type lines require receptor tyrosine kinases (RTKs) to recruit GEFs for nucleotide exchange of RAS (Lito et al., 2016; Patricelli et al., 2016; Young et al., 2013). Members of this canonical signaling axis thus represent potential targets for the KRAS-subtype. Conversely, RSK-type lines express mesenchymal markers and depend on the RSK-MTOR axis, which correlates with increased expression of oxidative phosphorylation genes and may supplement glycolysis in energy production and/or anabolic metabolism (Figure 3F).
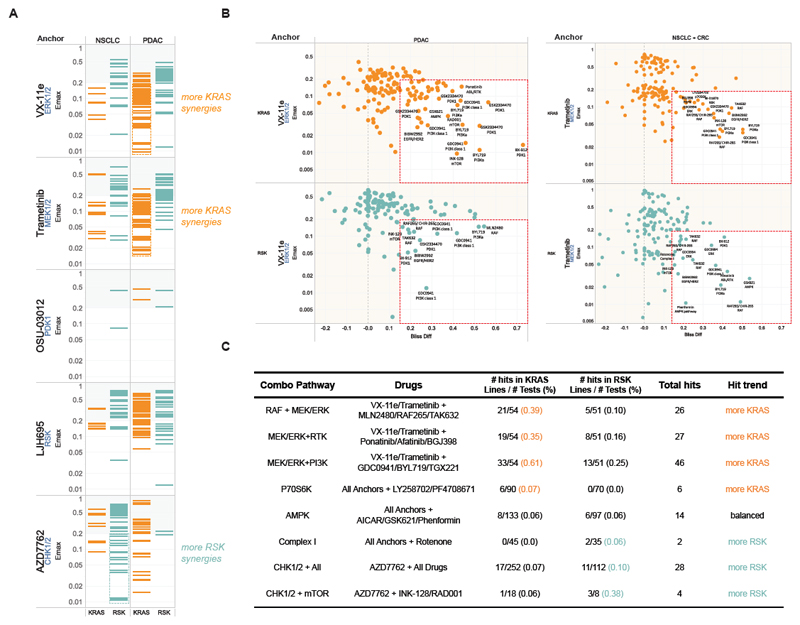
To further probe the potential difference in drug response of the KRAS and RSK-types we conducted a combinatorial drug screen on 20 cell lines (10 KRAS and 10 RSK) (Table S5). We focused on combinations that targeted the most differentiating phenotypes revealed by the siREN assay. Inhibitors of MEK1/2, ERK1/2, PDK1, RSK and CHK1/2 were used in combination with 28 drugs (Table S5). We found that the KRAS subtype presented with more effective synergies (Emax<20%, Synergy >15%) than the RSK subtype when drugs were combined with an ERK or MEK inhibitor particularly in the PDAC lines (Figure 4A). In contrast, combinations involving a CHK1/2 inhibitor resulted in more effective synergies in the RSK subtype. Consistent with previous findings (Engelman et al., 2008), combining PI3K inhibitors with MEK and ERK inhibitors frequently resulted in synergy (Figure 4B, Table S5), and vertical combinations of MEK and RAF inhibitors were recurrently synergistic across cell lines, especially in the KRAS subtype (p=0.005, Fisher Exact Test) (Figure 4B). Interestingly, inhibitors of p70S6K were engaged in more synergies in KRAS-type lines than RSK-type lines (p=0.035, Fisher Exact Test). Although statistically underpowered, pathway analysis of effective synergistic events showed enrichment of MEK/ERK inhibition with receptor tyrosine kinase inhibition in KRAS type lines (p=0.06, Fisher Exact Test), consistent with higher EGFR dependency and recent reports of synergy with FGFR (Kitai et al., 2016; Manchado et al., 2016). The RSK subtype showed some enrichment in synergistic events involving the Complex I inhibitor, rotenone, and CHK1/2 inhibitors particularly in combination with mTOR inhibitors (Figure 4C). Overall, these results confirm that subtype specific drug combinations can be identified and that siREN-defined phenotypes can inform the design of effective drug combinations.
Figure 4.
Combination drug screen identifies efficient synergies in KRAS- and RSK-type lines. (A) Synergistic events identified with different Anchor Drugs across the drug library within each tissue type and KRAS subtype. Only combinations achieving 15% more activity than predicted by the Bliss hypothesis are shown, and the resulting minimum viability (compared to untreated cells) is plotted (Emax: Maximum combination effect). All lines were run in biological duplicates, with most lines passing quality control (see Supplemental Information). (B) Effective Synergies (x-axis: viability loss over predicted viability, y-axis: viability outcome) for combinations including ERK (left) or MEK (right) inhibitor found in different tissues and for KRAS and RSK subtypes: Hits are in the lower right box of each panel and the most dramatic hits labeled by drug name and target. (C) Pathway and target analysis of effective synergies found in KRAS- and RSK-type lines.
Predicting effector dependence
In order to predict effector engagement in cell lines and patient samples, we performed Elastic Net Regression (EN) to identify genomic biomarkers of effector dependency (AUC) from a set of 20,771 features, including gene expression, mutation and copy number variation (Table S4). Our previous results suggested that KRAS- and RSK-type lines diverge most strikingly in their morphological and metabolic properties. KRAS-type lines are epithelial while RSK-type lines are mesenchymal, and our data suggest that the epithelial fate is driven by engagement of KRAS with the RAC and ROCK effectors. KRAS-type lines are also glycolytic while RSK-type lines utilize oxidative phosphorylation, and our data indicate that engagement of RSK, MTOR and TCA Cycle is the driver of oxidative metabolism. We thus identified strong biomarkers of KRAS, RSK, RAC, ROCK, MTOR and TCA Cycle dependency. We generated three concise expression signatures to understand which phenotype best stratifies samples into the KRAS or RSK subtype (Table S4).
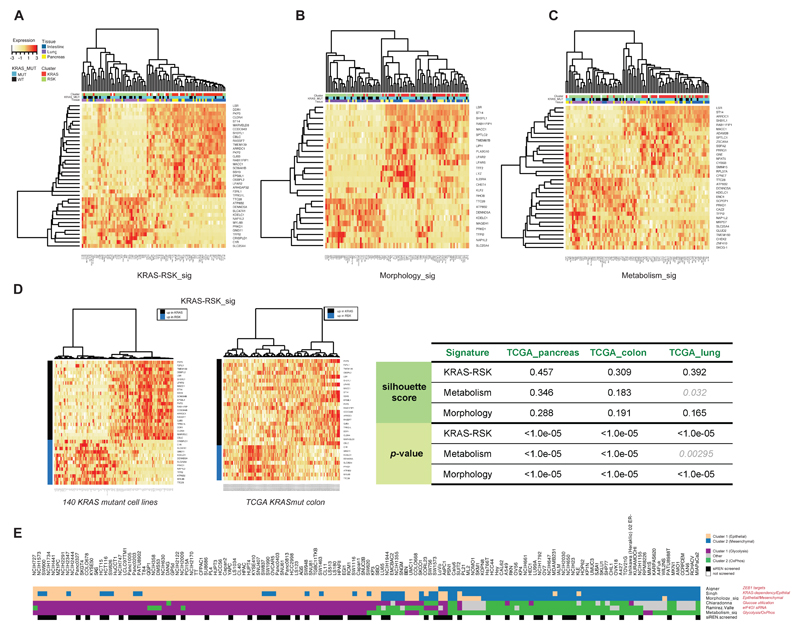
KRAS-RSK_sig is a signature comprised of all genes that predict KRAS or RSK dependence. Morphology_sig is enriched for genes predictive of RAC and ROCK engagement and specifically differentiates between epithelial and mesenchymal samples. Genes in this signature include RHOB, a small GTPase involved in cell adhesion, and SH3YL1, which interacts with DOCK4 and WASL, regulators of adherens junctions (Kobayashi et al., 2014). Metabolism_sig is enriched for genes predictive of MTOR and TCA Cycle dependency, thus differentiating between glycolytic and oxidative metabolism. This signature includes genes such as MRPS7, which encodes a mitochondrial ribosomal protein, and GLUD2, which shuttles glutamate into the TCA cycle through conversion to alpha-ketoglutarate (Shashidharan and Plaitakis, 2014). All three signatures quite accurately stratified cell lines into the KRAS or RSK subtype as determined by the siREN assay, indicating that in cell lines, metabolic or morphologic state are both good predictors of differential effector engagement (Figure 5A-C).
Figure 5.
Gene expression signatures stratify cell lines and patient samples into the KRAS and RSK subtypes. Elastic Net-derived genomic features of KRAS and RSK sensitivity were used to compile signatures of KRAS/RSK dependence (KRAS-RSK_sig, A), EMT status (Morphology_sig, B) and glycolysis or oxidative phosphorylation (Metabolism_sig, C). KRAS-RSK_sig was projected onto 140 KRAS mutant lines and TCGA patient samples. The ability to robustly stratify TCGA samples across 3 indications using KRAS-RSK_sig was scored using silhouette (D). (E) Six independently derived signatures of EMT status and metabolic dependency similarly classified 140 KRAS mutant lines, showing a strong correlation between the epithelial-glycolytic state and the mesenchymal-oxidative state.
We projected the signatures onto 140 KRAS mutant lines, including 76 that were not screened by siREN assay and thus not included in the EN training set. Unsupervised hierarchical clustering robustly stratified all lines into either the KRAS or RSK subtypes using any of the three signatures (Figure 5D, Figure S4A). We next projected the signatures onto KRAS mutant patient samples in TCGA and used Silhouette (Rousseeuw, 1987) to score how well each signature separated these samples into two distinct classes (Figure 5D, right panel, Figure S4B-C). With the exception of one signature in one indication, our signatures were able to robustly stratify KRAS mutant PAAD, COAD and LUAD TCGA samples into the KRAS- and RSK- subtypes. Samples that did not stratify well reveal that additional subtypes exist in patients that are not well-represented in cell lines. Lastly, we used KRAS-RSK_sig to stratify all lines screened in a pharmacological landscape study (Iorio et al., 2016) into either the KRAS or RSK subtype. We then showed that these predictions did in fact accurately predict for differential sensitivity to multiple drugs targeting the MAPK, MTOR and DNA repair pathways, as predicted by our drug screening results (Figure S3B, Figure 4). These data demonstrate that genomic biomarkers of effector engagement can predict morphological and metabolic phenotypes, either of which is sufficient to stratify KRAS mutant samples into the KRAS or RSK subtype.
LKB1/AMPK activity differentiates KRAS mutant subtypes
The striking correlation between morphology and metabolism in KRAS mutant cell lines prompted us to test the strength of this relationship using additional independently derived signatures. In addition to Morphology_sig and Metabolism_sig, we used Aigner_ZEB1_Targets (MSigDB: M14590) and Singh_KRAS_Dependency (MSigDB: M2851), two signatures of EMT (Aigner et al., 2007; Singh et al., 2009), and Chiaradonna_Neoplastic_Transformation_KRAS (MSigDB: M10381) and siRNA_EIF4GI_DN (MSigDB: M2846), two metabolic signatures (Chiaradonna et al., 2006; Ramirez-Valle et al., 2008) to stratify 140 KRAS mutant cell lines. There was excellent agreement using all six signatures (Figure 5E, Figure S4D-E), demonstrating a strong relationship between the epithelial and glycolytic states and between the mesenchymal and oxidative states in KRAS mutant cell lines.
To explore the mechanistic connection between morphology and metabolism, we applied expression data to a reverse causal network perturbation analysis, using a database of gene expression signatures assembled from thousands of genetic and chemical perturbation experiments. Biomarkers of KRAS and RSK dependency (as measured by AUC) were then correlated with inferred mechanism activity values. Figure S5A shows an example of such a mechanism (STK11 kinase activity) and its associated gene signature or “footprint”. Table S6 provides the footprints of 6 other relevant mechanisms. We identified many inferred mechanisms that correlated or anti-correlated with KRAS or RSK dependence (FDR < 0.1 by Pearson correlation, Supplemental Methods, Table S6). By integrating these mechanisms with the results of the siREN and drug screen, we assembled a core mechanistic model that provides a granular biological explanation for the phenotypic differences observed between KRAS- and RSK-types lines (Figure 6A, and see Figure S5B and Table S6 for more details).
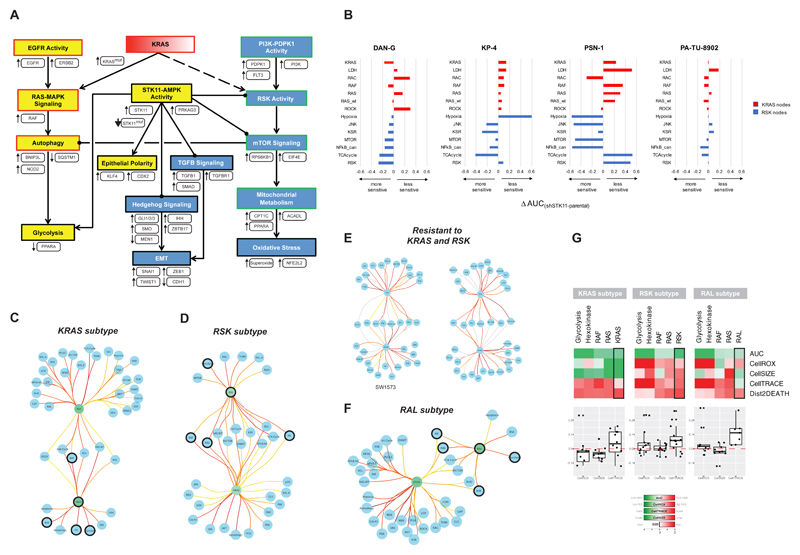
Figure 6.
A network linking morphology and metabolism and a model of oncogene addiction. (A) Summarized mechanistic model linking morphology and metabolism that explains features of KRAS and RSK phenotypes. Evidence is indicated as follows: Yellow nodes – processes that are more active in the KRAS subtype; Blue nodes – processes that are more active in the RSK subtype; White boxes – mechanisms identified by genomic features and/or correlations with KRAS/RSK sensitivity; Red and green borders – KRAS- and RSK-specific processes, respectively, that were empirically observed in the siREN and/or drug assays. (B) The siREN assay was run on four STK11-knockdown lines and their parental controls. All lines were run in technical duplicates. The delta-AUC measures the change in dependency in the knockdown lines compared to parent. (C) KRAS-type lines have fewer edges between KRAS and downstream effector nodes than edges from RSK. (D) RSK-type lines have fewer edges between RSK and downstream effector nodes than edges from KRAS. (E) Two cell lines resistant to both KRAS and RSK have equivalent edges from KRAS and RSK. (F) RAL-type lines have fewer edges between RAL and downstream effectors than edges from KRAS. (G) Knockdown of the 4 common nodes controlled by the onconode in KRAS, RSK and RAL-type lines reveals different phenotypic outcomes. Means of all lines within a subtype are shown (top), and values from individual lines for CellROX, CellTRACE and CellSIZE are plotted (bottom).
In the model shown in Figure 5A, yellow processes were associated with the KRAS subtype and blue processes were associated in the RSK subtype. Individual mechanisms are shown in white boxes. We found that genomic predictors of the KRAS subtype are indicative of higher canonical signaling through RAF, which can be driven by oncogenic KRAS and/or signaling through EGFR/ERBB2. Other mechanisms, including inferred relative elevation of NOD2 and BNIP3L and a decrease in SQSTM1 activity, are indicative of elevated levels of autophagy, which along with repressed PPARA activity, may serve to drive glycolysis (Lock et al., 2011; Peeters and Baes, 2010; Ribet et al., 2010). The epithelial character of the KRAS subtype may be explained by elevated levels of KLF4 and CDX2, two inferred mechanisms linked to induction of epithelial polarity.
In the RSK-type cells, high inferred PDK1, PI3K and FLT3 activity are likely drivers of RSK activation. High MTOR activity, as inferred by elevated RPS6KB1 and EIF4E mechanisms, negatively regulates autophagy and may drive CPT1C, ACADL and PPARA activity to elevate oxidative phosphorylation and fatty acid oxidation (Jung et al., 2010). High levels of superoxides and elevated NFE2L2 activity were also inferred mechanisms that indicate high oxidative stress, a potential byproduct of increased mitochondrial metabolism. The process of EMT was also strongly supported by mechanistic inferences, such as SNAI1, ZEB1, and TWIST1 and a negative correlation with CDH1 activity, which may be supported by high TGFβ and hedgehog signaling, as evidenced by high activity of TGFB1, SMAD, GLI1/2/3 and SMO.
Strikingly, our analysis suggests that the activation state of STK11 (LKB1) is one of the major determining factors of KRAS/RSK fate. LKB1 regulates both metabolism and morphology, and accurately explains the directionality of these phenotypes in both the KRAS and RSK subtypes. High LKB1 activity favors the KRAS-state by promoting glycolysis and the establishment of epithelial polarity, whereas low activity favors the RSK-state by de-repressing MTOR and driving oxidative phosphorylation, Hedgehog, TGFβ, and EMT (Di Magno et al., 2016; Lin et al., 2015; Shackelford and Shaw, 2009). As such, inactivating mutations in STK11 were indeed significantly correlated with the RSK-subtype, and both STK11 mRNA and inferred biological activity strongly correlated with KRAS- and RSK-sensitivity (Figure S5C). In support of this model, KRAS;STK11 mutant lung adenocarcinomas have been shown to comprise a distinct molecular subtype exhibiting adaptation to oxidative stress and other subtype-specific vulnerabilities (Skoulidis et al., 2015). However, while all STK11 mutant lines we tested classified as RSK-type lines, 80% of RSK-type lines were in fact STK11 wildtype. This emphasizes that mutation calls are not sufficient to predict pathway activity. To test our model, we generated stable STK11-knockdown lines to modulate LKB1-AMPK activity and looked for a shift in dependency to RSK. Indeed we found that in three out of four lines, STK11 knockdown drives sensitivity to RSK-associated nodes and resistance to KRAS-associated nodes compared to parental lines (Figure 5B).
Modeling oncogene addiction to KRAS and RSK
We next sought to model effector engagement in the KRAS and RSK subtypes. The siREN assay generates a vector of 4 phenotypic responses in live cells upon node knockdown: 1) viability, 2) ROS, 3) proliferation and 4) cell size. This allowed us to correlate node-node pairs (903 total), where high correlations suggest phenotypic similarity and functional epistasis. We were thus able to generate RAS effector networks for every cell line (Table S7). To identify critical signaling hubs within KRAS- and RSK-type networks, we counted the number of edges (positive correlations between nodes) emanating from every node in all 92 lines. Not surprisingly, we found that the number of edges emanating from KRAS and RSK in mutant lines were most significantly different between subtypes (p-value = 3.33x10-4 and 1.55x10-4, respectively). Unexpectedly, we found that in KRAS-type lines, the average number of edges emanating from KRAS was lower than the number of edges emanating from RSK (13 compared to 32) (Figure 6C). Similarly, in RSK-type lines, the number of edges emanating from RSK was lower than the number of edges from KRAS (17 compared to 25) (Figure 6D).
This suggested that the driving “onconode” is linked to few downstream nodes and does not act as a central hub that controls many nodes, as might be expected. In support of this model, cell lines that are resistant to both KRAS and RSK have equivalently large numbers of edges emanating from both nodes (Figure 6E). A possible interpretation of these results is that oncogenes shed many “wildtype” connections to downstream nodes in order to signal maximally through a few select nodes that enhance survival. This model would thus suggest that KRAS selectively binds few effectors (such as RAF, but not not RalGDS or p110α) in KRAS-addicted cells in order to maximally activate the RAF-MAPK pathway. In non-KRAS-addicted cells (RSK-type cells), KRAS may bind many more effectors, effectively dampening the ability to maximally activate any single effector pathway to cause pathway addiction. In these lines, RSK appears to selectively and maximally activate few downstream effectors, such as MTOR. In both cases, adaptation to an onconode (KRAS or RSK) thus results in the inability of an “addicted” cellular system to compensate for the loss of the onconode.
Using this method of edge counting, we searched for other potential onconodes that lose edges while KRAS gains edges. We identified RAL as an onconode for a small subset of KRAS mutant lines (9 edges from RAL and 33 from KRAS, Figure 6F). We confirmed that the majority of these lines do indeed have a strong dependency on the RAL node (mean AUCRAL subtype = -0.628; mean AUCAll Others = -0.103) and a moderate dependency on KRAS. This suggests that KRAS engagement of RalGDS is important in the RAL subtype, analogous to KRAS engagement of RAF in the KRAS subtype. Figure 2A further shows that RAL clusters with PI3K and AKT dependencies, thus representing a third pattern of effector engagement in KRAS mutant cell lines.
Interestingly, we found that in all three subtypes, the onconode controlled a set of common nodes: Hexokinase, Glycolysis, RAS and RAF (Figure 6C-E; dark circles). These nodes comprise 4 of the top 5 nodes with the greatest AUC effect (mean AUCCommonNodes = -1.259; mean AUCAll Others = 0.158), suggesting that the onconode must maintain control of these key effectors of viability in KRAS mutant lines. Despite sharing these common dependencies, as predicted by Figure 2, the phenotypic outcome of node knockdown was strikingly different between subtypes (Figure 6G, Table S3). RAF knockdown, for instance, led to a decrease in ROS and a severe stall in proliferation in the KRAS subtype, indicative of a metabolic shutdown. Whereas RAF knockdown in the RSK subtype resulted in a slight increase in ROS that only slowed proliferation, indicative of a very different metabolic response.
Lastly, we showed that co-mutation of KRAS and STK11 correlates with RSK membership (KRASmut;STK11mut: EdgestoRSK = 15.8 and EdgestoKRAS = 23.2; KRASmut;STK11wt: EdgestoRSK = 22.1 and EdgestoKRAS = 20.4; p= 0.0019), which confirms our model in Figure 6A. Conversely, co-mutation of KRAS and CDKN2A correlates with RAL membership (KRASmut;CDKN2Amut: EdgestoRAL = 18.8 and EdgestoKRAS = 22.3; KRASmut;CDKNAwt: EdgestoRAL = 23.5 and EdgestoKRAS = 19.7; p= 0.002). These data suggest that secondary tumor suppressor mutations play a role in triggering a switch in effector engagement and thus oncogene addiction.
Discussion
The siREN assay employs combinatorial siRNA knockdown and multi-parameter readouts to identify differential patterns of KRAS-effector engagement. This large-scale integration of RNAi screening, small molecule screening and genomic analysis of cell lines and TCGA samples reveals that although KRAS has the potential to bind many effectors, specific cellular contexts, defined by lineage, secondary mutations and energy states, dictate which effectors are activated.
We describe two such cellular contexts, into which most KRAS mutant cancers stratify. The KRAS subtype maintains oncogene addiction to KRAS and maximally activates MAPK by engaging RAF and very few other effectors. Signal transduction in these lines subsequently maintains the epithelial phenotype and drives flux through glycolysis. The RSK subtype has switched oncogene addiction from KRAS to RSK, preserving many KRAS-effector interactions, but maximally activating MTOR through RSK activation. Signaling in these lines maintains the mesenchymal phenotype and drives oxidative phosphorylation. We also identify a third subtype, the RAL subtype, which represents a smaller fraction of KRAS mutant cell lines that is dependent on RAL and PI3K signaling.
This variety of KRAS mutant subtypes calls into question how such heterogeneity evolved. KRAS mutations are probably initiating events in lung adenocarcinoma and pancreatic cancer, though not in colorectal cancers. Deep sequencing of multiple primary and metastatic pancreatic tumors revealed that KRAS mutations are retained throughout tumor evolution, an observation consistent with an essential role at all stages of this disease. In lung adenocarcinoma, KRAS-independence may be acquired at later stages of tumorigenesis. We show evidence that secondary mutations in tumor suppressors, such as STK11 and CDKN2A, can contribute to a switch in oncogene addiction, which may suggest that clones harboring such mutations are selected for under changing micro-environmental pressures. Alternatively, RSK-type tumors may depend on KRAS functions that remain unknown and are not measured in our 2D cell-based assay system. For example, there is evidence that KRAS dependency increases in 3D culture systems (Fujita-Sato et al., 2015; Patricelli et al., 2016). This may be related to the expression of receptor tyrosine kinases in 2D and 3D, which would impact nucleotide exchange particularly of wildtype RAS isoforms. This could subsequently influence activation of downstream nodes. Further inquiry into node dependence in the 3D context is a critical next step and may reveal dependencies that are stronger in vivo and possibly more clinically relevant.
It also serves to ask how this knowledge lends to the design of effective therapies. We show that targeting vertical and orthogonal combinations of effectors achieves a greater depth of response compared to single agent inhibition. The efficacy of vertical combinations indicates that within-subtype dependencies (RAF-ERK) are hard-wired and frequently buffered by feedback mechanisms that require inhibition at multiple steps of a linear pathway. The efficacy of orthogonal or between-subtype dependencies (ERK-AMPK) may hint at an escape from the KRAS state to the RSK state and vice versa. If the transition from the KRAS to the RSK state is fluid or reversible, a tumor might be able to escape therapies that target either state alone. Our study provides a framework on which rational combinations can be further designed and tested.
Experimental Procedures
Cell Culture
All cell lines were obtained from the Genomics of Drug Sensitivity in Cancer (GDSC) cell line collection (Supplemental Table S2). Pancreatic and colorectal cell lines were tested for mycoplasma (no antibiotics) and authenticated before and at several points throughout the entire study. Lung cell lines were verified ahead of inclusion in the drug sensitivity panel.
siREN Assay
EGFP-positive cell lines were seeded based on doubling time (<24 – 36 h: 5 x 104 or >36h: 7 x 104 cells/well) in twelve-well plates. siRNAs were mixed with optimal RNAiMAX concentrations and added to the cells. CellTrace Violet (Thermo-Fisher) was added to the wells 24h post-siRNA transfection according to the manufacturer’s recommendations. Ninety-six hours post-transfection, floating (dead) cells were collected and CellROX Deep Red (Thermo Fisher) was added to the adherent cells according to the manufacturer’s recommendations. Adherent cells were trypsinized, collected, added to the floating (dead) cells and incubated with propidium iodide (Thermo Fisher) prior to flow cytometry analysis. Cells were transferred to 96-well plates in technical duplicates and analyzed by HTS flow cytometry using a BD Canto II or BD LSRFortessa and BD FACSDiva software. Gating for single cells was applied and ~30,000 cells per sample were acquired (see Supplemental Methods for a detailed protocol).
Drug Profiling
Cells were treated with the following drugs in an 8 point, 2-fold dilution series (highest concentration in parentheses): Bleomycin sulfate (64 μM), OSU-03012 (16 μM), PF4708671 (10.24 μM), VX-11e (5.12 μM), BX-795 (5.12 μM), 681640 (2.56 μM), AZD8055 (2 μM), AZD7762 (2 μM), BEZ-235 (0.05 μM), BI-D1870 (10.24 μM), Erlotinib (2 μM). Cells were seeded in 96-well plates and incubated with compound for 96h. Response to drug was assayed using Cell Titer-Glo (Promega) according to the manufacturer’s instructions.
Elastic Net Regression (EN)
The EN was performed on all siREN nodes, for both the AUC and D2D response measures, as previously described but with updated genomic features(Garnett et al., 2012). For each cell line, we used point mutation data from exome sequencing. For copy number variation, we selected subset of 1789 genes, which had a correlation of at least 0.5 between mRNA expression and copy number across all the 986 cell lines of the GDSC project. EN parameters alpha and lambda were selected by cross-validation and regression coefficients were determined for 100 independent runs. Frequency was computed for each genomic feature as the number of runs where the corresponding coefficient was non zero, divided by 100. A positive effect between a feature and a node indicates resistance to that node and a negative feature indicates sensitivity.
Statistical Procedures
Correlations across phenotypic parameters in KRAS- and RSK-type lines were measured by Pearson correlation coefficient and p-values were determined using the cor.mtest function in the R package corrplot. For drug screening data, p-values for significance of the difference between KRAS- and RSK-type lines were determined by two-sample t-test or Wilcoxon rank test. Synergy calculations were performed according to the Bliss independence hypothesis. Here we apply this hypothesis as follows: Vd xVa = Vd,a,where Vd is the relative viability observed in the presence of the library drug alone and Va the relative viability observed in the presence of the anchor drug alone. The causal mechanism analysis assessed the significance of univariate correlations between mechanism and node AUC and corrected for FDR with the Benjamini & Hochberg method. Differences in expression between KRAS- and RSK-type lines as measured by qPCR was assessed for significance using a two-sample t-test. Differential gene expression was assessed using the R package limma and corrected for FDR with the Benjamini & Hochberg method. Retrospective drug sensitivity predictions based on KRAS- and RSK- classification by expression were tested for significance using a two-sample t-test and corrected for FDR with the Benjamini & Hochberg method.
Supplementary Material
Acknowledgements
We thank Jacqueline Galeas and Cayde Ritchie for technical support, Brett Tomson for critical reading of the manuscript and Bin Zhang for reagents. This work was supported by funding from the NIH (a grant from the CTD2 network to FM and K99 CA194284-02 to TLY), Susan G. Komen Foundation (KG111338 to TLY), a grant from Daiichi Sankyo (to FM), Wellcome Trust (102696 to CB) and Stand up to Cancer (to CB and FM).
Footnotes
Author Contributions
TY, RB, CB and FM designed the experiments. TY, RB, SA, WB, NF, LG, KP, BS, AW, RL, LP, GS, EM and MB conducted the experiments. AA, MY, TY, RS, CB, DD, TT and PG computationally analyzed the data. CF, SL, JL, CL and LCL designed and validated the siRNA library. TY, CB and FM wrote the paper.
Declaration of Interests
The authors declare no competing interests.
References
- Aigner K, Dampier B, Descovich L, Mikula M, Sultan A, Schreiber M, Mikulits W, Brabletz T, Strand D, Obrist P, et al. The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene. 2007;26:6979–6988. doi: 10.1038/sj.onc.1210508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RH, Sarnecki C, Blenis J. Nuclear localization and regulation of erk- and rsk-encoded protein kinases. Molecular and cellular biology. 1992;12:915–927. doi: 10.1128/mcb.12.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaradonna F, Sacco E, Manzoni R, Giorgio M, Vanoni M, Alberghina L. Ras-dependent carbon metabolism and transformation in mouse fibroblasts. Oncogene. 2006;25:5391–5404. doi: 10.1038/sj.onc.1209528. [DOI] [PubMed] [Google Scholar]
- Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- Di Magno L, Basile A, Coni S, Manni S, Sdruscia G, D'Amico D, Antonucci L, Infante P, De Smaele E, Cucchi D, et al. The energy sensor AMPK regulates Hedgehog signaling in human cells through a unique Gli1 metabolic checkpoint. Oncotarget. 2016;7:9538–9549. doi: 10.18632/oncotarget.7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downward J. RAS Synthetic Lethal Screens Revisited: Still Seeking the Elusive Prize? Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21:1802–1809. doi: 10.1158/1078-0432.CCR-14-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, Maira M, McNamara K, Perera SA, Song Y, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nature medicine. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellmann C, Zuber J, McJunkin K, Chang K, Malone CD, Dickins RA, Xu Q, Hengartner MO, Elledge SJ, Hannon GJ, et al. Functional identification of optimized RNAi triggers using a massively parallel sensor assay. Molecular cell. 2011;41:733–746. doi: 10.1016/j.molcel.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita-Sato S, Galeas J, Truitt M, Pitt C, Urisman A, Bandyopadhyay S, Ruggero D, McCormick F. Enhanced MET Translation and Signaling Sustains K-Ras-Driven Proliferation under Anchorage-Independent Growth Conditions. Cancer research. 2015;75:2851–2862. doi: 10.1158/0008-5472.CAN-14-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P, Thompson IR, Luo X, Soares J, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM, Karantza V, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes & development. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Iorio F, Knijnenburg TA, Vis DJ, Bignell GR, Menden MP, Schubert M, Aben N, Goncalves E, Barthorpe S, Lightfoot H, et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell. 2016;166:740–754. doi: 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS letters. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissil JL, Walmsley MJ, Hanlon L, Haigis KM, Bender Kim CF, Sweet-Cordero A, Eckman MS, Tuveson DA, Capobianco AJ, Tybulewicz VL, et al. Requirement for Rac1 in a K-ras induced lung cancer in the mouse. Cancer research. 2007;67:8089–8094. doi: 10.1158/0008-5472.CAN-07-2300. [DOI] [PubMed] [Google Scholar]
- Kitai H, Ebi H, Tomida S, Floros KV, Kotani H, Adachi Y, Oizumi S, Nishimura M, Faber AC, Yano S. Epithelial-to-Mesenchymal Transition Defines Feedback Activation of Receptor Tyrosine Kinase Signaling Induced by MEK Inhibition in KRAS-Mutant Lung Cancer. Cancer discovery. 2016;6:754–769. doi: 10.1158/2159-8290.CD-15-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Harada K, Negishi M, Katoh H. Dock4 forms a complex with SH3YL1 and regulates cancer cell migration. Cellular signalling. 2014;26:1082–1088. doi: 10.1016/j.cellsig.2014.01.027. [DOI] [PubMed] [Google Scholar]
- Lin H, Li N, He H, Ying Y, Sunkara S, Luo L, Lv N, Huang D, Luo Z. AMPK Inhibits the Stimulatory Effects of TGF-beta on Smad2/3 Activity, Cell Migration, and Epithelial-to-Mesenchymal Transition. Molecular pharmacology. 2015;88:1062–1071. doi: 10.1124/mol.115.099549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lito P, Solomon M, Li LS, Hansen R, Rosen N. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science. 2016;351:604–608. doi: 10.1126/science.aad6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock R, Roy S, Kenific CM, Su JS, Salas E, Ronen SM, Debnath J. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Molecular biology of the cell. 2011;22:165–178. doi: 10.1091/mbc.E10-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchado E, Weissmueller S, Morris JPt, Chen CC, Wullenkord R, Lujambio A, de Stanchina E, Poirier JT, Gainor JF, Corcoran RB, et al. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature. 2016;534:647–651. doi: 10.1038/nature18600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Dooley AL, Feldser DM, Shen L, Turk E, Ouyang C, Jacks T. Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;462:104–107. doi: 10.1038/nature08462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Gravel SP, Chenard V, Sikstrom K, Zheng L, Alain T, Gandin V, Avizonis D, Arguello M, Zakaria C, et al. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell metabolism. 2013;18:698–711. doi: 10.1016/j.cmet.2013.10.001. [DOI] [PubMed] [Google Scholar]
- Patricelli MP, Janes MR, Li LS, Hansen R, Peters U, Kessler LV, Chen Y, Kucharski JM, Feng J, Ely T, et al. Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State. Cancer discovery. 2016;6:316–329. doi: 10.1158/2159-8290.CD-15-1105. [DOI] [PubMed] [Google Scholar]
- Peeters A, Baes M. Role of PPARalpha in Hepatic Carbohydrate Metabolism. PPAR research. 2010;2010 doi: 10.1155/2010/572405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelossof R, Fairchild L, Huang CH, Widmer C, Sreedharan VT, Sinha N, Lai DY, Guan Y, Premsrirut PK, Tschaharganeh DF, et al. Prediction of potent shRNAs with a sequential classification algorithm. Nature biotechnology. 2017;35:350–353. doi: 10.1038/nbt.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Valle F, Braunstein S, Zavadil J, Formenti SC, Schneider RJ. eIF4GI links nutrient sensing by mTOR to cell proliferation and inhibition of autophagy. The Journal of cell biology. 2008;181:293–307. doi: 10.1083/jcb.200710215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribet C, Montastier E, Valle C, Bezaire V, Mazzucotelli A, Mairal A, Viguerie N, Langin D. Peroxisome proliferator-activated receptor-alpha control of lipid and glucose metabolism in human white adipocytes. Endocrinology. 2010;151:123–133. doi: 10.1210/en.2009-0726. [DOI] [PubMed] [Google Scholar]
- Rousseeuw PJ. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math. 1987;20:53–65. [Google Scholar]
- Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nature reviews Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shashidharan P, Plaitakis A. The discovery of human of GLUD2 glutamate dehydrogenase and its implications for cell function in health and disease. Neurochemical research. 2014;39:460–470. doi: 10.1007/s11064-013-1227-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, Settleman J. A gene expression signature associated with "K-Ras addiction" reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, Behrens C, Kadara H, Parra ER, Canales JR, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer discovery. 2015;5:860–877. doi: 10.1158/2159-8290.CD-14-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sanchez N, Marchesini M, Carugo A, Green T, Seth S, Giuliani V, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628–632. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A, Lou D, McCormick F. Oncogenic and wild-type Ras play divergent roles in the regulation of mitogen-activated protein kinase signaling. Cancer discovery. 2013;3:112–123. doi: 10.1158/2159-8290.CD-12-0231. [DOI] [PubMed] [Google Scholar]
- Yuan TL, Fellmann C, Lee CS, Ritchie CD, Thapar V, Lee LC, Hsu DJ, Grace D, Carver JO, Zuber J, et al. Development of siRNA payloads to target KRAS-mutant cancer. Cancer discovery. 2014;4:1182–1197. doi: 10.1158/2159-8290.CD-13-0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.