Abstract
Circulating biomarkers available in clinical practice do not allow to stratify patients with coronary heart disease (CHD) prior the onset of a clinically relevant event. We evaluated the methylation status of specific genomic segments and gene expression in peripheral blood of patients undergoing Cardiac Computed Tomography (CCT) for CHD (n = 95).
We choose to investigate cholesterol metabolism. Methylation and gene expression of low density lipoprotein receptor (LDLR), sterol regulatory element-binding factor 2 (SREBF2) and ATP-binding cassette transporter 1 (ABCA1) were evaluated by qRT-PCR. Calcium score (CACS), stenosis degree, total plaque volume (TPV), calcified plaque volume (CPV), non-calcified plaque volume (NCPV) and plaque burden (PB) were assessed in all CHD patients (n = 65). The percentage of methylation at the specific analyzed segment of LDLR promoter was higher in CHD patients vs healthy subjects (HS) (n = 30) (p = 0.001). LDLR, SREBF2 and ABCA1 mRNAs were up-regulated in CHD patients vs HS (p = 0.02; p = 0.019; p = 0.008). SREBF2 was overexpressed in patients with coronary stenosis ≥50% vs subjects with stenosis <50% (p = 0.036). After adjustment for risk factors and clinical features, ABCA1 (p = 0.005) and SREBF2 (p = 0.010) gene expression were identified as independent predictors of CHD and severity. ROC curve analysis revealed a good performance of ABCA1 on predicting CHD (AUC = 0.768; p<0.001) and of SREBF2 for the prediction of disease severity (AUC = 0.815; p<0.001). Moreover, adjusted multivariate analysis demonstrated SREBF2 as independent predictor of CPV, NCPV and TPV (p = 0.022; p = 0.002 and p = 0.006) and ABCA1 as independent predictor of NCPV and TPV (p = 0.002 and p = 0.013).
CHD presence and characteristics are related to selected circulating transcriptional and epigenetic-sensitive biomarkers linked to cholesterol pathway. More extensive analysis of CHD phenotypes and circulating biomarkers might improve and personalize cardiovascular risk stratification in the clinical settings.
Introduction
Despite advances in diagnosis, treatment and prognosis, coronary heart disease (CHD) is still the most prevalent cause of mortality and morbidity worldwide [1].
The main pathophysiological process underlying the development of CHD is represented by coronary atherosclerosis whose pathogenesis involves an imbalanced lipid metabolism and impaired immune response. These phenomena contribute to endothelial dysfunction and chronic inflammation, with the consequent formation of the atherosclerotic plaque, erosion and unstable atheroma, and vessel lumen stenosis [2,3].
Several studies reported an implication of epigenetic modifications in the pathogenesis of multifactorial diseases such as CHD, focusing on the evaluation of global DNA methylation in atherosclerotic tissues, and in peripheral blood cells of CHD patients [4–9].
Blood gene expression profiling showed a differential transcriptional signature in CHD patients and healthy subjects (HS). Major alterations were detected in genes coding for biomolecules involved in oxidative stress, cell motility, metabolic pathways, and inflammation. Interestingly, the expression pattern was found to correlate with the severity of CHD and gene expression in vascular tissues, suggesting a mirroring between circulating cells and changes in the atherosclerotic coronary wall [10–14].
Lipid homeostasis plays a key role in the atherosclerotic process and genes actively involved are represented by low-density lipoprotein receptor (LDLR) that has a central role in regulating the internalization of plasma LDL-cholesterol (LDL-c); sterol regulatory element binding transcription factor 2 (SREBF2), involved at transcriptional level in cholesterol metabolism, and ATP binding cassette transporter1 (ABCA1), the main regulator of cholesterol cellular efflux [15–18].
Few studies have correlated circulating molecular patterns to quantitative imaging parameters derived by Cardiac Computed Tomography (CCT) [19–23].
CCT is a powerful diagnostic tool to rule out CHD thanks to its high negative predictive value [24–26] allowing characterization and quantification of atherosclerotic plaque burden (PB) and providing comprehensive information about the location, severity and features of coronary atherosclerotic plaques [27,28]. Although CCT represents the most promising tool for CHD assessment, mainly to avoid unnecessary invasive coronary angiography, it is challenging to improve the conventional risk scores by assessing new non-invasive biomarkers that could support patient stratification and clinical decision making toward personalized treatments.
Given the central role both of the epigenetics and immune system in the pathogenesis of atherosclerosis and CHD development through dynamic changes of molecular patterns [26,29,30], in this observational study we investigated the association between gene expression/epigenetic markers and CHD features. Indeed, we evaluated by quantitative realtime PCR (qRT-PCR) the methylation status of specific genomic segments and the relative expression of genes in peripheral blood mononuclear cells (PBMNCs) of patients with suspected CHD underwent to CCT aiming to find a possible screening methodology for non-invasive and non-radiation-utilizing detection of CHD. We analyzed epigenetic-sensitive genes involved in cholesterol bioactivity such as LDLR, SREBF2 and ABCA1 in correlation with CHD features and quantitative imaging parameters derived by CCT.
Methods
Study population
The study has been approved by the institutional ethics committee (IRCCS Fondazione SDN, protocol no. 7–13) on research on humans in accordance with the ethical guidelines of the 1975 Declaration of Helsinki. A written informed consent was obtained from all subjects enrolled.
During the experimental design we performed a power calculation analysis using G*Power software, obtaining an estimated total sample size of 81 with an effect size equal to 0.45. In a period of 36 months, between November 2014 and November 2017, 250 consecutive patients were enrolled in the study at IRCCS SDN (Naples, Italy) (see S1 Appendix for sample characterization).
Patients with known history of cancer (n = 51), active infections (n = 9), chronic or immune-mediated diseases (n = 15) were excluded from the study to avoid confounding effects due to other variables. Furthermore, subjects with cardiomyopathy, known CHD, previous percutaneous transluminal coronary angioplasty and coronary artery bypass grafting (n = 80), systemic atherosclerosis such as lower extremity peripheral arterial disease or supra-aortic arterial disease were not included in the study population. The remaining 95 subjects without a history of cardiovascular events and referred to our institution for suspected CHD were included in the study.
In detail, patients with Calcium Score (CACS) = 0 and uninjured coronaries were considered as HS (n = 30). Obstructive CHD was defined by the presence of a stenosis greater than or equal to 50% in one or more of the major coronary arteries detected by CCT.
Sample collection and molecular analysis
PBMNCs were isolated by Ficoll gradient using HISTOPAQUE-1077 (Sigma Diagnostics, USA) according to manufacturer’s instructions and frozen at -80°C at the IRCCS SDN Biobank [31]. (S1 Appendix for details).
Methylated DNA immunoprecipitation (MeDIP)
For DNA extraction and immunoprecipitation from isolated PBMNCs was used MagMeDIP kit (Diagenode, Belgium) (see S1 Appendix for the detailed protocol).
Genome Browser and Methbase [32] tools were used to select genetic regulatory regions and design methylation-specific primers for specific genomic sequences: LDLR promoter and intron 1, SREBF2 promoter and ABCA1 5’UTR (S1 Appendix for details).
RNA extraction and quantitative realtime PCR assay
Total RNAs were extracted from PBMNCs of patients and HS using TRIzol solution (Thermo Fischer Scientific, USA), according to the manufacturer’s instructions (S1 Appendix for details). Oligonucleotide sequences are reported in S1 Table. The relative expression levels of mRNA were measured by CFX96 Touch Real-Time PCR Detection System (BioRad Laboratories, Ltd, USA). Target gene expression levels were normalized using RPS18 as housekeeping gene [33] for each sample (S1 Appendix for details). Each sample was analyzed in triplicate and data expressed as mean ± standard error. For logistic regression analysis delta Ct (Δct) values of each gene were considered.
Cardiac computed tomography and image analysis
All patients underwent a CCT with a third-generation dual source multidetector computed tomography scanner (Somatom Force, Siemens Healthcare AG, Germany). Details on the imaging protocols are given in the S1 Appendix. CCT derived parameters such as CACS, stenosis degree (in percentage of lumen reduction), non-calcified plaque volume (NCPV), calcified plaque volume (CPV), total plaque volume (TPV) and PB were calculated as previously reported [34]. Plaque segmentations were performed by two independent technicians and averaged values for each derived imaging parameter were considered. Only three patients showed multivessel CHD at CCT for whom the most significant plaque was considered for the segmentation and statistical analysis. All scans were analyzed by 2 experienced radiologists. After independent evaluations were made, a consensus interpretation was achieved according to the international SCCT guidelines [35]. Patients were grouped according to imaging parameters. Two representative examples of plaque segmentation by CCT to calculate imaging parameters are reported in Fig 1 and Fig 2.
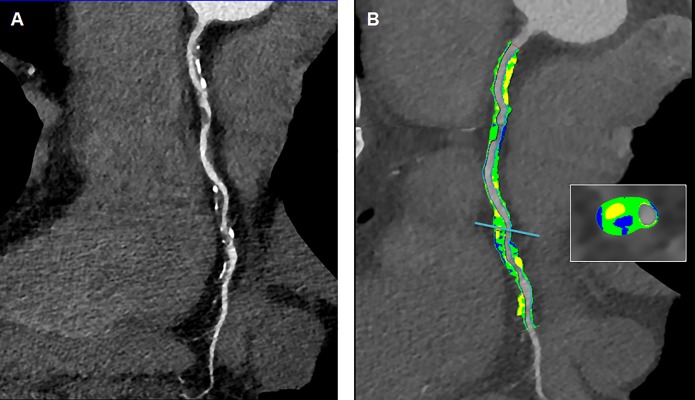
Fig 1.
(A) 3D volume rendering and (B) Curved multi planar reconstruction of a left anterior descending artery showing a proximal mostly non-calcified plaque with a spotty calcification (white arrow in (A) and (B) panels) and a distal calcified plaque. Semi-automatic segmentation of coronary vessel: calcified component (yellow), lipid component (green) and fibrotic (blue).
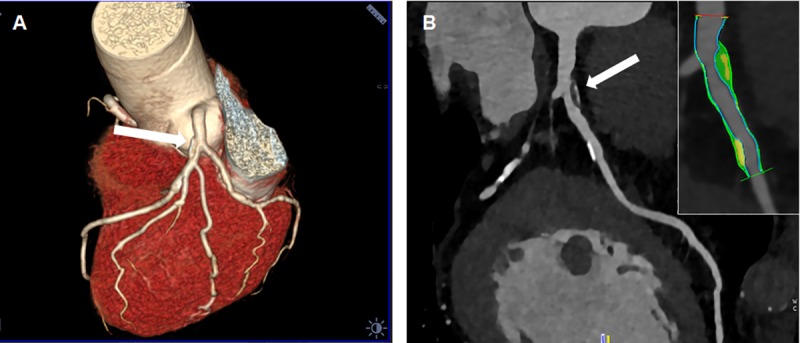
Fig 2.
(A) Curved multi planar reconstruction of a right coronary artery showing a severe atherosclerosis with both calcified and non-calcified plaques. (B) Semi-automatic segmentation of coronary vessel: calcified component (yellow), lipid component (green) and fibrotic (blue).
Statistical analysis
Statistical analysis was performed using R Core Team (version 3.03, Austria). Continuous variables were expressed as mean ± standard deviation or standard error. Data were tested for normality through the Shapiro-Wilk test. Unpaired Student’s t-test or Mann-Whitney U test, as required, were used for comparison between two groups. The one-way analysis of variance (ANOVA) or the Kruskal-Wallis test were used for comparison among three or four groups for parametric and non-parametric variables, respectively, with Bonferroni’s adjustment for multiple comparisons. Categorical variables were expressed as percentage and were compared using the Chi-Square test or the Fisher’s exact test.
Methylation statistical analysis revealed a significance only for LDLR promoter methylation. For this reason data reporting SREBF2 and ABCA1 promoter and LDLR intron 1 methylation were not reported. Molecular markers were tested in a univariate logistic regression analysis; then, significant variables were included in a multivariate logistic regression analysis (stepwise forward model) adjusted for the traditional cardiovascular risk factors and baseline clinical features [36]. In order to assess the possible effects of dyslipidemia treatments (e.g., statins) on the molecular marker expression, we used an unpaired Student’s t-test to compare treated vs untreated subgroups. Receiver operating characteristic (ROC) curves were subsequently generated using CHD and obstructive CHD as the events. Areas under the curve (AUC) were compared for each single molecular variable and the multivariate model. For all tests a p<0.05 was considered for statistical significance.
Results
Study population features
The baseline characteristics of CHD patients and HS are summarized in Table 1. The mean age was 57.87±9.5 years in HS compared to 63.12±10.97 years in CHD patients (p = 0.016). The percentage of male was significantly higher in patients with CHD (75.4%) compared to HS (43.3%) (p = 0.002).The mean body mass index (BMI), the pericardial fat, the heart rate (HR) and the left ventricular ejection fraction (LVEF) were comparable between two groups (p = 0.728, p = 0.327, p = 0.152, p = 0.974, respectively). Cardiovascular risk factors such as CHD familiarity, smoke, hypertension, diabetes and dyslipidemia were not significantly different between HS and CHD patients (p = 0.759; p = 0.684; p = 0.484; p = 0.163; p = 0.125, respectively). Moreover, total cholesterol, LDL- and HDL-cholesterol plasmatic concentrations did not significantly differ among the two groups (Table 1).
Table 1. Baseline characteristics of CHD patients and HS.
| Variables | CHD (n = 65) |
HS (n = 30) |
p value |
|---|---|---|---|
| Age (years)* | 63.12 ± 11.0 | 57.87 ± 9.60 | 0.016 |
| Men (%) | 49 (75.4%) | 13 (43.3%) | 0.002 |
| BMI (kg/m2)* | 27.86 ± 4.52 | 27.40 ± 3.92 | 0.728 |
| HR (bpm) | 64.45 ± 12.39 | 69.70 ± 17.12 | 0.152 |
| LVEF (%) | 68.30 ± 9.34 | 68.21 ± 11.04 | 0.974 |
| Pericardial fat (mL)* | 175.33 ± 90.2 | 150.90 ± 74.00 | 0.327 |
| CHD familiarity (%) | 39 (60%) | 17 (56.7%) | 0.759 |
| Smoke (%) | 20 (30.8%) | 8 (26.7%) | 0.684 |
| Hypertension (%) | 43 (66.2%) | 22 (73.3%) | 0.484 |
| Diabetes (%) | 9 (13.8) | 1 (3.3%) | 0.163 |
| Dyslipidemia (%) | 37 (56.9%) | 12 (40%) | 0.125 |
| Total cholesterol (mg/dL)* | 186.4 ± 46.4 | 189.3 ± 38.7 | 0.721 |
| LDL cholesterol (mg/dL)* | 103.8 ± 36.4 | 117.3 ± 31.5 | 0.103 |
| HDL cholesterol (mg/dL)* | 53.0 ±18.6 | 50.3 ± 10.6 | 0.372 |
| Physical activity (%) | 27 (41.5%) | 5 (16.7%) | 0.020 |
| Anti-hypertensive therapy (%) | 39 (60%) | 18 (60%) | 1.000 |
| Dyslipidemia treatment (%) | 29 (44.6%) | 5 (16.7%) | 0.007 |
*Data are represented as mean ± SD. Bold values were considered statistically significant with a p < 0.05.
CHD, coronary artery disease; BMI, body mass index; HDL, high density lipoprotein; HR, heart rate; HS, healthy subjects; LVEF, left ventricular ejection fraction; LDL, low density lipoprotein
Hypertensive subjects were in treatment with blood-pressure lowering drugs while dyslipidemic patients were in treatment with statins. All diabetic patients were treated with oral hypoglycemic agents.
CHD molecular features
A significantly higher methylation level in a promoter region of LDLR gene was found in PBMNCs of CHD patients (1.37%±0.25) as compared to HS (0.45%±0.09; p = 0.001).
In addition, gene expression analysis showed a significant up-regulation of LDLR (p = 0.021), SREBF2 (p = 0.019) and ABCA1 (p = 0.008) mRNA levels in CHD patients as compared to HS (Fig 3A, 3B and 3C and Table 2).
Fig 3.
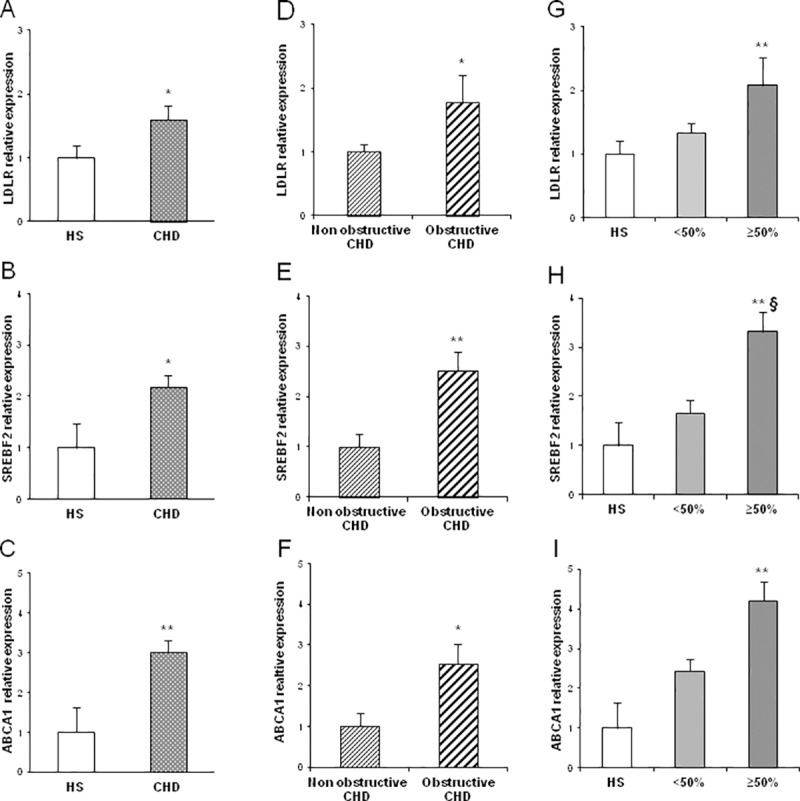
(A-C) LDLR, SREBF2 and ABCA1 mRNA relative expression in CHD patients (n = 65) and HS (n = 30); (D-F) LDLR, SREBF2 and ABCA1 mRNA relative expression in obstructive (n = 69) and non-obstructive CHD patients (n = 26); (G-I) LDLR, SREBF2 and ABCA1 mRNA relative expression in CHD patients with coronary stenosis <50% (n = 39) and coronary stenosis ≥50% (n = 26) compared to HS (n = 30) (*p value<0.05 vs HS; **p value<0.01 vs HS; §p value <0.05 coronary stenosis ≥50% vs coronary stenosis <50%).
Table 2. CHD features and molecular analysis.
| CHD | HS | CHD | p value |
| LDLR promoter methylation** | 0.45±0.09 | 1.37±0.25 | 0.001 |
| LDLR gene expression‡ | 9.31±0.19 | 8.63±0.21 | 0.021 |
| SREBF2 gene expression | 6.94±0.48 | 5.81±0.23 | 0.019 |
| ABCA1 gene expression | 7.92±0.64 | 6.32±0.27 | 0.008 |
| Obstructive CHD |
No CHD or CHD <50% |
Obstructive CHD | p value |
| LDLR gene expression | 9.07±0.12 | 8.24±0.43 | 0.015 |
| SREBF2 gene expression | 6.53±0.26 | 5.2±0.38 | 0.007 |
| ABCA1 gene expression | 7.19±0.33 | 5.84±0.48 | 0.026 |
| Stenosis degree | <50% | ≥50% | p value* |
| LDLR promoter methylation | 1.3±0.15 | 1.47±0.2 | 0.708 |
| LDLR gene expression | 8.89±0.15 | 8.24±0.43 | 0.079 |
| SREBF2 gene expression | 6.21±0.28 | 5.21±0.39 | 0.036 |
| ABCA1 gene expression | 6.63±0.3 | 5.84±0.49 | 0.239 |
*p value from multiple comparisons. Bold values were considered statistically significant with a p < 0.05.
** Methylation data are expressed as percentage of methylation/total input.
‡Gene expression data are expressed in terms of Δct derived from qRT-PCR experiments.
ABCA1, ATP binding cassette subfamily A member 1; CHD, coronary artery disease; HS, healthy subjects; LDLR, low density lipoprotein receptor; SREBF2, sterol regulatory element-binding factor 2.
Furthermore, molecular analysis showed that higher levels of LDLR (p = 0.015), SREBF2 (p = 0.007) and ABCA1 (p = 0.026) were found in subjects with coronary obstructive CHD as compared to HS and patients with non-obstructive CHD. (Fig 3D, 3E and 3F and Table 2).
Statistical differences were also found stratifying the study population according to stenosis degree. High levels of methylation were found in LDLR promoter region in patients with coronary stenosis ≥50% (1.47%± 0.20) and coronary stenosis <50% (1.3%± 0.15) compared to HS (p = 0.029 and p = 0.043, respectively) while no difference was found between CHD groups. Gene expression data demonstrated an up-regulation of LDLR (p = 0.007), SREBF2 (p = 0.003) and ABCA1 (p = 0.004) in CHD patients with stenosis degree ≥50% vs HS, with a significance for SREBF2 mRNA overexpression in the group with coronary stenosis ≥50% as compared to <50% (p = 0.036) (Fig 3G, 3H and 3I and Table 2).
The univariate regression analysis showed that LDLR promoter methylation was significantly associated to the presence of CHD (OR = 0.464 CI 95% = 1.049–1.480; p = 0.043) while LDLR, SREBF2 and ABCA1 gene expression were predictors of both CHD and critical stenosis (OR = 1.516 CI95% = 1.024–2.245, p = 0.038; OR = 1.281 CI 95% = 1.049–1.588, p = 0.024; OR = 1.249 CI95% = 1.034–1.480, p = 0.012; respectively) (Table 3).
Table 3. Univariate and multivariate logistic regression§ analysis to predict CHD and obstructive CHD.
| Univariate analysis | ||||
| Predictors | CHD | Obstructive CHD | ||
| OR (95% CI) | p value | OR (95% CI) | p value | |
| LDLR promoter methylation** | 0.464 (0.220–0.976) | 0.043 | 1.170 (0.921–1.485) | 0.198 |
| LDLR gene expression‡ | 1.516 (1.024–2.245) | 0.038 | 0.666 (0.456–0.972) | 0.035 |
| SREBF2 gene expression | 1.281 (1.049–1.588) | 0.024 | 0.701 (0.525–0.937) | 0.016 |
| ABCA1 gene expression | 1.249 (1.034–1.480) | 0.012 | 0.806 (0.659–0.986) | 0.036 |
| Multivariate analysis | ||||
| Predictors | CHD | |||
| OR (95% CI) | p value | |||
| Male | 0.121 (0.038–0.380) | <0.001 | ||
| ABCA1 gene expression | 1.352 (1.096–1.666) | 0.005 | ||
| Predictors | Obstructive CHD | |||
| OR (95% CI) | p value | |||
| Male | 25.757 (3.216–206.275) | 0.002 | ||
| SREBF2 gene expression | 0.652 (0.471–0.903) | 0.010 | ||
** Methylation data are expressed as percentage of methylation/total input.
‡Gene expression data are expressed in terms of Δct derived from qRT-PCR experiments.
§Multivariate analysis corrected for male gender
Bold values were considered statistically significant with a p < 0.05.
ABCA1, ATP binding cassette subfamily A member; CHD, coronary heart disease; CI, confidence interval; LDLR, low density lipoprotein receptor; OR, odds ratio; SREBF2, sterol regulatory element-binding factor 2.
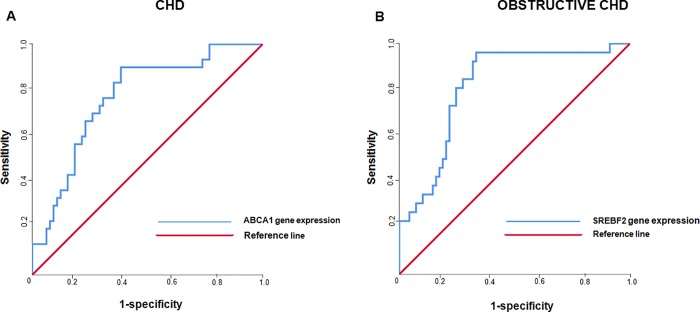
The multivariate regression models, adjusted for cardiovascular risk factors, baseline features and clinical characteristics, showed that gender (male) and ABCA1 gene expression (OR = 1.352 CI 95% = 1.096–1.666; p = 0.005) were independently associated with CHD as well as SREBF2 gene expression (OR = 0.652CI 95% = 0.471–0.903; p = 0.010) was found to be a predictor of obstructive CHD independently by male gender (Table 3). ROC curve analysis of the multivariate models revealed a good performance on predicting the presence of CHD and CHD severity. In detail, ABCA1 gene expression showed an AUC of 0.768 (p<0.001) in predicting CHD (Fig 4A), while mRNA expression levels of SREBF2 provided an AUC of 0.815 (p<0.001) for the prediction of obstructive CHD (Fig 4B). Subgroup analysis comparing polygenic dyslipidemia treated vs untreated patients showed no significant differences for the expression of selected molecular markers.
Fig 4.
(A) ROC curve analysis generated from the multivariate model for the presence of CHD and ABCA1 gene expression; (B) ROC curve analysis generated from the multivariate model for the presence of obstructive CHD and SREBF2 gene expression.
Plaque composition
According to CACS stratification (see S1 Appendix for details), molecular data showed high percentage of methylation in the analyzed CpG segment of LDLR promoter in severe group compared to normal, low and moderate groups (p<0.001; p = 0.017; p = 0.002, respectively) (S1A Fig). An up-regulation of SREBF2 and ABCA1 mRNA levels was observed in severe (p = 0.002 for both markers) and moderate (p = 0.038, and p = 0.008, respectively) compared to normal group. Moreover, SREBF2 gene was overexpressed in severe compared to low group (p = 0.038) (S1B and S1C Fig).
As regarding quantitative plaque parameters (see Supplementary material for details), LDLR promoter methylation was higher in CHD patients with CPV>50 compared to HS (p = 0.02) and to CPV<50 (p = 0.036) (Fig 5A). SREBF2 mRNA was up-regulated in CPV>50 group vs HS (p = 0.002) and in CPV>50 vs CPV<50 group (p = 0.026) (Fig 5B). ABCA1 relative expression was higher in CPV>50 respect to HS (p = 0.004) with no statistically significant difference between the two CHD subgroups (Fig 5C and Table 4).
Fig 5.
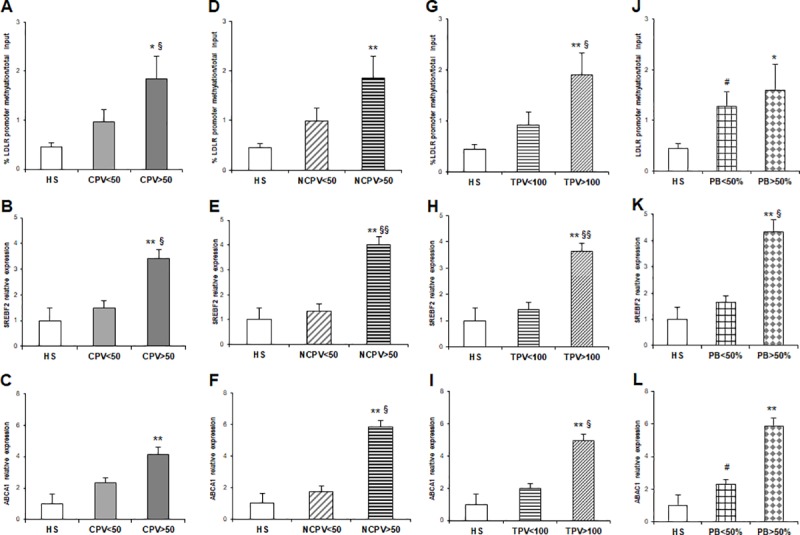
(A) % of LDLR promoter methylation/total input in CHD patients with CPV<50 (n = 35) and CPV>50 (n = 30) compared to HS (n = 30); (B-C) SREBF2 and ABCA1 mRNA relative expression in CHD patients with CPV<50 and CPV>50 vs HS; (D) % of LDLR promoter methylation/total input in CHD patients with NCPV<50 (n = 36) and NCPV>50 (n = 29) compared to HS (n = 30); (E-F) SREBF2 and ABCA1 mRNA relative expression in CHD patients with NCPV<50 and NCPV>50 vs HS; (G) % of LDLR promoter methylation/total input in CHD patients with TPV<100 (n = 35) and TPV>100 (n = 30) compared to HS (n = 30); (H-I) SREBF2 and ABCA1 mRNA relative expression in CHD patients with TPV<100and TPV>100 vs HS; (K-L) SREBF2 and ABCA1 mRNA relative expression in CAD patients with PB<50% (n = 46) and PB>50% (n = 19) vs HS (§p value<0.05 vs PB<50); (*p value<0.05 vs HS; **p value<0.01 vs HS for all comparisons).
Table 4. Plaque composition and molecular analysis.
| NCPV (mm3) | <50 | >50 | p value* |
| LDLR promoter methylation** | 0.99±0.26 | 1.85±0.45 | 0.104 |
| SREBF2 gene expression‡ | 6.51±0.28 | 4.94±0.33 | 0.008 |
| ABCA1 gene expression | 7.09±0.31 | 5.36±0.39 | 0.022 |
| CPV (mm3) | <50 | >50 | p value* |
| LDLR promoter methylation | 0.96±0.25 | 1.85±0.45 | 0.036 |
| SREBF2 gene expression | 6.36±0.29 | 5.17±0.35 | 0.026 |
| ABCA1 gene expression | 6.7±0.31 | 5.87±0.44 | 0.214 |
| TPV (mm3) | <100 | >100 | p value* |
| LDLR promoter methylation | 0.92±0.26 | 1.90±0.44 | 0.021 |
| SREBF2 gene expression | 6.44±0.29 | 5.08±0.33 | 0.010 |
| ABCA1 gene expression | 6.92±0.30 | 5.62±0.43 | 0.048 |
| PB | <50% | >50% | p value* |
| LDLR promoter methylation | 1.28±0.29 | 1.60±0.51 | 0.493 |
| SREBF2 gene expression | 6.22±0.25 | 4.82±0.44 | 0.015 |
| ABCA1 gene expression | 6.71±0.30 | 5.37±0.50 | 0.065 |
*p value from multiple comparisons. Bold values were considered statistically significant with a p < 0.05.
** Methylation data are expressed as percentage of methylation/total input.
‡Gene expression data are expressed in terms of Δct derived from qRT-PCR experiments.
ABCA1, ATP binding cassette subfamily A member 1; CPV, calcified plaque volume; LDLR, low density lipoprotein receptor; NCPV, non-calcified plaque volume; PB, plaque burden; SREBF2, sterol regulatory element-binding factor 2; TPV, total plaque volume.
Results from statistical analysis derived from the NCPV and the molecular data showed a differential methylation of LDLR promoter segment in CHD patients with NCPV>50 compared to HS (p = 0.005) but this epigenetic data was not able to discriminate patients with a prevalent non-calcified plaque composition (NCPV>50 vs NCPV<50 patients) (Fig 5D).
Gene expression analysis revealed an overexpression of SREBF2 and ABCA1. In detail, SREBF2 relative expression was higher in NCPV>50 patient category compared to HS (p = 0.001) with a differential overexpression as compared to NCPV<50 patients (p = 0.008). Significantly high expression levels of ABCA1 gene were found in NCPV>50 vs HS (p = 0.001) and NCPV>50 vs NCPV<50 (p = 0.022) (Fig 5E and 5F and Table 4).
MeDIP data showed an increased methylation in LDLR promoter of CHD patients with a TPV >100 (1.9%±0.44) compared to HS (p = 0.001) with a significant difference between TPV >100 and TPV <100 (0.92%±0.26; p = 0.021) groups (Fig 5G). SREBF2 and ABCA1 gene expression were significantly higher in TPV>100 compared to HS (p = 0.001 for both genes). Furthermore, SREBF2 and ABCA1 relative expressions were significantly higher in TPV>100 group compared to TPV<100 (p = 0.01 and p = 0.048, respectively) (Fig 5H and 5I and Table 4).
The analysis of molecular data in association to PB showed that LDLR promoter methylation, and the relative expression of SREBF2 and ABCA1 genes were overall associated with PB >50%. In particular, the percentage of methylation at the analyzed locus in LDLR promoter was high in CHD patients with PB>50% (1.60%±0.51; p = 0.024) compared to HS and in PB<50% patients (1.28%±0.29; p = 0.042) compared to HS. No statistical difference was found between the two CHD groups (Fig 5J).
SREBF2 gene was significantly upregulated in PB>50% patients compared to HS and to PB<50 patients (p = 0.001 and p = 0.015 respectively). ABCA1 was suggestive of a PB>50%, with high mRNA levels in CHD patients respect HS (p = 0.001) while border significances were obtained between PB>50% and PB<50% (p = 0.052) and between PB<50% and HS (p = 0.065) (Fig 5K and 5L and Table 4).
The univariate multinomial logistic regression analysis to predict coronary plaque features showed that SREBF2gene expression was able to predict/discriminate CPV<50 vs CPV>50 (OR = 0.702 CI 95%0.513–0.961; p = 0.027), NCPV<50 vs NCPV>50 (OR = 0.574 CI 95%0.391–0.845;p = 0.005) and TPV<100 vs TPV >100 (OR = 0.648 CI 95% 0.461–0.909; p = 0.012). Furthermore, ABCA1 gene expression was found to be associated with NCPV (ORNCPV<50vsNCPV>50 = 0.706 CI 95% 0.549–0.908; p = 0.007) and TPV (ORTPV<100vsTPV>100 = 0.784 CI 95% 0.624–0.984; p = 0.036) (Table 5). Adjusted multivariate analysis confirmed SREBF2 as independent predictor of CPV, NCP and TPV (p = 0.022; p = 0.002 and p = 0.006, respectively) and ABCA1 as independent predictor of NCPV and TPV (p = 0.002 and p = 0.013) (Table 6).
Table 5. Univariate multinomial logistic regression analysis to predict coronary plaque features.
| OR (95% CI) | p value | OR (95% CI) | p value | OR (95% CI) | p value | |
|---|---|---|---|---|---|---|
| Predictors | CPV<50 vs HS | CPV>50 vs HS | CPV<50 vs CPV>50 | |||
| LDLR promoter methylation | 1.922 (0.892–4.139) | 0.095 | 2.447 (1.135–5.274) | 0.022 | 1.273 (0.958–1.693) | 0.096 |
| LDLR gene expression | 0.660 (0.434–1.003) | 0.051 | 0.660 (0.430–1.011) | 0.056 | 1 (0.730–1.369) | 0.998 |
| SREBF2 gene expression | 0.881 (0.699–1.109) | 0.280 | 0.618 (0.447–0.855) | 0.004 | 0.702 (0.513–0.961) | 0.027 |
| ABCA1 gene expression | 0.851 (0.706–1.025) | 0.089 | 0.735 (0.588–0.917) | 0.007 | 0.863 (0.698–1.068) | 0.175 |
| Predictors | NCPV<50 vs HS | NCPV>50 vs HS | NCPV<50 vs NCPV>50 | |||
| LDLR promoter methylation | 1.940 (0.903–4.169) | 0.089 | 2.440 (1.133–5.257) | 0.023 | 1.258 (0.953–1.659) | 0.105 |
| LDLR gene expression | 0.710 (0.467–1.080) | 0.109 | 0.598 (0.383–0.932) | 0.023 | 0.842 (0.598–1.185) | 0.324 |
| SREBF2 gene expression | 0.910 (0.723–1.144) | 0.418 | 0.522 (0.352–0.775) | 0.001 | 0.574 (0.391–0.845) | 0.005 |
| ABCA1 gene expression | 0.895 (0.746–1.073) | 0.231 | 0.632 (0.485–0.823) | 0.001 | 0.706 (0.549–0.908) | 0.007 |
| Predictors | TPV<100 vs HS | TPV>100 vs HS | TPV<100 vs TPV>100 | |||
| LDLR promoter methylation | 1.893 (0.876–4.093) | 0.105 | 2.484 (1.149–5.372) | 0.021 | 1.312 (0.976–1.764) | 0.072 |
| LDLR gene expression | 0.675 (0.444–1.0124) | 0.065 | 0.642 (0.418–0.987) | 0.043 | 0.952 (0.695–1.306) | 0.762 |
| SREBF2 gene expression | 0.896 (0.712–1.128) | 0.351 | 0.580 (0.409–0.823) | 0.002 | 0.648 (0.461–0.909) | 0.012 |
| ABCA1 gene expression | 0.876 (0.729–1.053) | 0.159 | 0.687 (0.541–0.872) | 0.002 | 0.784 (0.624–0.984) | 0.036 |
Bold values were considered statistically significant with a p < 0.05.ABCA1, ATP binding cassette subfamily A member; CHD, coronary heart disease; CI, confidence interval; CPV, calcified plaque volume; LDLR, Low density lipoprotein receptor; NCPV, non-calcified plaque volume; OR, odds ratio; SREBF2, sterol regulatory element-binding factor 2; TPV, total plaque volume.
Table 6. Multivariate multinomial logistic regression analysis§ of risk factors associated with coronary plaque features.
| Predictors | CPV<50 vs HS | CPV>50 vs HS | CPV<50 vs CPV>50 | |||
| Model 1 | OR (95% CI) | p value | OR (95% CI) | p value | OR (95% CI) | p value |
| Male | 0.266 (0.078–0.909) | 0.035 | 0.080 (0.019–0.343) | 0.001 | 0.302 (0.082–1.116) | 0.073 |
| Dyslipidemia treatment | 0.164 (0.040–0.677) | 0.012 | 0.105 (0.023–0.0476) | 0.003 | 0.639 (0.208–1.964) | 0.434 |
| Physical activity | 0.162 (0.045–0.579) | 0.005 | 0.626 (0.145–2.709) | 0.531 | 3.878 (1.220–12.322) | 0.022 |
| SREBF2 gene expression | 0.867 (0.651–1.156) | 0.331 | 0.574 (0.390–0.846) | 0.005 | 0.662 (0.466–0.942) | 0.022 |
| NCPV<50 vs HS | NCPV>50 vs HS | NCPV<50 vs NCPV>50 | ||||
| Model 1 | OR (95% CI) | p value | OR (95% CI) | p value | OR (95% CI) | p value |
| Male | 0.365 (0.108–1.238) | 0.106 | 0.24 (0.004–0.156) | <0.001 | 0.067 (0.012–0.375) | 0.002 |
| Dyslipidemia treatment | 0.150 (0.038–0.588) | 0.007 | 0.140 (0.27–0.713) | 0.018 | 0.934 (0.269–3.246) | 0.914 |
| Physical activity | 0.174 (0.49–0.620) | 0.007 | 0.626 (0.134–2.929) | 0.552 | 3.603 (1.010–12.851) | 0.048 |
| SREBF2 gene expression | 0.900 (0.677–1.195) | 0.466 | 0.441 (0.274–0.710) | 0.001 | 0.490 (0.313–0.766) | 0.002 |
| Model 2 | OR (95% CI) | p value | OR (95% CI) | p value | OR (95% CI) | p value |
| Male | 0.328 (0.092–1.174) | 0.087 | 0.016 (0.002–0.115) | <0.001 | 0.049 (0.008–0.299) | 0.001 |
| Dyslipidemia treatment | 0.165 (0.043–0.639) | 0.009 | 0.161 (0.032–0.820) | 0.028 | 0.974 (0.276–3.430) | 0.967 |
| Physical activity | 0.201 (0.054–0.745) | 0.016 | 0.790 (0.166–3.770) | 0.768 | 3.926 (1.082–14.247) | 0.038 |
| ABCA1 gene expression | 0.899 (0.715–1.129) | 0.359 | 0.537 (0.379–0.762) | <0.001 | 0.598 (0.435–0.822) | 0.002 |
| TPV<100 vs HS | TPV>100 vs HS | TPV<100 vs TPV>100 | ||||
| Model 1 | OR (95% CI) | p value | OR (95% CI) | p value | OR (95% CI) | p value |
| Male | 0.263 (0.076–0.913) | 0.035 | 0.084 (0.020–0.348) | 0.001 | 0.320 (0.087–1.182) | 0.087 |
| Dyslipidemia treatment | 0.126 (0.031–0.516) | 0.004 | 0.162 (0.036–0.731) | 0.018 | 1.291 (0.408–4.079) | 0.664 |
| Physical activity | 0.144 (0.039–0.531) | 0.004 | 0.666 (0.154–2.875) | 0.586 | 4.638 (1.412–15.239) | 0.011 |
| SREBF2 gene expression | 0.900 (0.673–1.204) | 0.478 | 0.525 (0.348–0.794) | 0.002 | 0.584 (0.397–0.858) | 0.006 |
| Model 2 | OR (95% CI) | p value | OR (95% CI) | p value | OR (95% CI) | p value |
| Male | 0.236 (0.066–0.850) | 0.027 | 0.066 (0.015–0.284) | <0.001 | 0.278 (0.074–1.039) | 0.057 |
| Dyslipidemia treatment | 0.142 (0.035–0.575) | 0.006 | 0.184 (0.041–0.824) | 0.027 | 1.296 (0.411–4.084) | 0.658 |
| Physical activity | 0.171 (0.045–0.652) | 0.010 | 0.812 (0.186–3.546) | 0.781 | 4.741 (1.443–15.577) | 0.010 |
| ABCA1 gene expression | 0.884 (0.703–1.113) | 0.295 | 0.638 (0.479–0.848) | 0.002 | 0.721 (0.558–0.932) | 0.013 |
Bold values were considered statistically significant with a p < 0.05.
§Multivariate analysis corrected for male gender.
ABCA1, ATP binding cassette subfamily A member; CHD, coronary heart disease; CI, confidence interval; CPV, calcified plaque volume; NCPV, non-calcified plaque volume; OR, odds ratio; SREBF2, sterol regulatory element-binding factor 2; TPV, total plaque volume.
Discussion
Here, we evaluated in PBMNCs from suspected CHD patients and HS, undergoing CCT, the methylation status of specific regulatory regions in principal genes involved in cholesterol pathway. Furthermore, we have analyzed the mRNA levels of the selected genes.
Some studies associated mutations or polymorphisms in LDLR gene and impairment of its function with atherosclerosis burden and/or an increased incidence of CVDs and a high risk for both incident and recurrent cardiac acute events [37–40]. In particular, Ten Kate et al. performed an association study considering the mutational status of LDLR gene and the extent of subclinical coronary atherosclerosis, detected by CCT, expressed in terms of Diseased Segment Score (DSS) [40].
Our study investigated for the first time the methylation and expression levels of LDLR with quantitative features derived from CCT. A study by Liu et al., suggested that abnormal lipid homeostasis may contribute to the pathogenesis of vascular calcification. Indeed, authors performed an immunohistochemical analysis reporting an up-regulation of LDLR in atherosclerotic vascular tissues and a parallel increase in calcified plaque deposition [41]. Even if LDLR expression was evaluated at protein level and in vascular tissues, our results, obtained from PBMNCs, are in line with the above-mentioned data. We established in a non-invasive way that high levels of LDLR gene were associated with the presence of CHD and in particular to obstructive CHD. Furthermore, the degree of methylation in a promoter region of the same gene was more associated with severe CACS.
SREBF2 performs a fine control on the transcription of sterol-regulated genes such as LDLR and ABCA1. Previous studies reported an altered SREBF2 gene expression in atherosclerotic tissues as well as an association of specific genotypes with the risk of cardiovascular events and sudden cardiac death [42–45]. Shchelkunova et al., analyzed the expression levels of SREBF2 gene in human aortic samples. Results showed that SREBF2 mRNA was found comparable to non-diseased tissue in initial lesions. Otherwise SREBF2 transcript was found to be progressively up-regulated in fatty streaks and fibrous lipid plaques, supporting an active role of this biomolecule during the atherosclerosis progression [46]. The present study shows an association between the circulating expression levels of SREBF2 mRNA in PBMNCs and the presence of obstructive CHD. We have detected, in non-invasive manner, that SREBF2 gene expression was a predictor of critical stenosis independent by gender, so it could be a surrogate biomarker of CHD and disease severity.
Regarding the molecular analysis of ABCA1, literature data reported significant associations of dysregulation of this cholesterol transporter with the atherosclerotic process and CHD [47–52]. Indeed, an involvement of ABCA1 has been demonstrated in the formation and composition of carotid atherosclerotic plaques with an upregulation of ABCA1 levels between atherosclerotic and healthy tissues [51], as well as the significance of this biomarker in plaque rupture [52]. In our study we have shown from peripheral blood sample that ABCA1 mRNA was up-regulated in patients with CHD underwent to CCT, suggesting a possible implication in disease diagnosis and high-risk plaque detection. In light of our results, we hypothesize that ABCA1 could be a predictive biomarker of CHD while SREBF2 could be involved in the identification of obstructive stenosis. In addition, plaque feature results suggest SREBF2 gene expression as predictive of total plaque burden (both calcified and non-calcified plaque component) while ABCA1 could be related to non-calcified plaque composition hence allowing to identify high-risk plaque.
Considering that the selected molecular markers are all involved in the cholesterol metabolism, we firstly compared total, LDL, HDL, and cholesterol serum concentrations between CHD and HS group, reporting no significant alterations at the time of CCT. Following, in order to investigate the effects of dyslipidemia treatments on the expression of the selected molecular markers, we performed a subgroup analysis comparing treated vs untreated subjects, reporting no effects of medications on ABCA1 and SREBF2 gene expression and on the percentage of LDLR promoter methylation.
The present study has some limitations. We recognize that a genome-wide approach could have been the best strategy in identifying more molecular markers associated to CHD presence, severity and coronary plaque features. The epigenetic and analysis was performed considering only the methylation levels of limited portions in promoters and regulatory elements in a specific set of genes of interest already recognized to be related to atherogenesis and endothelial function in vascular tissues. Although GWAS and EWAS require huge amount of blood samples, further studies are needed to evaluate other epigenetic modified sequences and we cannot exclude that others post transcriptional events (i.e. microRNAs) may influence mRNA expression levels.
Another limitation is represented by the small sample size, moreover our analysis was conducted only in suspected CHD patients but an evaluation of patients with known CHD should be considered for future analysis. It could be of great interest to evaluate a possible association of these epigenetic-sensitive biomarkers with occurrence/recurrence of cardiac acute events. At the same time, a further analysis could investigate the association of such biomarkers with myocardial ischemia to assess the morphological-functional relationship in CHD patients.
Moreover, the prognostic value of the studied cholesterol/metabolic biomarkers for cardiovascular events and their contribution to the risk-stratification of the population undergoing diagnostic CCT could be tested in a larger cohort with prospective follow-up.
Finally, although the increasing technological widespread, the costs incurred to perform gene expression/methylation analysis have to be taken into account, although the high sensitivity, specificity and reproducibility of qRT-PCR technique.
Conclusions and future perspectives
Our study could have an important clinical impact, representing a possible initial epigenetic/molecular-sensitive approach in the diagnosis and prediction of CHD through a non-invasive approach. Therefore, the evaluation of reproducible and non-invasive techniques for assessing endothelial function should enable screening of large populations and may guide interventions designed specifically to reduce the individual vascular risk.
Supporting information
(DOCX)
(DOCX)
(A) % of LDLR promoter methylation/total input in CHD patients categorized according to CACS levels in: Normal (CACS = 0) (n = 32), Low (CACS = 1–100) (n = 21), Moderate (CACS = 101–400) (n = 18), and Severe (CACS>400) (n = 24). (B) SREBF2 mRNA relative expression in CHD patients categorized according to CACS levels; (C) ABCA1 gene relative expression in CHD patients categorized according to CACS levels.
(TIF)
Abbreviations
- ABCA1
ATP binding cassette subfamily A member 1
- CCT
cardiac computed tomography
- CHD
coronary heart disease viruses
- CPV
calcified plaque volume
- LDLR
low density lipoprotein receptor
- NCPV
non-calcified plaque volume
- PB
plaque burden
- SREBF2
sterol regulatory element-binding factor 2
- TPV
total plaque volume
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This study was supported by Italian Ministry of Health grants “Giovani Ricercatori 2011-12”, project code GR-2011-02349436 (TI) and “Ricerca Corrente 2013-15” (CN). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Dalen JE, Alpert JS, Goldberg RJ, Weinstein RS. The epidemic of the 20(th) century: coronary heart disease. Am J Med. 2014; 127(9):807–12. 10.1016/j.amjmed.2014.04.015 . [DOI] [PubMed] [Google Scholar]
- 2.Weber C, Noels H. Atherosclerosis: current pathogenesis and therapeutic options. Nat Med. 2011; 17(11):1410–22. 10.1038/nm.2538 . [DOI] [PubMed] [Google Scholar]
- 3.Libby P, Ridker PM, Hansson GK. Progress and challenges in translating the biology of atherosclerosis. Nature. 2011; 473(7347):317–25. 10.1038/nature10146 . [DOI] [PubMed] [Google Scholar]
- 4.Grimaldi V, Vietri MT, Schiano C, Picascia A, De Pascale MR, Fiorito C,et al. Epigenetic reprogramming in atherosclerosis. Curr Atheroscler Rep. 2015; 17 (2):476 10.1007/s11883-014-0476-3 . [DOI] [PubMed] [Google Scholar]
- 5.Schiano C, Vietri MT, Grimaldi V, Picascia A, De Pascale MR, Napoli C. Epigenetic-related therapeutic challenges in cardiovascular disease. Trends Pharmacol Sci. 2015; 36(4):226–35. 10.1016/j.tips.2015.02.005 . [DOI] [PubMed] [Google Scholar]
- 6.Napoli C, Crudele V, Soricelli A, Al-Omran M, Vitale N, Infante T, et al. Primary prevention of atherosclerosis: a clinical challenge for the reversal of epigenetic mechanisms? Circulation. 2012; 125(19):2363–73. 10.1161/circulationaha.111.085787 . [DOI] [PubMed] [Google Scholar]
- 7.Napoli C, Grimaldi V, De Pascale MR, Sommese L, Infante T, Soricelli A. Novel epigenetic-based terapies useful in cardiovascular medicine. World J Cardiol. 2016; 8(2):211–19. 10.4330/wjc.v8.i2.211 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duan L, Hu J, Xiong X, Liu Y, Wang J. The role of DNA methylation in coronary artery disease. Gene. 2018; 646:91–97. 10.1016/j.gene.2017.12.033 . [DOI] [PubMed] [Google Scholar]
- 9.de Nigris F, Cacciatore F, Mancini FP, Vitale DF, Mansueto G, D'Armiento FP, et al. Epigenetic Hallmarks of Fetal Early Atherosclerotic Lesions in Humans. JAMA Cardiol. 2018; 3(12):1184–1191. 10.1001/jamacardio.2018.3546 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen HH, Stewart AF. Transcriptomic Signature of Atherosclerosis in the Peripheral Blood: Fact or Fiction? Curr Atheroscler Rep. 2016; 18(12):77 10.1007/s11883-016-0634-x . [DOI] [PubMed] [Google Scholar]
- 11.Wingrove JA, Daniels SE, Sehnert AJ, Tingley W, Elashoff MR, Rosenberg S, et al. Correlation of peripheral-blood gene expression with the extent of coronary artery stenosis. Circ Cardiovasc Genet. 2008;1:31–8. 10.1161/CIRCGENETICS.108.782730 . [DOI] [PubMed] [Google Scholar]
- 12.Sinnaeve PR, Donahue MP, Grass P, Seo D, Vonderscher J, Chibout SD, et al. Gene expression patterns in peripheral blood correlate with the extent of coronary artery disease. PLoS One. 2009;4:e7037 10.1371/journal.pone.0007037 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kontaraki JE, Kochiadakis GE, Marketou ME, Chlouverakis G, Igoumenidis NE, Saloustros IG, et al. Early cardiac gene transcript levels in peripheral blood mononuclear cells reflect severity in stable coronary artery disease. Hellenic J Cardiol. 2014;55:119–25. . [PubMed] [Google Scholar]
- 14.Mozzini C, Fratta Pasini A, Garbin U, Stranieri C, Pasini A, Vallerio P, et al. Increased endoplasmic reticulum stress and Nrf2 repression in peripheral blood mononuclear cells of patients with stable coronary artery disease. Free Radic Biol Med. 2014;68:178–85. 10.1016/j.freeradbiomed.2013.12.017 . [DOI] [PubMed] [Google Scholar]
- 15.Goldstein JL, Brown MS. The LDL receptor. Arterioscler Thromb Vasc Biol. 2009; 29(4):431–38. 10.1161/atvbaha.108.179564 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eberlé D, Hegarty B, Bossard P, Ferré P, Foufelle F. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie. 2004; 86(11):839–48. 10.1016/j.biochi.2004.09.018 . [DOI] [PubMed] [Google Scholar]
- 17.Sato R. Sterol metabolism and SREBP activation. Arch Biochem Biophys. 2010; 501(2):177–81. 10.1016/j.abb.2010.06.004 . [DOI] [PubMed] [Google Scholar]
- 18.Westerterp M, Bochem AE, Yvan-Charvet L, Murphy AJ, Wang N, et al. ATP-binding cassette transporters, atherosclerosis, and inflammation. Circ Res. 2014; 114(1):157–70. 10.1161/circresaha.114.300738 . [DOI] [PubMed] [Google Scholar]
- 19.Voros S, Elashoff MR, Wingrove JA, Budoff MJ, Thomas GS, Rosenberg S. A peripheral blood gene expression score is associated with atherosclerotic plaque burden and stenosis by cardiovascular CT-angiography: results from the PREDICT and COMPASS studies. Atherosclerosis. 2014;233(1):284–90. 10.1016/j.atherosclerosis.2013.12.045 . [DOI] [PubMed] [Google Scholar]
- 20.Harada K, Amano T, Uetani T, Yoshida T, Kato B, Kato M, et al. Association of inflammatory markers with the morphology and extent of coronary plaque as evaluated by 64-slice multidetector computed tomography in patients with stable coronary artery disease. Int J Cardiovasc Imaging. 2013; 29(5):1149–58. 10.1007/s10554-013-0181-2 . [DOI] [PubMed] [Google Scholar]
- 21.Canpolat U, Aytemir K, Yorgun H, Hazırolan T, Kaya EB, Şahiner L, et al. Association of serum total bilirubin levels with the severity, extent and subtypes of coronary atherosclerotic plaques detected by coronary CT angiography. Int J Cardiovasc Imaging. 2013; 29(6):1371–79. 10.1007/s10554-013-0209-7 . [DOI] [PubMed] [Google Scholar]
- 22.Kunita E, Yamamoto H, Kitagawa T, Ohashi N, Utsunomiya H, Oka T,et al. Association between plasma high molecular-weight adiponectin and coronary plaque characteristics assessed by computed tomography angiography in conditions of visceral adipose accumulation. Circ J. 2012; 76(7):1687–96. 10.1253/circj.cj-66-0044 . [DOI] [PubMed] [Google Scholar]
- 23.Bamberg F, Truong QA, Koenig W, Schlett CL, Nasir K, Butler J, et al. Differential associations between blood biomarkers of inflammation, oxidation, and lipid metabolism with varying forms of coronary atherosclerotic plaque as quantified by coronary CT angiography. Int J Cardiovasc Imaging. 2012; 28(1):183–92. 10.1007/s10554-010-9773-2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forte E, Aiello M, Inglese M, Infante T, Soricelli A, Tedeschi C, et al. Coronary artery aneurysms detected by computed tomography coronary angiography. Eur Heart J Cardiovasc Imaging. 2017; 18(11):1229–35. 10.1093/ehjci/jew218 . [DOI] [PubMed] [Google Scholar]
- 25.Forte E, Inglese M, Infante T, Schiano C, Napoli C, Soricelli A, et al. Anomalous left main coronary artery detected by CT angiography. Surg Radiol Anat. 2016; 38(8):987–90. 10.1007/s00276-016-1634-9 . [DOI] [PubMed] [Google Scholar]
- 26.Infante T, Forte E, Schiano C, Cavaliere C, Tedeschi C, Soricelli A, et al. An integrated approach to coronary heart disease diagnosis and clinical management. Am J Transl Res. 2017; 9(7):3148–66. . [PMC free article] [PubMed] [Google Scholar]
- 27.Rizvi A, Hartaigh BÓ, Danad I, Han D, Lee JH, GransarH, et al. Diffuse coronary artery disease among other atherosclerotic plaque characteristics by coronary computed tomography angiography for predicting coronary vessel-specific ischemia by fractional flow reserve. Atherosclerosis. 2017; 258:145–51. 10.1016/j.atherosclerosis.2017.01.018 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cademartiri F, Nistri S, Tarantini G, Maffei E. Management of coronary artery disease with cardiac CT beyond gatekeeping. Heart. 2017; 103(13):975–76. 10.1136/heartjnl-2016-310473 . [DOI] [PubMed] [Google Scholar]
- 29.Bobryshev YV, Ivanova EA, Chistiakov DA, Nikiforov NG, Orekhov AN. Macrophages and Their Role in Atherosclerosis: Pathophysiology and Transcriptome Analysis. Biomed Res Int. 2016; 2016:9582430 10.1155/2016/9582430 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tabas I, Lichtman AH. Monocyte-Macrophages and T Cells in Atherosclerosis. Immunity. 2017; 47(4):621–34. 10.1016/j.immuni.2017.09.008 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mirabelli P, Incoronato M, Coppola L, Infante T, Parente CA, Nicolai E,et al. SDN Biobank: Bioresource of Human Samples Associated with Functional and/or Morphological Bioimaging Results for the Study of Oncological, Cardiological, Neurological, and Metabolic Diseases. Open Journal of Bioresources 2017; 4:p.2 10.5334/ojb.26. [DOI] [Google Scholar]
- 32.Song Q, Decato B, Hong EE, Zhou M, Fang F, Qu J, et al. A reference methylome database and analysis pipeline to facilitate integrative and comparative epigenomics. PLoS One. 2013; 8(12):e81148 10.1371/journal.pone.0081148 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rienzo M, Schiano C, Casamassimi A, Grimaldi V, Infante T, Napoli C. Identification of valid reference housekeeping genes for gene expression analysis in tumor neovascularization studies. Clin Transl Oncol. 2013; 15(3):211–18. 10.1007/s12094-012-0904-1 . [DOI] [PubMed] [Google Scholar]
- 34.Infante T, Forte E, Aiello M, Salvatore M, Cavaliere C. In Vivo and In Vitro Analysis in Coronary Artery Disease Related to Type 2 Diabetes. Front Endocrinol (Lausanne). 2017; 8:209 10.3389/fendo.2017.00209 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leipsic J, Abbara S, Achenbach S, Cury R, Earls JP, Mancini GJ,et al. SCCT guidelines for the interpretation and reporting of coronary CT angiography: A report of the Society of Cardiovascular Computed Tomography Guidelines Committee. J Cardiovasc Comput Tomogr. 2014; 8(5):342–58. 10.1016/j.jcct.2014.07.003 . [DOI] [PubMed] [Google Scholar]
- 36.Guaricci AI, Pontone G, Fusini L, De Luca M, Cafarelli FP, Guglielmo M,et al. Additional value of inflammatory biomarkers and carotid artery disease in prediction of significant coronary artery disease as assessed by coronary computed tomography angiography. Eur Heart J Cardiovasc Imaging. 2017; 18(9):1049–56. 10.1093/ehjci/jew173 . [DOI] [PubMed] [Google Scholar]
- 37.Romano M, Di Taranto MD, D'Agostino MN, Marotta G, Gentile M, Abate G,et al. Identification and functional characterization of LDLR mutations in familial hypercholesterolemia patients from Southern Italy. Atherosclerosis. 2010; 210(2):493–96. 10.1016/j.atherosclerosis.2009.11.051 . [DOI] [PubMed] [Google Scholar]
- 38.De Castro-Orós I, Pérez-López J, Mateo-Gallego R, Rebollar S, Ledesma M, León M,et al. A genetic variant in the LDLR promoter is responsible for part of the LDL-cholesterol variability in primary hypercholesterolemia. BMC Med Genomics. 2014; 7:17 10.1186/1755-8794-7-17 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mega JL, Stitziel NO, Smith JG, Chasman DI, Caulfield M, Devlin JJ,et al. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials. Lancet. 2015; 385(9984):2264–71. 10.1016/s0140-6736(14)61730-x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ten Kate GJ, Neefjes LA, Dedic A, Nieman K, Langendonk JG, Galema-Boers AJ, et al. The effect of LDLR-negative genotype on CT coronary atherosclerosis in asymptomatic statin treated patients with heterozygous familial hypercholesterolemia. Atherosclerosis. 2013; 227(2):334–41. 10.1016/j.atherosclerosis.2012.12.016 . [DOI] [PubMed] [Google Scholar]
- 41.Liu J, Ma KL, Gao M, Wang CX, Ni J, Zhang Y, et al. Inflammation disrupts the LDL receptor pathway and accelerates the progression of vascular calcification in ESRD patients. PLoS One. 2012; 7(10):e47217 10.1371/journal.pone.0047217 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fan YM, Karhunen PJ, Levula M, Ilveskoski E, Mikkelsson J, Kajander OA, et al. Expression of sterol regulatory element-binding transcription factor (SREBF) 2 and SREBF cleavage-activating protein (SCAP) in human atheroma and the association of their allelic variants with sudden cardiac death. Thromb J. 2008; 6:17 10.1186/1477-9560-6-17 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Friedlander Y, Schwartz SM, Durst R, Meiner V, Robertson AS, Erez G, et al. SREBP-2 and SCAP isoforms and risk of early onset myocardial infarction. Atherosclerosis. 2008; 196(2):896–904. 10.1016/j.atherosclerosis.2007.02.006 . [DOI] [PubMed] [Google Scholar]
- 44.Miserez AR, Muller PY, Barella L, Barella S, Staehelin HB, Leitersdorf E,et al. Sterol-regulatory element-binding protein (SREBP)-2 contributes to polygenic hypercholesterolaemia. Atherosclerosis. 2002; 164(1):15–26. 10.1016/s0021-9150(01)00762-6 . [DOI] [PubMed] [Google Scholar]
- 45.Muller PY, Miserez AR. Identification of mutations in the gene encoding sterol regulatory element binding protein (SREBP)-2 in hypercholesterolaemic subjects. J Med Genet. 2002;39(4):271–75. 10.1136/jmg.39.4.271 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shchelkunova TA, Morozov IA, Rubtsov PM, Bobryshev YV, Sobenin IA, Orekhov AN,et al. Lipid regulators during atherogenesis: expression of LXR, PPAR, and SREBP mRNA in the human aorta. PLoS One. 2013; 8(5):e63374 10.1371/journal.pone.0063374 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strang AC, Hovingh GK, Stroes ES, Kastelein JJ. The genetics of high-density lipoprotein metabolism: clinical relevance for therapeutic approaches. Am J Cardiol. 2009; 104(10 Suppl):22E–31E. 10.1016/j.amjcard.2009.09.016 . [DOI] [PubMed] [Google Scholar]
- 48.Clee SM, Kastelein JJ, van Dam M, Marcil M, Roomp K, Zwarts KY,et al. Age and residual cholesterol efflux affect HDL cholesterol levels and coronary artery disease in ABCA1 heterozygotes. J Clin Invest. 2000; 106(10):1263–70. 10.1172/jci10727 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Demina EP, Miroshnikova VV, Rodygina TI, Kur'ianov PS, Vinogradov AG, Denisenko AD,et al. ABCA1 gene expression in peripheral blood lymphocytes and macrophages in patients with atherosclerosis. Mol Biol (Mosk). 2011; 45(2):289–93. 10.1134/s0026893310061019 . [DOI] [PubMed] [Google Scholar]
- 50.Maiwald S, Zwetsloot PP, Sivapalaratnam S, Dallinga-Thie GM. Monocyte gene expression and coronary artery disease. Curr Opin Clin Nutr Metab Care. 2013; 16(4):411–17. 10.1097/mco.0b013e32836236f9 . [DOI] [PubMed] [Google Scholar]
- 51.Liu HF, Cui KF, Wang JP, Zhang M, Guo YP, Li XY, et al. Significance of ABCA1 in human carotid atherosclerotic plaques. Exp Ther Med. 2012; 4(2):297–302. 10.3892/etm.2012.576 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heo SH, Lee EH, Park HH, Kim BJ, Youn HC, Kim YS, et al. Differences between the Molecular Mechanisms Underlying Ruptured and Non-Ruptured Carotid Plaques, and the Significance of ABCA1. J Stroke. 2018; 20(1):80–91. 10.5853/jos.2017.02390 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(A) % of LDLR promoter methylation/total input in CHD patients categorized according to CACS levels in: Normal (CACS = 0) (n = 32), Low (CACS = 1–100) (n = 21), Moderate (CACS = 101–400) (n = 18), and Severe (CACS>400) (n = 24). (B) SREBF2 mRNA relative expression in CHD patients categorized according to CACS levels; (C) ABCA1 gene relative expression in CHD patients categorized according to CACS levels.
(TIF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.