Abstract
Background and Aims
Although dioecy, which characterizes only 6 % of angiosperm species, has been considered an evolutionary dead end, recent studies have demonstrated that this is not necessarily the case. Moraceae (40 genera, 1100 spp., including Ficus, 750 spp.) are particularly diverse in breeding systems (including monoecy, gynodioecy, androdioecy and dioecy) and thus represent a model clade to study macroevolution of dioecy.
Methods
Ancestral breeding systems of Ficus and Moraceae were inferred. To do so, a new dated phylogenetic tree of Ficus and Moraceae was first reconstructed by combining a revised 12-fossil calibration set and a densely sampled molecular data set of eight markers and 320 species. Breeding system evolution was then reconstructed using both parsimony and model-based (maximum likelihood and Bayesian) approaches with this new time scale.
Key Results
The crown group ages of Ficus and Moraceae were estimated in the Eocene (40.6–55.9 Ma) and Late Cretaceous (73.2–84.7 Ma), respectively. Strong support was found for ancestral dioecy in Moraceae. Although the ancestral state of Ficus remained particularly sensitive to model selection, the results show that monoecy and gynodioecy evolved from dioecy in Moraceae, and suggest that gynodioecy probably evolved from monoecy in Ficus.
Conclusions
Dioecy was found not to be an evolutionary dead end in Moraceae. This study provides a new time scale for the phylogeny and a new framework of breeding system evolution in Ficus and Moraceae.
Keywords: Breeding system evolution, dioecy, evolutionary dead end, molecular dating, ancestral state reconstruction, fossil calibration, Ficus, Moraceae
INTRODUCTION
Flowers are the reproductive structures of angiosperms. The approx. 350 000 species of angiosperms are highly diverse in floral morphological traits, including breeding systems, ranging from bisexuality (hermaphroditism) to separate sex on distinct plants (dioecy) through several presumed intermediate states between these two ends (monoecy, andromonoecy, gynomonoecy, androdioecy and gynodioecy) (Renner, 2014). Although a recent study demonstrated that bisexual flowers are ancestral in angiosperms and evolved many times independently to unisexual flowers (Sauquet et al., 2017), the exact number and the context of these transitions remain to be characterized. Dioecy is rare in angiosperms (only approx. 6 %) (Renner, 2014) and has been suggested to be an evolutionary dead end (Bull and Charnov, 1985; Heilbuth, 2000). Transitions from hermaphroditism to dioecy have been suggested to occur through three alternative pathways: the dimorphic pathway (through gynodioecy, androdioecy or polygamodioecy) (Dufay et al., 2014); the monomorphic pathway (through monoecy, andromonoecy, gynomonoecy or polygamomonoecy) (Renner and Ricklefs, 1995); and the direct pathway (Barrett, 2002; Goldberg et al., 2017). However, the view of dioecy as an evolutionary dead end has gradually changed over the last decade after it was found that transitions from dioecy to other breeding systems are possible and that the flexibility of breeding system transitions may be caused by different selective pressures and constraints (Barrett, 2013; Goldberg et al., 2017). Thus, it remains unclear why dioecy is in fact so rare in angiosperms (Käfer et al., 2017).
The mulberry family (Moraceae) consists of approx. 40 genera and 1100 species, and represents a good model system to study dioecy transitions (Clement and Weiblen, 2009). Four breeding systems are observed in the family: monoecy, androdioecy, gynodioecy and dioecy (Datwyler and Weiblen, 2004). Ficus, the largest genus in Moraceae, contains almost 75 % of the species in the family (750 spp.). About half of the species of Ficus are monoecious, while the other half are gynodioecious (Cook and Rasplus, 2003). In monoecious species of Ficus, both functionally male and female flowers coexist in the same inflorescence (syconium); female flowers vary in style length. Fig wasp pollinators (Agaonidae) lay eggs in part of these female flowers and pollinate the others (Cook and Rasplus, 2003). Gynodioecious species of Ficus are characterized by two kinds of plants and inflorescences: female individuals bear syconia that contain only functionally female flowers (i.e. flowers that can develop seeds) and functionally male individuals bear syconia that contain both functionally male and gall flowers (female flowers that develop only wasps) (Cook and Rasplus, 2003). Therefore, structurally gynodioecious species of figs are functionally dioecious.
A previous study, based on parsimony reconstruction sampling 46 Ficus species (but no outgroup), suggested that monoecy is ancestral in Ficus, and that gynodioecy originated at least once or twice within the genus (Weiblen, 2000). A subsequent family-level parsimony study, sampling 83 Moraceae species (including 11 species of Ficus) suggested monoecy and dioecy to be ancestral in Ficus and Moraceae, respectively (Datwyler and Weiblen, 2004). However, the accuracy and precision of ancestral state reconstruction depend on the reliability of the phylogenetic tree and sampling density, and important progress has been made to reconstruct phylogenetic relationships in Ficus and Moraceae since these two studies were published (Zerega et al., 2005; Clement and Weiblen, 2009; Zerega et al., 2010; Cruaud et al., 2012; Williams et al., 2017). In addition, new probabilistic approaches for ancestral state reconstruction, taking into account phylogenetic uncertainty and divergence times, are now available (Pagel et al., 2004). Parsimony, maximum likelihood (ML) and Bayesian approaches today are the most commonly used methods for reconstructing ancestral states. The accuracy of branch length estimation may be important in model-based approaches (ML and Bayesian), which rely on them for computing the likelihood of evolutionary change along the phylogeny (Pagel and Meade, 2006). However, to date only a few attempts have been made to estimate divergence times in Moraceae (Datwyler and Weiblen, 2004; Zerega et al., 2005). Although more studies have addressed the time scale of Ficus evolution (Rønsted et al., 2005; Zerega et al., 2005; Cruaud et al., 2012), partly inconsistent results have been obtained, for instance with crown group age estimates for Ficus ranging from 40.1 to 101.9 million years ago (Mya). In addition to providing a framework to reconstruct ancestral states, estimating divergence times in Ficus and Moraceae is also important for other evolutionary questions, including the evaluation of the co-diversification and biogeographic history of the genus (Cruaud et al., 2012).
Here, we investigate two key questions. (1) What are the crown group ages of Ficus and Moraceae? (2) What are the ancestral breeding systems of Ficus and Moraceae, and how many times did monoecy and dioecy evolve in Ficus and Moraceae? To do so, we reconstruct a new phylogenetic tree for Moraceae using a densely sampled molecular data set, estimate divergence times using a relaxed molecular clock calibrated with a revised set of 12 fossil age constraints, and reconstruct ancestral breeding systems in Ficus and Moraceae with state-of-the-art model-based approaches. We estimate the age of the crown group of Ficus and Moraceae in the Eocene and Late Cretaceous, respectively. Our results suggest that dioecy is not an evolutionary dead end in Moraceae and the transitions from dioecy to the other breeding systems (androdioecy, gynodioecy and monoecy) occurred several times during the evolutionary history of the family. This study sheds light on the evolution of dioecy into other breeding systems and more generally improves our understanding of breeding system evolution in angiosperms.
MATERIALS AND METHODS
Taxonomic sampling and molecular data set assembly
We selected GenBank sequences from 320 species belonging to 65 genera and eight families of Rosales, including 272 species and 36 genera (three-quarters of the approx. 40 genera) of Moraceae (Supplementary Data Table S1). Our efforts include a comprehensive sample of outgroups in order to estimate accurately divergence times in the family while taking advantage of the rich fossil record of the order (see below). We used eight markers from two genomes: three chloroplast genes (matK, rbcL and ndhF) and five nuclear markers including two non-coding regions [internal transcibed spacer (ITS) and external transcribed spacer (ETS)] and three genes (G3pdh, ncpGS and the waxy region). This combination of coding and non-coding markers was selected to resolve both deep- and shallow-level relationships. MatK and rbcL have been proposed as standard barcoding regions for land plants (Hollingsworth et al., 2009) and ndhF proved to be very useful in previous phylogenetic studies of Moraceae (Datwyler and Weiblen, 2004; Zerega et al., 2005; Clement and Weiblen, 2009). ITS, ETS, G3pdh, ncpGS and the waxy region were used in the latest and most densely sampled phylogenetic study of Ficus (Cruaud et al., 2012). Considering the comparatively fast molecular rates of ITS and ETS, these markers may be difficult to align and may introduce more noise than signal at the family level. Therefore, ITS and ETS sequences were here only used for species of Ficus. All sequences were aligned using Muscle v3.7 (Edgar, 2004) as implemented on CIPRES (Miller et al., 2010); alignments were then checked and adjusted by hand. In the ITS alignment, we deleted three short regions (approx. 10 bp) because of the uncertainty of gap length and position of base pairs in these regions. Combined multimarker alignments were assembled with Mesquite v3.04 (Maddison and Maddison, 2016).
Phylogenetic reconstruction
To identify and exclude problematic GenBank sequences (e.g. incorrectly identified), separate gene analyses were conducted first with RAxML v8.2.9 (Stamatakis, 2014) on CIPRES (Miller et al., 2010), using the GTRCAT model and 100 bootstrap replicates. We then compared single-marker phylogenetic trees with each other and with the latest published phylogenies of Ficus (Cruaud et al., 2012), Moraceae (Clement and Weiblen, 2009) and other families of Rosales (Wiegrefe et al., 1998; Potter et al., 2007; Zhang et al., 2011; Wu et al., 2013; Yang et al., 2013; Onstein et al., 2015; Hauenschild et al., 2016). A sequence was excluded when its position in one tree conflicted strongly (i.e. with high support) with those in other trees or published phylogenies. After excluding problematic sequences (Celtis: L12638, AY263941, AY263961, AY263925, AY263899; Morus: L01933) and confirming that no supported conflict remained among single-marker trees, combined phylogenetic analyses of chloroplast genes (matK, rbcL and ndhF), nuclear markers (ITS, ETS, G3pdh, ncpGS the waxy region) and of all eight markers were conducted with ML and Bayesian approaches using RAxML and BEAST on CIPRES. For ML analyses, the data set was divided into eight partitions according to marker, each with the GTRCAT model and 1000 bootstrap replicates. All trees are presented rooted on the most external outgroup, Rosaceae, which have been shown to be the sister group of all remaining Rosales in all recent higher level phylogenetic analyses (e.g. Wang et al., 2009; Soltis et al., 2011). The phylogenetic reconstruction by Bayesian approach was done simultaneously with divergence time estimation (see below).
Fossil calibration
The fossil record of Moraceae is comparatively poor and in need of critical revision (Collinson, 1989), whereas unambiguous fossils exist for other families of Rosales (Burge and Manchester, 2008; Benedict et al., 2011; Friis et al., 2011). When reliable fossils are absent or scarce for the ingroup, outgroup calibration may provide more accurate estimates than secondary calibration (Sauquet et al., 2012; Hipsley and Müller, 2014). Therefore, we specifically designed the taxon sample of this study to include sufficient outgroup nodes to take advantage of the fossil record of Rosales. Our set of fossil age constraints includes four in Moraceae and eight distributed among the remaining families of Rosales (Supplementary Data Table S2). To revise fossil calibrations in Rosales, we proceeded as follows (Parham et al., 2012; Sauquet et al., 2012): (1) we started from lists of calibrations used in previous molecular dating studies (Zerega et al., 2005; Cruaud et al., 2012; Magallón et al., 2015) and completed this list with specific reviews (Collinson, 1989; Friis et al., 2011) and recently published fossil taxa (Manchester, 1999; Calvillo-Canadell and Cevallos-Ferriz, 2007; Manos et al., 2007; Benedict et al., 2011); (2) for each fossil, we critically assessed the phylogenetic assignment based on original descriptions and subsequent reviews, and using the latest reference phylogeny for each family (Manchester, 1999; Calvillo-Canadell and Cevallos-Ferriz, 2007; Manos et al., 2007; Benedict et al., 2011; Friis et al., 2011; Jud et al., 2017); (3) for each fossil, we also critically revised the absolute age or age range of the fossil using the latest stratigraphy and geological time scale from the International Commission on Stratigraphy (Cohen et al., 2017). Because none of these fossil taxa has been included in total evidence phylogenetic analyses, our assignments here are at best ‘apomorphy based’ and therefore we have been particularly conservative in both selecting our final calibrations and assigning them to clades. This implies that some taxa previously used as calibrations are typically used here to constrain the age of a more inclusive node than in previous studies (e.g. the fossil achenes we used to calibrate the stem group of Ficus were used to calibrate the crown group of Ficus in previous studies; Datwyler and Weiblen, 2004; Rønsted et al., 2005; Zerega et al., 2005; Cruaud et al., 2012).
Molecular dating analyses
We used BEAST v1.8.0 (Drummond et al., 2012) implemented on CIPRES (Miller et al., 2010) to estimate divergence times and topology simultaneously. To reduce computational burden, we used empirical base frequencies and divided the data set into three partitions: chloroplast DNA (matK, rbcL and ndhF), non-coding nuclear markers (ITS and ETS) and coding nuclear genes (G3pdh, ncpGS and the waxy region). The substitution model for each partition was selected using the Akaike information criterion (AIC) with jModelTest v2.1.6 (Darriba et al., 2012) as implemented on CIPRES. The best substitution model was TVM + I + G for the chloroplast (matK, rbcL and ndhF) and non-coding nuclear (ITS and ETS) partitions, and TPM3uf + I + G for the coding nuclear (G3pdh, ncpGS and the waxy region) partition.
For each calibrated node, we chose the oldest reliable fossil as a minimum age constraint. Because most fossils typically provide only minimum ages for the clade to which they can be safely attributed, a uniform prior distribution was used for each fossil calibration, using the upper (younger) boundary of the fossil oldest stratigraphic age range as the minimum bound and the maximum constraint for the root (see below) as the maximum bound. Molecular dating analyses usually require at least one maximum age constraint (Ho and Phillips, 2009; Sauquet, 2013). Here, we chose to set the maximum age bound for the root (i.e. the crown group node of Rosales) to 107 million years (Ma) based on the crown group age estimate of Rosales (98.96–106.94 Ma) in the latest, large-scale molecular dating study of angiosperms (Magallón et al., 2015). Although the phylogenetic relationships among sampled members of Rhamnaceae are inconsistent with previous work (Onstein et al., 2015; Hauenschild et al., 2016; see the Discussion), they are unlikely to have affected divergence times because we chose conservative fossil assignments in Rhamnaceae. Paliurus clarnensis has been proposed to be more closely related to extant species of Paliurus than any other member of Rhamnaceae (Burge and Manchester, 2008), thus providing a minimum age for the stem node of Paliurus. Because of conflict in the position of the genus, we decided to calibrate the crown group node of Rhamnaceae instead. Coahuilanthus belindae has been proposed to be a member of Rhamnaceae, but its position in the phylogeny remains unclear (Calvillo-Canadell and Cevallos-Ferriz, 2007), therefore we used it as a minimum age constraint for the stem node of the family.
The tree prior in BEAST was set as a Birth–Death process. To produce a starting tree that conforms with the hard age constraints enforced in this analysis, we transformed the best-known RAxML tree into an ultrametric tree using the bladj function of phylocom v4.2 (Webb et al., 2008). Four separate runs were conducted, each with 100 million generations, sampling trees and parameters every 10 000 generations. Chain convergence was checked in Tracer v1.5 (Rambaut and Drummond, 2009), with the first 10 % of chain length excluded as burn-in. After confirming that chains of these four runs had converged, we combined them using LogCombiner v 1.8.0, resampling every 100 000 generations and discarding the first 10 % (i.e. 10 million) generations as burn-in. Tree statistics were summarized on the Maximum Clade Credibility (MCC) tree by TreeAnnotator v1.8.0, using median ages as node heights.
In addition to our main dating analysis, we also conducted a series of sensitivity experiments. First, we tested the impact of calibrations on the estimated topology by conducting another run keeping the maximum age constraint on the root but without internal age constraints (Sauquet et al., 2012). Secondly, we produced another run by excluding the age constraint on the stem node of Ficus, but maintaining all other 11 age constraints to test the influence of the former on the estimated crown group age of Ficus. Last, we tested the impact of our root maximum age constraint on divergence time estimates for the ingroup. Indeed, there remains considerable uncertainty on the crown group age of angiosperms (Doyle, 2012; Magallón et al., 2015; Herendeen et al., 2017), and recent analyses that do not constrain this age consistently estimate it to be much older than the first accepted crown group fossils (Foster et al., 2017). Therefore, we also re-ran our analysis using an older maximum age constraint on the root, set to 112 Ma, based on the crown group age of Rosales (87.82–111.78 Ma) estimated in the unconstrained analysis of Magallón et al. (2015), in which the crown group age of angiosperms was estimated to be 160–256 Ma rather than 136–140 Ma (S. Magallón, pers. comm.).
Ancestral state reconstruction
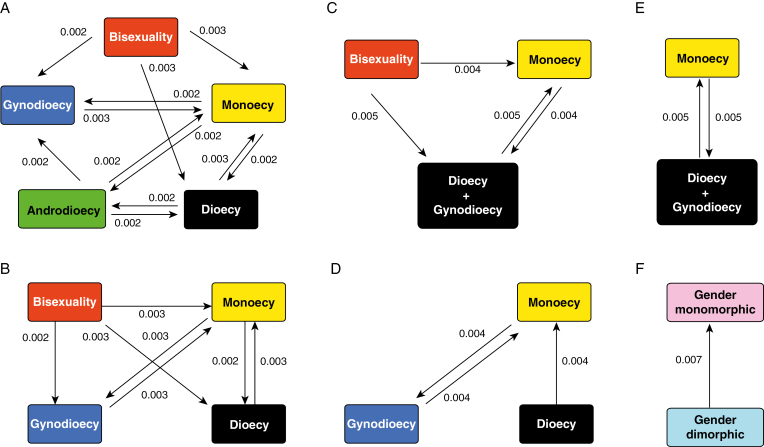
The breeding system state of each species was determined from the literature, and then recorded in the PROTEUS database (Sauquet, 2016). A list of all data records (each linked to an explicit reference) and the matrix are provided as Supplementary Data Table S3. For the main analysis, we reconstructed the ancestral state with all 320 species in the data set (i.e. including all outgroups). To characterize a reliable evolutionary scenario supported by different classification methods of breeding systems (e.g. Anger et al., 2017), we analysed six configurations of the same character and data, differing in the number of character states considered (Table 1; Fig. 1). Configuration A distinguishes among five breeding system states (as recorded and listed in Supplementary Data Table S3). Configuration B (four states) treats androdioecy as missing data. In our data set, there are only three androdioecious species, accounting for <1 % of all species sampled. Configuration C (three states) is similar to configuration B, but with gynodioecy and dioecy pooled as a single state (dioecy). In Ficus, gynodioecious species are functionally dioecious (Cook and Rasplus, 2003), and all gynodioecious species but one (Dryas octopetala) in this study belong to Ficus. Configuration D (three states) is similar to configuration B, but treats bisexual flowers as missing data. In Rosales, families Cannabaceae, Moraceae and Urticaceae all have unisexual flowers (Simpson, 2010); thus, maintaining the bisexual state is not essential to this study and could be detrimental by unnecessarily increasing the number of parameters to estimate. Configuration E (two states) combines the two transformations of configurations C and D: gynodioecy and dioecy pooled, bisexuality treated as missing. In configuration F (two states), we contrast gender monomorphic and gender dimorphic species. Gender monomorphic and gender dimorphic means that in one population there are one or two functional classes of sex (Lloyd, 1980). Therefore, bisexuality and monoecy are gender monomorphic, and androdioecy, gynodioecy and dioecy are gender dimorphic.
Table 1.
Summary of the six character state configurations used to reconstruct ancestral breeding systems in Moraceae
| Configuration | States | Justification |
|---|---|---|
| A | Bisexuality, gynodioecy, androdioecy, monoecy, dioecy | Original states recorded in species |
| B | Bisexuality, gynodioecy, monoecy, dioecy | Excluding state androdioecy for its low frequency in the data set of this study |
| C | Bisexuality, monoecy, dioecy + gynodioecy | Based on configuration B, gynodioecy combined with dioecy: all gynodioecious species (but one) in this study belong to Ficus, where they are functionally dioecious |
| D | Gynodioecy, monoecy, dioecy | Based on configuration B, excluding state bisexuality: only distant outgroups of Moraceae include species with bisexual flowers (e.g. Rosaceae, Elaeagnaceae and Rhamnaceae) |
| E | Monoecy, dioecy + gynodioecy | Based on configuration B, gynodioecy combined with dioecy (as in C) and bisexuality excluded (as in D) |
| F | Gender monomorphic, gender dimorphic | Recognition of bisexuality and monoecy as gender monomorphic, and gynodioecy, androdioecy and dioecy as gender dimorphic (Lloyd, 1980) |
Fig. 1.
Schematic representation of the six configurations for the breeding system character analysed in this study. The arrows denote transition rate parameters, as estimated in the reversible-jump Markov Chain Monte Carlo analyses, with mean rates reported. The absence of an arrow indicates a near-zero estimate for the corresponding parameter, suggesting that the model does not support direct transition between the two states.
For each configuration, we used and compared parsimony, ML and Bayesian approaches to reconstruct ancestral states (Sauquet et al., 2015, 2017). Parsimony analyses were conducted in Mesquite v3.04 (Maddison and Maddison, 2016); ML and Bayesian analyses were conducted in BayesTraits V2 (Pagel and Meade, 2013). Parsimony and ML analyses were conducted with the MCC tree produced from the BEAST analysis, whereas Bayesian analyses were conducted with 3600 trees randomly sampled from the posterior of the BEAST analysis. To test the influence of topological uncertainty on ancestral state reconstruction, we also conducted additional Bayesian analyses with a fixed (MCC) tree.
Maximum likelihood analyses presented here explored two models for each configuration: an equal rates (ER, or Mk1) model, assuming equal transition rates among all character states (Lewis, 2001), and an all rates different (ARD) model, allowing distinct (asymmetric) transition rates among character states (Pagel, 1994). The best model (equal or unequal rate) for each configuration was selected according to the AIC. We calculated ΔAIC between two models for each configuration, using a ΔAIC of ≥2 as a criterion for positive support of the best-fit model (Posada and Buckley, 2004).
We also reconstructed ancestral breeding system states using both a ‘common’ (i.e. fixed-model) Bayesian approach and a reversible-jump Markov chain Monte Carlo (rjMCMC) strategy. While both allowed us to take parameter and phylogenetic (including molecular dating) uncertainty into account in ancestral state reconstruction (Pagel et al., 2004), the rjMCMC approach allowed us to explore and visit multiple models of morphological evolution in proportion to their posterior probabilities (Pagel and Meade, 2006). Both the ER and ARD models were tested in the common Bayesian approach, and their relative fit was compared with the Bayes factor (Kass and Raftery, 1995) according to the criteria of Lodewyckx et al. (2011). Chain lengths were set to 10 million generations (or 2 million generations for fixed-tree analyses), and parameters and ancestral states were sampled every 1000 generations. Chain convergence was checked in Tracer v1.5 (Rambaut and Drummond, 2009), with the first 10 % of generations excluded as burn-in.
To investigate the impact of different taxonomic sampling strategies (with or without outgroups of Ficus) on ancestral state reconstruction of breeding systems in Ficus, we also conducted analyses with only Ficus species (same topology). Here, after excluding non-Ficus species in configuration A, we used Mesquite instead of BayesTraits for ML reconstructions because we wished to test the impact of different root states priors on ancestral state reconstruction. We applied both the equal rates (Mk1) and the asymmetric 2 rate (AsymmMk, equivalent to ARD for a binary character) models to reconstruct the ancestral state of Ficus. In addition, two different root states priors (equal and equilibrium frequencies) were applied for the asymmetric 2 rate (AsymmMk) model in Mesquite (whereas BayesTraits assumes an equal frequency root state prior for all states).
RESULTS
Phylogenetic reconstruction
Chloroplast markers were insufficient to resolve relationships within Ficus, as most internal nodes had low support (Supplementary Data Fig. S1A–D). We found three instances of conflict (F. trigonata clustered with F. tinctoria; F. religiosa clustered with F. benghalensis; F. pumila clustered with F. hirta, bootstrap probability 79, 80 and 57) between topologies reconstructed from nuclear and chloroplast sequences (Supplementary Data Fig. S1D, E). With the exception of these, we found no well-supported conflict between chloroplast and nuclear trees (Supplementary Data Fig. S1). We thus combined the eight markers and focus here on the results from this combined analysis (Fig. 2; Supplementary Data Fig. S2). Hereafter we only discuss the trees reconstructed from the whole data set. The topologies reconstructed with ML and Bayesian approaches were consistent. Differences were only observed in some weakly supported nodes. All families, all tribes of Moraceae except Moreae, and all genera were supported as monophyletic. Tribe Moreae was reconstructed as polyphyletic because Streblus smithii clustered with Maclureae (the other two species of Streblus sampled were instead found to be nested in Moreae; Supplementary Data Fig. S2). In Ficus, subgenera Phamacosycea, Urostigma and Ficus were found to be paraphyletic, and subgenus Synoecia nested in subgenus Ficus. In addition, some deep nodes (e.g. the most recent common ancestor of F. carica and Urostigma) were poorly supported in the Bayesian analysis (Supplementary Data Fig. S2). With or without fossil age constraints, the topologies obtained with BEAST were identical and the posterior probability values of each nodes were similar as well (result not shown). Excluding the crown group calibration of Ficus or using an older maximum root age constraint did not influence the topology either (Supplementary Data Table S4).
Fig. 2.
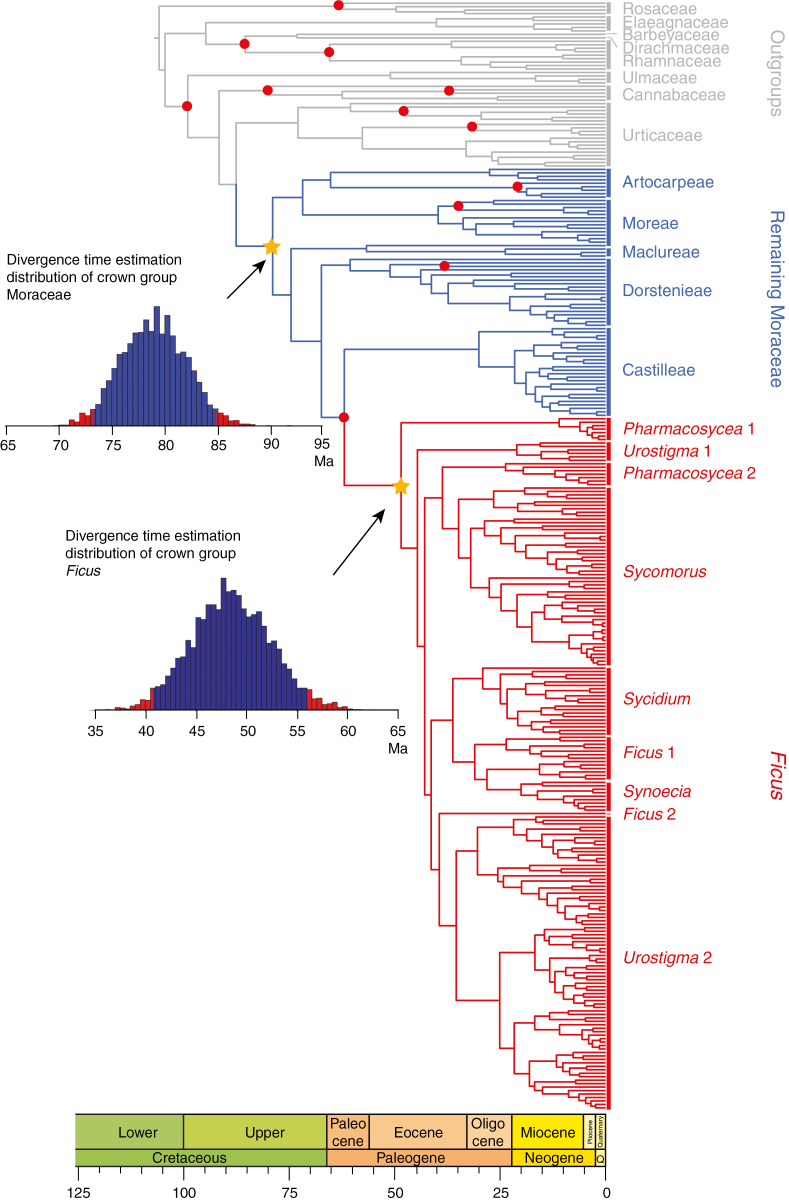
Dated phylogenetic tree of 320 species with family names in Rosales, tribe names in Moraceae and section names in Ficus. This is the Maximum Clade Credibility (MCC) from the BEAST analysis of eight molecular markers (see text). Fossil-calibrated nodes are indicated in red. The posterior distribution of estimated ages is shown for crown group Moraceae and Ficus (marked with a yellow star). For full details of this tree, see Supplementary Data Fig. S2.
Molecular dating
After combination of the four separate runs, the effective sample size (ESS) of each parameter was >100, with most of them over 200. The crown group ages of Ficus and Moraceae were estimated as Eocene (40.6–55.9 Ma) and Late Cretaceous (73.2–84.7 Ma), respectively (Table 2; Fig. 2). The oldest tribe was Artocarpeae (64.0–68.6 Ma), and Maclureae originated most recently (8.9–41.1 Ma). In Ficus, subgenus Sycomorus was the oldest (28.0–41.1Ma) (Table 2). Excluding the Ficus stem group calibration or using an older maximum root age constraint did not have strong impact on estimated ages (Supplementary Data Table S4).
Table 2.
Estimated divergence times for key nodes of Moraceae, following the tribal classification of Clement and Weiblen (2009), as shown in Fig. 1
| Nodes | Breeding systems | Support values (BP/PP)* | Mean age (95 % HPD)† (Ma) |
|---|---|---|---|
| CG‡ Rosales | Bisexuality, monoecy, androdioecy, gynodioecy, dioecy | – | 105.5 (102.8–107.0) |
| SG§ Moraceae | – | 100/1.00 | 87.5 (81.7–93.3) |
| CG Moraceae | Monoecy, androdioecy, gynodioecy, dioecy | 99/1.00 | 79.0 (73.2–84.7) |
| CG Artocarpeae | Monoecy, dioecy | 99/1.00 | 65.6 (64.0–68.6) |
| CG Moreae¶ | Monoecy, dioecy | 91/1.00 | 40.3 (34.9–46.8) |
| CG Maclureae | Dioecy | 98/1.00 | 24.7 (8.9–41.1) |
| CG Dorstenieae | Monoecy, dioecy | 98/1.00 | 60.5 (51.4–70.2) |
| CG Castilleae | Monoecy, androdioecy, dioecy | 100/1.00 | 31.2 (18.7–47.0) |
| SG Ficeae | – | 98/1.00 | 62.2 (56.0–68.6) |
| CG Ficeae | Monoecy, gynodioecy | 99/1.00 | 48.5 (40.6–55.9) |
| CG Pharmacosycea 1 | Monoecy | 58/1.00 | 11.6 (5.5–19.4) |
| CG Pharmacosycea 2 | Monoecy | 100/1.00 | 24.2 (13.2–36.7) |
| CG Sycomorus | Monoecy, gynodioecy | 77/1.00 | 38.7 (28.0–41.1) |
| CG Sycidium | Gynodioecy | 100/1.00 | 29.2 (22.6–36.4) |
| CG Synoecia | Gynodioecy | 100/1.00 | 20.1 (13.7–26.8) |
| CG Ficus | Gynodioecy | 82/1.00 | 31.0 (24.7–38.0) |
| CG Urostigma 1 | Monoecy | 100/1.00 | 21.5 (12.0–31.2) |
| CG Urostigma 2 | Monoecy | 99/1.00 | 35.5 (28.0–42.3) |
*PP, posterior probability; BP, bootstrap probability.
†95 % HPD, 95 % highest posterior density.
‡CG, crown group.
§SG, stem group.
¶ Streblus smithii was here excluded from crown group Moreae (see text for details).
Ancestral state reconstruction
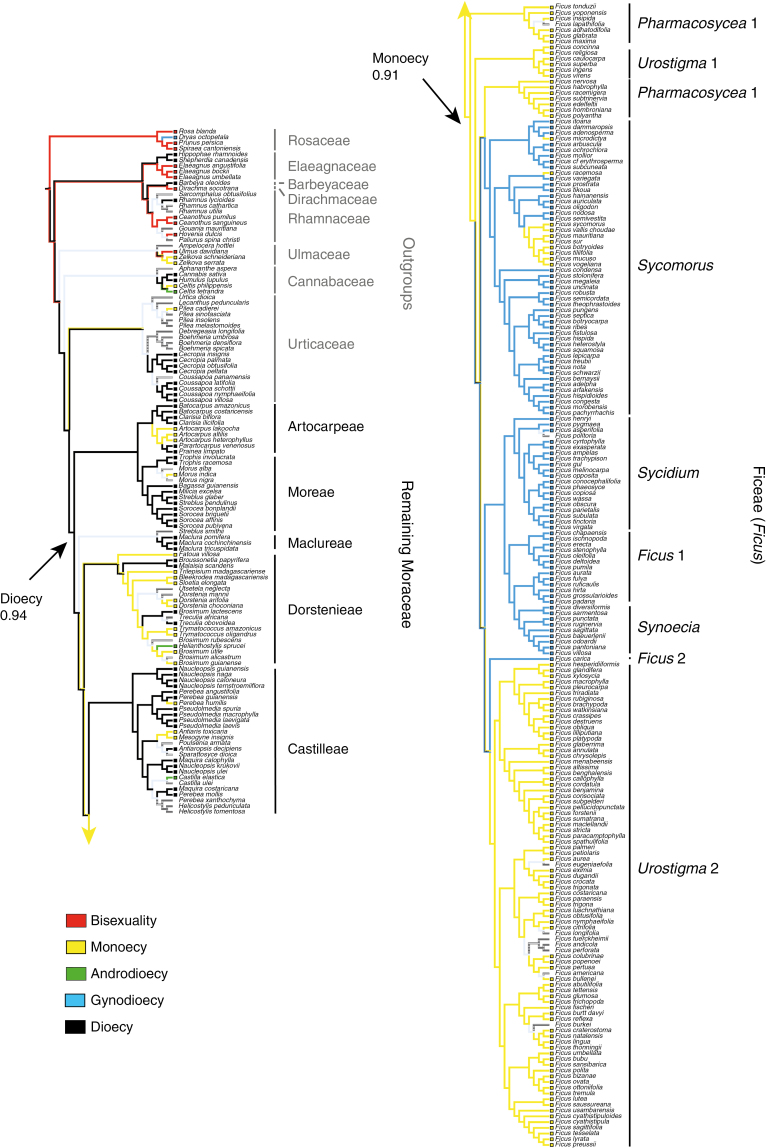
Parsimony, ML and Bayesian analyses of the full data set (320 species) reconstruct dioecy as ancestral for Moraceae with strong support in all six character state configurations (Table 3; Fig. 3; Supplementary Data Figs S3 and S4). Furthermore, we find that dioecy has evolved into monoecy at least five times in Moraceae. However, several alternative scenarios are possible due to uncertainty in the most recent common ancestor of Dorstenieae, Castilleae and Ficeae (Ficus) (Fig. 3). In one possible scenario, monoecy evolved independently once in Artocarpeae (Artocarpus); once in Moreae (Morus); once in Dorstenieae (with at least two subsequent reversals); twice in Castilleae (Perebea humilis; and the clade of Antiaris toxicaria and Mesogyne insignis); and at least once in Ficeae (Ficus). In another possible scenario, monoecy evolved independently in Artocarpeae and Moreae (as above), but shares a common origin in the clade of Dorstenieae, Castilleae and Ficus, with at least two reversals (to dioecy) in Dorstenieae, and one in the ancestor of Castilleae (followed by two subsequent gains again in the tribe as above). Within Ficus, no matter whether gynodioecy (functional dioecy) is ancestral (and therefore intermediate) or not (see below), at least three transitions from gynodioecy to monoecy are reconstructed (within section Sycomorus).
Table 3.
Summary results of ancestral state reconstruction for the complete (320 species) data set by the maximum likelihood approach
| CG Moraceae* | Prob† | CG Ficus‡ | Prob | Lh§ | No. of par¶ | AIC | |
|---|---|---|---|---|---|---|---|
| Equal rate | |||||||
| A | Dioecy | 0.91 | Monoecy | 0.92 | 121.6 | 10 | 263.2 |
| B | Dioecy | 0.90 | Monoecy | 0.91 | 102.7 | 6 | 217.4 |
| C | Gynodioecy + dioecy | 0.95 | Monoecy | 0.87 | 91.5 | 3 | 189 |
| D | Dioecy | 0.88 | Monoecy | 0.89 | 78.9 | 3 | 163.8 |
| E | Gynodioecy + dioecy | 0.92 | Monoecy | 0.80 | 63.1 | 1 | 128.2 |
| F | Gender dimorphic | 0.89 | Gender monomorphic | 0.76 | 76.1 | 1 | 154.2 |
| Unequal rate | |||||||
| A | Androdieocy; dioecy; gynodioecy | Each approx. 0.33 | Gynodioecy | 0.95 | 124.7 | 20 | 289.4 |
| B | Dioecy | 0.99 | Gynodioecy | 0.98 | 90.8 | 12 | 205.6 |
| C | Gynodioecy + dioecy | 0.99 | Gynodioecy + dioecy | 0.84 | 76.9 | 6 | 165.8 |
| D | Dioecy | 1.00 | Gynodioecy | 1.00 | 70.0 | 6 | 152 |
| E | Gynodioecy + dioecy | 1.00 | Gynodioecy + dioecy | 0.83 | 61.6 | 2 | 127.2 |
| F | Gender dimorphic | 1.00 | Gender dimorphic | 0.84 | 74.6 | 2 | 153.2 |
*CG Moraceae, breeding system state of crown group Moraceae.
†Prob, probability of support estimated by maximum likelihood.
‡CG Ficus, breeding system state of crown group Ficus.
§Lh, minus log likelihood.
¶No. of par, number of free parameters.
The best-fit model identified by AIC for each configuration is indicated in bold.
Fig. 3.
Ancestral state reconstruction of breeding systems in Moraceae. Here we show results from the maximum likelihood analysis of configuration A (five character states) with the equal rate model (for other results, see Supplementary Data Figs S3–S5). Clade labels: family names in Rosales, tribe names in Moraceae and section names in Ficus.
Conversely, the ancestral breeding system of Ficus could not be reconstructed with confidence. First, parsimony reconstructions suggested either monoecy (configurations A, B and D) or an equivocal state (monoecy or the combination of dioecy and gynodioecy; configurations C, E and F) as ancestral in Ficus (Supplementary Data Fig S4). Secondly, ML reconstructions with an equal rate model with all configurations supported monoecy (or gender monomorphic) as ancestral in Ficus, whereas the unequal rate model supported gynodioecy (or gynodioecy + dioecy or gender dimorphic) as ancestral in the genus (Table 3). The strength of support differed among different configurations, but the ancestral state did not change. In the ML approach, we compared the models in each configuration using the AIC. Only with configuration A was the equal rate model selected, whereas the unequal rate model better fit the data with configurations B–E. As a result, considering best-fit models only, gynodioecy (or gynodioecy + dioecy, or gender dimorphic) was found as ancestral in Ficus with five out of six configurations (Table 3). Thirdly, the Bayesian approach with rjMCMC weakly supported monoecy (or gender monomorphic) as ancestral in Ficus, but with a broad 95 % highest posterior density (HPD; 0–1.00), suggesting high uncertainty in the estimation (Table 4). Bayesian analyses with fixed model selected the unequal rate model in four out of six configurations (two with weak support), and monoecy as ancestral in Ficus in four out of six configurations.
Table 4.
Summary results of ancestral state reconstruction for the complete (320 species) data set obtained with the Bayesian approach (reversible-jump MCMC)
| Configuration | One MCC tree | 3600 posterior trees | ||||||
|---|---|---|---|---|---|---|---|---|
| CG Moraceae* | 95 % HPD† | CG Ficus‡ | 95 % HPD | CG Moraceae | 95 % HPD | CG Ficus | 95 % HPD | |
| A | Dioecy | 0.82–1.00 | Monoecy | 0.00–0.88 | Dioecy | 0.80–1.00 | Monoecy | 0–0.99 |
| B | Dioecy | 0.84–1.00 | Monoecy | 0.71–0.90 | Dioecy | 0.80–1.00 | Monoecy | 0.71–1.00 |
| C | Gynodioecy + dioecy | 0.86–1.00 | Monoecy | 0.00–0.84 | Gynodioecy + dioecy | 0.74–1.00 | Monoecy | 0.00–0.97 |
| D | Dioecy | 0.85–1.00 | Monoecy | 0.66–0.90 | Dioecy | 0.82–1.00 | Monoecy | 0.65–1.00 |
| E | Gynodioecy + dioecy | 0.87–1.00 | Monoecy | 0.00–0.85 | Gynodioecy + dioecy | 0.75–1.00 | Monoecy | 0.00–0.98 |
| F | Gender dimorphic | 0.85–1.00 | Gender dimorphic | 0.19–1.00 | Gender dimorphic | 0.76–1.00 | Gender dimorphic | 0.03–1.00 |
*CG Moraceae, breeding system state of crown group Moraceae.
†95 % HPD, 95 % highest posterior density.
‡CG Ficus, breeding system state of crown group Ficus.
Some of the transitions such as the direct transition between gynodioecy and dioecy in configurations A, B or D were estimated to be unlikely (i.e. transition rates were estimated to be zero) in the rjMCMC analyses (Fig 2). The 95 % HPD of transition rates in each configuration broadly overlapped. In the Bayesian analysis with the unequal rate model in configuration A (Supplementary Data Table S5), the chain did not converge. This remained true when fixing the tree (i.e. using the MCC tree) instead of using a sample of trees from the BEAST posterior. These results suggest that the unequal rate model is overparameterized for this configuration with five character states. Indeed, 20 free parameters (transition rates) are estimated in this model. In contrast, a four-state unequal rate model requires 12 parameters, a three-state model requires six parameters and a binary model requires only two parameters. The potential overparameterization of configuration A was also suggested by the results of rjMCMC analyses. For each configuration, the number of free parameters in the model was estimated to be between one and three.
Reconstructions with only species of Ficus also showed an uncertain result with respect to the ancestral state of the genus (Supplementary Data Fig. S5). Both parsimony and ML with the equal rate model strongly supported monoecy to be ancestral in Ficus, whereas the ML approach with the unequal rate model supported gynodioecy as ancestral in the genus. The support for ancestral gynodioecy differed with root state prior: with the equal frequencies root state prior, gynodioecy was highly supported (0.99 out of 1) whereas with the equilibrium frequencies root state prior, the probability for gynodioecy decreased sharply to 0.56, suggesting high uncertainty.
DISCUSSION
Phylogenetic relationships in Moraceae
The general topology of phylogenetic trees reconstructed in this study (both ML and Bayesian approach) is consistent with previous studies in Ficus (Cruaud et al., 2012), Moraceae (Clement and Weiblen, 2009) and Rosales (Zhang et al., 2011), except for relationships within Rhamnaceae, for which our results partly differ from those of two recent phylogenetic studies (Onstein et al., 2015; Hauenschild et al., 2016). This conflict may be due to the use of different molecular markers and to our sampling of this outgroup family with low density. Here we used matK, rbcL and ndhF, while Hauenschild et al. (2016) used ITS and trnL–F, and Onstein et al. (2015) used eight molecular markers (including all the markers we used and trnL–F, psbA, psbA–trnH, rpl16 and ITS). However, these differences are unlikely to affect our ancestral state reconstructions, given the distant positions of Rhamnaceae and Moraceae in the phylogeny of Rosales, and the fact that the breeding system state of most of the species in Rhamnaceae has been scored as missing data in this study. In our phylogenetic reconstruction, outgroup relationships are consistent with previous work (Wang et al., 2009; Soltis et al., 2011; Zhang et al., 2011).
Within Moraceae, phylogenetic relationships among genera are consistent with recent studies (Zerega et al., 2005, 2010; Clement and Weiblen, 2009; Williams et al., 2017) except for Moreae (Fig. 2; Supplementary Data Fig. S2). Tribe Moreae was found to be paraphyletic due to the position of Streblus smithii as sister to Maclureae. This relationship is not strongly supported (posterior probability 0.76) and may be caused by too few informative sites for this species. In our data set, S. smithii is only represented by the ndhF sequence and our separate analysis of the ndhF data set, in which all three species of Streblus were sampled, supports the same result as our combined analysis (Supplementary Data Fig. S1C). In the original paper from where the sequence came (Datwyler and Weiblen, 2004), the species was found in an uncertain phylogenetic position. However, in a later reconstruction (Clement and Weiblen, 2009) where ndhF, 26S and morphological data were combined, S. smithii clustered with the other two species in the genus, possibly because the nuclear (26S) and morphological signal overcame a divergent ndhF sequence in their combined analysis.
Within Ficus, subgenera Phamacosycea, Urostigma and Ficus were found to be paraphyletic, and subgenus Synoecia is nested within subgenus Ficus and some deep nodes remain poorly supported. These results are consistent with the most recent comprehensive phylogenetic study of the genus (Cruaud et al., 2012), while they are in conflict with a recent phylogenomic study of Ficus based on full chloroplast genomes, and this conflict could be caused by potential cyto-nuclear discordance (Bruun-Lund et al., 2017). Chloroplast markers performed poorly in Ficus as the informative sites they provide are too few (Cruaud et al., 2012). Almost no branch length can be observed in the chloroplast tree for the shallow nodes (Supplementary Data Fig. S1). In our analysis, Ficus species were represented not only by chloroplast markers but also by nuclear markers which were found informative enough in previous studies (Rønsted et al., 2008; Cruaud et al., 2012). In the Bayesian analyses, 3600 trees were taken into account, therefore the uncertainty of topology was considered.
A new time scale for Moraceae diversification
The new time scale presented here for Moraceae was estimated with 12 fossil calibrations (four ingroup and eight outgroup), more than in any previous study of the family so far. Here the crown group age of Moraceae was estimated as 73.3–84.9 Ma. This estimate is similar to that found by Zerega et al. (2005) (72.6–110.0 Ma), but with a narrower confidence interval. Our stem group age estimate for the family (81.7–93.3 Ma) is older than that reported by Magallón et al. (2015) (57.7–77.84 Ma), most probably because our increased sampling of the family and its outgroups allowed us to use more fossil calibrations in Rosales. The crown group age of Ficus was here estimated as 40.8–56.0 Ma, which is similar to the age reported by Zerega et al. (2005) (40.1–51.0 Ma), but younger than the age found by Rønsted et al. (2005) (51.4–78.0 Ma) and Cruaud et al. (2012) (60.0–101.9 Ma). These differences may be explained by different calibration strategies. When fossil species have morphological traits similar to extant species of a clade, these fossils are often used optimistically to calibrate the crown group of this clade. This practice is problematic because it rules out the possibility that such fossils are stem relatives of the clade (Ronquist et al., 2012; Sauquet, 2013). Here we used an apomorphy-based approach, using fossils as minimum age constraints modelled with uniform prior distributions to calibrate the stem node of the clade with which they share apomorphies (Sauquet, 2013) (Supplementary Data Table S2). For instance, the fossil achenes attributed to Ficus (Collinson, 1989) were here used to calibrate the stem node of Ficus, whereas Rønsted et al. (2005) and Cruaud et al. (2012) used the same fossils to calibrate the crown node of Ficus. We also used different prior distributions for fossil calibrations. Although some authors have suggested that priors such as lognormal or exponential distributions may help to improve the accuracy of divergence time estimation (Yang and Rannala, 2005; Ho and Phillips, 2009), the prior setting of parameters such as mean or standard deviation for these distributions is often arbitrary and the shape of these distributions implicitly assumes that the fossil diverged close to the node calibrated. Our strategy here was instead to use uniform distribution with fossil ages as the minimum bound and maximum root age as the maximum bound. The crown group ages of tribes Artocarpeae and Dorstenieae were estimated as 64.0–68.6 and 51.4–70.2 Ma, respectively, both of which are younger than the ages reported by Williams et al. (2017) for Artocarpeae (61.4–78.5 Ma) and by Misiewicz and Zerega (2012) for Dorstenia (84.8–132.0 Ma).
The BEAST analyses without internal calibrations showed identical topology and similar posterior support compared with our reference analysis using all calibrations, suggesting that here calibration did not influence the topology. Here, we used the maximum age estimated for crown group Rosales by Magallón et al. (2015) as the maximum age constraint on the root in our analyses, but also tested the influence of older root constraints. Consistent with previous work (Sauquet et al., 2009; Massoni et al., 2014; Foster et al., 2017), we found that alternative maximum root constraints had very little impact on estimated divergence times. In addition, the crown group age of Ficus was similar with or without the stem group Ficus calibration (Supplementary Data Table S4). These results suggest that our estimates are not an artefact of the calibration points near the nodes of interest.
Breeding system transitions in Moraceae and Ficus
It was found that increased taxon sampling density can be helpful in ancestral state reconstruction (Salisbury and Kim, 2001). Ancestral states of breeding systems in Moraceae have been previously reconstructed with parsimony and a phylogenetic tree of 46 species (Weiblen, 2000), 83 species (Datwyler and Weiblen, 2004) and 73 species (Clement and Weiblen, 2009). In this study, we reconstructed ancestral states with a more densely sampled phylogenetic tree (including 200 species of Ficus and 271 species from other genera of Moraceae), compared different model-based approaches and took into account phylogenetic uncertainty in our Bayesian analyses. Here, we found similar results for two focal nodes (Moraceae and Ficus) whether using a single tree or thousands of trees sampled from the posterior of BEAST analyses, suggesting that, in our analyses, topology did not affect the estimation. In all the approaches and models, dioecy was strongly supported to be ancestral for Moraceae, while the ancestral state for Ficus remains unclear.
Dioecy has been reported to correlate with several ecological traits, including abiotic pollination, fleshy fruits and woody growth form (Freeman et al., 1979; Renner and Ricklefs, 1995; Vamosi et al., 2003). During the history of Moraceae, dioecy evolved into monoecy at least six times (see above; Fig. 3; Supplementary Data Figs S3 and S4). Although a previous study suggested no strong statistical support for a relationship between breeding system and pollination syndrome in Moraceae (Clement and Weiblen, 2009), our results reinforce the general pattern that would be consistent with such a relationship: dioecious taxa tend to be wind pollinated, whereas monoecious taxa tend to be insect pollinated. For instance, we infer Artocarpeae as ancestrally dioecious (Fig. 3; Supplementary Data Figs S3 and S4), and Datwyler and Weiblen (2004) reconstructed the clade as ancestrally anemophilous. In our reconstruction, dioecy has evolved to monoecy in Artocarpus, which is characterized by inflorescences with an insect pollination syndrome. This relationship may also apply to tribe Dorstenieae as a whole, most species of which are monoecious and were hypothesized to be insect pollinated according to their inflorescence structure (Berg and Hijman, 1999). However, the relationship becomes less clear in Castilleae and Ficus (Datwyler and Weiblen, 2004), partly because all species of Ficus are insect pollinated (Cook and Rasplus, 2003), yet only half of them are monoecious. Unfortunately, the current lack of sufficient data on actual pollination modes in Moraceae (outside Ficus) precludes us from further testing this potential correlation at the family level.
Although the ancestral state of Ficus proved to be particularly difficult to reconstruct in this study due to our exploration of various models and character state configurations, most of the evidence supports monoecy rather than gynodioecy as the ancestral state in the genus. As highlighted in recent studies of floral traits using the same methods (Sauquet et al., 2015, 2017), our results suggest that great caution should be exercised when interpreting results from ML analyses exploring a limited set of models. Indeed, the models with highest posterior probability identified through our rjMCMC analyses corresponded to neither the equal rates nor the unequal rates models explored with ML. These best-fit models typically involved only one free parameter, but excluded some transitions (Fig. 1). In addition, Bayesian analyses present the advantage of taking into account phylogenetic and branch length uncertainty (Pagel et al., 2004). All our Bayesian analyses that took into account phylogenetic and branch length uncertainty suggested monoecy as ancestral in Ficus, whether models were allowed to vary (Table 4) or were fixed (Supplementary Data Table S5), except for configuration F. In addition, the model-averaged rates from the rjMCMC analyses of the three configurations where dioecy and gynodioecy were treated as separate states (A, B and F) were estimated to be zero, suggesting that direct transitions between the two states do not occur in Moraceae. Therefore, we argue that it is more probable that monoecy is ancestral in Ficus and represents an intermediate state between dioecy and gynodioecy in the genus (Fig. 3). From a functional point of view, it also seems more likely that the highly specialized pollination association between Ficus and fig wasps started in monoecious figs.
Assuming that monoecy is ancestral in Ficus, the phylogenetic distribution of monoecy and gynodioecy in the genus and the uncertainty remaining in the ancestral states of several deep nodes allow for various scenarios (Fig. 3; Supplementary Data Figs S3–S5 ). It is possible that gynodioecy evolved only once (after divergence of the Pharmacosycea 1 and Urostigma 1 lineages), followed by two main reversals to monoecy (Pharmacosycea 2 and Urostigma 2). Alternatively, gynodioecy may have evolved twice independently (once in Sycomorus and once in the ancestral lineage of Sycidium, Ficus 1 and Synoecia). In all scenarios, gynodioecy reverted to monoecy at least three times within Sycomorus (Fig. 3). The biological reasons for these fluctuations between monoecy and gynodioecy in Ficus remain unclear, but several hypotheses have been proposed. Gynodioecy in Ficus may be linked with an adaptation to seasonality (Kjellberg et al., 1987); reduction of non-pollinating wasps (Kerdelhué and Rasplus, 1996); persistence of pollinator populations when the host fig populations are small (Kameyama et al., 1999); seed protection (Greeff and Compton, 2002); disperser selection; de-coupling of wasp and seed size; survival from predation; non-pollinators and predator satiation; and chronic pollinator shortages, crop asynchrony and inbreeding depression (Harrison and Yamamura, 2003). In addition, breeding system evolution in Ficus may have a relationship with biogeographic distribution. For instance, there appear to be no gynodioecious species of figs in South America (Cruaud et al., 2012). Different climates and habitats in different continents may have accelerated breeding system evolution in Ficus. The genus has been inferred to have originated in Eurasia, then migrated and diversified in South America during the Miocene (Cruaud et al., 2012). Migration to a new continent may have led to breeding system reversal from gynodioecy to monoecy in the clade Urostigma 2 (Fig. 3; Supplementary Data Figs S3 and S4) under the scenario where gynodioecy originated only once in Ficus.
Dioecy is not an evolutionary dead end in Moraceae
Our results provide additional strong support for dioecy to be ancestral in Moraceae as a whole, regardless of the approach or model used for reconstructing ancestral states (Fig. 3; Tables 3 and 4), consistent with previous work (Weiblen, 2000; Datwyler and Weiblen, 2004; Clement and Weiblen, 2009). Furthermore, our extensive sample of outgroups shows that dioecy in Moraceae is ancestral to a larger clade including at least Cannabaceae, Urticaceae and Moraceae (Fig. 3).
These results add to a growing number of studies that have challenged the notion that dioecy is an irreversible trait in flowering plants. Dioecy was once suggested to be an evolutionary dead end (Bull and Charnov, 1985). It is found in only approx. 6 % of angiosperms and distributed widely in around 43 % of families of flowering plants (Renner, 2014). However, an ‘evolutionary dead end’ has been used to describe different patterns of macroevolution (Bromham et al., 2015), including: (1) irreversible evolution (Bull and Charnov, 1985; Glémin and Muyle, 2014); or (2) a state that presents more long-term disadvantages than advantages compared with other states, ultimately leading to probable extinction (Käfer et al., 2014, 2017). Dioecy used to be thought of as irreversible (Bull and Charnov, 1985), and high extinction rates were found on dioecious clades by a comparative approach (Heilbuth, 2000). However, it has been known for a long time that although dioecious angiosperms are exposed to a higher risk of failure to find a sex partner, they are also advantageous in having obligate cross-fertilization (Darwin, 1876). Various studies have shown that dioecious species may include individuals with other breeding systems and that dioecy in general may be more labile than previously thought (Pannell, 1997; Korpelainen, 1998; Ainsworth, 2000; Käfer et al., 2017). These transitions could be linked to ecological processes including changing plant–pollinator relationships and environments (Case et al., 2008); long-distance dispersal (Schaefer and Renner, 2010); and hybridization and polyploidy (Obbard et al., 2006). In addition, various studies have documented clades where dioecy is the ancestral rather than the derived state, challenging the assumption of irreversibility of dioecy evolution. For instance, dioecy has been reconstructed as ancestral in Myristicaceae, with at least four subsequent shifts to monoecy in the family (Sauquet, 2003) and appears to be a transition state between monoecy and polygamy in Arecaceae (Nadot et al., 2016). A recent analysis of macroevolutionary dynamics of breeding systems in 40 angiosperm genera supported transitions away from dioecy in Bursera (Burseraceae) and Dodonaea (Sapindaceae; Goldberg et al., 2017). Furthermore, two recent angiosperm-wide analyses showed that dioecy does not have a negative relationship with diversity: dioecy was shown to correlate with an increased diversification rate (Käfer et al., 2014) or not to have a significant relationship with the latter (Sabath et al., 2016).
Conclusion
Here we reconstructed divergence times and ancestral breeding systems in Moraceae. With 12 fossil calibrations, the crown group ages of Ficus and Moraceae were estimated as 40.6–55.9 Ma (Eocene) and 73.2–84.7 Ma (Late Cretaceous). Dioecy was supported as the ancestral state of Moraceae and we showed that it evolved into monoecy at least five times in the family, and subsequently reversed in some clades (e.g. Artocarpeae and Dorstenieae). In Ficus, which represents 75 % of species in Moraceae, the ancestral state was estimated to be monoecy with moderate support, and at least one transition from monoecy to gynodioecy followed by at least three reversals were estimated. In future work, to investigate the exact breeding system evolutionary scenario of Ficus, it would be important to sample basal lineages of Ficus more densely and improve phylogenetic resolution of the backbone of the genus. The new time scale of Moraceae and Ficus we provide here will also be useful for future analyses of biogeography and co-diversification in the family. Finally, our results lend further support to the growing idea that dioecy does not always represent an evolutionary dead end, shedding light on the understanding of breeding system evolution in angiosperms more generally.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1: list of GenBank accession numbers for the sequences used in this study. Table S2: detailed information for the fossil calibrations used in this study. Table S3: extraction of the PROTEUS database including a list of all data records and linked references, lists of characters and character states used in this study, and the matrix in ancestral state reconstruction analyses. Table S4: a list of divergence time estimates for tribes of Moraceae. Table S5: summary results of ancestral state reconstruction for the complete (320 species) data set by Bayesian approach with fixed model. Figure S1: phylogenetic trees reconstructed by maximum likelihood with different molecular marks or combinations in this study. Figure S2: detailed version of the dated phylogenetic tree of 320 species. Figure S3: ancestral state reconstruction with a 320-species data set by parsimony approach with tip names with all six configurations. Figure S4: ancestral state reconstruction with a 320-species data set by equal-rate maximum likelihood. Figure S5: ancestral state reconstruction with only Ficus species in the data set with tip names by different approaches.
ACKNOWLEDGEMENTS
We thank Jos Käfer for his constructive review of this paper; Sophie Nadot for comments on an earlier draft; Selena Smith for comments on fossils of Artocarpus; Susana Magallón for discussion of divergence time estimation; and Nyree Zerega and Elliot Gardner for discussion of breeding system evolution in Moraceae. This work was supported by a China Scholarship Council PhD grant [grant no. 201506140077] to Q.Z.
LITERATURE CITED
- Ainsworth C. 2000. Boys and girls come out to play: the molecular biology of dioecious plants. Annals of Botany 86: 211–221. [Google Scholar]
- Anger N, Fogliani B, Scutt CP, Gâteblé G. 2017. Dioecy in Amborella trichopoda: evidence for genetically based sex determination and its consequences for inferences of the breeding system in early angiosperms. Annals of Botany 119: 591–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett SCH. 2002. The evolution of plant sexual diversity. Nature Reviews. Genetics 3: 274–284. [DOI] [PubMed] [Google Scholar]
- Barrett SCH. 2013. The evolution of plant reproductive systems: how often are transitions irreversible?Proceedings of the Royal Society B: Biological Sciences 280: 20130913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict JC, DeVore ML, Pigg KB. 2011. Prunus and Oemleria (Rosaceae) flowers from the Late Early Eocene Republic Flora of Northeastern Washington State, U.S.A. International Journal of Plant Sciences 172: 948–958. [Google Scholar]
- Berg CC, Hijman MEE. 1999. The genus Dorstenia (Moraceae). Ilicifolia 2: 1–211. [Google Scholar]
- Bromham L, Hua X, Cardillo M. 2015. Detecting macroevolutionary self-destruction from phylogenies. Systematic Biology 65: 109–127. [DOI] [PubMed] [Google Scholar]
- Bruun-Lund S, Clement WL, Kjellberg F, Rønsted N. 2017. First plastid phylogenomic study reveals potential cyto-nuclear discordance in the evolutionary history of Ficus L. (Moraceae). Molecular Phylogenetics and Evolution 109: 93–104. [DOI] [PubMed] [Google Scholar]
- Bull JJ, Charnov EL. 1985. On irreversible evolution. Evolution 39: 1149–1155. [DOI] [PubMed] [Google Scholar]
- Burge DO, Manchester SR. 2008. Fruit morphology, fossil history, and biogeography of Paliurus (Rhamnaceae). International Journal of Plant Sciences 169: 1066–1085. [Google Scholar]
- Calvillo-Canadell L, Cevallos-Ferriz SRS. 2007. Reproductive structures of Rhamnaceae from the Cerro del Pueblo (Late Cretaceous, Coahuila) and Coatzingo (Oligocene, Puebla) Formations, Mexico. American Journal of Botany 94: 1658–1669. [DOI] [PubMed] [Google Scholar]
- Case AL, Graham SW, Macfarlane TD, Barrett SCH. 2008. A phylogenetic study of evolutionary transitions in sexual systems in Australasian Wurmbea (Colchicaceae). International Journal of Plant Sciences 169: 141–156. [Google Scholar]
- Clement WL, Weiblen GD. 2009. Morphological evolution in the mulberry family (Moraceae). Systematic Botany 34: 530–552. [Google Scholar]
- Cohen KM, Finney SC, Gibbard PL, Fan J-X. 2017. The ICS International Chronostratigraphic Chart (2013; updated). Episodes 36: 199–204. [Google Scholar]
- Collinson ME. 1989. The fossil history of the Moraceae, Urticaceae (including Cecropiaceae), and Cannabaceae. In: Crane PR, Blackmore S, eds. Evolution, systematics, and fossil history of the Hamamelidae, Vol. 2 Oxford: Clarendon Press, 319–339. [Google Scholar]
- Cook JM, Rasplus J-Y. 2003. Mutualists with attitude: coevolving fig wasps and figs. Trends in Ecology & Evolution 18: 241–248. [Google Scholar]
- Cruaud A, Ronsted N, Chantarasuwan B, et al. 2012. An extreme case of plant–insect codiversification: figs and fig-pollinating wasps. Systematic Biology 61: 1029–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. 1876. The effects of cross and self fertilisation in the vegetable kingdom. London: J. Murray. [Google Scholar]
- Datwyler SL, Weiblen GD. 2004. On the origin of the fig: phylogenetic relationships of Moraceae from ndhF sequences. American Journal of Botany 91: 767–777. [DOI] [PubMed] [Google Scholar]
- Doyle JA. 2012. Molecular and fossil evidence on the origin of angiosperms. Annual Review of Earth and Planetary Sciences 40: 301–326. [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution 29: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufay M, Champelovier P, Käfer J, Henry JP, Mousset S, Marais GAB. 2014. An angiosperm-wide analysis of the gynodioecy–dioecy pathway. Annals of Botany 114: 539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster CSP, Sauquet H, van der Merwe M, McPherson H, Rossetto M, Ho SYW. 2017. Evaluating the impact of genomic data and priors on Bayesian estimates of the angiosperm evolutionary timescale. Systematic Biology 66: 338–351. [DOI] [PubMed] [Google Scholar]
- Freeman DC, Harper KT, Kent Ostler W. 1979. Ecology of plant dioecy in the intermountain region of Western North America and California. Oecologia 44: 410–417. [DOI] [PubMed] [Google Scholar]
- Friis EM, Crane PR, Pedersen KR. 2011. Early flowers and angiosperm evolution. Cambridge: Cambridge University Press. [Google Scholar]
- Glémin S, Muyle A. 2014. Mating systems and selection efficacy: a test using chloroplastic sequence data in angiosperms. Journal of Evolutionary Biology 27: 1386–1399. [DOI] [PubMed] [Google Scholar]
- Goldberg EE, Otto SP, Vamosi JC, et al. 2017. Macroevolutionary synthesis of flowering plant sexual systems. Evolution 71: 898–912. [DOI] [PubMed] [Google Scholar]
- Greeff JM, Compton SG. 2002. Can seed protection lead to dioecy in Ficus?Oikos 96: 386–388. [Google Scholar]
- Harrison RD, Yamamura N. 2003. A few more hypotheses for the evolution of dioecy in figs (Ficus, Moraceae). Oikos 100: 628–635. [Google Scholar]
- Hauenschild F, Matuszak S, Muellner-Riehl AN, Favre A. 2016. Phylogenetic relationships within the cosmopolitan buckthorn family (Rhamnaceae) support the resurrection of Sarcomphalus and the description of Pseudoziziphus gen. nov. Taxon 65: 47–64. [Google Scholar]
- Heilbuth JC. 2000. Lower species richness in dioecious clades. American Naturalist 156: 221–241. [DOI] [PubMed] [Google Scholar]
- Herendeen PS, Friis EM, Pedersen KR, Crane PR. 2017. Palaeobotanical redux: revisiting the age of the angiosperms. Nature Plants 3: 17015. [DOI] [PubMed] [Google Scholar]
- Hipsley CA, Müller J. 2014. Beyond fossil calibrations: realities of molecular clock practices in evolutionary biology. Frontiers in Genetics 5: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SYW, Phillips MJ. 2009. Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times. Systematic Biology 58: 367–380. [DOI] [PubMed] [Google Scholar]
- Hollingsworth PM, Forrest LL, Spouge JL, et al. 2009. A DNA barcode for land plants. Proceedings of the National Academy of Sciences, USA 106: 12794–12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jud NA, Gandolfo MA, Iglesias A, Wilf P. 2017. Flowering after disaster: Early Danian buckthorn (Rhamnaceae) flowers and leaves from Patagonia. PLoS One 12: e0176164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käfer J, de Boer HJ, Mousset S, Kool A, Dufay M, Marais GAB. 2014. Dioecy is associated with higher diversification rates in flowering plants. Journal of Evolutionary Biology 27: 1478–1490. [DOI] [PubMed] [Google Scholar]
- Käfer J, Marais GAB, Pannell JR. 2017. On the rarity of dioecy in flowering plants. Molecular Ecology 26: 1225–1241. [DOI] [PubMed] [Google Scholar]
- Kameyama T, Harrison R, Yamamura N. 1999. Persistence of a fig wasp population and evolution of dioecy in figs: a simulation study. Researches on Population Ecology 41: 243–252. [Google Scholar]
- Kass RE, Raftery AE. 1995. Bayes factors. Journal of the American Statistical Association 90: 773–795. [Google Scholar]
- Kerdelhué C, Rasplus J-Y. 1996. The evolution of dioecy among Ficus (Moraceae): an alternative hypothesis involving non-pollinating fig wasp pressure on the fig–pollinator mutualism. Oikos 77: 163–166. [Google Scholar]
- Kjellberg F, Gouyon P-H, Ibrahim M, Raymond M, Valdeyron G. 1987. The stability of the symbiosis between dioecious figs and their pollinators: a study of Ficus carica L. and Blastophaga psenes L. Evolution 41: 693–704. [DOI] [PubMed] [Google Scholar]
- Korpelainen H. 1998. Labile sex expression in plants. Biological Reviews 73: 157–180. [Google Scholar]
- Lewis PO. 2001. A likelihood approach to estimating phylogeny from discrete morphological character data. Systematic Biology 50: 913–925. [DOI] [PubMed] [Google Scholar]
- Lloyd DG. 1980. Sexual strategies in plants III. A quantitative method for describing the gender of plants. New Zealand Journal of Botany 18: 103–108. [Google Scholar]
- Lodewyckx T, Kim W, Lee MD, Tuerlinckx F, Kuppens P, Wagenmakers E-J. 2011. A tutorial on Bayes factor estimation with the product space method. Journal of Mathematical Psychology 55: 331–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. 2016. Mesquite: a modular system for evolutionary analysis. Version 3.10.http://mesquiteproject.org. [Google Scholar]
- Magallón S, Gomez-Acevedo S, Sanchez-Reyes LL, Hernandez-Hernandez T. 2015. A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytologist 207: 437–453. [DOI] [PubMed] [Google Scholar]
- Manchester SR. 1999. Biogeographical relationships of North American tertiary floras. Annals of the Missouri Botanical Garden 86: 472–522. [Google Scholar]
- Manos PS, Soltis PS, Soltis DE, et al. 2007. Phylogeny of extant and fossil Juglandaceae inferred from the integration of molecular and morphological data sets. Systematic Biology 56: 412–430. [DOI] [PubMed] [Google Scholar]
- Massoni J, Forest F, Sauquet H. 2014. Increased sampling of both genes and taxa improves resolution of phylogenetic relationships within Magnoliidae, a large and early-diverging clade of angiosperms. Molecular Phylogenetics and Evolution 70: 84–93. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Gateway Computing Environments Workshop (GCE), 2010. IEEE, 1–8. [Google Scholar]
- Misiewicz TM, Zerega NC. 2012. Phylogeny, biogeography and character evolution of Dorstenia (Moraceae). Edinburgh Journal of Botany 69: 413–440. [Google Scholar]
- Nadot S, Alapetite E, Baker WJ, Tregear JW, Barfod AS. 2016. The palm family (Arecaceae): a microcosm of sexual system evolution. Botanical Journal of the Linnean Society 182: 376–388. [Google Scholar]
- Obbard DJ, Harris SA, Buggs RJA, Pannell JR. 2006. Hybridization, polyploidy, and the evolution of sexual systems in Mercurialis (Euphorbiaceae). Evolution 60: 1801–1815. [PubMed] [Google Scholar]
- Onstein RE, Carter RJ, Xing YW, Richardson JE, Linder HP. 2015. Do Mediterranean-type ecosystems have a common history? Insights from the buckthorn family (Rhamnaceae). Evolution 69: 756–771. [DOI] [PubMed] [Google Scholar]
- Pagel M. 1994. Detecting correlated evolution on phylogenies: a general method for the comparative analysis of discrete characters. Proceedings of the Royal Society B: Biological Sciences 255: 37–45. [Google Scholar]
- Pagel M, Meade A. 2006. Bayesian analysis of correlated evolution of discrete characters by reversible‐jump Markov chain Monte Carlo. American Naturalist 167: 808–825. [DOI] [PubMed] [Google Scholar]
- Pagel M, Meade A. 2013. BayesTraits V2. http://www.evolution.rdg.ac.uk/BayesTraits.html. [Google Scholar]
- Pagel M, Meade A, Barker D. 2004. Bayesian estimation of ancestral character states on phylogenies. Systematic Biology 53: 673–684. [DOI] [PubMed] [Google Scholar]
- Pannell J. 1997. Widespread functional androdioecy in Mercurialis annua L. (Euphorbiaceae). Biological Journal of the Linnean Society 61: 95–116. [Google Scholar]
- Parham JF, Donoghue PCJ, Bell CJ, et al. 2012. Best practices for justifying fossil calibrations. Systematic Biology 61: 346–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Buckley TR. 2004. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Systematic Biology 53: 793–808. [DOI] [PubMed] [Google Scholar]
- Potter D, Eriksson T, Evans RC, et al. 2007. Phylogeny and classification of Rosaceae. Plant Systematics and Evolution 266: 5–43. [Google Scholar]
- Rambaut A, Drummond A. 2009. Tracer v1.5. http://tree.bio.ed.ac.uk/software/tracer/. [Google Scholar]
- Renner SS. 2014. The relative and absolute frequencies of angiosperm sexual systems: dioecy, monoecy, gynodioecy, and an updated online database. American Journal of Botany 101: 1588–1596. [DOI] [PubMed] [Google Scholar]
- Renner SS, Ricklefs RE. 1995. Dioecy and its correlates in the flowering plants. American Journal of Botany 82: 596–606. [Google Scholar]
- Ronquist F, Klopfstein S, Vilhelmsen L, Schulmeister S, Murray DL, Rasnitsyn AP. 2012. A total-evidence approach to dating with fossils, applied to the early radiation of the Hymenoptera. Systematic Biology 61: 973–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rønsted N, Weiblen GD, Clement WL, Zerega NJC, Savolainen V. 2008. Reconstructing the phylogeny of figs (Ficus, Moraceae) to reveal the history of the fig pollination mutualism. Symbiosis (Rehovot) 45: 45. [Google Scholar]
- Rønsted N, Weiblen GD, Cook JM, Salamin N, Machado CA, Savolainen V. 2005. 60 million years of co-divergence in the fig–wasp symbiosis. Proceedings of the Royal Society B: Biological Sciences 272: 2593–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabath N, Goldberg EE, Glick L, et al. 2016. Dioecy does not consistently accelerate or slow lineage diversification across multiple genera of angiosperms. New Phytologist 209: 1290–1300. [DOI] [PubMed] [Google Scholar]
- Salisbury BA, Kim J. 2001. Ancestral state estimation and taxon sampling density. Systematic Biology 50: 557–564. [PubMed] [Google Scholar]
- Sauquet H. 2003. Androecium diversity and evolution in Myristicaceae (Magnoliales), with a description of a new Malagasy genus, Doyleanthus gen. nov. American Journal of Botany 90: 1293–1305. [DOI] [PubMed] [Google Scholar]
- Sauquet H. 2013. A practical guide to molecular dating. Comptes Rendus Palevol 12: 355–367. [Google Scholar]
- Sauquet H. 2016. PROTEUS: a database for recording morphological data and creating NEXUS matrices. Version 1.26. http://eflower.myspecies.info/proteus. [Google Scholar]
- Sauquet H, Weston PH, Barker NP, Anderson CL, Cantrill DJ, Savolainen V. 2009. Using fossils and molecular data to reveal the origins of the Cape proteas (subfamily Proteoideae). Molecular Phylogenetics and Evolution 51: 31–43. [DOI] [PubMed] [Google Scholar]
- Sauquet H, Ho SYW, Gandolfo MA, et al. 2012. Testing the impact of calibration on molecular divergence times using a fossil-rich group: the case of Nothofagus (Fagales). Systematic Biology 61: 289–313. [DOI] [PubMed] [Google Scholar]
- Sauquet H, Carrive L, Poullain N, Sannier J, Damerval C, Nadot S. 2015. Zygomorphy evolved from disymmetry in Fumarioideae (Papaveraceae, Ranunculales): new evidence from an expanded molecular phylogenetic framework. Annals of Botany 115: 895–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauquet H, von Balthazar M, Magallón S, et al. 2017. The ancestral flower of angiosperms and its early diversification. Nature Communications 8: 16047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer H, Renner SS. 2010. A three-genome phylogeny of Momordica (Cucurbitaceae) suggests seven returns from dioecy to monoecy and recent long-distance dispersal to Asia. Molecular Phylogenetics and Evolution 54: 553–560. [DOI] [PubMed] [Google Scholar]
- Simpson MG. 2010. Plant Systematics. Cambridge: Academic Press. [Google Scholar]
- Soltis DE, Smith SA, Cellinese N, et al. 2011. Angiosperm phylogeny: 17 genes, 640 taxa. American Journal of Botany 98: 704–730. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vamosi JC, Otto SP, Barrett SCH. 2003. Phylogenetic analysis of the ecological correlates of dioecy in angiosperms. Journal of Evolutionary Biology 16: 1006–1018. [DOI] [PubMed] [Google Scholar]
- Wang H, Moore MJ, Soltis PS, et al. 2009. Rosid radiation and the rapid rise of angiosperm-dominated forests. Proceedings of the National Academy of Sciences, USA 106: 3853–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CO, Ackerly DD, Kembel SW. 2008. Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics 24: 2098–2100. [DOI] [PubMed] [Google Scholar]
- Weiblen GD. 2000. Phylogenetic relationships of functionally dioecious Ficus (Moraceae) based on ribosomal DNA sequences and morphology. American Journal of Botany 87: 1342–1357. [PubMed] [Google Scholar]
- Wiegrefe SJ, Sytsma KJ, Guries RP. 1998. The Ulmaceae, one family or two? Evidence from chloroplast DNA restriction site mapping. Plant Systematics and Evolution 210: 249–270. [Google Scholar]
- Williams EW, Gardner EM, Harris R III, Chaveerach A, Pereira JT, Zerega NJC. 2017. Out of Borneo: biogeography, phylogeny and divergence date estimates of Artocarpus (Moraceae). Annals of Botany 119: 611–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z-Y, Monro AK, Milne RI, et al. 2013. Molecular phylogeny of the nettle family (Urticaceae) inferred from multiple loci of three genomes and extensive generic sampling. Molecular Phylogenetics and Evolution 69: 814–827. [DOI] [PubMed] [Google Scholar]
- Yang M-Q, van Velzen R, Bakker FT, Sattarian A, Li D-Z, Yi T-S. 2013. Molecular phylogenetics and character evolution of Cannabaceae. Taxon 62: 473–485. [Google Scholar]
- Yang Z, Rannala B. 2005. Bayesian estimation of species divergence times under a molecular clock using multiple fossil calibrations with soft bounds. Molecular Biology and Evolution 23: 212–226. [DOI] [PubMed] [Google Scholar]
- Zerega NJC, Clement WL, Datwyler SL, Weiblen GD. 2005. Biogeography and divergence times in the mulberry family (Moraceae). Molecular Phylogenetics and Evolution 37: 402–416. [DOI] [PubMed] [Google Scholar]
- Zerega NJC, Supardi N, Motley TJ. 2010. Phylogeny and recircumscription of Artocarpeae (Moraceae) with a focus on Artocarpus. Systematic Botany 35: 766–782. [Google Scholar]
- Zhang S-D, Soltis DE, Yang Y, Li D-Z, Yi T-S. 2011. Multi-gene analysis provides a well-supported phylogeny of Rosales. Molecular Phylogenetics and Evolution 60: 21–28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.