Abstract
Objective:
This investigation was performed to evaluate the registration accuracy between magnetic resonance imaging (MRI) and pathology using three-dimensional (3D) printed molds.
Methods:
Tissue-mimicking prostate phantoms were manufactured with embedded fiducials. The fiducials were used to measure and compare target registration error (TRE) between phantoms that were sliced by hand versus phantoms sliced within 3D-printed molds. Subsequently, 10 radical prostatectomy specimens were placed inside molds, scanned with MRI, and then sliced. The ex vivo scan was used to assess the true location of whole mount (WM) slides relative to in vivo MRI. The TRE between WM and in vivo MRI was measured using anatomic landmarks.
Results:
Manually sliced phantoms had a 4.1 mm mean TRE, whereas mold-sliced phantoms had a 1.9 mm mean TRE. Similarly, mold-assisted slicing reduced mean angular misalignment around the left-right (LR) anatomic axis from 10.7 to 4.5 degrees. However, ex vivo MRI revealed that excised prostates were misaligned within molds, including a mean 14-degree rotation about the LR axis. The mean in-plane TRE was 3.3 mm using molds alone, and 2.2 mm after registration was corrected with ex vivo MRI.
Conclusion:
Patient-specific molds improved accuracy relative to manual slicing techniques in a phantom model. However, the registration accuracy of surgically resected specimens was limited by their imperfect fit within molds. This limitation can be overcome with the addition of ex vivo imaging.
Significance:
The accuracy of 3D printed molds was characterized, quantifying their utility for facilitating MRI-pathology registration.
Keywords: Image registration, Magnetic resonance imaging, Pathology, Phantoms, Three-dimensional printing
I. Introduction
Multi-parametric magnetic resonance imaging (mpMRI) has become an important tool in the diagnosis and localization of prostate cancer (CaP) [1], though many tumors are missed or mischaracterized [2], [3]. In order to improve prostate mpMRI, imaging must be correlated with groundtruth histopathology. The true extent of CaP tumors can be observed on whole-mount (WM) prostatectomy slides, but existing MR-WM registration techniques introduce substantial errors [4]. These techniques rely on assumptions regarding the position, orientation, and shape of an excised gland relative to preoperative MRI, which may be erroneous.
One common assumption during MR-WM registration is that prostate specimens are grossed with uniform slice width and orientation. However, since excised glands are typically sliced by hand, the resulting slides are neither evenly spaced nor parallel to one another, and their correspondence with MRI is uncertain [5]. Furthermore, the sparse through-plane resolution of whole mount slides can cause tumors to be missed or mischaracterized [6].
Prostate geometry on WM slides can differ considerably from the gland’s size and shape observed on MRI, further confounding attempts at correlation. During MRI, the use of endorectal receiver coils can substantially deform the gland [7]. During prostatectomy, extraprostatic tissue is often resected beyond the capsule, while factors such as bladder neck dissection, vascular and urethral collapse, and tissue dehydration can reduce prostate volume by 10% [8]. The specimen is subject to further deformation during pathology processing, including shrinkage from formalin fixation and the tensile stresses of tissue slicing [9], [10].
Many efforts have been made to improve MR-WM registration accuracy. Manually annotated landmarks such as the urethra, capsule, and prostate nodules have been used to inform nonrigid MR-WM registrations [11]–[13]. Voxel intensities and image features have also been leveraged to perform registrations using machine vision and/or mutual information (MI) [13]–[16]. However, these efforts were limited by small sample sizes and often made specious assumptions regarding the spacing and orientation of WM slides. Since the true position and angle of manually acquired slices is unknown, registration errors likely persisted.
Others have employed a guide or template to regulate prostate slicing, ensuring that slides were acquired with uniform spacing and orientation [5], [17]–[19]. Though this approach enabled a reasonably accurate calculation of tumor volume, such systems had no way of ensuring that the gland was sliced at positions and orientations that matched MR images. They also had no prospective means of adjusting for patient-specific anatomy or compensating for gland deformation.
Ex vivo
MRI of excised glands has been employed by some groups to visualize the surgical specimen and register it with preoperative images [8]–[9], [12], [19]–[21]. Park et al. employed MI and nonrigid transformations to register WM with block-face photographs, ex vivo MRI, and in vivo MRI. Using an indirect measure of target registration error (TRE) in 2 specimens, they reported 2.3-3.7 mm mean error [12]. Reynolds et al. stabilized 6 specimens in agarose, performed ex vivo MRI, and nonrigidly registered them to the in vivo image series. An analysis of anatomic landmarks suggested that their approach conferred 3.3 mm TRE between in vivo MRI and WM [19]. Gibson et al. supplemented anatomic landmarks with gadolinium-soaked fiducials in 9 specimens. Since the fiducials were visible on both ex vivo MRI and WM slides, they could serve as the basis for an affine transformation that yielded sub-mm TRE [20]. These efforts have shown that ex vivo MRI can enable highly accurate registrations. However, ex vivo scanning studies have been limited by small sample sizes, since they tend to require specialized pathology processing, custom imaging, and additional analysis.
In recent years, 3D-printed patient-specific prostate molds have been introduced as a means of processing excised glands and facilitating MR-WM registration. Unlike other slicing templates, molds are manufactured to match each gland’s anatomy on MRI. If the specimen is effectively held in the same shape and orientation that was observed during in vivo imaging, each slide acquired should match a specific MRI slice. This approach was first described by the National Cancer Institute [22]–[24], which reported on the use of custom molds to obtain WM slides in 6-mm increments for 73 patients. Subsequently the University of California Los Angeles designed a patient-specific mold for obtaining slices in 4.5-mm increments, and reported on its use in 114 cases [3]. Patient-specific molds show promise, but to date few groups have reported on the errors associated with mold-based registration. When Elen et. al. used ex vivo MRI to evaluate 3 excised prostates within conventional 3D-printed molds, they observed substantial rotational misalignment [25]. This effect could greatly reduce registration accuracy and the utility of 3D-printed molds.
In order to substantiate the benefits of 3D-printed molds versus conventional slicing, we conducted a statistically powered study using tissue-mimicking prostate phantoms. This study helped to establish a lower bound for registration error. Subsequently, in order to quantify the errors resulting from the fit and alignment of actual specimens within 3D-printed molds, we performed ex vivo imaging and MR-WM registration for 10 excised prostates.
II. Materials and Methods
The design, manufacture, and use of 3D-printed patientspecific molds at our institution has been previously reported [3], [26], [27]. In order to generate a mold, patients received pre-surgical T2-weighted MRI (SPACE, FOV 170×170×90 mm3, TE/TR = 204/2230 ms, resolution 0.66×0.66×1.5 mm3, 2 averages, 7 min). The prostate capsule was contoured in 3D using segmentation software (Profuse, Eigen, Grass Valley CA), and the contours were used to create a cavity within a mold using computer-aided design software (Solidworks, Dassault Systémes, France). Slots with 4.5 mm spacing were aligned with MRI slice positions, allowing every third MRI slice to be sampled during pathology processing. Molds were manufactured using consumer-grade 3D printers (Makerbot, Makerbot Industries, Brooklyn NY) and printing software (Simplify3D, Simplify3D Inc., Cincinnati OH). Each mold cost approximately $3 USD, took approximately 5 hours to print (20% infill, 0.2 mm resolution), and was composed of ∼0.1 kg of polylactic acid, a biocompatible plastic.
A. Comparison of Mold-Assisted Slicing Vs. Conventional Slicing in Tissue-Mimicking Phantoms
In an IRB-approved protocol, tissue-mimicking prostate phantoms were produced to compare mold-assisted gland slicing with conventional techniques (N = 28 phantom pairs, sliced by 7 operators). Statistical analysis based on a test case indicated that 20 or more phantom pairs would yield greater than 95% power for detection of differences in slice angle and slice thickness.
Six MRI prostate contours were selected from a prior clinical study to serve as models for the phantoms. They were chosen to represent a variety of morphologies, tissue volumes (33-68 cc), and MR acquisition angles relative to the posterior capsule surface (1-22 degrees). The prostate contours were used to generate cavities within six templates, along with divots to indicate urethra location and channels for 12 vector fiducials (Fig 1A). The templates were designed and 3D-printed via the same procedure as patient-specific molds. They were then used to cast six unique phantoms (Fig 1B).
Fig. 1.
(A) Computer aided design of casting template, (B) 3D printed casting template for manufacture of tissue mimicking prostate phantoms with implanted fiducials, C) a digital slice plane from the phantom model showing color-coded fiducials, and D) a flatbed scan of a phantom with inked fiducials visible.
After sealing the casting template, 26-Ga needles were inserted through each planned fiducial trajectory. A 3% agarose solution was heated to a boil and allowed to cool below 50°C before filling the casting template. The cast phantom was cooled for three hours within a refrigerator, solidifying into an agar gel that approximated the mechanical properties of prostate tissue. Next, water-insoluble acrylic ink was injected through the needles as each was withdrawn, leaving behind 12 color-coded fiducials. Last, each phantom was inked externally to indicate urethra location and anatomic orientation.
Four of the 12 fiducials were inked blue and ran parallel to the inferior-superior (IS) axis, framing the corners of a 15×15 mm rectangular prism. Four other fiducials were inked red, and framed the corners of a tapering pyramid. Since they were tightly clustered near the phantom base but far apart near the apex, their relative positions were sensitive to slicing depth. The last four fiducials were inked green and assigned random trajectories, each intended to be uncorrelated with all other fiducials.
Fiducial positions were computed for simulated slice planes in each of the six phantom models (Fig 1C), sampled in 0.2 mm depth increments and 1 degree angle increments for rotations about the anterior-posterior (AP) and left-right (LR) anatomic axes. Fiducial coordinates were recorded that corresponded to 275,000 – 350,000 unique slice planes per model, depending on prostate size. The coordinates were projected into 2D and saved into a look-up table.
Seven operators (3 pathologists and 4 pathology assistants) were recruited for this study, all of whom had experience grossing prostate specimens in a surgical pathology laboratory. Each operator was assigned a set of 4 prostate phantom pairs. The first 3 phantom pairs were all derived from different prostate models, and were used to compare manual slicing versus mold-based slicing. The fourth phantom pair was a repeat of the first prostate model, and was used to assess intra-operator variability. Operators were also provided with 3D-printed slicing molds, the cavities of which were enlarged 5% relative to the agar phantom volume in order to simulate imprecise fit within the mold. The operators utilized sectioning knives, forceps, paper towels, gloves, and a cutting board when grossing the specimens.
Each operator was instructed to section each pair of phantoms, first by hand exactly as they would have processed a clinical specimen. Specifically, their goal was to place the prostate posterior-side down and then to section it from apex to base, with evenly spaced (∼5 mm thick) slices perpendicular to the cutting board and the apex-to-base axis. After manual slicing was complete, mold-assisted slicing was performed by first positioning the phantom within the mold, then cutting down through each mold slot serially from apex to base. Hand slicing always preceded mold-assisted slicing, so the operator would not be biased by the slice orientation imposed by the mold. This process was repeated four times by each operator, on each pair of phantoms.
After sectioning, each phantom slice was placed on a transparency and digitized via flatbed scanner with a resolution of 600 dots per inch (Fig 1D). The 2D positions of the color-coded fiducials on each slice were recorded. The mean squared errors (MSE) were then computed between the observed fiducial coordinates and all entries of the precomputed look-up table. The lowest MSE corresponded to the best estimate of the slice’s true 3D position within the specimen. The depths and angles of each slice were compared to the ‘ideal’ slices typically assumed, i.e. evenly spaced and parallel to the MR image plane. The difference between observed and ideal slice angle, depth, and thickness were recorded for each sample.
In total, the seven operators sectioned four pairs of phantoms each: 3 unique pairs assigned from the 6 available prostate models, and 1 repeated pair. This resulted in a total of 56 phantoms (28 matched-pairs), each sliced 5-11 times. Wilcoxon signed-rank tests were performed to evaluate differences between the mold-sliced and manually sliced phantom populations (alpha = 0.05). As a measure of intraoperator variability, the difference in slice position and angle for the repeated phantoms is also reported.
B. Mold Registration Accuracy With Ex Vivo Specimens
In an IRB-approved and biosafety-approved protocol, we measured the MR-WM registration error of 10 prostates sliced within patient-specific molds. Prior to radical prostatectomy each patient received mpMRI, which was used to design and 3D-print a prostate mold. After surgery, each prostate specimen was trimmed and placed within the mold (Fig 2A). The mold was locked into a 3D-printed frame that centered it within a cylindrical container. The cylinder was then filled with a perfluorocarbon solution (Fomblin, Solvay S.A., Belgium) chosen to match the magnetic susceptibility of tissue while minimizing background signal (Fig 2B) [12], [28]. The cylinder was sealed and placed inside a 15-channel knee coil, and it was positioned within a whole-body 3T MRI scanner (Prisma, Siemens, Erlangen Germany).
Fig. 2.
Setup for ex vivo scanning of a prostate specimen, including A) an inked prostate gland within a patient-specific mold, B) the specimen, mold, and frame immersed in perfluorocarbon solution, C) design of the fiducial cartridge with the agar-filled region shaded blue, and D) the fiducial cartridge as seen on sagittal T2-weighted MRI.
Ex vivo mpMRI was performed [29], including a highresolution T2-weighted sequence with field of view 75×75 mm2, in-plane resolution 0.29×0.29 mm2, slice thickness 1.5 mm, and 3 averages over 8 minutes. An external fiducial, containing a patterned cartridge with 2% agarose gel (Fig 2C-2D), was placed within the mold and visualized during ex vivo MRI. This fiducial indicated the alignment of the specimen with the intended WM slide locations. After scanning, the prostate was sliced within the mold, sampling every third ex vivo MR image plane. The mean time required for ex vivo MRI, including specimen preparation and transportation, was 116 minutes.
A variation of the registration methodology described by Fei et al. [30] was performed in order to maximize mutual information between the in vivo (V1) and ex vivo (VE) T2-weighted MR volumes. The formula for MI is
| (1) |
where pRF is the joint probability, pR is the marginal probability of the ex vivo reference volume (VE), and pF is the marginal probability of the in vivo floating volume (VI). These were estimated from the normalized joint and marginal intensity histograms, respectively.
Both VI and VE were resampled to have an isotropic 3D resolution of 0.375 mm, i.e. ¼ of the 1.5 mm through-plane resolution. T2 intensity values within the prostate were normalized, and a baseline alignment was performed with reference to the fiducial cartridge. Then, a standard matrix was used to rigidly transform VI in order to maximize MI, rotating θx, θy, and θz in 1° increments and translating Δx, Δy, and Δz in 0.375 mm increments.
First, MI was measured for a broad range (±20°, ±6 mm) of coarsely-sampled transformations in 3 dimensions (θxΔyΔz). After performing the transform that maximized MI, this was repeated for 2 additional dimension sets (θvΔxΔz and θzΔxΔy). This method broadly sampled the space while keeping computation time reasonable, and reliably transformed VI into rough alignment with VE.
Second, gradient descent optimization was performed along all 6 dimensions with fixed step size (1 voxel translation and 1 degree rotation). The convergence criterion was arrival at a local minimum, for which further changes along any dimension would decrease MI. The combination of rotations and translations observed to maximize MI between VI and VE were used to define the transformation matrix T0
| (2) |
such that any point in VI could be transformed to VE coordinates through matrix multiplication with T0. Application of this transform to every voxel in VI resulted in the postregistered in vivo volume VReg. In order to evaluate the accuracy of T0, a set of corresponding anatomic landmarks LVReg-VE was identified on VReg and VE. The LVReg-VE landmark centroids were recorded in 3D, and their coordinates were compared.
The VI to VE registration runtime was approximately 20 minutes. Once the in vivo and ex vivo volumes were aligned, the MR images most closely matching each WM prostatectomy slide were determined by consensus between a radiologist and two imaging scientists. Then, for every set of matched images, a second set of corresponding anatomic landmarks, LVReg-WM, was identified on VReg and WM. Between 1 and 4 (mean 3.6) anatomic landmarks were chosen for each pair of images, and the transform inverse to T0 was used to determine the VReg landmark coordinates in VI.
In order to register WM to MRI, a previously described nonrigid registration procedure was performed [3], [26]. First, each whole-mount specimen was contoured (CWM), encapsulating only prostatic tissue. Second, corresponding 2D MRI prostate contours (CMR) were traced on the MR image planes corresponding each WM slide. Third, in-plane rotational misalignment was corrected by rotating the WM slide until the mean distance between CMR and CWM was minimized. Fourth, lines were generated orthogonal to the slope of CMR at evenly spaced intervals. Corresponding control points on CWM were then assigned by computing the intersection of these lines with CWM. Fifth, in order to compensate for nonrigid gland deformation, these control points were used to perform a thin-plate spline transformation as described by Fei et. al [31]. Lastly, the WM slide was projected into 3D, now fully registered to MRI. The WM to MRI registration runtime was approximately 25 minutes.
This procedure was performed twice per specimen, to nonrigidly register: 1) WM to VI and 2) WM to VReg. The WM to VI registration (Fig 3) was performed to characterize TRE when blinded to ex vivo images, naively assuming that the mold had correctly aligned the surgical specimen with VI, i.e. assuming that each slide had been acquired from the planned location. The WM landmarks could then be compared to the corresponding MRI landmark coordinates. The WM to VReg registration (Fig 4) was used to characterize the LVReg-WM TRE when ex vivo information was available, i.e. when WM images were registered with the correct image slice, as determined using ex vivo MRI. Inter- and intra-patient mean TRE was then calculated, and in-plane TREs were compared using a Wilcoxon signed-rank test.
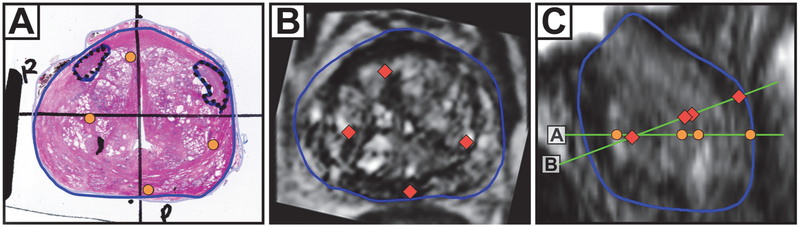
Fig. 3.
A) nonrigid registration of WM with in vivo MRI while blinded to ex vivo MRI results. B) In vivo MR image corresponding to true WM slide location, as determined via ex vivo registration. C) sagittal view of in vivo MRI, with the projected slice planes and fiducials from 4A and 4B, showing rotational misalignment. Prostate contours (blue), image planes (green), WM landmarks (orange circles), and MRI landmarks (red diamonds) are superimposed.
Fig. 4.
A) Nonrigid registration of WM with ex vivo MRI. B) Rigid intensity-based registration of in vivo MRI to ex vivo MRI. C) Ex vivo MRI. Prostate contours (blue), 15×15 mm gridlines (red), WM landmarks (orange circles), and MRI landmarks (red diamonds) are superimposed.
III. Results
A. Comparison of Mold-Assisted Slicing Vs. Conventional Slicing in Tissue-Mimicking Phantoms
Angle, depth, and 3D registration errors for the matchedpair phantom study are reported in Table 1. For the N=21 (non-repeated) pairs of sliced phantoms, the mean slice depth was farther from ideal (evenly-spaced from apex to base) for hand-sliced specimens than for mold-sliced specimens (2.1 vs 1.0 mm depth error, p<0.01). The mean angular misalignment about the LR anatomic axis was greater in hand-sliced specimens (10.7 vs. 4.5 degrees, p<0.01). Mean registration error was 4.1 mm in hand-sliced specimens, much larger than the 1.9 mm mean error in mold-sliced specimens (p<0.01). Similar trends were observed in the N=7 matched pairs of repeat-sliced specimens, though the study was underpowered for these comparisons and they did not reach significance.
Table 1:
Mean ± SD between mold-sliced and hand-sliced prostate phantoms
| Phantom Error Mean ± SD (N = 21) |
Mold- Sliced |
Hand- Sliced |
P-Value |
|---|---|---|---|
| LR-Axis Angle Error (°) | 4.5 ± 2.9 | 10.7 ± 6.7 | 0.001 |
| AP-Axis Angle Error (°) | 3.9 ± 3.6 | 3.7 ± 4.2 | 0.9 |
| Depth Error (mm) | 1.0 ± 0.4 | 2.1 ± 1.4 | 0.002 |
| 3D Registration Error (mm) | 1.9 ± 0.6 | 4.1 ± 1.3 | <0.001 |
| Repeat-Phantom Mean Difference ± SD (N = 7) |
Mold- Sliced |
Hand- Sliced |
P-Value |
|---|---|---|---|
| LR-Axis Angle (°) | 5.3 ± 5.3 | 10.6 ± 5.6 | 0.22 |
| AP-Axis Angle (°) | 3.1 ± 2.3 | 4.8 ± 4.5 | 0.46 |
| Depth Error (mm) | 1.2 ± 0.5 | 1.8 ± 0.7 | 0.29 |
B. Mold Registration Accuracy with Ex Vivo Specimens
The registration parameters associated with T0 (the transform between VI and VE) are reported in Table 2. Mean (absolute value) rotations of 14°, 4°, and 6° were performed about the LR, AP, and IS axis respectively. Rotation was followed by a mean 3D translation of 2.7 mm. Using the N=50 pairs of 3D anatomic landmarks that comprised LVReg-VE, the mean TRE was 4.5 mm for the baseline mold alignment and 1.7 mm after registration via T0.
Table 2:
MI-based transform parameters, prostate geometry changes, and in vivo MRI to ex vivo MRI mean TRE for 3D Landmarks (LVReg-VE)
| Mi-Based Rigid Registration Parameters | Prostate Geometry Changes | Mean 3D Landmark TRE | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Case No. | LR-Axis Rot. (°) |
AP-Axis Rot. (°) |
IS-Axis Rot. (°) |
Shift 3D (mm) |
In Vivo Vol. Missing |
Ex Vivo Vol. Extra |
Vol. Change cc (%) |
Pre-Reg. TRE (mm) |
Post-Reg. TRE (mm) |
| 1 | 31.8 | 8.0 | 8.0 | 1.1 | 12.7% | 10.5% | −0.9 (−2.5%) | 6.2 | 1.1 |
| 2 | 27.4 | −2.0 | −4.4 | 4.8 | 15.9% | 15.8% | −0.1 (−0.2%) | 8.3 | 3.1 |
| 3 | 11.8 | −4.8 | −4.6 | 2.3 | 9.5% | 9.9% | 0.3 (0.5%) | 2.8 | 1.1 |
| 4 | 2.1 | −1.3 | 6.7 | 2.1 | 11.6% | 6.4% | −3.0 (−5.5%) | 2.5 | 1.4 |
| 5 | −14.9 | 7.1 | 9.0 | 4.4 | 30.9% | 5.9% | −12.4(−26.6%) | 4.8 | 2.8 |
| 6 | 18.3 | 2.6 | 10.5 | 1.5 | 8.7% | 15.1% | 2.3 (7.5%) | 3.9 | 1.5 |
| 7 | −11.9 | 0.5 | −2.1 | 2.5 | 12.8% | 8.6% | −1.6 (−4.6%) | 3.4 | 1.2 |
| 8 | 12.0 | −7.0 | −3.5 | 2.5 | 14.3% | 11.6% | −2.6 (−3.1%) | 5.2 | 1.1 |
| 9 | 5.0 | 3.9 | 1.8 | 2.3 | 12.1% | 7.7% | −1.8 (−4.7%) | 3.6 | 1.9 |
| 10 | −8.5 | 4.4 | 9.6 | 3.3 | 18.5% | 13.1% | −2.2 (−6.3%) | 3.9 | 1.4 |
| Mean | 7.3 | 1.1 | 3.1 | 2.7 | 14.7% | 10.4% | −2.2 (−4.6%) | 4.5 | 1.7 |
| Mean(ABS) | 14.4 | 4.2 | 6.0 | ||||||
The in vivo prostate segmentation, with which the mold cavity was generated, was compared to a segmentation of the surgical specimen on ex vivo MRI. A mean volume reduction of 4.6% occurred relative to the in vivo segmentation, less shrinkage than the 10% reported by Orczyk et. al [8]. After registration via T0, on average 10.4% (±3.3%) of the excised specimen’s volume was considered extraprostatic relative to the in vivo segmentation. Conversely, 14.7% (±6.1%) of the tissue considered prostatic on in vivo imaging was not present after resection. Extraprostatic tissue was most commonly found on the gland’s lateral and posterior aspects, especially towards the prostate base. Relative to the preoperative segmentation, tissue was commonly missing from the base and far apex, particularly the anterior base adjacent to the bladder neck. Figure 5 shows post-registration contour overlays from an exemplary case.
Figure 5:
Post-registration 3D surfaces in A) axial, B) coronal, and C) sagittal perspective from a single case (no. 6) where 7% of in vivo tissue (blue) was missing and 13.6% of ex vivo tissue (yellow) was considered extraprostatic. Overlap is shown in green.
TREs for WM to MRI registration are reported in Table 3, based on N = 148 pairs of anatomic landmarks that comprised LVReg-WM. For the “mold-only” WM to VI registration, which was blinded to the specimen misalignment observed on ex vivo MRI, the mean TRE was 3.3 mm in-plane, 2.9 mm out-of-plane, and 4.8 mm in 3D. After ex vivo MRI was used to compensate for angular misalignment and the through-plane error component, the in-plane TRE was 2.2 mm, a significant improvement (P = 0.002).
Table 3:
In Vivo MRI to WM mean target registration errors for LReg-WM ± standard deviation (SD), with all measures reported in mm.
| WM to VReg |
WM to VI (Blinded to Ex Vivo) |
|||
|---|---|---|---|---|
| Case No. | In-Plane | In-Plane | Through- Plane (abs) |
3D |
| 1 | 2.5 ± 1.3 | 5.4 ± 2.5 | 4.6 ± 3.2 | 7.5 ± 3.2 |
| 2 | 3.0 ± 1.4 | 5.1 ± 1.7 | 3.4 ± 2.0 | 6.4 ± 1.7 |
| 3 | 2.6 ± 1.5 | 3.1 ± 1.2 | 3.5 ± 2.1 | 4.8 ± 2.0 |
| 4 | 1.6 ± 1.8 | 2.6 ± 1.6 | 3.0 ± 0.4 | 4.1 ± 1.3 |
| 5 | 2.2 ± 1.3 | 3.7 ± 1.9 | 2.9 ± 2.0 | 5.0 ± 2.1 |
| 6 | 2.3 ± 1.3 | 3.2 ± 1.2 | 3.7 ± 1.9 | 5.2 ± 1.3 |
| 7 | 1.7 ± 1.3 | 2.6 ± 1.5 | 2.1 ± 1.6 | 3.5 ± 1.6 |
| 8 | 2.4 ± 1.3 | 4.0 ± 1.7 | 2.1 ± 1.5 | 4.7 ± 1.7 |
| 9 | 2.0 ± 1.2 | 2.0 ± 1.2 | 1.2 ± 1.0 | 2.6 ± 1.2 |
| 10 | 1.9 ± 1.2 | 2.4 ± 1.4 | 2.9 ± 1.5 | 3.9 ± 1.7 |
| Mean ± SDoM* | 2.2 ± 0.4 | 3.3 ± 0.5 | 2.9 ± 1.7 | 4.8 ± 1.8 |
SDoM is the Standard Deviation of the Mean
IV. Discussion
The two studies described herein present a systematic evaluation of 3D-printed prostate molds for MR-pathology correlation. The phantom study enabled a controlled, repeatable comparison between specimens sliced manually and specimens sliced with patient-specific molds. The results indicate that molds improve registration accuracy relative to manual slicing techniques.
The clearest benefit appears be regulation of the LR-axis slicing angle, which deviated from the angle of MR images by an average of 4.5 degrees using molds. By contrast, manually sliced specimens had a mean LR-axis rotational misalignment of over 10 degrees, and some cases exceeded 20 degrees. This is likely due to the practice of manually slicing prostate specimens posterior-side down, whereas MR images are never exactly perpendicular to the gland’s posterior face. Interestingly, rotational about the AP axis was not reduced by use of a mold. This may be because, within a mold, the sectioning knife cuts down from right to left, exerting torque about the AP axis and increasing this aspect of rotational misalignment. Conversely, when sliced by hand, the sectioning knife cuts down from anterior to posterior, exerting torque about the LR axis.
Perhaps the most meaningful metric was registration accuracy, the consequence of both rotation and translation errors. The mean TRE of mold-sliced phantoms was under 2 mm, less than half that of manually sliced specimens. In addition to improving registration accuracy, patient-specific molds helped improve consistency and repeatability; the standard deviation for LR-rotation error, depth error, and registration error for mold-sliced specimens was less than half that of manually sliced specimens. Since patient-specific molds hold the prostate in a fixed position and regulate slice spacing, it stands to reason that they would reduce interspecimen and inter-operator variability. Even intra-operator variability appeared to diminish through use of patient-specific molds, though the study was not adequately powered to prove the significance these measures (N = 7).
The phantom study showed that registrations can be very accurate, if the cavity of a prostate mold is a good representation of the excised specimen. However, as observed in the ex vivo tissue study, differences between in vivo prostate segmentations and excised specimens can lead to larger registration errors. When registering VI to VE, the matrix T0 was necessary to transform in vivo data from its baseline position, correcting for mold misalignment. The mean 3D translation associated with T0 was 2.7 mm, a relatively minor adjustment. However, substantial rotations—sometimes over 30 degrees—were necessary to optimally register VI to VE. This indicates that the mold was not orienting specimens correctly with in vivo MRI, contributing to a mean 3D TRE of 4.5 mm for the baseline mold alignment. Ex vivo MRI was able to correct for this misalignment, as evidenced by the reduction of TRE to 1.7 mm after MI-based registration was performed.
Unlike the phantom study, where LR-axis misalignment was minimal, LR rotation error between ex vivo and in vivo volumes was 14 degrees on average. This discrepancy can largely be ascribed to the presence of extraprostatic tissue around the posterior base in combination with missing tissue towards the anterior base, together causing the ex vivo specimen to rotate relative to in vivo imaging. Conversely, rotations about the AP axis were very similar to phantom study predictions. The prostate’s bilateral symmetry ensured that discrepancies between in vivo segmentations and ex vivo specimens affected both hemispheres equally, resulting in very little AP and IS misalignment on average.
After the “mold-only” nonrigid registration blinded to the ex vivo scan, the mean TRE between whole mount and in vivo MRI was 3.3 mm in the axial plane and 4.8 mm in 3D. These registration errors are much higher than the TRE measured in the phantom study. However, since landmark identification is inherently subjective, some part of the registration error is likely attributable to fiducial localization variability. When registration was performed with Vreg instead of VI, the 2D TRE was significantly reduced to 2.2 mm, which is sufficient for most clinical purposes. Thus, the addition of ex vivo MRI substantially improved registration accuracy, though it necessitated more personnel and higher costs. It is noteworthy that TRE appeared to improve with time, suggesting that early cases were subject to operator error and mean accuracy may be higher for future work.
To our knowledge, this is the first published study on moldsliced vs. hand-sliced phantoms, and the first report of mold TRE with and without ex vivo MRI. Unlike most ex vivo prostate studies, we have integrated with our institution’s clinical workflow and processed specimens prior to formalin fixation. The 2.2 mm TRE using molds in combination with ex vivo MRI is consistent with the 1-4 mm error previously reported for ex vivo imaging studies [12], [19], [20], [25]. Only Gibson et. al and Elen et. al. reported substantially better accuracy (with mean TRE of 0.7 mm and 0.9 respectively), which they were able to achieve by registering with the aid of internal landmarks. In the absence of ex vivo MRI we observed TRE to be relatively large, a fact that should be acknowledged in any study relying exclusively on 3D-printed molds of this type.
In addition to the influence of landmark localization error, there are several limitations to the work presented here. First, for the phantom study, the agarose was an imperfect replica of prostate tissue mechanics and was more easily sliced than actual prostate specimens. Second, in our analysis of phantom slice location and orientation, each slice was assumed to be planar. Any curvature would have perturbed fiducial vector positions, resulting in a conformation that was not present in the lookup table of possible slice planes. This effect was present but fairly minimal, since the mean difference between observed fiducial coordinates and those of the matched slice planes was less than 0.2 mm. Third, key differences exist between in vivo prostate segmentations and ex vivo specimens, an effect that was observed in the ex vivo tissue study but not accounted for in the phantom study.
Fourth, for the ex vivo tissue study, the in vivo to ex vivo transformation was limited by its constraint of rigidity. For 2 of the 10 cases, the in vivo MR and ex vivo MR prostate shapes were not well matched and TRE was nearly double the 1.7 mm average. Since the comparison of landmark positions depended on an accurate in vivo to ex vivo registration, changes in prostate shape likely resulted in an inflation of the measured WM-to-MRI TRE as well. Fifth, during landmark annotation it was assumed that each whole mount slide corresponded to a single ex vivo image. However, the throughplane MRI resolution was 1.5 mm, and nominally 2D error measures were unavoidably influenced by a through-plane uncertainty of this magnitude. Sixth, landmarks were identified on Vreg instead of VI, an arrangement that can result in bias but was unavoidable due to large rotational misalignments between VE and VI. However, the anatomic correspondence of 3D landmarks was reviewed and verified after performing an inverse transformation to VI.
Several improvements can be made to the mold’s design in order to address its limitations and further improve registration accuracy. First, systematic differences between the in vivo prostate segmentation and ex vivo specimen should be compiled, quantified, and used to adjust mold cavities. For example, since tissue is usually missing from the bladder neck as a consequence of resection, the mold’s cavity can be adjusted to hold the specimen more snugly in that region. Conversely, where excess tissue is usually present in the posterior and lateral base, the mold’s cavity can be enlarged to confer the best mean alignment accuracy. Second, in order to avoid displacement during slicing of the specimen, it would be advantageous if the mold could be adjustably tightened, holding the gland firmly in place even if the cavity was oversized. Third, in order to avoid rotational error due to torque applied by the sectioning knife, it may be preferable to obtain multiple slices simultaneously. A multi-bladed device may serve this purpose [26], if gland compression can be minimized during slicing. Lastly, Elen et. al. showed that mold-based registration can be substantially improved by constraining mold alignment with an intubated prostatic urethra [25], an approach we intend to investigate. It is likely that, with these improvements, molds may be used to facilitate accurate registration without the time and expense necessitated by ex vivo MRI.
The phantoms, molds, and registration methods described herein have many potential applications for future work. The tissue-mimicking prostate phantoms enabled a repeatable, objective measure of tissue slice quality and uniformity, and can therefore serve as a tool to evaluate tissue-processing methods and operator skill. Furthermore, the ex vivo registration methodology presented here can be employed to aid research efforts including the refinement of prostate cancer detection and the development of new prostate MRI sequences. Since ex vivo scanning within a mold enabled measurement of discrepancies between pre-and post-operative specimen geometry, this procedure could be used to improve mold design, update MRI contouring technique, assess surgeon skill, and even evaluate margins.
Since 3D printers can also produce anatomic models based on 3D ultrasound or computed tomography scans [32]–[35], it is likely that these modalities could be used to generate patient-specific molds. Once the fiducial design and ex vivo scanning procedure are amended to accommodate another imaging modality, a similar registration procedure could be performed. Patient-specific molds theoretically have the potential to facilitate highly accurate registrations between pathology and any medical imaging modality, with numerous applications including resected kidney, heart, liver, and brain.
V. Conclusions
We have demonstrated in a phantom model that patient-specific molds reduce MR-WM registration error relative to conventional sectioning. However, ex vivo specimens tended to have extraprostatic tissue in the posterior base and missing prostatic tissue from the anterior base, resulting in rotational misalignment within the molds. The best registrations were achieved only after specimen misalignment was corrected using ex vivo MRI.
3D printed molds have the potential to improve prostate MRI-pathology correlations, and this approach may be applicable to other organs and imaging modalities. The accurate registration that molds help facilitate can be used to improve prostate MRI and contouring techniques. As mold design improves, its value will continue to grow as a registration platform between MRI and pathology.
Acknowledgments
This work was supported in part by the National Cancer Institute, via award number R01CA158627 and the Prostate Cancer SPORE at the University of California Los Angeles (P50CA092131). Additional support was provided by the UCLA Integrated Diagnostics program, the Beckman Coulter Foundation, the Jean Perkins Foundation, and the Steven C. Gordon Family Foundation.
Footnotes
This paper has supplementary downloadable material available at http://mrrl.ucla.edu/pages/prostate_molds, provided by the authors. This includes stereolithography files of the patient-specific mold, along with instructions detailing mold design, manufacture, and implementation for ex vivo scanning. This material is 451 KB in size.
Contributor Information
Alan Priester, Department of Bioengineering, UCLA, CA, USA. He is now with the Department of Urology, David Geffen School of Medicine, UCLA, CA, USA aprieste@ucla.edu)..
Holden Wu, Department of Radiological Sciences, David Geffen School of Medicine, UCLA, CA, USA..
Pooria Khoshnoodi, Department of Radiological Sciences, David Geffen School of Medicine, UCLA, CA, USA..
Douglas Schneider, Department of Ecology and Evolutionary Biology, UCLA, CA, USA..
Zhaohuan Zhang, Department of Radiological Sciences, David Geffen School of Medicine, UCLA, CA, USA..
Nazanin H. Asvadi, Department of Radiological Sciences, David Geffen School of Medicine, UCLA, CA, USA.
Anthony Sisk, Department of Pathology, David Geffen School of Medicine, UCLA, CA, USA..
Steven Raman, Department of Radiological Sciences, David Geffen School of Medicine, UCLA, CA, USA..
Robert Reiter, Department of Urology, David Geffen School of Medicine, Los Angeles, CA, USA..
Warren Grundfest, Department of Bioengineering, UCLA, CA, USA..
Leonard S. Marks, Department of Urology, David Geffen School of Medicine, Los Angeles, CA, USA.
Shyam Natarajan, Department of Urology, David Geffen School of Medicine, Los Angeles, CA, USA..
References
- [1].Hegde JV et al. , “Multiparametric MRI of prostate cancer: An update on state-of-the-art techniques and their performance in detecting and localizing prostate cancer,” J. Magn. Reson. Imaging, vol. 37, no. 5, pp. 1035–1054, May 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].De Rooij M et al. , “Accuracy of multiparametric MRI for prostate cancer detection: A meta-analysis,” Am. J. Roentgenol, vol. 202, no. 2, pp. 343–351, February 2014. [DOI] [PubMed] [Google Scholar]
- [3].Priester A et al. , “MRI Underestimation of Prostate Cancer Geometry: Use of Patient-Specific Molds to Correlate Images with Whole-Mount Pathology,” J. Urol, vol. 197, no. 2, pp. 320–326, February 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gibson E et al. , “3D prostate histology image reconstruction: Quantifying the impact of tissue deformation and histology section location,” J. Pathol. Inform, vol. 4, no. 1, October 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Yamamoto H et al. , “A workflow to improve the alignment of prostate imaging with whole-mount histopathology,” Acad. Radiol, vol. 21, no. 8, pp. 1009–1019, August 2014. [DOI] [PubMed] [Google Scholar]
- [6].Noguchi M et al. , “Assessment of Morphometric Measurements of Prostate Carcinoma Volume,” Am. Cancer Soc, vol. 89, no. 5, pp. 1056–1064, September 2000. [DOI] [PubMed] [Google Scholar]
- [7].Kim Y et al. , “Expandable and rigid endorectal coils for prostate MRI: impact on prostate distortion and rigid image registration,” Med. Rhys, vol. 32, no. 12, pp. 3569–78, December 2005. [DOI] [PubMed] [Google Scholar]
- [8].Orczyk C et al. , “Imaging of prostate cancer: a platform for 3D coregistration of in-vivo MRI ex-vivo MRI and pathology,” in Proc. of SPIE, 2012, pp 83162M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Meyer C et al. , “Challenges in accurate registration of 3-D medical imaging and histopathology in primary prostate cancer,” Eur. J. Nucl Med. Mol. Imaging, vol. 40, no. 1, pp. 72–78, March 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jonmarker S et al. , “Tissue shrinkage after fixation with formalin injection of prostatectomy specimens,” Virchows Arch, vol. 449, no. 3, pp. 297–301, September 2006. [DOI] [PubMed] [Google Scholar]
- [11].Kalavagunta C et al. , “Registration of in vivo prostate MRI and pseudo-whole mount histology using Local Affine Transformations guided by Internal Structures (LATIS),” J. Magn. Reson. Imaging, vol. 41, no. 4, pp. 1104–1114, April 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Park H et al. , “Registration Methodology for Histological Sections and In Vivo Imaging of Human Prostate,” Acad. Radiol, vol. 15, no. 8, pp. 1027–1039, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Orczyk C et al. , “Preliminary experience with a novel method of three-dimensional co-registration of prostate cancer digital histology and in vivo multiparametric MRI,” Clin. Radiol, vol. 68, no. 12, pp. e652–e658, December 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Meyer CR et al. , “A methodology for registration of a histological slide and in vivo MRI volume based on optimizing mutual information,”Mol. Imaging, vol. 5, no. 1, pp. 16–23, January 2006. [PMC free article] [PubMed] [Google Scholar]
- [15].Chappelow J et al. , “Elastic registration of multimodal prostate MRI and histology via multiattribute combined mutual information,”Med Phys, vol. 38, no. 4, pp. 2005–2018, April 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Xiao G et al. , “Determining histology-MRI slice correspondences for defining MRI-based disease signatures of prostate cancer,” Comput. Med. Imaging Graph, vol. 35, no. 7–8, pp. 568–578, October 2011. [DOI] [PubMed] [Google Scholar]
- [17].Drew B et al. , “Device for sectioning prostatectomy specimens to facilitate comparison between histology and in vivo MRI,” J. Magn. Reson. Imaging, vol. 32, no. 4, pp. 992–996, October 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chen LH et al. , “Optimum slicing of radical prostatectomy specimens for correlation between histopathology and medical images,” Int. J. Comput. Assist. Radiol. Surg, vol. 5, no. 5, pp. 471–487, 2010. [DOI] [PubMed] [Google Scholar]
- [19].H. M. Reynolds et al. , “Development of a registration framework to validate MRI with histology for prostate focal therapy,” Med. Phys, vol. 42, no. 12, pp. 7078–7089, December 2015. [DOI] [PubMed] [Google Scholar]
- [20].Gibson E et al. , “Registration of prostate histology images to ex vivo MR images via strand-shaped fiducials,” J. Magn. Reson. Imaging, vol. 36, no. 6, pp. 1402–1412, December 2012. [DOI] [PubMed] [Google Scholar]
- [21].Fan X et al. , “High-resolution MRI of excised human prostate specimens acquired with 9.4T in detection and identification of cancers: Validation of a technique,” J. Magn. Reson. Imaging, vol. 34, no. 4, pp. 956–961, October 2011. [DOI] [PubMed] [Google Scholar]
- [22].Shah V et al. , “A method for correlating in vivo prostate magnetic resonance imaging and histopathology using individualized magnetic resonance -based molds,” Rev. Sci. Instrum, vol. 80, no. 10, p. 104301, October 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Trivedi H et al. , “Use of patient-specific MRI-based prostate mold for validation of multiparametric MRI in localization of prostate cancer,” Urology, vol. 79, no. 1, pp. 233–239, January 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Turkbey B et al. , “Correlation of Magnetic Resonance Imaging Tumor Volume with Histopathology,” Juro, vol. 188, no. 4, pp. 1157–1163, October 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Elen A et al. , “Validation of an Improved Patient-Specific Mold Design for Registration of In-vivo MRI and Histology of the Prostate”, in Clin. Image-BasedProc, vol. 9958, pp 36–43, Springer, Cham, 2016. [Google Scholar]
- [26].Priester A et al. , “A system for evaluating magnetic resonance imaging of prostate cancer using patient-specific 3D printed molds,” Am. J. Clin. Exp. Urol, vol. 2, no. 2, pp. 127–35, July 2014. [PMC free article] [PubMed] [Google Scholar]
- [27].Priester A et al. , “Informing Focal Therapy Margins Through MRI-Pathology Correlation,” in American Urology Association Annual Meeting, 2016, p. e313. [Google Scholar]
- [28].Kung GL et al. , “The presence of two local myocardial sheet populations confirmed by diffusion tensor MRI and histological validation,” J. Magn. Reson. Imaging, vol. 34, no. 5, pp. 1080–1091, Nov. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wu HH et al. , “A New System to Spatially Align In Vivo MRI, Ex Vivo MRI, and Whole-Mount Histopathology Slides for Integrated Prostate Cancer Research,” in 103rd RNSA, Chicago: 2017. [Google Scholar]
- [30].Fei B et al. , “A comparative study of warping and rigid body registration for the prostate and pelvic MR volumes,” Comput. Med. Imaging Graph, vol. 27, no. 4, pp. 267–281, Jul. 2003. [DOI] [PubMed] [Google Scholar]
- [31].Fei B et al. , “Three-Dimensional Rigid and Non-Rigid Image Registration for the Pelvis and Prostate,” in Handbook of Biomedical Image Analysis Volume III: Registration Models, New York: Kluwer Academic / Plenum Publishers, 2005, Chapter 3, Section 2, pp. 122–125. [Google Scholar]
- [32].Werner H et al. , “Additive manufacturing models of fetuses built from three-dimensional ultrasound, magnetic resonance imaging and computed tomography scan data,” Ultrasound Obstet. Gynecol, vol. 36, no. 3, pp. 355–361, 2010. [DOI] [PubMed] [Google Scholar]
- [33].Miller MA and Hutchins GD, “Development of anatomically realistic PET and PET/CT phantoms with rapid prototyping technology,” IEEE Nucl. Sci. Symp. Conf. Rec., vol. 6, pp. 4252–4257, 2007. [Google Scholar]
- [34].Mitsouras D et al. , “Medical 3D Printing for the Radiologist,” RadioGraphics, vol. 35, no. 7, pp. 1965–1988, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Samuel BP et al. , “Ultrasound-Derived Three-Dimensional Printing in Congenital Heart Disease,” J. Digit. Imaging, vol. 28, no. 4, pp. 459–461, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]