Abstract
Introduction.
IDH1 mutation has been identified as a early genetic event driving low grade gliomas (LGGs) and it has been proven to exerts a powerful epigenetic effect. Cells containing IDH1 mutation are refractory to epigenetical reprogramming to iPSC induced by expression of Yamanaka transcription factors, a feature that we employed to study early genetic amplifications or deletions in gliomagenesis.
Methods.
We made iPSC clones from freshly surgically resected IDH1 mutant LGGs by forced expression of Yamanaka transcription factors. We sequenced the IDH locus and analyzed the genetic composition of multiple iPSC clones by array-based comparative genomic hybridization (aCGH).
Results.
We hypothesize that the primary cell pool isolated from LGG tumor contains a heterogeneous population consisting tumor cells at various stages of tumor progression including cells with early genetic lesions if any prior to acquisition of IDH1 mutation. Because cells containing IDH1 mutation are refractory to reprogramming, we predict that iPSC clones should originate only from LGG cells without IDH1 mutation, i.e. cells prior to acquisition of IDH1 mutation. As expected, we found that none of the iPSC clones contains IDH1 mutation. Further analysis by aCGH of the iPCS clones reveals that they contain regional chromosomal amplifications which are also present in the primary LGG cells.
Conclusions.
These results indicate that there exists a subpopulation of cells harboring gene amplification but without IDH1 mutation in the LGG primary cell pool. Further analysis of TCGA LGG database demonstrates that these regional chromosomal amplifications are also present in some cases of low grade gliomas indicating they are reoccurring lesions in glioma albeit at a low frequency. Taken together, these data suggest that regional chromosomal alterations may exist prior to the acquisition of IDH mutations in at least some cases of LGGs.
Keywords: induced pluripotent stem cells, low-grade glioma, IDH1, regional chromosomal amplification, gliomagenesis
Introduction
Glial tumors are among the most fatal human tumors [1]. Monoallelic R132 (Arginine 132) mutations of IDH1 or R172 (Arginine 172) mutations of IDH2, which encode isocitrate dehydrogenases (IDH) that convert isocitrate into α-ketoglutarate (αKG), have been detected in nearly all secondary glioblastoma multiforme (GBM) and 80% of low grade gliomas (LGGs) [2–4]. The mutant IDH, which forms a heterodimer with the wild-type enzyme, acquires neomorphic activity to catalyze αKG into D-2-Hydroxyglutarate (D2HG) [5,6]. Because of the structural similarity, D2HG effectively inhibits αKG-dependent enzymes including the TET-family DNA methyl hydroxylases, Jumonji-family histone demethylases, and the hydroxylases that regulate HIF1α stability [7]. The IDH mutations are therefore considered as drivers for gliomagenesis because of the acquired capacities to alter the epigenome and respond to hypoxia.
Clonal evolution is a major force to drive tumor progression, in which cancer cells accumulate genetic lesions stepwise, generate intratumoral heterogeneity, and selectively expand the fittest clones [8]. Besides the IDH mutations, progression of LGGs involves gradual accumulation of additional genetic alterations including mutations in TP53, ATRX, TERT and chromosome 1p/19q co-deletion [4,9–13]. Analyses of multiple LGG biopsies collected from the same patients indicated sequential acquisition of genetic alterations, in which IDH mutations occur invariably prior to the other aforementioned mutations/chromosomal deletions [14]. However, whether there are LGG-associated genetic lesions that occur even prior to the IDH mutations remains unknown. One approach to gain insight is to make induced pluripotent stem cell (iPSC) clones from pooled primary tumor cells and then examine their genetic composition including amplifications or deletions by aCGH. The iPSC clones collectively should reflect the primary tumor cell pool if each cell has similar potential to be induced to iPSCs. Interestingly, we observed that cells containing IDH1 mutation are refractory to reprogramming by forced expression of Yamanaka transcription factors Oct4, Sox2, Klf4, and c-Myc. Therefore, iPSC clones originated from primary cell pool of IDH1 mutant glioma should exclude all the cells harboring IDH mutations and thus amplify cells containing early genetic lesions.
In this study, we generated iPSC (LGG-iPSC) clones from primary cells from freshly surgically resected IDH1 mutant LGGs. Because cells with IDH mutation is blocked from forming iPCS, we hypothesized that analyzing the genome of LGG-iPSCs would reveal early genetic changes in gliomagenesis. We established multiple iPSC clones from IDH1 mutation-bearing primary LGGs. As expected, we found that none of the iPSC clones carried the IDH1 mutations suggesting that tumor cells cannot be reprogrammed as long as they contain IDH1 mutation. Importantly, we found that the LGG-iPSCs contained regional chromosomal amplifications and these amplifications are also present in the primary LGG cells. These data suggest that the LGG-iPSCs are originated from tumor cells not yet acquiring the IDH1 mutations. Furthermore, the observed chromosomal alterations were detected in a fraction of LGGs in the Cancer Genome Atlas (TCGA) database. Taken together, by reprogramming freshly resected LGG cells, we demonstrated that IDH1 mutation-bearing LGG cells are resistant to become iPSCs and that certain regional chromosomal alterations exist prior to the acquisition of IDH mutations at least in a fraction of LGGs. The functional role of the amplification await further study.
Materials and Methods
Preparation and culture of primary low-grade glioma cells
De-identified LGG samples were acquired in accordance with the policies of Institutional Review Board (IRB) of University of Alabama at Birmingham. Freshly resected tumors were minced with #11 scalpel blades and minced tumors were disaggregated via gentleMacs Dissociator per standard tumor protocol supplied by Tumor Dissociation Kit (Miltenyi Biotec, Auburn, CA). Collected cells were washed twice with serumless DMEM/F12 (300×G, 7 minutes, 4°C), counted in the presence of 0.04% trypan blue to enumerate viable cells. Dissociated LGG cells were cultured in cell culture plates coated with poly-L-ornithine (Sigma-Aldrich) and laminin (EMD-Millipore) in LGG medium (Neurobasal-A medium containing 1% N-2, 2% vitamin A-free B-27, 2 mM L-Glutamine [Thermo Fisher], 20 ng/ml basic FGF [Stemgent], and 20 ng/ml human EGF [Cell Signaling]) as described [15,16]. Medium was exchanged as needed, and cells grew adherently to the bottom of flasks. All cell cultures were maintained at 37°C, 20.8% O2, and 5% CO2.
Immunohistochemistry and DNA FISH
Immunohistochemistry was performed as described [17]. In brief, paraffin sections were deparaffinized in xylene and rehydrated in decreasing concentration of ethanol solutions. Antigen is retrieved by incubation in Target Retrieval Solution (Agilent) at 95 °C for 20 min and endogenous peroxidase and alkaline phosphatase activity were quenched using BLOXALL blocking solution (Vector Laboratories) at RT for 10 min. Slides were blocked by with horse serum, incubated with the IDH1 R132H antibody (DIA-H09, Dianova) at 4°C overnight, and secondary antibody at RT for 30 min. FISH was performed as per manufacturer instructions. Chromosome arms 1p and 19q status was determined by Vysis LSI 1p36/1q25 and LSI 19q13/19p13 Dual Color probe (Abbott Molecular).
Lentiviral preparation and iPSC derivation
STEMCCA lentivirus, which express all reprogramming factors from a polycistronic transcript [18], was prepared as described [19]. Primary LGG cells were transduced with the STEMCCA lentivirus at an MOI of 3 as described [20]. Three days after transduction, the LGG cells were reseeded onto cell culture plates-coated with hESC-qualified Geltrex basement membrane matrix (Thermo Fisher) at a density of 1−3×105 cells/cm2 in LGG medium. The cells were then cultured in E7 medium (E8 medium without TGF-β) from the following day for two weeks and in E8 medium for one to two weeks as described [20–22]. Independent clones of iPSCs were manually picked into Geltrex-coated 12-well tissue culture plates and maintained in E8 medium as described [23,24].
DNA isolation and Sanger sequencing
Genomic DNA of primary LGG cells and LGG-iPSCs were prepared using the Quick-gDNA miniPrep kit (Zymo Research). DNA fragments containing the R132 mutation of IDH1 and R172 mutation of IDH2 were PCR amplified. Primers used include IDH1f (5’ - AAT GAG CTC TAT ATG CCA TCA CTG – 3’), IDH1r (5’ - TTC ATA CCT TGC TTA ATG GGT GT – 3’), IDH2f (5’ - TTG TTG CTT GGG GTT CAA AT – 3’), and IDH2r (5’ - AAG GAA AGC CAC GAG ACA GA – 3’). PCR products were purified using the DNA Clean & Concentrator-5 kit (Zymo Research) and sequenced using the forward primer. Chromatograms were viewed using the Sequence Scanner v1.0 software (Applied Biosystems).
RNA isolation and quantitative RT-PCR analysis
Total RNA was isolated with the DirectZol RNA Kit (Zymo Research) and cDNA were synthesized by the Verso cDNA Synthesis Kit (Thermo Fisher Scientific). Quantitative PCR was performed using 2x Absoluate Blue Q-PCR Master Mix (Thermo Fisher Scientific) on a ViiA7 real-time PCR system (Thermo Fisher Scientific). The primers used to quantify REST include: REST-F 5’ – CAT ACA GGA GAA CGC CCA TAT AA – 3’; REST-R 5’ – ATG GCT TCT CAC CTG AAT GAG – 3’; bActin-F 5’ – TCA GAA GGA TTC CTA TGT GGG CGA – 3’; bActin-R 5’ -TTT CTC CAT GTC GTC CCA GTT GGT – 3’.
Stem cell marker analysis
For alkaline phosphatase staining, iPSCs were fixed in 4% paraformaldehyde (PFA) and stained using the Leukocyte Alkaline Phosphatase Kit (Sigma-Aldrich) per manufacturer’s instruction. For immunostaining, iPSCs were fixed in 4% PFA, blocked in Protein Block (Agilent), stained with primary antibodies at 4 °C overnight and secondary antibodies at room temperature for 1 hour as described [15,25]. Primary antibodies used were OCT4 (sc-5279, Santa Cruz Biotech), SOX2 (561469, BD Biosciences), and NANOG (AF1977, R&D Systems). Images were acquired by a Nikon Ti-S microscope and processed by the Adobe Photoshop CS6 software. For flow cytometry analysis, iPSCs were trypsinized and stained on ice with Tra-1–60 antibody (sc-21705, Santa Cruz Biotech) for 1 hour and FITC-conjugated anti-mouse-IgM secondary antibody (555988, BD Biosciences) for 30 min. Cells were analyzed on a BD Fortessa flow cytometer and data were analyzed by the Flowjo V10 software as described [20,26].
Teratoma formation assay
All experiments involving lab animals were conducted in accordance with guidelines from the University of Alabama at Birmingham (UAB) and National Institute of Health. Teratoma formation assay was performed as described [20,27]. In brief, 2−5×106 iPSCs were suspended in 100 μl of DMEM/F12 medium containing 30% of human ESC-qualified Geltrex membrane matrix (Thermo Fisher) and injected subcutaneously into 4–6 week-old NOD scid gamma (NSG) mice (005557, Jackson Laboratory). Teratomas were harvested, fixed in 10% Formalin (Sigma-Aldrich), and processed by the Comparative Pathology Laboratory at UAB.
Array-based Comparative Genomic Hybridization (aCGH) analysis
Frozen pellets or genomic DNA of primary LGG cells or iPSCs were submitted to Cell Line Genetics (Madison, WI) for aCGH analysis using the SurePrint G3 60K Human Genome CGH microarray (Agilent). Data were analyzed by the Agilent CytoGenomics software. Only amplifications and deletions not observed in the healthy population, as determined by referencing against the Database of Genomic Variants (DGV) [28], were considered as chromosomal aberrance.
TCGA data mining
TCGA low-grade glioma database was queried using the cBioportal tools [29,30]. Genetic regions of interest were analyzed for copy number variation in low-grade gliomas with and without IDH1 mutations. Tumors with regional chromosomal gain or loss were presented as a percentage of the total tumor samples.
Results
Characterization of primary LGGs
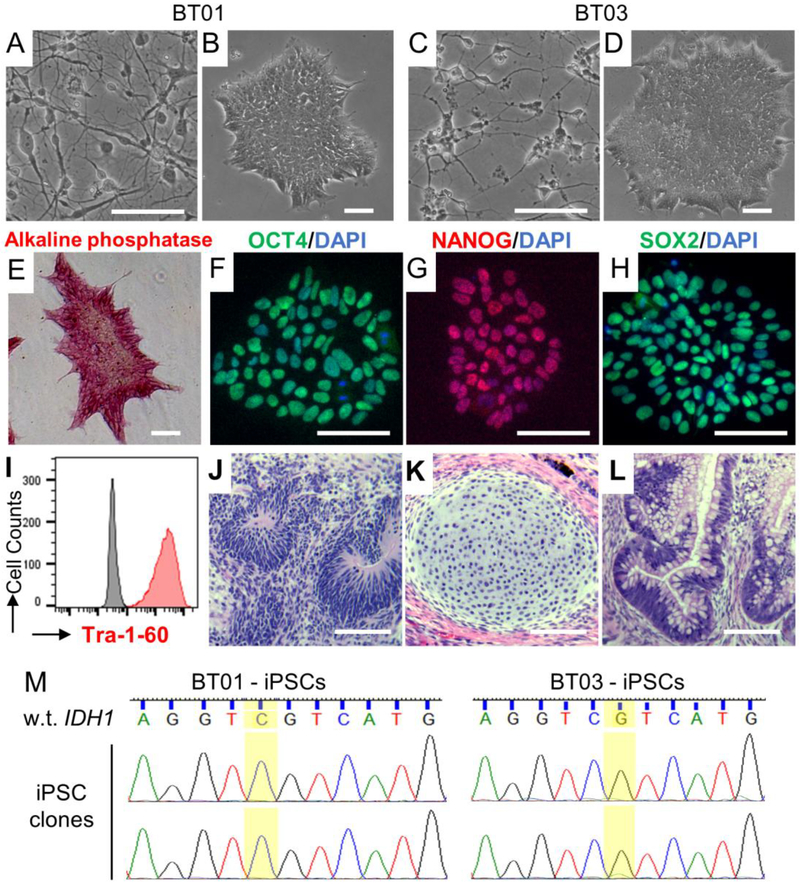
To investigate the early genetic alteration of LGGs, we acquired two samples of freshly surgically resected LGGs (BT01 and BT03). Both LGGs were from the frontal lobe in male patients (Fig. 1A–D). Histochemical analysis indicated that BT01 is an astrocytoma that is not reactive to the antibody specifically recognizing the IDH1 R132H mutation (Fig. 1E and 1F), while BT03 is an oligodendroglioma strongly expressing the IDH1 R132H mutation (Fig. 1G and 1H). Fluorescence in situ hybridization (FISH) revealed reduced copy number of chromosomes 1p and 19q in BT03 cells (Fig. 1I–K) but not in BT01 cells (data not shown), which demonstrated 1p/19q co-deletion in BT03 and is consistent with the molecular feature that 1p/19q co-deletion occurs in oligodendrogliomas [4,10,11,13]. Sequencing of the IDH1 gene demonstrated that BT01 carries an IDH1 R132C mutation while BT03 carries an IDH1 R132H mutation (Fig. 1L). Sequencing of IDH2 gene confirmed that both BT01 and BT03 have the wild type IDH2, which is consistent with the notion that mutations of IDH1 and IDH2 are mutually exclusive in LGGs [2–4].
FIG. 1.
Characterization of primary LGGs. (A-D) MRI images FLAIR (A and C) and T1 with contrast (B and D) of BT01 and BT03. (E-H) Analysis of primary tumor tissues. (E, G) H&E stain and (F, H) immunohistochemistry of the IDH1 R132H mutant of paraffin-embedded BT01 and BT03 tissue, respectively. (I-K) Fluorescence in situ hybridization (FISH) of chromosomes 1p and 19q of primary BT03 cells. (I, J) Representative FISH images of chromosome 1 and 19. Red, 1p and 19q, respectively; green, 1q and 19p (control), respectively. (K) Quantification of FISH analysis of 75 cells for 1p and 80 cells for 19q. (L) Sequencing of genomic DNA identified monoallelic IDH1 R132C and R132H mutations in BT01 and BT03, respectively. Shown are sequences of wild-type (w.t.) and mutant (mut.) IDH1 sequences and the chromatograms of BT01 (left) and BT03 (right) primary cells. (M) Sequencing of genomic DNA confirmed that BT01 and BT03 have wild type IDH2. Shown are wild-type IDH2 DNA sequences encoding the R172 residue (highlighted) and the chromatograms of BT01 (left) and BT03 (right) primary cells.
Generation of iPSC clones from primary LGG cells
To generate iPSCs from primary LGG cells, we transduced primary BT01 and BT03 cells with lentivirus expressing the reprogramming factors OCT4, SOX2, KLF4, and c-MYC [23,31]. Multiple clones of LGG-iPSCs, which have typical iPSC colony morphology and growth property, were isolated from both BT01 and BT03 cells (Fig. 2A–D). We confirmed that the LGG-iPSC clones are fully reprogrammed because they expressed the pluripotency-associated markers alkaline phosphatase, OCT4, NANOG, SOX2, and Tra-1–60 (Fig. 2E–I) and can differentiate into tissues from all the three embryonic germ layers (Fig. 2J–L). Sequencing analysis of the genomic DNA of at least 15 LGG-iPSC clones from each tumor revealed that none of the clones contained the IDH1 or IDH2 mutations (Fig. 2M and Fig. S1). These data indicate that the IDH1 mutations inhibit somatic cell reprogramming.
FIG. 2.
Generation of iPSCs from primary LGG cells. (A, C) Bright field images of primary BT01 and BT03 cells. (B, D) Images of representative iPSC colonies derived from BT01 and BT03 cells. (E) LGG-iPSCs express alkaline phosphatase. (F-H) Immunostaining demonstrated that LGG-iPSCs express pluripotency-associated transcription factors (F) OCT4, (G) NANOG, and (H) SOX2. (I) Flow cytometry analysis confirmed LGG-iPSCs expressing the pluripotency-specific surface marker Tra-1–60. (J-L) Teratoma formation assay demonstrated LGG-iPSCs can differentiated into (J) ectoderm (neuroepithelium), (K) mesoderm (cartilage), and (L) endoderm (glandular structure). Scale bars, 100 μm. (M) Representative sequencing results of the BT01- and BT03-iPSC clones. More than 15 independent clones of each LGG samples were sequenced. None carried the IDH1 mutations.
To further test whether IDH1 mutation inhibits somatic cell reprogramming, we attempted to generate iPSCs from the brain tumor cell line BT142 (ATCC #ACS-1018), which was originally established from a grade III oligoastrocytoma carrying the monoallelic IDH1 R132H mutation but subsequently became homozygous of the IDH1 mutation because of loss of heterozygosity [32]. Although we can propagate BT142 cells in culture, we were unable to make iPSCs (data not shown), suggesting that the IDH1 mutation and/or other genetic aberrations in BT142 cells are incompatible with iPSCs. Because the IDH1 mutations acquire the neomorphic activity to produce D2HG, which inhibits all αKG-dependent epigenetic modifying enzymes, we then tested whether human PSCs could tolerate D2HG. Our data discovered a deterioration of colonies of human H9 ESCs or BJ-iPSCs after a 24-hour treatment of D2HG (Fig. S2), which further supports the notion that iPSCs cannot tolerate D2HG and the IDH mutations.
To rule out the possibility that we may selectively eliminate LGG cells carrying the IDH1 mutations during tumor dissociation or cell culture, we compared IDH1 mutation status in freshly dissociated primary LGG cells and the same cells that have been cultured. As expected, we were unable to expand the primary LGG cells because in vitro culture these cells remains as a major challenge to the entire field. However, we found that the proportion of IDH1 mutation-carrying cells, as judged by the relative peak heights on the DNA sequencing chromatograms, remained relatively stable over a 4-month period for the BT01 cells or a 14-day period for the BT03 cells (Fig. S3). Therefore, our observation of none of the LGG-iPSCs carrying the IDH1 mutations is unlikely explained by selective loss of mutation-bearing cells during sample processing and the subsequent culture.
Because REST is known as a transcriptional regulator that plays critical roles in stem cells, neural differentiation, and glioblastoma aggressiveness [33–37], we examined how REST is expressed in LGG-iPSCs. Quantitative RT-PCR data confirmed that REST is expressed at similar levels in LGG-iPSCs as in human H9 ESCs and in the skin fibroblast-derived BJ-iPSCs (Fig. S4). These data suggest that REST expression in iPSCs is not affected by their somatic tissue origins. These data also support the notion that once reprogrammed iPSCs are indistinguishable from ESCs [38].
LGG-iPSCs contain regional chromosomal alterations
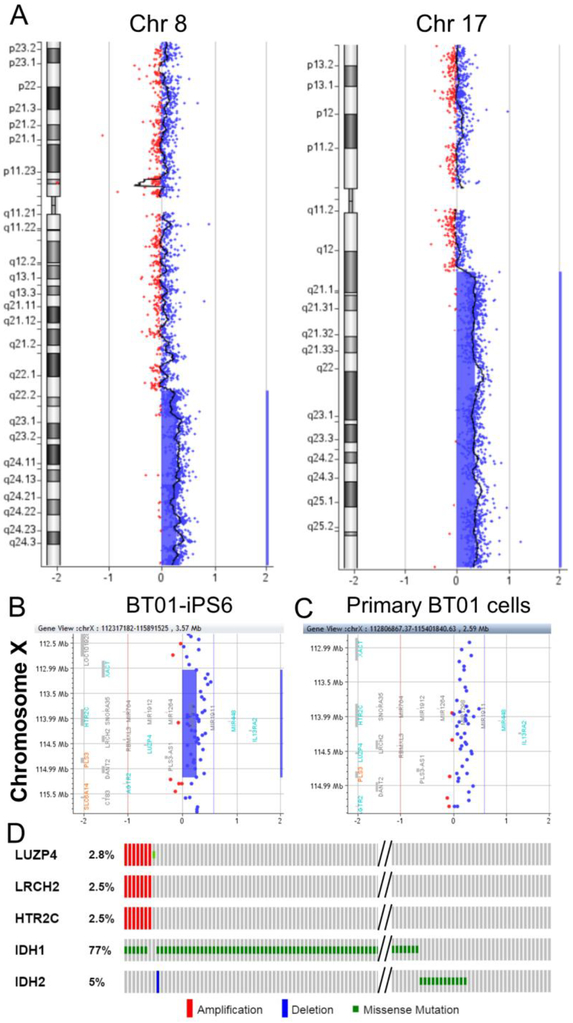
Because primary tumors likely consist of normal brain cells and a mixture of tumor cells at various stages of tumor progression, the LGG-iPSCs could originate from normal cells or tumor cells prior to acquiring the IDH1 mutations. To distinguish these possibilities and to investigate whether the LGG-iPSCs carry other genomic abnormalities, we performed array-based comparative genomic hybridization (aCGH) analysis on primary LGG cells and the derived LGG-iPSCs. The BT01 primary cells contained large amplifications on chromosomes 8q and 17q (Fig. 3A). An 8q gain is a common chromosomal abnormality reported in IDH1 mutated gliomas [39]. However, these large chromosomal amplifications were not detected in BT01 LGG-iPSC clones. One of the BT01-iPSC clones, BT01-iPS16, contains no chromosomal abnormality (data not shown), suggesting that the BT01-iPS16 cells originated from a normal cell in the tumor mass. The other clones contained a 2.1 Mb-amplification on chromosome Xq23 and a representative clone BT01-iPS6 is shown (Fig. 3B). Importantly, we found that the observed Xq23 amplification is present in the primary BT01 cells (Fig. 3C) but absent from over 22,300 genomes of healthy individuals, as determined by referencing the Database of Genomic Variants (DGV) [28]. To examine whether the Xq23 amplification is recurrent in LGGs, we analyzed TCGA LGG database [29,30]. We found that 2.5% of the LGGs (7 of 283) showed the Xq23 amplification, as determined by co-amplification of genes within this region such as LUZP4, LRCH2, and HTR2C (Fig. 3D and Table 1). Therefore, frequency of the Xq23 amplification in LGGs is significantly higher than that in healthy genomes (p = 4.5×10−14, Fisher’s exact test). Furthermore, among the subset LGGs carrying the Xq23 amplification, most (6 of 7) also have the IDH1 mutation (Fig. 3D). Taken together, these data demonstrated that the BT01-iPSCs originated either from normal cell or tumor cells carrying the Xq23 amplification but not the IDH1 mutation. Because tumor cells gradually accumulate multiple genetic lesions during tumorigenesis, these data suggest that the Xq23 amplification occurred before the IDH1 mutation in BT01.
FIG. 3.
aCGH analysis of primary BT01 cells and the derived iPSCs. (A) aCGH analysis of primary BT01 cells demonstrated amplification of chromosome 8q and 17q. (B-C) aCGH analysis revealed that (B) BT01-iPS6 cells contained a 2.1 Mb-amplification on chromosome Xq23, which was also detected in (C) primary BT01 cells. Red, deletion; blue, amplification. (D) Analysis of TCGA low-grade glioma database revealed that 2.5% of LGGs (7 of 283) were amplified at the 2.1 Mb region and most of them (6 of 7) also have the IDH1 mutation. LUZP4, LRCH2, and HTR2C are genes within the amplified region on chromosome Xq23.
Table 1.
All genes that are within the amplified chromosomal regions
| LGG sample | Amplified chromosomal region | Genes |
|---|---|---|
| BT01 |
Xq23 Start (bp) - 113,032,051 Stop (bp) - 115,176,657 |
XACT, HTR2C, SNORA35, MIR764, MIR1912, MIR1264, MIR1298, MIR1911, MIR448, IL13RA2, LRCH2, RBMXL3, LUZP4, PLS3-AS1, PLS3, DANT2 |
| BT03 |
7q31 Start (bp) - 121,187,838 Stop (bp) - 122,635,001 |
PTPRZ1, AASS, FEZF1, FEZF1-AS1, CADPS2,RNF133, RNF148, TAS2R16 |
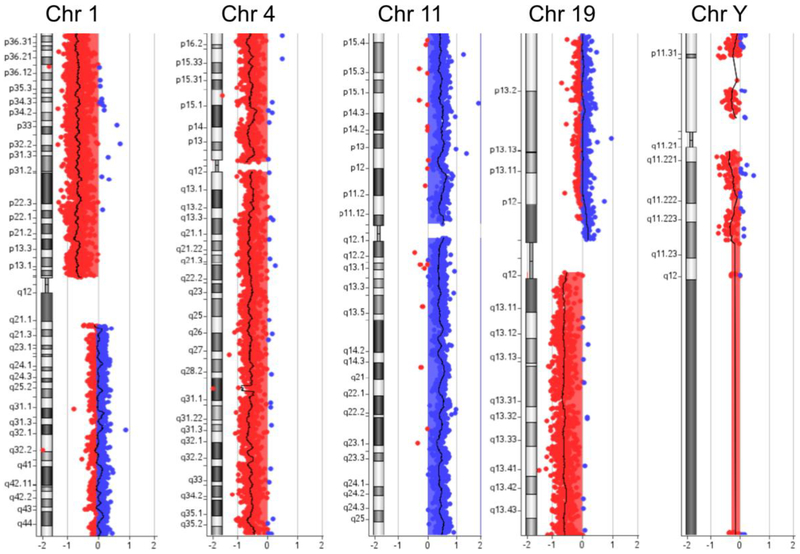
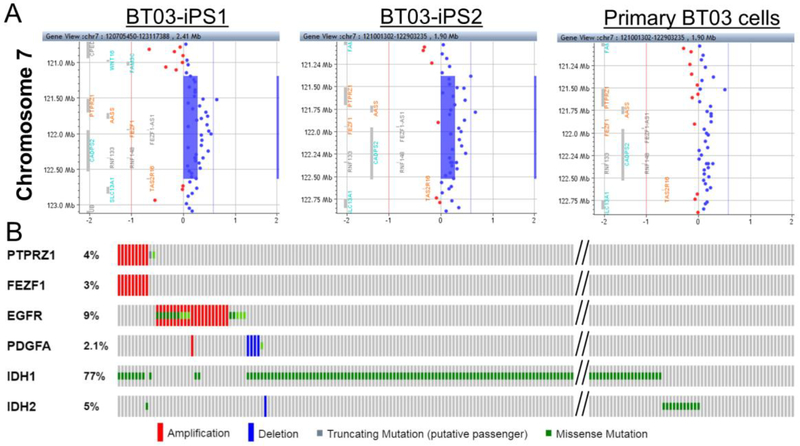
Similarly, aCGH analysis of the primary BT03 cells revealed amplification of chromosome 11 and deletion of chromosome 1p, 4, 19q, and Y (Fig. 4), which was consistent with the FISH data showing 1p/19q co-deletion of this tumor (Fig. 1I–K). However, analysis of randomly selected clones of iPSCs revealed that none of these major chromosomal aberrations were present in iPSCs. The aCGH data demonstrated that BT03-iPSC clones amplified a 1.4 Mb region on chromosome 7q31, which is also present in the primary BT03 cells (Fig. 5A) but absent from the over 22,300 genomes of healthy individuals in DGV [28]. A search of TCGA LGG database revealed that 3.2% of LGGs (9 of 283) showed the 7q31 amplification, as demonstrated by co-amplification of genes within this region such as PTPRZ1 and FEZF1 (Fig. 5B and Table 1). Similarly, frequency of the 7q31 amplification in LGGs is significantly higher than that in healthy genomes (p = 6.7×10−18, Fisher’s exact test). Among the nine LGGs carrying the 7q31 amplification, all carried mutations in IDH1 (8 of 9) or IDH2 (1 of 9) (Fig. 5B). Furthermore, genes on chromosome 7 such as PDGFA and EGFR that are known drivers of primary GBM were not amplified in this subset of LGG samples (Fig. 5B). Because the 7q31 amplification presents in both iPSCs and the primary tumor cells, these data indicated that the BT03-iPSC cells originated from tumor cells. Because these cells do not carry the IDH1 mutations, the 7q31 amplification in BT03 is mostly likely an early genetic amplification exist before the IDH1 mutations. Taken together, we conclude that the regional amplifications on chromosome Xq23 (BT01) and 7q31 (BT03) are early genetic lesions and is likely acquired prior to the IDH1 mutations (Fig. 6). These lesions could represent predisposing conditions for IDH1 mutations and/or be cooperating with the IDH1 mutations in gliomagenesis.
FIG. 4.
aCGH analysis of primary BT03 cells revealed amplification of chromosome 11 and deletion of chromosome 1p, 4, 19q, and Y. The results are consistent with a diagnosis of oligodendroglioma. Red, deletion; Blue, amplification.
FIG. 5.
aCGH analysis of BT03-iPSCs. (A) aCGH analysis revealed BT03-iPS1 (left) and BT03-iPS2 cells (center) shared a common 1.4 Mb-amplification on chromosome 7, which was also detected in the primary BT03 cells (right). (B) Analysis of the TCGA data revealed that 3% of LGGs (9 of 283) have amplification at the 1.4 Mb region on chromosome 7 and all of them also have the IDH1 (8 of 9) or IDH2 (1 of 9) mutation. PTPRZ1 and FEZF1 are genes within the amplified region. Note that genes on chromosome 7 that known to drive brain tumors, such as PDGFA and EGFR, were not amplified in this subset of tumor cells.
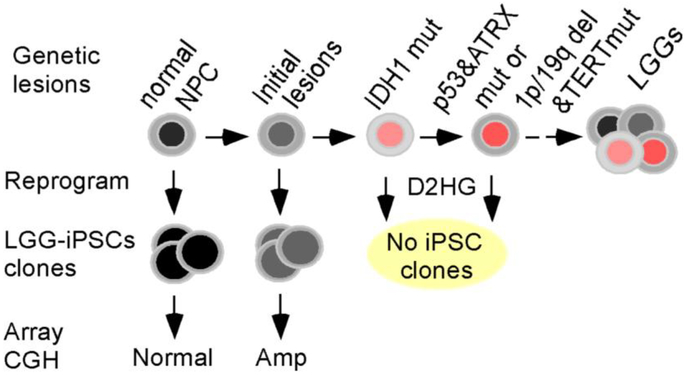
FIG. 6.
A model illustrating sequential acquisition of genetic lesions. Previous studies have established that IDH1 mutations are the key drivers for gliomagenesis and precede other major genetic alterations such mutations in p53, ATRX, and TERT and co-deletion of chromosome 1p/19q. We discovered the presence of regional chromosomal alterations in at least a subset of LGGs before acquiring the IDH1 mutations, which may represent predisposing conditions for IDH1 mutations and/or be cooperating with the IDH1 mutations in gliomagenesis.
Discussion
Clonal evolution is a major force to drive tumor progression and an improved understanding to the process is critical to novel target identification and cancer treatment. Cancer cell-derived iPSCs, which involves epigenetic reprogramming of cancer cells at various stages of cancer progression, have recently been used to deconvolute the phylogenetic history of leukemia and to test stage-specific responses to cancer therapies [40]. Here, we applied a similar method to LGG research by reprograming primary LGG cells. Although IDH1 mutations can be clearly detected in primary BT01 and BT03 cells (Fig. 1A), none of the derived iPSC clones carried the mutations. Because reprogramming of iPSCs involves global erasure of epigenetic marks of the differentiated cells and establishment of the marks of pluripotent cells [41], it is likely that the IDH mutations, which produce high levels of D2HG and inhibit epigenetic remodeling enzymes such as the TET-family DNA demethylases and Jumonji-family histone demethylases, completely block the transition of brain tumor cells into iPSCs. Furthermore, it is likely that pluripotent cells rely on the activities of these αKG-dependent enzymes to maintain its status because treatment with D2HG caused rapid death of PSCs (Figure S2). Consistent with this notion, human patients carrying the neomorphic IDH1 R132 germline mutations have never been reported, suggesting that the IDH1 mutations are detrimental to early embryonic cells.
The monoallelic mutations of IDH1 or IDH2 have been identified in most LGGs and secondary GBMs [2–4]. Comparing to the IDH2 mutations, the IDH1 mutations (e.g., R132H and R132C) are more prevalent and have been identified in over 90% of all patients carrying IDH mutations [2–4]. Furthermore, mutations in IDH1 or IDH2 are mutually exclusive in patients [2–4], suggesting that they play similar roles in driving gliomagenesis. Our data agree with the previous findings because BT01 and BT03 carry the IDH1 mutations but they have wild type IDH2 (Fig. 1L and 1M).
Besides the IDH mutations, additional genetic alterations have been identified to be closely associated with subtypes of LGGs. The recurrent genetic changes include mutations in TP53 and ATRX in over 80% of IDH mutation-bearing astrocytomas, and TERT promoter mutation and chromosome 1p/19q co-deletion in over 80% of oligodendrogliomas [4,11]. The IDH1 mutations are considered as an early event acquired prior to these other major genetic changes [14]. Our data agree with this conclusion because the LGG-iPSCs contained neither the IDH1 mutations nor the major chromosomal gains/losses detected in primary LGG cells (Fig. 3A and 4). It has been reported that the IDH1 mutations could be deleted in later stage of gliomagenesis [42,43], which raises the possibility that the LGG-iPSCs may originate from these cells. However, we argue this is unlikely because we did not detect loss or amplification of large regions of multiple chromosomes presented in the primary LGG cells (Fig. 3 and 5). Instead, we detected small regional chromosomal amplifications in BT01 and BT03-derived iPSCs (Fig. 3B–C and 5). Because these regional amplifications were found in the primary tumor cells, we conclude the LGG-iPSCs must originate from tumor cells at early stage of gliomagenesis before acquisition of the IDH1 mutations.
Several lines of evidences demonstrated that the observed chromosomal aberration is unlikely resulted from lentiviral transfection or other experimental artifact. First, we found no chromosomal aberration in BT01-iPS16 (data not shown), which was established by identical experimental conditions as all other iPS cell lines. Secondly, we found the primary LGG cells also contain amplifications in the same chromosomal regions (Fig. 3B–C and 5A), demonstrating that the observed chromosomal aberration is not generated by lentiviral transduction or other experimental manipulation. Third, amplifications in the same chromosomal regions were also identified in a fraction of TCGA LGG samples (Fig. 3D and Fig. 5B), suggesting that these amplifications are recurrent events and functionally important to LGGs. Consistent with this notion, several recent studies implicated potential roles of the genes within the amplified regions, such as FEZF1 [44], PTPRZ1 [45,46], AASS and TAS2R16 [47], in gliomagenesis (Table 1). Therefore, more research would be necessary to fully appreciate the functional consequence of these regional chromosomal amplifications in gliomagenesis.
Previous genome-wide association studies (GWAS) have identified rare inherited variants on chromosome 8q24 and 11q23 that may increase the risk of IDH-mutant gliomas [48,49]. In a similar way, the regional chromosomal amplifications of Xq23 and 7q31 may represent early genetic aberrations, which serve as predisposing conditions for gliomagenesis. It is also possible that these regional chromosomal changes represent early genetic events in gliomagenesis that can cooperate with IDH1 mutations. Indeed, there are examples of chromosomal alteration proceeding acquisition of driver mutations. In primary glioblastoma (GBM), it has been reported that the earliest genetic alterations are polysomy of chromosome 7 and monosomy of chromosome 10, which occur in 86% and 90% of GBM, respectively [50]. Although this is clearly different from the regional chromosomal amplifications reported in this study, which were only observed in a small fraction of LGGs (Fig. 3D and 5B), it is possible that many different genetic lesions may exist to facilitate the acquisition of IDH1 mutations and/or cooperate with the IDH mutations to initiate LGGs. The method established in current study enables identification of genetic lesions that occur prior to acquisition of the IDH mutations in gliomagenesis, which would provide novel insights into conditions leading to the initiation of LGGs.
Supplementary Material
Acknowledgements
This project is supported by the UAB CCC Neuro-oncology Research Acceleration Fund. X.H. and R.Z. are supported by the NIH grant R21NS106430. X.H. is supported by the NIH grant 1R01NS095626. R.Z. is supported by is supported by Institutional Research Grant number 128599-IRG-15–56-IRG from the American Cancer Society, Faculty Development Grant Program award, and UAB CFRC Pilot & Feasibility Grant (ROWE15R0). K.K. is supported by NIH (5R00HL093212–04), TriStem-Star Foundation (2013–049), Louis V. Gerstner, Jr. Young Investigators awards, Geoffrey Beene Junior Chair Award, Sidney Kimmel Scholar Award, Alfred W. Bressler Scholars Endowment Fund, and MSKCC Society Fund.
Footnotes
Conflict of interest statement
The authors indicate no potential conflicts of interest.
References
- 1.Ostrom QT, Gittleman H, Liao P, Rouse C, Chen Y, Dowling J, Wolinsky Y, Kruchko C and Barnholtz-Sloan J. (2014). CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2007–2011. Neuro Oncol 16 Suppl 4:iv1–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B and Bigner DD. (2009). IDH1 and IDH2 mutations in gliomas. N Engl J Med 360:765–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A, Felsberg J, Wolter M, Mawrin C, Wick W, Weller M, Herold-Mende C, Unterberg A, Jeuken JW, Wesseling P, Reifenberger G and von Deimling A. (2009). Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol 118:469–74. [DOI] [PubMed] [Google Scholar]
- 4.Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A, Felsberg J, Wolter M, Mawrin C, Wick W, Weller M, Herold-Mende C, Unterberg A, Jeuken JW, Wesseling P, Reifenberger G and von Deimling A. (2009). Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol 118:469–74. [DOI] [PubMed] [Google Scholar]
- 5.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG and Su SM. (2009). Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462:739–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pietrak B, Zhao H, Qi H, Quinn C, Gao E, Boyer JG, Concha N, Brown K, Duraiswami C, Wooster R, Sweitzer S and Schwartz B. (2011). A tale of two subunits: how the neomorphic R132H IDH1 mutation enhances production of alphaHG. Biochemistry 50:4804–12. [DOI] [PubMed] [Google Scholar]
- 7.Waitkus MS, Diplas BH and Yan H. (2016). Isocitrate dehydrogenase mutations in gliomas. Neuro Oncol 18:16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greaves M and Maley CC. (2012). Clonal evolution in cancer. Nature 481:306–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, Verhaak RG, Hoadley KA, Hayes DN, Perou CM, Schmidt HK, Ding L, Wilson RK, Van Den Berg D, Shen H, Bengtsson H, Neuvial P, Cope LM, Buckley J, Herman JG, Baylin SB, Laird PW, Aldape K and N Cancer Genome Atlas Research. (2010). Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17:510–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, Louis DN, Rozenblatt-Rosen O, Suva ML, Regev A and Bernstein BE. (2014). Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344:1396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA Jr., Friedman AH, Friedman H, Gallia GL, Giovanella BC, Grollman AP, He TC, He Y, Hruban RH, Jallo GI, Mandahl N, Meeker AK, Mertens F, Netto GJ, Rasheed BA, Riggins GJ, Rosenquist TA, Schiffman M, Shih Ie M, Theodorescu D, Torbenson MS, Velculescu VE, Wang TL, Wentzensen N, Wood LD, Zhang M, McLendon RE, Bigner DD, Kinzler KW, Vogelstein B, Papadopoulos N and Yan H. (2013). TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A 110:6021–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES and Getz G. (2014). Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Venteicher AS, Tirosh I, Hebert C, Yizhak K, Neftel C, Filbin MG, Hovestadt V, Escalante LE, Shaw ML, Rodman C, Gillespie SM, Dionne D, Luo CC, Ravichandran H, Mylvaganam R, Mount C, Onozato ML, Nahed BV, Wakimoto H, Curry WT, Iafrate AJ, Rivera MN, Frosch MP, Golub TR, Brastianos PK, Getz G, Patel AP, Monje M, Cahill DP, Rozenblatt-Rosen O, Louis DN, Bernstein BE, Regev A and Suva ML. (2017). Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe T, Nobusawa S, Kleihues P and Ohgaki H. (2009). IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol 174:1149–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Z, Zhang C, Khodadadi-Jamayran A, Dang L, Han X, Kim K, Li H and Zhao R. (2017). Canonical microRNAs Enable Differentiation, Protect Against DNA Damage, and Promote Cholesterol Biosynthesis in Neural Stem Cells. Stem Cells Dev 26:177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han X, Zhang W, Yang X, Wheeler CG, Langford CP, Wu L, Filippova N, Friedman GK, Ding Q, Fathallah-Shaykh HM, Gillespie GY and Nabors LB. (2014). The role of Src family kinases in growth and migration of glioma stem cells. Int J Oncol 45:302–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rana U,Liu Z, Kumar SN, Zhao B, Hu W, Bordas M, Cossette S, Szabo S, Foeckler J, Weiler H, Chrzanowska-Wodnicka M, Holtz ML, Misra RP, Salato V, North PE, Ramchandran R and Miao QR. (2016). Nogo-B receptor deficiency causes cerebral vasculature defects during embryonic development in mice. Dev Biol 410:190–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Somers A, Jean JC, Sommer CA, Omari A, Ford CC, Mills JA, Ying L, Sommer AG, Jean JM, Smith BW, Lafyatis R, Demierre MF, Weiss DJ, French DL, Gadue P, Murphy GJ, Mostoslavsky G and Kotton DN. (2010). Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells 28:1728–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao R, Deibler RW, Lerou PH, Ballabeni A, Heffner GC, Cahan P, Unternaehrer JJ, Kirschner MW and Daley GQ. (2014). A nontranscriptional role for Oct4 in the regulation of mitotic entry. Proc Natl Acad Sci U S A 111:15768–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Z and Zhao R. (2016). Generation of HEXA-deficient hiPSCs from fibroblasts of a Tay-Sachs disease patient. Stem Cell Res 17:289–291. [DOI] [PubMed] [Google Scholar]
- 21.Beers J, Gulbranson DR, George N, Siniscalchi LI, Jones J, Thomson JA and Chen G. (2012). Passaging and colony expansion of human pluripotent stem cells by enzyme-free dissociation in chemically defined culture conditions. Nat Protoc 7:2029–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park IH, Lerou PH, Zhao R, Huo H and Daley GQ. (2008). Generation of human-induced pluripotent stem cells. Nat Protoc 3:1180–6. [DOI] [PubMed] [Google Scholar]
- 23.Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, PH Lerou, Lensch MW and Daley GQ. (2008). Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451:141–6. [DOI] [PubMed] [Google Scholar]
- 24.Kim K, Zhao R, Doi A, Ng K, Unternaehrer J, Cahan P, Huo H, Loh YH, Aryee MJ, Lensch MW, Li H, Collins JJ, Feinberg AP and Daley GQ. (2011). Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat Biotechnol 29:1117–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Z, Skamagki M, Kim K and Zhao R. (2015). Canonical MicroRNA Activity Facilitates but May Be Dispensable for Transcription Factor-Mediated Reprogramming. Stem Cell Reports 5:1119–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woolnough JL, Atwood BL, Liu Z, Zhao R and Giles KE. (2016). The Regulation of rRNA Gene Transcription during Directed Differentiation of Human Embryonic Stem Cells. PLoS One 11:e0157276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Z, Zhang C, Skamagki M, Khodadadi-Jamayran A, Zhang W, Kong D, Chang CW, Feng J, Han X, Townes TM, Li H, Kim K and Zhao R. (2017). Elevated p53 Activities Restrict Differentiation Potential of MicroRNA-Deficient Pluripotent Stem Cells. Stem Cell Reports 9:1604–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacDonald JR, Ziman R, Yuen RK, Feuk L and Scherer SW. (2014). The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res 42:D986–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C and Schultz N. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C and Schultz N. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K and Yamanaka S. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–72. [DOI] [PubMed] [Google Scholar]
- 32.Luchman HA, Stechishin OD, Dang NH, Blough MD, Chesnelong C, Kelly JJ, Nguyen SA, Chan JA, Weljie AM, Cairncross JG and Weiss S. (2012). An in vivo patient-derived model of endogenous IDH1-mutant glioma. Neuro Oncol 14:184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gopalakrishnan V (2009). REST and the RESTless: in stem cells and beyond. Future Neurol 4:317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lunyak VV and Rosenfeld MG. (2005). No rest for REST: REST/NRSF regulation of neurogenesis. Cell 121:499–501. [DOI] [PubMed] [Google Scholar]
- 35.Kamal MM, Sathyan P, Singh SK, Zinn PO, Marisetty AL, Liang S, Gumin J, El-Mesallamy HO, Suki D, Colman H, Fuller GN, Lang FF and Majumder S. (2012). REST regulates oncogenic properties of glioblastoma stem cells. Stem Cells 30:405–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conti L, Crisafulli L, Caldera V, Tortoreto M, Brilli E, Conforti P, Zunino F, Magrassi L, Schiffer D and Cattaneo E. (2012). REST controls self-renewal and tumorigenic competence of human glioblastoma cells. PLoS One 7:e38486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang J, Meng Q, Zhao W, Tong P, Li P, Zhao Y, Zhao X and Li H. (2016). An expression based REST signature predicts patient survival and therapeutic response for glioblastoma multiforme. Sci Rep 6:34556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi J, Lee S, Mallard W, Clement K, Tagliazucchi GM, Lim H, Choi IY, Ferrari F, Tsankov AM, Pop R, Lee G, Rinn JL, Meissner A, Park PJ and Hochedlinger K. (2015). A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nat Biotechnol 33:1173–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cohen A, Sato M, Aldape K, Mason CC, Alfaro-Munoz K, Heathcock L, South ST, Abegglen LM, Schiffman JD and Colman H. (2015). DNA copy number analysis of Grade II-III and Grade IV gliomas reveals differences in molecular ontogeny including chromothripsis associated with IDH mutation status. Acta Neuropathol Commun 3:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cancer Genome Atlas Research N and Brat DJ and Verhaak RG and Aldape KD and Yung WK and Salama SR and Cooper LA and Rheinbay E and Miller CR and Vitucci M and Morozova O and Robertson AG and Noushmehr H and Laird PW and Cherniack AD and Akbani R and Huse JT and Ciriello G and Poisson LM and Barnholtz-Sloan JS and Berger MS and Brennan C and Colen RR and Colman H and Flanders AE and Giannini C and Grifford M and Iavarone A and Jain R and Joseph I and Kim J and Kasaian K and Mikkelsen T and Murray BA and O'Neill BP and Pachter L and Parsons DW and Sougnez C and Sulman EP and Vandenberg SR and Van Meir EG and von Deimling A and Zhang H and Crain D and Lau K and Mallery D and Morris S and Paulauskis J and Penny R and Shelton T and Sherman M and Yena P and Black A and Bowen J and Dicostanzo K and Gastier-Foster J and Leraas KM and Lichtenberg TM and Pierson CR and Ramirez NC and Taylor C and Weaver S and Wise L and Zmuda E and Davidsen T and Demchok JA and Eley G and Ferguson ML and Hutter CM and Mills Shaw KR and Ozenberger BA and Sheth M and Sofia HJ and Tarnuzzer R and Wang Z and Yang L and Zenklusen JC and Ayala B and Baboud J and Chudamani S and Jensen MA and Liu J and Pihl T and Raman R and Wan Y and Wu Y and Ally A and Auman JT and Balasundaram M and Balu S and Baylin SB and Beroukhim R and Bootwalla MS and Bowlby R and Bristow CA and Brooks D and Butterfield Y and Carlsen R and Carter S and Chin L and Chu A and Chuah E and Cibulskis K and Clarke A and Coetzee SG and Dhalla N and Fennell T and Fisher S and Gabriel S and Getz G and Gibbs R and Guin R and Hadjipanayis A and Hayes DN and Hinoue T and Hoadley K and Holt RA and Hoyle AP and Jefferys SR and Jones S and Jones CD and Kucherlapati R and Lai PH and Lander E and Lee S and Lichtenstein L and Ma Y and Maglinte DT and Mahadeshwar HS and Marra MA and Mayo M and Meng S and Meyerson ML and Mieczkowski PA and Moore RA and Mose LE and Mungall AJ and Pantazi A and Parfenov M and Park PJ and Parker JS and Perou CM and Protopopov A and Ren X and Roach J and Sabedot TS and Schein J and Schumacher SE and Seidman JG and Seth S and Shen H and Simons JV and Sipahimalani P and Soloway MG and Song X and Sun H and Tabak B and Tam A and Tan D and Tang J and Thiessen N and Triche, Jr. T and Van Den Berg DJ and Veluvolu U and Waring S and Weisenberger DJ and Wilkerson MD and Wong T and Wu J and Xi L and Xu AW and Yang L and Zack TI and Zhang J and Aksoy BA and Arachchi H and Benz C and Bernard B and Carlin D and Cho J and DiCara D and Frazer S and Fuller GN and Gao J and Gehlenborg N and Haussler D and Heiman DI and Iype L and Jacobsen A and Ju Z and Katzman S and Kim H and Knijnenburg T and Kreisberg RB and Lawrence MS and Lee W and Leinonen K and Lin P and Ling S and Liu W and Liu Y and Liu Y and Lu Y and Mills G and Ng S and Noble MS and Paull E and Rao A and Reynolds S and Saksena G and Sanborn Z and Sander C and Schultz N and Senbabaoglu Y and Shen R and Shmulevich I and Sinha R and Stuart J and Sumer SO and Sun Y and Tasman N and Taylor BS and Voet D and Weinhold N and Weinstein JN and Yang D and Yoshihara K and Zheng S and Zhang W and Zou L and Abel T and Sadeghi S and Cohen ML and Eschbacher J and Hattab EM and Raghunathan A and Schniederjan MJ and Aziz D and Barnett G and Barrett W and Bigner DD and Boice L and Brewer C and Calatozzolo C and Campos B and Carlotti, Jr. CG and Chan TA and Cuppini L and Curley E and Cuzzubbo S and Devine K and DiMeco F and Duell R and Elder JB and Fehrenbach A and Finocchiaro G and Friedman W and Fulop J and Gardner J and Hermes B and Herold-Mende C and Jungk C and Kendler A and Lehman NL and Lipp E and Liu O and Mandt R and McGraw M and McLendon R and McPherson C and Neder L and Nguyen P and Noss A and Nunziata R and Ostrom QT and Palmer C and Perin A and Pollo B and Potapov A and Potapova O and Rathmell WK and Rotin D and Scarpace L and Schilero C and Senecal K and Shimmel K and Shurkhay V and Sifri S and Singh R and Sloan AE and Smolenski K and Staugaitis SM and Steele R and Thorne L and Tirapelli DP and Unterberg A and Vallurupalli M and Wang Y and Warnick R and Williams F and Wolinsky Y and Bell S and Rosenberg M and Stewart C and Huang F and Grimsby JL and Radenbaugh AJ and Zhang J. (2015). Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med 372:2481–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao R and Daley GQ. (2008). From fibroblasts to iPS cells: induced pluripotency by defined factors. J Cell Biochem 105:949–55. [DOI] [PubMed] [Google Scholar]
- 42.Ichimura K, Pearson DM, Kocialkowski S, Backlund LM, Chan R, Jones DT and Collins VP. (2009). IDH1 mutations are present in the majority of common adult gliomas but rare in primary glioblastomas. Neuro Oncol 11:341–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin G, Reitman ZJ, Duncan CG, Spasojevic I, Gooden DM, Rasheed BA, Yang R, Lopez GY, He Y, McLendon RE, Bigner DD and Yan H. (2013). Disruption of wild-type IDH1 suppresses D-2-hydroxyglutarate production in IDH1-mutated gliomas. Cancer Res 73:496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu M, Yu S, Xue Y, Yu H, Chen D, Wei X and Liu Y. (2018). Over-Expressed FEZF1 Predicts a Poor Prognosis in Glioma and Promotes Glioma Cell Malignant Biological Properties by Regulating Akt-ERK Pathway. J Mol Neurosci 65:411–419. [DOI] [PubMed] [Google Scholar]
- 45.Azar S, Leventoux N, Ripoll C, Rigau V, Goze C, Lorcy F, Bauchet L, Duffau H, Guichet PO, Rothhut B and Hugnot JP. (2018). Cellular and molecular characterization of IDH1-mutated diffuse low grade gliomas reveals tumor heterogeneity and absence of EGFR/PDGFRalpha activation. Glia 66:239–255. [DOI] [PubMed] [Google Scholar]
- 46.Bao ZS, Chen HM, Yang MY, Zhang CB, Yu K, Ye WL, Hu BQ, Yan W, Zhang W, Akers J, Ramakrishnan V, Li J, Carter B, Liu YW, Hu HM, Wang Z, Li MY, Yao K, Qiu XG, Kang CS, You YP, Fan XL, Song WS, Li RQ, Su XD, Chen CC and Jiang T. (2014). RNA-seq of 272 gliomas revealed a novel, recurrent PTPRZ1-MET fusion transcript in secondary glioblastomas. Genome Res 24:1765–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y, Wang D, Wang L, Yu J, Du D, Chen Y, Gao P, Wang DM, Yu J, Zhang F and Fu S. (2013). Distinct genomic aberrations between low-grade and high-grade gliomas of Chinese patients. PLoS One 8:e57168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rice T, Zheng S, Decker PA, Walsh KM, Bracci P, Xiao Y, McCoy LS, Smirnov I, Patoka JS, Hansen HM, Hsuang G, Wiemels JL, Tihan T, Pico AR, Prados MD, Chang SM, Berger MS, Caron A, Fink S, Kollmeyer T, Rynearson A, Voss J, Kosel ML, Fridley BL, Lachance DH, Eckel-Passow JE, Sicotte H, O’Neill BP, Giannini C, Wiencke JK, Jenkins RB and Wrensch MR. (2013). Inherited variant on chromosome 11q23 increases susceptibility to IDH-mutated but not IDH-normal gliomas regardless of grade or histology. Neuro Oncol 15:535–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jenkins RB, Xiao Y, Sicotte H, Decker PA, Kollmeyer TM, Hansen HM, Kosel ML, Zheng S, Walsh KM, Rice T, Bracci P, McCoy LS, Smirnov I, Patoka JS, Hsuang G, Wiemels JL, Tihan T, Pico AR, Prados MD, Chang SM, Berger MS, Caron AA, Fink SR, Halder C, Rynearson AL, Fridley BL, Buckner JC, O’Neill BP, Giannini C, Lachance DH, Wiencke JK, Eckel-Passow JE and Wrensch MR. (2012). A low-frequency variant at 8q24.21 is strongly associated with risk of oligodendroglial tumors and astrocytomas with IDH1 or IDH2 mutation. Nat Genet 44:1122–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ozawa T, Riester M, Cheng YK, Huse JT, Squatrito M, Helmy K, Charles N, Michor F and Holland EC. (2014). Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell 26:288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.