The Red Sea is an attractive environment for bioprospecting. There are 25 brine-filled deeps in the Red Sea. The Atlantis II brine pool is the biggest and hottest of such hydrothermal ecosystems. We generated an environmental mercuric reductase library from the lowermost layer of the Atlantis II brine pool, in which we identified two variants of the mercuric reductase enzyme (MerA). One is the previously described halophilic and thermostable ATII-LCL MerA and the other is a nonhalophilic relatively less thermostable enzyme, designated ATII-LCL-NH MerA. We used the ATII-LCL-NH enzyme as a parent molecule to locate the amino acid residues involved in the noticeably higher thermotolerance of the homolog ATII-LCL MerA. Moreover, we designed a novel enzyme with superior thermal stability. This enzyme might have strong potential in the bioremediation of mercuric toxicity.
KEYWORDS: Atlantis II, bioprospecting, MerA, protein engineering, Red Sea, brine pools, extreme environments, mercuric reductase, site-directed mutagenesis, thermostable
ABSTRACT
The lower convective layer (LCL) of the Atlantis II brine pool of the Red Sea is a unique environment in terms of high salinity, temperature, and high concentrations of heavy metals. Mercuric reductase enzymes functional in such extreme conditions could be considered a potential tool in the environmental detoxification of mercurial poisoning and might alleviate ecological hazards in the mining industry. Here, we constructed a mercuric reductase library from Atlantis II, from which we identified genes encoding two thermostable mercuric reductase (MerA) isoforms: one is halophilic (designated ATII-LCL) while the other is not (designated ATII-LCL-NH). The ATII-LCL MerA has a short motif composed of four aspartic acids (4D414–417) and two characteristic signature boxes that played a crucial role in its thermal stability. To further understand the mechanism behind the thermostability of the two studied enzymes, we mutated the isoform ATII-LCL-NH and found that the substitution of 2 aspartic acids (2D) at positions 415 and 416 enhanced the thermal stability, while other mutations had the opposite effect. The 2D mutant showed superior thermal tolerance, as it retained 81% of its activity after 10 min of incubation at 70°C. A three-dimensional structure prediction revealed newly formed salt bridges and H bonds in the 2D mutant compared to the parent molecule. To the best of our knowledge, this study is the first to rationally design a mercuric reductase with enhanced thermal stability, which we propose to have a strong potential in the bioremediation of mercurial poisoning.
IMPORTANCE The Red Sea is an attractive environment for bioprospecting. There are 25 brine-filled deeps in the Red Sea. The Atlantis II brine pool is the biggest and hottest of such hydrothermal ecosystems. We generated an environmental mercuric reductase library from the lowermost layer of the Atlantis II brine pool, in which we identified two variants of the mercuric reductase enzyme (MerA). One is the previously described halophilic and thermostable ATII-LCL MerA and the other is a nonhalophilic relatively less thermostable enzyme, designated ATII-LCL-NH MerA. We used the ATII-LCL-NH enzyme as a parent molecule to locate the amino acid residues involved in the noticeably higher thermotolerance of the homolog ATII-LCL MerA. Moreover, we designed a novel enzyme with superior thermal stability. This enzyme might have strong potential in the bioremediation of mercuric toxicity.
INTRODUCTION
The Red Sea is a marine environment that is attractive for bioprospecting owing to its characteristic features. It is isolated, and its water is relatively warmer and more saline than other seas. The water temperature ranges from approximately 26°C at the north part to 30°C at the south part, with only 2°C variation between the winter and summer average temperatures (1). Its average salinity is 4.1%, compared to the average of 3.5% in other seas (2, 3).
There are 25 brine-filled deeps that have been discovered so far in the Red Sea (4–7) that were formed by the shift of the Arabian and African tectonic plates (5, 8). The Atlantis II (ATII) brine pool is considered the biggest, hottest, and largest ore deposit of any known hydrothermal system (6, 9). This deep is a harsh environment harboring diverse microbial communities adapted to its extreme conditions (10). The lower convective layer (LCL) is the deepest, 2,200 m below sea surface, with salinity 7.5 times that of seawater (2). It endures extreme hypoxic conditions and has a low pH of 5.3, high concentrations of heavy metals, and a temperature of 68°C (8, 9, 11) that has gradually increased over the past few decades as a result of the mounting volcanic activity in the vicinity (5).
Because of technical difficulties, studying the microbial communities in these deep sea brines has not attracted sufficient attention until recently (7, 10). Among the abundant stresses existing in this marine environment that challenge the microbial community is the mercurial toxicity.
Mercury is one of the most toxic heavy metals on earth (12). It is ranked the sixth among the ten most toxic elements in the universe (13). Its toxicity to organisms lacking mercury resistance mechanisms stems from its affinity to sulfhydryl groups (-SH) of proteins and enzymes (12, 13), altering their structure and function (14) and rendering them ineffective (15). Mercury abundance is correlated with the activity of hydrothermal systems (16), such as the ATII hydrothermal vent, but its measurement is elusive, since emissions from anthropogenic sources are variable and surpass emissions from natural origins (17). The total concentration of mercury in the Red Sea is remarkably higher than those of other seas: mercury levels reach up to 2,000 ng/liter in the Red Sea (18), compared to 0.6 ng/liter in the Baltic Sea (19) and 5.5 ng/liter in the Yellow Sea (20). Moreover, mercury content in the ATII sediments is remarkably higher than in seawater and reaches up to 30 × 106 ng/liter (9).
The mercuric reductase enzyme (MerA) is a homodimeric protein belonging to the family of flavin dinucleotide oxidoreductases (21, 22). MerA detoxifies inorganic Hg2+ by reducing the divalent mercuric ion into volatile Hg0. MerA is characterized by two active sites located at the dimer interface (see Fig. S1 in the supplemental material) comprising two pairs of cysteines at the catalytic core (C136 and C141), which are redox active, and a pair of cysteines (C558 and C559) each at the C termini of the two subunits, serving as both mercury-binding and -presenting sites to be further reduced by the enzyme active sites (23, 24).
Thermozymes are enzymes produced by thermophilic or hyperthermophilic organisms. The enhancement of their thermostability/thermophilicity is attributed to either a single amino acid or a group of amino acid changes in specific sites and critical locations on the polypeptide chain (25–28). However, there is no definitive method to predict such sites or amino acid residues to be substituted, because each protein behaves in a distinct manner (29).
A mercuric reductase enzyme from ATII-LCL was previously characterized (30). This enzyme is activated by sodium chloride in a salt-dependent manner to reach its maximum activity at a concentration of 4 M sodium chloride. The enzyme is also thermostable, as it retains 70% of its activity after 10 min of incubation at 70°C. Moreover, the ATII-LCL enzyme shows an abundance of acidic amino acids on its surface, and it has two distinguished short amino acid motifs of four and seven amino acids, named Box1 and Box2, respectively (30). In the vicinity of these boxes comes a stretch of four aspartic acids (4D; Asp414 to 417). Interestingly, one of them, Asp417, can potentially form a salt bridge with the first lysine residue (Lys 432) of Box1 (Fig. 1).
FIG 1.

Diagrammatic representation of the salt bridge between Lys432 of Box1 and Asp417 in the ATII-LCL MerA enzyme model. (A) A MerA monomer in which Box1 (random coiled loop), Box2 (beta sheet), and 4D are shown in purple. (B) Enlargement of the area containing Box1 and the 4D residues. (C) The two amino acid residues involved in the potential salt bridge in the ATII-LCL MerA enzyme.
In a previous study, when the Box1 motif was replaced with another set of amino acids in a soil ortholog of the enzyme, the salt bridge was lost, minimizing its thermal stability (30). On the other hand, replacing the Box2 motif further reduced the temperature resistance. Collectively, the removal of both boxes and replacing them with the corresponding residues in the soil ortholog drastically lowered the heat stability. Since the substitution of these two boxes affected neither the enzyme’s kinetic parameters nor its salinity profile, the two boxes were suggested to be directly involved in the heat stability of the enzyme (30).
These observations, which emphasize the importance of the boxes and the 4D motif in stabilizing the enzyme, initiated the current study, which aimed to engineer an enzyme with enhanced thermal stability.
For this purpose, we created a mercuric reductase library from the ATII LCL of the Red Sea, in which we identified an enzyme (ATII-LCL-NH) that is devoid of all the acidic substitutions of the ATII-LCL enzyme and does not have the two signature boxes. Subsequently, we implemented a rational design approach to generate a more thermostable enzyme by substituting its two signature boxes and the four aspartic acid residues 414 to 417 of the ATII-LCL enzyme with the corresponding motifs in ATII-LCL-NH. One of the mutants was more thermostable than both enzymes, as it retained 81% of its activity after 10 min of incubation at 70°C, relative to 44% and 70% in the cases of ATII-LCL-NH and ATII-LCL, respectively.
This study presents a novel isoform that is catalytically superior to several mercuric reductases previously described, rendering it a potentially effective catalyst for the bioremediation of environmental lethal mercury toxicity.
RESULTS
Establishment of a mercuric reductase PCR library from Atlantis II LCL.
Using a single pair of oligonucleotide primers homologous to a highly conserved area in merA, we amplified Atlantis II LCL DNA to generate a PCR library of candidate genes. The sequences of agricultural soil merA (NCBI accession number AEV57255.1) and Tn501 merA (NCBI accession number CAA77323.1) and the merA consensus sequence of assembled reads (CSAR) from the Atlantis II data set were used to generate oligonucleotide primers for PCR amplification. A single discrete band of approximately 1.7 kb was obtained, as expected from the merA gene length of 1,686 bp that potentially codes for full-length MerA of 561 amino acid residues (see Fig. S2 and S3 in the supplemental material).
The sequencing of the inserted DNA of the forty isolated recombinant plasmids from the merA library resulted in eight full-length nonredundant mercuric reductase sequences (see Fig. S4).
Very few amino acid differences (ranging from 1 to 4 substitutions) were detected upon translating the DNA sequences. The sequence designated ATII-LCL-NH has a high similarity to the well-characterized mercuric reductase Tn501, with only one amino acid difference at position 386: an alanine residue instead of a valine in Tn501. This sequence was also previously obtained from a separate library. The observed few substitutions were not in the redox active or mercury-binding cysteine residues. Therefore, one of them, ATII-LCL-NH, was chosen to be the backbone for mounting all the mutants generated in this work.
Generation of MerA mutants using site-directed mutagenesis.
The ATII-LCL-NH protein was found to be different by just one amino acid residue from the well-characterized Tn501 MerA. Its sequence is missing all the acidic amino acids, including the two boxes responsible for the thermostability of the MerA ATII-LCL (30) (Fig. 2). The ATII-LCL-NH sequence was therefore selected to introduce sequences from the ATII-LCL MerA that were shown to affect, or have the potential to affect, the thermostability of the protein.
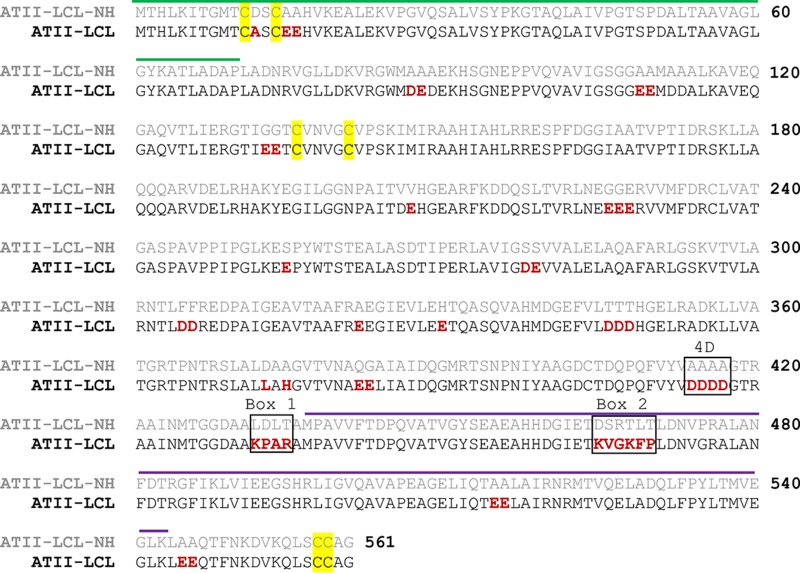
FIG 2.
Pairwise alignment of MerA ATII-LCL and ATII-LCL-NH. The amino acids different in ATII-LCL compared with ATII-LCL-NH are in red. The NmerA domain (55) is overlined in green. The dimerization domain (61) is overlined in purple. The cysteine pairs 11/14 and 558/559, which are responsible for Hg2+ binding, and the cysteine pair 136/141 involved in the catalytic site are highlighted in yellow. Positions of the amino acids involved in the mutations performed in this work are in black boxes.
Three mutants were generated by site-directed mutagenesis. All involved the four aspartic acids at positions 414 to 417 and the two boxes (Fig. 3). The substituted amino acid of each mutant and its corresponding residue in ATII-LCL-NH are shown in Table 1.
FIG 3.
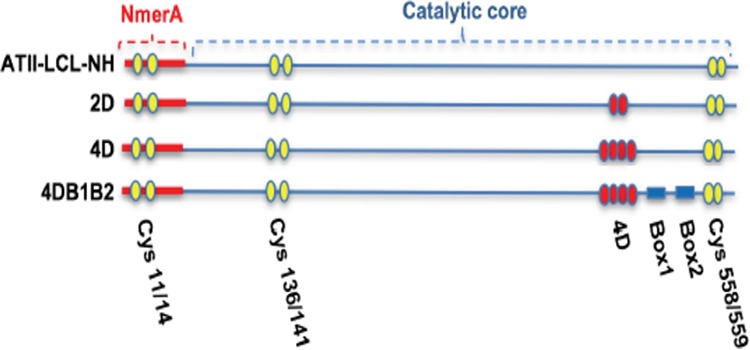
Diagram of the mutations shown in Table 1. Yellow spheres represent the cysteine residues involved in binding and reduction of Hg2+; red spheres represent aspartic acid residues; blue rectangles are Box1 and Box2. For simplicity, the mutants ATII-LCL-NH/2D, ATII-LCL-NH/4D, and ATII-LCL-NH/4DB1B2 are referred to as 2D, 4D, and 4DB1B2, respectively.
TABLE 1.
Mutations to replace residues from ATII-LCL-NH with their corresponding amino acids in the ATII-LCL enzyme
| Mutation | Amino acid residues in: |
|
|---|---|---|
| ATII-LCL-NH | ATII-LCL | |
| 2D | 415AA416 | 415DD416 |
| 4D | 414AAAA417 | 414DDDD417 |
| 4DB1B2 | 414AAAA417/432LDLT435/465DSRTLT470 | 414DDDD417/432KPAR435/465KVGKFP470 |
Expression and purification of ATII-LCL-NH and its mutants.
The ATII-LCL-NH gene was expressed in Escherichia coli BL21 cells under the control of the T7 promoter in the pET-SUMO expression vector at 37°C. Similar levels of MerA protein yield were obtained by inducting the recombinant protein at different IPTG (isopropyl-β-d-thiogalactopyranoside) concentrations: 0.1, 0.2, or 0.5 mM IPTG (see Fig. S5). Moreover, the maximum level of expression of the recombinant protein did not change with the timing of induction.
The protein was expressed in the soluble cellular fraction of the cell lysate. However, mutant enzymes (4D and 4DB1B2) were only expressed in the soluble fraction after an overnight induction with 0.1 or 0.2 mM IPTG at 24°C. Their expression for 2 h at 37°C resulted in the formation of inclusion bodies. This might be explained in the context of their thermal stability behavior. Those mutants precipitate and become inactivated by heat (see Fig. 4). The purification procedure was applied successfully with MerA ATII-LCL-NH and its mutants.
FIG 4.
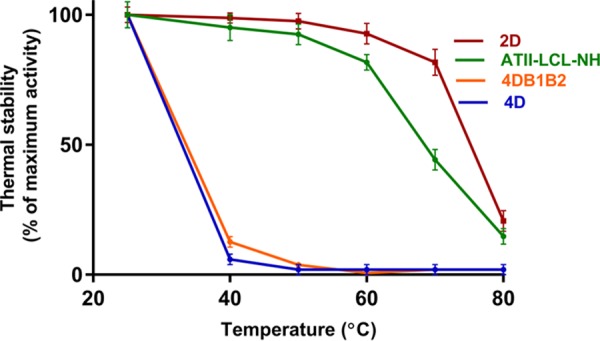
Thermostability of ATII-LCL-NH and its mutants. The enzymes were incubated at the indicated temperatures for 10 min, and the residual activities were measured under standard assay conditions (see Materials and Methods for details).
Two column chromatography methods were used in the purification process, namely, HisTag affinity chromatography and size exclusion on Superdex 75. The elution profiles obtained from the two column chromatography methods were similar for MerA ATII-LCL-NH and its mutants. Therefore, only the results from the purification of MerA ATII-LCL-NH are presented in Fig. S6 to S9. The process yielded 10 to 12 mg pure protein/liter of induced bacterial culture.
Effect of NaCl on the activity and kinetic parameters of ATII-LCL-NH and its mutants.
The ATII-LCL-NH enzyme and its mutants were strongly inhibited by NaCl. Although all the mutants were inhibited by NaCl, a slight difference in the degree of inhibition was noticed between the different mutants (Table 2).
TABLE 2.
Residual activities of ATII-LCL-NH and its mutants at 0.5 M NaCl
| MerA isoform | Residual activity at 0.5 M NaCl (%) |
|---|---|
| ATII-LCL-NH | 55 ± 4.6 |
| 2D | 56 ± 3.9 |
| 4D | 27 ± 6.5 |
| 4DB1B2 | 29 ± 5.5 |
The kinetic parameters of ATII-LCL-NH, 2D, 4DB1B2, 4D, and 4DB1B2 mutants were determined. The specific activities, the affinity to the substrate (Hg), and the catalytic efficiency—measured as kcat/Km—were very similar for the ATII-LCL-NH, 2D, 4D, and 4DB1B2 enzymes (Table 3). The 2D mutant showed a specific activity of 10.95 μmol/min/mg, Km of 10.96 μM, kcat of 12 s−1, and a kcat/Km of 1.1.
TABLE 3.
Kinetics data of ATII-LCL-NH, 2D, 4D, and 4DB1B2 mutantsa
| MerA isoform | Sp act (μmol/min/mg) | Km (μM) | Vmax (μmol/min/mg) | kcat (s−1) | kcat/Km |
|---|---|---|---|---|---|
| ATII-LCL-NH | 10.96 ± 0.74 | 11.25 ± 0.71 | 14.21 ± 0.81 | 12.25 ± 1.2 | 1.1 |
| 2D | 10.95 ± 0.58 | 10.96 ± 0.82 | 14.05 ± 0.73 | 12 ± 1 | 1.1 |
| 4D | 11.1 ± 0.6 | 13.9 ± 1.35 | 15.09 ± 0.93 | 12.6 ± 1.4 | 0.9 |
| 4DB1B2 | 10.9 ± 0.68 | 12.8 ± 0.93 | 14.4 ± 0.78 | 12.4 ± 0.98 | 0.96 |
Data were determined by Lineweaver-Burk (double reciprocal plot) and GraphPad Prism software (GraphPad, La Jolla, CA, USA).
Thermal stability of ATII-LCL-NH merA and its mutants.
Although the ATII-LCL-NH enzyme was missing the two boxes and all the acidic residues present in its ATII-LCL homolog, it still retained 81% residual activity at 60°C. The residual activities of the enzymes were measured after incubating for 10 min at the corresponding temperature. The values (residual activity) were relative to those obtained after incubating at 25°C for 10 min.
However, the ATII-LCL-NH enzyme was remarkably less stable at 70°C than ATII-LCL, as it retained 44% of its activity relative to 70% residual activity in the case of ATII-LCL. The 4D mutation caused severe instability of the mutant toward heat treatment. The 4DB1B2 mutant, unexpectedly, was also as severely unstable to heat as the 4D mutant (Fig. 4).
Surprisingly, in comparison with both ATII-LCL-NH and ATI-LCL, the 2D (aspartic acid 415 and 416) mutation alone increased the thermostability of the molecule, as it retained 81% of its activity after 10 min of incubation at 70°C. Interestingly, the catalytic efficiency (kcat/Km) of the heat-stable 2D mutant was much higher than that of the recently reported Metallosphaera sedula MerA (31) and Lysinibacillus sphaericus MerA (32), which makes it a better candidate for mercuric bioremediation.
We therefore computationally analyzed the hydrogen and ionic bonds present in the ATII-LCL-NH dimer and compared them with the same bonds in the 2D, 4D, and 4DB1B2 mutants to determine the effects of the 2D mutation on increasing the stability, the 4D mutation on the destabilization of the molecule to heat, and the two boxes together with the 4D mutation.
The hydrogen bonds between the amino acid side chains, within the MerA dimer molecules, were computed using the Hydrogen Bond Calculator version 1.1 web server (http://cib.cf.ocha.ac.jp/bitool/HBOND/) (33, 34).
We did not observe a direct relation between the total hydrogen bonds in the molecules and the observed stabilities of the mutants compared to those of the ATII-LCL-NH protein. A clear difference that correlated with the heat stabilities was observed by comparing the loss and gain of hydrogen bonds between the ATII-LCL-NH enzyme and its mutants (see Fig. S10).
The 2D mutant lost 26 hydrogen bonds present in ATII-LCL-NH and gained 42 new hydrogen bonds, whereas the 4D mutant, which is unstable to heat, lost 69 hydrogen bonds and gained 90 new hydrogen bonds. In the case of the 4DB1B2 mutant, the loss and gain of hydrogen bonds were very prominent, since 174 bonds were lost and the molecule gained 150 newly established hydrogen bonds (see Fig. S11).
The salt bridges in ATII-LCL-NH and its mutants were also analyzed using the web server for evaluating salt bridges in proteins (http://bioinformatica.isa.cnr.it/ESBRI/introduction.html) (35). In comparison with ATII-LCL-NH, the introduction of the 2D or the 4D mutation increased the salt bridges by 6 or 7 bridges, respectively (Fig. S11).
A ConSurf (36) analysis of the evolutionary conservation of the mutated amino acid positions indicated that Ala414 is the most conserved among homologous mercuric reductases, while Ala416 is more variable (see Fig. S12).
DISCUSSION
The Atlantis II LCL of the Red Sea is a unique environment characterized by high temperature, salinity of 4.4 M, very low oxygen concentration, low pH of 5.3, and high concentrations of toxic heavy metals. Biocatalysts isolated from such an environment could have valuable applications in industry and bioremediation. In a previous work (30), a thermostable mercuric reductase that is activated by NaCl, MerA ATII-LCL, was isolated from the ATII LCL environment. It is characterized by the abundance of acidic amino acids, most notably on the surface of the molecule, and has two signature boxes in close proximity to a four-aspartic-acid motif that might form a potential salt bridge with Box1. Substitution of the two boxes was shown to drastically lower the enzyme’s thermal stability.
Towards engineering an enzyme variant with enhanced thermal stability, we constructed a mercuric reductase library from the Atlantis II brine LCL. Of the 40 clones sequenced, we obtained 8 full-length nonredundant sequences, yet very few differences between the 8 sequences were observed. We selected an ortholog from the same environment as a template to study the effects of the two signature boxes and the short aspartic acid motif.
The selected template lacks the acidic amino acids and the signature boxes found in the ATII-LCL MerA enzyme. It is also one amino acid different than the well-studied Tn501 MerA. We used site-directed mutagenesis to replace the two boxes, the four-alanine motif of the ATII-LCL-NH with the corresponding signature boxes, and the four-aspartic-acid motif of the ATII-LCL MerA. The ATII-LCL-NH enzyme and its mutants were purified on a HisTrap affinity column followed by a Superdex75 size exclusion column. A yield of 10 to 12 mg of pure protein was obtained per liter of induced bacterial culture. We noticed that induction at 37°C of the 4D and 4DB1B2 mutants resulted in the precipitation of the proteins and the formation of inclusion bodies. This might be explained by the heat sensitivity of these two mutants. Therefore, all the mutants in this work were induced at 24°C overnight.
A previous work with MerA ATII-LCL revealed that the enzyme has distinguished properties, is activated by NaCl in a salt-dependent manner, and attains its highest activity in the presence of 4 M sodium chloride (30). This high molarity of sodium chloride is similar to the salinity of the ATII LCL environment. In contrast to the ATII-LCL enzyme, the ATII-LCL-NH homolog described in this work did not show an increase in acidic and polar uncharged side chains at the expense of hydrophobic amino acids, a characteristic of nonhalophilic proteins (30, 37–40).
To examine the responses of the ATII-LCL-NH protein and its mutants to NaCl, we assayed the enzymes’ activity in the presence of increasing concentrations of NaCl. As expected from their amino acid compositions, the ATII-LCL-NH enzyme and its mutants were strongly inhibited by NaCl. The kinetic parameters of the NaCl-inhibited ATII-LCL-NH protein and the 2D, 4D, and 4DB1B2 mutants were determined. The specific activities, affinity to the substrate (Hg), and efficiency (measured as kcat/Km) were very similar for the ATII-LCL-NH, 2D, and 4D enzymes. This result indicates that the different degrees of NaCl inhibition of the three MerA enzymes are not due to changes of the affinity of the enzymes to the substrate or the efficiency of the catalytic process. Of note, activity assays and kinetic experiments for ATII-LCL MerA were performed at concentrations of 0.5 M and 4 M NaCl (30) and thus cannot be compared to our assays in the absence of inhibitor NaCl, while the temperature stability analyses for all our mutants, and the previously described ATII-LCL, were performed in the absence of salt and could thus be compared.
Interestingly, the 2D MerA mutant had superior catalytic properties to numerous reported mercuric reductase enzymes. The 2D mutant had a kcat/Km of 1.1 μM−1 · s−1, compared to 0.6 μM−1 · s−1 for Lysinibacillus sphaericus MerA and 0.15 μM−1 · s−1 for Metallosphaera sedula MerA (31). The specific activity of the 2D mutant was 10.95 μmol/min/mg, compared to 1.9 μmol/min/mg for M. sedula MerA (31) and 9 μmol/min/mg for Klebsiella pneumoniae MerA (41) under similar assay conditions.
It is worth noting that Box1 and Box2 each have one proline residue. Proline is known to reduce the structural flexibility of polypeptide regions containing it (42–44). It is proposed that certain proline substitutions at critical sites on a protein backbone account for the rigidity and restriction of polypeptide flexibility and therefore improve protein thermal stability in a cumulative manner (43, 44). Conversely, proline substitutions at undesirable sites result in a disruption of conformation and lowered protein thermal stability (45); thus, site-specific mutation and/or replacement has to be taken into consideration when engineering a thermostable enzyme (46).
Salt bridge formation or disruption in site-directed mutagenesis studies may improve or lower thermal stability depending on the location of the bridge and its geometry (35, 47). In fact, modeling the three-dimensional structure of the MerA ATII-LCL indicated the presence of a salt bridge between Lys432 in Box1 (a random coiled loop) and Asp417 located in the stretch of the 4D residues (Asp414 to 417). The presence of this salt bridge suggests a possible involvement of this interaction in the thermal stability of ATII-LCL. To examine if the two boxes and the 4D residues increase the thermostability of ATII-LCL-NH, we examined the thermostabilities of the established set of mutants in which the two boxes (Box1 and Box2) and the 4D substituted for their corresponding residues in the ATII-LCL-NH. Interestingly, in comparison with ATII-LCL-NH, the 2D mutation (Asp415 and 416) alone increased the thermostability of the molecule, whereas the 4D mutation caused severe instability of the mutant toward heat treatment. The 4DB1B2 mutant, however, was as severely unstable to heat as the 4D mutant.
Modeling the three-dimensional structure of the 4DB1B2 mutant indicated that the salt bridge between Lys432 in Box1 and aspartic acid 417 of the 4D was indeed established. This suggests that this salt bridge did not change or alter the negative behavior of the 4DB1B2 mutant molecule toward heat stability. We computationally analyzed the hydrogen and ionic bonds present in the ATII-LCL-NH dimer and compared it with those in the 2D, 4D, and 4DB1B2 mutants to determine the effects of the 2D mutation on increasing the stability, the 4D mutation on its destabilization of the molecule to heat, and the two boxes together with the 4D. The hydrogen bonds and salt bridges helped stabilize the protein molecules by optimizing the secondary structure interactions and subunit associations to form more stable tertiary and quaternary structures (48). There is no direct relation between the total hydrogen bonds in the molecules and the observed stabilities of the mutants compared to that of the ATII-LCL-NH. A clear difference that correlated with the heat stabilities was observed by comparing the loss and gain of hydrogen bonds between the ATII-LCL-NH and its mutants.
Analyzing the Venn diagram of these results showed that the 45 hydrogen bonds present in the ATII-LCL-NH and the 2D mutant are missing in the 4D heat-labile molecule. In addition, of the 42 newly formed hydrogen bonds in the 2D mutant, 28 were shared with the 4D heat-labile mutant and 14 were specific to the 2D heat-stable mutant. Moreover, 62 hydrogen bonds were specific to the 4D mutant. These results nominate the 45 hydrogen bonds lost from the 4D mutants and the 62 newly formed hydrogen bonds in the 4D mutant as potential candidates for destabilizing the molecule, turning it into a heat-labile structure. In support of this potential proposition, 52 of the 62 hydrogen bonds are still present in the 4DB1B2 thermolabile mutant, and the 45 hydrogen bonds shared between the ATII-LCL-NH and the 2D mutant are absent in both 4DB1B2 and 4D mutants.
In comparison with ATII-LCL-NH, the introduction of the 2D or the 4D residues increased the salt bridges by six or seven bridges, respectively. Note that the six new salt bridges in the 2D mutant are also shared with the 4D mutant. Since the introduction of the 2D residues did not cause a loss of the molecule’s thermostability, those six salt bridges most probably did not contribute to the dramatic decreases of the thermostability in the 4D mutant. The only difference that potentially explains the extreme heat sensitivity of the 4D mutant is the one salt bridge that is specific for this mutant. Salt bridge formation or disruption in site-directed mutagenesis studies may improve or lower thermal stability depending on the location of the bridge and its geometry (35, 47). Although the 4DB1B2 mutant did not gain this unique salt bridge, it gained 16 salt bridges that are not present in the thermostable ATII-LCL-NH molecule and the 2D mutant.
A ConSurf analysis (36) highlighted the correlation between evolutionary conservation and the positions of each amino acid in homologous mercuric reductase proteins. We observed that Ala414 of the ATII-LCL-NH MerA enzyme is the most conserved (Fig. S12 in the supplemental material, dark purple in the alignment), while Ala415 is less conserved (Fig. S12, light purple), and Ala416 is average (Fig. S12, white). Given that Ala415 and 416 are the residues of the 2D mutant which were replaced with aspartic acids and showed the highest heat stability, this might explain, together with the salt bridges and hydrogen bond analysis, the elevated heat stability of the 2D mutant and the noticeable temperature sensitivity of the 4D mutant.
In conclusion, both mercuric reductases isolated from the same environment demonstrated the complexity by which an enzyme adapts to several abiotic stressors simultaneously. The disruption of the geometry and balanced interactions of the ATII-LCL-NH enzyme and its mutants might explain the remarkable alteration of heat stability.
A molecule behaves as a unit and requires favorable conformation and interactions with the solvent to impart stability toward heat. Accordingly, we here report the generation of a novel heat-stable mercuric reductase with superior catalytic properties compared to those of previously characterized enzymes. This enzyme could be used in the bioremediation of mercurial toxicity.
MATERIALS AND METHODS
Sample collection and DNA extraction.
Water samples were collected from the ATII brine pool of the Red Sea at a depth of 2,200 m. The Atlantis II brine pool is located at 21°20.760′N, 38°04.68′E (49) between Saudi Arabia and Sudan (50). According to the oxygen content, salinity, and temperature, the ATII depression is divided into four layers: three upper convective layers (UCL-1 to UCL-3) and a lower convective layer (LCL) (2). Samples were collected in March/April 2010 during the expedition of the research vessel Aegaeo as part of the collaboration between King Abdullah University for Science and Technology (KAUST) and Woods Hole Oceanographic Institution (WHOI). A rosette of Niskin bottles connected to a conductivity, temperature, and depth device was lowered to the lower convective layer of the Atlantis II brine pool. Water samples from this layer were serially filtered through a 0.3-μm filter, a 0.8-μm filter, and a 0.1-μm cellulose-ester filter (Millipore) and stored in sucrose buffer. Collective DNA was extracted in our laboratory using a marine DNA isolation kit (Epicenter) as described in detail in reference 30.
Establishment of mercuric reductases library from ATII LCL.
To design primers for amplification, we performed multiple sequence alignment of agricultural soil MerA (NCBI accession number AEV57255.1), Tn501 MerA (NCBI accession number CAA77323.1), and a consensus sequence of assembled reads (CSAR) using Multalin (51). The CSAR was obtained by screening an established metagenomics library from the lower convective layer of Atlantis II for mercuric reductase sequences. The designed primer pairs started with the ATG start codon of the forward primer and ended with the TGA stop codon of the reverse primer. Two perfectly matching primers were synthesized accordingly: MerA-F, 5′-ATGACCCATCTAAAAATCACCGGCATGACTTG-3′, and MerA-R, 5′-TCACCCGGCGCAGCAGGAAAGCTGCTTC-3′. PCR was performed on environmental DNA isolated from Atlantis II LCL with Phusion high-fidelity DNA polymerase (Thermo Scientific, MA, USA) at a concentration of 1 U/50 μl reaction mixture as follows: 5× HF reaction buffer (containing MgCl2 7.5 mM) to a final concentration of 1×, 0.3 μM primers, 0.3 mM deoxynucleoside triphosphates (dNTPs), 100 ng DNA, and 3% dimethyl sulfoxide (DMSO). Thirty cycles were performed in a Veriti thermal cycler (Applied Biosystems, CA, USA) as follows: 30 s for denaturing at 95°C, 30 s for annealing at 70°C, 1.5 min for extension at 72°C, followed by a final extension step of 7 min at 72°C. The band of interest was excised from the gel and purified by the QIAquick PCR purification kit (Qiagen, MD, USA). The purified PCR fragments were ligated overnight at 15°C into the pGEM vector (Promega, WI, USA) according to the manufacturer’s manual.
Plasmids were transformed into XL-1 Blue cells and plated on Luria Bertani (LB) plates containing 100 μg/ml ampicillin, which were incubated overnight at 37°C. Forty random colonies were picked, and each was grown in 5 ml LB broth supplemented with 100 μg/ml ampicillin and shaken at 220 rpm overnight at 37°C. Glycerol stocks were made and stored at −80°C. Plasmids were extracted with a PureYield plasmid miniprep kit (Promega, WI, USA). Sanger dideoxy sequencing from both ends was performed using the T7 and SP6 pGEM universal primers (Macrogen Korea Laboratory). This custom-made gene was synthesized at GenScript (NJ, USA) in order to optimize codon usage and GC content and to increase expression levels in the host Escherichia coli. The synthesized gene was cloned into Champion pET SUMO (Invitrogen, CA, USA) and transformed into BL21(DE3) cells for expression. This gene was used to generate all the mutants in this work. NCBI BLASTp (52) shows that its closest match is Aquabacterium parvum (53). It is a Gram-negative Betaproteobacteria.
Generation of MerA mutants using site-directed mutagenesis.
The codon-optimized ATII-LCL-NH mercuric reductase gene was used as the template to generate the mutants described in this work. Mutagenic primers were generated using the web-based QuikChange primer design software (http://www.agilent.com/genomics/qcpd). The QuikChange II site-directed mutagenesis kit (Agilent, CA, USA) was used to create the various mutants addressed in this paper as recommended in the instruction manual. The annealing temperatures (Ta) were calculated to be 77.5°C for the Box1 primer, 77.2°C for Box2, 75.4°C for 2D, and 75.1°C for 4D. The mutant genes were cloned into the pGEM vector (Promega, WI, USA) for propagation and further sequencing and into the pET SUMO vector (Invitrogen, CA, USA) for expression. The primer sequences used to generate each of the mutants are as follows: 4D sense, 5′-GACCAACCGCAGTTTGTCTACGTGGATGATGATGATGGTACCCGTGCAGCAATCAACATG-3′; 4D antisense, 5′-CATGTTGATTGCTGCACGGGTACCATCATCATCATCCACGTAGACAAACTGCGGTTGGTC-3′; 2D antisense, 5′-TTGCTGCACGGGTACCAGCATCATCTGCCACGTAGACAAACTG-3′; 2D sense, 5′-CAGTTTGTCTACGTGGCAGATGATGCTGGTACCCGTGCAGCAA-3′; Box1 sense, 5′-CAACATGACGGGCGGTGATGCAGCTAAGCCCGCGAGGGCAATGCCGGCTGTGGTTTTC-3′; Box1 antisense, 5′-GAAAACCACAGCCGGCATTGCCCTCGCGGGCTTAGCTGCATCACCGCCCGTCATGTTG-3′; Box2 antisense, 5′-GGGCACGCGGCACGTTATCCAGGGGGAACTTACCGACCTTGGTTTCAATGCCATCGTGATGCGC-3′; Box2 sense, 5′-GCGCATCACGATGGCATTGAAACCAAGGTCGGTAAGTTCCCCCTGGATAACGTGCCGCGTGCCC-3′.
Expression and purification of recombinant MerA isoforms.
An overnight culture of transformed E. coli BL21(DE3) cells harboring the recombinant plasmid (carrying the merA isoform) was diluted 50 times in fresh LB medium supplemented with 20 μM flavin adenine dinucleotide (FAD) and 50 μg/ml kanamycin. The expression of recombinant enzymes was induced at an optical density at 600 nm (OD600) of 0.6 by the addition of IPTG to a final concentration 0.2 mM overnight at 24°C. Cultures were centrifuged, resuspended in lysis buffer (20 mM phosphate buffer [pH 7.4] containing 1 mM EDTA, 2 mM β-mercaptoethanol, 50 μM FAD, 1 mM protease inhibitor phenylmethylsulfonyl fluoride [PMSF], 1% Triton X-100 [vol/vol], 40 mM imidazole, and 0.2 mg/ml lysozyme [31]), and incubated for 30 min at room temperature. The sample was then sonicated three times for 30 s on ice in a Branson Sonifier 150 at an output power of 5. Cellular debris was removed by centrifugation at 14,000 × g for 45 min at 4°C.
The clear supernatant was filtered through a 0.2-μm syringe filter and applied to a preequilibrated 1-ml His-Trap HP column (GE Healthcare, IL, USA) at a flow rate of 1 ml/min using the AKTA purifier instrument (UNICORN version 4.0, Pump P-950, and Frac-950). The wavelength detector was adjusted to 280 and 450 nm. The protein was then eluted from the column using 500 mM imidazole at a rate of 0.5 ml/min. The elution continued until no detectable reading was observed at 280 and 450 nm, and the fractions were collected. SDS-PAGE was used to analyze the eluted fractions for the presence of the desired band on a 10% gel.
Fractions containing the desired band were pooled and slowly passed on a preequilibrated 24-ml Superdex 75 10/30 gel filtration column (GE Healthcare, IL, USA) at a flow rate of 0.5 ml/min to remove excess FAD and further purify the protein. The protein was eluted with 1× phosphate-buffered saline (PBS) (2 mM KCl, 137 mM NaCl, 10 mM Na2HPO4, 1.8 mM KH2PO4). Fractions were collected and analyzed on a 10% SDS-PAGE gel, and the fractions containing the desired band were pooled. The protein sample was concentrated in Amicon Ultra-4 10K centrifugal filters (Millipore, MA, USA) to a final concentration of 30 mg/ml and stored in aliquots at −80°C until further use. The protein concentration was determined by a Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Scientific, MA, USA).
Mercuric reductase activity assay.
Routine enzyme assays were performed in triplicates at 25°C in the assay mixture (80 mM phosphate buffer, 1 mM β-mercaptoethanol, 1 mM EDTA, 50 μM HgCl2). Enzymes were used at 100 μg in a final volume of 1 ml. The reaction was started by the addition of 100 μM NADPH. The enzyme activity was monitored by observing the initial rate of substrate-dependent NADPH consumption at 340 nm using a Shimadzu UV-1800 spectrophotometer. The unit of activity was defined as the amount of enzyme that catalyzes Hg2+-dependent oxidation of 1 μmol of NADPH/min (54). Kinetic and statistical parameters were calculated using GraphPad Prism software (GraphPad, La Jolla, CA, USA) with a confidence interval of 95%.
Heat stability analysis.
Replicate samples of each enzyme were incubated at the designated temperatures for 10 min in a Veriti thermal cycler (Applied Biosystems, CA, USA) and then dipped immediately on ice. The vials were spun down to separate any precipitated protein, and the clear supernatant was taken. The residual activity was then measured by the addition of the heated enzyme to the reaction mixture as previously described. The results are expressed relative to the activity observed after incubating for 10 min at 25°C (30).
Salt tolerance analysis.
Replicate samples of each enzyme were added to the standard reaction mixture containing the selected concentrations of NaCl. The reaction was started by the addition of 100 μM NADPH. The initial rate of NADPH oxidation was measured. The percentage of inactivation by NaCl was calculated relative to the enzyme activity in the absence of NaCl. The enzyme activity in the absence of NaCl was designated 100%.
Modeling of the three-dimensional structure of the wild-type ATII-LCL-NH enzyme.
The amino acid sequences of the ATII-LCL-NH and other mutants were used to obtain the Protein Data Bank (PDB) file using the free online software SWISS-MODEL. The three-dimensional structure of ATII-LCL-NH MerA was built by homology modeling against structures of the N-terminal domain of Tn501 mercuric reductase (PDB code 2kt2) (55) and the C-terminal core (amino acids 95 to 561; PDB code 1zk7) (56) domain of the Tn501 mercuric reductase, using SWISS-MODEL (57). Manual inspection of the output was performed in PyMOL (PyMOL Molecular Graphics System, version 1.5.0.1; Schrödinger, LLC). Rendering of the final three-dimensional models was performed using PyMOL (58).
Salt bridge and hydrogen bond prediction.
The PDB file of the ATII-LCL-NH enzyme and all the mutants was used as the input file to predict the putative salt bridges between oppositely charged residues in the entire dimer. A cutoff of 4 Å was used. Potential salt bridges were predicted using the online software ESBRI (35, 59, 60), available from http://bioinformatica.isa.cnr.it/ESBRI/input.html. Hydrogen bonds were predicted with the online software tool available at http://cib.cf.ocha.ac.jp/bitool/HBOND/ (33, 34), using the default parameters.
Accession number(s).
All nucleotide sequences obtained from the merA library were deposited in NCBI GenBank with accession numbers MF363130 to MF363137. Of note, the accession number of the gene encoding the main enzyme used in this paper (ATII-LCL-NH) is MF363137.
Supplementary Material
ACKNOWLEDGMENTS
We declare no conflict of interest.
M.M. and E.R. conceived and designed the experiments; M.M. and E.R. performed the experiments; M.M., A.E.H., R.K.A., and M.K.S. analyzed the data; A.E.H. and R.K.A. performed computational analyses; M.M., R.K.A., and E.R. wrote the manuscript. All authors read and approved the final manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02387-18.
REFERENCES
- 1.Anonymous. 2015. Red Sea New World Encyclopedia. http://www.newworldencyclopedia.org/p/index.php?title=Red_Sea&oldid=989212.
- 2.Bower A. 2009. R/V Oceanus Voyage 449-6 Red Sea Atlantis II Deep Complex Area, 19 October–1 November 2008. Woods Hole Oceanographic Institute Technical Report. Woods Hole Oceanographic Institute (WHOI) and King Abdullah University of Science and Technology, Woods Hole, MA: http://www.whoi.edu/science/PO/people/www-abower/abower/techreps/KAUST_CTR0901_press.pdf. [Google Scholar]
- 3.Botz R, Schmidt M, Wehner H, Hufnagel H, Stoffers P. 2007. Organic-rich sediments in brine-filled Shaban- and Kebrit deeps, northern Red Sea. Chem Geol 244:520–553. doi: 10.1016/j.chemgeo.2007.07.004. [DOI] [Google Scholar]
- 4.Speth DR, Lagkouvardos I, Wang Y, Qian P-Y, Dutilh BE, Jetten MSM. 2017. Draft genome of Scalindua rubra, obtained from the interface above the Discovery Deep brine in the Red Sea, sheds light on potential salt adaptation strategies in Anammox bacteria. Microb Ecol 74:1–5. doi: 10.1007/s00248-017-0929-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Cao H, Zhang G, Bougouffa S, Lee OO, Al-Suwailem A, Qian PY. 2013. Autotrophic microbe metagenomes and metabolic pathways differentiate adjacent Red Sea brine pools. Sci Rep 3:1748. doi: 10.1038/srep01748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartmann M, Scholten JC, Stoffers P, Wehner F. 1998. Hydrographic structure of brine-filled deeps in the Red Sea—new results from the Shaban, Kebrit, Atlantis II, and Discovery Deep. Mar Geol 144:311–330. doi: 10.1016/S0025-3227(97)00055-8. [DOI] [Google Scholar]
- 7.Wang Y, Yang J, Lee OO, Dash S, Lau SC, Al-Suwailem A, Wong TY, Danchin A, Qian PY. 2011. Hydrothermally generated aromatic compounds are consumed by bacteria colonizing in Atlantis II Deep of the Red Sea. ISME J 5:1652–1659. doi: 10.1038/ismej.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Li JT, He LS, Yang B, Gao ZM, Cao HL, Batang Z, Al-Suwailem A, Qian PY. 2015. Zonation of microbial communities by a hydrothermal mound in the Atlantis II Deep (the Red Sea). PLoS One 10:e0140766. doi: 10.1371/journal.pone.0140766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laurila TE, Hannington MD, Leybourne M, Petersen S, Devey CW, Garbe-Schönberg D. 2015. New insights into the mineralogy of the Atlantis II Deep metalliferous sediments, Red Sea. Geochem Geophys Geosyst 16:4449–4478. doi: 10.1002/2015GC006010. [DOI] [Google Scholar]
- 10.Antunes A, Ngugi DK, Stingl U. 2011. Microbiology of the Red Sea (and other) deep-sea anoxic brine lakes. Environ Microbiol Rep 3:416–433. doi: 10.1111/j.1758-2229.2011.00264.x. [DOI] [PubMed] [Google Scholar]
- 11.Siam R, Mustafa GA, Sharaf H, Moustafa A, Ramadan AR, Antunes A, Bajic VB, Stingl U, Marsis NG, Coolen MJ, Sogin M, Ferreira AJ, Dorry HE. 2012. Unique prokaryotic consortia in geochemically distinct sediments from Red Sea Atlantis II and discovery deep brine pools. PLoS One 7:e42872. doi: 10.1371/journal.pone.0042872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nascimento AM, Chartone-Souza E. 2003. Operon mer: bacterial resistance to mercury and potential for bioremediation of contaminated environments. Genet Mol Res 2:92–101. [PubMed] [Google Scholar]
- 13.Mathema VB, Thakuri BC, Sillanpaa M. 2011. Bacterial mer operon-mediated detoxification of mercurial compounds: a short review. Arch Microbiol 193:837–844. doi: 10.1007/s00203-011-0751-4. [DOI] [PubMed] [Google Scholar]
- 14.Moller AK, Barkay T, Hansen MA, Norman A, Hansen LH, Sorensen SJ, Boyd ES, Kroer N. 2014. Mercuric reductase genes (merA) and mercury resistance plasmids in High Arctic snow, freshwater and sea-ice brine. FEMS Microbiol Ecol 87:52–63. doi: 10.1111/1574-6941.12189. [DOI] [PubMed] [Google Scholar]
- 15.Zheng R, Wu S, Ma N, Sun C. 2018. Genetic and physiological adaptations of marine bacterium Pseudomonas stutzeri 273 to mercury stress. Front Microbiol 9:682. doi: 10.3389/fmicb.2018.00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Varekamp JC, Buseck PR. 1984. The speciation of mercury in hydrothermal systems, with applications to ore deposition. Geochim Cosmochim Acta 48:177–185. doi: 10.1016/0016-7037(84)90359-4. [DOI] [Google Scholar]
- 17.Gworek B, Bemowska-Kałabun O, Kijeńska M, Wrzosek-Jakubowska J. 2016. Mercury in marine and oceanic waters—a review. Water Air Soil Pollut 227:371. doi: 10.1007/s11270-016-3060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ay N. 1962. Content of mercury in some natural waters. Chem Abstr 57:16336. [Google Scholar]
- 19.Pempkowiak J, Cossa D, Sikora A, Sanjuan J. 1998. Mercury in water and sediments of the southern Baltic sea. Sci Total Environ 213:185–192. [DOI] [PubMed] [Google Scholar]
- 20.Ci Z, Zhang X, Wang Z, Niu Z. 2011. Phase speciation of mercury (Hg) in coastal water of the Yellow Sea, China. Mar Chem 126:250–255. doi: 10.1016/j.marchem.2011.06.004. [DOI] [Google Scholar]
- 21.Brown NL, Ford SJ, Pridmore RD, Fritzinger DC. 1983. Nucleotide sequence of a gene from the Pseudomonas transposon Tn501 encoding mercuric reductase. Biochemistry 22:4089–4095. [DOI] [PubMed] [Google Scholar]
- 22.Schiering N, Kabsch W, Moore MJ, Distefano MD, Walsh CT, Pai EF. 1991. Structure of the detoxification catalyst mercuric ion reductase from Bacillus sp. strain RC607. Nature 352:168–172. doi: 10.1038/352168a0. [DOI] [PubMed] [Google Scholar]
- 23.Miller SM, Moore MJ, Massey V, Williams CH Jr, Distefano MD, Ballou DP, Walsh CT. 1989. Evidence for the participation of Cys558 and Cys559 at the active site of mercuric reductase. Biochemistry 28:1194–1205. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Freedman Z, Lu-Irving P, Kaletsky R, Barkay T. 2009. An initial characterization of the mercury resistance (mer) system of the thermophilic bacterium Thermus thermophilus HB27. FEMS Microbiol Ecol 67:118–129. doi: 10.1111/j.1574-6941.2008.00603.x. [DOI] [PubMed] [Google Scholar]
- 25.Li WF, Zhou XX, Lu P. 2005. Structural features of thermozymes. Biotechnol Adv 23:271–281. doi: 10.1016/j.biotechadv.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Vieille C, Burdette DS, Zeikus JG. 1996. Thermozymes. Biotechnol Annu Rev 2:1–83. [DOI] [PubMed] [Google Scholar]
- 27.Huang R, Yang Q, Feng H. 2015. Single amino acid mutation alters thermostability of the alkaline protease from Bacillus pumilus: thermodynamics and temperature dependence. Acta Biochim Biophys Sin (Shanghai) 47:98–105. doi: 10.1093/abbs/gmu120. [DOI] [PubMed] [Google Scholar]
- 28.Trivedi S, Gehlot HS, Rao SR. 2006. Protein thermostability in Archaea and Eubacteria. Genet Mol Res 5:816–827. [PubMed] [Google Scholar]
- 29.Suzuki Y. 1989. A general principle of increasing protein thermostability. Proc Jpn Acad Ser B Phys Biol Sci 65:146–148. [Google Scholar]
- 30.Sayed A, Ghazy MA, Ferreira AJ, Setubal JC, Chambergo FS, Ouf A, Adel M, Dawe AS, Archer JA, Bajic VB, Siam R, El-Dorry H. 2014. A novel mercuric reductase from the unique deep brine environment of Atlantis II in the Red Sea. J Biol Chem 289:1675–1687. doi: 10.1074/jbc.M113.493429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Artz JH, White SN, Zadvornyy OA, Fugate CJ, Hicks D, Gauss GH, Posewitz MC, Boyd ES, Peters JW. 2015. Biochemical and structural properties of a thermostable mercuric ion reductase from Metallosphaera sedula. Front Bioeng Biotechnol 3:97. doi: 10.3389/fbioe.2015.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bafana A, Khan F, Suguna K. 2017. Structural and functional characterization of mercuric reductase from Lysinibacillus sphaericus strain G1. Biometals 30:809–819. doi: 10.1007/s10534-017-0050-x. [DOI] [PubMed] [Google Scholar]
- 33.Baker EN, Hubbard RE. 1984. Hydrogen bonding in globular proteins. Prog Biophys Mol Biol 44:97–179. [DOI] [PubMed] [Google Scholar]
- 34.Petsko GA, Ringe D. 2004. Bonds that stabilize folded proteins, p 10–11. Protein structure and function. New Science Press, London, United Kingdom. [Google Scholar]
- 35.Costantini S, Colonna G, Facchiano AM. 2008. ESBRI: a web server for evaluating salt bridges in proteins. Bioinformation 3:137–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Celniker G, Nimrod G, Ashkenazy H, Glaser F, Martz E, Mayrose I, Pupko T, Ben-Tal N. 2013. ConSurf: using evolutionary data to raise testable hypotheses about protein function. Isr J Chem 53:199–206. doi: 10.1002/ijch.201200096. [DOI] [Google Scholar]
- 37.Madern D, Ebel C, Zaccai G. 2000. Halophilic adaptation of enzymes. Extremophiles 4:91–98. [DOI] [PubMed] [Google Scholar]
- 38.Mevarech M, Frolow F, Gloss LM. 2000. Halophilic enzymes: proteins with a grain of salt. Biophys Chem 86:155–164. doi: 10.1016/S0301-4622(00)00126-5. [DOI] [PubMed] [Google Scholar]
- 39.Fukuchi S, Yoshimune K, Wakayama M, Moriguchi M, Nishikawa K. 2003. Unique amino acid composition of proteins in halophilic bacteria. J Mol Biol 327:347–357. doi: 10.1016/S0022-2836(03)00150-5. [DOI] [PubMed] [Google Scholar]
- 40.Siglioccolo A, Paiardini A, Piscitelli M, Pascarella S. 2011. Structural adaptation of extreme halophilic proteins through decrease of conserved hydrophobic contact surface. BMC Struct Biol 11:50. doi: 10.1186/1472-6807-11-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeroual YMA, Dzairi FZ, Talbi M, Chung PU, Lee K. 2003. Purification and characterization of cytosolic mercuric reductase from Klebsiella pneumoniae. Ann Microbiol 53:149–160. [Google Scholar]
- 42.Takano K, Higashi R, Okada J, Mukaiyama A, Tadokoro T, Koga Y, Kanaya S. 2009. Proline effect on the thermostability and slow unfolding of a hyperthermophilic protein. J Biochem 145:79–85. doi: 10.1093/jb/mvn144. [DOI] [PubMed] [Google Scholar]
- 43.Watanabe K, Masuda T, Ohashi H, Mihara H, Suzuki Y. 1994. Multiple proline substitutions cumulatively thermostabilize Bacillus cereus ATCC7064 oligo-1,6-glucosidase. Irrefragable proof supporting the proline rule. Eur J Biochem 226:277–283. [DOI] [PubMed] [Google Scholar]
- 44.Watanabe K, Kitamura K, Suzuki Y. 1996. Analysis of the critical sites for protein thermostabilization by proline substitution in oligo-1,6-glucosidase from Bacillus coagulans ATCC 7050 and the evolutionary consideration of proline residues. Appl Environ Microbiol 62:2066–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hardy F, Vriend G, Veltman OR, van der Vinne B, Venema G, Eijsink VG. 1993. Stabilization of Bacillus stearothermophilus neutral protease by introduction of prolines. FEBS Lett 317:89–92. doi: 10.1016/0014-5793(93)81497-N. [DOI] [PubMed] [Google Scholar]
- 46.Imanaka T. 2011. Molecular bases of thermophily in hyperthermophiles. Proc Jpn Acad Ser B Phys Biol Sci 87:587–602. doi: 10.2183/pjab.87.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar S, Ma B, Tsai CJ, Nussinov R. 2000. Electrostatic strengths of salt bridges in thermophilic and mesophilic glutamate dehydrogenase monomers. Proteins 38:368–383. [DOI] [PubMed] [Google Scholar]
- 48.Xu D, Tsai CJ, Nussinov R. 1997. Hydrogen bonds and salt bridges across protein-protein interfaces. Protein Eng 10:999–1012. [DOI] [PubMed] [Google Scholar]
- 49.Kamanda Ngugi D, Blom J, Alam I, Rashid M, Ba-Alawi W, Zhang G, Hikmawan T, Guan Y, Antunes A, Siam R, El Dorry H, Bajic V, Stingl U. 2015. Comparative genomics reveals adaptations of a halotolerant thaumarchaeon in the interfaces of brine pools in the Red Sea. ISME J 9:396–411. doi: 10.1038/ismej.2014.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sonbol SA, Ferreira AJ, Siam R. 2016. Red Sea Atlantis II brine pool nitrilase with unique thermostability profile and heavy metal tolerance. BMC Biotechnol 16:14. doi: 10.1186/s12896-016-0244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Corpet F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16:10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mount DW. 2007. Using the Basic Local Alignment Search Tool (BLAST). CSH Protoc 2007:pdb.top17. doi: 10.1101/pdb.top17. [DOI] [PubMed] [Google Scholar]
- 53.Kalmbach S, Manz W, Wecke J, Szewzyk U. 1999. Aquabacterium gen. nov., with description of Aquabacterium citratiphilum sp. nov., Aquabacterium parvum sp. nov. and Aquabacterium commune sp. nov., three in situ dominant bacterial species from the Berlin drinking water system. Int J Syst Bacteriol 49:769–777. doi: 10.1099/00207713-49-2-769. [DOI] [PubMed] [Google Scholar]
- 54.Fox B, Walsh CT. 1982. Mercuric reductase. Purification and characterization of a transposon-encoded flavoprotein containing an oxidation-reduction-active disulfide. J Biol Chem 257:2498–2503. [PubMed] [Google Scholar]
- 55.Ledwidge R, Hong B, Dotsch V, Miller SM. 2010. NmerA of Tn501 mercuric ion reductase: structural modulation of the pKa values of the metal binding cysteine thiols. Biochemistry 49:8988–8998. doi: 10.1021/bi100537f. [DOI] [PubMed] [Google Scholar]
- 56.Ledwidge R, Patel B, Dong A, Fiedler D, Falkowski M, Zelikova J, Summers AO, Pai EF, Miller SM. 2005. NmerA, the metal binding domain of mercuric ion reductase, removes Hg2+ from proteins, delivers it to the catalytic core, and protects cells under glutathione-depleted conditions. Biochemistry 44:11402–11416. doi: 10.1021/bi050519d. [DOI] [PubMed] [Google Scholar]
- 57.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, Schwede T. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holec PV, Hackel BJ. 2016. PyMOL360: multi-user gamepad control of molecular visualization software. J Comput Chem 37:2667–2669. doi: 10.1002/jcc.24489. [DOI] [PubMed] [Google Scholar]
- 59.Sarakatsannis JN, Duan Y. 2005. Statistical characterization of salt bridges in proteins. Proteins 60:732–739. doi: 10.1002/prot.20549. [DOI] [PubMed] [Google Scholar]
- 60.Kumar S, Nussinov R. 2002. Relationship between ion pair geometries and electrostatic strengths in proteins. Biophys J 83:1595–1612. doi: 10.1016/S0006-3495(02)73929-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y, Bond CS, Bailey S, Cunningham ML, Fairlamb AH, Hunter WN. 1996. The crystal structure of trypanothione reductase from the human pathogen Trypanosoma cruzi at 2.3 Å resolution. Protein Sci 5:52–61. doi: 10.1002/pro.5560050107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.