Multiple gene modifications of specific fungal strains are required for achieving industrial-scale production of enzymes and secondary metabolites. In the present study, we developed an efficient multiple genetic engineering technique for the filamentous fungus Aspergillus oryzae. The approach is based on a clustered regulatory interspaced short palindromic repeats (CRISPR)/Cas9 system and recycling of an AMA1-based autonomous replicating plasmid. Because the plasmid harbors a drug resistance marker (ptrA), the approach does not require the construction of auxotrophic industrial strains prior to genome editing and allows for forced recycling of the gene-editing plasmid. The established plasmid-recycling technique involves an Aoace2-conditional expression cassette, whose induction severely impairs fungal growth. We used the developed genetic engineering techniques for highly efficient marker-free multiple gene deletion/integration in A. oryzae. The genome-editing approaches established in the present study, which enable unlimited repeatable genetic engineering, will facilitate multiple gene modification of industrially important fungal strains.
KEYWORDS: Aspergillus oryzae, CRISPR/Cas9, marker-free engineering, multiplex genome editing, plasmid recycling
ABSTRACT
Filamentous fungi are used for food fermentation and industrial production of recombinant proteins. They also serve as a source of secondary metabolites and are recently expected as hosts for heterologous production of useful secondary metabolites. Multiple-step genetic engineering is required to enhance industrial production involving these fungi, but traditional sequential modification of multiple genes using a limited number of selection markers is laborious. Moreover, efficient genetic engineering techniques for industrial strains have not yet been established. We have previously developed a clustered regulatory interspaced short palindromic repeats (CRISPR)/Cas9-based mutagenesis technique for the industrial filamentous fungus Aspergillus oryzae, enabling mutation efficiency of 10 to 20%. Here, we improved the CRISPR/Cas9 approach by including an AMA1-based autonomously replicating plasmid harboring the drug resistance marker ptrA. By using the improved mutagenesis technique, we successfully modified A. oryzae wild and industrial strains, with a mutation efficiency of 50 to 100%. Conditional expression of the Aoace2 gene from the AMA1-based plasmid severely inhibited fungal growth. This enabled forced recycling of the plasmid, allowing repeated genome editing. Further, double mutant strains were successfully obtained with high efficiency by expressing two guide RNA molecules from the genome-editing plasmid. Cotransformation of fungal cells with the genome-editing plasmid together with a circular donor DNA enabled marker-free multiplex gene deletion/integration in A. oryzae. The presented repeatable marker-free genetic engineering approach for mutagenesis and gene deletion/integration will allow for efficient modification of multiple genes in industrial fungal strains, increasing their applicability.
IMPORTANCE Multiple gene modifications of specific fungal strains are required for achieving industrial-scale production of enzymes and secondary metabolites. In the present study, we developed an efficient multiple genetic engineering technique for the filamentous fungus Aspergillus oryzae. The approach is based on a clustered regulatory interspaced short palindromic repeats (CRISPR)/Cas9 system and recycling of an AMA1-based autonomous replicating plasmid. Because the plasmid harbors a drug resistance marker (ptrA), the approach does not require the construction of auxotrophic industrial strains prior to genome editing and allows for forced recycling of the gene-editing plasmid. The established plasmid-recycling technique involves an Aoace2-conditional expression cassette, whose induction severely impairs fungal growth. We used the developed genetic engineering techniques for highly efficient marker-free multiple gene deletion/integration in A. oryzae. The genome-editing approaches established in the present study, which enable unlimited repeatable genetic engineering, will facilitate multiple gene modification of industrially important fungal strains.
INTRODUCTION
Filamentous fungi efficiently secrete proteins and have been used for food fermentation and industrial production of recombinant proteins (1). In addition, they serve as a source of secondary metabolites and are recently expected as hosts for heterologous production of useful secondary metabolites (2, 3). Development of transformation methods and several selection markers allowed for homologous recombination-based genetic engineering of many filamentous fungal species. Generation of deletion mutants of genes related to nonhomologous end joining (NHEJ), such as ku70-ku80 and ligD, greatly enhances the engineering efficiency (4). These efforts facilitated many attempts to increase the production of proteins and secondary metabolites, e.g., overexpression of chaperone proteins and deletion of multiple protease-encoding genes for increased production of recombinant proteins (1), and introduction of multiple biosynthetic genes for the production of secondary metabolites (3), whereas multiple steps of genetic engineering involving a limited number of selection markers are required for such improvements. In addition, although the full potential of industrially exploited strains has possibly not yet been unlocked, genetic engineering of such strains remains laborious because of the difficulty in application of advanced genetic technologies in these strains.
A bacterial and archaeal immune mechanism, clustered regulatory interspaced short palindromic repeats (CRISPR)/Cas9, has been developed into a powerful genetic engineering system (5). Two components, Cas9 nuclease and a single chimeric guide RNA (sgRNA), required for the recruitment of Cas9 to a specific genomic locus, allow for the introduction of targeted DNA double-strand breaks (DSBs). This, in turn, enables genome editing, including mutagenesis by NHEJ repair or gene deletion/integration by homology-directed repair (HDR) (5). The CRISPR/Cas9 system has been applied to various organisms, such as mammalian cells, plants, fish, and yeast (5). In addition, genetic engineering using this system was also performed in industrial microbes (6).
Mutagenesis and gene deletion/integration using the CRISPR/Cas9 system have recently been reported in filamentous fungi (6, 7). In the case of mutagenesis, mutants containing deletion or insertion of nucleotide sequences of various lengths were obtained by introducing Cas9 and sgRNA expression cassettes (8–16) or a Cas9 expression cassette and synthesized sgRNA (17, 18). However, the mutation efficiencies of such systems are low: 10 to 20% in Aspergillus oryzae (10), 25 to 67% in Aspergillus fumigatus (9, 12), and 8% in Phytophthora sojae (14). Such efficiencies are lower than that of genetic engineering in mutants lacking NHEJ-related genes (4). In addition, genes, whose deletion affects the phenotype of transformant, such as pigment, growth, and drug resistance, have been selected as the target genes (7). Therefore, mutagenesis that does not result in phenotypic changes is probably more difficult to screen. The CRISPR/Cas9 system involving a donor DNA integrated into the recipient genome via HDR was also used for the introduction of mutation (19, 20), gene deletion (8, 12, 14, 15, 18, 21–25), and gene integration (26–28). In these studies, positively selectable markers, such as pyrG (URA5) (8, 18, 21, 22) and amdS (24), and drug-resistant selection markers, such as the pyrithiamine resistance marker ptrA (19), the hygromycin resistance marker hph (12, 22, 25, 26), and the G418 resistance marker neo (NPTII) (14, 15, 27), were inserted into the donor DNA, resulting in higher efficiencies of genetic engineering. In contrast, Pohl et al. reported marker-free gene deletion using a donor DNA without any selection markers in Penicillium chrysogenum (24). Recently, marker-free introduction of a stop-codon triplet was efficiently performed by using a single-stranded oligonucleotide as the template in the NHEJ-deficient Aspergillus strains (20).
Genetic engineering of multiple genes requires multiplex engineering or reintroduction of sgRNA expression cassette. Simultaneous deletion of up to four genes using a single selection marker has been reported in filamentous fungi (15, 18). However, increased number of target genes leads to reduced efficiency of multiplex engineering (15), indicating that the number of genes that can be simultaneously engineered is limited. Therefore, a method to recycle the sgRNA expression cassettes is required for efficient engineering of multiple genes.
Plasmids containing the AMA1 sequence from Aspergillus nidulans (29), which allows for autonomous plasmid replication, are often used for the expression of Cas9 and sgRNA (8, 12, 20–22, 24–26). Since multiple copies of the AMA1-based plasmid are present in a cell (29), this would contribute to increasing the CRISPR/Cas9-mediated mutation efficiency due to high expression of these factors. In addition, several rounds of subculturing under nonselective conditions results in the removal of the AMA1-based plasmid from transformants (30). Weyda et al. reported natural loss of the AMA1-based plasmid from Aspergillus carbonarius transformants by three rounds of subculturing under nonselective conditions (22). Forced recycling of the genome-editing plasmid would theoretically facilitate infinite repeatability of genetic engineering steps. Finally, drug resistance markers are preferred for genetic modifications of industrial strains, since obtaining auxotrophic mutants is laborious. Nonetheless, forced recycling of an AMA1-based plasmid containing a drug resistance marker had not been reported in filamentous fungi.
Although numerous A. oryzae strains are used industrially for Japanese traditional food fermentation and the production of recombinant proteins, an efficient genetic engineering method has not been established for such strains. In the present study, we performed a highly efficient mutagenesis of A. oryzae wild and industrial strains using AMA1-based genome-editing plasmids. We also successfully performed simultaneous double mutagenesis of these strains by expressing two sgRNAs from a single genome-editing plasmid. Further, we developed a forced recycling method for the AMA1-based genome-editing plasmid harboring the drug resistance marker and performed efficient gene deletion/integration using a donor DNA without inserting any selection markers, allowing for repeatable genetic engineering.
RESULTS
Highly efficient mutagenesis involving AMA1-based genome-editing plasmids.
We previously performed CRISPR/Cas9-mediated mutagenesis by integrating a genome-editing plasmid into the genome of A. oryzae; however, the mutation efficiencies of this approach were 10 to 20% (10). The strategy for mutagenesis by integrating the cas9 and sgRNA expression cassettes into the genome also resulted in low efficiency (8%) in P. sojae (14). In contrast, use of the AMA1-based genome-editing plasmid allowed for efficient mutagenesis (8), possibly due to high expression of cas9 and sgRNA from multiple copies. In the present study, we therefore constructed a new genome-editing vector, ppAsAcas9, containing the AMA1 sequence for autonomous plasmid replication to improve the efficiency of CRISPR/Cas9-mediated mutagenesis (Fig. 1A). Since truncated AMA1 is also functional (26, 29), we used half of the AMA1 sequence (half AMA1), and the pyrithiamine resistance marker ptrA was used for screening. Further, sgRNA expression cassettes were inserted into ppAsACas9, yielding plasmids ppAsAC9gwA, ppAsAC9gyA, and ppAsAC9gpG, for the mutagenesis of the wA, yA, and pyrG genes, respectively (Fig. 1A). When the plasmids were introduced into the A. oryzae wild strain RIB40, we successfully obtained transformants exhibiting the expected phenotypes, i.e., white conidium formation (wA mutants), yellow conidium formation (yA mutants), and uridine/uracil auxotrophy (pyrG mutants) (Fig. 1B). Mutation efficiencies in RIB40 were 87.5, 100, and 55.6% for wA, yA, and pyrG genes, respectively (Fig. 1C). Various mutation patterns were observed, and most mutants contained short deletions of up to 26 bp (see Fig. S1 in the supplemental material). In some yA mutants, the regions adjacent to the target sequences were largely deleted, and large fragments from other regions of the genome were inserted. The plasmid ppAsAC9gwA for wA mutagenesis was also introduced into A. oryzae industrial strains RIB128 and RIB915, which are used for sake and soy sauce fermentation, respectively (http://www.nrib.go.jp/data/asp/strain.html). Transformants exhibiting white conidium formation were obtained with the mutation efficiencies of 94.1 and 58.3% for RIB128 and RIB915, respectively (Fig. 1C and D). These results indicate that the AMA1-based genome-editing plasmid enabled highly efficient mutagenesis in A. oryzae wild and industrial strains.
FIG 1.
Mutagenesis of A. oryzae wild (RIB40) and industrial (RIB128 and RIB915) strains using AMA1-based genome-editing plasmids. (A) Construction of AMA1-based genome-editing plasmid. (B) Phenotypes of wA, yA, and pyrG mutants generated from the wild strain. A conidial suspension (104 conidia in 5 μl) of each strain was spotted onto potato dextrose (PD) agar medium for the wA and yA mutants and on PD either with uridine and uracil (+Uri/Ura) or without uridine and uracil (−Uri/Ura) for the pyrG mutant, followed by incubation for 5 days at 30°C. (C) Efficiencies of mutation using the AMA1-based genome-editing plasmids. (D) Phenotypes of the wA mutants generated from the industrial strains. A conidial suspension (104 conidia in 5 μl) of each strain was spotted onto PD agar medium, followed by incubation for 5 days at 30°C.
Multiple mutagenesis by recycling of genome-editing plasmids.
The AMA1-based plasmid is reportedly removed from transformants by continuous subculturing on a nonselective medium (30). Efficient plasmid removal can be achieved using counterselective markers, such as pyrG and niaD, but obtaining the corresponding auxotrophic strains is laborious. Therefore, we attempted to design an efficient plasmid recycling system when a drug resistance marker is used. In a transcription factor screen, we have previously identified AO090003000678 as a gene whose deletion leads to hyperproduction of sclerotia (H. Nakamura and J. Maruyama, unpublished data). Since the orthologues of AO090003000678 are conserved in filamentous fungi, including Aspergillus fumigatus, of which Ace2 was identified as an orthologue to Ace2 of Schizosaccharomyces pombe (31), we designated AO090003000678 Aoace2 (Fig. S2A). When Aoace2 was expressed under the control of amyB promoter (PamyB), which is induced strongly by dextrin, induced intermediately by glucose, and repressed by glycerol (32), the harboring strain exhibited severe growth defects on a medium containing dextrin but not glycerol as the sole carbon source (Fig. S2B). As the induction of Aoace2 expression resulted in cell lysis (Fig. S2C), growth defect by overexpression of Aoace2 is possibly due to its involvement in cell wall maintenance, which was suggested in A. fumigatus and S. pombe (31, 33). Thus, conditional expression of Aoace2 from the AMA1-based plasmid could have enabled plasmid removal. To test this, the Aoace2 expression cassette under the control of PamyB was inserted into the genome-editing plasmid, yielding plasmid ppAsATC9a2. Therein, the cas9 gene was placed under the control of promoter of AO090120000080 (Aotef1) encoding a translation elongation factor EF-1-α (Fig. 2A). An sgRNA expression cassette was then inserted into ppAsATC9a2 (Fig. 2A).
FIG 2.
Generation of wA niaD double mutant of the A. oryzae wild strain RIB40 by recycling the genome-editing plasmid. (A) Construction of the genome-editing plasmid containing the Aoace2 conditional expression cassette. (B) Pyrithiamine sensitivity test of transformants before and after plasmid removal. A conidial suspension (104 conidia in 5 μl) of each strain was spotted onto the Czapek-Dox (CD) agar medium with (+) or without (−) pyrithiamine, followed by incubation for 5 days at 30°C. (C) Nucleotide sequence analysis of the target sequence within the wA gene in the indicated strains. Gray letters indicate the deleted nucleotides in the generated mutants. (D) Southern blot analysis to detect the removal of the genome-editing plasmid. The indicated probe detected a 6.8-kb genomic DNA fragment in the three indicated strains and a 4.9-kb fragment of the genome-editing plasmid in the plasmid-containing strain RIB40wA7C9-1. (E) Inability to assimilate nitrate by the wA niaD double mutant. A conidial suspension (104 conidia in 5 μl) of each strain was spotted onto the indicated agar medium, followed by incubation for 5 days at 30°C. (F) Nucleotide sequence analysis of the target sequence within the niaD gene in the indicated strains. Gray letters indicate the deleted nucleotide in the generated mutants.
For multiple mutagenesis by plasmid recycling, we performed three steps: first-gene mutagenesis, removal of the plasmid for first-gene mutagenesis, and second-gene mutagenesis (Fig. S3). The wA and niaD genes were selected as target genes. First, the genome-editing plasmid for wA mutagenesis ppAsATC9a2gwA7, containing the Aoace2 expression cassette, was introduced into the strain RIB40 under PamyB-repressing conditions to retain the plasmid. As expected, a wA mutant RIB40wA7C9-1 that formed white conidia was obtained; a 17-bp deletion in the wA gene was confirmed in the mutant (Fig. 2B and C). The mutant exhibited pyrithiamine resistance, indicating that the genome-editing plasmid was retained (Fig. 2B). The mutant was then incubated under a PamyB-inducing condition to remove the genome-editing plasmid, yielding a strain RIB40wA7-1. The strain exhibited pyrithiamine sensitivity (Fig. 2B), and a band specific for the genome-editing plasmid was not detected by Southern blot analysis (Fig. 2D). These results indicated that the genome-editing plasmid was absent in RIB40wA7-1. In addition, the 17-bp deletion in wA was retained after plasmid removal (Fig. 2C). Subsequently, the genome-editing plasmid for niaD mutagenesis ppAsATC9a2gniaD was introduced into the plasmid-devoid wA mutant RIB40wA7-1. The transformant was grown under PamyB-inducing conditions to remove the genome-editing plasmid, yielding RIB40wAnD-1. The resulting strain did not grow on the Czapek-Dox (CD) minimal medium, in which NaNO3 is the sole nitrogen source (Fig. 2E). Indeed, RIB40wAnD-1 harbored a 1-bp deletion in the target sequence within the niaD gene (Fig. 2F). We also attempted to use plasmid recycling to generate a wA niaD double mutant derived from the A. oryzae industrial strain RIB128, and we successfully obtained the double mutant containing 1-bp insertion and 1-bp deletion within the target sequences of wA and niaD, respectively (Fig. S4). These findings indicated that the recycling of the genome-editing plasmid enabled multiple gene mutagenesis in A. oryzae strains.
Double mutagenesis in a single transformation step using a genome-editing plasmid expressing two sgRNA molecules.
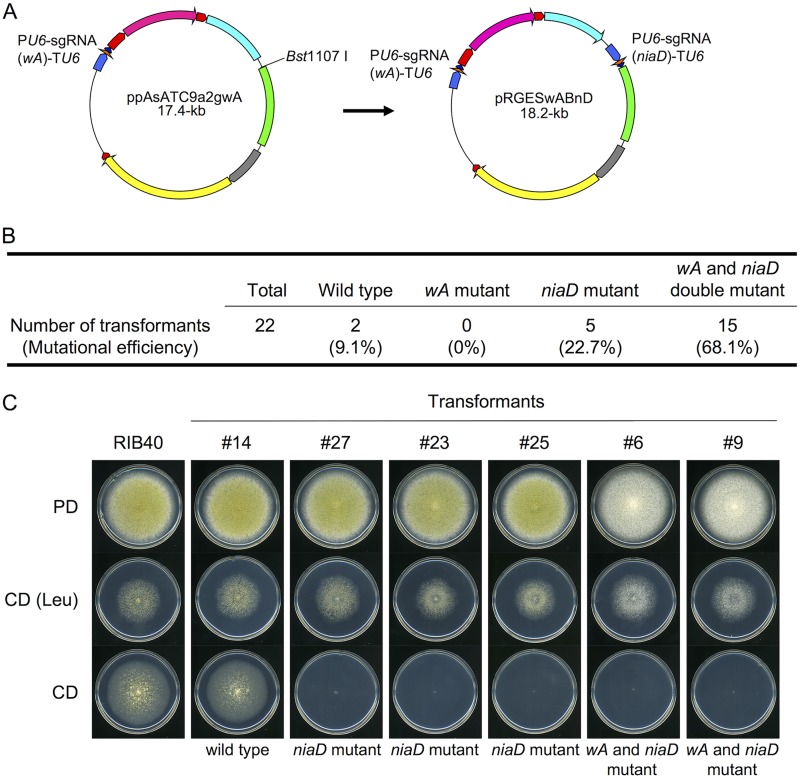
In the discussion above, we presented a method for repeatable genetic engineering by recycling of a genome-editing plasmid, while construction of double mutants using this strategy relied on tandem transformation. Multiplex mutagenesis in a single transformation step would increase the efficiency of genetic engineering. When the strain RIB40 was cotransformed with two genome-editing plasmids, ppAsATC9a2gwA7 and ppAsATC9a2gniaD, for multiplex mutagenesis, only single mutants of either wA or niaD, and not wA niaD double mutants, were obtained (M. B. Arnaud, T. Katayama, and J. Maruyama, data not shown). Therefore, we next attempted to simultaneously express two sgRNAs from a single genome-editing plasmid. Consequently, the genome-editing plasmid pRGESwABnD for simultaneous mutagenesis of wA and niaD was constructed (Fig. 3A) and introduced into the strain RIB40. As target sequences that included specific restriction enzyme sites were chosen in the experiment, successful mutagenesis could be detected by restriction enzyme digestion of amplified PCR products from genomic DNA (Fig. S5A). Among 22 transformants, 15 transformants were wA niaD double mutants (68.1%), 5 were niaD single mutants (22.7%), and 2 were not mutated (9.1%) (Fig. 3B and C; see also Fig. S5B). The majority of transformants exhibited the expected phenotypes: white conidium formation (wA mutants) and the inability to assimilate nitrate (niaD mutants) (Fig. S5B). As an exception, transformant 27 appeared to be the wild type, as determined by the digestion of PCR products (Fig. S5B), but it exhibited the phenotype of a niaD mutant (Fig. 3C). This was probably due to a newly generated restriction site after mutagenesis at the niaD locus. We also investigated the feasibility of multiplex mutagenesis in the industrial strain RIB128. When the plasmid pRGESpGBwA for wA pyrG double mutagenesis was introduced into the strain, three of six transformants exhibited the phenotypes of a double mutant, white conidium formation and uridine/uracil auxotrophy (50%) (Fig. S6). These findings indicated that the genome-editing plasmid for simultaneous expression of two sgRNA molecules enabled efficient multiplex mutagenesis of A. oryzae strains.
FIG 3.
Multiplex mutagenesis of wA and niaD in the A. oryzae wild strain RIB40. (A) Construction of the genome-editing plasmid containing two sgRNA expression cassettes. (B) Efficiencies of mutation of multiplex mutagenesis of wA and niaD. (C) Phenotypes of transformants obtained by multiplex mutagenesis. The conidia of each strain were spotted onto the indicated agar medium, followed by incubation for 5 days at 30°C.
Marker-free deletion of target genes using genome-editing plasmids and donor DNA.
In filamentous fungi, the CRISPR/Cas9 system can be used for efficient gene deletion/integration by HDR using a donor DNA that contains a selection marker (7). However, multiple rounds of genetic engineering require marker-free deletion/integration. Therefore, we constructed the donor plasmid pwAupDN for the deletion of the 5′ region of the wA gene and introduced it into the strain RIB40, together with the genome-editing plasmid pRGE-gwAup for DSB, upstream of wA (Fig. 4A). Since circular donor DNA is more frequently integrated into the target locus than linear donor DNA (8), we used circular donor DNA. In all 10 transformants, the 5′ region of wA was deleted and white conidium formation was observed (Fig. 4A to C). We also constructed the donor plasmid pnDdownDN for the deletion of the 3′ region of the niaD gene and introduced it into the strain RIB40 together with the genome-editing plasmid pRGE-gnDdown for DSB downstream of niaD (Fig. S7A). In all 10 transformants, the 3′ region of niaD was deleted, and inability to assimilate nitrate was observed (Fig. S7B and C). These results indicate that cointroduction of donor and genome-editing plasmids enabled efficient marker-free gene deletion in A. oryzae strains.
FIG 4.
Gene deletion in the A. oryzae wild strain RIB40 by cotransformation with a genome-editing plasmid, together with a circular donor DNA. (A) Strategy for deletion of the 5′ region of the wA gene by using the CRISPR/Cas9 system. (B) PCR analysis of genomic DNA using the primers indicated in panel A for partial deletion of wA. (C) Phenotype of transformants resulting from partial deletion of wA. A conidial suspension (104 conidia in 5 μl) of each strain was spotted onto PD agar medium, followed by incubation for 5 days at 30°C. (D) PCR analysis of genomic DNA using the primers indicated in panel A and Fig. S7A for double deletion of wA and niaD. (E) Phenotypes of transformants resulting from double deletion of wA and niaD. A conidial suspension (104 conidia in 5 μl) of each strain was spotted onto the indicated agar medium, followed by incubation for 5 days at 30°C.
In addition, for double deletion, we constructed a genome-editing plasmid expressing two sgRNAs, pRGE-gwAupgnDdown, for DSB upstream of wA and downstream of niaD. The genome-editing plasmid was then introduced, together with the two donor plasmids, pwAupDN and pnDdownDN, into the strain RIB40. Both the 5′ region of wA and the 3′ region of niaD were deleted in 13 of 21 transformants (61.9%) (Fig. 4D). The selected transformants exhibited the expected phenotypes, i.e., white conidium formation and inability to assimilate nitrate (Fig. 4E). These results indicate that marker-free double gene deletion can be efficiently performed in A. oryzae.
Marker-free integration of exogenous genes into the genome.
To integrate exogenous genes into the genome, we inserted the EGFP expression cassette under the control of PamyB into plasmid pwAupDN, yielding the donor plasmid pwAupEGFP, and introduced it together with the genome-editing plasmid pRGE-gwAup into the strain RIB40. In all 15 transformants, the EGFP expression cassette was integrated into the wA locus (Fig. 5A and B). The selected transformants exhibited white conidium formation and cytoplasmic enhanced green fluorescent protein (EGFP) fluorescence (Fig. 5C and D). We also constructed the donor plasmid pnDdownDR for the expression of mDsRed under the control of PamyB at the niaD locus and introduced it, together with the genome-editing plasmid pRGE-gnDdown, into strain RIB40. In all 10 transformants, the mDsRed expression cassette was integrated into the niaD locus (Fig. S8A and B). The selected transformants were unable to assimilate nitrate and exhibited cytoplasmic mDsRed fluorescence (Fig. S8C and D). These results demonstrated that cotransformation with donor and genome-editing plasmids enabled efficient integration of exogenous genes at the target locus.
FIG 5.
Gene integration in the A. oryzae wild strain RIB40 by cotransformation with the genome-editing plasmid together with a circular donor DNA. (A) Strategy for integration of the EGFP expression cassette into the wA locus by using the CRISPR/Cas9 system. (B) PCR analysis of genomic DNA and restriction enzyme digestion analysis of the integration of the EGFP expression cassette at the wA locus. Amplified DNA fragments using the primers indicated in panel A were digested with SmaI and analyzed by electrophoresis. (C) Phenotype of transformants resulting from gene integration. A conidial suspension (104 conidia in 5 μl) of each strain was spotted onto the PD medium, followed by incubation for 5 days at 30°C. (D) Fluorescent microscopic analysis of transformants expressing EGFP. Scale bar, 10 μm. (E) PCR analysis of genomic DNA using the primers indicated in panel A and in Fig. S8A for double integration of the EGFP and mDsRed expression cassettes into the wA and the niaD loci, respectively. (F) Phenotype of transformants resulting from the double integration. A conidial suspension (104 conidia in 5 μl) of each strain was spotted onto the indicated agar medium, followed by incubation for 5 days at 30°C. (G) Fluorescent microscopic analysis of transformants expressing EGFP and mDsRed. Scale bar, 10 μm.
For double integration of exogenous DNA sequences into the wA and niaD loci, we cotransformed the strain RIB40 with the genome-editing plasmid pRGE-gwAup-gnDdown and two donor plasmids, pwAupEGFP and pnDdownDR. Integration of both fragments was confirmed in 11 of 13 transformants (84.6%) (Fig. 5E). The selected transformants exhibited the expected phenotypes, white conidium formation and inability to assimilate nitrate (Fig. 5F), and cytoplasmic EGFP and mDsRed fluorescence (Fig. 5G). These observations indicated that marker-free double gene integration of exogenous genes was indeed efficiently performed in A. oryzae.
To investigate whether the amount of a donor plasmid influences its integration pattern into the genome, we cointroduced 1 μg of genome-editing plasmid along with with 2 or 3 μg of donor plasmid into strain RIB40. In all transformants, a single band was detected by Southern blot analysis (Fig. S9A and B), indicating that the donor DNA was integrated only into the target locus. A single copy was integrated in four and two of five transformants with 2 and 3 μg of donor plasmid, respectively, containing the EGFP expression cassette (Fig. S9A) and in two and four of five transformants with 2 and 3 μg of donor plasmid, respectively, containing the mDsRed expression cassette (Fig. S9B). These results suggest that the donor plasmid amounts tested do not affect the ratio of single-copy integration. In the transformants, except for those with single-copy integration, longer DNA fragments were detected (Fig. S9A and B), and multiple-copy integration of the donor DNA was demonstrated by quantitative PCR (Fig. S9C and D), indicating that multiple donor DNAs were tandemly integrated. In the experiment using 3 μg of donor plasmid, we obtained transformants containing multiple copies of the donor DNA, e.g., 5 copies of EGFP expression cassette and 30 copies of mDsRed expression cassette (Fig. S9C and D). In accordance, the higher production level of EGFP was detected in the transformant containing five copies of EGFP expression cassette (Fig. S9E). Hence, increased amount of multiple-copy integration of donor DNA possibly enhances the heterologous protein production.
DISCUSSION
Filamentous fungi are used for the production of industrially desirable enzymes and are expected as hosts for heterologous production of useful secondary metabolites (1, 3). Although multistep genetic engineering of recipient strains is required for efficient production of these enzymes and compounds, sequential (single) engineering of multiple genes using traditional methods is time-consuming. Previously, we developed a CRISPR/Cas9 approach for the mutagenesis of A. oryzae, but the efficiency of the approach was low (10). In the present study, we improved the mutation efficiency of A. oryzae wild and industrial strains up to 50 to 100% using autonomous replicating genome-editing plasmids harboring half AMA1 (Fig. 1C). The improved mutation efficiency was probably associated with the increased expression of Cas9 and sgRNA because the AMA1-bearing plasmid is present in multiple copies per cell (29).
For repeatable mutagenesis, we inserted the Aoace2 gene conditionally controlled by PamyB into the genome-editing plasmid (Fig. 2A) and successfully removed the plasmid from the resulting mutants (Fig. 2B and D). It was previously reported that a genome-editing plasmid harboring AMA1 was removed from A. carbonarius transformants by continuous subculturing in the absence of selective pressure, but further genetic engineering was not performed (22). Therefore, it was unclear whether the plasmid was completely removed. Forced recycling of the AMA1-based plasmid containing a drug resistance marker had not been reported in filamentous fungi. We performed further mutagenesis and obtained double mutants (Fig. 2E and F), indicating that the constructed genome-editing plasmid that harbored the Aoace2 expression cassette was indeed removed. In addition, since the overexpression of Aoace2 under the control of PamyB led to a severe growth defect of the harboring strain (Fig. S2), induction of Aoace2 enabled complete removal of the plasmid (Fig. 2D). We also demonstrated that plasmid recycling based on Aoace2 expression was feasible in A. oryzae industrial strain RIB128 (Fig. S4). Therefore, the genome-editing plasmid with conditionally expressed Aoace2 allowed for repeatable genetic engineering of A. oryzae industrial strains even if no auxotrophic strains are generated.
To perform simultaneous double mutagenesis, we inserted two sgRNA expression cassettes into a single genome-editing plasmid. This resulted in the mutation efficiencies of 68.1% for wA niaD double mutagenesis in the wild strain RIB40 and 50% for wA pyrG double mutagenesis in the industrial strain RIB128 (Fig. 3A and B; see also Fig. S6). Simultaneous deletion of up to four genes by HDR using individual donor DNAs with a single selection marker has been reported (15, 18). However, increased number of target genes results in reduced homologous recombination efficiency (15), and therefore the number of simultaneously modifiable genes is thought to be limited in filamentous fungi. In contrast, the genome-editing plasmids used in double mutagenesis in the present study are recyclable since they harbor the Aoace2 expression cassette (Fig. 3A; see also Fig. S6A).
In most previous attempts of CRISPR/Cas9-mediated HDR-based engineering in filamentous fungi, the selection markers were inserted into the donor DNAs, resulting in highly efficient genetic engineering. Recently, the marker-free HDR-based mutagenesis by using a single-stranded oligonucleotide as the template was performed with high efficiency in NHEJ-deficient Aspergillus strains (20). In the present study, we succeeded in efficient marker-free gene deletion/integration by cointroduction of genome-editing plasmids and circular donor DNA even in the A. oryzae strain with a functional NHEJ pathway (Fig. 4; see also Fig. S7). In addition, we also succeeded simultaneous double gene deletion without inserting any selection markers into the target loci (Fig. 4; see also Fig. S7). These findings indicate that marker-free genetic engineering can indeed be efficiently performed in filamentous fungi with functional NHEJ pathway. Furthermore, if the drug-resistant marker is used, the genetic engineering method developed in this study can be directly applied to filamentous fungal industrial strains without any NHEJ deficiency and auxotrophies. Many strategies for increased production of recombinant proteins using filamentous fungi have been reported (1). Among these, deletion of protease-encoding genes is an effective strategy for some Aspergillus species (34–36), and combinatorial deletion of up to 10 genes enhances recombinant protein production (35, 37, 38). For multiple gene deletion, the pyrG selection marker was recycled as the selected multiple genes were deleted one at a time in the NHEJ-deficient strain (37, 38). Consequently, generation of an NHEJ-deficient strain and multiple deletion mutants was time-consuming. The repeated multiplex gene deletion technique developed in the present study will facilitate generation of multiple gene deletion mutants in filamentous fungi.
We also demonstrated that gene integration could be efficiently performed, by inserting the EGFP and mDsRed expression cassettes into the donor DNAs without any selection marker (Fig. 5; see also Fig. S8). Filamentous fungi are used for the production of secondary metabolites such as antibiotics (2, 3). Since multiple enzymes, which are often encoded in a gene cluster on the genome, are related to a biosynthetic pathway of secondary metabolites (39), introduction of many genes is required for heterologous production in filamentous fungi (40). However, it is difficult to introduce many genes using the limited selection markers. Therefore, the technique for repeated marker-free gene integration developed in the present study would facilitate efficient generation of strains producing various secondary metabolites.
In conclusion, in the present study, we developed highly efficient techniques of CRISPR/Cas9-mediated genetic engineering, i.e., mutagenesis and gene deletion/integration, and for repeatable genetic engineering by plasmid recycling in A. oryzae wild and industrial strains. Theoretically, the combination of these techniques would allow infinite rounds of genetic engineering. Since, in various filamentous fungi, including not only Aspergillus species but also P. chrysogenum, Talaromyces atroroseus, Trichoderma reesei, and Gaemannomyces graminis, the AMA1 sequence was shown to be functional (8, 24, 25, 41, 42) and orthologues of Aoace2 are also conserved (Fig. S2A) (31), repeatable genetic engineering approaches developed in the present study can be applied to other filamentous fungal species. These techniques can be used for molecular breeding of high-level heterologous production of proteins and secondary metabolites in filamentous fungi.
MATERIALS AND METHODS
A. oryzae strains, growth conditions, and transformation.
A. oryzae strains used in the present study are listed in Table 1 . A. oryzae strains were grown in CD medium (0.3% NaNO3, 0.2% KCl, 0.1% KH2PO4, 0.05% MgSO4⋅7H2O, 0.002% FeSO4⋅7H2O, and 2% glucose [pH 5.5]) and PD medium (Nissui, Tokyo, Japan). GP medium (0.5% yeast extract, 1% Hipolypeptone [Fujifilm Wako Pure Chemical Corporation, Osaka, Japan], 2% glucose, 0.5% KH2PO4, 0.05% MgSO4⋅7H2O) was used for preculture during transformation. To grow the pyrG mutants, the CD medium was supplemented with 0.5% uridine and 0.2% uracil. CD(Leu) medium containing 10 mM leucine instead of NaNO3 as the sole nitrogen source was used to grow the niaD mutants. For microscopic analysis, strains were grown in the CD medium containing 1% Casamino Acids. For the induction and repression of PamyB, 2% dextrin and 2% glycerol, respectively, instead of glucose were included in the CD medium as the sole carbon source. To repress PamyB, CD medium containing 1.2 M sorbitol was also used.
TABLE 1.
A. oryzae strains used in this study
| Strain | Genotype or description | Source or reference |
|---|---|---|
| RIB40 | Wild strain | 45 |
| RIB128 | Industrial strain for sake fermentation | NRIBa |
| RIB915 | Industrial strain for soy sauce fermentation | NRIB |
| SlD1 | niaD− sC− adeA− argB::adeA− ΔligD::argB ΔpyrG::adeA pgEpG[pyrG] pNR10[niaD] | 46 |
| NSlD1 | niaD− sC− adeA− argB−::adeA− ΔligD::argB ΔpyrG::adeA pgEpG[pyrG] | 47 |
| SlD-UtAoace2 | niaD−::niaD-PamyB-Aoace2-TamyB sC− adeA− ΔargB::adeA− ΔligD::argB ΔpyrG::adeA pgEpG[pyrG] | This study |
| RIB40wA7C9-1 | wA1 ppAsATC9a2gwA7 | This study |
| RIB40wA7-1 | wA1 | This study |
| RIB40wAnD-1 | wA1 niaD1 | This study |
| RIB128wA7C9-1 | wA2 ppAsATC9a2gwA7 | This study |
| RIB128wA7-1 | wA2 | This study |
| RIB128wAnD-1 | wA2 niaD1 | This study |
NRIB, National Research Institute of Brewing, Higashihiroshima, Hiroshima, Japan.
Transformation of A. oryzae strains.
Transformation of A. oryzae was performed as previously described, with some modifications (43). The GP medium was used for preculture to prepare protoplasts. To select the transformants, 0.1 μg/ml pyrithiamine was added to the medium. The transformants by using the genome-editing plasmids without the Aoace2 expression cassette were subcultured twice on the CD or CD(Leu) medium containing pyrithiamine. The DNA fragment around the target sequence was amplified by genomic PCR, and the DNA sequence was analyzed by nucleotide sequencing. The transformants by using the genome-editing plasmids bearing the Aoace2 expression cassette were subcultured once on the CD or CD(Leu) medium containing 1.2 M sorbitol and pyrithiamine. They were further shifted onto the CD or CD(Leu) containing dextrin as the sole carbon source to remove the genome-editing plasmid, and then the DNA fragment around the target sequence was amplified by genomic PCR to investigate mutation and gene deletion/integration.
DNA manipulation.
Escherichia coli DH5α was used for DNA manipulation. Primers used in this study are listed in Table 2 . The PCR (PCR) was performed using PrimesSTAR HS DNA polymerase (TaKaRa, Otsu, Japan). An In-Fusion HD cloning kit (Clontech, Palo Alto, CA) was used for plasmid construction. Plasmids were extracted by the alkaline lysis method or using a plasmid midikit (Qiagen, Hilden, Germany). Nucleotide sequencing analysis of target genes was performed commercially by Fasmac Co., Ltd.
TABLE 2.
Primers used in this study
Construction of autonomously replicating plasmids for genome editing.
The autonomously replicating plasmid ppAsAcas9 was constructed as follows: a DNA fragment containing the ptrA marker and a half part of AMA1 was amplified from the plasmid pPTRII (TaKaRa) using a primer set, 19IF-ptrA-F and 19IF-mAMA1-R, and was ligated with BamHI-digested pUC19 (TaKaRa), yielding ppAsAMA. A DNA fragment containing the amyB promoter, cas9, and the amyB terminator was amplified from pUNAFNC9gwA1 (10) using a primer set, amyB(p)-F and amyB(t)-R, and was ligated with HindIII-digested ppAsA, yielding ppAsAcas9. The sgRNA expression cassettes were then inserted into ppAsAcas9 as described in the supplemental material.
Construction of recycling plasmids for genome editing.
The plasmid pUtNAa2N for Aoace2-conditional expression was constructed as follows: a DNA fragment containing the 3′ region and the terminator of niaD, downstream of niaD, and the amyB terminator were amplified from the genome DNA of A. oryzae wild strain RIB40 using the primer sets pUC19-niaD3_F and niaD3-PaB_R, niaDd-TaB-F and pUC19nDAADnD_R, and TaB-PaB_F and TaB-niaDd_R, respectively. The amyB promoter was amplified from pUNA (44) using the primer set PaB-niaD3_F and PaB-TaB_R. These four fragments were fused using the primer set pUC19-niaD3_F and pUC19nDAADnD_R and ligated with BamHI-digested pUC19, yielding pUtNAN. The Aoace2 ORF was amplified from the genome DNA of strain RIB40 using a primer set (pUNA′Aoace2ORF_F and pUNA′Aoace2ORF+TGA_R) and ligated with SmaI-digested pUtNAN, yielding pUtNAa2N.
The recycling plasmid ppAsATC9a2 for genome editing was constructed as follows. A DNA fragment containing cas9 and the amyB terminator was amplified from the plasmid ppAsAcas9 using the primer set Ptef-FNC9F and amyB(t)-R. The Aotef1 promoter was amplified from the genome DNA of the strain RIB40 using the primer set IF-Ptef1F and Ptef1R. These two DNA fragments were fused using the primer set IF-Ptef1F and amyB(t)-R, and the fused fragment was ligated with SmaI-digested ppAsAMA, yielding ppAsATC9. A DNA fragment containing the amyB promoter and the 5′ half of Aoace2 was amplified from the plasmid pUtNAa2N using the primer set sApAIF-PamyB and Aoace2-mR. A DNA fragment containing 3′ half of Aoace2 and the amyB terminator was amplified from pUtNAa2N using the primer set Aoace2-mF and sApAIF-TamyB. These two DNA fragments were fused using the primer set sApAIF-PamyB and sApAIF-TamyB, and the fused fragment was ligated with SmaI-digested ppAsATC9, yielding ppAsATC9a2. The sgRNA expression cassettes were then inserted into ppAsATC9a2 as described in the supplemental material.
Construction of the Aoace2-expressing strain.
NotI-digested pUtNAa2N was introduced into the strain NSlD1, yielding strain SlD-UtAoace2 (Fig. S10A). The integration of the Aoace2 expression cassette downstream of niaD was confirmed by PCR analysis of genome DNA (Fig. S10B).
Extraction of A. oryzae genome DNA and Southern blot analysis.
Genomic DNA of A. oryzae was extracted as previously described (43). For Fig. 2D and Fig. S4C, genomic DNA was digested with PvuII, and a DNA fragment amplified from pUtNAa2N using the primer set ace2_probeF and ace2_probeR was used as a probe in Southern blot analysis. For Fig. S9A, genomic DNA was digested with BglII and DNA fragment amplified from pwAupEGFP using the primer set 19IF-wADN-F and wADN-5R. For Fig. S9B, genomic DNA was digested with EcoRV and DNA fragment amplified from pnDdownDR using the primer set 19IF-nDdownDN5F and nDdownDN5R. Southern blot analysis was performed with an ECL detection kit (GE Healthcare, IL) according to the manufacturer’s instructions, and the nucleic acid bands were detected using a LAS-4000 miniEPUV luminescent image analyzer (Fuji Photo Film, Tokyo, Japan).
Fluorescence microscopy and image analysis.
Conidia (1 × 104) were inoculated into 150 μl of the CD liquid medium containing Casamino Acids in a glass-bottomed dish and incubated at 30°C for 18 h. The cultures were observed by confocal laser scanning microscopy using an IX71 inverted microscope (Olympus, Tokyo, Japan) equipped with a ×40 Neofluor lens objective (1.00 numerical aperture), 488-nm (Furukawa Electric, Tokyo, Japan) and 561-nm (Melles Griot, Rochester, NY) semiconductor lasers, GFP and DsRed filters (Nippon Roper, Tokyo, Japan), a CSU22 confocal scanning system (Yokogawa Electronics, Tokyo, Japan), and an Andor iXon cooled digital CCD camera (Andor Technology PLC, Belfast, UK). The captured images were analyzed using Andor iQ 1.9 software (Andor Technology PLC).
Construction of plasmids, quantitative PCR, and Western blot analysis.
The construction of the genome-editing plasmids containing sgRNA expression cassettes and of the donor plasmids is described in detail in the supplemental material, as well as the methods used for the quantitative PCR and Western blot analyses.
Supplementary Material
ACKNOWLEDGMENTS
This study was financially supported by a Grant-in-Aid for Scientific Research on Innovative Areas (17H05431) and by Grants-in-Aid for Scientific Research (18H02123 and 17K15242) from the Ministry of Education, Culture, Sports, Science, and Technology (Japan). We thank Yasuji Koyama and Masahiro Ogawa (Noda Institute for Scientific Research, Japan) for technical support in analysis of transcription factors.
T.K., H.N., Y.Z., A.P., W.F., and J.M. conceived and designed the experiments. T.K., H.N., Y.Z., and A.P. performed the experiments. T.K., H.N., Y.Z., A.P., and J.M. analyzed the data. T.K. and J.M. wrote the manuscript. All the authors reviewed the manuscript.
There are no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01896-18.
REFERENCES
- 1.Ward OP. 2012. Production of recombinant proteins by filamentous fungi. Biotechnol Adv 30:1119–1139. doi: 10.1016/j.biotechadv.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 2.Hoffmeister D, Keller NP. 2007. Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat Prod Rep 24:393–416. doi: 10.1039/b603084j. [DOI] [PubMed] [Google Scholar]
- 3.Alberti F, Foster GD, Bailey AM. 2017. Natural products from filamentous fungi and production by heterologous expression. Appl Microbiol Biotechnol 101:493–500. doi: 10.1007/s00253-016-8034-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang D, Zhu W, Wang Y, Sun C, Zhang KQ, Yang J. 2013. Molecular tools for functional genomics in filamentous fungi: recent advances and new strategies. Biotechnol Adv 31:1562–1574. doi: 10.1016/j.biotechadv.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Doudna JF, Charpentier E. 2014. The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 6.Donohoue PD, Barrangou R, May AP. 2017. Advances in industrial biotechnology using CRISPR-Cas systems. Trends Biotechnol S0167-7799:30187–30187. doi: 10.1016/j.tibtech.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 7.Shi TQ, Liu GN, Ji RY, Shi K, Song P, Ren LJ, Huang H, Ji XJ. 2017. CRISPR/Cas9-based genome editing of the filamentous fungi: the state of the art. Appl Microbiol Biotechnol 101:7435–7443. doi: 10.1007/s00253-017-8497-9. [DOI] [PubMed] [Google Scholar]
- 8.Nødvig CS, Nielsen JB, Kogle ME, Mortensen UH. 2015. A CRISPR-Cas9 system for engineering of filamentous fungi. PLoS One 10:e0133085. doi: 10.1371/journal.pone.0133085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuller KK, Chen S, Loros JJ, Dunlap JC. 2015. Development of the CRISPR/Cas9 system for targeted gene disruption in Aspergillus fumigatus. Eukaryot Cell 14:1073–1080. doi: 10.1128/EC.00107-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katayama T, Tanaka Y, Okabe T, Nakamura H, Fujii W, Kitamoto K, Maruyama J. 2016. Development of a genome editing technique using the CRISPR/Cas9 system in the industrial filamentous fungus Aspergillus oryzae. Biotechnol Lett 38:637–642. doi: 10.1007/s10529-015-2015-x. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura H, Katayama T, Okabe T, Iwashita K, Fujii W, Kitamoto K, Maruyama J. 2017. Highly efficient gene targeting in Aspergillus oryzae industrial strains under ligD mutation introduced by genome editing: strain-specific differences in the effects of deleting EcdR, the negative regulator of sclerotia formation. J Gen Appl Microbiol 63:172–178. doi: 10.2323/jgam.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Zhang C, Meng X, Wei X, Lu L. 2016. Highly efficient CRISPR mutagenesis by microhomology-mediated end joining in Aspergillus fumigatus. Fungal Genet Biol 86:47–57. doi: 10.1016/j.fgb.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 13.Wenderoth M, Pinecker C, Voss B, Fischer R. 2017. Establishment of CRISPR/Cas9 in Alternaria alternata. Fungal Genet Biol 101:55–60. doi: 10.1016/j.fgb.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Fang Y, Tyler BM. 2016. Efficient disruption and replacement of an effector gene in the oomycete Phytophthora sojae using CRISPR/Cas9. Mol Plant Pathol 17:127–139. doi: 10.1111/mpp.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Q, Gao R, Li J, Lin L, Zhao J, Sun W, Tian C. 2017. Development of a genome-editing CRISPR/Cas9 system in thermophilic fungal Myceliophthora species and its application to hyper-cellulase production strain engineering. Biotechnol Biofuels 10:1. doi: 10.1186/s13068-016-0693-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schuster M, Schweizer G, Reissmann S, Kahmann R. 2016. Genome editing in Ustilago maydis using the CRISPR-Cas9 system. Fungal Genet Biol 89:3–9. doi: 10.1016/j.fgb.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Qin H, Xiao H, Zou G, Zhou Z, Zhong JJ. 2017. CRISPR-Cas9 assisted gene disruption in the higher fungus Ganoderma species. Process Biochem 56:57–61. doi: 10.1016/j.procbio.2017.02.012. [DOI] [Google Scholar]
- 18.Liu R, Chen L, Jiang Y, Zhou Z, Zou G. 2015. Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discov 1:15007. doi: 10.1038/celldisc.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber J, Valiante V, Nødvig CS, Mattern DJ, Slotkowski RA, Mortensen UH, Brakhage AA. 2017. Functional reconstruction of a fungal natural product gene cluster by advanced genome editing. ACS Synth Biol 6:62–68. doi: 10.1021/acssynbio.6b00203. [DOI] [PubMed] [Google Scholar]
- 20.Nødvig CS, Hoof JB, Kogle ME, Jarczynska ZD, Lehmbeck J, Klitgaard DK, Mortensen UH. 2018. Efficient oligonucleotide-mediated CRISPR-Cas9 gene editing in aspergilli. Fungal Genet Biol 115:78–89. doi: 10.1016/j.fgb.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Kuivanen J, Wang YMJ, Richard P. 2016. Engineering Aspergillus niger for galactaric acid production: elimination of galactaric acid catabolism by using RNA sequencing and CRISSPR/Cas9. Microb Cell Fact 15:210. doi: 10.1186/s12934-016-0613-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weyda I, Yang L, Vang J, Ahring B, Lübeck M, Lübeck PS. 2017. A comparison of Agrobacterium-mediated transformation and protoplast-mediated transformation with CRISPR-Cas9 and bipartite gene targeting substrate, as effective gene targeting tools for Aspergillus carbonarius. J Microbiol Methods 135:26–34. doi: 10.1016/j.mimet.2017.01.015. [DOI] [PubMed] [Google Scholar]
- 23.Arazoe T, Miyoshi K, Yamato T, Ogawa T, Ohsato S, Arie T, Kuwata S. 2015. Tailor-made CRISPR/Cas system for highly efficient targeted gene replacement in the rice blast fungus. Biotechnol Bioeng 112:2543–2549. doi: 10.1002/bit.25662. [DOI] [PubMed] [Google Scholar]
- 24.Pohl C, Kiel JAKW, Driessen AJM, Bovenberg RAL, Nygård Y. 2016. CRISPR/Cas9-based genome editing of Penicillium chrysogenum. ACS Synth Biol 5:754–764. doi: 10.1021/acssynbio.6b00082. [DOI] [PubMed] [Google Scholar]
- 25.Nielsen ML, Isbrandt T, Rasmussen KB, Thrane U, Hoof JB, Larsen TO, Mortensen UH. 2017. Genes linked to production of secondary metabolites in Talaromyces atroroseus revealed using CRISPR-Cas9. PLoS One 12:e0169712. doi: 10.1371/journal.pone.0169712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarkari P, Marx H, Blumhoff ML, Mattanovich D, Sauer M, Steiger MG. 2017. An efficient tool for metabolic pathway construction and gene integration in Aspergillus niger. Bioresour Technol 245:1327–1333. doi: 10.1016/j.biortech.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 27.Zheng YM, Lin FL, Gao H, Zou G, Zhang JW, Wang GQ, Chen GD, Zhou ZH, Yao XS, Hu D. 2017. Development of a versatile and conventional technique for gene disruption in filamentous fungi based on CRISPR-Cas9 technology. Sci Rep 7:9250. doi: 10.1038/s41598-017-10052-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsu-Ura T, Baek M, Kwon J, Hong C. 2015. Efficient gene editing in Neurospora crassa with CRISPR technology. Fungal Biol Biotechnol 2:4. doi: 10.1186/s40694-015-0015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gems D, Johnstone IL, Clutterbuck AJ. 1991. An autonomously replicating plasmid transforms Aspergillus nidulans at high frequency. Gene 98:61–67. doi: 10.1016/0378-1119(91)90104-J. [DOI] [PubMed] [Google Scholar]
- 30.Aleksenko A, Clutterbuck AJ. 1997. Autonomous plasmid replication in Aspergillus nidulans: AMA1 and MATE elements. Fungal Genet Biol 21:373–387. doi: 10.1006/fgbi.1997.0980. [DOI] [PubMed] [Google Scholar]
- 31.Ejzykowicz DE, Cunha MM, Rozental S, Solis NV, Gravelat FN, Sheppard DC, Filler SG. 2009. The Aspergillus fumigatus transcription factor Ace2 governs pigment production, conidiation, and virulence. Mol Microbiol 72:155–169. doi: 10.1111/j.1365-2958.2009.06631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tada S, Gomi K, Kitamoto K, Takahashi K, Tamura G, Hara S. 1991. Construction of a fusion gene comprising the Taka-amylase A promoter and the Escherichia coli β-glucuronidase gene and analysis of its expression in Aspergillus oryzae. Mol Gen Genet 229:301–306. doi: 10.1007/BF00272170. [DOI] [PubMed] [Google Scholar]
- 33.Bähler J. 2005. A transcriptional pathway for cell separation in fission yeast. Cell Cycle 4:39–41. doi: 10.4161/cc.4.1.1336. [DOI] [PubMed] [Google Scholar]
- 34.van den Hombergh JPTW, Gelpke MDS, van de Vondervoort PJI, Buxton FP, Visser J. 1997. Disruption of three acid proteases in Aspergillus niger: effect on protease spectrum, intracellular proteolysis, and degradation of target proteins. Eur J Biochem 247:605–613. doi: 10.1111/j.1432-1033.1997.00605.x. [DOI] [PubMed] [Google Scholar]
- 35.Jin FJ, Watanabe T, Juvvadi PR, Maruyama J, Arioka M, Kitamoto K. 2007. Double disruption of the proteinase genes, tppA and pepE, increases the production level of human lysozyme by Aspergillus oryzae. Appl Microbiol Biotechnol 76:1059–1068. doi: 10.1007/s00253-007-1088-4. [DOI] [PubMed] [Google Scholar]
- 36.Moralejo FJ, Cardoza RE, Gutierrez S, Lombraña M, Fierro F, Martín JF. 2002. Aspergillopepsin (pepB) gene of Aspergillus awamori by antisense RNA expression or protease removal by gene disruption results in a large increase in thaumatin production. Appl Environ Microbiol 68:3550–3559. doi: 10.1128/AEM.68.7.3550-3559.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoon J, Kimura S, Maruyama J, Kitamoto K. 2009. Construction of quintuple protease gene disruptant for heterologous protein production in Aspergillus oryzae. Appl Microbiol Biotechnol 82:691–701. doi: 10.1007/s00253-008-1815-5. [DOI] [PubMed] [Google Scholar]
- 38.Yoon J, Maruyama J, Kitamoto K. 2011. Disruption of ten protease genes in the filamentous fungus Aspergillus oryzae highly improves production of heterologous proteins. Appl Microbiol Biotechnol 89:747–759. doi: 10.1007/s00253-010-2937-0. [DOI] [PubMed] [Google Scholar]
- 39.Inglis DO, Binkley J, Skrzypek MS, Arnaud MB, Cerqueira GC, Shah P, Wymore F, Wortman JR, Sherlock G. 2013. Comprehensive annotation of secondary metabolite biosynthetic genes and gene clusters of Aspergillus nidulans, A. fumigatus, A. niger, and A. oryzae. BMC Microbiol 13:91. doi: 10.1186/1471-2180-13-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alberti F, Khairudin K, Venegas ER, Davies JA, Hayes PM, Willis CL, Bailey AM, Foster GD. 2017. Heterologous expression reveals the biosynthesis of the antibiotic pleuromutilin and generates bioactive semi-synthetic derivatives. Nat Commun 8:1831. doi: 10.1038/s41467-017-01659-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kubodera T, Yamashita N, Nishimura A. 2002. Transformation of Aspergillus sp. and Trichoderma reesei using the pyrithiamine resistance gene (ptrA) of Aspergillus oryzae. Biosci Biotechnol Biochem 66:404–406. doi: 10.1271/bbb.66.404. [DOI] [PubMed] [Google Scholar]
- 42.Ruiz-Díez B. 2002. Strategies for the transformation of filamentous fungi. J Appl Microbiol 92:189–195. doi: 10.1046/j.1365-2672.2002.01516.x. [DOI] [PubMed] [Google Scholar]
- 43.Maruyama J, Kitamoto K. 2011. Targeted gene disruption in koji mold Aspergillus oryzae. Methods Mol Biol 765:447–456. doi: 10.1007/978-1-61779-197-0_27. [DOI] [PubMed] [Google Scholar]
- 44.Wada R, Jin FJ, Koyama Y, Maruyama J, Kitamoto K. 2014. Efficient formation of heterokaryotic sclerotia in the filamentous fungus Aspergillus oryzae. Appl Microbiol Biotechnol 98:325–334. doi: 10.1007/s00253-013-5314-y. [DOI] [PubMed] [Google Scholar]
- 45.Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto K, Arima T, Akita O, Kashiwagi Y, Abe K, Gomi K, Horiuchi H, Kitamoto K, Kobayashi T, Takeuchi M, Denning DW, Galagan JE, Nierman WC, Yu J, Archer DB, Bennett JW, Bhatnagar D, Cleveland TE, Fedorova ND, Gotoh O, Horikawa H, Hosoyama A, Ichinomiya M, Igarashi R, Iwashita K, Juvvadi PR, Kato M, Kato Y, Kin T, Kokubun A, Maeda H, Maeyama N, Maruyama J, Nagasaki H, Nakajima T, Oda K, Okada K, Paulsen I, Sakamoto K, Sawano T, Takahashi M, Takase K, Terabayashi Y, et al. 2005. Genome sequencing and analysis of Aspergillus oryzae. Nature 438:1157–1161. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- 46.Nakamura H, Kikuma T, Jin FJ, Maruyama J, Kitamoto K. 2016. AoRim15 is involved in conidial stress tolerance, conidiation and sclerotia formation in the filamentous fungus Aspergillus oryzae. J Biosci Bioeng 121:365–371. doi: 10.1016/j.jbiosc.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 47.Yoon J, Aishan T, Maruyama J, Kitamoto K. 2010. Enhanced production and secretion of heterologous proteins by the filamentous fungus Aspergillus oryzae via disruption of vacuolar protein sorting receptor gene Aovps10. Appl Environ Microbiol 76:5718–5727. doi: 10.1128/AEM.03087-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.