Natural diversification of CTXФ and ctxAB genes certainly influences disease severity and shifting patterns in major etiological agents of cholera, e.g., the overwhelming emergence of hybrid El Tor variants, replacing the prototype El Tor strains of V. cholerae. This report, showing the occurrence of CTXET comprising a novel variant of ctxAB in V. mimicus, points out a previously unnoticed evolutionary event that is independent of the evolutionary event associated with the El Tor strains of V. cholerae. Identification and cluster analysis of the newly discovered alleles of tcpA and toxT suggest their horizontal transfer from an uncommon clone of V. cholerae. The genomic contents of ToxT regulon and of tandemly arranged multiple pre-CTXФEnv and of a CTXФET in V. mimicus probably act as salient raw materials that induce natural recombination among the hallmark virulence genes of hybrid V. cholerae strains. This report provides valuable information to enrich our knowledge on the evolution of new variant CT and ToxT regulons.
KEYWORDS: CTXФ, El Tor biotype, Vibrio cholerae, Vibrio mimicus, cholera toxin, classical biotype, tcpA, toxT
ABSTRACT
Atypical El Tor strains of Vibrio cholerae O1 harboring variant ctxB genes of cholera toxin (CT) have gradually become a major cause of recent cholera epidemics. Vibrio mimicus occasionally produces CT, encoded by ctxAB on CTXФ genome; toxin-coregulated pilus (TCP), a major intestinal colonization factor; and also the CTXФ-specific receptor. This study carried out extensive molecular characterization of CTXФ and ToxT regulon in V. mimicus ctx-positive (ctx+) strains (i.e., V. mimicus strains containing ctx) isolated from the Bengal coast. Southern hybridization, PCR, and DNA sequencing of virulence-related genes revealed the presence of an El Tor type CTX prophage (CTXET) carrying a novel ctxAB, tandem copies of environmental type pre-CTX prophage (pre-CTXEnv), and RS1 elements, which were organized as an RS1-CTXET-RS1-pre-CTXEnv-pre-CTXEnv array. Additionally, novel variants of tcpA and toxT, respectively, showing phylogenetic lineage to a clade of V. cholerae non-O1 and to a clade of V. cholerae non-O139, were identified. The V. mimicus strains lacked the RTX (repeat in toxin) and TLC (toxin-linked cryptic) elements and lacked Vibrio seventh-pandemic islands of the El Tor strains but contained five heptamer (TTTTGAT) repeats in ctxAB promoter region similar to those seen with some classical strains of V. cholerae O1. Pulsed-field gel electrophoresis (PFGE) analysis showed that all the ctx+ V. mimicus strains were clonally related. However, their in vitro CT production and in vivo toxigenicity characteristics were variable, which could be explainable by differential transcription of virulence genes along with the ToxR regulon. Taken together, our findings strongly suggest that environmental V. mimicus strains act as a potential reservoir of atypical virulence factors, including variant CT and ToxT regulons, and may contribute to the evolution of V. cholerae hybrid strains.
IMPORTANCE Natural diversification of CTXФ and ctxAB genes certainly influences disease severity and shifting patterns in major etiological agents of cholera, e.g., the overwhelming emergence of hybrid El Tor variants, replacing the prototype El Tor strains of V. cholerae. This report, showing the occurrence of CTXET comprising a novel variant of ctxAB in V. mimicus, points out a previously unnoticed evolutionary event that is independent of the evolutionary event associated with the El Tor strains of V. cholerae. Identification and cluster analysis of the newly discovered alleles of tcpA and toxT suggest their horizontal transfer from an uncommon clone of V. cholerae. The genomic contents of ToxT regulon and of tandemly arranged multiple pre-CTXФEnv and of a CTXФET in V. mimicus probably act as salient raw materials that induce natural recombination among the hallmark virulence genes of hybrid V. cholerae strains. This report provides valuable information to enrich our knowledge on the evolution of new variant CT and ToxT regulons.
INTRODUCTION
Vibrio mimicus is genetically and ecologically very similar to Vibrio cholerae, the cholera bacterium, and shares similar environmental niches in freshwater and estuarine ecosystems, particularly in tropical regions such as the Bengal delta. V. mimicus is known to be associated with sporadic cholera-like diarrhea cases. Despite extensive efforts at hygiene promotion and therapeutic advances, cholera continues to pose as a major health problem worldwide, accounting for millions of episodes and thousands of deaths, with ca. 132,000 cases in 2016 reported to the World Health Organization (http://www.who.int/gho/epidemic_diseases/cholera/en/). The principal pathogenic factor instigating the disease is the cholera toxin (CT), encoded by the ctxAB operon, predominantly found in V. cholerae strains belonging to the O1 and O139 serogroups and occasionally in a few non-O1/non-O139 serogroups. Among the seven known cholera pandemics, the El Tor biotype of V. cholerae O1 is associated with the current, seventh pandemic since 1961 whereas the counterpart classical biotype was associated with the sixth pandemic. In Bangladesh, the classical form of cholera reemerged in 1983, was later detected at diminishing levels as a consequence of the rise in El Tor cholera, and is believed to have been extinct since 1993 (1). However, since the last decade, hybrid El Tor strains producing classical CT have been the dominant cause of epidemic and endemic cholera, replacing the prototype El Tor strains that produce El Tor CT (1). Occurrences of such variant El Tor strain types have also been reported to have spread in many countries in Asia and Africa and in Haiti (2–4). This indicates a cryptic existence of the variant or classical ctxB gene and variant CTXΦ in environmental reservoirs that has so far been left mostly unexplored. In vitro experiments have shown that CTXΦ can infect certain V. mimicus strains (5). In line with this, although rarely isolated, the occurrence of the ctxAB gene among V. mimicus strains in Bangladesh, India, Japan, and the United States attests to the hypothesis of inter-species genetic exchange (6–9).
The ctxAB operon encoding the A and B subunits of CT is a part of the genome of CTXΦ, a filamentous bacteriophage. The precursor form of the CTXΦ, pre-CTXΦ, does not carry the ctxAB genes (10). Before this study, a total of 13 genotypes of ctxB have been distinguished based on single nucleotide polymorphisms (SNPs) at 10 loci of this toxigenic factor (see Table 2). Notably, ctxB genotypes 1 and 2 are typical of all classical strains and El Tor strains from Australia, respectively, while genotypes 3 and 7 are featured among the pandemic El Tor and the Haitian variant strains. V. cholerae O1 El Tor strains are also characterized by the presence of TLC (toxin-linked cryptic) element and RTX (repeat in toxin) genes in the flanking region of CTX prophage and of two large genomic islands termed Vibrio seventh pandemic island I (VSP-I) and VSP-II (11). Other known virulence factors of V. cholerae, particularly of the non-O1/non-O139 strains, include heat-stable enterotoxin/NAG-ST (encoded by stn), the type III secretion system (TTSS) (vcsN2), and cytotoxic cholix toxin (chxA) (12, 13). Natural recombination events, compounded with the integration of phages, contribute to the evolution of genes, especially those related to virulence and ecological fitness (14). While persisting in the aquatic environment, V. cholerae and V. mimicus interact with diverse phages, and a portion of their populations, harboring a selective receptor, can integrate toxigenic phages into their genome (6).
TABLE 2.
Comparative diversity levels in the ctxAB gene among V. mimicus and V. cholerae strains
| Strain (GenBank accession no.)a | Isolation |
ctxA (aa position)b |
ctxB (aa position)c |
Designated genotype |
Reference | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Country(ies) | Yr(s) | 46 | 190 | 198 | 226 | 255 | 20 | 24 | 28 | 34 | 36 | 39 | 46 | 55 | 67 | 68 | |||
| V. cholerae O1, CL, O395 (CP000627) | India | 1948 | S | R | I | V | K | H | Q | D | H | T | H | F | K | A | T | 1 | 1 |
| V. cholerae O1, ET, Australia | Australia | H | Q | D | H | T | H | L | K | A | T | 2 | 1 | ||||||
| V. cholerae O1, ET, N16961 (NC_002505) | Bangladesh | 1975 | S | R | I | V | K | H | Q | D | H | T | Y | F | K | A | I | 3 | 1 |
| V. cholerae O139 (FJ821557) | Bangladesh | 1998 | H | Q | D | H | T | Y | F | K | A | T | 4 | 1 | |||||
| V. cholerae O139 (FJ821556) | Bangladesh | 2005 | H | Q | A | H | T | H | F | K | A | T | 5 | 1 | |||||
| V. cholerae O139 (FJ821581) | Bangladesh | 2007 | H | Q | D | P | T | Y | F | K | A | T | 6 | 1 | |||||
| V. cholerae O1 (EU496273, NC_016445) | India, Haiti | 2007, 2010 | N | Q | D | H | T | H | F | K | A | T | 7 | 1 | |||||
| V. cholerae O27 (AF390572) | Japan | 1996 | S | R | I | V | E | H | H | A | H | T | H | F | K | A | T | 8 | 1 |
| V. cholerae O37 (D30052) | Sudan | 1968 | N | R | I | V | K | H | Q | D | H | T | H | L | N | A | T | 9 | 1 |
| V. cholerae O1 (EU932878) | Zambia | 1996 | H | Q | D | P | T | Y | F | K | A | I | 10 | 54 | |||||
| V. cholerae O1 (EU932881) | Zambia | 2003 | H | Q | D | P | T | H | F | K | A | T | 11 | 54 | |||||
| V. mimicus (ACYV01000039) | USA | 1990 | N | I | I | I | K | H | Q | D | H | T | H | L | K | A | T | 12 | 55 |
| V. cholerae O1 (KC754370) | China | 1965 | H | Q | D | H | A | Y | L | N | A | T | 13 | 56 | |||||
| V. mimicus (LC427969) | Bangladesh | 2000 | N | I | Vd | V | K | H | Q | D | H | T | H | L | K | Ed | T | 14 | This study |
Known serogroups of V. cholerae strains are shown. Accession numbers of the gene sequences are given in parentheses.
The deduced amino acid (aa) positions are indicated by the given numbers; only the variable amino acids deduced from ctxA gene sequences available from 7 of 14 representative strains of V. cholerae and V. mimicus are shown.
The deduced amino acid (aa) positions are indicated by the given numbers; only the variable amino acids deduced from the ctxB gene among the representative strains of V. cholerae and V. mimicus are shown. Boldface data for positions 39 and 68 indicate the amino acid markers, differentiating classical and El Tor type ctxB genes.
Unique change in deduced amino acid of ctxAB in V. mimicus strains of this study.
The CTXФ genome (∼6.9 kb) contains core and RS2 regions. The core region includes genes involved in phage morphogenesis and CT production, including ctxAB, zot, and orfU (or “gIIICTX”). The RS2 region contains genes required for replication (rstA), integration (rstB), and regulation (rstR) of CTXФ (15). Moreover, the upstream promoter of ctxAB possesses heptamer repeats, considered an evolutionary signature, while its downstream intergenic region (ig) contains a site for CTXФ integration, mediated by XerC and XerD recombinases (16). In El Tor strains, the prophage DNA is flanked by a genetic element known as RS1, which is a satellite phage (17). In comparison to RS2, the RS1 additionally contains rstC, which encodes an antirepressor of rstR and promotes transmission of RS1 and CTXФ (18). In V. cholerae strains, the presence of both CTX prophage and the RS1 element, in the form of solitary and multiple copies with diverse arrays of genetic organization, has been documented (19, 20). On the basis of nucleotide sequence polymorphism in its several genes, including rstR and orfU, the CTX prophage can be differentiated into several types such as the classical, El Tor, Kolkata, and environmental types (20). Among the El Tor variant or hybrid strains, different ctxB genotypes, including classical ctxB1 and Haitian ctxB7 and occurring predominantly in classical and El Tor types of CTX prophages, have been reported (2, 3, 19, 21). Although extensive investigations have revealed nucleotide sequence polymorphism and diversity in the array of CTX prophages in the V. cholerae genome (19–22), little is known of those V. mimicus strains.
The transmission of CTXФ into a Vibrio strain relies on the presence of a specific cell surface type IV pilus receptor, termed the toxin-coregulated pilus (TCP), which also plays a vital role, aiding colonization of V. cholerae in human or animal intestine (23). The TCP is located on the Vibrio pathogenicity island (VPI) and is produced by the action of a cluster of genes, termed the TCP island. The major structural subunit of TCP is encoded by tcpA (14). The expression of CT and TCP is activated by ToxT, present on the TCP island, and is under the control of the ToxR regulon, comprising toxR, toxS, tcpP, and tcpH (24). On the basis of the nucleotide sequence polymorphism in tcpA, the TCP can be differentiated into several types, e.g., El Tor, classical, Nandi, and Novais (25). CTX/pre-CTX prophages and genes of VPIs are found scattered throughout environmental isolates of V. cholerae (22). Despite the absence of the classical biotype strains along with the classical CTX phage (1), the increasing occurrence of hybrid El Tor strains of V. cholerae O1 harboring variant ctxB genes is intriguing and requires detailed exploration for determination of their environmental reservoirs. Being genetically and ecologically the two most closely related species, there is a high probability of genetic exchange between V. mimicus and V. cholerae, particularly among their populations interacting closely in the environment. Therefore, some V. mimicus strains may act as a potential reservoir of virulence genes associated with cholera and diarrhea epidemics. However, our knowledge of the traits and diversity of genetic determinants of virulence, particularly that associated with cholera-like diarrhea, in V. mimicus, and their similarity to those of epidemic strains of V. cholerae is very limited. In this study, several V. mimicus ctx-positive (ctx+) strains (i.e., V. mimicus strains containing ctx) isolated from estuarine surface waters in Bangladesh were analyzed to ascertain whether they can act as reservoirs of the CTXΦ-carrying ctxAB variant present in V. cholerae strains associated with recent epidemics. The objectives were to investigate (i) the molecular diversity of genetic elements within CTX prophage and TCP islands, (ii) in vitro CT production, (iii) in vivo fluid accumulation using a suckling mouse model, and (iv) differential expression of the ToxT regulon in these environmental V. mimicus strains. Comparison of these phenotypic and genetic traits to those of toxigenic V. cholerae would aid in improving understanding the evolution of new variant CT and ToxT regulon.
RESULTS
Antimicrobial susceptibility.
With regard to the increasing threat of multidrug resistance, V. mimicus strains were tested for antimicrobial susceptibility, which also partly demonstrated their phenotypic variations. Among the 11 antimicrobials tested, polymyxin B is also known as a phenotypic signature of the El Tor and corresponding hybrid strains of V. cholerae (1). All ctx+ V. mimicus strains showed resistance to ampicillin (10 μg) and cephalothin (30 μg) and intermediate resistance to erythromycin (15 μg). However, two types of antimicrobial resistance pattern were observed based on the resistance to polymyxin B (50 μg) and gentamicin (10 μg) (Table 1). Three of six V. mimicus strains showed resistance to polymyxin B (50 μg), while the other three strains showed intermediate resistance to gentamicin (10 μg).
TABLE 1.
Antimicrobial susceptibility, cholera toxin production, PFGE patterns, and enterotoxigenicity in ctx+ V. mimicus strains
| Strain IDa |
Date of isolation |
Antimicrobial resistance (μg)b |
PFGE pattern |
CT productiond (ng ml−1) |
Suckling mouse assaye (n = 5) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PB (50) |
CF (30) |
EM (15) |
ABPC (10) |
GM (10) |
Othersc | SfiI | NotI | LB | AKI | FA ratio | Diarrhea | ||
| V. mimicus DCT1 | 17 July 2000 | R | R | I | R | S | S | I | a | 0.3 | 0.1 | nd | nd |
| V. mimicus DCT2 | 05 August 2000 | R | R | I | R | S | S | II | a | 0.4 | 0.2 | 0.068 | 0/5 |
| V. mimicus DCT3 | 22 August 2000 | R | R | I | R | S | S | I | b | 0.2 | 0.1 | nd | nd |
| V. mimicus DCT4 | 11 September 2000 | S | R | I | R | I | S | III | b | 0.2 | 0.1 | nd | nd |
| V. mimicus DCT5 | 03 October 2000 | S | R | I | R | I | S | II | b | 110 | 30 | 0.087 | 5/5 |
| V. mimicus DCT6 | 27 October 2000 | S | R | I | R | I | S | II | a | 0.5 | 0.4 | nd | nd |
| V. cholerae 0395 | 1948 | S | I | S | R | S | 270 | 150 | 0.098 | 5/5 | |||
| V. cholerae N16961 | 1975 | R | S | S | I | S | 1.6 | 2.5 | nd | nd | |||
“DCT1” to “DCT6” represent V. mimicus strains examined in this study; “O395” and “N16961” represent V. cholerae reference strains of classical and El Tor biotypes, respectively. ID, identifier. Boldface highlighting indicates data from low-CT-producing V. mimicus strain DCT2 and from high-CT-producing V. mimicus strain DCT5.
“PB,” “CF,” “EM,” “ABPC,” and “GM” indicate polymyxin B, cephalothin, erythromycin, ampicillin, and gentamicin, respectively. S, R, and I designate susceptible, resistant, and intermediate patterns, respectively.
Others, other antibiotics (e.g., furazolidone, trimethoprim-sulfamethoxazole, nalidixic acid, ciprofloxacin, and tetracycline) were also used at standard doses (i.e., 100, 1.25/23.75, 10, 5, and 30 μg, respectively).
CT production was measured by bead-ELISA after cells were cultured (4 h static plus 4 h shaking, 120 revolutions min−1) in Luria-Bertani broth and AKI medium, representing inducible conditions for the classical and El Tor strain types, respectively. Mean values given are based on three experiments performed for each strain.
A fluid accumulation (FA) ratio of >0.08 in the suckling mouse assay indicates enterotoxigenicity. Mean values (n = 5) are given. nd, not done.
PFGE-based screening for genomic relatedness.
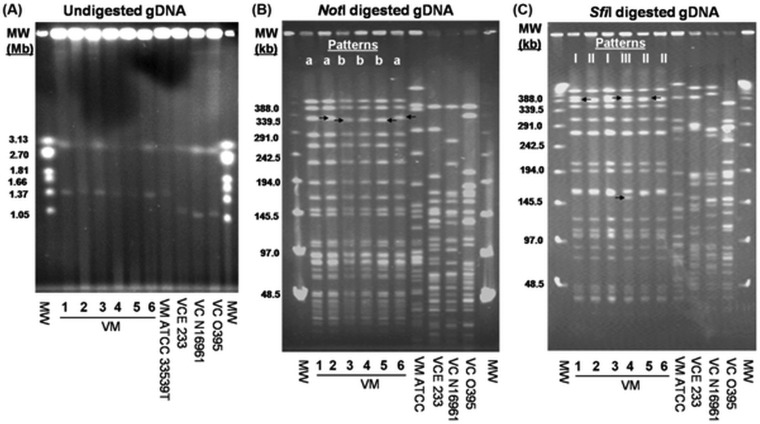
Pulsed-field gel electrophoresis (PFGE) of the undigested genomic DNA (gDNA) showed that the ctx+ V. mimicus strains possessed ca. 2.9 and 1.3 Mb of large and small chromosomes, respectively, which were similar to sizes seen with the V. mimicus type strain (ATCC33539T). Interestingly, the small chromosome in V. mimicus strains was larger (ca. 0.2 Mb) than those in the classical (O395), El Tor (N16961), and non-O1/non-O139 strains of V. cholerae E233 (VCE233) (Fig. 1). PFGE analysis of NotI- and SfiI-digested gDNA of ctx+ V. mimicus strains showed 1-to-2-band differences that resulted in two (a and b) and three (I, II, and III) subtypes, respectively (Fig. 1 and Table 1). According to criteria previously specified by Tenover et al. (26), these strains are clonal, since they did not differ by more than 6 bands. However, combining the NotI and SfiI patterns resulted in categorization of these six strains into four subtypes (Ia, Ib, IIb, and IIIb).
FIG 1.
PFGE analysis of environmental ctx+ and reference (ATCC) strains of V. mimicus (VM) and of ctx+ non-O1/non-O139 (VCE 233), O1 El Tor (VC N16961), and O1 classical (VC O395) strains of V. cholerae. (A) Gel image of PFGE profiles of undigested gDNA showing similar sizes of the two chromosomes of ctx+ V. mimicus and ATCC V. mimicus strains. (B and C) Gel images showing PFGE patterns of NotI-digested (B) and SfiI-digested (C) gDNA of ctx+ V. mimicus and the reference strains. The ctx+ V. mimicus strains were clonal but differed in 1 to 2 bands, indicated by arrows. NotI- and SfiI-digested gDNA of ctx+ V. mimicus strains generated two (a and b) and three (I, II, and III) PFGE profiles, respectively. “MW” represents the molecular weight standard of the Hansenula wingei chromosomes (Bio-Rad) for undigested gDNA (A) and lambda ladder (Bio-Rad) for digested gDNA (B and C).
Occurrence of the major virulence factors.
Among the hallmark genes associated with the toxigenic V. cholerae strains, several related to CT production were detected in the V. mimicus strains used in this study. Colony blot hybridization showed the presence of ctxA and zot of CTXФ, rstC of the RS1 element, and tcpA of the VPI. However, these strains did not harbor genes representing VSP-I and VSP-II. They were also negative for TLC and RTX elements, which are commonly present in the flanking region of CTXФ in V. cholerae El Tor and corresponding hybrid strains. No other important virulence genes of V. cholerae, namely, vcsN2, chxA, and stn, encoding type III secretion system (ATPase), cholix toxin, and heat-stable enterotoxin (NAG-ST), respectively, were detected in the ctx+ V. mimicus strains.
Characteristics of ctxAB, orfU, and CTXФ-associated genetic elements.
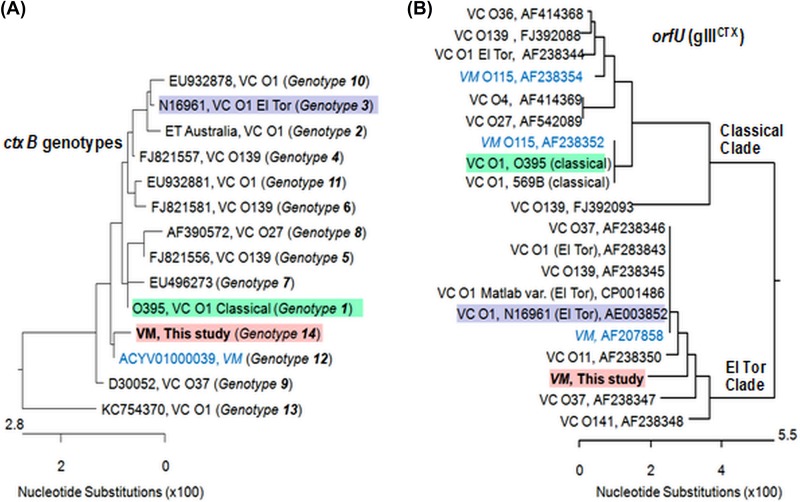
On the basis of the results of the mismatch amplification mutation assay (MAMA)-PCR, all the V. mimicus strains contained the classical type of the ctxB gene. Sequencing of the entire ctxB gene revealed that all instances of the gene were identical in all six V. mimicus strains. Although showing signature changes at positions 39 (tyrosine to histidine) and 68 (isoleucine to threonine), similarly to classical ctxB genotype 1, the V. mimicus ctxB gene had additional nonsynonymous substitutions conferring subtle changes in the deduced amino acids at positions 46 (phenylalanine/F to leucine/L) and 67 (alanine/A to glutamic acid/E) (Table 2). Comparative analyses performed with other known ctxAB genotypes reported among V. mimicus and V. cholerae strains revealed the existence of a new ctxB gene, which was designated genotype 14. This genotype was almost identical to genotype 12, reported from a V. mimicus strain which also contains leucine at position 46 but contains alanine at position 67 (Table 2) (Fig. 2). Notably, in all other genotypes of ctxB identified so far, alanine is present at position 67 (Table 2).
FIG 2.
Genetic relatedness among ctxB and orfU genes of V. mimicus and V. cholerae strains. (A) Novel ctxB genotype 14 of V. mimicus showed the highest affiliation with genotype 12, reported to occur in a V. mimicus strain isolated from the United States. However, ctxB genotype 14 showed more closeness with both ctxB genotype 1 and ctxB genotype 7, representing classical and Haitian strains, respectively, in comparison to genotype 3, usually found in the typical El Tor strains of V. cholerae O1. (B) The orfU gene of V. mimicus of this study showed close affiliation to the strains grouped into the El Tor clade of V. cholerae.
Sequencing analysis of ctxA of V. mimicus strains also showed alterations from the canonical ctxA sequences. This novel ctxA sequence differed from that of the reference El Tor and classical strains at three amino acid positions, namely, positions 46, 190, and 198, while the highest similarity was observed with ctxA of a V. mimicus strain with ctxB genotype 12 (Table 2). A unique change from isoleucine to valine at position 198 of ctxA in the V. mimicus strains was noteworthy.
Because the orfU gene is important in studying the diversity in the core region of CTX prophage, this gene was also sequenced and phylogenetic analysis was done. Interestingly, complete sequence homology among all the V. mimicus strains and also within CTXΦET and pre-CTXΦEnv prophages was observed. Comparative analyses of OrfU (pIIICTX) sequences performed with the reference El Tor strain and classical strains of V. cholerae and a recently studied V. mimicus strain indicated that the V. mimicus strains of this study did not have 100% sequence similarity with any of them but rather they possessed nine unique changes with a total of 31 polymorphic sites. The data showed a high level of similarity of V. mimicus OrfU to that of V. cholerae El Tor strain N16961, with 11 polymorphic sites, whereas with V. cholerae classical strain O395 and the other V. mimicus strain, there were 27 amino acid substitutions (see Fig. S1 in the supplemental material). Thus, sequencing analysis indicated that these V. mimicus strains carried a variant OrfU in their CTXФ genome. In phylogenetic analysis, the orfU sequences of these V. mimicus strains did not cluster with partial (702 of 1,083 nucleotides) orfU sequences of classical V. cholerae available in GenBank database; instead, they grouped with El Tor, El Tor variant, and O139 strains and with several non-O1/non-O139 strains of V. cholerae (Fig. 2).
The presence of RS1 element was confirmed by rstC gene-based PCR (Fig. S1), followed by sequencing analysis. The rstC gene in the V. mimicus strains was identical to that in the reference El Tor strain. PCR-based genotyping of rstR showed the presence of two different alleles, the El Tor type (rstRET) and the environmental type (rstREnv), indicating the occurrence of multiple prophages, i.e., CTXФET and CTXФEnv.
Genetic organization of CTXΦ-associated elements.
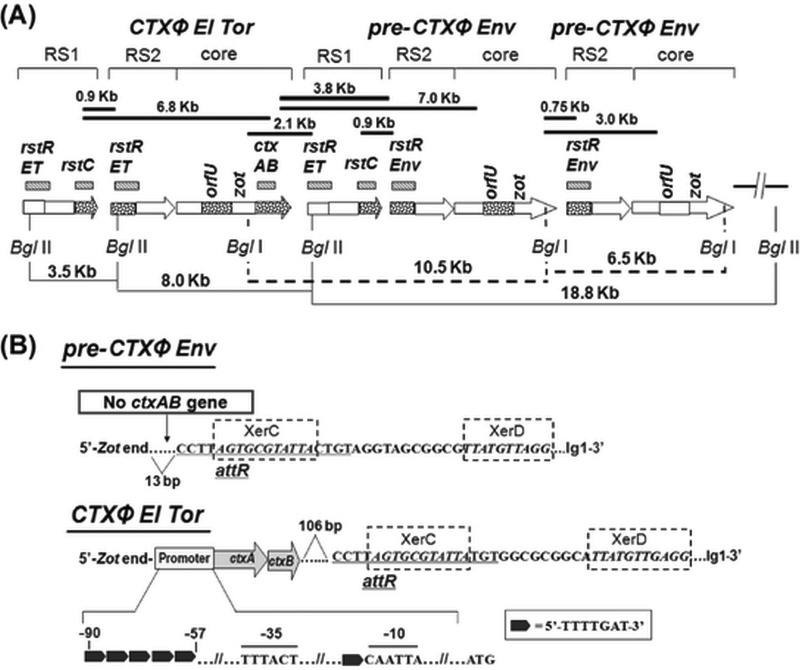
Southern hybridization of chromosomal DNA digested with BglI and BglII, which have single cutting sites in CTXΦ at rstRET/rstREnv and zot, respectively, showed identical RFLP patterns for all V. mimicus strains. Size-wise comparative analyses of the bands detected by hybridization, using different probes, of the enzyme-digested gDNA revealed similar results, i.e., the presence of two copies of pre-CTXΦEnv prophages and a single CTXΦET prophage, in all six strains of ctx+ V. mimicus. Probing with ctxA and rstREnv of the BglI-digested gDNA generated a single positive band (ca. 10.5 kb) and two positive bands (ca. 6.5 and 10.5 kb), respectively. These results indicated the presence of one copy of CTXΦ containing ctxAB, followed by a pre-CTXΦEnv (lacking ctxAB) with an adjacent RS1 and pre-CTXΦEnv without RS1 (Fig. 3 and Fig. S3). Probing with rstREnv and rstRET of the BglII-digested gDNA resulted in one (ca. 18.8 kb) and three (ca. 3.5, 8.0, and 18.8 kb) positive bands, justifying the adjacent locations of one RS1 followed by two pre-CTXΦEnv prophages, along with the preceding occurrence of another RS1 and El Tor type full-length CTXΦ containing ctxAB. Hybridization with rstC probe justified the presence of two copies of the RS1 element, one before the CTXΦET and the other preceding the adjacent pre-CTXΦEnv. However, there was no RS1 element between the two adjacent CTXΦEnv prophages. Taking the data together, an array of RS1–CTXΦET–RS1–pre-CTXΦEnv–pre-CTXΦEnv was deduced from the Southern hybridization analysis.
FIG 3.
Organization of CTXΦEl Tor, RS1, and pre-CTXΦEnv in V. mimicus strains. (A) Filled bars indicate the PCR arrays used to check probable locations of genes, and the sizes of PCR products are given at the top. Hashed bars indicate the genetic regions (names mentioned at the top) used as probes for Southern hybridization after restriction digestion with BglI or BglII enzymes; arrows indicate RS1, RS2, or core prophage data where dotted regions were analyzed by sequencing. Lines (filled and dotted) at the bottom show the distances between specific genetic locations determined by Southern hybridization analysis of the BglI- or BglII-digested genomic DNA performed using specific probes. (B) Signature sequences in the flanking region of pre-CTXΦEnv and CTXΦEl Tor in V. mimicus. The ctxAB promoter of CTXΦEl Tor contains 5 heptamer (TTTTGAT) repeats, shown by filled black arrows, which is characteristic of classical type ctxAB. The phage integration site of the CTXΦEl Tor in V. mimicus, the attR sequence, followed by XerC and XerD binding sites, starts at 106 bp downstream of ctxAB gene, which is similar to the reference El Tor strain, N16961, of V. cholerae. However, the phage integration site, characterized by these signature sequences, of pre-CTXΦEnv in V. mimicus starts at 13 bp downstream of the zot gene.
To verify the hybridization results, PCR arrays were performed using the allele-specific forward and reverse primers with multiple combinations of rstC, rstR, ctxAB, and orfU genes (Fig. 3). All six ctx+ V. mimicus strains yielded similar amplicons after PCR performed using primers specific for different regions of the CTX element. Size-wise comparisons of the PCR amplicons were in concordance with the published genetic organization of El Tor strains of V. cholerae. The results of PCR walking were in concordance with hybridization results, confirming the presence of the RS1 element rstRET in RS2 of the CTXΦ carrying ctxAB and of the rstREnv allele in RS2 of a pre-CTXΦ element(s), which lacked the ctxAB operon (Fig. S2; see also Fig. S3). The flanking sequences of CTXΦET harbored both the RS1 and RS2 elements, similarly to the reference V. cholerae O1 El Tor strain N16961. Sequencing analysis of several core and intergenic regions of the CTX element (Fig. 3), confirmed the tandem presence of three copies of the CTX element, including one intact CTXΦET copy and two pre-CTXΦEnv prophages lacking ctxAB.
Genomic signatures in ctxAB promoter and intergenic sequences.
The sequential integration of pre-CTXΦEnv (lacking ctxAB) and CTXΦET in V. mimicus strains prompted sequencing analysis of intergenic regions, particularly of the ctxAB promoter and the prophage flanking regions, to reveal genomic signatures associated with their lysogenic transformation. Forward and reverse primers of the adjacent genes, i.e., the zot, ctxA, ctxB, and rstRET genes for El Tor CTX prophage and the zot and rstREnv genes for environmental pre-CTX prophage, amplified the desired portions of ctxAB promoter and intergenic regions. Sequencing analysis of the El Tor CTX prophage showed that the promoter at the 5ʹ upstream region of ctxAB contained 5-heptamer (TTTTGAT) repeat spanning the sequence from −90 to −57 bp, which is a characteristic of the classical type ctxAB promoter, while the RNA polymerase binding sites at −35 bp (TTTACT) and −10 bp (CAATTA) were conserved (Fig. 3). The 3ʹ end of ctxAB was characterized by attR sequences coupled with the XerC and XerD binding sites for CTXФ integration, which started 106 bp downstream, similarly to N16961, the reference El Tor strain of V. cholerae. On the other hand, the 3ʹ end of pre-CTXEnv prophage was characterized by a 13-bp gap between the zot (the last gene of the phage core region) and attR sequences, followed by XerC and XerD binding sites (Fig. 3).
PCR, sequencing, and phylogenetic analysis of tcpA and toxT.
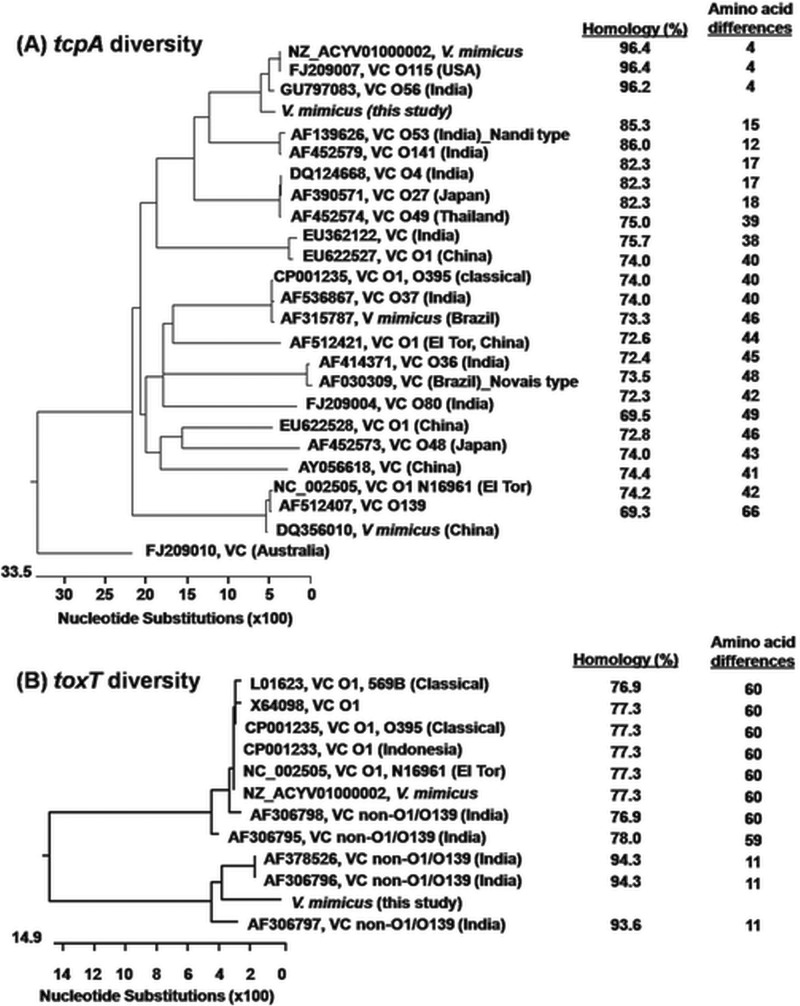
PCR performed using a forward primer (primer tcpA-F [Table 3]) from the starting part of the 5′-terminal conserved region and a reverse primer for the El Tor type or classical type of tcpA did not yield any amplicon. However, using the forward primer tcpA-F and a reverse primer for tcpQ (tcpQ-R) yielded a 2.1-kb product, reconfirming the results of a previous study (8), for all of the ctx+ V. mimicus strains. DNA sequencing showed that all of the strains had identical tcpA genes, and BLAST search analysis revealed the occurrence of a new tcpA allele, designated tcpAEnv_Vm. Phylogenetic analysis revealed that this gene had homology of between 69.3% and 96.4% to other reported tcpA sequences and could be categorized into a novel cluster clearly separated from other major tcpA clusters, including the classical, El Tor, Nandi, and Novais types. The novel tcpAEnv_Vm gene found in V. mimicus strains showed the closest similarity to a couple of V. cholerae non-O1/non-O139 strains isolated in India and the United States and also to a V. mimicus strain (GenBank accession no. NZ_ACYV01000002) isolated from the USA (Fig. 4). Most of the diversities observed among the tcpA alleles were in the carboxy-terminal half, but the amino-terminal region was almost completely conserved among the compared sequences. Comparative sequence analysis performed with the reference classical and El Tor strains of V. cholerae O1 showed that the tcpAEnv_Vm gene had 74% homology at the DNA level to that of the El Tor (N16961) and classical (O395) tcpA genes, with 40 and 43 substitutions, respectively, among 224 deduced amino acids of the tcpA gene (Fig. 4; see also Fig. S4). The Nandi and Novais TcpA types differed by 15 and 45 amino acid residues in comparison to that of the V. mimicus of this study. Phylogenetic analysis also observed high TcpA sequence homology of one V. mimicus strain isolated from Brazil and another strain from China to the canonical TcpA of classical and El Tor O1 V. cholerae, respectively. However, those classical and El Tor TcpA types of V. mimicus had 40 and 42 differences in amino acids, respectively, from TcpAEnv_Vm.
TABLE 3.
Primers and probes used in this study
| Targeta | Primer IDb | Sequence (5′–3′) | Amplicon (bp)/special fragment(s) in Fig. 3 (kb) |
Purpose(s)c | Reference or source |
|---|---|---|---|---|---|
| ctxA | ctxA-F | ACAGAGTGAGTACTTTGACC | 307 bp | PCR/CBH/Seq | 57 |
| ctxA-R | ATACCATCCATATATTTGGGAG | 307 bp/6.8 kb | PCR/CBH/Seq | 57 | |
| ctxB | ctxB-F | GATACACATAATAGAATTAAGGAT | 399 bp/3.8 kb, 7.0 kb | PCR/Seq | 58 |
| ctxB-R | TTGCCATACTAATTGCGG | PCR/Seq | This study | ||
| ctxB-com5-F (common) | ACTATCTTCAGCATATGCACATGG | MAMA PCR | This study | ||
| ctxB-cla3-R (classical) | CCTGGTACTTCTACTTGAAACG | 366 bp | MAMA PCR | 47 | |
| ctxB-e13-R (El Tor) | CCTGGTACTTCTACTTGAAACA | 366 bp | MAMA PCR | 47 | |
| orfU | orfU-F | CGTCACACCAGTTACTTTTCG | 1,072 bp | PCR/Seq | 43 |
| orfU-R | AGAATGTACGCCATCGC | PCR/Seq | 43 | ||
| orfU-R1 | CGAAAAGTAACTGGTGTGACG | 7.0 kb, 3.0 kb | PCR/Seq | This study | |
| zot | zot-1 | GGCTTAAACCTTGAACGC | 1,036 bp | PCR | This study |
| zot-2 | AACCCCGTTTCACTTCTAC | PCR | This study | ||
| zot-F1 | ATGAGTTTGAAACCATACACTTT | 2.1 kb, 0.75 kb, 3.0 kb | PCR/Seq | This study | |
| rstC | rstC-F | ATGAGTTTGAAACCATACACTTT | 225 bp/0.9 kb, 6.8 kb | PCR/CBH/Seq | 17 |
| rstC-R | TTACAGTGATGGATCAGTCAAT | 225 bp | PCR/CBH/Seq | 17 | |
| rstR | rstR-R (common) | TCGAGTTGTAATTCATCAAGAGTG | PCR/CBH/Seq | 20 | |
| rstR (classical)-F | CTTCTCATCAGCAAAGCCTCCATC | 474 bp | PCR | 20 | |
| rstR (El Tor)-F | GCACCATGATTTAAGATGCTC | 501 bp | PCR/CBH/Seq | 20 | |
| rstR (Calcutta)-F | CTGTAAATCTCTTCAATCCTAGG | 313 bp | PCR | 20 | |
| rstR (Environment)-F | GTTAACGCTTCAAGCCTG | 372 bp | PCR/CBH/Seq | This study | |
| rstR (El Tor)-R1 | GAGCATCTTAAATCATGGTGC | 0.9 kb, 2.1 kb | PCR/Seq | This study | |
| rstR (Environment)-R1 | CAGGCTTGAAGCGTTAAC | 0.9 kb, 3.8 kb, 0.75 kb | PCR/Seq | This study | |
| tcpA | tcpH1-F | AGCCGCCTAGATAGTCTGTG | 1,324 bp | PCR/Seq | 45 |
| tcpA4-R | TCGCCTCCAATAATCCGAC | PCR/Seq | 45 | ||
| tcpA-F | CACGATAAGAAAACCGGTCAAGAG | 2,108 bp | PCR/Seq | 25 | |
| tcpQ-R | GAGGACTGTTCTGCAAATCTGCTCAT | PCR | 34 | ||
| tcpA (classical)-R | TTACCAAATGCAACGCCGAATG | Seq | This study | ||
| tcpA (El Tor)-R | CGAAAGCACCTTCTTTCACACGTTG | Seq | This study | ||
| toxT | toxT-R | CCATGAATGTAGCACCAAG | PCR/Seq | This study | |
| toxT-F | CTTGTCTATTGTTCGTAAAGTG | 834 bp | PCR/Seq | This study | |
| toxT-F2 | CTGATGATCTTGATGCTATG | 298 bp | PCR/Seq | This study | |
| TLC | tlc-F | GGGAATGTTGAGTTCTCAGTG | 1,548 bp | PCR | 6 |
| tlc-R | GTTGCGAAGTGGATTTTGTG | PCR | 6 | ||
| RTX | rtx-F | CACTCATTCCGATAACCAC | 1,366 bp | PCR | 6 |
| rtx-R | GCGATTCTCAAAGAGATGC | PCR | 6 | ||
| T3SS-2 | vcsN2-F | CAACACCTTCAAAGCCTTG | 848 bp | PCR/CBH | This study |
| vcsN2-R | GCGAGCTCCAATTGAAAC | PCR/CBH | This study | ||
| NAG-ST | stn-F | GAGAAACCTATTCATTGC | 216 bp | PCR/CBH | 59 |
| stn-R | GCAAGCTGGATTGCAAC | PCR/CBH | 59 | ||
| VSP-I | VC0175F2 | TGAGGGCTGCTGAATAAG | 689 bp | PCR/CBH | This study |
| VC0175R2 | TTGGCTCTGGTGTAAAGG | PCR/CBH | This study | ||
| VC0183F2 | AATACCACAAAGGACGCAC | 687 bp | PCR/CBH | This study | |
| VC0183R2 | TGGCCTAACCATAAGCAG | PCR/CBH | This study | ||
| VSP-II | VSPII LA F2 | CGCACATACTCTTTGGTGGCATCAG | 10,091 bp | PCR/CBH | This study |
| VSP LA R2 | ACTCTCTCTGATGGTGATGGCCTTC | PCR/CBH | This study | ||
| chxA | chxAF1 | GTCGAAGATGAGTTAAACAT | 1,904 bp | PCR/CBH | 13 |
| chxAR1 | TTATTTCAGTTCATCTTTTCGC | PCR/CBH | 13 | ||
| VSP-I | VC0175F2 | TGAGGGCTGCTGAATAAG | 689 bp | PCR/CBH | This study |
| VC0175R2 | TTGGCTCTGGTGTAAAGG | PCR/CBH | This study | ||
| VC0183F2 | AATACCACAAAGGACGCAC | 687 bp | PCR/CBH | This study | |
| VC0183R2 | TGGCCTAACCATAAGCAG | PCR/CBH | This study | ||
| VSP-II | VSPII LA F2 | CGCACATACTCTTTGGTGGCATCAG | 10,091 bp | PCR/CBH | This study |
| VSP LA R2 | ACTCTCTCTGATGGTGATGGCCTTC | PCR/CBH | This study | ||
| chxA | chxAF1 | GTCGAAGATGAGTTAAACAT | 1,904 bp | PCR/CBH | 13 |
| chxAR1 | TTATTTCAGTTCATCTTTTCGC | PCR/CBH | 13 | ||
| recA | Vm-recA-F | CTGGGTGTTAATATCGATGAG | 73 bp | RT-PCR | This study |
| Vm-recA-R | TCACAAATTTCTAGTGCTTG | RT-PCR | This study | ||
| Vm-recA-P | CTACTGGTTTCTCAGCCAGATACCGGTGA | RT-PCR | This study | ||
| toxR | Vm-toxR-F | TCCGATTAGGCAGTAACGAAA | 83 bp | RT-PCR | This study |
| Vm-toxR-R | ATGCAAATCGTTGCGAGAAAC | RT-PCR | This study | ||
| Vm-toxR-P | TCGAATTCTTTGGCTGCTTGCTCAGC | RT-PCR | This study | ||
| toxS | Vm-ToxS-F | TGGCAATCGAAGATGGTTTCA | 76 bp | RT-PCR | This study |
| Vm-ToxS-R | ACATCAACACGGCTCAATGG | RT-PCR | This study | ||
| Vm-ToxS-P | TGATCAAAACCAAGAGTAACAGTCCAGCA | RT-PCR | This study | ||
| hns | Vm-hns-F | GCTTATTTCTGCACTTTCAGG | 85 bp | RT-PCR | This study |
| Vm-hns-R | GTCGATGTACTTGTATTTCGC | RT-PCR | This study | ||
| Vm-hns-P | ACTAAAGCAAAAGGCAAACGTGCTCCTCG | RT-PCR | This study | ||
| tcpA | Vm-tcpA-F | GCTCAGCGTGCGATTGACT | 76 bp | RT-PCR | This study |
| Vm-tcpA-R | GAGTAAGCGACACTTGGATTG | RT-PCR | This study | ||
| Vm-tcpA-P | ATGACCAAGGCTGCGCAAAATCTCA | RT-PCR | This study | ||
| toxT | Vm-toxT-F | CGAGATTCTAGTTCTTTTTTCAAAACC | 123 bp | RT-PCR | This study |
| Vm-toxT-R | TGATGCTATGGAGAAAATATCATGTT | RT-PCR | This study | ||
| Vm-toxT-P | TAATTCATCACAAACATCAGACCAACGCCA | RT-PCR | This study | ||
| ctxA | ctxA-F | GGAGGGAAGAGCCGTGGAT | 66 bp | RT-PCR | 49 |
| ctxA-R | CATCGATGATCTTGGAGCATTC | RT-PCR | 49 | ||
| ctxA-P | CATCATGCACCGCCGGGTTG | RT-PCR | 49 | ||
| tcpP | tcpP-F | TGGTACACCAAGCATAATACAGACTAAG | 100 bp | RT-PCR | 49 |
| tcpP-R | AGGCCAAAGTGCTTTAATTATTTGA | RT-PCR | 49 | ||
| tcpP-P | TACTCTGTGAATATCATCCTGCCCCCTGTC | RT-PCR | 49 | ||
| tcpH | tcpH-F | GCCGTGATTACAATGTGTTGAGTAT | 82 bp | RT-PCR | 49 |
| tcpH-R | TCAGCCGTTAGCAGCTTGTAAG | RT-PCR | 49 | ||
| tcpH-P | TCAACTCGGCAAAGGTTGTTTTCTCGC | RT-PCR | 49 | ||
“T3SS-2” and “NAG-ST” indicate type III secretion system 2 and heat-stable enterotoxin of V. cholerae, respectively.
“F,” “R,” and “P” indicate forward primer, reverse primer and probe, respectively. Gene names are indicated in the primer designations.
“CBH” and “Seq” indicate colony blot hybridization and sequencing analysis, respectively.
FIG 4.
Genetic relatedness of tcpA and toxT genes among strains of V. cholerae and V. mimicus of different serogroups. (A) The novel tcpA gene of V. mimicus of this study did not cluster in classical strains, El Tor strains, or other types of strains but formed a separate clade showing closeness to strains of serogroups O56 and O115 of V. cholerae. (B) The novel toxT gene of V. mimicus of this study did not group into the major cluster comprising the toxT genes of V. cholerae O1 classical, El Tor, O139, and non-O1/non-O139 strains or of other V. mimicus strains but grouped into a separate cluster with atypical toxT genes reported in a few non-O1/non-O139 strains of V. cholerae.
Similarly to the results seen with tcpA amplification, PCR using the conventional primers for toxT did not produce any amplicon for the environmental V. mimicus strains of this study. However, application of newly designed primers (Table 3), considering variations in classical, El Tor, and environmental types of toxT, successfully yielded a specific amplicon of this gene in the study strains. Sequencing results showed identical levels of sequence homology of toxT in all the environmental V. mimicus strains. Comparative analysis identified the presence of a new allele with several unique substitutions and 76.9% to 78.0% homology among the deduced amino acid residues in comparison to the canonical toxT gene of the classical and El Tor V. cholerae strains (Fig. S5). Higher diversity was observed in the amino-terminal half of ToxT sequences than was seen in those of the environmental V. mimicus and V. cholerae O1 strains. Phylogenetic analysis clearly differentiated the toxT genes into two major clusters, one including the usual toxT gene commonly found in epidemic V. cholerae O1 strains and the other comprising the variant toxT gene identified in this study and in several V. cholerae non-O1/non-O139 strains from India (Fig. 4). However, the variant ToxT present in environmental V. mimicus strains was novel in terms of the acquired differences in 11 amino acid residues in comparison to that of the non-O1/non-O139 V. cholerae strain in the same phylogenetic cluster and to 59 to 60 amino acid residues with the canonical ToxT found in classical and El Tor V. cholerae O1.
CT production capacity and virulence potential.
All the environmental V. mimicus strains showed identical patterns for the major virulence-related genes, including those of the predicted amino acid sequences in CTX prophages and the TCP island, which indicated their probable functional capability to produce CT and virulence-related proteins. Therefore, bead enzyme-linked immunosorbent assays (bead-ELISA) were carried out under established conditions for both the El Tor and classical strains of V. cholerae O1 to check the functional CT production capacity and its variations, if any, among the environmental V. mimicus strains. Results showed that the CT production capacities differed among the environmental V. mimicus strains and were higher under the in vitro conditions favorable for the classical strains (LB medium, pH 6.6, 30°C) than under El Tor conditions for V. cholerae strains (AKI medium [1.5% Bacto peptone, 0.4% yeast extract, 0.5% NaCl, 0.3% NaHCO3], pH 7.4, 37°C). Of the six V. mimicus strains, one strain (V. mimicus DCT5) showed high CT production capacity (110 and 30 ng ml−1 in LB and AKI, respectively) whereas the toxin production was very low (0.1 to 0.5 ng ml−1) in others (Table 1).
Suckling mouse assay (SMA)-based in vivo experiments produced results in congruence with the CT production capacity for the V. mimicus strains (Table 1). Both live cells (106 to 107 CFU) and culture filtrates of V. mimicus strain DCT5 producing high CT levels induced fluid accumulation and diarrhea in all the experimental mice and hence were revealed to be enterotoxigenic. The SMA score, representing the fluid accumulation ratio, ranged between 0.083 and 0.090 (0.087 ± 003) for the high-CT-producing V. mimicus strain DCT5. However, none of the experimental mice exhibited diarrhea when low-CT-producing V. mimicus strain DCT2 was administered at a normal dose (107 CFU) or even when it was administered at a higher dose (>5 × 109 CFU). The fluid accumulation ratio shown by this strain with attenuated CT production ranged between 0.065 and 0.070 (0.0068 ± 002), which was similar to the range seen with the negative control (0.062 ± 002) (Table S1).
Transcriptional analysis of genes associated with CT production.
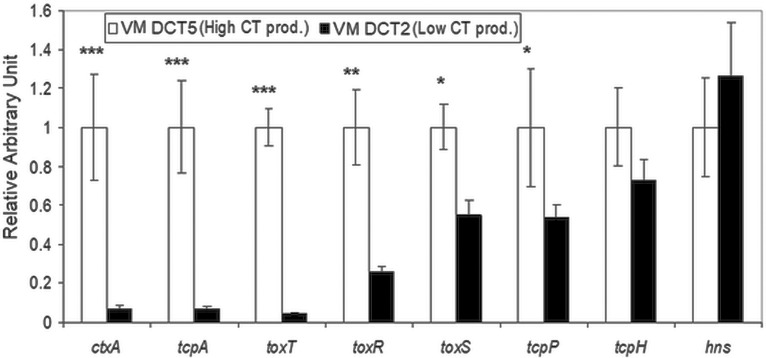
According to the results of bead-ELISA, the optimum culture condition for CT production, i.e., the classical type condition using LB medium, was selected for transcriptional analysis of ctxAB and its known regulatory genes by quantitative reverse transcription-PCR (qRT-PCR) in the high-CT-producing (V. mimicus DCT5) and low-CT-producing (V. mimicus DCT2) environmental V. mimicus strains used in SMA in vivo experiments. As expected, a significantly lower level of transcription of ctxA was observed in the low-CT-producing strain than in the high-CT producer. Similarly, significant low-level transcription of tcpA and toxT, which are known to directly interact with CT production, was also observed. In checking the transcription of other genes in the ToxR regulon influencing CT production, high and low levels of transcription of ctxA were observed to be correlated with the mRNA transcription of toxR, toxS, and tcpP (Fig. 5). In the low-CT-producing strain, the levels of transcription of the ctxA, tcpA, and toxT genes were significantly lower by about 15-fold to 25-fold (P < 0.005) than those of the high-CT-producing strains. Similarly, toxR expression was also significantly (about 4-fold [P <0.01]) lower, while both toxS and tcpP showed about 2-fold (P < 0.05)-lower transcription. In the low-CT-producing strain, tcpH transcription was about 1.4-fold lower but the difference was not significant in comparison to the high-CT producer. On the other hand, an opposite trend was observed for hns transcription; the high-CT-producing strain showed about 1.3-fold-lower hns transcription than the low CT producer, although the difference was not significant.
FIG 5.
Variation in mRNA transcription levels of virulence genes and regulated genes between high-CT-producing and low-CT-producing strains of Vibrio mimicus. Transcriptional levels of various virulence-related genes were analyzed by qRT-PCR. The relative transcriptional level of each gene was normalized with the housekeeping recA gene. The mRNA transcription level of each gene in a low-CT-producing strain was compared with that of a high-CT-producing strain. The transcriptional level of each virulence-related gene of the high-CT-producing strain was arbitrarily assigned a value of 1 (relative arbitrary unit). Statistically significant differences were calculated using the two-sample t test. A P value of <0.05 was considered significant (***, P <0.005; **, P <0.01; *, P <0.05).
DISCUSSION
Understanding the adaptive evolutionary mechanism of the CTXФ and ctxAB genes encoding the cholera toxin (CT) is highly important because of its direct relation to severe diarrheal diseases such as cholera, which causes health hazards throughout the world. In this aspect, through acquisition of toxigenic ctxAB genes, V. mimicus might play a salient role in its maintenance and propagation in the natural environment. A previous investigation (8), suggesting a possible occurrence of atypical tcpA in these ctx+ V. mimicus strains, prompted us to perform detailed characterizations (e.g., genotyping, sequence diversity, and transcriptional analysis) of genes of CTX prophage and ToxR regulon and to compare the characteristics of genomic relatedness, CT production in vitro, and virulence potential in vivo of these strains.
Novel ctxAB allele and multiple CTX prophages in environmental V. mimicus strains.
Comparing the core and flanking regions of CTX prophage in V. mimicus with those of V. cholerae strains showed a lot of previously unidentified variation in amino acid residues. Remarkably, the novel ctxB variant (Table 2), designated genotype 14, in the V. mimicus strains is distantly related to El Tor genotype 3 but is more closely related to the classical genotype 1 and Haitian genotype 7, the predominant traits of V. cholerae hybrid strains associated with the recent cholera epidemics (2–4). The functional implications of these changes in ctxAB in V. mimicus are required to be investigated in detail, particularly in relation to virulence potential, e.g., the crystal structure of the variant CT (27), constructing mutant strains and verifying the possible impacts of the changes by in vivo experiments. On the other hand, comparison of the orfU genes of CTX prophage in V. mimicus strains could identify its close homology with that of the El Tor type, which is inconsistent with its reported affiliation with the classical clade of V. cholerae O1 (7). Nonetheless, several unique changes in the amino acid residues within the first two domains (D1 and D2) of three domains of orfU (28) indicated the ongoing evolution of CTX prophage in V. mimicus in parallel to those of the epidemic V. cholerae O1. These polymorphic residues in orfU (gIIICTX) most likely interact with the periplasmic protein, TolA, and with “adsorption” domains, associated with phage penetration (28).
The occurrence of a single-copy CTX prophage in the studied V. mimicus strains was claimed in a previous study, based on hybridization of gDNA restriction digestions with ctxA probe (8). However, our results obtained by genome walking through a series of hybridization and PCR assays demonstrated that the V. mimicus strains actually contain one mature El Tor type prophage (CTXФET) with ctxAB and two environmental type pre-CTX prophages (pre-CTXФEnv) without ctxAB. The existence of pre-CTXΦ has been reported for only a very few strains of V. cholerae, including the epidemic O1/O139 strain and also non-O1/O139 strains (19, 22, 25, 29). Integration of at least two types of CTXФ (El Tor and environmental) within the genome of V. mimicus is an interesting and novel observation. In V. cholerae strain N16961, the reference El Tor strain, the phage integration site, characterized by the attR sequence followed by XerC and XerD binding sites, starts at 106 bp downstream of ctxAB (30). That study observed a similar location for the integration site of CTXΦET but a different intergenic location of attR-XerC-XerD for pre-CTXΦEnv. The occurrence of the RS1 element in the V. mimicus strains follows the evolutionary trait of the El Tor strains of V. cholerae. Acquisition of RS1 may not only increase CTXФ production, with a possible influence on ctxAB transcription, but also promote genomic diversity by the loss of CTX prophage and lysogenic immunity to make room for new CTX prophage to be integrated (18, 29, 31). In comparison to the canonical El Tor type strains, the hybrid El Tor strains with the classical ctxB genotype are considered to be more virulent than the El Tor CT producer (32). The acquisition of hybrid CTXΦET by the V. mimicus strains might have equipped them with greater evolutionary fitness, since these pathogenic strains can utilize the chance of becoming selectively enriched in the intestine of humans and animals (33).
Diverse TCP and ToxT alleles in V. mimicus strains.
Not only the CTX elements but also the TCP genes in V. cholerae can be mobilized by generalized transduction (34). The sequence of the tcpA locus in the TCP element is known to be more divergent than those of other loci in the VPI (35). Similarly, we have observed a high diversity in tcpA sequences by phylogenetic analysis, which is in congruence with previous studies (36, 37). Notably, the environmental V. mimicus strains contain a novel allele of tcpA, identified as tcpAEnv_Vm, with ca. 70% to 74% homology at nucleotide level compared to the reference El Tor and classical strains of V. cholerae O1. The progenitors of these V. mimicus strains might have acquired this tcpA genotype from V. cholerae non-O1/non-O139 strains of a particular phylogenetic clade, including the O56 and O115 serogroups, and/or vice versa (Fig. 4). This observation is not coherent with the previously reported V. mimicus tcpA genotypes, clustered to the phylogenetic clades comprising the canonical classical and El Tor strains of V. cholerae O1 (36). Most likely, this diversity is a reflection of diversifying selection with respect to CTXΦ susceptibility as part of the ongoing adaptive evolution among potentially pathogenic Vibrio populations of this kind. Similar reasons may explain the occurrence in studied V. mimicus strains of a novel toxT allele, affiliating with the atypical toxT gene of certain non-O1/non-O139 strains but not the canonical toxT of epidemic O1 classical and El Tor strains of V. cholerae (25).
Comparative analysis of tcpA sequences has clearly identified a high substitution rate in the carboxy-terminal half, encoding the exposed part of the TCP pilus on the cell surface. Among the many differences between the novel tcpAEnv_Vm gene and the tcpACla gene is the ca. 187 V>K substitution, which may influence pilus-mediated autoagglutination (38). In the case of toxT, the diversity in amino acid residuals in comparison to the reference strain was higher in the amino-terminal half, which is in accordance with a previous study (25). The relatively conserved carboxy-terminal half is known to determine the specificity of ToxT protein binding to DNA regulatory sites (25). Apart from acting as a virulence factor, TCP may also aid in environmental persistence, e.g., of biofilm formation on aquatic particles and of organisms, particularly, chitinous zooplankton (39). The occurrence of a new variant tcpA gene, with possible alterations in cell surface epitopes, among the toxigenic V. mimicus strains of the present study might be have been a consequence of an adaptive evolutionary response to the changes in environmental niche.
Variation in CT production and virulence potential and its regulatory framework in V. mimicus.
In the case of V. cholerae, strains producing a concentration of CT of at least ∼20 ng ml−1 are known to cause fluid accumulation or diarrhea in experimental animal (32). Most of the V. mimicus strains did not cause fluid accumulation in sucking mice, in concordance with the very low level of CT production (<0.5 ng ml−1) seen under in vitro growth conditions. Yet these strains are potentially toxigenic, as reflected in one of the V. mimicus strains, with identical virulence genotypes but producing a high amount (>100 ng ml−1) of CT and diarrhea in vivo. We also cannot rule out the possibility that the experimental conditions used in this study may not be optimal for CT production in V. mimicus strains and need to be further investigated using different culture conditions and growth media. The absence of any other potential virulence factors, e.g., the TTSS, NAG-ST, ChxA, and RTX, indicates that the observed enterotoxicity in mice intestine is due to CT produced by the environmental V. mimicus strains. However, detailed investigations, including understanding the role of intestinal receptors (other than GM1) of CT and the intestinal colonization capacity of these V. mimicus strains containing a new variant of tcpA, are required to decipher the virulence mechanism triggering diarrhea in mouse (40, 41).
Despite the presence of CTXΦ and the TCP element, the variation in CT production in V. mimicus strains is likely influenced by other genetic factors; e.g., the expression of CT and TCP in V. cholerae is activated by ToxT, which is regulated by the TcpP-TcpH-ToxR-ToxS complex (24). In the case of the high-CT-producing V. mimicus strain, higher transcription of ctxA in conjunction with that of tcpA and toxT and the upstream-regulatory genes toxR and toxS corroborates the known ToxT-ToxR-mediated genetic regulation influencing CT production in V. cholerae. In addition, the histone-like nucleoid structuring protein (H-NS) encoded by hns, a global prokaryotic gene regulator, has been shown to repress the transcription of virulence genes, including toxT, ctxAB, and tcpA in V. cholerae (42). However, the variation in CT production in V. mimicus strains is not influenced by the H-NS since its transcription did not show a considerable change in parallel to that of ctxAB. The ToxR regulon is thought to be controlled by environmental stimuli, such as temperature, pH, and osmolarity (24). Hence, understanding the precise genetic and physiological mechanisms behind the variations in the level of CT production in V. mimicus requires more-extensive research beyond the scope of this study.
Probable role of V. mimicus in the evolution of CTXФ.
In the V. mimicus strains, the presence of the recombinase XerC and XerD binding sequences at both ends of the pre-CTXФEnv and CTXФET prophages support the concept of phage-mediated integration events of these external genetic elements (23). Pathogenic V. cholerae strains are thought to have evolved from their nonpathogenic progenitors first through acquisition of tcpA and other genes of VPI and then through integration of CTXФ (43). On the other hand, special forms of the CTXΦ family, designated pre-CTXΦ, do not carry ctxAB but contain other genes considered to be CTXΦ precursors (6). The occurrence of RS1–CTXET–RS1–pre-CTXEnv–pre-CTXEnv in V. mimicus suggests their integration through stepwise horizontal gene transfers (HGTs) and recombination of phage genes, and these strains may be considered to be among the missing evolutionary links for the diverse arrays of multiple alleles of CTXET, pre-CTXEnv, and RS1 reported to occur in the O1 and non-O1/O139 strains of V. cholerae (19–22, 29). The presence of tandem copies of CTX and RS1 phages along with the novel types of ctxAB, tcpA, toxT, and orfU indicates that CTX prophages in the V. mimicus genome might have evolved independently of the seventh-pandemic El Tor clones, probably through independent integration of pre-CTXФEnv, in duplicate, and then of a primeval CTXФET (19). This proposition is in congruence with the absence of other hallmark factors of the El Tor strains, including VSP-I and VSP-II gene clusters, and also the RTX and TLC elements (11, 21), in these V. mimicus strains. Several studies comparing genome sequences have also indicated horizontal transfer of virulence-related genes in V. mimicus from an uncommon clone of V. cholerae rather than from the seventh-pandemic strains (9, 44). This is further supported by the existence in the ctxAB promoter region of V. mimicus strains of five heptamer (TTTTGAT) repeats, a characteristic genomic signature of the classical O1 strain isolated during the 1960s, while the El Tor O1 strains, isolated from the 1970s onward, contained four heptamer repeats (30).
The evolutionary success or failure of a new allele depends on numerous factors, e.g., the consecutive direction of gene flow, precursor-product relationships and influences of regulatory molecular-genetic processes during evolution, and the ecological fitness of the host bacterium, which is very difficult to predict or decipher. Nonetheless, the adaptive survival of particular ctx+ V. mimicus strains in the estuarine environment is indicated in their isolation during different months of a given year. These strains have been observed to be clonally related, which suggests their probable origin from a common ancestor. However, as reflected by their variable antimicrobial resistance traits and differentiation into several PFGE subtypes, V. mimicus strains of this kind are prone to genomic modifications, some of which may aid in their ecological fitness. The probable influence of environmental V. mimicus on the ongoing population shift of typical El Tor strains to hybrid El Tor strains carrying classical and variant type ctxB cannot be dismissed. Through acquisition of pre-CTXФ, particular tcp+ strains of V. mimicus probably serve as a cryptic but important natural reservoir of the CTXФ and TCP and related virulence genes.
Conclusion.
It can be inferred that certain clonally related environmental V. mimicus strains can act as reservoir of variant ctxB genes, designated genotype 14, which is phylogenetically closer to currently predominant genotypes 1 and 7 associated with cholera outbreaks worldwide. This report provides molecular insight into the virulence potential of ctx+ V. mimicus strains, which could potentially serve as reservoirs of novel or variant types of not only ctxAB but also tcpA, toxT, and orfU. The genomic contents of tandemly arranged multiple pre-CTXФEnv types and a CTXФET with novel classical type ctxAB probably act as salient raw materials for the natural recombination events driving the evolution of virulence genes related to CT production. Though CT production in some examples of environmental V. mimicus strains of this kind can be naturally attenuated, the strains might be toxigenic under favorable conditions and might be able to instigate cholera-like diarrhea. The variation in CT production capacity in V. mimicus is partially explained by the coordinated differential transcription of several genes within the ToxR-ToxT regulatory cascade. The cryptic existence of the virulence genes related to CT production in V. mimicus genome points out an unnoticed event in the evolutionary pathway of CTXΦ ecology and cholera epidemiology. Systematic environmental surveillance of nonepidemic strains, including V. mimicus and V. cholerae, and associated detailed investigations of the genetic and functional diversities of genes influencing CT production and virulence potential in different hosts and under different environmental conditions would allow us to improve understanding the evolution of new variant ctx elements and of CTXФ as well as of the genes that regulate them.
MATERIALS AND METHODS
Bacterial strains and their antimicrobial susceptibility.
Six ctx+ V. mimicus strains (Table 1) were obtained from the culture collection of the Environmental Microbiology Laboratory of icddr,b, Dhaka. These strains were isolated during the postmonsoon and early winter months in 2000 from the Karnaphuli River estuary, Bangladesh. The ctx+ strains were screened from 1,600 presumptive V. mimicus colonies, grown on thiosulfate citrate bile salts sucrose (TCBS) agar after enrichment of environmental samples in alkaline-peptone-water (APW) (pH 8.0). All strains were grown in Luria-Bertani (LB) broth, and their identity was verified according to a standard protocol (45). Strains stored as glycerol stock at −80°C were grown in APW and subsequently on TCBS agar (Difco Laboratories, MI, USA), gelatin agar (Difco), and LB at 37°C whenever needed. Several reference strains of V. cholerae (i.e., N16961 and O395, representing the El Tor and classical biotypes, respectively; VCE233 and AS522, non-O1/non-O139 strains containing environmental and Kolkata type CTX prophages; SG6, a type III secretion system [TTSS]-positive non-O1/non-O139 strain; GP156, a stn-positive O1 El Tor strain; and C9, a chxA-positive non-O1/non-O139 strain) and V. mimicus ATCC 33653T were used as controls. Each of the ctx+ V. mimicus strains was examined for resistance to some commonly used antimicrobials (Table 1) to observe related phenotypic variations, if any. Antimicrobial resistance was determined by the disc diffusion method according to the Clinical and Laboratory Standards Institute (http://www.clsi.org) using Mueller-Hinton agar (Difco) and commercially available discs (Oxoid, Hampshire, England).
Pulsed-field gel electrophoresis (PFGE).
PFGE was performed according to the Pulse Net USA protocol (https://www.cdc.gov/pulsenet/pathogens/pfge.html) with slight modifications. Briefly, freshly grown V. mimicus strains were embedded into 1% Seakem Gold agarose (Sigma-Aldrich, MO, USA) followed by lysis of the cells with 0.5 mg ml−1 proteinase K (P8044-5G; Sigma) and 1% sarcosine (Sigma) at 54°C for 1 h. Agarose blocks containing genomic DNA were digested with NotI and SfiI (TaKaRa Bio Inc., Otsu, Japan) (30 and 40 U, respectively) using appropriate buffer at 37°C for 3 h. DNA fragments were electrophoresed in 1% pulsed-field-certified agarose gel (Bio-Rad Laboratories, CA, USA) using a Chef Mapper (Bio-Rad). Gels were stained for 30 min and destained twice for 15 min each time, and images were captured using a Gel-Doc 2000 system (Bio-Rad). A lambda ladder (Bio-Rad) was used as a molecular mass standard. The PFGE fingerprints were analyzed by the use of Fingerprinting II software (Bio-Rad).
Colony blot hybridization of virulence-related genes.
DNA probes for colony blot hybridization included the major toxigenic factors, e.g., cholera toxin (ctxA), zonula occludens toxin (zot; part of CTX phage), RS1 element (rstC), Vibrio seventh-pandemic island (including VSP-I and VSP-II, markers of the present pandemic O1 El Tor biotype), the TLC element, and other known virulence genes of V. cholerae, namely, vcsN2, chxA, stn, and rtxA, encoding the type III secretion system, cytotoxic cholix toxin, heat stable enterotoxin, and repeat in toxin, respectively. DNAs of reference V. cholerae strains and gene-specific primers were used as a template for PCR amplification of these genes (Table 3). PCR products were purified by standard methods and labeled by random priming with [α-32P]dCTP (370 MBq mmol−1) using a Multi-Prime DNA labeling system (GE Healthcare, Buckinghamshire, United Kingdom). Environmental V. mimicus strains were grown on nitrocellulose membrane, overlaid on LB agar at 37°C for 4 to 6 h, and subjected to colony blot hybridization following the procedure described by Yamasaki et al. (46). Radioactivity in the hybridized membrane was detected using a BAS FLA-3000 system (Fuji Film, Tokyo, Japan).
PCR-based typing of virulence genes and CTX phage element.
Template DNA was prepared by the use of the standard boiling method and was stored at −30°C until use. MAMA-PCR (47) was employed to detect ctxB genotypes in V. mimicus strains to define their potential for classical or El Tor type CT production. The presence of the RS1 element was determined by rstC gene-based PCR (18). Genotypes of rstR, namely, classical, El Tor, Kolkata, and environmental genotypes, were also determined by PCR using a newly designed primer set for the environmental type and previously established protocols for others (20). The genotypes of tcpA, belonging to the VPI, were also checked by PCR using previously established methods (25). Details of the primers and PCR conditions for screening these genes are mentioned in Table 3.
Southern hybridization and PCR arrays to understand the genetic organization of CTXФ.
Southern hybridization analyses of BglI-digested and BglII-digested gDNA of V. mimicus strains were carried out with probes for selected virulence-related genes, including ctxAB, rstRET, rstREnv, and rstC. Briefly, 5-μg aliquots of total gDNA were digested with the restriction enzymes using appropriate buffer and electrophoresed in 0.8% pulsed-field-certified agarose gel (Bio-Rad) using a Chef Mapper (Bio-Rad). Once separated, the gDNA fragments were subjected to Southern hybridization transfer and blotted onto nylon membranes (Hybond-N+; Amersham, GE Healthcare, Buckinghamshire, United Kingdom). The genomic blots were hybridized with the gene probes, labeled by random priming with [α-32P]dCTP (370 MBq mmol−1), and autoradiographed as described previously (13). In order to verify the genetic organization of CTXФ and associated elements, a series of PCR arrays were performed using the forward and reverse allelic primers, with multiple combinations of genes rstC, rstRET, rstREnv, ctxAB, and orfU as shown in Fig. 3 and the respective primers (Table 3).
Nucleotide sequencing and phylogenetic analysis of diversity of virulence genes.
V. mimicus strains were subjected to nucleotide sequencing analysis for several target virulence genes of CTXФ, including ctxAB, orfU, and rstR, and for associated flanking regions comprising zot, intergenic region 1 (ig-1) and ig-2, and TCP elements, namely, tcpA and toxT. Briefly, PCRs using primers (Table 3) targeting these virulence genes and flanking regions were conducted following standard protocols. The amplified products were purified using a QIAquick purification kit (Qiagen GmbH, Hilden, Germany), and then cycle sequencing was carried out using a BigDye Terminator v3.1 cycle sequencing kit according to the instructions of the manufacturer (Applied Biosystems, CA, USA). Afterwards, a further purification was done using CleanSEQ (Agencourt Bioscience, MA, USA), and nucleotide sequences were determined by the use of an ABI Prism 3100 Avant Genetic Analyzer (Applied Biosystems). The obtained gene sequences were assembled and aligned by the use of DNA Lasergene software (DNASTAR, WI, USA). Homology searching was performed using the BLAST program (http://blast.ncbi.nlm.nih.gov/Blast.cgi), and the nucleotide and deduced amino acid sequences were compared with those of published genes. A phylogenetic tree was constructed using the ClustalW algorithm to understand the genetic lineage and sequence diversity within the study strains and other representative sequences of the target genes of V. mimicus and V. cholerae published in GenBank. Representative sequences of the novel ctxAB, tcpA, toxT, and orfU genes in the V. mimicus strains of this study can be obtained from NCBI/DDBJ under accession no. LC427969, LC427970, LC427971, and LC427972, respectively.
Measuring CT production by bead enzyme-linked immunosorbent assay (bead-ELISA).
The ctx-positive V. mimicus strains were grown in AKI medium (pH 7.4) and LB medium (L-broth) (Difco, KS, USA) (pH 6.6) for 12 h at 37 and 30°C, respectively, to compare their CT production levels under conditions favorable for the El Tor and classical strains of V. cholerae (48). Subsequently, the optical density at 600 nm (OD) nm of the bacterial cultures was adjusted to 1.0, followed by 100-fold dilution in the respective media and incubation under stationary and shaking conditions, for 4 h each, at 180 rpm (48, 49). The cell-free supernatant (CFS) of each culture was prepared by centrifugation at 12,000 × g for 10 min followed by filtration through a 0.22-μm-pore-size filter (Iwaki, Tokyo, Japan). The CFS from each culture was diluted 10, 100, and 500 times in 10 mM sodium phosphate buffer containing 100 mM NaCl (pH 8.0), and the level of CT produced was measured by bead-ELISA. Purified CT was obtained following the methodology described by Uesaka et al. (50) and was used as controls for known concentrations. Preparation of polyclonal rabbit antisera against CT, conjugation of Fab′ of antitoxin IgG with horseradish peroxidase, and estimation of the CT level secreted by each strain by bead-ELISA were done according to the method previously described by Oku et al. (51). All experiments were done in triplicate.
Detection of pathogenic potential in vivo.
Two strains of ctx+ V. mimicus showing similar PFGE pulsotypes but producing high and low CT levels, as detected by ELISA, were selected for evaluating enterotoxigenic potential in vivo. A suckling mouse assay (SMA) was performed using 3-day-old Swiss albino suckling mice according to standard procedures (52). Briefly, an aliquot (0.1 ml) of bacterial culture freshly grown in LB medium (as well as its filtrate [obtained using a 0.2-μm-pore-size filter]) was mixed with Evans blue (0.01% [wt/vol]) and intragastrically inoculated into each suckling mouse. A volume approximately equal to 107 CFU was inoculated as the normal dose; however, higher and lower doses were also administered for low-CT and high-CT producers, respectively. After 6 h of incubation, the intestines of the mice were removed, pooled, and weighed. The fluid accumulation score from the SMA was expressed as the ratio of the weight of the intestine to the remaining body weight, and a ratio of ≥0.08 was considered to represent a positive result. Culture filtrates of the reference strains of ctx+ V. cholerae O1 (O395) and of V. mimicus lacking ctx (ATCC 33653T) were used as positive and negative controls, respectively. The pathogenic potential of each strain was verified using five and three mice for live cells and culture filtrates, respectively.
RNA isolation and qRT-PCR assay.
The ctx+ V. mimicus strains, expressing high and low CT levels, were freshly grown to the mid-logarithmic phase (∼108 CFU ml−1) in LB medium under the classical condition (30°C) of CT production (48). Total RNA was extracted and purified using TRIzol reagent (Gibco-BRL, NY, USA) according to the manufacturer’s instructions. The qRT-PCR assay was carried out with the primers and probes for the appropriate genes, namely, ctxA, tcpA, toxT, toxR, toxS, tcpP, tcpH, and hns, which are known to regulate CT production, and a housekeeping recA gene as an internal control (Table 3) following the TaqMan probe method. Each probe was labeled with 6-carboxyfluorescein (FAM) and 6-carboxytetramethylrhodamine (TAMRA) as the 5ʹ reporter and 3ʹ quencher dyes, respectively. Reverse transcription was carried out for cDNA synthesis from an RNA template (1 μg) using a quick RNA-cDNA kit (Applied Biosystems) according to the manufacturer’s instructions. Real-time PCR was carried out using the amplified cDNA and TaqMan Gene Expression master mix containing each set of primer and probe (Applied Biosystems). PCR conditions were 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min in an ABI Prism 7000 sequence detection system (Applied Biosystems). The relative levels of transcription in comparison with the internal control were analyzed according to the method of Hagihara et al. (53).
Statistical analysis.
Statistica (ver. 10.0; StatSoft, OK, USA) was used to explore the differences between the mean values by applying Student’s two-sample t test. A P value of <0.05 was considered significant.
Accession number(s).
Newly determined sequence data have been deposited in NCBI/DDBJ under accession numbers LC427969, LC427970, LC427971, and LC427972.
Supplementary Material
ACKNOWLEDGMENTS
We appreciate the technical support of the environmental surveillance team of icddr,b. The thoughtful suggestions received from Prodyot Kumar Basu Neogi, formerly a scientist of icddr,b, are gratefully remembered.
This research was supported by the Osaka Prefecture University under the Monbukagakusho:MEXT scholarship and JASSO fellowship programs. icddr,b is thankful to the governments of Bangladesh, Canada, Sweden, and the United Kingdom for providing core/unrestricted support.
S.B.N. and N.C. designed and performed laboratory experiments and participated in data analysis. S.Y. and G.B.N. coordinated the experiments and analyzed the data. Z.H.M. and M.S.I. performed field studies. S.P.A., M.A., K.O., and A.H. helped design the study and participated in laboratory experiments. S.B.N., N.C., and S.Y. wrote the draft of the manuscript. All of us read and approved the final manuscript.
We declare that we have no conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01977-18.
REFERENCES
- 1.Safa A, Nair GB, Kong RY. 2010. Evolution of new variants of Vibrio cholerae O1. Trends Microbiol 18:46–54. doi: 10.1016/j.tim.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Rashed SM, Mannan SB, Johura FT, Islam MT, Sadique A, Watanabe H, Sack RB, Huq A, Colwell RR, Cravioto A, Alam M. 2012. Genetic characteristics of drug-resistant Vibrio cholerae O1 causing endemic cholera in Dhaka, 2006–2011. J Med Microbiol 61:1736–1745. doi: 10.1099/jmm.0.049635-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saidi SM, Chowdhury N, Awasthi SP, Asakura M, Hinenoya A, Iijima Y, Yamasaki S. 2014. Prevalence of Vibrio cholerae O1 El Tor variant in a cholera-endemic zone of Kenya. J Med Microbiol 63:415–420. doi: 10.1099/jmm.0.068999-0. [DOI] [PubMed] [Google Scholar]
- 4.Adewale AK, Pazhani GP, Abiodun IB, Afolabi O, Kolawole OD, Mukhopadhyay AK, Ramamurthy T. 2016. Unique clones of Vibrio cholerae O1 El Tor with Haitian type ctxB allele implicated in the recent cholera epidemics from Nigeria, Africa. PLoS One 11:e0159794. doi: 10.1371/journal.pone.0159794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faruque SM, Rahman MM, Asadulghani NKMI, Mekalanos JJ. 1999. Lysogenic conversion of environmental Vibrio mimicus strains by CTXPhi. Infect Immun 67:5723–5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd EF, Moyer KE, Shi L, Waldor MK. 2000. Infectious CTXФ and the vibrio pathogenicity island prophage in Vibrio mimicus: evidence for recent horizontal transfer between V. mimicus and V. cholerae. Infect Immun 68:1507–1513. doi: 10.1128/IAI.68.3.1507-1513.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bi K, Miyoshi SI, Tomochika KI, Shinoda S. 2001. Detection of virulence associated genes in clinical strains of Vibrio mimicus. Microbiol Immunol 45:613–616. doi: 10.1111/j.1348-0421.2001.tb01292.x. [DOI] [PubMed] [Google Scholar]
- 8.Islam MS, Rahman MZ, Khan SI, Mahmud ZH, Ramamurthy T, Nair GB, Sack RB, Sack DA. 2005. Organization of the CTX prophage in environmental isolates of Vibrio mimicus. Microbiol Immunol 49:779–784. doi: 10.1111/j.1348-0421.2005.tb03668.x. [DOI] [PubMed] [Google Scholar]
- 9.Wang D, Wang H, Zhou Y, Zhang Q, Zhang F, Du P, Wang S, Chen C, Kan B. 2011. Genome sequencing reveals unique mutations in characteristic metabolic pathways and the transfer of virulence genes between Vibrio mimicus and Vibrio cholerae. PLoS One 6:e21299. doi: 10.1371/journal.pone.0021299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyd EF, Heilpern AJ, Waldor MK. 2000. Molecular analysis of a putative CTXΦ precursor and evidence for independent acquisition of distinct CTXΦs by toxigenic Vibrio cholerae. J Bacteriol 182:5530–5538. doi: 10.1128/JB.182.19.5530-5538.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dziejman M, Balon E, Boyd D, Fraser CM, Heidelberg JF, Mekalanos JJ. 2002. Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc Natl Acad Sci U S A 99:1556–1561. doi: 10.1073/pnas.042667999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chatterjee S, Ghosh K, Raychoudhuri A, Chowdhury G, Bhattacharya MK, Mukhopadhyay AK, Ramamurthy T, Bhattacharya SK, Klose KE, Nandy RK. 2009. Incidence, virulence factors, and clonality among clinical strains of non-O1, non-O139 Vibrio cholerae isolates from hospitalized diarrheal patients in Kolkata, India. J Clin Microbiol 47:1087–1095. doi: 10.1128/JCM.02026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Awasthi SP, Asakura M, Chowdhury N, Neogi SB, Hinenoya A, Golbar HM, Yamate J, Arakawa E, Tada T, Ramamurthy T, Yamasaki S. 2013. Novel cholix toxin variants, ADP-ribosylating toxins in Vibrio cholerae non-O1/non-O139 strains, and their pathogenicity. Infect Immun 81:531–541. doi: 10.1128/IAI.00982-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faruque SM, Mekalanos JJ. 2003. Pathogenicity islands and phages in Vibrio cholerae evolution. Trends Microbiol 11:505–510. doi: 10.1016/j.tim.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 15.Waldor MK, Rubin EJ, Pearson GD, Kimsey H, Mekalanos JJ. 1997. Regulation, replication, and integration functions of the Vibrio cholerae CTXФ are encoded by region RS2. Mol Microbiol 24:917–926. doi: 10.1046/j.1365-2958.1997.3911758.x. [DOI] [PubMed] [Google Scholar]
- 16.Moyer KE, Kimsey HH, Waldor MK. 2001. Evidence for a rolling-circle mechanism of phage DNA synthesis from both replicative and integrated forms of CTXФ. Mol Microbiol 41:311–323. doi: 10.1046/j.1365-2958.2001.02517.x. [DOI] [PubMed] [Google Scholar]
- 17.Faruque SM, Asadulghani KM, Nandi RK, Ghosh AN, Nair GB, Mekalanos JJ, Sack DA. 2002. RS1 element of Vibrio cholerae can propagate horizontally as a filamentous phage exploiting the morphogenesis genes of CTXФ. Infect Immun 70:163–170. doi: 10.1128/IAI.70.1.163-170.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis BM, Kimsey HH, Kane AV, Waldor MK. 2002. A satellite phage-encoded antirepressor induces repressor aggregation and cholera toxin gene transfer. EMBO J 21:4240–4249. doi: 10.1093/emboj/cdf427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nandi S, Maiti D, Saha A, Bhadra RK. 2003. Genesis of variants of Vibrio cholerae O1 biotype El Tor: role of the CTXФ array and its position in the genome. Microbiology 149:89–97. doi: 10.1099/mic.0.25599-0. [DOI] [PubMed] [Google Scholar]
- 20.Bhattacharya T, Chatterjee S, Maiti D, Bhadra RK, Takeda Y, Nair GB, Nandy RK. 2006. Molecular analysis of the rstR and orfU genes of the CTX prophages integrated in the small chromosomes of environmental Vibrio cholerae non-O1, non-O139 strains. Environ Microbiol 8:526–534. doi: 10.1111/j.1462-2920.2005.00932.x. [DOI] [PubMed] [Google Scholar]
- 21.Faruque SM, Tam VC, Chowdhury N, Diraphat P, Dziejman M, Heidelberg JF, Clemens JD, Mekalanos JJ, Nair GB. 2007. Genomic analysis of the Mozambique strain of Vibrio cholerae O1 reveals the origin of El Tor strains carrying classical CTX prophage. Proc Natl Acad Sci U S A 104:5151–5156. doi: 10.1073/pnas.0700365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mantri CK, Mohapatra SS, Colwell RR, Singh DV. 2010. Sequence analysis of Vibrio cholerae orfU and zot from pre-CTXΦ and CTXΦ reveals multiple origin of pre-CTXΦ and CTXΦ. Environ Microbiol Rep 2:67–75. doi: 10.1111/j.1758-2229.2009.00085.x. [DOI] [PubMed] [Google Scholar]
- 23.Waldor MK, Mekalanos JJ. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 24.Skorupski K, Taylor RK. 1997. Control of the ToxR virulence regulon in Vibrio cholerae by environmental stimuli. Mol Microbiol 25:1003–1009. doi: 10.1046/j.1365-2958.1997.5481909.x. [DOI] [PubMed] [Google Scholar]
- 25.Mukhopadhyay AK, Chakraborty S, Takeda Y, Nair GB, Berg DE. 2001. Characterization of VPI pathogenicity island and CTXФ prophage in environmental strains of Vibrio cholerae. J Bacteriol 183:4737–4746. doi: 10.1128/JB.183.16.4737-4746.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33:2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heilpern AJ, Waldor MK. 2003. pIIICTX, a predicted CTXФ minor coat protein, can expand the host range of coliphage fd to include Vibrio cholerae. J Bacteriol 185:1037–1044. doi: 10.1128/JB.185.3.1037-1044.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ford CG, Kolappan S, Phan HT, Waldor MK, Winther-Larsen HC, Craig L. 2012. Crystal structures of a CTX pIII domain unbound and in complex with a Vibrio cholerae Tol A domain reveal novel interaction interfaces. J Biol Chem 287:36258–36272. doi: 10.1074/jbc.M112.403386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H, Pang B, Xiong L, Wang D, Wang X, Zhang L, Kan B. 2015. The hybrid pre-CTXФ-RS1 prophage genome and its regulatory function in environmental Vibrio cholerae O1 strains. Appl Environ Microbiol 81:7171–7177. doi: 10.1128/AEM.01742-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Halder K, Das B, Nair GB, Bhadra RK. 2010. Molecular evidence favouring step-wise evolution of Mozambique Vibrio cholerae O1 El Tor hybrid strain. Microbiology 156:99–107. doi: 10.1099/mic.0.032458-0. [DOI] [PubMed] [Google Scholar]
- 31.Kamruzzaman M, Robins WP, Bari SM, Nahar S, Mekalanos JJ, Faruque SM. 2014. RS1 satellite phage promotes diversity of toxigenic Vibrio cholerae by driving CTX prophage loss and elimination of lysogenic immunity. Infect Immun 82:3636–3643. doi: 10.1128/IAI.01699-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghosh-Banerjee J, Senoh M, Takahashi T, Hamabata T, Barman S, Koley H, Mukhopadhyay AK, Ramamurthy T, Chatterjee S, Asakura M, Yamasaki S, Nair GB, Takeda Y. 2010. Cholera toxin production by the El Tor variant of Vibrio cholerae O1 compared to prototype El Tor and classical biotypes. J Clin Microbiol 48:4283–4286. doi: 10.1128/JCM.00799-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tikoo A, Singh DV, Sanyal SC. 1994. Influence of animal passage on haemolysin and enterotoxin production in Vibrio cholerae O1 biotype El Tor strains. J Med Microbiol 40:246–251. doi: 10.1099/00222615-40-4-246. [DOI] [PubMed] [Google Scholar]
- 34.O’Shea YA, Boyd EF. 2002. Mobilization of the Vibrio pathogenicity island between Vibrio cholerae isolates mediated by CP-T1 generalized transduction. FEMS Microbiol Lett 214:153–157. doi: 10.1111/j.1574-6968.2002.tb11339.x. [DOI] [PubMed] [Google Scholar]
- 35.Sarkar A, Nandy RK, Nair GB, Ghose AC. 2002. Vibrio pathogenicity island and cholera toxin genetic element-associated virulence genes and their expression in non-O1 non-O139 strains of Vibrio cholerae. Infect Immun 70:4735–4742. doi: 10.1128/IAI.70.8.4735-4742.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar P, Thulaseedharan A, Chowdhury G, Ramamurthy T, Thomas S. 2011. Characterization of novel alleles of toxin co-regulated pilus A gene (tcpA) from environmental isolates of Vibrio cholerae. Curr Microbiol 62:758–763. doi: 10.1007/s00284-010-9774-3. [DOI] [PubMed] [Google Scholar]
- 37.Li F, Du P, Li B, Ke C, Chen A, Chen J, Zhou H, Li J, Morris JG Jr, Kan B, Wang D. 2014. Distribution of virulence-associated genes and genetic relationships in non-O1/O139 Vibrio cholerae aquatic isolates from China. Appl Environ Microbiol 80:4987–4992. doi: 10.1128/AEM.01021-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirn TJ, Lafferty MJ, Sandoe CM, Taylor RK. 2000. Delineation of pilin domains required for bacterial association into microcolonies and intestinal colonization by Vibrio cholerae. Mol Microbiol 35:896–910. doi: 10.1046/j.1365-2958.2000.01764.x. [DOI] [PubMed] [Google Scholar]
- 39.Reguera G, Kolter R. 2005. Virulence and the environment: a novel role for Vibrio cholerae toxin-coregulated pili in biofilm formation on chitin. J Bacteriol 187:3551–3555. doi: 10.1128/JB.187.10.3551-3555.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boyd EF, Waldor MK. 2002. Evolutionary and functional analyses of variants of the toxin-coregulated pilus protein TcpA from toxigenic Vibrio cholerae non-O1/non-O139 serogroup isolates. Microbiology 148:1655–1666. doi: 10.1099/00221287-148-6-1655. [DOI] [PubMed] [Google Scholar]
- 41.Cervin J, Wands AM, Casselbrant A, Wu H, Krishnamurthy S, Cvjetkovic A, Estelius J, Dedic B, Sethi A, Wallom K-L, Riise R, Bäckström M, Wallenius V, Platt FM, Lebens M, Teneberg S, Fändriks L, Kohler JJ, Yrlid U. 2018. GM1 ganglioside-independent intoxication by cholera toxin. PLoS Pathog 14:e1006862. doi: 10.1371/journal.ppat.1006862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nye MB, Pfau JD, Skorupski K, Taylor RK. 2000. Vibrio cholerae H-NS silences virulence gene expression at multiple steps in the ToxR regulatory cascade. J Bacteriol 182:4295–4303. doi: 10.1128/JB.182.15.4295-4303.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Shea YA, Reen FJ, Quirke AM, Boyd EF. 2004. Evolutionary genetic analysis of the emergence of epidemic Vibrio cholerae isolates on the basis of comparative nucleotide sequence analysis and multilocus virulence gene profiles. J Clin Microbiol 42:4657–4671. doi: 10.1128/JCM.42.10.4657-4671.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hasan NA, Grim CJ, Haley BJ, Chun J, Alam M, Taviani E, Hoq M, Munk AC, Saunders E, Brettin TS, Bruce DC, Challacombe JF, Detter JC, Han CS, Xie G, Nair GB, Huq A, Colwell RR. 2010. Comparative genomics of clinical and environmental Vibrio mimicus. Proc Natl Acad Sci U S A 107:21134–21139. doi: 10.1073/pnas.1013825107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis BR, Fanning GR, Madden JM, Steigerwalt AG, Bradford HB, Smith HL, Brenner DJ. 1981. Characterization of biochemically atypical Vibrio cholerae strains and designation of a new pathogenic species, Vibrio mimicus. J Clin Microbiol 14:631–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamasaki S, Garg S, Nair GB, Takeda Y. 1999. Distribution of Vibrio cholerae O1 antigen biosynthesis genes among O139 and other non-O1 serogroups of Vibrio cholerae. FEMS Microbiol Lett 179:115–121. doi: 10.1111/j.1574-6968.1999.tb08716.x. [DOI] [PubMed] [Google Scholar]
- 47.Morita M, Ohnishi M, Arakawa E, Bhuiyan NA, Nusrin S, Alam M, Siddique AK, Qadri F, Izumiya H, Nair GB, Watanabe H. 2008. Development and validation of a mismatch amplification mutation PCR assay to monitor the dissemination of an emerging variant of Vibrio cholerae O1 biotype El Tor. Microbiol Immunol 52:314–317. doi: 10.1111/j.1348-0421.2008.00041.x. [DOI] [PubMed] [Google Scholar]
- 48.Iwanaga M, Yamamoto K, Higa N, Ichinose Y, Nakasone N, Tanabe M. 1986. Culture conditions for stimulating cholera toxin production by Vibrio cholerae O1 El Tor. Microbiol Immunol 30:1075–1083. doi: 10.1111/j.1348-0421.1986.tb03037.x. [DOI] [PubMed] [Google Scholar]
- 49.Chatterjee S, Asakura M, Chowdhury N, Neogi SB, Sugimoto N, Haldar S, Awasthi SP, Hinenoya A, Aoki S, Yamasaki S. 2010. Capsaicin, a potential inhibitor of cholera toxin production in Vibrio cholerae. FEMS Microbiol Lett 306:54–60. doi: 10.1111/j.1574-6968.2010.01931.x. [DOI] [PubMed] [Google Scholar]
- 50.Uesaka Y, Otsuka Y, Lin Z, Yamasaki S, Yamaoka J, Kurazono H, Takeda Y. 1994. Simple method of purification of Escherichia coli heat-labile enterotoxin and cholera toxin using immobilized galactose. Microb Pathog 16:71–76. doi: 10.1006/mpat.1994.1007. [DOI] [PubMed] [Google Scholar]
- 51.Oku Y, Uesaka Y, Hirayama T, Takeda Y. 1988. Development of a highly sensitive bead-ELISA to detect bacterial protein toxins. Microbiol Immunol 32:807–816. doi: 10.1111/j.1348-0421.1988.tb01442.x. [DOI] [PubMed] [Google Scholar]
- 52.Dean AG, Ching TC, Williams RG, Harden LB. 1972. Test for Escherichia coli enterotoxin in infant mice: application in a study of diarrhea in children in Honolulu. J Infect Dis 125:407–411. doi: 10.1093/infdis/125.4.407. [DOI] [PubMed] [Google Scholar]
- 53.Hagihara K, Nishikawa T, Isobe T, Song J, Sugamata Y, Yoshizaki K. 2004. IL-6 plays a critical role in the synergistic induction of human serum amyloid (SAA) gene when stimulated with proinflammatory cytokines as analyzed with an SAA isoform real-time quantitative RT-PCR assay system. Biochem Biophys Res Commun 314:363–369. doi: 10.1016/j.bbrc.2003.12.096. [DOI] [PubMed] [Google Scholar]
- 54.Marin MA, Vicente AC. 2012. Variants of Vibrio cholerae O1 El Tor from Zambia showed new genotypes of ctxB. Epidemiol Infect 140:1386–1387. doi: 10.1017/S0950268811001944. [DOI] [PubMed] [Google Scholar]
- 55.Thompson CC, Vicente AC, Souza RC, Vasconcelos AT, Vesth T, Alves N, Ussery DW, Iida T, Thompson FL. 2009. Genomic taxonomy of vibrios. BMC Evol Biol 9:258. doi: 10.1186/1471-2148-9-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang P, Zhou H, Kan B, Wang D. 2013. Novel ctxB variants of Vibrio cholerae O1 isolates, China. Infect Genet Evol 20:48–53. doi: 10.1016/j.meegid.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 57.Hoshino K, Yamasaki S, Mukhopadhyay AK, Chakraborty S, Basu A, Bhattacharya SK, Nair GB, Shimada T, Takeda Y. 1998. Development and evaluation of a multiplex PCR assay for rapid detection of toxigenic Vibrio cholerae O1 and O139. FEMS Immunol Med Microbiol 20:201–207. doi: 10.1111/j.1574-695X.1998.tb01128.x. [DOI] [PubMed] [Google Scholar]
- 58.Olsvik Ø, Wahlberg J, Petterson B, Uhlen M, Popovic T, Wachsmuth IK, Fields PI. 1993. Use of automated sequencing of polymerase chain reaction-generated amplicons to identify three types of cholera toxin subunit B in Vibrio cholerae O1 strains. J Clin Microbiol 31:22–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singh DV, Matte MH, Matte GR, Jiang S, Sabeena F, Shukla BN, Sanyal SC, Huq A, Colwell RR. 2001. Molecular analysis of Vibrio cholerae O1, O139, non-O1, and non-O139 strains: clonal relationships between clinical and environmental isolates. Appl Environ Microbiol 67:910–921. doi: 10.1128/AEM.67.2.910-921.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.