PAH contamination has become a worldwide environmental issue because of the potential toxic effects on natural ecosystems and human health. Biotransformation and biodegradation are considered the main natural elimination forms of PAHs from contaminated sites. Therefore, the knowledge of the degradation potential of the microbial community in contaminated sites is crucial for PAH pollution bioremediation. However, the nonspecificity of pahAc as a functional marker of PAH-degrading bacteria has resulted neither in a reliable prediction of PAH degradation potential nor an accurate assessment of degradation. Here, we introduced pahE encoding the PAH hydratase-aldolase as a new and better functional marker gene of PAH-degrading bacteria. This study provides a powerful molecular tool to more effectively explore the ecological role and degradation potential of PAH-degrading bacteria in ecosystems, which is significant to the bioremediation of PAH pollution.
KEYWORDS: amplicon Illumina sequencing, functional marker gene, hydratase-aldolase (PahE), PAH ring-hydroxylating dioxygenase, polycyclic aromatic hydrocarbons (PAHs)
ABSTRACT
The characterization of native polycyclic aromatic hydrocarbon (PAH)-degrading bacteria is significant for understanding the PAH degradation process in the natural environment and developing effective remediation technologies. Most previous investigations of PAH-degrading bacteria in environmental samples employ pahAc, which encodes the α-subunit of PAH ring-hydroxylating dioxygenase, as a functional marker gene. However, the poor phylogenetic resolution and nonspecificity of pahAc result in a misestimation of PAH-degrading bacteria. Here, we propose a PAH hydratase-aldolase-encoding gene, pahE, as a superior biomarker for PAH-degrading bacteria. Comparative phylogenetic analysis of the key enzymes involved in the upper pathway of PAH degradation indicated that pahE evolved dependently from a common ancestor. A phylogenetic tree constructed based on PahE is largely congruent with PahAc-based phylogenies, except for the dispersion of several clades of other non-PAH-degrading aromatic hydrocarbon dioxygenases present in the PahAc tree. Analysis of pure strains by PCR confirmed that pahE can specifically distinguish PAH-degrading bacteria, while pahAc cannot. Illumina sequencing of pahE and pahAc amplicons showed more genotypes and higher specificity and resolution for pahE. Novel reads were also discovered among the pahE amplicons, suggesting the presence of novel PAH-degrading populations. These results suggest that pahE is a more powerful biomarker for exploring the ecological role and degradation potential of PAH-degrading bacteria in ecosystems, which is significant to the bioremediation of PAH pollution and environmental microbial ecology.
IMPORTANCE PAH contamination has become a worldwide environmental issue because of the potential toxic effects on natural ecosystems and human health. Biotransformation and biodegradation are considered the main natural elimination forms of PAHs from contaminated sites. Therefore, the knowledge of the degradation potential of the microbial community in contaminated sites is crucial for PAH pollution bioremediation. However, the nonspecificity of pahAc as a functional marker of PAH-degrading bacteria has resulted neither in a reliable prediction of PAH degradation potential nor an accurate assessment of degradation. Here, we introduced pahE encoding the PAH hydratase-aldolase as a new and better functional marker gene of PAH-degrading bacteria. This study provides a powerful molecular tool to more effectively explore the ecological role and degradation potential of PAH-degrading bacteria in ecosystems, which is significant to the bioremediation of PAH pollution.
INTRODUCTION
Polycyclic aromatic hydrocarbons (PAHs) are formed by two or more fused aromatic rings, and a portion of them have been recognized as priority pollutants by the US Environmental Protection Agency due to their recalcitrance and potentially deleterious effects on the water ecosystem and human health (1, 2). Though bioremediation is regarded as the most cost-effective and sustainable PAH remediation technology in natural environments (3, 4), its use is still constrained by limited knowledge of PAH-degrading bacteria populations in situ. Identification and detection of indigenous PAH degraders are therefore necessary to better understand natural biodegradation processes and for the successful application of bioremediation technologies.
Culture-independent methods targeting the 16S rRNA gene can effectively reveal the microbial diversity of many PAH-polluted systems (5–7). However, 16S rRNA gene methods cannot directly identify which organisms are responsible for the degradation of PAH or have degradation potential (8, 9). Specific metabolic genes provide us with a powerful molecular tool to infer the metabolic capacity of microbial communities directly. PAH ring-hydroxylating dioxygenases (PAH-RHD) are multicomponent enzymes that catalyze the initial oxidation of PAH via adding dioxygen to aromatic ring structures, which is thought to be the rate-limiting step in PAH degradation (10, 11). A gene encoding the α-subunit of PAH-RHD (pahAc) is commonly used as a functional marker gene of PAH-degrading bacteria (8), because pahAc has been implicated in substrate specificity and is more conserved than genes encoding other components of PAH-RHDs (12).
In the last decades, PCR amplification and clone library sequencing using specific or degenerate primers targeting pahAc have been extensively applied to estimate the diversity and abundance of PAH-RHD genes in PAH-degrading isolates and multiplex systems (11, 13–17). In these studies, specific primers are designed to target a rather narrow range of sequences, e.g., nahAc-, phnAc-, nagAc-, and nidA-type sequences (13, 18), which are useful for describing enriched cultures or isolates but may be less reliable for detecting the complete diversity of PAH degraders. Variations in pahAc sequences in PAH-degrading bacteria hinder inclusive targeting of PAH degraders with such highly specific primers, resulting in an underestimation of PAH degraders at contaminated sites (19, 20). To circumvent this problem, Cébron and her colleagues (10) used a two-degenerate-primer system targeting PAH-RHDα genes of Gram-positive and Gram-negative bacteria, simultaneously detecting many pahAc-like genes (including pahAc-, nahAc-, phnAc-, nagAc-, nidA3-, and pdoA2-like genes) in contaminated soils and sediments. Similarly, an investigation of PAH-degrading bacteria in marine sediments from Patagonia using degenerate primers (21) revealed the nahAc-like and phnAc-like genes identified in Alcaligenes faecalis AFK2, the phnA1-like genes from Cycloclasticus spp., and five novel PAH-RHDαs. The abundance of these genes was later found to be correlated with contamination level (22). The major drawback of degenerate primers is their lack of specificity for PAH-RHDα. PAH-RHDαs are typically closely related to the α-subunits of other aromatic-ring-hydroxylating dioxygenase (other-ArhAc) which cannot degrade PAH (20, 23). Degenerate primers targeting pahAc are known to cross-react with other-ArhAc, especially when used for multicontaminated environmental samples containing complex microbial communities (12, 24–26). For example, Ní Chadhain et al. (24) found that the degenerate primer pair targeting the Rieske gene fragment from PAH-RHDα might also target the polychorobiphenyl-, benzene-, toluene-, and xylene-dioxygenases, along with other sequences that are not closely related to any RHDα genes present in PAH-spiked soil. Such cross-reactions can occur even when using specific PAH-RHDα primers, as observed in a study that found operational protein families (OPFs) affiliated with homogentisate 1,2-dioxygenase (HgmA) from Brevibacillus brevis in contaminated Antarctic soils by using the primer pairs PAH-RHDα-GN-F/PAH-RHDα-GN-R and PAH-RHDα-GP-F/PAH-RHDα-GP-R (25). All these cross-reactions would lead to a misestimation of PAH-RHD diversity and abundance and hinder the identification and characterization of novel PAH dioxygenase genes.
Effective biomarkers were usually thought to be able to characterize microbial degradation rate and potential. Previous studies have found that there is a good positive correlation between the abundance of nahAc and degradation of naphthalene (27, 28), and the degradation of pyrene is usually positively related to the abundance and expression of nidA (29, 30). For other PAH substrates, the relationship between specific PAH degradation and pahAc is irregular (positive, negative, or no correlation), and correlations are generally not strong (31–33). Hence, pahAc can neither reliably predict PAH degradation potential nor accurately assess degradation rate, although it catalyzes the degradation rate-limited step, a more specific functional biomarker of PAH-degrading bacteria is urgently needed.
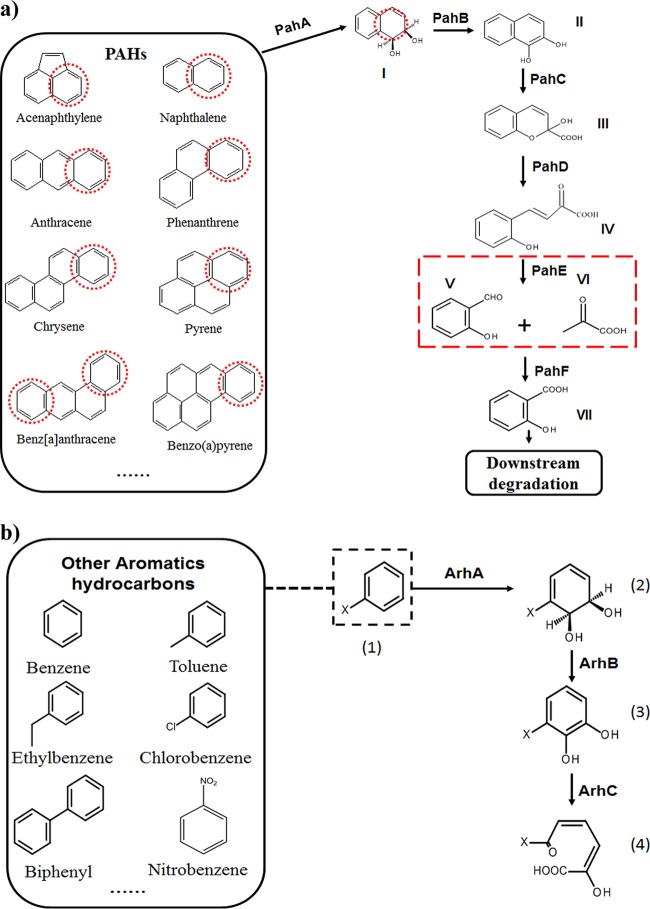
The objective of this study is to use comparative analyses of functional genes to identify a better biomarker for PAH-degrading bacteria and evaluate different biomarkers on specific metrics. As a good functional marker should be associated with a functional trait, have specificity, and provide fine phylogenetic resolution of closely related populations (34), we collected the protein sequences of key enzymes (including PahA, PahB, PahC, PahD, PahE, and PahF) responsible for the upper pathway of PAH metabolism (Fig. 1) and analyzed their phylogenies with the closely related members in their protein family. PahE catalyzes the fifth step of the PAH aerobic degradation process, converting analogues of trans-o-hydroxybenzylidenepyruvate (tHBPA) to aldehydes and pyruvic acid (Fig. 1) (35). The gene encoding PAH hydratase-aldolase (pahE) showed the greatest potential as a suitable functional marker gene of PAH-degrading bacteria. To evaluate the potential of pahE as a functional marker, degenerate primers specific for pahE gene sequences in PAH-degrading bacteria and a PCR-based assay were developed. The specificity and the target range of pahE and pahAc as functional markers of PAH degradation were then compared by testing their ability to identify PAH degraders from pure cultures and environmental samples.
FIG 1.
The upper consensus metabolic pathway of PAHs, with naphthalene as an example (a), and other aromatic hydrocarbons, with x-benzene as an example (b). Chemicals are I, cis-1,2-dihydroxy-1,2-dihydronaphthalene; II, 1,2-dihydroxynaphthalene; III, 2-hydroxy-4-(2′-oxo-3,5-cyclohexadienyl)-buta-2,4-dienoate; IV, trans-o-hydroxybenzylidenepyruvate; V, salicylaldehyde; VI, pyruvate; VII, salicylate; (1), x-benzene; (2), cis-5,6-dihydroxy-5,6-dihydro-1-x-benzene; (3), 5,6-dihydroxy-1-x-benzene; (4), 2-hydroxyl-6-oxo-6-x-2,4-diadienic acid. Enzymes are PahA, naphthalene dioxygenase; PahB, cis-1,2-dihydroxy-1,2-dihydronaphthalene dehydrogenase; PahC, 1,2-dihydroxynaphthalene dioxygenase; PahD, 2-hydroxychromene-2-carboxylate isomerase; PahE, trans-o-hydroxy- benzylidene pyruvate hydratase-aldolase; PahF, salicylaldehyde dehydrogenase; ArhA, aromatic hydrocarbon ring-hydroxylating dioxygenase; ArhB, aromatic hydrocarbon dihydrodiol dehydrogenase; ArhC, aromatic hydrocarbon extradiol dioxygenase. Red dotted circle, activated aromatic ring; red dotted box, the reaction catalyzed by PahE; black dotted box, x-benzene as an example.
RESULTS
pahE is a better functional marker of PAH-degrading bacteria.
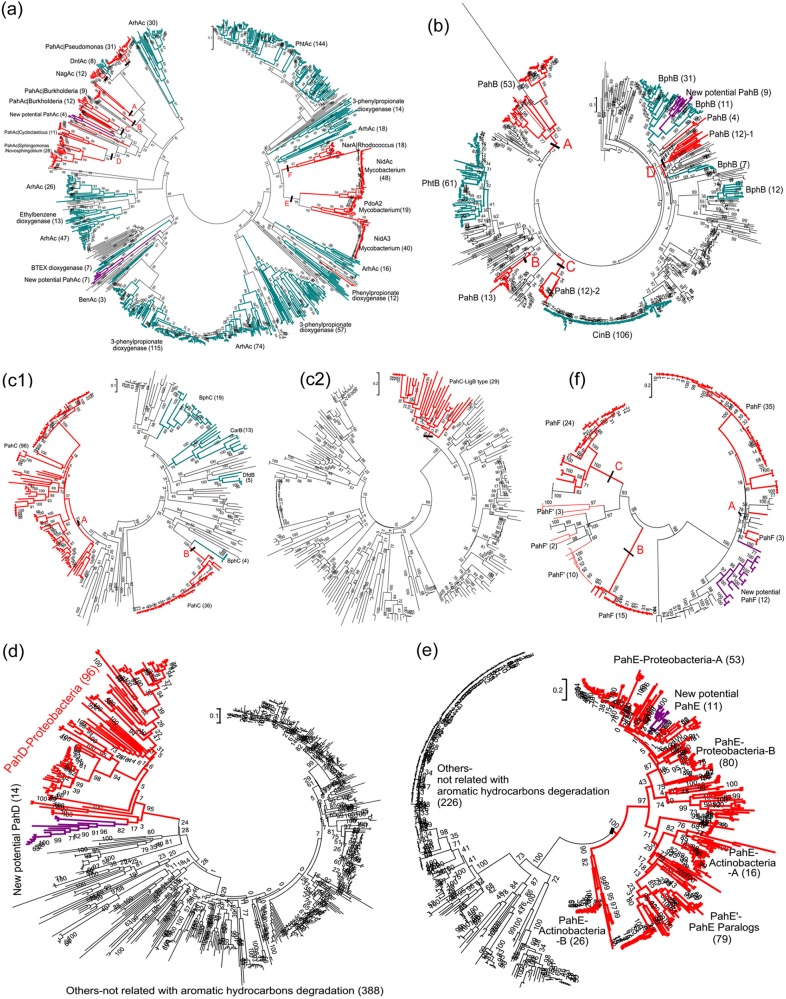
To select a suitable functional marker gene for PAH-degrading bacteria, the phylogenies of the key enzymes, including PahA, PahB, PahC, PahD, PahE, and PahF, responsible for the upper pathway of PAH metabolism, were analyzed for their similarity with other members of the same protein family (Fig. 2; see also Table S2 in the supplemental material).
FIG 2.
Evolutionary relationship of PahAc (a), PahB (b), PahC (vicinal chelate protein family) (c1), PahC (LigAB protein family) (c2), PahD (d), PahE (e), and PahF (f) based on amino acid sequences. The detailed information of each tree is provided in corresponding Table S2a, b, c1, c2, and d to f. Bootstrap support is indicated at individual branches. Parentheses in labels represent the number of branches in the clade. The branches or clades belonging to Pah are indicated by red, while those belonging to Arh are indicated by lake blue. The new potential Pah are indicated by purple branches or clades. The scale bar indicates 0.1 branch distance.
The phylogenetic clustering of the genes encoding these different functional enzymes shows that pahE has the greatest potential as a biomarker for PAH-degrading bacteria. All the PahE enzymes (hydratase-aldolases) cluster in an independent clade distinct from other subfamilies that are not related to aromatic hydrocarbon degradation (Fig. 2e and Table S2e) present in the dihydrodipicolinate synthase (DHDPS) family phylogenetic tree. The distinct deep branching of the pahE genes is favorable for the identification of PAH hydratase-aldolase through sequence alignment and phylogenetic analysis. The genes encoding the PahA, PahB, PahC, and PahF enzymes cluster in multiple clades separated by clades unrelated to PAH degradation (Fig. 2a and b, c1, c2, and f and Table S2a, b, c1, c2, and f). Genes encoding PahD also cluster into a single clade (Fig. 2d and Table S2d). However, since it remains unclear whether Gram-positive PAH-degrading bacteria require PahD for PAH degradation (36), the pahE gene is more likely to capture both Gram-positive and Gram-negative PAH degraders when used as a biomarker.
Based on PahE as a functional marker, a new group of potential PAH-degrading proteobacteria (indicated by purple branches in each tree) were inferred as their phylogenetic association with other PAH-degrading bacteria. PahAc/PahB/PahC/PahD/PahF sequences in the genomes of this new group clustered within the clades defined by these enzymes, providing evidence that this new group is likely capable of PAH degradation. These results indicate that pahE has great potential as a functional gene for identifying PAH-degrading bacteria.
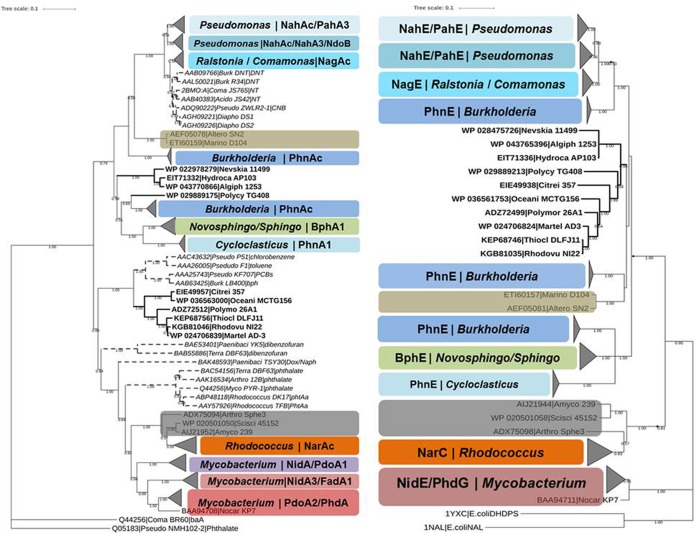
To further assess the potential of pahE as a new functional marker of PAH-degrading bacteria, the pahE and pahAc trees were directly compared. Curated reference databases of both genes were built based on the same criteria to avoid sampling artifacts, and the phylogenetic trees of both genes were calculated by two different classic treeing algorithms to ameliorate problems from individual treeing algorithms, so the results can be considered reliable phylogenetic estimates. The topological structures of the two trees for each gene are similar, so only the neighbor-joining consensus trees are shown for comparison (Fig. 3 and S2).
FIG 3.
Comparison of PahE and PahAc-based phylogenies of PAH-degrading bacteria. Both panels show majority rule consensus trees based on the neighbor-joining (NJ) method. Bootstrap support is indicated at individual branches, and only values greater than 0.5 are shown. Different types of both genes are marked by different colors. The same color in both trees indicates a one-to-one correspondence in both genes and that the two genes are from same degrading bacteria. The bold text in both trees indicates members of the group of newly defined potential PAH degraders, which also correspond to each other. The other aromatic ring-hydroxylating dioxygenase clades (other-ArhAc, which could not degrade PAH) in PahAc trees are indicated by dashed branches. The size of node triangles indicates the number of sequences in each clade. The scale bar indicates 0.1 branch distance. Accession numbers are shown to the left of each organism name.
The topological structure of the pahE tree is largely congruent with that of pahAc (Fig. 3 and S2), demonstrating that there is a specific one-to-one correspondence between the pahE and pahAc genes of PAH-degrading bacteria. Moreover, the presence of corresponding pahAc and pahE genes (Fig. 3, bold branches) further suggests that the newly identified potential PAH-degrading taxa have PAH-degrading capacity. The pahAc tree shows major topological incongruences in other-ArhAc lineages (Fig. 3, dashed branches), but these taxa have no corresponding pahE genes in the pahE tree. This indicates that pahE genes are more specific for PAH-degrading bacteria. Two other minor incongruences are notable within the Marinomonas sp. strain D104 and Alteromonas sp. strain SN2 clades and within Burkholderia clades, potentially indicating that the pahAc and pahE in these groups have been acquired through different horizontal gene transfer events, as they are different genera but share similar pahAc or pahE. These comparisons confirmed that pahE is more specific and is therefore more predictive as a functional marker for PAH-degrading bacteria than pahAc.
Determination of pahE specificity.
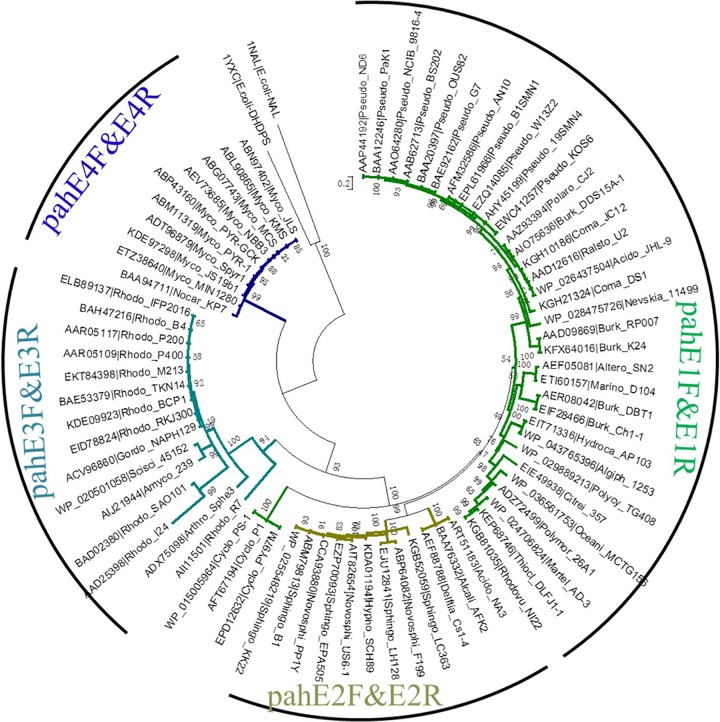
To test the specificity of pahE genes for PAH-degrading bacteria, a highly specific PCR-based assay detection system was developed. Four primer sets were designed to specifically amplify partial stretches of the pahE gene sequence from different clades (Table 1 and Fig. 4). The coverage and specificity of each primer set were tested in silico. Of the 73 reference pahE sequences, 68 sequences are specifically targeted (Table S5). Almost every pahE genotype is targeted except those of the Cycloclasticus genus, which are the dominant PAH-degrading bacteria in marine environments (37).
TABLE 1.
Primer sets designed for pahE genes and the primer sets chosen for pahAc genes
| Primer | Sequencea | Length (nt) | Degeneracy | Length of PCR products (bp) | Tm (°C)b | Reference |
|---|---|---|---|---|---|---|
| pahE1F | TGCGGCGGGTGTNAAYGGNAT | 21 | 32 | 377 | 57 | This study |
| pahE1R | CCTGAGGAATCTCGGACATYTSTGCCCARAA | 31 | 8 | |||
| pahE2F | AGCATGGGAACKYTKGGNGA | 20 | 32 | 366 | 52 | This study |
| pahE2R | TTTGGCGGTVACVACYTG | 18 | 18 | |||
| pahE3F | GACGGCGTSGACGGVATCAT | 20 | 6 | 323 | 54 | This study |
| pahE3R | TCAGGGTTGTCRTARAKSA | 19 | 16 | |||
| pahE4F | TGGTGCGYGAYGGBGYCGA | 19 | 24 | 324 | 54 | This study |
| pahE4R | GGCGTGCGGGTTSTSRTARAYCA | 23 | 32 | |||
| PAH-RHDα-396F | ATTGCGCTTAYCAYGGBTGG | 20 | 12 | 320 | 57 | 16 |
| PAH-RHDα-696R | ATAGGTGTCTCCAACRAARTT | 21 | 4 |
The pahE primers consisted of a 5′ consensus clamp region and a 3′ degenerate core region (italicized in the primer sequences).
Tm, melting temperature.
FIG 4.
Four different degenerate primer pair sets designed based on pahE phylogeny. The coverage and specificity of pahE primers designed in this study are indicated by colored branches. Green branches, pahE of PAH-degrading bacteria of Betaproteobacteria and Gammaproteobacteria; olive branches, pahE of PAH-degrading bacteria of Alphaproteobacteria; teal branches, pahE of PAH-degrading bacteria of Rhodococcus; blue, pahE of PAH-degrading bacteria of Mycobacterium. The scale bar indicates 0.1 branch distance. Accession numbers are shown to the left of each organism name.
The performance of the pahE and pahAc PCR assays was tested using pure cultured strains, including reference PAH-degrading bacteria, other aromatic hydrocarbon-degrading bacteria (those that could not grow on PAHs as the sole carbon source), and control bacteria (Table S3). The pahAc assays resulted in an ∼320-bp amplicon stretch of the pahAc gene from all of the reference PAH-degrading bacteria, as well as the homologous stretch of ArhAc from other aromatic hydrocarbon-degrading bacteria (Table 2). For example, neither Pseudomonas stutzeri ZWLR2-1 nor Comamonas sp. strain CNB1 degrade PAH, but their dioxygenases are closely homologous with PAH-RHDα and produced amplicons. Compared with the results of the pahAc PCR assay, the pahE assay only amplified the specific stretches of pahE from the referenced PAH-degrading bacteria. Neither nonspecific nor specific products were generated from other non-PAH-degrading strains. These results demonstrate that the pahE assay could specifically discriminate the PAH-degrading bacteria from closely related bacteria, while pahAc could not.
TABLE 2.
Comparisons of the amplification of pahAc and pahE from reference strains by pahAc and pahE primer sets, respectively
| Reference strain | Substrate(s)a | Amplification results with primer sets forb: |
|
|---|---|---|---|
| pahAc | pahE | ||
| PAH-degrading bacteriac | |||
| Pseudomonas stutzeri 1-5 | Phn/Pyr | + | + |
| Delftia sp. strain Cs4-1 | Phn | + | + |
| Marinomonas profundimaris D104 | Nah/Ant/Phn/Pyr | + | + |
| Sphingomonas sp. strain 1-1 | Phn | + | + |
| Novosphingobium pentaromativorans US6-1 | Nah/Ant/Phn/Pyr/BaP | + | + |
| Rhodococcus sp. strain B4 | Nah | + | + |
| Mycobacterium vanbaalenii PYR-1 | Nah/Ant/Phn/Pyr/Fla | + | + |
| Thioclava dalianensis DLFJ1-1 | New potential degrader | + | + |
| Other aromatic hydrocarbon-degrading bacteriad | |||
| Pseudomonas stutzeri ZWLR2-1 | 2-CNB | + | − |
| Comamonas sp. strain CNB-1 | 3-CNB | + | − |
| Other strains | |||
| E. coli DH5α | NO | − | − |
| Bacillus subtilis 168 | NO | − | − |
Phn, phenanthrene; Pyr, pyrene; Nah, naphthalene; Ant, anthracene; BaP, benzopyrene; Fla, fluoranthene; 2-CNB, 2-chloronitrobenzene; 3-CNB, 3-chloronitrobenzene; NO, not capable of degrading aromatic hydrocarbons.
+, correct sequence was amplified. −, no amplicon.
Can mineralize PAHs or grow on PAHs as the only carbon source.
Cannot mineralize PAHs or grow on PAHs as the only carbon source.
More genotypes and higher specificity detected by pahE.
To further evaluate the specificity and applicability of pahE as a functional marker for PAH-degrading bacteria in ecological studies, the diversity of pahE and pahAc genes in a selection of different environmental samples (including a chronically crude oil contaminated soil [ODS], an activated sludge of urban wastewater treatment plant [BAS], and an enrichment culture of PAH-degrading bacteria [ASE]; details are described in Tables S3 and S4) was compared by Illumina amplicon sequencing. In particular, this test was conducted to determine whether the primers would retrieve only the specific targeted genes (pahE or pahAc) of PAH-degrading bacteria and whether the approach could detect novel genotypes of the targets. Additionally, Illumina amplicon sequencing could provide insight into the diversity of pahE in PAH-degrading bacteria, a topic which has never been investigated by a specific deep-sequencing approach.
Our quality screening resulted in 308,549 high-quality reads with lengths ranging from 320 bp to 350 bp for pahAc and 806,682, 2,163, and 255,306 reads, with lengths ranging from 320 bp to 380 bp for pahE1F/pahE1R amplicons, pahE2F/pahE2R amplicons, and pahE3F/pahE3R amplicons, respectively (Table 3). Phylogenetic classification of reads from the long-term contaminated soil and enrichment culture revealed that an average of 98.10% and 93.32% of reads are affiliated with pahAc and pahE (mean value of pahE1 and pahE2 amplicons), respectively. The large fraction clearly shows the high selectivity of these primers for pahAc and pahE under the applied PCR conditions. However, in activated sludge from a wastewater treatment plant (WWTP), the primer sets for both genes resulted in very low selectivity; only 1.98% and 2.27% of reads were affiliated with pahAc and pahE, respectively. This result may be unique to the WWTP sludge used in this study, as it is operated solely for treating domestic sewage, which has low PAH pollution and hence has few PAH-degrading microorganisms and some matters leading to nonspecificity. As suggested by Good’s coverage parameter (38), which was above 0.99 for all the sequenced samples (Table 3), the sequencing depth is sufficient to cover a large fraction of the PAH-degrading bacterial richness at the approximate species level (90% identity of pahAc or pahE) in these environmental samples. At the 90% nucleic acid identity clustering level, 28 pahAc-affiliated operational taxonomic units (OTUs) and 42 pahE-affiliated OTUs are represented in total (Table 3).
TABLE 3.
Illumina sequencing results and observed numbers of OTUs based on reads that were affiliated with pahAc or pahE of PAH-degrading bacteria
| Primer/primer set | Sample typea | No. of high-quality reads | No. of PAH-degrading bacterial pahAc or pahE reads (over 50% identity) | Good’s coverageb | No. of observed OTUs (90% identity) |
|---|---|---|---|---|---|
| pahAc | ODS | 75,979 | 75,363 | 0.999 | 19 |
| ASE | 172,319 | 167,140 | 0.999 | 6 | |
| BAS | 60,251 | 1,194 | 1 | 11 | |
| pahE1F/pahE1R | ODS | 330,283 | 329,093 | 0.999 | 11 |
| ASE | 463,897 | 416,736 | 0.999 | 10 | |
| BAS | 12,502 | 560 | 0.999 | 7 | |
| pahE2F/pahE2R | ODS | 2,163 | 1,957 | 0.999 | 14 |
| pahE3F/pahE3R | ODS | 62,147 | 35,071 | 0.999 | 13 |
| ASE | 60,771 | 6 | 0.999 | 2 | |
| BAS | 132,388 | 77 | 0.999 | 6 |
ODS, contaminated-oilfield soil; ASE, phenanthrene and pyrene enrichment culture; BAS, activated sludge of Bei Xiaohe urban sewage treatment plant.
Calculated from the number of OTUs (at 90% identity level) represented by only one quality-controlled Illumina read (N1) and the total number of quality-controlled Illumina reads (N) as 1 – (N1/N).
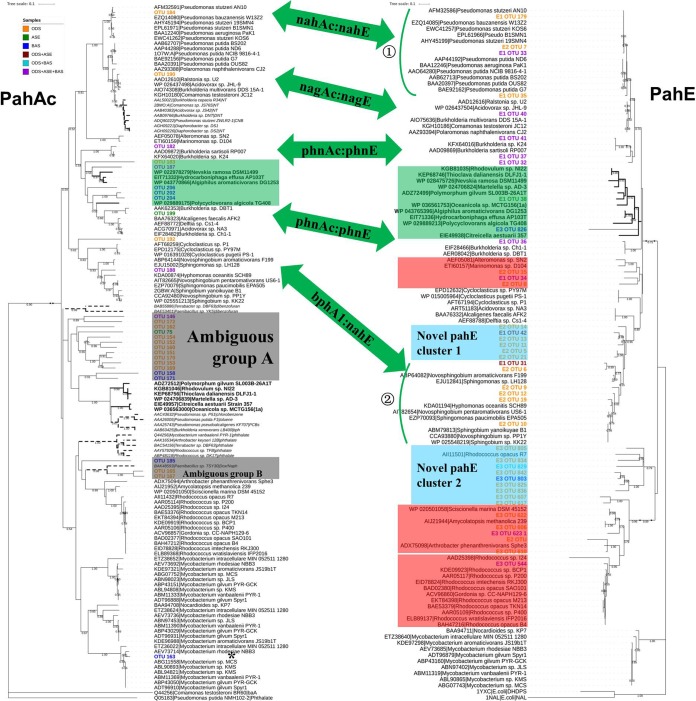
A large fraction of the pahAc OTUs matched the previously defined pahAc lineages of PAH-degrading bacteria, such as nahAc of Pseudomonas spp., nagAc of Ralstonia sp. strain U2, dntAc and phnAc of Burkholderia spp., bphA1 of Sphingomonas spp., nidA3 of Mycobacterium spp., and even pahAc of newly defined potential PAH degraders (Fig. 5, light-green box). The remaining pahAc reads fell between the defined pahAc lineages of PAH-degrading bacteria and other non-PAH-degrading aromatic dioxygenase lineages. These reads clustered into two groups, which were defined as ambiguous groups A and B (Fig. 5, gray box). Ambiguous group A contains 13 reads that show approximately 50 to 51.3% identity to the dibenzofuran dioxygenase of Terrabacter sp. strain DBF63 and approximately 50 to 53.7% identity to a new potential PAH dioxygenase (Table S7). The phylogenetic proximity and poor identities to disparate groups in these make it hard to determine whether they are PAH-RHDα genes or not. Ambiguous group B contains 3 reads (51.2 to ∼67.8% identity), which are most closely related to the putative naphthalene dioxygenase of Paenibacillus sp. strain TSY30 (Table S7) (39).
FIG 5.
Comparison of phylogenetic analysis of Illumina sequencing amplicons classified as pahAc and pahE in the environmental samples. The consensus trees were based on the translated amino acid of sequenced amplicons of pahAc and pahE by neighbor-joining (NJ) method. Bootstrap support is indicated at individual branches, and only those over 0.5 were shown. The environmental amplicons are indicated as OTU number, such as “OTU 190,” or “primer OTU number,” such as “E1/E2/E3 OTU 19,” for pahAc and pahE, respectively. Different color indicates amplicons from different samples according to the color legend. Corresponding pahE and pahAc are indicated by a green double-sided arrow. Members of the group of newly defined potential PAH degraders are marked by a light-green box. The gray boxes in the pahAc tree indicate ambiguous groups, while the red boxes in the pahE tree are the additional detected pahE genotypes with no corresponding pahAc detected. The detected novel pahE genes are indicated by light-sky-blue boxes. The detected pahAc with no corresponding pahE are labeled by an asterisk. The scale bar indicates 0.1 branch distance. Accession numbers are shown to the left of each organism name.
In contrast, most of the pahE sequence OTUs obtained fall into the previously defined pahE lineages of PAH-degrading bacteria (Fig. 5). The pahE OTUs were not only affiliated with the corresponding pahE of the PAH-degrading bacteria whose pahAc is also detected (the corresponding relationship between the detected pahAc and pahE are indicated by the green two-way arrow, Fig. 5 and Table S8), but also grouped with pahE of Marinomonas spp., aldolase of other Actinobacteria (Pseudarthrobacter spp. and Pseudonocardiaceae), and narC of Rhodococcus (Fig. 5, rose-red box). Moreover, some novel and unknown pahE genes were detected, which fell between the characterized pahE clades but do not exhibit significant similarity to the characterized pahE (indicated by “novel pahE light-blue box,” Fig. 5). In addition, pahE has a more precise resolution as a functional marker of PAH-degrading bacteria. Based on the same 90% identity nucleotide acid, pahE could be used to discriminate the different pahE genotypes of the Pseudomonas genus (Fig. 5, green arc ①; Table S8) and to distinguish the pahE genes of Sphingopyxis spp., Sphingobium spp., and Hyphomonas oceanitis (Fig. 5, green arc ②, and Table S8). Conversely, pahAc defined only one genotype of nahAc in Pseudomonas and only bphA1 of Sphingomonadaceae.
DISCUSSION
In this study, we propose the pahE gene as a new functional marker for analyzing the microbial ecology of PAH-degrading bacteria. Based on the PAH degradative pathways (Fig. 1) and our phylogenetic analyses (Fig. 2), it can be concluded that pahAc, pahB, and pahC are not specific to PAH degradation and have evolved closely with ArhAc, ArhB (aromatic hydrocarbon dihydrodiol dehydrogenases), and ArhC (aromatic hydrocarbon extradiol dioxygenases) (40–43). pahD and pahE are unique to PAH degradation and have evolved from a dependent ancestor, but PahD is not necessarily present in Gram-positive PAH degraders. In 1994, Eaton (44) proposed that PahD and PahE are unique to the biodegradation of polycyclic aromatic hydrocarbons, and the genes that encode these enzymes should be valuable as relatively specific probes for the identification or enumeration of bacteria that degrade polycyclic aromatic compounds. PahE catalyzes the fifth step of the PAH aerobic degradation process, converting analogues of trans-o-hydroxybenzylidenepyruvate (tHBPA) to aldehydes and pyruvic acid (35). This reaction is a significant step in PAH degradation, as degrading bacteria start to truly obtain energy from PAH degradation at this point. This is why many other aromatic hydrocarbon-degrading bacteria with dioxygenase but without PahE can add oxygen to PAH but cannot grow solely on PAH. For example, Comamonas sp. strain JS756, which lacks PahE, can oxidize naphthalene but does not grow on naphthalene, although its dioxygenase shows high similarity with the naphthalene dioxygenase of Pseudomonas spp. (45). In addition, phylogenetic analysis of PahE (Fig. 2) was used to identify a new group of potential PAH degraders in the phylum Proteobacteria. Members of this group were not previously known to contain corresponding PahA, PahB, PahC, PahD, and PahF, but pahA, pahB, pahD, and pahF, and possibly pahC, were subsequently identified by genome comparison.
Further phylogenetic analyses showed that pahE genes are more specific for PAH-degrading bacteria and provide better differentiation of closely related populations than pahAc (Fig. 3 and S2). pahAc sequences tend to be highly similar to other-ArhAc (20, 40, 43), and the two are hard to distinguish. For example, Ní Chadhain and his colleagues (24) designed a degenerate primer set to amplify 78 bp of the Rieske iron sulfur center to study the diversity of pahAc in soil exposed to different PAHs, detecting nahAc, nagAc, and phnAc, many other-ArhAc genes (including dbtA1, bphA1a, dbfA1, akbA1a, and carAa), and three new equivocal groups. The researchers proposed that these results are indicative of the wide range of the primers. However, separate studies using specific primer sets unexpectedly targeted the dinitrotoluene dioxygenase gene dntAc (approximately 84 to 92% identity to nahAc or nagAc) attempting to specifically target nahAc expression in contaminated groundwater (46) and to demonstrate the relationship of nagAc-like gene copies with naphthalene concentrations in coal-tar-contaminated freshwater sediments (18). In this study, we also found that pahAc primers amplified homologous stretches in dioxygenase genes from chloro-nitrobenzene-degrading bacteria (Table 2) and produced ambiguous reads in environmental samples (Fig. 5). Nonspecificity and poor resolution could result in misidentification of PAH-degrading bacteria when using pahAc as a functional marker and overestimation of degradation potential in practice. Moreover, it is worth noting that there are three different genotypes of pahAc but a specific pahE genotype in Mycobacterium spp. (Fig. 3 and S2). Previous studies found that many PAH-degrading genera have several copies or genotypes of pahAc, especially Mycobacterium, and these paralogues usually do not function simultaneously (47, 48). Therefore, when using pahAc as a target gene for quantitative PCR (qPCR) analyses of PAH functional gene abundance or expression, the range of pahAc copies in different PAH-degrading bacteria genomes must be taken into account.
Universality in ecosystems is important in using marker genes to identify the ecological role of target organisms. In this study, direct comparison of the performance of pahE and pahAc in environmental samples showed more genotypes detected, higher specificity, and better resolution of pahE than of pahAc. Consistent with other studies, we observed nahAc, nagAc, phnAc, and nidA3, which are found extensively in different environments (10, 17, 19, 24), among the read set amplified by pahAc primers. nahAc, nagAc, and phnAc are usually related to low-molecular-weight PAH degradation, while nidA3 genes are responsible for high-molecular-weight PAH degradation (8, 47). Sphingomonas species are versatile degraders and can use both monocyclic aromatic hydrocarbon and PAH as carbon sources in different environments (49, 50). pahAc reads from the potential PAH-degrading bacteria identified in this study were confirmed by alignment clusters of PAH degradation-associated genes and were identified in environmental samples. The pahE genes corresponding to pahAc in the identified potential PAH degraders were similarly found in environmental samples. These results confirm that the newly identified potential PAH-degrading genera are in these environments.
For pahE reads, aside from the pahE genes corresponding to the detectable pahAc, pahE from Marinomonas, narC of Rhodococcus, and aldolase genes of other Actinobacteria (Pseudarthrobacter and Pseudonocardiaceae) were also detected (Fig. 5), suggesting that these PAH-degrading genera are also in these systems. In previous studies, narAc of Rhodococcus and fadA2 of Arthrobacter were also found in contaminated soil and sediments (25), respectively. A possible, but unlikely, cause of the observed more pahE genotypes than pahAc could be the differences in the coverage and specificity of the primer sets used. The primer sets for pahAc were designed by Ding et al. (16) and had been confirmed to specifically target pahAc of both Gram-negative and Gram-positive PAH-degrading bacteria. Similarly, the four pahE primer pairs targeting different clades of pahE together cover all of pahE genes in PAH-degrading bacteria (Fig. 4). Amplification bias may also explain at least a portion of the observed differences in diversity from the two different marker genes (reviewed by von Wintzingerode et al. [51]). However, it is difficult to quantify PCR biases, and all PCRs were performed with the same mixtures and on the same equipment machines, except that the primers and thermal cycling conditions were different. The ambiguous groups and unspecific targets of pahAc are mainly the result of cross-reaction with other ArhAc clades (20, 43). It is particularly notable that novel pahE sequences were found, suggesting that there are unknown potential PAH degraders in these samples and that the approach detailed here could detect novel genotypes.
The only observed drawback of pahE as a target was that no pahE gene was detected corresponding to the detectable pahAc of Mycobacterium (nidA3). However, the detected Mycobacterium nidA3 genes were only found in the activated sludge and in low abundance (0.045%) (9, 52). Mycobacterium species are known to be prevalent in high-molecular-weight PAH-polluted sites (8, 53). Low abundance could explain the failure of the pahE4F/pahE4R primer pair to amplify the specific genes from this environmental system. Another controversial issue may be that four primer pair sets were used for detecting pahE, while only one primer set was used for detecting pahAc. However, nonspecificity and poor resolution of pahAc cannot be avoided even by multiple primer pair sets, as pahAc genes have evolved closely with other-ArhAc genes (Fig. 2a). The four pahE primer pairs only target all pahE genes in PAH-degrading bacteria (Fig. 4). This investigation of pahE as a functional marker utilized four primer pair sets for detecting pahE, which may complicate analyses and can confound estimate of the relative abundance based on Illumina sequencing. However, in order to ensure adequate coverage and specificity and to better understand the biology behind gene diversity, multiple primer pair sets are sometimes used to compensate for limitations in both the number of available sequences and conserved regions in functional genes (54). Similar to all PCR-based approaches, our approach should be assumed to have primer bias and cannot be comprehensive or reflect actual quantification. However, these primers reveal much more sequence information about the genes recovered and hence provide additional insight natural sequence variation in pahE genes. Moreover, as a greater diversity of full-length pahE sequences is resolved, a more highly conserved region could be identified, requiring fewer primers for achieving the same resolution. Thus, a crucial task might be the identification and discovery of additional pahE genes. A good functional marker of PAH degradation was usually expected to be able to characterize microbial degradation rate and potential. Actually, the degradation of PAHs in the environment is controlled by many factors, such as the features of PAHs itself, the species, activity, and quantity of microbes, environmental factors, and so on (55). However, a better relationship could be expected, as pahE is more specific for PAH-degrading bacteria than pahAc. The relationship between pahE and PAH degradation rate will be further established in future studies. The use of pahE as a functional marker of PAH-degrading bacteria will allow us to explore PAH-degrading bacteria and their degradation potential in ecosystems more precisely and effectively.
MATERIALS AND METHODS
Phylogenetic analyses of enzymes for upper pathway of PAH degradation.
To reconstruct the phylogenies of the key enzymes responsible for the upper pathway of PAH metabolism, we collected the protein sequences of each enzyme according to Fig. S1. Briefly, the reference protein sequences of each enzyme were collected by searching publicly referenced databases for known or functionally characterized sequences. Then the top x = 1,000 bit score hits were collected from a BLAST search of the NCBI nonredundant (NR) protein database (excluding uncultured and environmental sequences; other BLASTp parameters were defaults) with the reference protein sequences as queries. Retrieved sequences were filtered by length and aligned with MUSCLE (56), and a tentative phylogenetic tree was constructed using the neighbor-joining method in MEGA 5.0 (57). Then, based on whether the tree covered all seed clades for each enzyme and whether too many distant sequences were retrieved, x was manually curated until a sufficient protein sequence database was created for each enzyme. As arhAc genes, which code for the large subunit of oxygenase, are conserved among all ring-hydroxylating dioxygenase (RHOs) and are generally used to study the phylogenetic relationship among all RHOs (23, 58, 59), the phylogenies of PahA were indicated by PahAc.
For the phylogeny analysis, multiple-protein-sequence alignments were constructed using MUSCLE (56), and positions containing >95% gaps were removed. Phylogenetic trees were built using the neighbor-joining method within Poisson correction and maximum likelihood method within Poisson correction in MEGA 5.0 (57). In general, the topologies of trees built by these two methods were consistent, and hence, only the neighbor-joining trees were presented. For readability and clarity, the topological structures of the trees for each different enzyme are shown in Fig. 2, and detailed sequence information for each tree is listed in Table S2. As the original tree for PahF was too big, only a subtree is displayed here.
Reference databases of PAH-RHDα and PAH hydratase-aldolase.
For further analyses of PahE and PahAc, reference databases of PAH-RHDα (PahAc) and PAH hydratase-aldolase (PahE) were built by BLASTp analysis, according to previous studies (60, 61). First, we constructed a seed database for both protein sequences by searching for all well-known (i.e., functionally characterized and fully sequenced) sequences. To define a bit score threshold for the specific retrieval of PahAc or PahE sequences, each entry of both seed databases was used as a Basic Local Alignment Search Tool (BLAST) query against all other seed PahAc or PahE sequences and a set of corresponding outgroup sequences. The highest bit score of the outgroup entries +10% (to make the search more conservative) was then used as the bit score threshold for the BLASTp search. To account for the sequence divergence among the seed PahAc or PahE sequences, the bit score threshold was determined separately for each entry from both seed databases. Thereafter, each PahAc or PahE entry was used individually as a query for screening the NCBI nonredundant database, excluding uncultured and environmental sequences with individual bit score thresholds. Then, all PahAc or PahE sequences were filtered by length, aligned using MUSCLE (56), and compared using a percent identity matrix to eliminate 100% identical sequences to reduce sequence redundancy. PahE sequences with 100% identical identity were retained if the PahAc sequence in the corresponding bacterium was retained, and vice versa. In final curation, only functionally characterized sequences or those from PAH-degrading bacteria were used as reference PahAc or PahE sequences and for further analysis. The reference PahAc and PahE sequences are listed in Table S1. The nucleotide sequences of the reference PahAc and PahE sequences were also collected from the NCBI database. Further phylogenetic analyses of pahAc and pahE gene sequences were performed using full-length unambiguously aligned amino acid sequences. Amino acid sequences were preferred over nucleic acid sequences because of their higher functional conservation. The reference PahAc and PahE sequences were aligned by MUSCLE (56). Phylogenetic trees were reconstructed using the neighbor-joining method with Poisson model method and the maximum likelihood method with the Poisson correction model with MEGA 5.0 (57). Bootstrap support for neighbor-joining and maximum likelihood trees was determined using 1,000 resamplings. The topological structures of the trees for each gene were similar, so only the neighbor-joining trees were edited by iTOL online tool (http://itol.embl.de/) and shown.
Cultured strains and environmental samples.
All cultures and environmental samples used for the examination of PCR assay specificity and screened for pahE and pahAc of PAH-degrading bacteria are listed together with sampling details and DNA extraction methods in Table S3 in the supplemental material. Briefly, DNA extraction from bacterial pure cultures was performed using the TIANamp bacteria DNA kit (Tiangen Biotech [Beijing] Co., Ltd.), according to the manufacturer’s protocol. Genomic DNA of environmental samples was extracted using the FastDNA Spin kit for soil (MP Biomedicals, LLC), with modifications to the manufacturer’s instructions (details described in the supplemental material). Quadruplicate extractions were performed for each sample and then pooled. The DNA extracts were kept at −20°C until analyzed. Moreover, the degradation capacity of each environmental sample was analyzed (Table S4, and other details described in the supplemental material).
PCR primers.
Primers for PCR amplification of pahE genes were designed based on the PahE reference database using Codehop (COnsensus-DEgenerate-Hybrid Oligonucleotide Primer) (62) and manual curations (details described in the supplemental material). The specificity and coverage of each primer were tested against the pahE reference database and the GenBank database using NCBI Primer BLAST (Table S5). The annealing temperature for each clade-specific primer set was optimized using genomic DNA extracted from the selected reference bacterial strains by gradient temperature PCR.
Two previously published primer sets for amplification PAH-RHDα genes from both Gram-positive and Gram-negative bacteria were chosen by specificity and coverage. The degenerated primer set PAH-RHDα-396F/PAH-RHDα-696R (∼320-bp amplicon) can target all 40 referenced PAH-RHDα genes (16), while the primer pairs PAH-RHDα-GN-F/PAH-RHDα-GN-R (∼306-bp amplicon) and PAH-RHDα-GP-F/PAH-RHDα-GP-R (∼292-bp amplicon) target the dioxygenase genes specific for the Gram-positive and Gram-negative PAH-degrading bacteria, respectively (10). The PAH-RHDα-396F/PAH-RHDα-696R primer set amplified the exact fragment from more reference bacteria (data not shown) and so was selected for further analysis.
Specific PCR assays for recognition of PAH-degrading bacteria.
To test the specificity of the pahE genes and pahAc genes for PAH-degrading bacteria, pahAc and pahE were amplified with the primers described above. PCR was carried out in a 50-μl reaction mixture containing 25 μl 2× Es Taq (Beijing ComWin Biotech Co., Ltd.), 0.2 μM both forward and reverse primers, and 20 ng bacterial genomic DNA. For the pahE primer sets, the thermal cycling conditions were as follows: initial denaturation at 94°C for 5 min, followed by 35 cycles of 94°C for 30 s, 52 to ∼57°C (annealing temperature for each clade-specific primer set) for 45 s, and 72°C for 30 s, with a final extension at 72°C for 10 min. The thermal cycling conditions for pahAc were according to Ding et al. (16). Amplicons from each culture were cloned into Escherichia coli DH5α and then sequenced by Sanger sequencing (details described in the supplemental material). The obtained sequences were compared against the reference database using BLAST to confirm that the amplicon sequences matched target gene sequences.
Amplicon Illumina sequencing and data analysis.
pahE and pahAc amplicons for Illumina sequencing were generated using the four pahE primer sets designed and the primer set PAH-RHDα-396F/PAH-RHDα-696R described above, respectively. The only modifications included a barcoded reverse primer for each primer pair to discriminate among environment samples, a smaller cycle number (25) to reduce potential PCR biases (51), and a lower annealing temperature for some primer sets to account for possible base mismatches between the primer and unknown pahE or pahAc in the environmental samples. Although the lower annealing temperature resulted in increased nonspecific amplification in some samples, the expected PCR product was the dominant product in all samples. Not all clades of pahE were amplified from every environmental sample; only the positive amplicons were further sequenced (Table S6). The PCR products were extracted from 2% agarose gels and purified using the QIAquick gel extraction kit (Qiagen). After quantification using the Qubit double-stranded DNA (dsDNA) HS assay kit (Invitrogen, Life Technologies), amplicons from different samples were pooled in equal concentrations. The pooled library was quantified with a 2100 Bioanalyzer instrument (Agilent Technologies) before sequencing on an Illumina HiSeq 2500 platform.
Raw sequences were quality screened and trimmed using FastQC 0.11.5 (63). The filtered paired-end reads were assembled using PANDAseq 2.8 (64), with default parameters. Assembled reads with mismatches in barcodes or primer sequences were discarded, and the remaining sequences were filtered for appropriate length (300 to 400 nucleotides [nt]). The remaining high-quality sequences were dereplicated and clustered at 97% identity and then 90% identity using UPARSE (65), not excluding clusters of size 1 to avoid omitting correct sequences. Representative sequences of each cluster were screened for chimeras using UCHIME (66) by searching against the pahAc or pahE reference database, and identified chimeras were discarded. Remaining sequences were binned into OTUs and used for phylogenetic assignation. Phylogenetic assignment was performed by aligning OTU representatives individually to the pahAc or pahE reference database using tblastx. Only OTUs with >50% identity (amino acid) to a reference database entry with no frameshifts were regarded as pahAc- or pahE-affiliated sequences and translated into a deduced amino acid sequence. The phylogenetic positions of representative deduced amino acid sequences of pahAc or pahE OTUs within the consensus trees were inferred independently by both the distance matrix (neighbor-joining) and maximum likelihood methods within MEGA 5.0 (57). Bootstrap support for both trees was 1,000. The topological structures of the trees for each gene were similar, so only the neighbor-joining trees were edited using the iTOL online tool (http://itol.embl.de/) and shown.
Data availability.
The raw sequence data were submitted to the NCBI Sequence Read Archive (SRA) with accession numbers SRR6287136 to SRR6287145.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge all colleagues (William Hickey, Kae Kyoung Kwon, Zongze Shao, Ning-Yi Zhou, and Shuang-Jiang Liu) who sent us the reference bacterial strains. We are also very grateful to Zuotao Zhang for his assistance in environmental sample collection.
This work was financially supported by the National Natural Science Foundation of China (grants 41573065 and 41773082).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02399-18.
REFERENCES
- 1.Menzie CA, Potocki BB, Santodonato J. 1992. Exposure to carcinogenic PAHs in the environment. Environ Sci Technol 26:1278–1284. doi: 10.1021/es00031a002. [DOI] [Google Scholar]
- 2.Nikolaou A, Kostopoulou M, Lofrano G, Meric S. 2009. Determination of PAHs in marine sediments analytical methods and environmental concerns. Glob Nest J 11:391–405. [Google Scholar]
- 3.Gillespie IM, Philp JC. 2013. Bioremediation, an environmental remediation technology for the bioeconomy. Trends Biotechnol 31:329–332. doi: 10.1016/j.tibtech.2013.01.015. [DOI] [PubMed] [Google Scholar]
- 4.Vila J, Tauler M, Grifoll M. 2015. Bacterial PAH degradation in marine and terrestrial habitats. Curr Opin Biotechnol 33:95–102. doi: 10.1016/j.copbio.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 5.Martin F, Torelli S, Le Paslier D, Barbance A, Martin-Laurent F, Bru D, Geremia R, Blake G, Jouanneau Y. 2012. Betaproteobacteria dominance and diversity shifts in the bacterial community of a PAH-contaminated soil exposed to phenanthrene. Environ Pollut 162:345–353. doi: 10.1016/j.envpol.2011.11.032. [DOI] [PubMed] [Google Scholar]
- 6.Meynet P, Hale SE, Davenport RJ, Cornelissen G, Breedveld GD, Werner D. 2012. Effect of activated carbon amendment on bacterial community structure and functions in a PAH impacted urban soil. Environ Sci Technol 46:5057–5066. doi: 10.1021/es2043905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singleton DR, Jones MD, Richardson SD, Aitken MD. 2013. Pyrosequence analyses of bacterial communities during simulated in situ bioremediation of polycyclic aromatic hydrocarbon-contaminated soil. Appl Microbiol Biotechnol 97:8381–8391. doi: 10.1007/s00253-012-4531-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeBruyn JM, Chewning CS, Sayler GS. 2007. Comparative quantitative prevalence of Mycobacteria and functionally abundant nidA, nahAc, and nagAc dioxygenase genes in coal tar contaminated sediments. Environ Sci Technol 41:5426–5432. doi: 10.1021/es070406c. [DOI] [PubMed] [Google Scholar]
- 9.Wu P, Wang YS, Sun FL, Wu ML, Peng YL. 2014. Bacterial polycyclic aromatic hydrocarbon ring-hydroxylating dioxygenases in the sediments from the Pearl River estuary, China. Appl Microbiol Biotechnol 98:875–884. doi: 10.1007/s00253-013-4854-5. [DOI] [PubMed] [Google Scholar]
- 10.Cébron A, Norini MP, Beguiristain T, Leyval C. 2008. Real-time PCR quantification of PAH-ring hydroxylating dioxygenase (PAH-RHDα) genes from Gram positive and Gram negative bacteria in soil and sediment samples. J Microbiol Methods 73:148–159. doi: 10.1016/j.mimet.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 11.Martin F, Malagnoux L, Violet F, Jakoncic J, Jouanneau Y. 2013. Diversity and catalytic potential of PAH-specific ring-hydroxylating dioxygenases from a hydrocarbon-contaminated soil. Appl Microbiol Biotechnol 97:5125–5135. doi: 10.1007/s00253-012-4335-2. [DOI] [PubMed] [Google Scholar]
- 12.Zhou HW, Guo CL, Wong YS, Tam NF. 2006. Genetic diversity of dioxygenase genes in polycyclic aromatic hydrocarbon-degrading bacteria isolated from mangrove sediments. FEMS Microbiol Lett 262:148–157. doi: 10.1111/j.1574-6968.2006.00379.x. [DOI] [PubMed] [Google Scholar]
- 13.Lloyd-Jones G, Laurie AD, Hunter DW, Fraser R. 1999. Analysis of catabolic genes for naphthalene and phenanthrene degradation in contaminated New Zealand soils. FEMS Microbiol Ecol 29:69–79. doi: 10.1111/j.1574-6941.1999.tb00599.x. [DOI] [Google Scholar]
- 14.Stach JE, Burns RG. 2002. Enrichment versus biofilm culture: a functional and phylogenetic comparison of polycyclic aromatic hydrocarbon-degrading microbial communities. Environ Microbiol 4:169–182. doi: 10.1046/j.1462-2920.2002.00283.x. [DOI] [PubMed] [Google Scholar]
- 15.Flocco CG, Gomes NC, Mac Cormack W, Smalla K. 2009. Occurrence and diversity of naphthalene dioxygenase genes in soil microbial communities from the Maritime Antarctic. Environ Microbiol 11:700–714. doi: 10.1111/j.1462-2920.2008.01858.x. [DOI] [PubMed] [Google Scholar]
- 16.Ding GC, Heuer H, Zuhlke S, Spiteller M, Pronk GJ, Heister K, Kogel-Knabner I, Smalla K. 2010. Soil type-dependent responses to phenanthrene as revealed by determining the diversity and abundance of polycyclic aromatic hydrocarbon ring-hydroxylating dioxygenase genes by using a novel PCR detection system. Appl Environ Microbiol 76:4765–4771. doi: 10.1128/AEM.00047-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia X, Xia N, Lai Y, Dong J, Zhao P, Zhu B, Li Z, Ye W, Yuan Y, Huang J. 2015. Response of PAH-degrading genes to PAH bioavailability in the overlying water, suspended sediment, and deposited sediment of the Yangtze River. Chemosphere 128:236–244. doi: 10.1016/j.chemosphere.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 18.Dionisi HM, Chewning CS, Morgan KH, Menn FM, Easter JP, Sayler GS. 2004. Abundance of dioxygenase genes similar to Ralstonia sp. strain U2 nagAc is correlated with naphthalene concentrations in coal tar-contaminated freshwater sediments. Appl Environ Microbiol 70:3988–3995. doi: 10.1128/AEM.70.7.3988-3995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bordenave S, Goni-Urriza M, Vilette C, Blanchard S, Caumette P, Duran R. 2008. Diversity of ring-hydroxylating dioxygenases in pristine and oil contaminated microbial mats at genomic and transcriptomic levels. Environ Microbiol 10:3201–3211. doi: 10.1111/j.1462-2920.2008.01707.x. [DOI] [PubMed] [Google Scholar]
- 20.Iwai S, Johnson TA, Chai B, Hashsham SA, Tiedje JM. 2011. Comparison of the specificities and efficacies of primers for aromatic dioxygenase gene analysis of environmental samples. Appl Environ Microbiol 77:3551–3557. doi: 10.1128/AEM.00331-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lozada M, Mercadal JPR, Guerrero LD, Di Marzio WD, Ferrero MA, Dionisi HM. 2008. Novel aromatic ring-hydroxylating dioxygenase genes from coastal marine sediments of Patagonia. BMC Microbiol 8:50. doi: 10.1186/1471-2180-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcos MS, Lozada M, Di Marzio WD, Dionisi HM. 2012. Abundance, dynamics, and biogeographic distribution of seven polycyclic aromatic hydrocarbon dioxygenase gene variants in coastal sediments of Patagonia. Appl Environ Microbiol 78:1589–1592. doi: 10.1128/AEM.06929-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kweon O, Kim SJ, Baek S, Chae JC, Adjei MD, Baek DH, Kim YC, Cerniglia CE. 2008. A new classification system for bacterial Rieske non-heme iron aromatic ring-hydroxylating oxygenases. BMC Biochem 9:11. doi: 10.1186/1471-2091-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ní Chadhain SM, Norman RS, Pesce KV, Kukor JJ, Zylstra GJ. 2006. Microbial dioxygenase gene population shifts during polycyclic aromatic hydrocarbon biodegradation. Appl Environ Microbiol 72:4078–4087. doi: 10.1128/AEM.02969-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jurelevicius D, Alvarez VM, Peixoto R, Rosado AS, Seldin L. 2012. Bacterial polycyclic aromatic hydrocarbon ring-hydroxylating dioxygenases (PAH-RHD) encoding genes in different soils from King George Bay, Antarctic Peninsula. Appl Soil Ecol 55:1–9. doi: 10.1016/j.apsoil.2011.12.008. [DOI] [Google Scholar]
- 26.Yang Y, Wang J, Liao J, Xie S, Huang Y. 2014. Distribution of naphthalene dioxygenase genes in crude oil-contaminated soils. Microb Ecol 68:785–793. doi: 10.1007/s00248-014-0457-7. [DOI] [PubMed] [Google Scholar]
- 27.Tuomi PM, Salminen JM, Jørgensen KS. 2004. The abundance of nahAc genes correlates with the 14C-naphthalene mineralization potential in petroleum hydrocarbon-contaminated oxic soil layers. FEMS Microbiol Ecol 51:99–107. doi: 10.1016/j.femsec.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 28.Nyyssönen M, Piskonen R, Itavaara M. 2006. A targeted real-time PCR assay for studying naphthalene degradation in the environment. Microb Ecol 52:533–543. doi: 10.1007/s00248-006-9082-4. [DOI] [PubMed] [Google Scholar]
- 29.Zhou HW, Luan TG, Zou F, Tam NFY. 2008. Different bacterial groups for biodegradation of three- and four-ring PAHs isolated from a Hong Kong mangrove sediment. J Hazard Mater 152:1179–1185. doi: 10.1016/j.jhazmat.2007.07.116. [DOI] [PubMed] [Google Scholar]
- 30.Peng JJ, Cai C, Qiao M, Li H, Zhu YG. 2010. Dynamic changes in functional gene copy numbers and microbial communities during degradation of pyrene in soils. Environ Pollut 158:2872–2879. doi: 10.1016/j.envpol.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 31.Cébron A, Beguiristain T, Faure P, Norini MP, Masfaraud JF, Leyval C. 2009. Influence of vegetation on the in situ bacterial community and polycyclic aromatic hydrocarbon (PAH) degraders in aged PAH-contaminated or thermal-desorption-treated soil. Appl Environ Microbiol 75:6322–6330. doi: 10.1128/AEM.02862-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yergeau E, Arbour M, Brousseau R, Juck D, Lawrence JR, Masson L, Whyte LG, Greer CW. 2009. Microarray and real-time PCR analyses of the responses of high-arctic soil bacteria to hydrocarbon pollution and bioremediation treatments. Appl Environ Microbiol 75:6258–6267. doi: 10.1128/AEM.01029-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sawulski P, Clipson N, Doyle E. 2014. Effects of polycyclic aromatic hydrocarbons on microbial community structure and PAH ring hydroxylating dioxygenase gene abundance in soil. Biodegradation 25:835–847. doi: 10.1007/s10532-014-9703-4. [DOI] [PubMed] [Google Scholar]
- 34.Rotthauwe JH, Witzel KP, Liesack W. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63:4704–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eaton RW. 2000. trans-o-Hydroxybenzylidenepyruvate hydratase-aldolase as a biocatalyst. Appl Environ Microbiol 66:2668–2672. doi: 10.1128/AEM.66.6.2668-2672.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kweon O, Kim SJ, Cerniglia CE. 2010. Genomic view of mycobacterial high molecular weight polycyclic aromatic hydrocarbon degradation, p 1165–1178. In Timmis KN. (ed), Handbook of hydrocarbon and lipid microbiology, Springer Press, Berlin, Germany. [Google Scholar]
- 37.Niepceron M, Portet-Koltalo F, Merlin C, Motelay-Massei A, Barray S, Bodilis J. 2010. Both Cycloclasticus spp. and Pseudomonas spp. as PAH-degrading bacteria in the Seine estuary (France). FEMS Microbiol Ecol 71:137–147. doi: 10.1111/j.1574-6941.2009.00788.x. [DOI] [PubMed] [Google Scholar]
- 38.Good IJ. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237–264. doi: 10.2307/2333344. [DOI] [Google Scholar]
- 39.Kaiya S, Utsunomiya S, Suzuki S, Yoshida N, Futamata H, Yamada T, Hiraishi A. 2012. Isolation and functional gene analyses of aromatic-hydrocarbon-degrading bacteria from a polychlorinated-dioxin-dechlorinating process. Microb Environ 27:127–135. doi: 10.1264/jsme2.ME11283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Butler CS, Mason JR. 1996. Structure-function analysis of the bacterial aromatic ring-hydroxylating dioxygenases. Adv Microb Physiol 38:47–84. doi: 10.1016/S0065-2911(08)60155-1. [DOI] [PubMed] [Google Scholar]
- 41.Eltis LD, Bolin JT. 1996. Evolutionary relationships among extradiol dioxygenases. J Bacteriol 178:5930–5937. doi: 10.1128/jb.178.20.5930-5937.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barriault D, Vedadi M, Powlowski J, Sylvestre M. 1999. cis-2,3-Dihydro-2,3-dihydroxybiphenyl dehydrogenase and cis-1,2-dihydro-1,2-dihydroxynaphathalene dehydrogenase catalyze dehydrogenation of the same range of substrates. Biochem Biophys Res Commun 260:181–187. doi: 10.1006/bbrc.1999.0706. [DOI] [PubMed] [Google Scholar]
- 43.Meynet P, Head IM, Werner D, Davenport RJ. 2015. Re-evaluation of dioxygenase gene phylogeny for the development and validation of a quantitative assay for environmental aromatic hydrocarbon degraders. FEMS Microbiol Ecol 91:fiv049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eaton RW. 1994. Organization and evolution of naphthalene catabolic pathways: sequence of the DNA encoding 2-hydroxychromene-2-carboxylate isomerase and trans-o-hydroxybenzylidenepyruvate hydratase-aldolase from the NAH7 plasmid. J Bacteriol 176:7757–7762. doi: 10.1128/jb.176.24.7757-7762.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lessner DJ, Johnson GR, Parales RE, Spain JC, Gibson DT. 2002. Molecular characterization and substrate specificity of nitrobenzene dioxygenase from Comamonas sp. strain JS765. Appl Environ Microbiol 68:634–641. doi: 10.1128/AEM.68.2.634-641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson MS, Bakermans C, Madsen EL. 1999. In situ, real-time catabolic gene expression: extraction and characterization of naphthalene dioxygenase mRNA transcripts from groundwater. Appl Environ Microbiol 65:80–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krivobok S, Kuony S, Meyer C, Louwagie M, Willison JC, Jouanneau Y. 2003. Identification of pyrene-induced proteins in Mycobacterium sp. strain 6PY1: evidence for two ring-hydroxylating dioxygenases. J Bacteriol 185:3828–3841. doi: 10.1128/JB.185.13.3828-3841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DeBruyn JM, Mead TJ, Sayler GS. 2012. Horizontal transfer of PAH catabolism genes in Mycobacterium: evidence from comparative genomics and isolated pyrene-degrading bacteria. Environ Sci Technol 46:99–106. doi: 10.1021/es201607y. [DOI] [PubMed] [Google Scholar]
- 49.Pinyakong O, Habe H, Omori T. 2003. The unique aromatic catabolic genes in sphingomonads degrading polycyclic aromatic hydrocarbons (PAHs). J Gen Appl Microbiol 49:1–19. doi: 10.2323/jgam.49.1. [DOI] [PubMed] [Google Scholar]
- 50.Stolz A. 2009. Molecular characteristics of xenobiotic-degrading sphingomonads. Appl Microbiol Biotechnol 81:793–811. doi: 10.1007/s00253-008-1752-3. [DOI] [PubMed] [Google Scholar]
- 51.von Wintzingerode F, Göbel UB, Stackebrandt E. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev 21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 52.Jones MD, Crandell DW, Singleton DR, Aitken MD. 2011. Stable-isotope probing of the polycyclic aromatic hydrocarbon-degrading bacterial guild in a contaminated soil. Environ Microbiol 13:2623–2632. doi: 10.1111/j.1462-2920.2011.02501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnsen AR, de Lipthay JR, Sorensen SJ, Ekelund F, Christensen P, Andersen O, Karlson U, Jacobsen CS. 2006. Microbial degradation of street dust polycyclic aromatic hydrocarbons in microcosms simulating diffuse pollution of urban soil. Environ Microbiol 8:535–545. doi: 10.1111/j.1462-2920.2005.00935.x. [DOI] [PubMed] [Google Scholar]
- 54.Iwai S, Chai B, Sul WJ, Cole JR, Hashsham SA, Tiedje JM. 2010. Gene-targeted-metagenomics reveals extensive diversity of aromatic dioxygenase genes in the environment. ISME J 4:279–285. doi: 10.1038/ismej.2009.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haritash AK, Kaushik CP. 2009. Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): a review. J Hazard Mater 169:1–15. doi: 10.1016/j.jhazmat.2009.03.137. [DOI] [PubMed] [Google Scholar]
- 56.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Capyk JK, Eltis LD. 2012. Phylogenetic analysis reveals the surprising diversity of an oxygenase class. J Biol Inorg Chem 17:425–436. doi: 10.1007/s00775-011-0865-9. [DOI] [PubMed] [Google Scholar]
- 59.Gibson DT, Parales RE. 2000. Aromatic hydrocarbon dioxygenases in environmental biotechnology. Curr Opin Biotechnol 11:236–243. doi: 10.1016/S0958-1669(00)00090-2. [DOI] [PubMed] [Google Scholar]
- 60.Pester M, Rattei T, Flechl S, Gröngröft A, Richter A, Overmann J, Reinhold-Hurek B, Loy A, Wagner M. 2012. amoA-based consensus phylogeny of ammonia-oxidizing archaea and deep sequencing of amoA genes from soils of four different geographic regions. Environ Microbiol 14:525–539. doi: 10.1111/j.1462-2920.2011.02666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pester M, Maixner F, Berry D, Rattei T, Koch H, Lücker S, Nowka B, Richter A, Spieck E, Lebedeva E, Loy A, Wagner M, Daims H. 2014. NxrB encoding the beta subunit of nitrite oxidoreductase as functional and phylogenetic marker for nitrite-oxidizing Nitrospira. Environ Microbiol 16:3055–3071. doi: 10.1111/1462-2920.12300. [DOI] [PubMed] [Google Scholar]
- 62.Rose TM, Henikoff JG, Henikoff S. 2003. CODEHOP (COnsensus-DEgenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucleic Acids Res 31:3763–3766. doi: 10.1093/nar/gkg524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Andrews S. 2010. FastQC: a quality control tool for high-throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 64.Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. 2012. PANDAseq: paired-end assembler for Illumina sequences. BMC Bioinformatics 13:31. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 66.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence data were submitted to the NCBI Sequence Read Archive (SRA) with accession numbers SRR6287136 to SRR6287145.