In the oligotrophic environment of the Mariana Trench, alkanes as carbohydrates are important for the ecosystem, but their spatial and periodic spreading in deep waters has never been reported. Alkane-degrading bacteria such as Alcanivorax spp. are biological signals of the alkane distribution. In the present study, Alcanivorax was abundant in some waters, at depths of up to 6,000 m, in the Mariana Trench. Genomic, transcriptomic, and chemical analyses provide evidence for the presence and activities of Alcanivorax jadensis in deep sea zones. The periodic spreading of alkanes, probably from the subductive plates, might have fundamentally modified the local microbial communities, as well as perhaps the deep sea microenvironment.
KEYWORDS: Alcanivorax, alkanes, deep sea, subduction zone
ABSTRACT
In subduction zones, serpentinization and biological processes may release alkanes to the deep waters, which would probably result in the rapid spread of Alcanivorax. However, the timing and area of the alkane distribution and associated enrichment of alkane-degrading microbes in the dark world of the deep ocean have not been explored. In this study, we report the richness (up to 17.8%) of alkane-degrading bacteria, represented by Alcanivorax jadensis, in deep water samples obtained at 3,000 to 6,000 m in the Mariana Trench in two cruises. The relative abundance of A. jadensis correlated with copy numbers of functional almA and alkB genes, which are involved in alkane degradation. In these water samples, we detected a high flux of alkanes, which probably resulted in the prevalence of A. jadensis in the deep waters. Contigs of A. jadensis were binned from the metagenomes for examination of alkane degradation pathways and deep sea-specific pathways, which revealed a lack of nitrate and nitrite dissimilatory reduction in our A. jadensis strains. Comparing the results for the two cruises conducted close to each other, we suggest periodic release of alkanes that may spread widely but periodically in the trench. Distribution of alkane-degrading bacteria in the world’s oceans suggests the periodic and remarkable contributions of Alcanivorax to the deep sea organic carbon and nitrogen sources.
IMPORTANCE In the oligotrophic environment of the Mariana Trench, alkanes as carbohydrates are important for the ecosystem, but their spatial and periodic spreading in deep waters has never been reported. Alkane-degrading bacteria such as Alcanivorax spp. are biological signals of the alkane distribution. In the present study, Alcanivorax was abundant in some waters, at depths of up to 6,000 m, in the Mariana Trench. Genomic, transcriptomic, and chemical analyses provide evidence for the presence and activities of Alcanivorax jadensis in deep sea zones. The periodic spreading of alkanes, probably from the subductive plates, might have fundamentally modified the local microbial communities, as well as perhaps the deep sea microenvironment.
INTRODUCTION
Away from the eutrophic zone, deep sea organisms thriving in lower mesopelagic to hadal realms confront a nutrient-deprived environment (1). Given the fact that euphotic export and chemolithoautotrophic biosynthesis in the water column can contribute organic carbon to these deep layers (1–3), there is still discrepancy in the carbon budget in the dark ocean. It is controversial how much the puzzle will be resolved by extra organic carbon input from subsurface marine sediments and even the crust of the Earth. Movement of tectonic plates generates geothermal gases represented by methane, which is accumulating in marginal slopes and being released from deep sea vents, mud volcanoes, and pockmarks (4). There are also considerable amounts of CH4 and CO2 in the hydrothermal solution seeping out from deep ocean ridges and rifts (5, 6). The inorganic carbon is assimilated for microbial autotrophic growth. Therefore, tectonic activities beneath the seafloor contribute dramatically to the carbon budgets (7, 8). The role of subduction zones between convergent plates has not been fully evaluated, although special microbial lineages resembling those in early life were discovered in the 10-km-deep mantle, mud volcano, and cold seep in serpentinized sites of the Izu-Bonin-Mariana forearc and Lost City hydrothermal field (9–11). In addition to high concentrations of hydrogen and methane in serpentinized seepage fluids, there were also n-alkanes such as butane and propane (12). In experiments under simulation conditions, the serpentinization process may yield alkanes up to C27, which has been detected in fluid samples from the Rainbow hydrothermal field (13). However, alkanes were not detected in most of the previous studies (12), and how far alkanes can spread in deep sea waters was not investigated. Therefore, the contribution of alkanes to the deep ocean carbon budget is probably underestimated, as subduction zones are widely distributed along the convergent plates in the oceans.
Aside from thermogenic alkanes, decomposition of fatty acids from bacteria and algae also produces alkanes, particularly in anoxic sediments (14). Alkanes may be anaerobically degraded by nitrate- or sulfate-reducing bacteria but are more efficiently oxidized by aerobic microorganisms in open oceans (15). A large number of studies have shown the explosive prevalence of alkane-degrading bacteria in waters after oil pollution (16, 17). For example, Alcanivorax borkumensis SK2 and Alcanivorax jadensis T9 are able to utilize n-alkanes ranging from C5 to C32, depending on the alk and alm gene families, which are specialized for intermediate- and high-molecular-weight alkanes, respectively (18, 19). Their presence is a microbial signal for leakage of alkanes in waters. For deep sea zones, the alkane degradation mediated by microbes has rarely been reported (20). There is no direct evidence for the association between microbial degradation of alkanes and the serpentinization process. Probably alkanes are anaerobically degraded in sediments or injected into water columns. However, Alcanivorax species and other alkane-degrading bacteria were not detected in serpentinization sediments, fluids, and adjacent waters. Complete consumption of alkanes by microbes is occasionally limited by insufficient nitrogen and phosphate supplies (21). Depletion of oxygen may also hinder the removal of heavy alkanes from oil-polluted areas. The slow degradation permits long-distance spread of alkanes to form a plume after strong seepage of thermogenic or biogenic alkanes. The hypothesis of the formation of alkane plumes in dark oceans has not been substantiated to date.
The Mariana Trench was formed by subduction of the western North Pacific plate under the Philippine plate, which resulted in the deepest site on Earth, Challenger Deep. Recently, a cold seep was discovered on the forearc slope of the trench (10). The ecosystem was likely fueled by serpentinized fluids of high pH (10), which is supported by the evidence of serpentinization in the drilling core obtained in the nearby forearc (11). Alkane-degrading microbes have not been examined in the vents and approximate bottom waters. Therefore, the potential influence of alkanes produced by serpentinization was not evaluated. In this study, we sampled the water columns in the Mariana Trench, aiming to answer the questions regarding the spreading of alkanes in the subduction zone and the contribution of hydrocarbons to the ecosystem of deep waters. Alkanes up to C32 were detected in water samples at depths ranging from 1,000 m to 6,200 m. The high concentrations of alkanes remarkably correlated with the richness of A. jadensis, indicating the spreading of alkanes in the formation of plumes.
RESULTS
Microbial community structures.
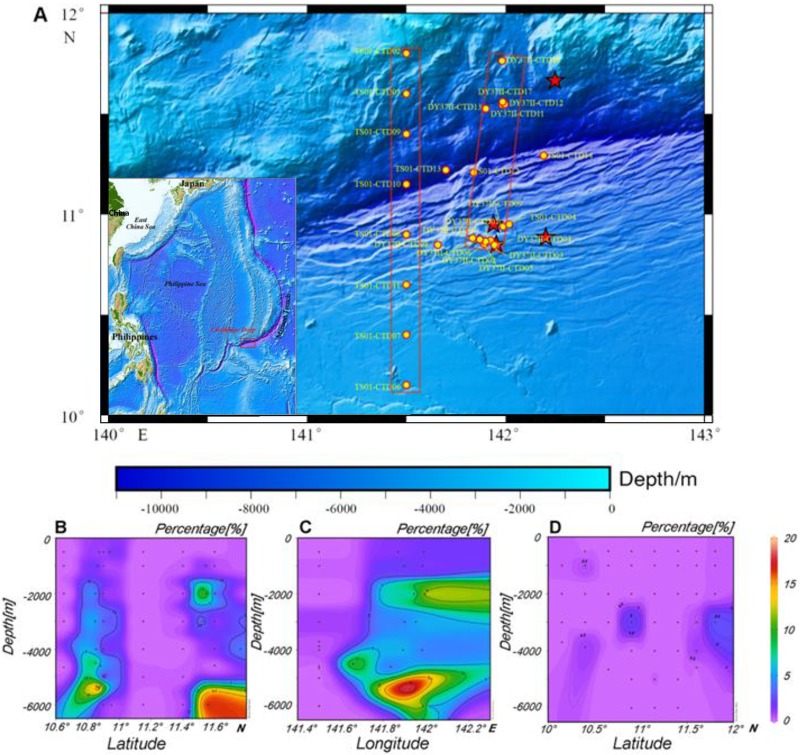
Along different water columns in the southern Mariana Trench, microbial communities were deciphered by sequencing 16S rRNA gene amplicons. The dominant bacterial groups in water samples were the Proteobacteria (mainly Gammaproteobacteria and Deltaproteobacteria), followed by the Thaumarchaeota and Euryarchaeota (see Fig. S1 in the supplemental material). The operational taxonomic units (OTUs) assigned to Alcanivorax were collected, to calculate their percentages at different depths (Table S1). The results showed that the relative abundance of Alcanivorax peaked at 17.8% in the 3,000-m water layer of the R/V Dayang no. 37-II (DY37II) conductivity, temperature, and depth 07 (CTD07) site (Fig. 1B). A prevalence of Alcanivorax (above 10%) was revealed in the water layers of 2,000 to 6,000 m at four sampling sites of the DY37II cruise (CTD01, CTD07, CTD12, and CTD17) (Fig. S1). In contrast, only a small percentage of OTUs for Alcanivorax was identified in the upper layers (<1,000 m). The distribution of Alcanivorax in bottom water (1 m above the bottom) and surface sediments was also examined. At four diving sites (5,460-m to 6,500-m depth) of the Jiaolong submersible (Fig. 1A), Alcanivorax was present only in the bottom waters and not in surface sediments (Fig. S2). The Alcanivorax in the bottom waters accounted for up to 5%, based on the 16S rRNA gene amplicon sequencing. For the samples from the R/V Tansuo no. 01 (TS01) cruise, the relative abundance of Alcanivorax was less than 1%. The TS01 samples were collected ∼2 months later than and ∼30 miles away from the DY37II samples, indicating spatial and periodic distribution of the Alcanivorax species in the Mariana deep waters.
FIG 1.
Collection of samples from the Mariana Trench. The water samples were collected with Niskin bottles (yellow dots) and a manned submersible (red stars) in two cruises (R/V TS01 and DY37II) (A). Multiple sampling sites were selected to illustrate the vertical distribution of Alcanivorax. Both latitude (B) and longitude (C) distributions are depicted for the sites of the DY37II cruise, and the latitude distribution is shown for the samples collected in the TS01 cruise (D). The percentages of Alcanivorax in the mesopelagic and abyssal layers (500 to 6,000 m) were based on sequencing of the 16S rRNA gene amplicons (65). The inset map was created with Generic Mapping Tools (GMT) (66).
Genome binning of an Alcanivorax genome.
Metagenomes that were enriched in Alcanivorax were sequenced for DY37II CTD13 and DY37II CTD17. Genome binning obtained two genome bins for Alcanivorax, as indicated by the classification of the 16S rRNA genes (Fig. S3). The genome bin of Alcanivorax from DY37II CTD17 was 3.2 Mbp and comprised 88 contigs; that from DY37II CTD13 was 3.9 Mbp and comprised 159 contigs (Fig. S3). Assessment of the genome bins resulted in 95.5% and 85.6% completeness for those from DY37II CTD17 and DY37II CTD13, respectively, with low levels (<1.8%) of contaminants in the bins. The similarity of the two genome bins was >99% in reference to the average nucleotide identity (ANI), indicating that they belonged to the same species.
Phylogenetic and correlation analyses.
A 16S rRNA gene sequence was extracted from each of the genome bins. A phylogenetic tree based on the 16S rRNA sequences showed that the Alcanivorax species with the binned genomes were clustered with Alcanivorax jadensis strain T9 (Fig. 2A) (22). Since the 16S rRNA sequences in the Mariana Trench were almost identical to that of Alcanivorax jadensis strain T9 (99.9%), the binned Alcanivorax genomes were very likely derived from A. jadensis; their strain names were designated C13 and C17. Representative reads of 5 major OTUs classified as Alcanivorax were also examined for their phylogenetic positions. The 2 most abundant OTUs, i.e., OTUs 6268 and 35507, were almost identical to the 16S rRNA genes of A. jadensis strains C13 and C17. Two of the remaining 3 OTUs were close to Alcanivorax xenomutans and Alcanivorax venustensis. The two genome bins of A. jadensis strains C13 and C17 were compared with those of A. jadensis strain T9, Alcanivorax borkumensis strain SK2, and Alcanivorax dieselolei strain B5. Again, A. jadensis strains C13 and C17 were close relatives of A. jadensis strain T9, as indicated by the presence of a large number of homologous regions with high levels of similarity between their genomes (Fig. 2B).
FIG 2.
Phylogenetic relationships of major Alcanivorax species and plotting of genomic homologous regions. The genomes of A. jadensis strains C13 and C17 were binned from the metagenomes, and the OTUs for Alcanivorax were picked at 97% similarity. Reference 16S rRNA genes were pooled by BLASTn search of the NCBI databases, to reconstruct a phylogenetic tree based on a maximum-likelihood algorithm (A). Bootstrap values (displayed as percentages of 1,000 replications) are shown at the branches of the maximum-likelihood tree. The homologous regions between the five genomes are demonstrated by lines of different colors (B). The outer circle represents the contigs, and the inner circle depicts the predicted CDSs.
Metabolism specialization.
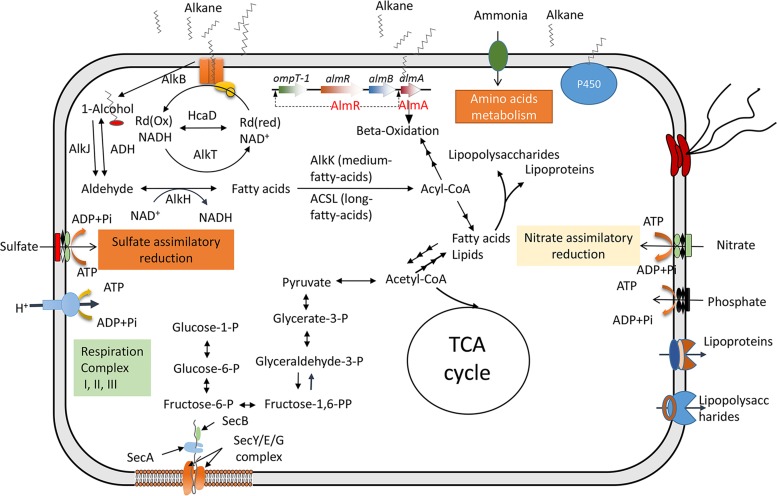
The A. jadensis C13 and C17 genome bins contained three known alkane utilization pathways, mediated by cytochrome P450 and the Alk and Alm alkane monooxygenase systems (Fig. 3). There was a complete set of genes involved in lipid production and degradation in the A. jadensis C13 and C17 genome bins. Sulfate and nitrate might be imported for assimilatory reduction, as indicated by the identification of the related genes. We also compared the Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations for five Alcanivorax genomes (Fig. S4). The A. dieselolei B5 genome contained more functional genes than the other four genomes. A. jadensis C13 and C17 genomes did not harbor the genes for periplasmic nitrogen reduction, such as narGHI, nirK, and norBZ, that were present in the other Alcanivorax genomes, particularly that of A. dieselolei strain B5 (Fig. S4). Probably nitrate was imported by the NrtABC cross-membrane complex and then was assimilated into amino acids directly by NirBD. Moreover, A. jadensis C13 differed from the other genomes by having nuoABFKMN genes, which encode subunits of respiratory chain complex I with proton-pumping function. However, several subunit-coding genes were missing from the genome bin, which leaves a question regarding the proper function of complex I in A. jadensis C13.
FIG 3.
Schematic model of metabolism and cross-membrane transporters. The model was predicted based on the predicted genes in the genome bins of Alcanivorax jadensis strains C13 and C17. TCA, tricarboxylic acid.
Copies of functional genes in water samples.
The almA and alkB genes are the functional genes for alkane degradation by Alcanivorax. To validate the association between the prevalence of Alcanivorax and the abundance of alkanes, copy numbers of the functional almA and alkB genes in the samples were examined by quantitative PCR (qPCR). Differences in the community structure of hydrocarbon-degrading microbial populations were assessed via absolute quantification of the genes. A total of 16 samples from different depths and sites were examined. Results showed that the gene copy numbers positively varied with the percentage of Alcanivorax (Fig. 4). The samples for DY37II CTD17 at 6,000 m and DY37II CTD12 at 2,000 m harbored the most abundant almA and alkB genes, which is in accord with the highest percentages of Alcanivorax in the microbial communities. There were many more almA gene copies than alkB gene copies in most of the samples, while copy numbers of alkB were higher than those of almA in the two bottom waters (DIVE114W and DIVE117W) (Fig. 4).
FIG 4.

Vertical profiling of Alcanivorax, copy numbers of functional genes, and amounts of alkanes in water samples. The abundance of Alcanivorax is an estimate based on sequencing of 16S rRNA gene amplicons. The copy numbers of functional almA and alkB genes of Alcanivorax along the water column were inferred from qPCR results. GC-MS was used to detect alkanes in the water samples. Alkanes were categorized into medium-chain (C11 to C16) and long-chain (C17 to C32) groups (see Table S2 in the supplemental material for details).
Composition of alkanes in water samples.
To explain the spatial distribution pattern of A. jadensis in the Mariana Trench, alkane contents in the water samples were analyzed with gas chromatography-mass spectrometry (GC-MS). From the results, pentadecanoic acid (C17) was the most abundant long-chain alkane, followed by hexanedioic acid (C16), in all samples. These fatty acids may be used to synthesize the storage lipids in Alcanivorax (23). In the DY37II CTD10 sample, there were 10 known alkane species, accounting for the greatest amounts of alkane content (Table S2). The waters from DY37II CTD13 and DY37II CTD17 contained the greatest amounts of long-chain alkanes such as C19, C21, and C26. The results were roughly consistent with the relative abundance of Alcanivorax in microbial communities (Fig. 4). DY37II CTD02 and DY37II CTD10 were sampled at the south and north slopes of the trench, respectively. Although both were at a depth of 4,000 m, they differed in the abundance of alkanes. For the two samples from the TS01 cruise, linolelaidic acid was detected in TS01 CTD09-7 (depth of 6,000 m) but not in TS01 CTD08-7 (Fig. 4). The alkane contents in the near-bottom water samples also correlated with the relative abundance of Alcanivorax in the community structure. DIVE114W was a bottom water sample collected by a submersible dive from the southern slope of the trench. GC-MS analysis revealed the greatest amount of long-chain (C32) alkanes in DIVE114W. TS01 CTD09-7 was close to DY37II CTD17 and at the same sampling depth. The GC-MS results displayed disparities in the contents and abundance of alkanes in the two samples, which was also an indicator of spatial and periodic spreading of alkanes in the Mariana Trench.
Transcriptional activity.
A total of 24 Gbp of metatranscriptomic data for seven aqueous samples was obtained and subjected to quality control. The 16S rRNA genes were then predicted in the data. Those assigned to A. jadensis were the most abundant (11.1%) in the DY37II CTD01 sample, which coincides with the enrichment of A. jadensis in the sample (Fig. 5). The transcriptional level of A. jadensis in the other layers was relatively lower, in agreement with the relative abundance in the communities. The almA and alkB genes of A. jadensis C13 and C17 were identified in the TS01 CTD07 metatranscriptome (Tables S3 and S4). Totals of 213 and 147 genes were transcribed for A. jadensis C13 and C17, respectively, in the TS01 CTD07 metatranscriptome. The number of the genes identified in the metatranscriptome accounted for ∼6.7% of the gene contents of A. jadensis C13 and C17.
FIG 5.

Relative abundance of Alcanivorax 16S rRNA in transcripts and amplicons. The 16S rRNA gene transcripts in seven metatranscriptomes were extracted and classified. The percentages assigned to A. jadensis among all of the 16S rRNA gene transcripts are shown. The relative abundance of A. jadensis in the microbial communities of these samples was estimated based on the percentage in sequenced 16S rRNA gene amplicons.
Global deep sea distribution.
Using public data on 16S rRNA gene amplicons and metagenomes, we investigated the distribution of Alcanivorax in deep waters of different oceans, particularly along subduction zones (Fig. 6). Overall, Alcanivorax was ubiquitous and accounted for less than 5% of the microbial communities in the collected data. At a depth of 10,200 m in the Mariana Trench, Alcanivorax accounted for ∼25% of the total. When subduction zones were compared with the other sites, there was not an obviously greater percentage of Alcanivorax there. At different depths, several cases of layer-specific Alcanivorax richness were observed.
FIG 6.
Global distribution of Alcanivorax in deep sea layers. The results were based on BLASTN searches of 16S rRNA fragments in 16S rRNA amplicons and metagenomes from public databases. The solid red lines show the locations of subduction zones. The map was created with Generic Mapping Tools (GMT) (66).
DISCUSSION
Previous studies on Alcanivorax focused on its role in ecological restoration, with very few studies on its spread in deep oceans. Our results showed that it may also be abundant in the depths but not dominant, which is perhaps attributable to the extreme environment of deep seas, with low temperature and high pressure (24). The richness of Alcanivorax in certain samples was perhaps due to the large amounts of alkanes released from sediments. In the Mariana Trench, pockmarks were observed by the Jiaolong submersible during the DY37II cruise (Fig. S6). The development of pockmarks is often related to the proliferation of deep hydrocarbon gases (25). A growing body of evidence shows that pockmarks are mainly formed by fluid escaping from under the seabed sediment with the diffusion of oil and gas (26). For the terrain of the Mariana Trench (27), the collision of plates resulted in mass flow from serpentinite-hosted systems, generating hydrocarbons under hydrothermal conditions. In this process, carbon and hydrogen elements accumulate to abiogenically form methane or short-chain and low-molecular-weight alkanes through the interaction of water and rock (28, 29). Undergoing long-term chemical reactions, the short-chain alkanes form medium- and long-chain alkanes. The hydrothermal gases probably fueled the origin of life on Earth (30). In subduction zones such as the Mariana Trench, methane and alkanes might be discharged to the deep sea waters through cracks in rocks and oceanic energy vents (31). Aside from the alkane-producing serpentinization process, alkanes are also generated biologically by organisms, through thermogenesis, and from rotting plants (32, 33). The microbial community structures of the samples collected by the two cruises in this study differed remarkably. Alcanivorax species not only were highly abundant in some sites but also were transcriptionally active at different levels in the DY37 samples. The low relative abundance of Alcanivorax was consistent with the small amounts of alkanes in the TS01 samples. The unstable supply of alkanes was a possible explanation for the spatial and periodic prevalence of Alcanivorax. Along the subduction zones of the world’s oceans, it was not evident that alkane-degrading Alcanivorax bloomed in the deep waters, as shown by our results. More sampling sites and time points may contribute to the tight link between approximation to subduction zones and the richness of Alcanivorax. In addition, A. jadensis strains C13 and C17 may be considered to be pressure adapted, which is different from previous reports (34, 35). More follow-up studies, such as separations of pure cultures, are required to verify the pressure tolerance.
Degradation of alkanes by Alcanivorax is highly dependent on oxygen, which prohibits the spread of Alcanivorax in zones and sediments with minimal oxygen (36). In this study, we compared the microbial community structures in bottom water samples and sediments. The dramatic difference in Alcanivorax abundance was probably attributable to the low oxygen flux in the sediments. In this regard, alkanes likely could be consumed only in waters. The slow degradation of long-chain alkanes may spread long distances with the effects of deep seawater currents. It was reported that, with the effects of physicochemical factors, spilled oil formed an oil plume containing gaseous and monoaromatic compounds, and a hydrocarbon-dependent microbial community in the plume was proposed (34). However, our study did not detect Colwellia, Cycloclasticus, and Pseudoalteromonas, which may multiply to form dominant bacterial groups in the plumes (34). Catching the plume is technically difficult, because it is rather dynamic under the influence of the water flow. This may partially explain the incongruent GC-MS and Alcanivorax relative abundance results, as well as the lack of consistency between the distribution of subduction zones and the relative abundance of Alcanivorax.
Although the related regulatory genes were identified in Alcanivorax genomes, the expression-regulating mechanism was unknown (37, 38). The presence of the three known alkane-degrading pathways indicates that A. jadensis in the present study might utilize alkanes of different sizes. Strains with the alkB gene were abundant in the bottom waters, probably due to the rapid consumption of short- and medium-chain alkanes. In the water column, the almA genes outcompeted the alkB genes in copy number, indicating that the Alcanivorax inhabitants there were prone to degradation of medium- and long-chain alkanes. This allows the proposition that the alkanes were released from the sediments and small alkanes were degraded first by the Alcanivorax strains bearing more alkB genes in the bottom waters.
Most alkane-degrading bacteria, including Colwellia, Cycloclasticus, and Pseudoalteromonas, were reported in shallow and coastal waters (39). In deep waters of open oceans, Alcanivorax is the predominant species for the degradation of alkanes of various sizes (40). The degradation of long-chain alkanes may also provide a carbon source for heterotrophic bacteria in the deep sea. In the microbial communities revealed for the DY37 cruise, the richness of Alcanivorax was associated with that of Alteromonas, a heterotrophic bacterium (41) (see Fig. S5 in the supplemental material). In contrast, there was distinct richness of Novosphingobium and Sphingobium in the samples collected by the TS01 cruise. The prevalence of polycyclic aromatic hydrocarbon (PAH) degraders (42) indicates a nutrient-poor deep sea environment at the TS01 sampling sites. The assimilatory nitrate reduction process carried out by the deep sea A. jadensis strains in this study likely could rapidly produce organic nitrogen sources such as amino acids and vitamins, which are important for the development and maintenance of deep sea ecosystems. There is some dissimilatory nitrate reduction in other Alcanivorax species, but the A. jadensis strains in this study seem to be independent of the periplasmic nitrate as an election acceptor. This was perhaps accounted for by sufficient oxygen flux for the deep inhabitants in the water column. A previous study reported a dissolved oxygen concentration of 172 μM at a depth of 6,000 m in the Mariana Trench (24), which allows rapid growth of Alcanivorax.
How the three alkane-degrading pathways are regulated in Alcanivorax is still an open question at present. More experiments, in situ cultivation, and transcriptomic work may provide clues to the puzzle. In sum, the microbial communities are more dynamic than previously expected, due to rapid changes in deep sea environments. Sampling efforts at more sites, time points, and depths with advanced techniques will cast light on the metabolic activities and adaptation strategies of deep sea microbial lineages with important ecological contributions.
MATERIALS AND METHODS
Sample collection and nucleotide extraction.
Two research cruises were carried out in the Mariana Trench in the summer of 2016, i.e., DY37II (from 4 June to 13 July 2016) and TS01 (from 22 June to 12 August 2016). About 70 liters of water was obtained for each sample, at different depths and sites, with Niskin bottles on the CTD sensors (Fig. 1A; also see Table S1 in the supplemental material). Push cores and 10-liter samples of bottom waters were also obtained, by four Jiaolong submersible dives (Fig. 1A). Water samples were filtered through 0.22-μm polycarbonate membranes (Millipore, Bedford, MA, USA). After filtration, the membranes were immediately frozen at −80°C for metagenomic work, and 500-ml samples of filtered water were stored at 4°C for chemical component analysis. A thin layer of the push cores was removed, and the remaining sediment was sliced into 2-cm layers.
DNA was extracted from the polycarbonate membranes and sediment layers from 0 to 2 cm below the seafloor (cmbsf) using the Mo Bio PowerSoil DNA isolation kit (Mo Bio, Carlsbad, CA, USA). RNA was extracted using the Mo Bio PowerSoil total RNA isolation kit, and DNA was degraded with the Turbo DNA-free kit (Ambion, Carlsbad, CA, USA). Reverse transcription was completed with the Ovation RNA-sequencing system v2 kit (NuGEN, San Carlos, CA, USA). Nucleotide concentrations were estimated with a Qubit 2.0 fluorometer (Invitrogen, Carlsbad, CA, USA). The quality of nucleotides was checked with gel electrophoresis.
Sequencing of 16S rRNA gene amplicons and data analyses.
A pair of universal primers (U341F, 5′-CCTAYGGGRBGCASCAG-3′; U802R, 5′-TACNVGGGTATCTAATCC-3′) was used to amplify the V3 to V4 region of the 16S rRNA gene, in triplicate (43). Six-nucleotide barcodes were appended to the primers for subsequent separation of different samples. The PCR was prepared according to the protocol for PrimeSTAR HS DNA polymerase (TaKaRa, Dalian, China), with 2 μl of forward and reverse primers (10 μM) and 1 ng of template DNA. The PCR was performed on a thermal cycler (Bio-Rad, Hercules, CA, USA), with the following program: (i) 10 s at 98°C to unwind the DNA, (ii) 30 cycles of 10 s at 98°C, 15 s at 50°C, and 30 s at 72°C, and (iii) 5 min at 72°C for extension. The quality of the PCR products was checked with gel electrophoresis, and the products were purified using the Cycle Pure kit (Omega, Norcross, GA, USA). All of the PCR products were mixed together, in the same amounts, for preparation of the Illumina sequencing library. The amplicons were sequenced on the Illumina MiSeq platform.
The adaptors, low-quality reads, and ambiguous nucleotides were trimmed from the raw data with the NGS QC Toolkit (44). The quality-filtered reads were assigned to OTUs at a 97% similarity level with UCLUST (45). The longest read of each OTU was selected as the representative for subsequent taxonomic classification. Taxonomic assignment was conducted using the Ribosomal Database Project (RDP) classifier v2.2 (46), by referring to the Silva128 database (47), with a confidence level of 80%. Calculation of diversity indices and OTU clustering were performed with QIIME v1.9.1 (48).
Metagenomic analyses.
A total of 200 ng of genomic DNA was fragmented to ∼500 bp by ultrasonication. Genomic libraries were arranged using the TruSeq Nano DNA Library kit (Illumina). The final library containing all of the pooled samples was sequenced on the MiSeq platform (Illumina). The raw data were evaluated using FastQC (49); low-quality reads that included N or were shorter than 50 bp were excluded. The quality of the paired-end reads (3 Gbp) was assessed using the NGS QC Toolkit (44), which resulted in filtration of low-quality reads with a minimum value of Q20. Clean data were assembled into scaffolds by SPAdes v3.6.2 (50). Genome binning was accomplished as reported previously (51). The read coverage and G+C contents of the contigs were used for binning of the genome bins, followed by a correspondence analysis of the tetranucleotide frequencies of their respective contigs (51). The 16S rRNA genes in the contigs were identified using rRNA_HMM (52). The completeness and contamination rate were assessed by CheckM v1.0.5 (53).
Coding sequences (CDSs) and proteins in the binned genomes were predicted by Prodigal v2.6.2 (54). Annotation of CDSs was carried out via BLASTP against databases including the NCBI nonredundant protein database, the KEGG database (55), and the Clusters of Orthologous Groups (COG) database (56), with a cutoff E-value of 1e−05.
Metatranscriptome sequencing and data analysis.
cDNA libraries were prepared with the Ovation Ultralow v2 1–16 kit (NuGEN, San Carlos, CA, USA) and sequenced on the MiSeq platform (Illumina). The raw data were evaluated using FastQC (49). The 16S rRNA genes in the metatranscriptomic data were predicted by SortMeRNA v2.1 (57) and classified by referring to the Silva128 database (47), with a confidence level of 90%. The quality-filtered reads were assembly by Trinity v2.3.2 (58). The genes that resembled the open reading frames (ORFs) predicted for the A. jadensis genome bins were identified by BLASTN, with a sequence identity threshold value of 90% (59).
Phylogenetic and correlation analyses.
The 16S RNA genes were pooled with the representative OTUs assigned to Alcanivorax and those of the relatives identified in the NCBI nucleotide database. All of the sequences were aligned with DNAMAN v6.0 (Lynnon Biosoft, San Ramon, CA, USA) and adjusted manually with MEGA v5.0 (60). Using a maximum-likelihood algorithm, a phylogenetic tree was constructed using raxmlGUI v1.5, with 1,000 replicates (61). The regions with homology between the binned genomes and the references were detected by a pairwise BLASTn search. The homologous regions were displayed by Circos v0.69-2 (62).
GC-MS analysis of hydrocarbons.
The water samples filtered with 0.22-μm polycarbonate membranes (Millipore) were shaken gently and extracted twice with 100 ml of methylene chloride and subsequently with 100 ml of hexane (63). The extracts was mixed and rotary-evaporated to 1 ml. A column filled with activated silica gel (mesh size of 200 to 325) was used for chromatography of the hexane extract. Twenty milliliters of hexane/methylene chloride (70:30 [vol/vol]) was used to elute the hydrocarbons (64). The collected distillate was blown dry with a nitrogen evaporator (Allsheng, Hangzhou, China). Before being injected into an Agilent 7890A/5975C GC-MS system (Agilent, Santa Clara, CA, USA), the samples were dissolved with 100 μl hexane. The resulting patterns were searched in the NIST14.L library. The area of the peaks was calculated as an estimate of amount of certain alkane content. The chemical components present in blank control that possibly resulted from containers, environment and solutions were then excluded from the GC-MS results of the water samples.
Quantification of functional genes.
Two pairs of primers were designed for the almA and alkB genes (almA-F, 5′-CGTGACAACGAAGACTGCATTAC-3′; almA-R, 5′-CGTGACAACGAAGACTGCATTAC-3′; alkB-F, 5′-TTGCGTTTGAAAAAGTGGGG-3′; alkB-R, 5′-CGGTAGCTCTTGTCCTGGGA-3′), based on the gene predictions for the genome. PCR amplification of the genes was performed with the 2× Taq PCR Master Mix kit (TianGen, Dalian, China). The fragments were cloned into the pMD18-T vector (TaKaRa) and then transformed into competent JM109 cells (TaKaRa). Colony PCR was performed by direct PCR with the M13F and M13R primers. After bacterial culture, the plasmid was extracted using a plasmid midi kit (Omega, Norcross, GA, USA). The DNA was serially diluted 10-fold, as were the standard PCR products. The gene copy numbers were estimated by qPCR in triplicate, using the StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA, USA) with SYBR green I (TaKaRa). The qPCR was performed in a 20-μl reaction mixture with 2 μl of template DNA (1 ng), 0.8 μl of each primer (10 nM), 0.4 μl of ROX reference dye, and 6 μl of sterile water. The thermal cycling conditions were 95°C for 30 s, 40 cycles of 95°C for 5 s and 60°C for 30 s, and a melting curve stage of 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s.
Data availability.
Data supporting the results of this article have been deposited in GenBank under BioProject no. PRJNA421240. The genome accession numbers are QAZC00000000 and QAZB00000000. SRA accession of sequence reads for 16S rRNA amplicons are SRR6466501, SRR6466502, SRR6466503, and SRR6466504.
Supplementary Material
ACKNOWLEDGMENTS
We thank the DY37II and TS01 cruise members for their efforts in the sampling cruises. We thank J. Li, J. Chen, and D. S. Cai for their help in the treatment of water samples.
This study was supported by the National Science Foundation of China (grants 31460001 and 41476104), Strategic Priority Research Program B of the Chinese Academy of Sciences (grant XDB06010201), and the National Key Research and Development Program of China (grant 2016YFC0302500).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02089-18.
REFERENCES
- 1.Herndl GJ, Reinthaler T. 2013. Microbial control of the dark end of the biological pump. Nat Geosci 6:718–724. doi: 10.1038/ngeo1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swan BK, Martinez-Garcia M, Preston CM, Sczyrba A, Woyke T, Lamy D, Reinthaler T, Poulton NJ, Masland EDP, Gomez ML, Sieracki ME, DeLong EF, Herndl GJ, Stepanauskas R. 2011. Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science 333:1296–1300. doi: 10.1126/science.1203690. [DOI] [PubMed] [Google Scholar]
- 3.Pachiadaki MG, Sintes E, Bergauer K, Brown JM, Record NR, Swan BK, Mathyer ME, Hallam SJ, Lopez-Garcia P, Takaki Y, Nunoura T, Woyke T, Herndl GJ, Stepanauskas R. 2017. Major role of nitrite-oxidizing bacteria in dark ocean carbon fixation. Science 358:1046–1050. doi: 10.1126/science.aan8260. [DOI] [PubMed] [Google Scholar]
- 4.Chen SC, Hsu SK, Tsai CH, Ku CY, Yeh YC, Wang YS. 2010. Gas seepage, pockmarks and mud volcanoes in the near shore of SW Taiwan. Mar Geophys Res 31:133–147. doi: 10.1007/s11001-010-9097-6. [DOI] [Google Scholar]
- 5.Connelly DP, Copley JT, Murton BJ, Stansfield K, Tyler PA, German CR, Dover CLV, Amon D, Furlong M, Grindlay N. 2012. Hydrothermal vent fields and chemosynthetic biota on the world's deepest seafloor spreading centre. Nat Commun 3:620. doi: 10.1038/ncomms1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pimenov NV, Lein AY, Sagalevich AM, Ivanov MV. 2000. Carbon dioxide assimilation and methane oxidation in various zones of the rainbow hydrothermal field. Microbiology 69:689–697. doi: 10.1023/A:1026662426126. [DOI] [PubMed] [Google Scholar]
- 7.Foley SF, Fischer TP. 2017. An essential role for continental rifts and lithosphere in the deep carbon cycle. Nat Geosci 10:897–902. doi: 10.1038/s41561-017-0002-7. [DOI] [Google Scholar]
- 8.Deming JW, Baross JA. 1993. Deep-sea smokers: windows to a subsurface biosphere? Geochim Cosmochim Acta 57:3219–3230. doi: 10.1016/0016-7037(93)90535-5. [DOI] [PubMed] [Google Scholar]
- 9.Brazelton WJ, Mehta MP, Kelley DS, Baross JA. 2011. Physiological differentiation within a single-species biofilm fueled by serpentinization. mBio 2:(4):e00127-11 10.1128/mBio.00127-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohara Y, Reagan MK, Fujikura K, Watanabe H, Michibayashi K, Ishii T, Stern RJ, Pujana I, Martinez F, Girard G, Ribeiro J, Brounce M, Komori N, Kino M. 2012. A serpentinite-hosted ecosystem in the Southern Mariana Forearc. Proc Natl Acad Sci U S A 109:2831–2835. doi: 10.1073/pnas.1112005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plümper O, King HE, Geisler T, Liu Y, Pabst S, Savov IP, Rost D, Zack T. 2017. Subduction zone forearc serpentinites as incubators for deep microbial life. Proc Natl Acad Sci U S A 114:4324–4329. doi: 10.1073/pnas.1612147114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schrenk MO, Brazelton WJ, Lang SQ. 2013. Serpentinization, carbon, and deep life. Rev Mineral Geochem 75:575–606. doi: 10.2138/rmg.2013.75.18. [DOI] [Google Scholar]
- 13.Holm NG, Charlou JL. 2001. Initial indications of abiotic formation of hydrocarbons in the Rainbow ultramafic hydrothermal system. Earth Planet Sci Lett 191:1–8. doi: 10.1016/S0012-821X(01)00397-1. [DOI] [Google Scholar]
- 14.Claypool GE, Kvenvolden KA. 1983. Methane and other hydrocarbon gases in marine sediment. Annu Rev Earth Planet Sci 11:299–327. doi: 10.1146/annurev.ea.11.050183.001503. [DOI] [Google Scholar]
- 15.So CM, Young LY. 1999. Initial reactions in anaerobic alkane degradation by a sulfate reducer, strain AK-01. Appl Environ Microbiol 65:5532–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Head IM, Jones DM, Roling WFM. 2006. Marine microorganisms make a meal of oil. Nat Rev Microbiol 4:173–182. doi: 10.1038/nrmicro1348. [DOI] [PubMed] [Google Scholar]
- 17.Brooijmans RJ, Pastink MI, Siezen RJ. 2009. Hydrocarbon-degrading bacteria: the oil-spill clean-up crew. Microb Biotechnol 2:587–594. doi: 10.1111/j.1751-7915.2009.00151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bredemeier R, Hulsch R, Metzger JO, Berthe-Corti L. 2003. Submersed culture production of extracellular wax esters by the marine bacterium Fundibacter jadensis. Mar Biotechnol (NY) 5:579–583. doi: 10.1007/s10126-003-0012-x. [DOI] [PubMed] [Google Scholar]
- 19.Rojo F. 2009. Degradation of alkanes by bacteria. Environ Microbiol 11:2477–2490. doi: 10.1111/j.1462-2920.2009.01948.x. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Li TG, Wang MY, Lai QL, Li JT, Gao ZM, Shao ZZ, Qian PY. 2016. Archive of bacterial community in anhydrite crystals from a deep-sea basin provides evidence of past oil-spilling in a benthic environment in the Red Sea. Biogeosciences 13:6405–6417. doi: 10.5194/bg-13-6405-2016. [DOI] [Google Scholar]
- 21.Schneiker S, Martins dos Santos VA, Bartels D, Bekel T, Brecht M, Buhrmester J, Chernikova TN, Denaro R, Ferrer M, Gertler C, Goesmann A, Golyshina OV, Kaminski F, Khachane AN, Lang S, Linke B, McHardy AC, Meyer F, Nechitaylo T, Puhler A, Regenhardt D, Rupp O, Sabirova JS, Selbitschka W, Yakimov MM, Timmis KN, Vorholter FJ, Weidner S, Kaiser O, Golyshin PN. 2006. Genome sequence of the ubiquitous hydrocarbon-degrading marine bacterium Alcanivorax borkumensis. Nat Biotechnol 24:997–1004. doi: 10.1038/nbt1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruns A, Berthe-Corti L. 1999. Fundibacter jadensis gen. nov., sp. nov., a new slightly halophilic bacterium, isolated from intertidal sediment. Int J Syst Bacteriol 49:441. doi: 10.1099/00207713-49-2-441. [DOI] [PubMed] [Google Scholar]
- 23.Kalscheuer R, Stoveken T, Malkus U, Reichelt R, Golyshin PN, Sabirova JS, Ferrer M, Timmis KN, Steinbuchel A. 2007. Analysis of storage lipid accumulation in Alcanivorax borkumensis: evidence for alternative triacylglycerol biosynthesis routes in bacteria. J Bacteriol 189:918–928. doi: 10.1128/JB.01292-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nunoura T, Takaki Y, Hirai M, Shimamura S, Makabe A, Koide O, Kikuchi T, Miyazaki J, Koba K, Yoshida N, Sunamura M, Takai K. 2015. Hadal biosphere: insight into the microbial ecosystem in the deepest ocean on Earth. Proc Natl Acad Sci U S A 112:E1230–E1236. doi: 10.1073/pnas.1421816112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gay A, Lopez M, Cochonat P, Séranne M, Levaché D, Sermondadaz G. 2006. Isolated seafloor pockmarks linked to BSRs, fluid chimneys, polygonal faults and stacked Oligocene-Miocene turbiditic palaeochannels in the Lower Congo Basin. Mar Geol 226:25–40. doi: 10.1016/j.margeo.2005.09.018. [DOI] [Google Scholar]
- 26.Judd AG, Hovland M. 2007. Seabed fluid flow: the impact on geology, biology, and the marine environment, p 492 Cambridge University Press, Cambridge, UK. [Google Scholar]
- 27.Oakley AJ, Taylor B, Fryer P, Moore GE, Goodliffe AM, Morgan JK. 2007. Emplacement, growth, and gravitational deformation of serpentinite seamounts on the Mariana forearc. Geophys J Int 170:615–634. doi: 10.1111/j.1365-246X.2007.03451.x. [DOI] [Google Scholar]
- 28.Lancet HS, Anders E. 1970. Carbon isotope fractionation in the Fischer-Tropsch synthesis and in meteorites. Science 170:980–982. doi: 10.1126/science.170.3961.980. [DOI] [PubMed] [Google Scholar]
- 29.McCollom TM, Ritter G, Simoneit BR. 1999. Lipid synthesis under hydrothermal conditions by Fischer-Tropsch-type reactions. Orig Life Evol Biosph 29:153–166. doi: 10.1023/A:1006592502746. [DOI] [PubMed] [Google Scholar]
- 30.Suda K, Gilbert A, Yamada K, Yoshida N, Ueno Y. 2017. Compound- and position-specific carbon isotopic signatures of abiogenic hydrocarbons from on-land serpentinite-hosted Hakuba Happo hot spring in Japan. Geochim Cosmochim Acta 206:201–215. doi: 10.1016/j.gca.2017.03.008. [DOI] [Google Scholar]
- 31.Lollar BS, Westgate TD, Ward JA, Slater GF, Lacrampe-Couloume G. 2002. Abiogenic formation of alkanes in the Earth’s crust as a minor source for global hydrocarbon reservoirs. Nature 416:522–524. doi: 10.1038/416522a. [DOI] [PubMed] [Google Scholar]
- 32.Volkman JK. 2006. Lipid markers for marine organic matter. Handb Environ Chem 2:27–70. [Google Scholar]
- 33.McCollom TM. 2013. Laboratory simulations of abiotic hydrocarbon formation in Earth’s deep subsurface. Rev Mineral Geochem 75:467–494. doi: 10.2138/rmg.2013.75.15. [DOI] [Google Scholar]
- 34.Scoma A, Yakimov MM, Boon N. 2016. Challenging oil bioremediation at deep-sea hydrostatic pressure. Front Microbiol 7:1203. doi: 10.3389/fmicb.2016.01203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scoma A, Barbato M, Borin S, Daffonchio D, Boon N. 2016. An impaired metabolic response to hydrostatic pressure explains Alcanivorax borkumensis recorded distribution in the deep marine water column. Sci Rep 6:31316. doi: 10.1038/srep31316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Genovese M, Crisafi F, Denaro R, Cappello S, Russo D, Calogero R, Santisi S, Catalfamo M, Modica A, Smedile F, Genovese L, Golyshin PN, Giuliano L, Yakimov MM. 2014. Effective bioremediation strategy for rapid in situ cleanup of anoxic marine sediments in mesocosm oil spill simulation. Front Microbiol 5:162. doi: 10.3389/fmicb.2014.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang WP, Shao ZZ. 2013. Enzymes and genes involved in aerobic alkane degradation. Front Microbiol 4:116. doi: 10.3389/fmicb.2013.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang WP, Shao ZZ. 2014. The long-chain alkane metabolism network of Alcanivorax dieselolei. Nat Commun 5:5755. doi: 10.1038/ncomms6755. [DOI] [PubMed] [Google Scholar]
- 39.Lamendella R, Strutt S, Borglin S, Chakraborty R, Tas N, Mason OU, Hultman J, Prestat E, Hazen TC, Jansson JK. 2014. Assessment of the Deepwater Horizon oil spill impact on Gulf Coast microbial communities. Front Microbiol 5:130. doi: 10.3389/fmicb.2014.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hara A, Syutsubo K, Harayama S. 2003. Alcanivorax which prevails in oil-contaminated seawater exhibits broad substrate specificity for alkane degradation. Environ Microbiol 5:746–753. doi: 10.1046/j.1468-2920.2003.00468.x. [DOI] [PubMed] [Google Scholar]
- 41.Weyman PD, Smith HO, Xu Q. 2011. Genetic analysis of the Alteromonas macleodii [NiFe]-hydrogenase. FEMS Microbiol Lett 322:180–187. doi: 10.1111/j.1574-6968.2011.02348.x. [DOI] [PubMed] [Google Scholar]
- 42.Maeda AH, Nishi S, Hatada Y, Ozeki Y, Kanaly RA. 2014. Biotransformation of the high-molecular weight polycyclic aromatic hydrocarbon (PAH) benzo[k]fluoranthene by Sphingobium sp. strain KK22 and identification of new products of non-alternant PAH biodegradation by liquid chromatography electrospray ionization. Microb Biotechnol 7:114–129. doi: 10.1111/1751-7915.12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Filardo S, Di Pietro M, Porpora MG, Recine N, Farcomeni A, Latino MA, Sessa R. 2017. Diversity of cervical microbiota in asymptomatic Chlamydia trachomatis genital infection: a pilot study. Front Cell Infect Microbiol 7:321. doi: 10.3389/fcimb.2017.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patel RK, Jain M. 2012. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7:e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 46.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuczynski J, Stombaugh J, Walters WA, Gonzalez A, Caporaso JG, Knight R. 2011. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Bioinformatics 36:10.7.1–10.7.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andrews S. 2014. FastQC: a quality control tool for high through-put sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 50.Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, Prjibelski AD, Pyshkin A, Sirotkin A, Sirotkin Y, Stepanauskas R, Clingenpeel SR, Woyke T, McLean JS, Lasken R, Tesler G, Alekseyev MA, Pevzner PA. 2013. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol 20:714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. 2013. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31:533–538. doi: 10.1038/nbt.2579. [DOI] [PubMed] [Google Scholar]
- 52.Huang Y, Gilna P, Li W. 2009. Identification of ribosomal RNA genes in metagenomic fragments. Bioinformatics 25:1338–1340. doi: 10.1093/bioinformatics/btp161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hyatt D, LoCascio PF, Hauser LJ, Uberbacher EC. 2012. Gene and translation initiation site prediction in metagenomic sequences. Bioinformatics 28:2223–2230. doi: 10.1093/bioinformatics/bts429. [DOI] [PubMed] [Google Scholar]
- 55.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tatusov RL, Galperin MY, Natale DA, Koonin EV. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kopylova E, Noe L, Touzet H. 2012. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 58.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, MacManes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, LeDuc RD, Friedman N, Regev A. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Silvestro D, Michalak I. 2012. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol 12:335–337. doi: 10.1007/s13127-011-0056-0. [DOI] [Google Scholar]
- 62.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cincinelli A, Stortini AM, Perugini M, Checchini L, Lepri L. 2001. Organic pollutants in sea-surface microlayer and aerosol in the coastal environment of Leghorn (Tyrrhenian Sea). Mar Chem 76:77–98. doi: 10.1016/S0304-4203(01)00049-4. [DOI] [Google Scholar]
- 64.Reddy CM, Quinn JG. 1999. GC-MS analysis of total petroleum hydrocarbons and polycyclic aromatic hydrocarbons in seawater samples after the North Cape oil spill. Mar Pollut Bull 38:126–135. doi: 10.1016/S0025-326X(98)00106-4. [DOI] [Google Scholar]
- 65.Schlitzer R. 2017. Ocean Data View. https://odv.awi.de.
- 66.Wessel P, Walter S. 1998. New, improved version of Generic Mapping Tools released. Eos Trans AGU 79:579. doi: 10.1029/98EO00426. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the results of this article have been deposited in GenBank under BioProject no. PRJNA421240. The genome accession numbers are QAZC00000000 and QAZB00000000. SRA accession of sequence reads for 16S rRNA amplicons are SRR6466501, SRR6466502, SRR6466503, and SRR6466504.