Bifidobacteria are natural inhabitants of the human intestinal tract. Some bifidobacterial strains are used as probiotics in fermented dairy production because of their health-promoting effects. Following consumption, bifidobacteria colonize the lower intestinal tract, where the concentrations of bile salts remain nearly 0.05% to 2.0%. Bile salts, as detergent-like antimicrobial compounds, can cause cellular membrane disruption, protein misfolding, and DNA damage. Therefore, tolerance to physiological bile stress is indeed essential for bifidobacteria to survive and to exert probiotic effects in the gastrointestinal tract. In B. longum BBMN68, the MarR-type regulator BmrR was involved in the bile stress response by autoregulating the bmrRAB operon, and ox bile as an inducer could increase the expression of the BmrAB transporter to enhance the bile tolerance of BBMN68. Our study represents a functional analysis of the bmrRAB operon in the bile stress response, which will provide new insights into bile tolerance mechanisms in Bifidobacterium and other bacteria.
KEYWORDS: ABC transporter, B. longum BBMN68, BmrR, MarR-type regulator, bile stress
ABSTRACT
In order to colonize the human gastrointestinal tract and exert their beneficial effects, bifidobacteria must effectively cope with toxic bile salts in the intestine; however, the molecular mechanism underlying bile tolerance is poorly understood. In this study, heterologous expression of a MarR family transcriptional regulator, BmrR, significantly reduced the ox bile resistance of Lactococcus lactis NZ9000, suggesting that BmrR might play a role in the bile stress response. In silico analysis combined with reverse transcription-PCR assays demonstrated that bmrR was cotranscribed with bmrA and bmrB, which encoded multidrug resistance (MDR) ABC transporters. Promoter prediction and electrophoretic mobility shift assays revealed that BmrR could autoregulate the bmrRAB operon by binding to the bmr box (ATTGTTG-6nt-CAACAAT) in the promoter region. Moreover, heterologous expression of bmrA and bmrB in L. lactis yielded 20.77-fold higher tolerance to 0.10% ox bile, compared to the wild-type strain. In addition, ox bile could disrupt the DNA binding activity of BmrR as a ligand. Taken together, our findings indicate that the bmrRAB operon is autoregulated by the transcriptional regulator BmrR and ox bile serves as an inducer to activate the bile efflux transporter BmrAB in response to bile stress in Bifidobacterium longum BBMN68.
IMPORTANCE Bifidobacteria are natural inhabitants of the human intestinal tract. Some bifidobacterial strains are used as probiotics in fermented dairy production because of their health-promoting effects. Following consumption, bifidobacteria colonize the lower intestinal tract, where the concentrations of bile salts remain nearly 0.05% to 2.0%. Bile salts, as detergent-like antimicrobial compounds, can cause cellular membrane disruption, protein misfolding, and DNA damage. Therefore, tolerance to physiological bile stress is indeed essential for bifidobacteria to survive and to exert probiotic effects in the gastrointestinal tract. In B. longum BBMN68, the MarR-type regulator BmrR was involved in the bile stress response by autoregulating the bmrRAB operon, and ox bile as an inducer could increase the expression of the BmrAB transporter to enhance the bile tolerance of BBMN68. Our study represents a functional analysis of the bmrRAB operon in the bile stress response, which will provide new insights into bile tolerance mechanisms in Bifidobacterium and other bacteria.
INTRODUCTION
Bifidobacteria are natural inhabitants of the human gastrointestinal tract, constituting approximately 60 to 90% of the total gut microbiome during early stages of life (1, 2). Some bifidobacteria are considered probiotics and are used as active ingredients in functional dairy-based products (3, 4). The health benefits are exerted mainly by inhibiting pathogens, preventing diarrhea, stimulating the immune response, and reducing serum cholesterol levels (5, 6). Upon ingestion, bifidobacteria inevitably need to cope with several stress conditions, such as the low pH in the stomach and bile salts in the intestine (7, 8). As detergent-like biological substances with strong antimicrobial activities, bile salts can disrupt the lipid bilayer structure of cellular membranes, induce protein misfolding, and cause DNA damage (9). Bifidobacteria have been reported to develop tolerance response to bile stress, but the comprehensive mechanism of bile resistance remains elusive (10–14).
Among the bile resistance mechanisms employed by bifidobacteria, bile salt hydrolase (BSH) and bile efflux transporter are well documented. BSHs are responsible for deconjugation of glycine- or taurine-conjugated bile salts, therefore decreasing the toxicity of conjugated bile salts (15). The bile efflux system is mediated by a multidrug resistance (MDR) transporter located on the cell membrane, such as Ctr in Bifidobacterium longum NCIMB 702259T (16), BetA in B. longum NCC2705 (14), and BbmAB in Bifidobacterium breve UCC2003 (17). Several studies showed that bifidobacteria modulated the cell envelope, including the fatty acid composition and membrane proteins, to decrease membrane permeability in response to bile salts (18, 19). In addition, the hemolysin-like protein TlyC1 functions as a barrier to protect the strain from bile toxicity and provides resistance to sodium taurocholate and sodium taurodeoxycholate in B. longum BBMN68 (20). The two-component system senX3-regX3 was reported to promote the expression of the pstS gene to maintain a high level of Pi uptake and to produce more ATP to resist bile stress in B. longum BBMN68 (11).
B. longum BBMN68 was isolated from healthy centenarians in Bama longevity villages of Guangxi province in China. This strain has been reported to enhance innate and adaptive immunity, to alleviate allergic responses, and to improve intestinal function in mice (21, 22). In our study, transcriptomic RNA sequencing analysis showed that the BBMN68_1796 gene encoding a MarR-type transcriptional regulator was 1.85-fold upregulated under bile stress in BBMN68 (NCBI accession no. GSE113993). Moreover, secondary structure prediction for BBMN68_1796 revealed high structural homology with other MarR family members used in the alignment (see Fig. S1 in the supplemental material). It has been reported that the MarR family transcriptional regulators are involved in regulation in response to diverse environmental signals, such as synthesis of virulence factors and antibiotic stress (23, 24). Martin and Rosner found that transcription of multidrug resistance operon marORAB was repressed by the MarR protein (25). Furthermore, another MarR-type repressor, EmrR, was reported to control the EmrAB efflux transporter in Escherichia coli (26). In the present work, we investigated the regulatory mechanism of protein BBMN68_1796, designated BmrR (Bifidobacterium multidrug resistance regulator), in the ox bile stress response in B. longum BBMN68. The data suggest that BmrR autoregulates the transcription of the bmrRAB operon and ox bile serves as a ligand of BmrR to attenuate this binding, which enhances the expression of efflux transporter genes to export ox bile in B. longum BBMN68.
RESULTS
Heterologous expression of bmrR in Lactococcus lactis NZ9000 increases its sensitivity to bile stress.
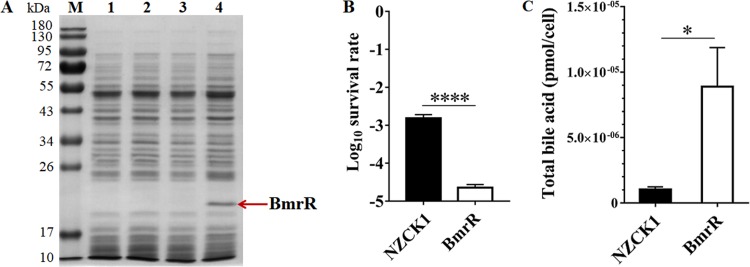
DNA sequencing showed that the length of the amplified gene bmrR was 534 bp, with 100% identity to the bmrR gene from B. longum BBMN68 (BBMN68_1796) (GenBank accession no. NC_014656.1). SDS-PAGE analysis revealed the production of an expected 20-kDa protein in L. lactis BmrR after nisin induction (Fig. 1A, lane 4), indicating the successful expression of bmrR in L. lactis NZ9000. The recombinant strains were incubated with 0.10% (wt/vol) ox bile for 1 h, and the survival of L. lactis BmrR was 57-fold lower than that of L. lactis NZCK1 (P < 0.0001) (Fig. 1B). Moreover, the bile acid level inside L. lactis BmrR was 8.90 × 10−6 ± 0.30 × 10−6 pmol per cell, which was 8.5-fold higher than that of L. lactis NZCK1 (P < 0.05) (Fig. 1C). These results showed that the heterologous expression of bmrR in L. lactis NZ9000 significantly reduced its resistance to ox bile, indicating that BmrR played a critical role in the bile stress response.
FIG 1.
Heterologous expression of BmrR with nisin induction detected by SDS-PAGE and the survival of L. lactis BmrR and L. lactis NZCK1 after ox bile challenge. (A) Soluble extracts were analyzed on 12% denaturing SDS-PAGE gels. Lane M, dual-color-prestained broad-molecular-size protein markers (10 to 180 kDa); lane 1, L. lactis NZCK1 without nisin induction; lane 2, L. lactis BmrR without nisin induction; lane 3, L. lactis NZCK1 with nisin (10 ng·ml−1) induction; lane 4, L. lactis BmrR with nisin (10 ng·ml−1) induction. The red arrow indicates the overexpressed BmrR. (B) The survival rate was calculated as the ratio of the number of colonies obtained on GM17 plates after and before ox bile treatment. (C) The bile acid content inside L. lactis BmrR was determined after ox bile treatment. Data are reported as means ± standard deviations (SDs) from at least three independent experiments and were analyzed by an unpaired, two-tailed Student's t test. *, P < 0.05; ****, P < 0.0001.
In silico analysis of the bmrR gene and determination of the bmrRAB operon.
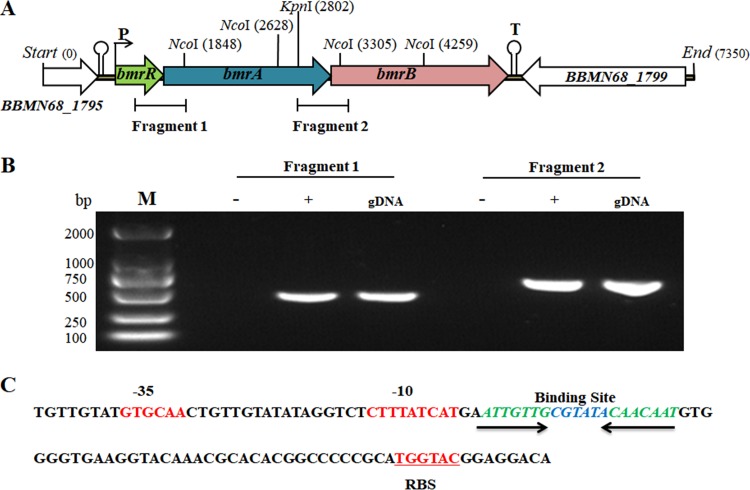
We noticed that the start codons of the bmrR, bmrA, and bmrB genes overlapped the stop codon of the proceeding gene. A putative promoter sequence was found 64 bp upstream of the potential bmrR start codon by the online promoter prediction tools NNPP and BPROM (27, 28), but no other promoter was predicted upstream of the bmrA or bmrB gene. The first gene, bmrR, possessed a putative ribosome binding site (TGGTAC) 8 bp upstream of its start codon, while the third gene, bmrB, was followed by a transcription-terminator-like sequence (Fig. 2A). Based on these observations, we hypothesized that these three genes were cotranscribed in the same cluster. A reverse transcription (RT)-PCR assay with cDNA as the template confirmed that genes from bmrR to bmrB formed a polycistronic operon, designated as bmrRAB (Fig. 2B). MarR family regulators were reported to bind recognizable palindromic sequences within the promoter region upstream of the target genes (29). Bioinformatic analysis revealed that an inverted repeat (IR) sequence (ATTGTTG-6nt-CAACAAT) was also found within the bmrRAB promoter in BBMN68 (Fig. 2C).
FIG 2.
In silico analysis and RT-PCR assays to verify the cotranscription of bmrR to bmrB. (A) Linear map of bmrR, bmrA, and bmrB with the genomic DNA flanking these genes in BBMN68. (B) RT-PCR assays to verify the cotranscription of bmrR to bmrB. gDNA, genomic DNA of wild-type BBMN68; + and −, cDNA and RNA, respectively, used as the template for PCR amplification; M, DNA marker. (C) Sequence analysis of the promoter region upstream of the bmrR gene. The putative −35 and −10 sequences and the ribosome binding site (RBS) are marked in red. The putative binding site is shown in italics.
Identification of the DNA binding specificity of BmrR by EMSA.
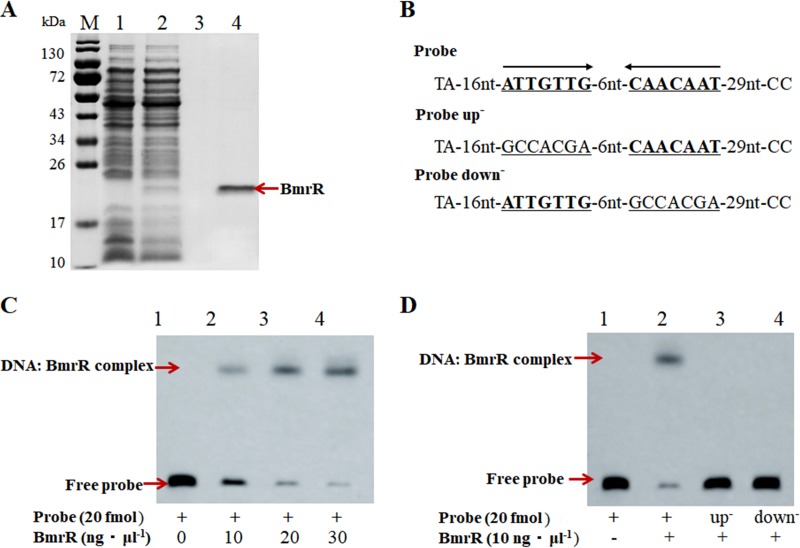
In order to confirm the DNA binding specificity of BmrR with its promoter, a 69-bp DNA probe was synthesized and labeled with biotin at the 3′ end for an electrophoretic mobility shift assay (EMSA). The BmrR with a C-terminal His tag was expressed in L. lactis NZ9000 and purified by affinity chromatography. SDS-PAGE revealed a single protein band with a molecular mass of approximate 20 kDa, indicating that the recombinant BmrRHis had been successfully expressed and purified for subsequent EMSAs (Fig. 3A, lane 4). The EMSA results indicated that the purified BmrRHis bound to the biotin-labeled bmrR probe and retarded its mobility (Fig. 3C, lane 2). Moreover, the quantity of DNA-protein-binding bands was enhanced with increasing concentrations of BmrR (Fig. 3C, lane 2 to lane 4). BmrRHis could not bind to either mutated probe up− or probe down− (Fig. 3D), indicating that the IR sequence (ATTGTTG-6nt-CAACAAT), designated a bmr box in the bmrRAB promoter region, was essential for BmrR binding. These findings indicated that BmrR could bind specifically to the bmr box upstream of the bmrRAB operon.
FIG 3.
SDS-PAGE analysis of the purified BmrRHis and specific binding of BmrRHis to its own promoter. (A) Lane M, dual-color-prestained broad-molecular-size protein markers (10 to 180 kDa); lane 1, L. lactis NZCK1 with 10 ng·ml−1 nisin induction; lane 2, L. lactis BmrRHis with 10 ng·ml−1 nisin induction; lane 3, protein sample from NZCK1 after purification; lane 4, purified recombinant BmrRHis from L. lactis BmrRHis. (B) DNA probes containing an intact palindromic sequence in the BmrR binding site or a mutated sequence. (C) Lane 1, 20 fmol labeled probes alone; lane 2 to lane 4, 20 fmol probes and 10, 20, and 30 ng·μl−1 BmrRHis, respectively. (D) Lane 1, 20 fmol labeled probes alone; lane 2 to lane 4, 10 ng·μl−1 BmrRHis with 20 fmol probes, 20 fmol probe up−, and 20 fmol probe down−, respectively.
Heterodimer ABC transporter BmrAB involvement in ox bile tolerance.
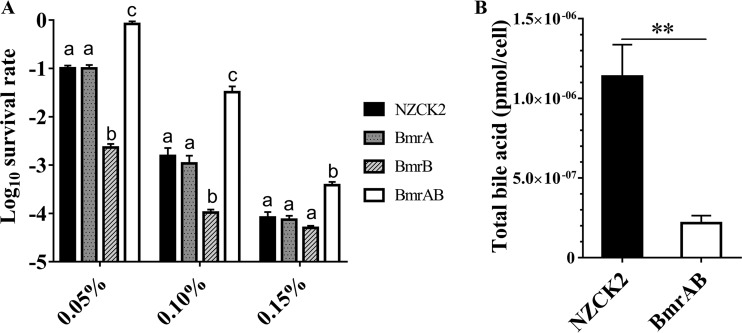
In order to determine whether the ABC transporter BmrAB was involved in the ox bile resistance, BmrA and BmrB were overexpressed, either singly or together, in the heterologous host L. lactis NZ9000. Survival assays showed that there was no significant difference between the recombinant strain L. lactis BmrA and the control strain L. lactis NZCK2 under bile stress (P > 0.05) (Fig. 4A), but the survival rate of L. lactis BmrB was 16-fold lower than that of L. lactis NZCK2 in the presence of 0.10% (wt/vol) ox bile. It is noteworthy that the survival rate of the BmrA- and BmrB-coexpressing strain L. lactis BmrAB was significantly increased, being 20.77-fold higher than that of the control strain in M17 medium with glucose (GM17) supplemented with 0.10% (wt/vol) ox bile (P < 0.05) (Fig. 4A). Moreover, the bile acid content of L. lactis BmrAB was 2.17 × 10−7 ± 0.47 × 10−7 pmol per cell, which was significantly lower than that of L. lactis NZCK2 (P < 0.001) (Fig. 4B). These results indicated that BmrA and BmrB together can enhance the bile resistance of the host strain, probably by forming a heterodimer ABC transporter to pump out the intracellular bile.
FIG 4.
Heterologous expression of genes bmrA, bmrB, and bmrAB in L. lactis affecting the survival of the host strain after bile stress. (A) The survival rate was calculated as the ratio of the number of colonies obtained on GM17 plates after and before ox bile treatment. Data are reported as means ± SDs from at least three independent experiments and were analyzed by one-way ANOVA with Tukey’s post hoc test. Bars with different letters are statistically significantly different (P < 0.05). (B) The bile acid content inside L. lactis BmrAB cells was determined after ox bile treatment. **, P < 0.001.
BmrR dissociates from DNA in the presence of ox bile.
The DNA binding activity of some transcriptional regulators from the MarR family was reported to be affected by specific ligands that dissociate the regulator from DNA, with consequent modulation of gene expression (30). In this study, the RT-quantitative PCR (qPCR) results showed that the mRNA level for bmrR was increased 2.73 ± 0.18-fold under ox bile stress in BBMN68. The bmrA and bmrB genes were upregulated 1.79 ± 0.23-fold and 1.54 ± 0.05-fold, respectively, in response to 0.075% (wt/vol) ox bile (Fig. 5). Therefore, we hypothesized that ox bile might be a ligand of BmrR and affect the interaction between BmrR and its binding site. To verify this hypothesis, different concentrations of ox bile were applied to EMSA reactions. The addition of 0.15% (wt/vol) ox bile led to complete dissociation of BmrR from its target DNA (Fig. 6, lane 5). These results indicated that ox bile was an effector for BmrR and could disrupt the DNA binding activity of BmrR in BBMN68.
FIG 5.
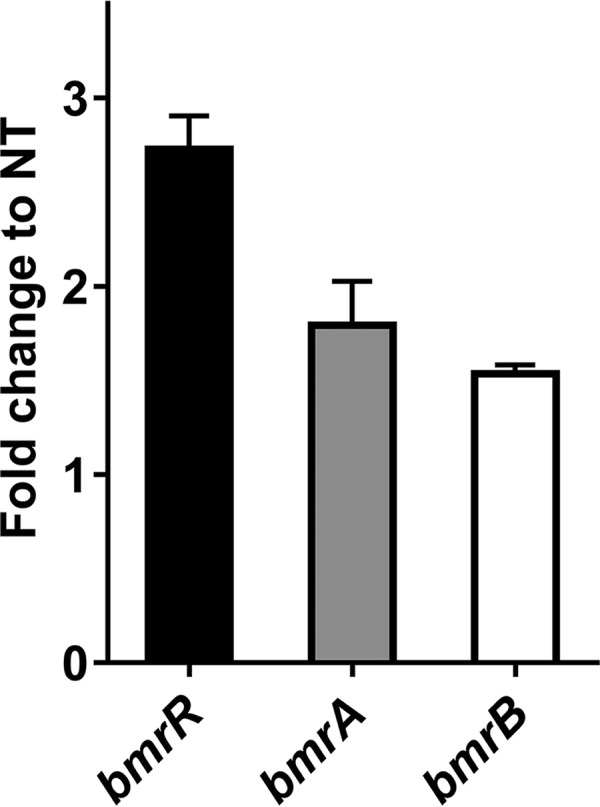
Fold changes in the relative expression of bmrR, bmrA, and bmrB genes after bile stress, as determined by RT-qPCR. The fold changes calculated were relative to the transcript levels under bile-treated conditions, compared with the nontreated (NT) control. Values were normalized using the 16S rRNA gene as an internal control. Data are reported as means ± SDs from three independent experiments.
FIG 6.
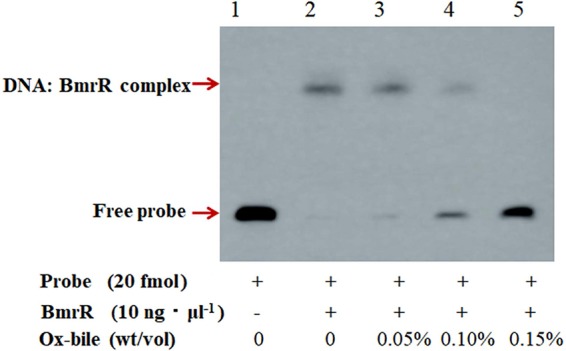
Effect of ox bile on the DNA binding activity of BmrR. The EMSA was performed with 20 fmol probe and 10 ng·μl−1 purified BmrRHis in the presence of 0.05%, 0.10%, or 0.15% (wt/vol) ox bile.
DISCUSSION
In bacteria, MarR family proteins constitute a diverse group of transcriptional regulators that modulate the expression of genes encoding proteins involved in metabolic pathways, stress responses, virulence, and degradation or efflux of harmful chemicals (31). Some MarR family transcription factors involved in bile stress responses have been identified in multiple bacterial species, including Salmonella enterica serovar Typhimurium, L. lactis, and Enterococcus faecalis (32–34). In S. Typhimurium, marRAB was activated in the presence of bile, and the marRAB mutant was more sensitive to bile stress than was the wild-type strain (32). SlyA is a MarR family transcriptional regulator identified in Enterococcus faecalis, and the growth of a slyA mutant strain was significantly impaired in the presence of bile salts (34). However, the paucity of efficient transformation methods and effective molecular tools for gene inactivation limits direct functional identification of BmrR in B. longum BBMN68. Consequently, heterologous expression in L. lactis NZ9000 was employed to explore the function of BmrR in bile stress responses, as described previously (11, 20, 35). In this study, heterologous expression of the bmrR gene in L. lactis NZ9000 decreased the bile tolerance of the host strain, suggesting that the bmrR gene might play a role in the bile stress response. The bmrR gene was cotranscribed with bmrA and bmrB, which encoded the MDR ABC transporters. The ABC transporter BmrAB was observed to enhance the bile tolerance of the host strain when the bmrAB gene was expressed in L. lactis NZ9000. Therefore, we supposed that the bmrRAB operon in bifidobacteria played a critical role in enhancing the resistance to bile stress.
The bmrA and bmrB genes were predicted by a database enquiry (BLASTP) to encode 652- and 671-amino-acid proteins, respectively, as putative ABC transporters. The hydropathic profile analysis demonstrated that both proteins possessed a transmembrane domain (TMD) with six putative helices, followed by cytoplasmically localized nucleotide binding domain (NBD) with a putative ATP binding domain and the ABC signature sequence (36, 37). The ATP-hydrolyzing domains are characterized by two short sequence motifs in their primary structures (Walker site A and Walker site B) that constitute a nucleotide binding fold (see Fig. S2 in the supplemental material). These analyses suggested that BmrA and BmrB might also serve as ABC half-transporters. The ABC transporter utilizes the free energy of ATP hydrolysis to drive substrate transport across lipid bilayers. It has been proved that several prokaryotic ABC transporters act as dimers. Homodimeric ABC transporters have been experimentally identified, such as LmrA in L. lactis (38) and MsbA in E. coli (39). Heterodimeric ABC transporters are also found in some species, such as LmrCD in L. lactis (40) and BbmAB in B. breve UCC2003 (17). BbmA and BbmB were further reported to be induced 3.21 ± 1.3-fold and 5.00 ± 0.9-fold, respectively, in the presence of bile salts (14). In agreement with these MDR transporters, the expression of BmrA and BmrB was found to be upregulated by ox bile in B. longum BBMN68, according to our transcriptome and RT-qPCR data. We found that the heterologous expression of BmrA did not affect the bile resistance of L. lactis. Moreover, the overexpression of BmrB increased the sensitivity of the host strain to ox bile. We speculated that the overexpressed BmrB might affect the membrane integrity (30, 41) or serve as a docking molecule for bile to increase its concentration directly at the membrane. Notably, the coexpression of BmrA and BmrB can enhance the bile resistance of the host strain, suggesting that BmrA and BmrB are essential for forming a functional heterodimer ABC transporter and then pumping out the intracellular bile.
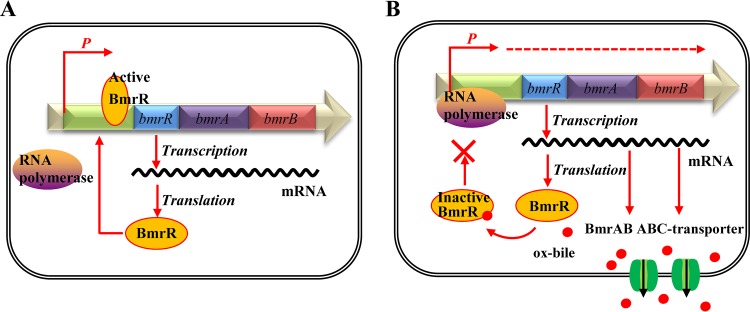
In the present study, we observed that the MarR family regulator BmrR could interact with the promoter region of the bmrRAB operon to regulate the transcription of these three genes. In addition, the three-dimensional structure of BmrR was generated using the SWISS-MODEL server (http://www.expasy.ch/swissmod/SWISS-MODEL.html), which indicated that BmrR is able to form a homodimer like other MarR proteins (Fig. S3). Generally, MarR family regulators were reported to bind recognizable palindromic sequences within the promoter region, producing attenuation of gene expression by sterically hindering the binding of RNA polymerase to the promoter. In addition, the MarR family transcription factors can respond to a variety of effector molecules (23, 31). When the ligand binds to a MarR family transcription factor, DNA binding is attenuated, resulting in derepression of transcription (23). In this study, we observed that the formation of a BmrR-DNA complex was impaired in the presence of ox bile (Fig. 6). Based on these results and the regulatory mechanism of other MarR-type regulators (31, 42), we proposed a bile-sensing and adaptive regulation model of the bmrRAB operon in B. longum (Fig. 7). Under normal growth conditions, ligand-free BmrR binds to the bmr box in the bmrRAB promoter region and prevents the RNA polymerase from initiating further transcription of bmrRAB. When ox bile enters the cell, BmrR interacts with ox bile and then causes significant conformational changes in the DNA binding domains, resulting in release of the BmrR repressor from the bmrRAB promoter. This modification allows the RNA polymerase to initiate the transcription of bmrRAB. Newly synthesized BmrAB ABC transporters are embedded in the membrane and mediate the efflux of the ox bile from the cell. Our study represents a functional analysis of the bmrRAB operon in the bile stress response, which is of great importance for exploring novel bile tolerance mechanisms in Bifidobacterium and other bacteria.
FIG 7.
Illustration of the proposed BmrR regulation mechanism. (A) Under normal growth conditions, the active form of BmrR binds to the bmr box and represses transcription of the BmrRAB operon. (B) In the presence of ox bile, the DNA binding activity of BmrR is disrupted by ox bile. This modification results in the transcription of BmrAB ABC transporters to pump out ox bile.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. B. longum BBMN68 was grown anaerobically (5% CO2, 5% H2, and 90% N2) at 37°C in de Man-Rogosa-Sharpe (MRS) broth supplemented with 0.05% (vol/vol) l-cysteine (MRSc). Lactococcus lactis NZ9000 was routinely grown at 30°C in M17 medium (Oxoid, Unipath, Basingstoke, UK) containing 0.5% (wt/vol) glucose (GM17). When necessary, medium was supplemented with 10 μg·ml−1 chloramphenicol for L. lactis.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant phenotype or genotypea | Source or reference |
|---|---|---|
| Bacterial strains | ||
| B. longum BBMN68 | Wild-type strain, isolated from feces from healthy centenarian | Hao et al. (46) |
| L. lactis NZ9000 | L. lactis MG1363 pepN::nisRK | de Ruyter et al. (43) |
| L. lactis BmrR | L. lactis NZ9000 harboring pNZBmrR | This work |
| L. lactis BmrRHis | L. lactis NZ9000 harboring pNZBmrRHis | This work |
| L. lactis BmrA | L. lactis NZ9000 harboring pNZBmrA | This work |
| L. lactis BmrB | L. lactis NZ9000 harboring pNZBmrB | This work |
| L. lactis BmrAB | L. lactis NZ9000 harboring pNZBmrAB | This work |
| L. lactis NZCK1 | L. lactis NZ9000 harboring pNZ8148 | This work |
| L. lactis NZCK2 | L. lactis NZ9000 harboring pNZ8147 | This work |
| Plasmids | ||
| pNZ8148 | Gene expression vector with PnisA; Cmr | de Ruyter et al. (43) |
| pNZ8147 | pNZ8148 derivative with modified MCS containing XbaI, SacI, and HindIII sites | This work |
| pNZBmrR | pNZ8148 derivative containing bmrR gene | This work |
| pNZBmrRHis | pNZ8148 derivative containing bmrRHis gene | This work |
| pNZBmrA | pNZ8147 derivative containing bmrA gene | This work |
| pNZBmrB | pNZ8147 derivative containing bmrB gene | This work |
| pNZBmrAB | pNZ8147 derivative containing bmrAB gene | This work |
PnisA, nisA promoter; Cmr, chloramphenicol resistance; MCS, multiple cloning site.
DNA manipulation techniques.
Genomic DNA from B. longum BBMN68 was prepared by lysozyme pretreatment and use of a genomic DNA extraction kit (Tiangen, Beijing, China). Mini-prep plasmid isolations from L. lactis were performed using the E.Z.N.A. plasmid mini kit I, according to the manufacturer’s instructions (Omega Bio-tek Inc., Doraville, GA, USA). Standard PCR was carried out using Q5 high-fidelity DNA polymerase, following the manufacturer’s instructions (NEB, Beijing, China). DNA digestion with restriction endonucleases and DNA ligation were performed according to the manufacturer’s instructions (NEB). The electroporation of L. lactis NZ9000 was performed according to previously described procedures (43). All primers and probes used in this study were designed using PRIMER v5 software (Premier Biosoft International, Palo Alto, CA, USA) and were synthesized by Sangon Biotech (Beijing, China). DNA sequencing was performed by Sangon Biotech, and the results were further analyzed with the DNAMAN software package (Lynnon Biosoftware, Vaudreuil, QC, Canada).
Construction of the L. lactis BmrR, BmrA, BmrB, and BmrAB recombinant strains.
The bmrR gene was amplified from genomic DNA from B. longum BBMN68 using the primer pair bmrR-F and bmrR-R (Table 2). The PCR product digested with NcoI and XbaI was inserted into the corresponding sites of pNZ8148 downstream of the nisA promoter. Subsequently, the ligation mixture was transformed into L. lactis NZ9000 to generate the recombinant strain L. lactis BmrR. Meanwhile, a control strain (L. lactis NZCK1) was constructed by introducing the empty vector pNZ8148 into L. lactis NZ9000. SDS-PAGE analysis was used to investigate the expression of bmrR in L. lactis.
TABLE 2.
Oligonucleotides and primers used in this study
aRestriction enzyme cutting sites are underlined (NcoI, CCATGG; XbaI, TCTAGA; HindIII, AAGCTT). Lowercase letters indicate the predicted binding sites of BmrR; italic boldface letters indicate the mutated binding sites.
The bmrA and bmrB genes were amplified from B. longum BBMN68 using the primer pairs bmrA-F/bmrA-R and bmrB-F/bmrB-R, respectively (Table 2). The bmrA and bmrB genes were then coamplified using bmrA-F and bmrB-R. Because both bmrA and bmrB contain two internal NcoI sites, bmrA, bmrB, and bmrAB digested with XbaI and HindIII were inserted into another nisin-inducible vector, pNZ8147. The ligation mixture was transformed into L. lactis NZ9000, resulting in recombinant strains L. lactis BmrA, L. lactis BmrB, and L. lactis BmrAB, respectively. Meanwhile, a L. lactis NZ9000 strain with the empty pNZ8147 vector was constructed as a control.
Survival assay and quantification of intracellular bile acid contents.
Overnight cultures of recombinant strains were inoculated into 10 ml of fresh GM17 supplemented with 10 μg·ml−1 chloramphenicol (1% inocula). When the cell density reached an optical density at 600 nm (OD600) of 0.3, nisin (final concentration, 10 ng·ml−1) was added; the cells were then incubated further for 2 h at 30°C. Aliquots of 1 ml of culture were collected and suspended in 1 ml of fresh GM17 containing 0.05%, 0.10%, or 0.15% (wt/vol) ox bile (Sigma, St. Louis, MO, USA). After incubation at 30°C for 1 h, the number of CFU per milliliter was determined by plating 10-fold serial dilutions on GM17 plates containing 10 μg·ml−1 chloramphenicol and incubating the plates at 30°C for 16 h. Survival rates were calculated by dividing the number of CFU per ml after ox bile incubation by the value obtained immediately after resuspension. In order to quantify intracellular bile acid, 10 ml of nisin-induced culture was collected and suspended in 1 ml of GM17 containing 0.85 μmol·ml−1 bile acid. The total bile acid contents in GM17 before and after 1 h of incubation were determined enzymatically with a commercial kit from Jiancheng Institute of Biotechnology (Nanjing, China), as described previously (44). The bile acid content inside each cell was normalized to total cell numbers after bile stress treatment. All results were obtained in at least three independent experiments, with each performed in triplicate.
Validation of bmrR operon by RT-PCR.
B. longum BBMN68 cells were immediately harvested, at an OD600 of 0.6, by centrifugation at 6,000 × g for 10 min. Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions, and digested with RNase-free DNase I (Tiangen). RNA concentrations were quantified using a NanoDrop 2000 spectrophotometer (Thermo Scientific). RNA quality was assessed with a 2100 bioanalyzer (Agilent Technologies, Amstelveen, Netherlands). Subsequently, RT was carried out with the PrimeScript II 1st strand cDNA synthesis kit (TaKaRa, Beijing, China), with 1 μg of total RNA as the template. Specific primers (operon 1-F/R and operon 2-F/R) are listed in Table 2. Standard PCR was carried out using Q5 high-fidelity DNA polymerase (NEB), with cDNA as the template, RNA as the negative control, and BBMN68 genomic DNA as the positive control.
Transcriptional analysis of bmrRAB under bile stress.
Overnight-cultured B. longum BBMN68 was inoculated (1% [vol/vol]) into MRSc medium with or without 0.075% (wt/vol) ox bile. After 24 h of incubation at 37°C, BBMN68 cells were harvested by centrifugation at 6,000 × g for 10 min. Total RNA was isolated using TRIzol reagent (Invitrogen). Then, cDNA was obtained using the PrimeScript II 1st strand cDNA synthesis kit (TaKaRa), with 1 μg of total RNA as the template. Real-time qPCR was performed using the SuperReal PreMix Plus SYBR green kit (Tiangen) on a LightCycler 480 real-time thermocycler (Roche Diagnostics, Meylan, France). The primers used for qPCR are listed in Table 2. Primer specificity was assessed by examination of the melting curve at the end of amplification. All reactions were performed in triplicate. The results were obtained in three independent experiments. The statistical analysis (unpaired t test) was performed to check the qPCR data quality. Data were analyzed with the 2−ΔΔCt method, using the untreated group mean as the reference condition (45). The internal control 16S rRNA gene was used for transcript normalization.
Purification of recombinant BmrR and EMSA.
The bmrR gene was amplified by PCR using the primer pair bmrRHis-F and bmrRHis-R (Table 2), which introduced a six-histidine tag at the C-terminal end of this protein, immediately prior to the stop codon, to simplify protein purification by affinity chromatography using a nickel column. The PCR product, digested with NcoI and XbaI, was ligated into pNZ8148 at the corresponding restriction sites, resulting in recombinant plasmid pBmrRHis. This plasmid was then introduced into L. lactis NZ9000, and the transformant harboring the correct construct was designated L. lactis BmrRHis. The BmrR protein with a C-terminal His tag (designated BmrRHis) was purified with Ni Sepharose 6 Fast Flow resin (GE Healthcare, Uppsala, Sweden), according to the manufacturer’s recommendations. Subsequently, purified BmrRHis was concentrated by ultrafiltration (10-kDa cutoff; Millipore, Bedford, MA, USA) and centrifugation at 13,000 × g for 30 min at 4°C. The purified BmrRHis was analyzed by SDS-PAGE, and the protein concentration was quantified using the Qubit protein assay kit and a Qubit 2.0 fluorometer (Invitrogen). Purified protein was used immediately or stored at −80°C for subsequent experiments.
EMSAs were performed using the LightShift chemiluminescent EMSA kit (Thermo Scientific, Rockford, IL, USA). To obtain biotin 3′-end-labeled probes, two complementary oligonucleotides (listed in Table 2) were synthesized and annealed at 95°C for 5 min, with the temperature decreasing by 1°C per minute thereafter until holding at 4°C. EMSAs were performed according to the manufacturer’s instructions, the binding reaction mixtures (20 μl) contained 1× binding buffer, 50 ng·μl−1 poly(dI-dC), 2.5% (vol/vol) glycerol, 0.05% (vol/vol) NP-40, 20 fmol labeled probe, 5 mM MgCl2, and 10 ng·μl−1 BmrRHis, and the reactions were performed for 20 min at room temperature. To determine whether the IR structure of the predicted binding site was essential, conserved binding site IR1 (ATTGTTG) was changed to GCCACGA and IR2 (CAACAAT) was changed to GCCACGA, as shown in Fig. 3B. In addition, different concentrations of BmrRHis (10 ng·μl−1, 20 ng·μl−1, and 30 ng·μl−1) and ox bile (0.05%, 0.10%, and 0.15% [wt/vol]) were applied to determine the dose effects on the binding activity of BmrR. The subsequent steps were carried out following the manufacturer’s instructions. Chemiluminescence signals of biotinylated probes were captured using a charge-coupled device (CCD) camera imaging system (UVP, Upland, CA, USA).
Statistical analysis.
Data were analyzed using GraphPad Prism 6 software for Windows (GraphPad Software, Inc., La Jolla, CA, USA). When two groups were compared, an unpaired Student's t test with Welch’s correction was used to calculate P values. When three or more groups were compared, one-way analysis of variance (ANOVA) was used, followed by an appropriate post hoc test.
Accession number(s).
Newly determined sequence data were deposited in GenBank under accession number GSE113993.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Sciences Foundation of China (contracts 21676294 and 21476250) and the International Postdoctoral Exchange Fellowship Program (grant 20150027).
We thank Willem M. de Vos (Wageningen University) for the gift of L. lactis NZ9000 and plasmid pNZ8148. We also thank Kuanqing Liu (UT Southwestern Medical Center) for helpful comments.
Q.X., Z.Z., H.A., and Y.H. designed the research, Q.X., Z.Z., and H.A. performed the research, Y.Y., J.Y., and G.W. contributed new reagents or analytic tools, and Q.X., Z.Z., F.R., and Y.H. analyzed the data and wrote the paper. All authors read and approved the final manuscript.
We declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02453-18.
REFERENCES
- 1.Roger LC, Costabile A, Holland DT, Hoyles L, McCartney AL. 2010. Examination of faecal Bifidobacterium populations in breast-and formula-fed infants during the first 18 months of life. Microbiology 156:3329–3341. doi: 10.1099/mic.0.043224-0. [DOI] [PubMed] [Google Scholar]
- 2.Collado MC, Cernada M, Baüerl C, Vento M, Pérez-Martínez G. 2012. Microbial ecology and host-microbiota interactions during early life stages. Gut Microbes 3:352–365. doi: 10.4161/gmic.21215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Granato D, Branco GF, Cruz AG, Faria JAF, Shah NP. 2010. Probiotic dairy products as functional foods. Compr Rev Food Sci Food Saf 9:455–470. doi: 10.1111/j.1541-4337.2010.00120.x. [DOI] [PubMed] [Google Scholar]
- 4.Kabeerdoss J, Devi RS, Mary RR, Prabhavathi D, Vidya R, Mechenro J, Mahendri N, Pugazhendhi S, Ramakrishna BS. 2011. Effect of yoghurt containing Bifidobacterium lactis BB12 on faecal excretion of secretory immunoglobulin A and human beta-defensin 2 in healthy adult volunteers. Nutr J 10:138. doi: 10.1186/1475-2891-10-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tannock GW. (ed). 2002. Probiotics and prebiotics: where are we going? Caister Academic Press, Wymondham, UK. [Google Scholar]
- 6.Pagnini C, Saeed R, Bamias G, Arseneau KO, Pizarro TT, Cominelli F. 2010. Probiotics promote gut health through stimulation of epithelial innate immunity. Proc Natl Acad Sci U S A 107:454–459. doi: 10.1073/pnas.0910307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noriega L, Gueimonde M, Sánchez B, Margolles A, de los Reyes-Gavilán CG. 2004. Effect of the adaptation to high bile salts concentrations on glycosidic activity, survival at low pH and cross-resistance to bile salts in Bifidobacterium. Int J Food Microbiol 94:79–86. doi: 10.1016/j.ijfoodmicro.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Sánchez B, Champomier-Vergès M-C, Stuer-Lauridsen B, Ruas-Madiedo P, Anglade P, Baraige F, Clara G, Johansen E, Zagorec M, Margolles A. 2007. Adaptation and response of Bifidobacterium animalis subsp. lactis to bile: a proteomic and physiological approach. Appl Environ Microbiol 73:6757–6767. doi: 10.1128/AEM.00637-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernstein C, Bernstein H, Payne CM, Beard SE, Schneider J. 1999. Bile salt activation of stress response promoters in Escherichia coli. Curr Microbiol 39:68–72. doi: 10.1007/s002849900420. [DOI] [PubMed] [Google Scholar]
- 10.Ruiz L, Margolles A, Sánchez B. 2013. Bile resistance mechanisms in Lactobacillus and Bifidobacterium. Front Microbiol 4:396. doi: 10.3389/fmicb.2013.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.An H, Douillard FP, Wang G, Zhai Z, Yang J, Song S, Cui J, Ren F, Luo Y, Zhang B, Hao Y. 2014. Integrated transcriptomic and proteomic analysis of the bile stress response in a centenarian-originated probiotic Bifidobacterium longum BBMN68. Mol Cell Proteomics 13:2558–2572. doi: 10.1074/mcp.M114.039156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawasaki S, Watanabe M, Fukiya S, Yokota A. 2017. Stress responses of Bifidobacteria: oxygen and bile acid as the stressors, p 131–143. In Mattarelli P, Biavati B, Holzapfel W, Wood BJB (ed), The Bifidobacteria and related organisms. Elsevier, New York, NY. [Google Scholar]
- 13.Sánchez B, Champomier-Vergès M-C, Anglade P, Baraige F, Clara G, Margolles A, Zagorec M. 2005. Proteomic analysis of global changes in protein expression during bile salt exposure of Bifidobacterium longum NCIMB 8809. J Bacteriol 187:5799–5808. doi: 10.1128/JB.187.16.5799-5808.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gueimonde M, Garrigues C, van Sinderen D, de los Reyes-Gavilan CG, Margolles A. 2009. Bile-inducible efflux transporter from Bifidobacterium longum NCC2705, conferring bile resistance. Appl Environ Microbiol 75:3153–3160. doi: 10.1128/AEM.00172-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grill J, Perrin S, Schneider F. 2000. Bile salt toxicity to some bifidobacteria strains: role of conjugated bile salt hydrolase and pH. Can J Microbiol 46:878–884. doi: 10.1139/w00-066. [DOI] [PubMed] [Google Scholar]
- 16.Price CE, Reid SJ, Driessen AJ, Abratt VR. 2006. The Bifidobacterium longum NCIMB 702259T ctr gene codes for a novel cholate transporter. Appl Environ Microbiol 72:923–926. doi: 10.1128/AEM.72.1.923-926.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Margolles A, Florez AB, Moreno JA, van Sinderen D, de los Reyes-Gavilan CG. 2006. Two membrane proteins from Bifidobacterium breve UCC2003 constitute an ABC-type multidrug transporter. Microbiology 152:3497–3505. doi: 10.1099/mic.0.29097-0. [DOI] [PubMed] [Google Scholar]
- 18.Ruiz L, Sánchez B, Ruas-Madiedo P, De Los Reyes-Gavilán CG, Margolles A. 2007. Cell envelope changes in Bifidobacterium animalis ssp. lactis as a response to bile. FEMS Microbiol Lett 274:316–322. doi: 10.1111/j.1574-6968.2007.00854.x. [DOI] [PubMed] [Google Scholar]
- 19.Ruiz L, Coute Y, Sanchez B, de los Reyes-Gavilan CG, Sanchez J-C, Margolles A. 2009. The cell-envelope proteome of Bifidobacterium longum in an in vitro bile environment. Microbiology 155:957–967. doi: 10.1099/mic.0.024273-0. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y, An H, Zhang J, Zhou H, Ren F, Hao Y. 2014. Functional role of tlyC1 encoding a hemolysin-like protein from Bifidobacterium longum BBMN68 in bile tolerance. FEMS Microbiol Lett 360:167–173. doi: 10.1111/1574-6968.12601. [DOI] [PubMed] [Google Scholar]
- 21.Yang H, Liu A, Zhang M, Ibrahim SA, Pang Z, Leng X, Ren F. 2009. Oral administration of live Bifidobacterium substrains isolated from centenarians enhances intestinal function in mice. Curr Microbiol 59:439–445. doi: 10.1007/s00284-009-9457-0. [DOI] [PubMed] [Google Scholar]
- 22.Yang J, Zhang H, Jiang L, Guo H, Luo X, Ren F. 2015. Bifidobacterium longum BBMN68-specific modulated dendritic cells alleviate allergic responses to bovine β-lactoglobulin in mice. J Appl Microbiol 119:1127–1137. doi: 10.1111/jam.12923. [DOI] [PubMed] [Google Scholar]
- 23.Wilkinson SP, Grove A. 2006. Ligand-responsive transcriptional regulation by members of the MarR family of winged helix proteins. Curr Issues Mol Biol 8:51. [PubMed] [Google Scholar]
- 24.Wagner A, Segler L, Kleinsteuber S, Sawers G, Smidt H, Lechner U. 2013. Regulation of reductive dehalogenase gene transcription in Dehalococcoides mccartyi. Philos Trans R Soc Lond B Biol Sci 368:20120317. doi: 10.1098/rstb.2012.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin RG, Rosner JL. 1995. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc Natl Acad Sci U S A 92:5456–5460. doi: 10.1073/pnas.92.12.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lomovskaya O, Lewis K, Matin A. 1995. EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J Bacteriol 177:2328–2334. doi: 10.1128/jb.177.9.2328-2334.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reese MG. 2001. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput Chem 26:51–56. doi: 10.1016/S0097-8485(01)00099-7. [DOI] [PubMed] [Google Scholar]
- 28.Salamov VSA, Solovyevand A. 2011. Automatic annotation of microbial genomes and metagenomic sequences, p 61–78. In Li RW. (ed), Metagenomics and its applications in agriculture. Nova Science Publishers, Hauppauge, NY. [Google Scholar]
- 29.Deochand DK, Grove A. 2017. MarR family transcription factors: dynamic variations on a common scaffold. Crit Rev Biochem Mol Biol 52:595–613. doi: 10.1080/10409238.2017.1344612. [DOI] [PubMed] [Google Scholar]
- 30.Grkovic S, Brown MH, Skurray RA. 2002. Regulation of bacterial drug export systems. Microbiol Mol Biol Rev 66:671–701. doi: 10.1128/MMBR.66.4.671-701.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perera IC, Grove A. 2010. Molecular mechanisms of ligand-mediated attenuation of DNA binding by MarR family transcriptional regulators. J Mol Cell Biol 2:243–254. doi: 10.1093/jmcb/mjq021. [DOI] [PubMed] [Google Scholar]
- 32.Prouty AM, Brodsky IE, Falkow S, Gunn JS. 2004. Bile-salt-mediated induction of antimicrobial and bile resistance in Salmonella typhimurium. Microbiology 150:775–783. doi: 10.1099/mic.0.26769-0. [DOI] [PubMed] [Google Scholar]
- 33.Zaidi AH, Bakkes PJ, Lubelski J, Agustiandari H, Kuipers OP, Driessen AJ. 2008. The ABC-type multidrug resistance transporter LmrCD is responsible for an extrusion-based mechanism of bile acid resistance in Lactococcus lactis. J Bacteriol 190:7357–7366. doi: 10.1128/JB.00485-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michaux C, Martini C, Hanin A, Auffray Y, Hartke A, Giard JC. 2011. SlyA regulator is involved in bile salts stress response of Enterococcus faecalis. FEMS Microbiol Lett 324:142–146. doi: 10.1111/j.1574-6968.2011.02390.x. [DOI] [PubMed] [Google Scholar]
- 35.Ruiz L, Zomer A, O'Connell-Motherway M, van Sinderen D, Margolles A. 2012. Discovering novel bile protection systems in Bifidobacterium breve UCC2003 through functional genomics. Appl Environ Microbiol 78:1123–1131. doi: 10.1128/AEM.06060-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider E, Hunke S. 1998. ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol Rev 22:1–20. doi: 10.1111/j.1574-6976.1998.tb00358.x. [DOI] [PubMed] [Google Scholar]
- 37.Walker JE, Saraste M, Runswick MJ, Gay NJ. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J 1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Veen HW, Margolles A, Muller M, Higgins CF, Konings WN. 2000. The homodimeric ATP-binding cassette transporter LmrA mediates multidrug transport by an alternating two-site (two-cylinder engine) mechanism. EMBO J 19:2503–2514. doi: 10.1093/emboj/19.11.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang G, Roth CB. 2001. Structure of MsbA from E. coli: a homolog of the multidrug resistance ATP binding cassette (ABC) transporters. Science 293:1793–1800. doi: 10.1126/science.293.5536.1793. [DOI] [PubMed] [Google Scholar]
- 40.Lubelski J, Mazurkiewicz P, van Merkerk R, Konings WN, Driessen AJ. 2004. ydaG and ydbA of Lactococcus lactis encode a heterodimeric ATP-binding cassette-type multidrug transporter. J Biol Chem 279:34449–34455. doi: 10.1074/jbc.M404072200. [DOI] [PubMed] [Google Scholar]
- 41.Grkovic S, Brown MH, Skurray RA. 2001. Transcriptional regulation of multidrug efflux pumps in bacteria. Semin Cell Dev Biol 12:225–237. doi: 10.1006/scdb.2000.0248. [DOI] [PubMed] [Google Scholar]
- 42.Prajapat MK, Jain K, Saini S. 2015. Control of MarRAB operon in Escherichia coli via autoactivation and autorepression. Biophys J 109:1497–1508. doi: 10.1016/j.bpj.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Ruyter PG, Kuipers OP, de Vos WM. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl Environ Microbiol 62:3662–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu PL, Yuan YH, Yue TL, Guo CF. 2018. Bile acid patterns in commercially available oxgall powders used for the evaluation of the bile tolerance ability of potential probiotics. PLoS One 13:e0192964. doi: 10.1371/journal.pone.0192964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 46.Hao Y, Huang D, Guo H, Xiao M, An H, Zhao L, Zuo F, Zhang B, Hu S, Song S, Chen S, Ren F. 2011. Complete genome sequence of Bifidobacterium longum subsp. longum BBMN68, a new strain from a healthy Chinese centenarian. J Bacteriol 193:787–788. doi: 10.1128/JB.01213-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.