Abstract
Background
We aim to analyze the ability to detect epithelial growth factor receptor (EGFR) mutations on chest CT images of patients with lung adenocarcinoma using radiomics and/or multi-level residual convolutionary neural networks (MCNNs).
Methods
We retrospectively collected 1,010 consecutive patients in Shanghai Chest Hospital from 2013 to 2017, among which 510 patients were EGFR-mutated and 500 patients were wild-type. The patients were randomly divided into a training set (810 patients) and a validation set (200 patients) according to a balanced distribution of clinical features. The CT images and the corresponding EGFR status measured by Amplification Refractory Mutation System (ARMS) method of the patients in the training set were utilized to construct both a radiomics-based model (MRadiomics) and MCNNs-based model (MMCNNs). The MRadiomics and MMCNNs were combined to build the ModelRadiomics+MCNNs (MRadiomics+MCNNs). Clinical data of gender and smoking history constructed the clinical features-based model (MClinical). MClinical was then added into MRadiomics, MMCNNs, and MRadiomics+MCNNs to establish the ModelRadiomics+Clinical (MRadiomics+Clinical), the ModelMCNNs+Clinical (MMCNNs+Clinical) and the ModelRadiomics+MCNNs+Clinical (MRadiomics+MCNNs+Clinical). All the seven models were tested in the validation set to ascertain whether they were competent to detect EGFR mutations. The detection efficiency of each model was also compared in terms of area under the curve (AUC), sensitivity and specificity.
Results
The AUC of the MRadiomics, MMCNNs and MRadiomics+MCNNs to predict EGFR mutations was 0.740, 0.810 and 0.811 respectively. The performance of MMCNNs was better than that of MRadiomics (P=0.0225). The addition of clinical features did not improve the AUC of the MRadiomics (P=0.623), the MMCNNs (P=0.114) and the MRadiomics+MCNNs (P=0.058). The MRadiomics+MCNNs+Clinical demonstrated the highest AUC value of 0.834. The MMCNNs did not demonstrate any inferiority when compared with the MRadiomics+MCNNs (P=0.742) and the MRadiomics+MCNNs+Clinical (P=0.056).
Conclusions
Both of the MRadiomics and the MCNNs could predict EGFR mutations on CT images of patients with lung adenocarcinoma. The MMCNNs outperformed the MRadiomics in the detection of EGFR mutations. The combination of these two models, even added with clinical features, is not significantly more efficient than MMCNNs alone.
Keywords: Adenocarcinoma of lung, epithelial growth factor receptor mutation (EGFR mutation), radiomics, neural networks
Introduction
Tyrosine kinase inhibitors (TKIs) are today the first-line standard modality for the treatment of stage IV non-small cell lung cancer (NSCLC) with epithelial growth factor receptor (EGFR) mutations (1). Data suggest that the EGFR mutations are found in approximately in 10% Caucasian patients and about 50% Asian-Pacific patients with NSCLC (2,3). First-line treatment with TKIs would provide longer progression-free survival than chemotherapy. The detection of EGFR mutations before treatment in those NSCLC is the prerequisite for TKIs treatment (4). Biopsies through endoscope or fine needle aspiration (FNA) usually provide the specimens for the detection of EGFR mutations. However, these methods come with several limitations in practice. Patients with low Karnofsky performance scores (KPS) are less likely to tolerate such invasive procedures repeatedly and not all tumors with various sizes or locations are appropriate for biopsy. Most importantly, specimens acquired by biopsies are unable to demonstrate the intra-tumor and inter-tumor heterogeneity and provide us relatively limited information about the genotype and phenotype of tumors (5,6). Repeated biopsies alongside the whole treatment process to monitor genetic change or biopsies on each metastatic lesion to reflect inter-tumor heterogeneity may not be practical. Therefore, new technologies recently attempt to address these problems, among which liquid biopsy and image analysis are currently the most promising ones. Image analysis is the technology utilized to extract and analyze indiscernible information in medical images to acquire biological information of pathologies. Radiomics and convolutional neural networks (CNNs) are now the most frequently utilized methods in medical images analysis.
Radiomics is defined as the conversion of images to higher dimensional mineable data and the subsequent mining of these data for improved decision support. The main steps of radiomics include image acquisition, segmentation of region of interests (ROIs), features extraction and qualification, and classifier modeling (7). Given the hypothesis that imaging phenotypes may reflect the effects of genotypes, radiomics has been applied to detect EGFR mutations and achieved good results (8-10). However, many factors may influence the quantification of radiomic features, including acquisition modes (11), reconstruction parameters (12), and smoothing (13), and segmentation thresholds (14,15). Therefore, the reproducibility, repeatability and robustness of radiomic results are relatively unsatisfactory (16,17).
Another currently prevalent technology to analyze medical images are CNNs, which requires only a set of data with minor preprocessing and then discovers the informative representations in a self-taught manner (18,19). Therefore, CNNs may be likely more reproducible than radiomics. CNNs have also demonstrated perfect diagnostic ability in retinal diseases and skin cancer, which even outperformed experienced experts (20,21). Possessing these advantages, CNNs may serve as a useful tool in the detection of EGFR mutations. But so far, no studies have been available to indicate whether CNNs could be utilized in the detection of EGFR mutations and to compare its efficacy with that of radiomics. To address these two issues, we aim to implement a CNN and perform radiomic analysis to detect EGFR mutations and to explore whether these two methods are mutually complementary.
Methods
Clinical data collection
We retrospectively collected data from patients in Shanghai Chest Hospital from 2013 to 2017. The study was approved by Shanghai Chest Hospital, Shanghai Jiao Tong University. Ethical approval (ID: KS 1716) was obtained for use of the CT images and information of EGFR mutations. Because of its retrospective nature, informed consents were waived. The inclusion criteria for the data in this study were as follows: (I) All patients were pathologically diagnosed with lung adenocarcinoma regardless of their clinical or pathological stages; (II) patients should take CT scanning in our hospital before any treatments; (III) the pulmonary lesions for EGFR mutation tests should be solid nodules not ground glass opacities (GGO); the margins of these solid nodules were well-defined on CT images with the longest dimension equal to or larger than 0.8 cm; (IV) there was only one lesion in bilateral lung, rather than multiple lesions; (V) complete clinical data including gender, age, smoking history, staging images and EGFR status should be available for all patients. As EGFR mutations mainly exist in exon 19 and 21, we collected patients harboring exon 19 and 21 mutations only to ensure enough sample size. All patients were split into the training set and the validation set randomly with a balanced distribution of clinical features including gender, age, smoking history, clinical or pathological stage and EGFR status.
Image data collection
Contrast and non-contrast CT scanning were undertaken before any treatments using Philips Brilliance 64 scanner and GE Discovery CT750 HD scanner. The parameters used were as follows: Tube voltage, tube current, pitch and thickness are 120 kV, 250 mA/s, 0.641 and 5 mm for Philips Brilliance 64 scanner and 120 kV, 400 mA/s, 0.984 and 5 mm for GE Discovery CT750 HD scanner.
EGFR mutations test
EGFR mutation tests by fluorescence PCR (ARMS) were conducted on the specimens acquired from surgeries and biopsies through FNA or endoscope. The PCR machine (Stratagene Mx3000PTM) was provided by Agilent. The Human EGFR Gene Mutation Detection Kit was manufactured by Amoy Diagnostics Co., Ltd. All the EGFR gene mutation tests were accomplished using the same test system and protocol.
Tumor segmentation
CT images were introduced into the treatment planning system (Pinnacle3 Version 9.10). Pulmonary lesions were delineated on non-contrast images with the window level of −400 and width of 1,600. The delineation of the lesions was performed by two experienced radiation oncologists. They reviewed each other’s delineation. Discrepancies about delineation were solved by discussion until consensus was reached.
The framework of our models building
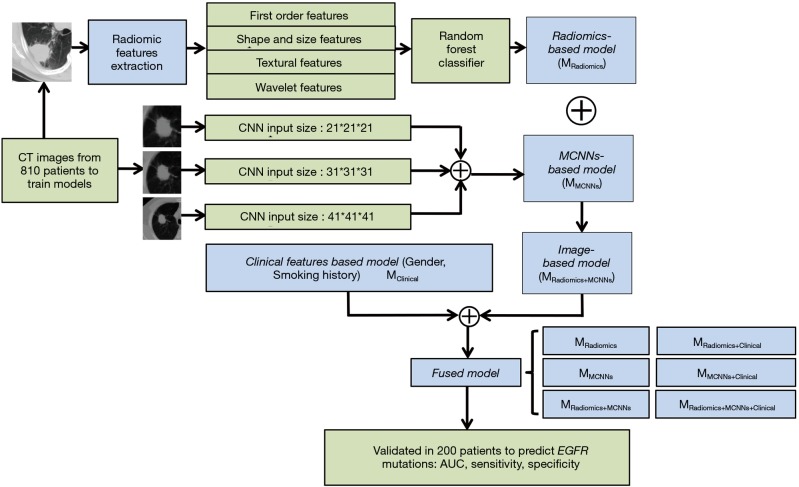
Patients with EGFR mutations were defined as positive samples (label is 1) and others were defined as negative samples (label is 0). Our study consisted of two models. The major model was CT images-based model, which includes radiomics-based model and the multi-level residual CNNs (MCNNs) based model. Another model is clinical features-based model. The performance was validated on the validation set using area under the receiver operating characteristic curve (AUC), sensitivity and specificity (Figure 1).
Figure 1.
The framework of this study. Radiomics-based model (MRadiomics) and MCNNs-based model (MMCNNs) were constructed on the CT-images from the training set. Then radiomics-based model and MCNNs-based model were combined to build the image-based model (MRadiomics+MCNNs). Clinical features (MClinical) were added into the image-based model to establish the fused model (MRadiomics+Clinical, MMCNNs+Clinical, MRadiomics+MCNNs+Clinical). All these seven models were tested in the validation set to calculate each AUC, sensitivity and specificity. MCCNs, multi-level residual CNNs; CNN, convolutional neural network; AUC, area under the receiver operating characteristic curve; EGFR, epithelial growth factor receptor.
Radiomics-based model (MRadiomics)
Quantitative radiomic features of four categories were extracted from the ROIs: 14 first-order features, 8 shape-based and size-based features, 34 textural features, and 384 wavelet features. A total of 440 features were obtained from CT images of one patient for each ROI. We computed the P value for each feature by performing an independent test between positive and negative samples. The grid search method was used to tune the value of P value from 0.00 to 1.00 with step size 0.01. The optimal threshold was finally set as 0.76. Therefore, several features with the corresponding P values lower than 0.76 were selected as discriminative radiomic features. We used a random forest (RF) classification method to combine the merits while ignoring the weaknesses of the selected features. The inputs to the RF were the discriminative radiomic features and the outputs were the EGFR status.
Multi-level residual CNNs based model (MMCNNs)
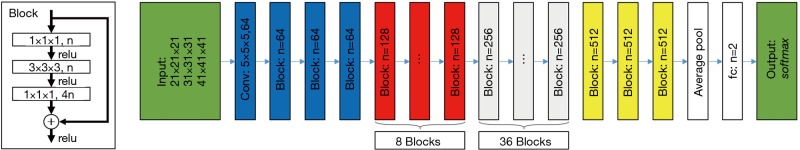
The MMCNNs contained three residual CNNs with each input patches of 21×21×21 voxels (42 mm × 42 mm × 42 mm), 31×31×31 voxels (62 mm × 62 mm × 62 mm), and 41×41×41 (82 mm × 82 mm × 82 mm) voxels, respectively. Each residual CNN had 152 layers. The structure of the MCNNs is presented in Figure 2. The input patches were augmented by random rotation, translation, and flipping before each and every training epoch. Data augmentation is well known to the overcome overfitting of training data and to improve the robustness of the model. The output of each residual CNN was the probabilities of EGFR mutation and wild type.
Figure 2.
The structure of the MCNNs used in this study. The MMCNNs contained three residual CNNs with each input patches of 21×21×21 voxels (42 mm × 42 mm × 42 mm), 31×31×31 voxels (62 mm × 62 mm × 62 mm), and 41×41×41 (82 mm × 82 mm × 82 mm) voxels, respectively. Each residual CNN had 152 layers. MCCNs, multi-level residual CNNs; CNN, convolutional neural network.
The Fusion of MRadiomics and MMCNNs
The ModelRadiomics+MCNNs (MRadiomics+MCNNs) consisting of the MRadiomics and the MMCNNs was defined as follows:
MRadiomics+MCNNs = wRadiomicsMRadiomics+ wMCNNsMMCNNs
wRadiomics and wMCNNs were weighs that determined the contribution of each sub-model to the fused model.
Clinical features-based model (MClinical)
Among all relevant clinical features, there was significant difference only in gender (P<0.0001) and smoking history (P<0.0001) between patients with EGFR mutations or not (Table 1). Therefore, we built a simple clinical feature-based model based on gender and smoking history. Logistic regression was utilized to test whether these two clinical features were indeed associated with EGFR mutations. Thereafter we assigned scores to each sample according to the clinical feature: female non-smokers given 1.00, female smokers and male non-smokers were both given 0.50, and male smokers were given 0.00.
Table 1. Characteristics of total patients.
| Characteristic | Overall (n=1,010) | Mutation (n=510) | Wild type (n=500) | Statistical method and P value |
|---|---|---|---|---|
| Gender, n (%) | Pearson χ2 test, P<0.0001 | |||
| Male | 553 (54.8) | 209 (41.0) | 344 (68.8) | |
| Female | 457 (45.2) | 301 (59.0) | 156 (31.2) | |
| Age (years) | Independent-samples t-test, P=0.352 | |||
| Median age | 63 | 62 | 61 | |
| Range | 25–88 | 30–88 | 25–85 | |
| Smoking history, n (%) | Pearson χ2 test, P<0.0001 | |||
| Smoking | 261 (25.8) | 69 (13.5) | 193 (38.6) | |
| No smoking | 749 (74.2) | 441 (86.5) | 307 (61.4) | |
| Pathological stage, n (%) | Mann-Whitney test, P=0.062 | |||
| I | 307 (30.4) | 181 (35.5) | 126 (25.2) | |
| II | 49 (4.9) | 24 (4.7) | 25 (5.0) | |
| III | 380 (37.6) | 161 (31.6) | 219 (43.8) | |
| IV | 274 (27.1) | 144 (28.2) | 130 (26.0) | |
| Sample type, n (%) | Pearson χ2 test, P=0.023 | |||
| Biopsy | 386 (38.2) | 177 (34.7) | 209 (41.8) | |
| Surgery | 624 (61.8) | 333 (65.3) | 291 (58.2) |
The Fusion of image-based model and clinical features-based model
The fused model was constructed by image-based model and clinical features-based model as follows:
MRadiomics+Clinical = wRadiomicsMRadiomics + wClinicalMClinical
MMCNNs+Clinical = wMCNNsMMCNNs + wClinicalMClinical
MRadiomics+MCNNs+Clinical = wRadiomics+MCNNsMRadiomics+MCNNs + wClinicalMClinical
wRadiomics, wMCNNs and wRadiomics+MCNNs were weighs that determined the contribution of each sub-model to the fused model. The weight parameters mentioned above were decided by using grid search method in the training set.
Statistical analysis
Receiver operating characteristic (ROC) curve was performed on the validation set to evaluate the performance of the seven models (trained by the training set) in detecting the EGFR mutation statues, and the AUC was calculated. The P value of paired z-test were conducted to compare the AUC of each model and the significance level was set at P<0.05. The sensitivity and specificity were obtained from the best diagnostic decision point of ROC. The weight parameters mentioned above were decided by using grid search method in the training set.
Results
Patients’ characteristics
CT images from 1,010 consecutive patients who met eligibility criteria from 2013 to 2017 with matching EGFR status were retrospectively collected, among which 510 patients were EGFR-mutated and 500 patients were wild-type. The patients’ demographic and clinical characteristics were presented in Table 1. There were 553 males and 457 females with a median age of 63 years old (25 to 88 years). Two hundred and sixty-one patients (25.8%) were smokers and 749 patients (74.2%) were not. Pathological stages were distributed as follows: stage I in 307 patients (30.4%), stage II in 49 patients (4.9%), stage III in 380 patients (37.6%) and stage IV in 274 patients (27.1%). The 1,010 patients were randomized into training set (810 patients) and validation set (200 patients). There was no significant statistical difference of the patients’ characteristics in these two sets as seen in Table 2.
Table 2. The comparison of patients’ characteristics between training set and validation set. There was no significant difference of patients’ characteristics between training set and validation set, including gender, age, smoking history, pathological stage, EGFR status and sample type.
| Characteristic | Training set (n=810) | Validation set (n=200) | Statistical method and P value |
|---|---|---|---|
| Gender, n (%) | Pearson χ2 test, P=0.813 | ||
| Male | 442 (54.6) | 111 (55.5) | |
| Female | 368 (45.4) | 89 (44.5) | |
| Age (years) | Independent-samples t-test, P=0.108 | ||
| Median age | 61 | 62 | |
| Range | 25–88 | 39–85 | |
| Smoking history, n (%) | Pearson χ2 test, P=0.954 | ||
| Smoking | 209 (25.8) | 53 (26.5) | |
| No smoking | 601 (74.2) | 147 (73.5) | |
| Pathological stage, n (%) | Mann-Whitney test, P=0.149 | ||
| I | 253 (31.2) | 54 (27.0) | |
| II | 40 (4.9) | 9 (4.5) | |
| III | 304 (37.5) | 76 (38.0) | |
| IV | 213 (26.3) | 61 (30.5) | |
| Genetic status, n (%) | Pearson χ2 test, P=0.364 | ||
| Wild type | 407 (50.2) | 93 (46.5) | |
| Exon 19 mutation | 192 (23.7) | 57 (28.5) | |
| Exon 21 mutation | 211 (26.0) | 50 (25.0) | |
| Sample type, n (%) | Pearson χ2 test, P=0.81 | ||
| Biopsy | 308 (38.0) | 78 (39.0) | |
| Surgery | 502 (62.0) | 122 (61.0) |
EGFR, epithelial growth factor receptor.
Models performances in the validation set
We utilized the CT images with corresponding EGFR status from the 810 patients to train MRadiomics and MMCNNs. The weights parameters were 0.16, 0.20, 0.64, 0.84 for wClinical, wRadiomics, wMCNNs and wRadiomics+MCNNs, respectively. All the models were tested individually in the validation set including 200 patients and the results were presented in Figures 3,4, Tables 3,4. According to the P value of independent test, we ultimately selected 388 radiomic features to be put into RF classifier including 14 first-order features, 7 shape-based and size-based features, 33 textural features, and 334 wavelet features. The AUC of the MRadiomics to predict EGFR mutations was 0.740 [95% confidence interval (CI), 0.670–0.811, P<0.0001] with specificity of 0.677 and sensitivity of 0.794. The MMCNNs achieved an AUC of 0.810 (95% CI, 0.748–0.872, P<0.0001) to predict EGFR mutations with specificity of 0.753 and sensitivity of 0.813, which outperformed the MRadiomics (P=0.0225). After combining the MRadiomics and the MMCNNs model, the MRadiomics+MCNNs could predict EGFR mutations with an AUC of 0.811 (95% CI, 0.749–0.873, P<0.0001) with specificity of 0.763 and sensitivity of 0.804. The MRadiomics+MCNNs did better than MRadiomics (P=0.009), but not MMCNNs (P=0.742).
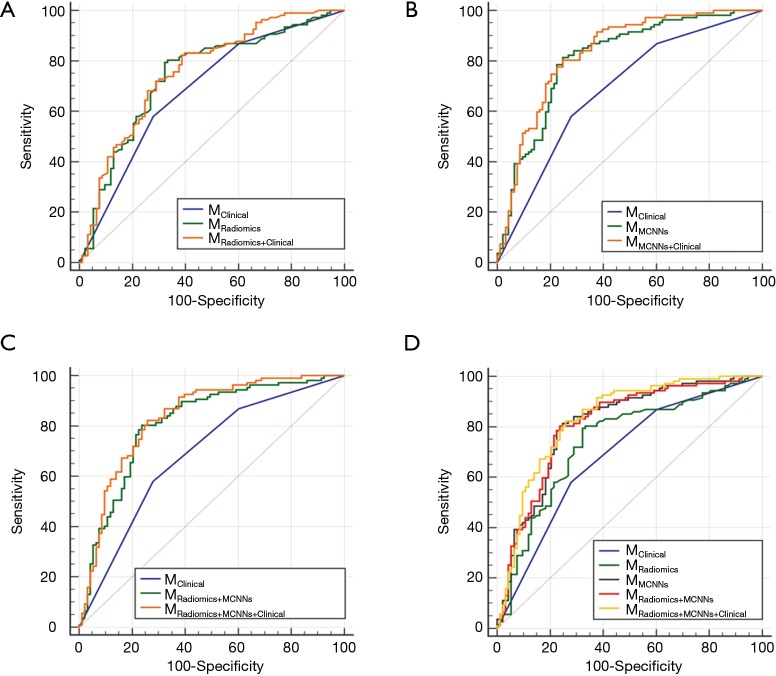
Figure 3.
The AUC of each model to predict EGFR mutations. (A) The AUC of MRadiomics and MRadiomics+Clinical is 0.740 (95% CI, 0.670–0.811) and 0.758 (95% CI, 0.690–0.825); (B) the AUC of MMCNNs and MMCNNs+Clinical is 0.810 (95% CI, 0.748–0.872) and 0.831 (95% CI, 0.773–0.890); (C) the AUC of MRadiomics+MCNNs and MRadiomics+MCNNs+Clinical is 0.811 (95% CI, 0.749–0.873) and 0.834 (95% CI, 0.776–0.892); (D) the AUC of MClinical is 0.686 (95% CI, 0.617–0.756) and the lowest one. MRadiomics is less efficient than other models except MClinical. There is no significant different between MMCNNs, MRadiomics+MCNNs and MRadiomics+MCNNs+Clinical. MCCNs, multi-level residual CNNs; CNN, convolutional neural network; AUC, area under the receiver operating characteristic curve; EGFR, epithelial growth factor receptor.
Figure 4.
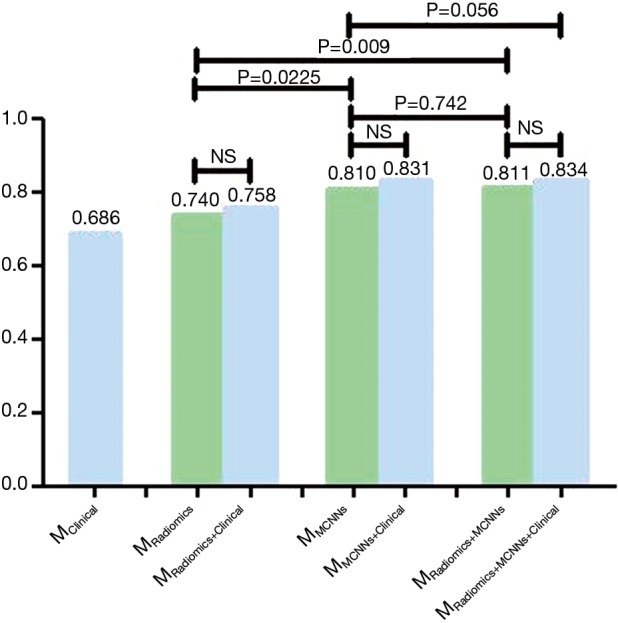
The addition of clinical features did not improve the AUC of the MRadiomics (P=0.623), the MMCNNs (P=0.114) and the MRadiomics+MCNNs (P=0.058). The MMCNNs outperformed the MRadiomics (P=0.0225). The MMCNNs+Radiomics did better than the MRadiomics (P=0.009), but not the MMCNNs (P=0.742). The MMCNNs did not demonstrate inferiority compared with the MRadiomics+MCNNs (P=0.742) and the MRadiomics+MCNNs+Clinical (P=0.056) in terms of AUC. MCCNs, multi-level residual CNNs; CNN, convolutional neural network; AUC, area under the receiver operating characteristic curve; EGFR, epithelial growth factor receptor.
Table 3. The specificity and sensitivity of these seven models at best decision point.
| Model | Specificity | Sensitivity |
|---|---|---|
| MClinical | 0.730 | 0.579 |
| MRadiomics | 0.677 | 0.794 |
| MRadiomics+Clinical | 0.613 | 0.822 |
| MMCNNs | 0.753 | 0.813 |
| MMCNNs+Clinical | 0.688 | 0.832 |
| MRadiomics+MCNNs | 0.763 | 0.804 |
| MRadiomics+MCNNs+Clinical | 0.742 | 0.822 |
Table 4. The AUC of each model validated in different pathological stages. Because sample size (n=9) of stage II in validation set is too small to evaluate, we combine stage I and II for analysis.
| Model | AUC in stage I and II (n=63) | AUC in stage III (n=76) | AUC in stage IV (n=61) |
|---|---|---|---|
| MClinical | 0.630 | 0.800 | 0.617 |
| MRadiomics | 0.755 | 0.742 | 0.736 |
| MRadiomics+Clinical | 0.713 | 0.838 | 0.718 |
| MMCNNs | 0.767 | 0.815 | 0.848 |
| MMCNNs+Clinical | 0.800 | 0.860 | 0.847 |
| MRadiomics+MCNNs | 0.771 | 0.818 | 0.839 |
| MRadiomics+MCNNs+Clinical | 0.803 | 0.861 | 0.854 |
MCCNs, multi-level residual CNNs; CNN, convolutional neural network; AUC, area under the receiver operating characteristic curve.
Clinical features like gender [odds ratio (OR) =2.1, 95% CI, 1.57–2.84, P<0.0001] and smoking (OR =0.39, 95% CI, 0.27–0.55, P<0.0001) were significantly associated with EGFR mutations. The MClinical acquired the lowest AUC of 0.686 (95% CI, 0.617–0.756, P<0.0001) with specificity of 0.730 and sensitivity of 0.579. The MClinical did worse than MMCNNs (P=0.005), but showed no significant difference with MRadiomics (P=0.256). Finally, we added these two clinical features to the image-based model to build the fused model. The AUC of MRadiomics+Clinical, MMCNNs+Clinical and MRadiomics+MCNNs+Clinical is 0.758 (95% CI, 0.690–0.825, P<0.0001), 0.831 (95% CI, 0.773–0.890, P<0.0001), and 0.834 (95% CI, 0.776–0.892, P<0.0001) respectively. But the addition of clinical features did not show significant improvement than MRadiomics (P=0.623) and MMCNNs (P=0.114). There was an increasing trend for the MRadiomics+MCNNs (P=0.058), but still without significance. The MRadiomics+MCNNs+Clinical demonstrated the highest AUC to predict EGFR mutations than other models, but exhibited significant difference only with MRadiomics (P=0.0009) rather than MRadiomics+MCNNs (P=0.058) and MMCNNs (P=0.056).
Discussion
The value of image analysis to reveal biological information is by no means a replacement of pathological biopsy and liquid biopsy. Compared with pathological biopsy, the most promising advantage of image analysis lies in that the biological information acquired by images could describe the genotype and phenotype of the whole tumor and even project the biological information onto each pixel of images to reflect intra-tumor heterogeneity. Liquid biopsy could reveal the genetic mutations via peripheral blood but such systemic information is unable to disclose the different molecular changes of each lesion due to inter-tumor heterogeneity. Image analysis could complement this shortcoming and instruct more delicate combination between systemic treatment of TKIs and local treatment like radiotherapy or mini-invasive surgery. Therefore, it is worthwhile to develop image analysis to complement pathological biopsy and liquid biopsy for more precise systemic treatment and local therapy.
Due to the development and application of targeted therapy, examination of patients’ genetic profile is recommended to gauge the tumor progression for some patients. A single examination before the start of the targeted therapy is insufficient for an effective treatment nowadays. Multiple examinations, for example by performing repeated biopsies, are needed. However, some thorny clinical scenarios do not allow for these procedures. Medical imaging analysis, as a non-invasive method to complement biopsies, has been studied to detect genetic mutations. The underlying hypothesis of imaging analysis is that advanced imaging technology could capture genomic and proteomic patterns expressed in terms of macroscopic image-based features (22). Difference of protein expression patterns within tumors has been demonstrated to be correlated to radiographic findings (23,24). Currently there have been several studies on the detection of EGFR mutations with the utilization of semantic features, radiomic features and CNNs.
Semantic features refer to the manual assessment of the tumor phenotype by an expert radiologist, like pleural attachment, poorly-defined margin or strong enhancement. They are all quantities that vary wildly between radiologists as no standards for their definitions exist. Although studies utilizing semantic features have shown excellent AUC values approaching 0.9, these results would be difficult to reproduce (25,26).
Radiomic features are calculated by algorithms from the defined ROIs to extract indiscernible biological information from images. These features include tumor intensity histogram-based features, shape-based features, texture-based features, and other higher-order features (10). These features have been demonstrated to predict EGFR mutations with AUC values ranging from 0.7 to 0.9 (8,9,15). But some of these results were acquired without external independent validation. In our study, the AUC of radiomic features was achieved in validation set and demonstrated that radiomic features are able to predict EGFR mutations. To achieve reproducible and best predictive performance, factors including scanning parameters, reconstruction algorithm and segmentation of ROIs should be standardized and universalized, which would be hard to be realized in practice. Radiomic features are often case-specific which means that the same set of features may not perform optimally on different image segmentation problems (27,28). In contrast, CNNs need relatively low requirements for producing reproducible results. Additionally, CNNs automatically extract the features that optimally represent the data for the specific problem at hand (19). The utilization of CNNs on chest imaging currently emphasized on the detection of malignant pulmonary nodules with their AUC values ranging from 0.7 to 0.9 (29-31). So far there have been no studies utilizing CNNs to predict EGFR mutations and let alone studies comparing CNNs and radiomic features in a same sample.
Our study utilized a CNN and radiomic analysis to detect EGFR mutations with an aim to explore their differences and mutual complementarities. The AUC of the CNN to detect EGFR mutations was 0.81 on independent validation set, which is comparable to the results of the above-mentioned studies to detect malignant pulmonary nodules. With specificity maintaining 0.753, sensitivity of CNN is 0.813. These results indicate that the CNN is competent to detect EGFR mutations tentatively. Radiomic analysis in our study achieves an AUC of 0.74, which is achieved under strict external validation and higher than that of previous studies (8,9,15). Nevertheless, the specificity of the radiomic features was only 0.677 with a sensitivity of 0.794. Therefore, considering AUC, specificity and sensitivity, radiomic analysis performs worse than the CNN. Thereafter we fused the clinical, radiomics, and MCNN models to improve the detection accuracy by giving each model a voting weight. A higher weight was given to a model with better performance. Then the final possibility of EGFR mutations was figured out. However, the AUC has not been improved significantly compared to the CNN alone after the combination. Given that automatically generated features by the CNN could generate the same performance of hand-crafted features (32), this phenomenon maybe due to that the CNN has already extracted enough features to support the detection and there has left little room for improvement with the addition of radiomic features. Another explanation of this phenomenon may attribute to our relatively tentative combination method. It was just a simple algorithm that assigned voting weights to the predictive result of each submodel. This combination method did not involve the MCNN structure. The addition of clinical features including gender and smoking history into all these three image-based models (MRadiomics, MMCNNs, MRadiomics+MCNNs) did not improve the AUC with statistical significance despite the increasing tendency of the MMCNNs+Clinical. This phenomenon also implicates that CNN alone is competent to predict EGFR mutations without the help of radiomics or clinical features in spite of the close association between Asian female non-smokers and EGFR mutations.
Compared with previous studies utilizing semantic (25,26) or radiomic features (8,9,15) to predict EGFR mutations, our study recruited the largest sample size and achieved a satisfactory result under strict external validation. We compared the predictive efficacy between CNNs and radiomic features in a same sample, and explored the possibility of the combination of these two methods. Clinical features of gender and smoking history were also proposed to complement CNNs or radiomic features. Our result is equivalent to or even better than that of previous studies. The predictive ability of all the model studies, however, is far from clinical demands. But there are several issues with our study entailing further improvement: (I) The CT-images were acquired by different scanners in our hospital and the scanning parameters have not been standardized. The scanning thickness of 5 mm may lead to loss of some radiomic information. The radiomic features extracted have not been validated by a test-retest in RIDER dataset to ensure reproducibility (33). All the above-mentioned flaws may compromise the results of radiomics-based model. More quality control measures are needed. (II) The structure of our MCNNs is relatively preliminary and the specificity of the models is unsatisfactory, which currently cannot be utilized in clinical practice to predict EGFR mutations. (III) The combination method we used was relatively simple, which only combined the result of each submodel through assigning voting weights. (IV) As female non-smokers are strongly associated with EGFR mutations, separate analysis of male and female patients may produce more reliable results. (V) The generalizability of our findings should be evaluated in other institutes because all the patients were enrolled only in our hospital. This study is a tentative exploration about whether CNNs could predict EGFR mutations on CT-images. Further improvement of the predictive ability and broader validation of CNN would be warranted. Other uncommon mutation sites like exon 18 and 20 of EGFR would be included as well. Among the forthcoming work, the emphasis would be the improvement the predictive ability of MMCNNs and development of an advanced method combining MMCNNs and MRadiomics. It includes the utilization of transfer learning (34), multi-instance deep learning (35), aggregated residual transformations for deep neural networks (36) and other methods. We hope that the refined MMCNNs could satisfy clinical requirements to detect EGFR mutations and furthermore provide information about other biological processes.
Conclusions
Both of the MRadiomics and the MMCNNs could predict EGFR mutations on CT images of patients with lung adenocarcinoma. The MMCNNs outperformed the MRadiomics in the detection of EGFR mutations. The combination of these two models, even added with clinical features, is not significantly more efficient than MMCNNs alone. Therefore, MMCNNs would be the main modality for future exploration of detecting EGFR mutations by image analysis.
Acknowledgements
Funding: This study was supported by the Shanghai Jiao Tong University Medical Engineering Cross Research Funds (No. YG2017ZD10).
Ethical Statement: This study was approved by Shanghai Chest Hospital, Shanghai Jiao Tong University. Ethical approval (ID: KS 1716) was obtained for use of the CT images and information of EGFR mutations.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Fukuoka M, Wu YL, Thongprasert S, et al. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS). J Clin Oncol 2011;29:2866-74. 10.1200/JCO.2010.33.4235 [DOI] [PubMed] [Google Scholar]
- 2.Han B, Tjulandin S, Hagiwara K, et al. EGFR mutation prevalence in Asia-Pacific and Russian patients with advanced NSCLC of adenocarcinoma and non-adenocarcinoma histology: The IGNITE study. Lung Cancer 2017;113:37-44. 10.1016/j.lungcan.2017.08.021 [DOI] [PubMed] [Google Scholar]
- 3.Hirsch FR, Bunn PA., Jr EGFR testing in lung cancer is ready for prime time. Lancet Oncol 2009;10:432-3. 10.1016/S1470-2045(09)70110-X [DOI] [PubMed] [Google Scholar]
- 4.Gridelli C, Ciardiello F, Gallo C, et al. First-line erlotinib followed by second-line cisplatin-gemcitabine chemotherapy in advanced non-small-cell lung cancer: the TORCH randomized trial. J Clin Oncol 2012;30:3002-11. 10.1200/JCO.2011.41.2056 [DOI] [PubMed] [Google Scholar]
- 5.Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer 2012;12:323-34. 10.1038/nrc3261 [DOI] [PubMed] [Google Scholar]
- 6.Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med 2017;376:2109-21. 10.1056/NEJMoa1616288 [DOI] [PubMed] [Google Scholar]
- 7.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016;278:563-77. 10.1148/radiol.2015151169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rios Velazquez E, Parmar C, Liu Y, et al. Somatic Mutations Drive Distinct Imaging Phenotypes in Lung Cancer. Cancer Res 2017;77:3922-30. 10.1158/0008-5472.CAN-17-0122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, Kim J, Balagurunathan Y, et al. Radiomic Features Are Associated With EGFR Mutation Status in Lung Adenocarcinomas. Clin Lung Cancer 2016;17:441-448.e6. 10.1016/j.cllc.2016.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aerts HJ, Grossmann P, Tan Y, et al. Defining a Radiomic Response Phenotype: A Pilot Study using targeted therapy in NSCLC. Sci Rep 2016;6:33860. 10.1038/srep33860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mackin D, Fave X, Zhang L, et al. Measuring Computed Tomography Scanner Variability of Radiomics Features. Invest Radiol 2015;50:757-65. 10.1097/RLI.0000000000000180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim H, Park CM, Lee M, et al. Impact of Reconstruction Algorithms on CT Radiomic Features of Pulmonary Tumors: Analysis of Intra- and Inter-Reader Variability and Inter-Reconstruction Algorithm Variability. PLoS One 2016;11:e0164924. 10.1371/journal.pone.0164924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao B, Tan Y, Tsai WY, et al. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci Rep 2016;6:23428. 10.1038/srep23428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parmar C, Rios Velazquez E, Leijenaar R, et al. Robust Radiomics feature quantification using semiautomatic volumetric segmentation. PLoS One 2014;9:e102107. 10.1371/journal.pone.0102107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Q, Lu L, Dercle L, et al. Interobserver variability in tumor contouring affects the use of radiomics to predict mutational status. J Med Imaging (Bellingham) 2018;5:011005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balagurunathan Y, Gu Y, Wang H, et al. Reproducibility and Prognosis of Quantitative Features Extracted from CT Images. Transl Oncol 2014;7:72-87. 10.1593/tlo.13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalpathy-Cramer J, Mamomov A, Zhao B, et al. Radiomics of Lung Nodules: A Multi-Institutional Study of Robustness and Agreement of Quantitative Imaging Features. Tomography 2016;2:430-7. 10.18383/j.tom.2016.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen D, Wu G, Suk HI. Deep Learning in Medical Image Analysis. Annu Rev Biomed Eng 2017;19:221-48. 10.1146/annurev-bioeng-071516-044442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60-88. 10.1016/j.media.2017.07.005 [DOI] [PubMed] [Google Scholar]
- 20.Kermany DS, Goldbaum M, Cai W, et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 2018;172:1122-1131.e9. 10.1016/j.cell.2018.02.010 [DOI] [PubMed] [Google Scholar]
- 21.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115-8. 10.1038/nature21056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441-6. 10.1016/j.ejca.2011.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hobbs SK, Shi G, Homer R, et al. Magnetic resonance image-guided proteomics of human glioblastoma multiforme. J Magn Reson Imaging 2003;18:530-6. 10.1002/jmri.10395 [DOI] [PubMed] [Google Scholar]
- 24.Grossmann P, Stringfield O, El-Hachem N, et al. Defining the biological basis of radiomic phenotypes in lung cancer. Elife 2017;6:e23421. 10.7554/eLife.23421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gevaert O, Echegaray S, Khuong A, et al. Predictive radiogenomics modeling of EGFR mutation status in lung cancer. Sci Rep 2017;7:41674. 10.1038/srep41674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rizzo S, Petrella F, Buscarino V, et al. CT Radiogenomic Characterization of EGFR, K-RAS, and ALK Mutations in Non-Small Cell Lung Cancer. Eur Radiol 2016;26:32-42. 10.1007/s00330-015-3814-0 [DOI] [PubMed] [Google Scholar]
- 27.Yip SS, Aerts HJ. Applications and limitations of radiomics. Phys Med Biol 2016;61:R150-66. 10.1088/0031-9155/61/13/R150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar V, Gu Y, Basu S, et al. Radiomics: the process and the challenges. Magn Reson Imaging 2012;30:1234-48. 10.1016/j.mri.2012.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masood A, Sheng B, Li P, et al. Computer-Assisted Decision Support System in Pulmonary Cancer detection and stage classification on CT images. J Biomed Inform 2018;79:117-28. 10.1016/j.jbi.2018.01.005 [DOI] [PubMed] [Google Scholar]
- 30.Shen W, Zhou M, Yang F, et al. Multi-crop Convolutional Neural Networks for lung nodule malignancy suspiciousness classification. Pattern Recognition 2017;61:663-73. 10.1016/j.patcog.2016.05.029 [DOI] [Google Scholar]
- 31.Tajbakhsh N, Suzuki K. Comparing two classes of end-to-end machine-learning models in lung nodule detection and classification: MTANNs vs. CNNs. Pattern Recognition 2017;63:476-86. 10.1016/j.patcog.2016.09.029 [DOI] [Google Scholar]
- 32.Sun W, Zheng B, Qian W. Automatic feature learning using multichannel ROI based on deep structured algorithms for computerized lung cancer diagnosis. Comput Biol Med 2017;89:530-9. 10.1016/j.compbiomed.2017.04.006 [DOI] [PubMed] [Google Scholar]
- 33.Balagurunathan Y, Kumar V, Gu Y, et al. Test-retest reproducibility analysis of lung CT image features. J Digit Imaging 2014;27:805-23. 10.1007/s10278-014-9716-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin HC, Roth HR, Gao M, et al. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Trans Med Imaging 2016;35:1285-98. 10.1109/TMI.2016.2528162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhennan Y, Yiqiang Z, Zhigang P, et al. Multi-Instance Deep Learning: Discover Discriminative Local Anatomies for Bodypart Recognition. IEEE Trans Med Imaging 2016;35:1332-43. 10.1109/TMI.2016.2524985 [DOI] [PubMed] [Google Scholar]
- 36.Xie S, Girshick R, Dollár P, et al. Aggregated Residual Transformations for Deep Neural Networks. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, Hawaii, USA 2017:5987-95. [Google Scholar]