Abstract
Although Aspergillus fumigatus is the major agent of invasive aspergillosis, an increasing number of infections are caused by its cryptic species, especially A. lentulus and the A. viridinutans species complex (AVSC). Their identification is clinically relevant because of antifungal drug resistance and refractory infections. Species boundaries in the AVSC are unresolved since most species have uniform morphology and produce interspecific hybrids in vitro. Clinical and environmental strains from six continents (n = 110) were characterized by DNA sequencing of four to six loci. Biological compatibilities were tested within and between major phylogenetic clades, and ascospore morphology was characterised. Species delimitation methods based on the multispecies coalescent model (MSC) supported recognition of ten species including one new species. Four species are confirmed opportunistic pathogens; A. udagawae followed by A. felis and A. pseudoviridinutans are known from opportunistic human infections, while A. felis followed by A. udagawae and A. wyomingensis are agents of feline sino-orbital aspergillosis. Recently described human-pathogenic species A. parafelis and A. pseudofelis are synonymized with A. felis and an epitype is designated for A. udagawae. Intraspecific mating assay showed that only a few of the heterothallic species can readily generate sexual morphs in vitro. Interspecific mating assays revealed that five different species combinations were biologically compatible. Hybrid ascospores had atypical surface ornamentation and significantly different dimensions compared to parental species. This suggests that species limits in the AVSC are maintained by both pre- and post-zygotic barriers and these species display a great potential for rapid adaptation and modulation of virulence. This study highlights that a sufficient number of strains representing genetic diversity within a species is essential for meaningful species boundaries delimitation in cryptic species complexes. MSC-based delimitation methods are robust and suitable tools for evaluation of boundaries between these species.
Keywords: Aspergillus felis, Aspergillus fumigatus, invasive aspergillosis, mating-type genes, multispecies coalescence model, Neosartorya udagawae, scanning electron microscopy, soil fungi
INTRODUCTION
Aspergillus is a speciose genus with almost 400 species classified into six subgenera and approximately 25 sections (Samson et al. 2014, Jurjević et al. 2015, Hubka et al. 2016a, 2017, Chen et al. 2016a, b, 2017, Kocsubé et al. 2016, Sklenář et al. 2017, Tanney et al. 2017). The species are widely distributed in nature and have a significant economic impact in human and animal health (causative agents of aspergillosis; allergies and respiratory problems associated with presence of fungi in the indoor environment), the food industry (source of enzymes and organic acids for fermentation, food and feed spoilage, production of hazardous mycotoxins), biotechnology and pharmacology (production of bioactive substances, heterologous proteins) (Pitt & Hocking 2009, Meyer et al. 2011, Frisvad & Larsen 2015b, Sugui et al. 2015, Gautier et al. 2016).
Aspergillus sect. Fumigati includes approximately 60 species occurring predominantly in soil (Hubka et al. 2017). Many are of considerable medical importance as they cause human and animal infections (Balajee et al. 2005b, 2009, Katz et al. 2005, Yaguchi et al. 2007, Hubka et al. 2012, Talbot & Barrs 2018). Aspergillus fumigatus is usually reported as both the most common member of the section in soil worldwide and the most common cause of aspergillosis (Klich 2002, Domsch et al. 2007, Mayr & Lass-Flörl 2011). A series of recent studies highlighted the high prevalence (11–19 %) of so-called cryptic Aspergillus species in clinical samples (Balajee et al. 2009, Alastruey-Izquierdo et al. 2013, Negri et al. 2014, Sabino et al. 2014). Their identification is clinically relevant since many demonstrate drug resistance to commonly used antifungals, thus their recognition influences therapeutic management. Reliable identification of clinical isolates to the species level and susceptibility testing by reference methods is thus warranted (Lyskova et al. 2018). Many of these less common pathogens belong to sect. Fumigati and the highest numbers of infections are attributed to A. lentulus, A. thermomutatus (syn. Neosartorya pseudofischeri) and species from A. viridinutans species complex (AVSC) (Balajee et al. 2005a, 2006, Sugui et al. 2010, 2014, Barrs et al. 2013, Talbot & Barrs 2018).
Homothallism is a predominant reproductive mode in sect. Fumigati and many species readily produce ascomata (neosartorya-morph) in culture, while others are heterothallic or have an unknown sexual morph (Hubka et al. 2017). Homothallic species are infrequently pathogenic, although A. thermomutatus is a notable exception. The majority of clinically relevant species belong to the A. fumigatus clade (Balajee et al. 2005b, 2009, Yaguchi et al. 2007, Alcazar-Fuoli et al. 2008) or the AVSC (Sugui et al. 2010, 2014, Barrs et al. 2013, Nováková et al. 2014) and are heterothallic. A cryptic sexual cycle of several of these opportunistic pathogens, including A. fumigatus (O’Gorman et al. 2009), A. lentulus (Swilaiman et al. 2013) and A. felis (Barrs et al. 2013), was discovered recently by crossing opposite mating type isolates in vitro.
Molecular methods are routinely used for identification of species from sect. Fumigati due to overlapping morphological features of their asexual morph. In contrast, the morphology of the sexual morph, especially of ascospores, is amongst the most informative of phenotypic characteristics in sect. Fumigati. The taxonomy of AVSC has developed rapidly since eight of the currently 11 recognized species were described in the last four years (Barrs et al. 2013, Eamvijarn et al. 2013, Nováková et al. 2014, Sugui et al. 2014, Matsuzawa et al. 2015, Talbot et al. 2017). The species boundaries delimitation was usually based on comparison of single-gene phylogenies and principles of genealogical concordance. In addition, some studies supported the species concept by results of in vitro mating experiments between opposite mating type strains. With the increasing number of species, available isolates and new mating experiment data, the species boundaries in AVSC became unclear as pointed out by Talbot et al. (2017) who used the designation ‘A. felis clade’ for A. felis and related species. Importantly, Sugui et al. (2014) and Talbot et al. (2017) identified that interpretation of in vitro mating assays in sect. Fumigati may be problematic because different phylogenetic species in the AVSC were able to produce fertile ascomata when crossed between themselves. Some even mated successfully with A. fumigatus s.str.
Here we present a critical re-evaluation of species boundaries in the AVSC. We examined a large set of clinical and environmental strains collected worldwide. We did not use classical phylogenetic methods or genealogical concordance phylogenetic species recognition rules (GCPSR) for species delimitation due to their unsatisfactory results in previous AVSC studies. Such methods, based predominantly on analysis of concatenated DNA sequence data or comparison of single-gene phylogenies are frequently prone to species over-delimitation or are affected by subjective judgements of species boundaries. Instead, we used recently introduced delimitation techniques based on coalescent theory and the multispecies coalescent model (MSC) (Flot 2015). We followed the approach recommended by Carstens et al. (2013) that combines species delimitation, species tree estimation and species validation steps. Although these methods have already been applied to other groups of organisms such as animals and plants their use in fungi is scarce (Stewart et al. 2014, Singh et al. 2015, Liu et al. 2016, Sklenář et al. 2017, Hubka et al. 2018). Here, the results of MSC methods were taken as a basic hypothesis for species delimitation and then further verified by analysis of intra- and interspecific biological compatibilities, as well as ascospore dimensions and ornamentation.
MATERIAL AND METHODS
Fungal strains
A total of 110 isolates were examined including new isolates and isolates obtained from previously published studies (Katz et al. 2005, Vinh et al. 2009, Coelho et al. 2011, Shigeyasu et al. 2012, Barrs et al. 2013, 2014, Eamvijarn et al. 2013, Nováková et al. 2014, Sugui et al. 2014, Matsuzawa et al. 2015, Talbot et al. 2017) and culture collections. The set comprised 38 clinical strains and 72 environmental isolates, including 67 from soil, four from cave environments and one from plant material. The provenance of isolates is detailed in Table 1. Newly isolated strains were deposited into the Culture Collection of Fungi at the Department of Botany, Charles University, Prague, Czech Republic (CCF). Dried herbarium specimens were deposited into the herbaria of the Medical Mycology Research Center, Chiba University, Japan (IFM) and Mycological Department of the National Museum, Prague, Czech Republic (PRM).
Table 1.
List of Aspergillus strains, information on isolation source and reproductive strategy.
| Species / Culture collection nos.1,2 | Locality, substrate, year of isolation3 | MAT locus4,5 |
|---|---|---|
| Aspergillus acrensis | ||
| IFM 57291T = CCF 4670T (01-BA-462-5) | Brazil, Acre, Xapuri, grassland soil in cattle farm, 2001 | MAT1-1-1 |
| IFM 57290 = CCF 4666 (01-BA-666-5) | Brazil, Amazonas, Manaus, tropical rain forest soil, 2001 | MAT1-2-1 |
| CCF 4959 (S973) | Romania, Movile cave, above the Lake Room, cave sediment, 2014 | MAT1-2-1 |
| CCF 4960 (S974) | Romania, Movile cave, cave sediment, 2014 | MAT1-2-1 |
| CCF 4961 (S975) | Romania, Movile cave, Lake Room, cave sediment, 2014 | MAT1-1-1 |
| A. arcoverdensis | ||
| IFM 61334T = JCM 19878T = CCF 4900T (6-2-32) | Brazil, Pernambuco, near Arcoverde, semi-desert soil in a caatinga area, 2011 | MAT1-1-1 |
| IFM 61333 = CCF 4899 (10-2-3) | Brazil, Pernambuco, near Arcoverde, semi-desert soil in a caatinga area, 2011 | MAT1-1-1 |
| IFM 61337 = JCM 19879 = CCF 4901 (1-1-34) | Brazil, Pernambuco, near Arcoverde, semi-desert soil in a caatinga area, 2011 | MAT1-1-1 |
| IFM 61338 = JCM 19880 = CCF 4902 (6-2-3) | Brazil, Pernambuco, near Arcoverde, semi-desert soil in a caatinga area, 2011 | MAT1-2-1 |
| IFM 61339 = CCF 4903 (2-1-11) | Brazil, Pernambuco, near Arcoverde, semi-desert soil in a caatinga area, 2011 | MAT1-1-1 |
| IFM 61340 = CCF 4904 (7-2-33) | Brazil, Pernambuco, near Arcoverde, semi-desert soil in a caatinga area, 2011 | MAT1-1-1 |
| IFM 61345 = CCF 5633 (3-2-2) | Brazil, Pernambuco, near Arcoverde, semi-desert soil in a caatinga area, 2011 | MAT1-2-1 |
| IFM 61346 = CCF 4906 (4-2-14) | Brazil, Pernambuco, near Arcoverde, semi-desert soil in a caatinga area, 2011 | MAT1-2-1 |
| IFM 61349 = CCF 4907 (4-2-9) | Brazil, Pernambuco, near Arcoverde, semi-desert soil in a caatinga area, 2011 | MAT1-2-1 |
| IFM 61362 = CCF 4908 (5-2-2) | Brazil, Pernambuco, near Arcoverde, semi-desert soil in a caatinga area, 2011 | MAT1-2-1 |
| IFM 59922 = CCF 4560 (08-SA-2-2) | China, soil, 2008 | MAT1-1-1 |
| IFM 59923 = CCF 4569 (08-SA-2-1) | China, soil, 2008 | MAT1-1-1 |
| FRR 1266 = CBS 121595 = DTO 019-F2 = CCF 4574 | Australia, New South Wales, Warrumbungle National Park, sandy soil, 1971 | MAT1-1-1 |
| A. aureolus | ||
| IFM 47021T = IFM 46935T = IFM 53589T = CBS 105.55T = NRRL 2244T = IMI 06145T = KACC 41204T = KACC 41095T = CCF 4644T = CCF 4646T = CCF 4648T | Ghana, Tafo, soil, 1950 | homothallic |
| IFM 46584 = IFM 46936 = CBM-FA-0692 = CCF 4645 = CCF 4647 | Brazil, São Paulo State, Botucatú, soil, 1993 | homothallic |
| IFM 53615 = CBM-FA-934 = CCF 4571 (ex-type of A. indohii) | Brazil, Acre, Cruzeiro do Sul, soil in a grassland in a tropical rain forest, 2001 | homothallic |
| IHEM 22515 (RV 71215) | Peru, Lima, human cornea, < 1995 | homothallic |
| A. felis | ||
| CBS 130245T = DTO 131-F4T = CCF 5620 | Australia, Sydney, retrobulbar mass, sino-orbital aspergillosis in a 3.5-year-old DSH cat, MN, 2008 | MAT1-2-1 (KC797620) |
| NRRL 62900 = CM-3147 = CCF 4895 (ex-type of A. parafelis) | Spain, human oropharyngeal exudate, 2004 | MAT1-2-1 (KJ858505) |
| NRRL 62903 = CM-6087 = CCF 4897 (ex-type of A. pseudofelis) | Spain, human sputum, 2010 | MAT1-2-1 (KJ858507) |
| NRRL 62901 = CM-5623 = CCF 4896 = CCF 4557 (Viridi-Pinh) | Portugal, bronchoalveolar lavage, chronic invasive aspergillosis in a 56-year-old male, 2007 | MAT1-1-1 (KJ858506) |
| IFM 59564 = CCF 5612 | Japan, human, sputum, 2011 | MAT1-2-1 |
| IFM 60053 = CCF 4559 | Japan, abscess near thigh bone, 40-year-old man with osteomyelitis, 2012 | MAT1-1-1 (HF937392) |
| IFM 54303 = CCF 4570 | Japan, human, clinical material, < 2007 | MAT1-1-1 |
| FRR 5679 = CCF 5613 (MK246) | Australia, thoracic mass in a cat, < 2005 | MAT1-2-1 |
| FRR 5680 = CCF 5615 (MK284) | Australia, retrobulbar mass, sino-orbital aspergillosis in a cat, < 2005 | MAT1-2-1 |
| CCF 2937 | Czech Republic, near Kladno, soil of spoil-bank, 1993 | MAT1-2-1 (LT796767) |
| CCF 4002 (AK 196/07) | Czech Republic, Markovičky, near Kutná Hora, old silver mine waste dump, 2007 | MAT1-2-1 |
| CCF 4003 (AK 27/07) | Czech Republic, Chvaletice, soil crust, abandoned tailing pond, 2007 | MAT1-2-1 |
| CCF 4171 = CMF ISB 2162 = IFM 60852 (F39) | USA, Wyoming, Glenrock, soil from coal mine dump, 2010 | MAT1-2-1 (LT796766) |
| CCF 4172 (F47) | Spain, Andalusia, Aracena, Gruta de la Maravillas, cave air, 2010 | ND |
| CCF 4148 = CMF ISB 1975 = IFM 60868 (F22) | USA, Wyoming, Glenrock, soil from coal mine dump, 2010 | MAT1-1-1 (LT796760) |
| CCF 4376 (AK 102/11) | Czech Republic, Krušné hory, near Abertamy, soil from old dump, 2011 | MAT1-1-1 |
| CCF 4497 = CMF ISB 1936 (F6) | USA, Wyoming, Glenrock, soil from coal mine dump, 2010 | MAT1-2-1 |
| CCF 4498 = IFM 60853 (F49) | USA, Wyoming, Glenrock, soil from coal mine dump, 2010 | MAT1-2-1 |
| DTO 131-E4 = CCF 5609 (2384/07) | Australia, Brisbane, retrobulbar mass, sino-orbital aspergillosis, 7-year-old DSH cat, FN, 2007 | MAT1-2-1 (KC797622) |
| DTO 131-E5 = CCF 5610 (4091/09) | Australia, Brisbane, retrobulbar mass, sino-orbital aspergillosis, 3-year-old Himalayan cat, FN, 2009 | MAT1-1-1 (KC797627) |
| DTO 131-G1 = CCF 5611 (834/07) | Australia, Sydney, retrobulbar mass, sino-orbital aspergillosis, 2-year-old Himalayan cat, MN, 2007 | MAT1-2-1 (KC797625) |
| CCF 5614 (14/4138) | Australia, Sydney, retrobulbar mass, sino-orbital aspergillosis, 5-year-old cat, Ragoll, MN, 2013 | ND |
| CCF 5616 (Felix H. D) | Australia, Canberra, retrobulbar mass, sino-orbital aspergillosis, 8-year-old domestic longhair cat | ND |
| DTO 131-F1 = CCF 5617 (66/10) | Australia, Brisbane, retrobulbar mass, sino-orbital aspergillosis, 5-year-old DSH cat, FN, 2010 | MAT1-1-1 (KC797629) |
| CCF 5618 (Luigi C.) | Australia, Sydney, retrobulbar mass, sino-orbital aspergillosis, 2-year-old BSH cat, MN, 2012 | MAT1-2-1 |
| CBS 130248 = DTO 131-G3 = CCF 5619 (1767/10) | Australia, Brisbane, retrobulbar mass, sino-orbital aspergillosis, 4-year-old DSH cat, FN, 2010 | MAT1-2-1 (KC797621) |
| CBS 130249 = DTO 155-G3 = CCF 5621 (1207/05) | Australia, Sydney, vitreous humor, disseminated invasive apsergillosis 9-year-old Old English Sheepdog, MN, 2005 | MAT1-2-1 |
| DTO 131-F2 = CCF 5622 (3532/09) | Australia, Brisbane, retrobulbar mass, sino-orbital aspergillosis, 4.5-year-old Ragdoll cat, MN, 2009 | MAT1-2-1 |
| CBS 130247 = DTO 131-G2 = CCF 5623 (1020/07) | Australia, Sydney, retrobulbar mass, sino-orbital aspergillosis, 2-year-old DSH cat, FN, 2007 | MAT1-1-1 (KC797632) |
| DTO 131-E9 = CCF 5624 (1848/08) | Australia, Brisbane, retrobulbar mass, sino-orbital aspergillosis, 1.5-year-old DSH cat, MN, 2008 | MAT1-1-1 (KC797628) |
| DTO 131-E3 = CCF 5625 (3008/08 D) | Australia, Brisbane, retrobulbar mass, sino-orbital aspergillosis, 8-year-old Persian cat, FN, 2008 | MAT1-1-1 (KC797634) |
| DTO 131-F6 = CCF 5626 (8651/09) | Australia, Brisbane, retrobulbar mass, sino-orbital aspergillosis, 8-year-old DSH cat, MN, 2009 | MAT1-2-1 (KC797624) |
| CBS 130244 = DTO 131-E6 = CCF 5627 (4067/09D) | Australia, Sydney, retrobulbar mass, sino-orbital aspergillosis, 5-year-old Cornish Rex cat, FN, 2009 | MAT1-1-1 (KC797630) |
| DTO 131-F3 = CCF 5628 (2188/08) | Australia, Brisbane, retrobulbar mass, sino-orbital aspergillosis, 7-year-old DSH cat, FN, 2008 | MAT1-2-1 |
| CBS 130246 = DTO 131-F9 = CCF 5629 (448/08) | Australia, Sydney, nasal cavity, sino-nasal aspergillosis 13-year-old DLH cat, MN, 2008 | MAT1-1-1 (KC797631) |
| A. frankstonensis | ||
| CBS 142233T = IBT 34172T = DTO 341-E7T = CCF 5799T | Australia, Victoria, Frankston, woodland soil, 2015 | MAT1-2-1 |
| CBS 142234 = IBT 34204 = DTO 341-F3 = CCF 5798 | Australia, Victoria, Frankston, woodland soil, 2015 | MAT1-2-1 |
| A. pseudoviridinutans | ||
| NRRL 62904T = CCF 5631 (NIHAV1, 1720) | USA, U.S. National Institutes of Health, mediastinal lymph node, 14-year-old boy with chronic granulomatous disease, 2004 | MAT1-1-1 (KJ858509) |
| CBS 458.75 = KACC 41203 = IHEM 9862 (ex-type of A. fumigatus var. sclerotiorum) | India, Lucknow, Mohanlalganj, soil, < 1971 | MAT1-2-1 |
| IMI 182127 = KACC 41614 = CCF 5630 | Srí Lanka, Pinus caribea, < 1974 | MAT1-2-1 |
| IFM 55266 = CCF 5644 | Japan, human, lung, 2004 | MAT1-1-1 |
| IFM 57289 = CCF 4665 | Brazil, Mato Grosso, soil | MAT1-2-1 |
| IFM 59502 = CCF 4561 | Japan, cornea, keratomycosis, 26-year-old woman, 2011 | MAT1-1-1 |
| IFM 59503 = CCF 4562 | Japan, cornea, keratomycosis, 26-year-old woman, 2011 | MAT1-1-1 |
| CCF 5632 (NIHAV2, 2594) | USA, lung biopsy, 8-year-old boy with hyperimmunoglobulin-E syndrome, 2004 | MAT1-1-1 (LT796761) |
| A. siamensis | ||
| IFM 59793T = KUFC 6349T = CCF 4685T | Thailand, Chonburi Province, Samaesarn Island, coastal forest soil, 2008 | homothallic |
| IFM 61157 = KUFC 6397 = CCF 4686 | Thailand, Chiang Mai, termite nest soil, 2009 | homothallic |
| A. udagawae | ||
| IFM 46972T = CBS 114217T = DTO 157-D7T = CBM-FA 0702T = KACC 41155T = CCF 4558T | Brazil, São Paulo State, Botucatú, Lagoa Seka Avea, plantation soil, 1993 | MAT1-1-1 |
| IFM 46973 = CBS 114218 = DTO 157-D8 = CBM-FA 0703 = KACC 41156 = CCF 5672 | Brazil, São Paulo State, Botucatú, Lagoa Seka Avea, plantation soil, 1993 | MAT1-2-1 |
| IFM 5058 = CCF 4662 | Japan, human, eye | MAT1-1-1 |
| IFM 51744 = CCF 4671 | Japan, human, clinical material, 2002 | MAT1-1-1 |
| IFM 53868 = CCF 4667 | Japan, human, clinical material, 2004 | MAT1-2-1 |
| IFM 54131 = CBM-FA-0697 = CCF 4663 | China, Shaanxi, soil, 1994 | MAT1-1-1 |
| IFM 54132 = CBM-FA-0698 = CCF 4664 | China, Shaanxi, soil, 1994 | MAT1-2-1 |
| IFM 54745 = CBM-FA-694 = CCF 4661 | China, Shaanxi, soil, 1994 | MAT1-1-1 |
| IFM 55207 = NBRC 31952 = CCF 4660 | Russia, soil, 1985 | MAT1-2-1 |
| IFM 62155 = CCF 4668 | Brazil, soil, 2008 | MAT1-1-1 |
| CCF 4475 (F2) | USA, Wyoming, Glenrock, prairie soil, 2010 | MAT1-2-1 |
| CCF 4476 (F32) | USA, Wyoming, Glenrock, soil, mine waste dump, 2010 | MAT1-1-1 |
| CCF 4478 = CMF ISB 2193 (F66) | USA, Wyoming, Gilette, soil, mine waste dump, 2011 | MAT1-2-1 |
| CCF 4479 = CMF ISB 2189 (F70) | USA, Illinois, soil, mine waste dump, 2011 | MAT1-2-1 |
| CCF 4481 = CMF ISB 2191 (F83) | USA, Wyoming, Gilette, soil, mine waste dump, 2011 | MAT1-2-1 |
| CCF 4491 = CMF ISB 1971 (F3) | USA, Wyoming, Glenrock, prairie soil, 2010 | MAT1-2-1 |
| CCF 4492 (F21) | USA, Wyoming, Glenrock, soil, mine waste dump, 2010 | MAT1-2-1 (HF937389) |
| CCF 4494 (F44) | USA, Wyoming, Glenrock, prairie soil, 2010 | MAT1-2-1 |
| CMF ISB 1972 = CCF 4502 (F11) | USA, Wyoming, Glenrock, soil, mine waste dump, 2010 | MAT1-2-1 |
| CMF ISB 2190 = CCF 5635 (F76) | USA, Indiana, soil, mine waste dump, 2011 | MAT1-1-1 |
| CMF ISB 2509 = CCF 5636 (F20) | USA, Wyoming, Glenrock, soil, mine waste dump, 2010 | MAT1-2-1 |
| CCF 5637 (F37) | USA, Wyoming, Gilette, soil, mine waste dump, 2008 | MAT1-1-1 |
| CCF 5638 (3C8) | USA, Philadelphia, retrobulbar mass, sino-orbital aspergillosis, 4-year-old Persian cat, MN, 2012 | MAT1-1-1 |
| DTO 166-D6 = CCF 5639 (11.3356, Milo) | Australia, Sydney, retrobulbar mass, sino-orbital aspergillosis 2-year-old DSH cat, MN, 2011 | ND |
| CCF 5634 (B3) | Czech Republic, Hostěradice, earthworm casts, 2012 | MAT1-2-1 |
| A. viridinutans | ||
| IFM 47045T = IFM 47046T = IMI 367415T = IMI 062875T = NRRL 4365T = NRRL 576T = CBS 127.56T = KACC 41142T = CCF 4382T = CCF 4568T | Australia, Victoria, Frankston, rabbit dung, 1954 | MAT1-1-1 (HF937390) |
| A. wyomingensis | ||
| CCF 4417T = CMF ISB 2494T = CBS 135456T (F30) | USA, Wyoming, Glenrock, soil, mine waste dump, 2010 | MAT1-1-1 (HF937391) |
| CCF 4169 = CMF ISB 2486 (F24) | USA, Wyoming, Glenrock, soil, 2010 | MAT1-1-1 |
| CCF 4170 = CMF ISB 2485 (F12) | USA, Wyoming, Glenrock, soil, 2010 | MAT1-2-1 (LT796765) |
| CCF 4411 = CMF ISB 1977 = IFM 60854 (F5) | USA, Wyoming, Glenrock, soil, mine waste dump, 2010 | MAT1-2-1 |
| CCF 4412 (F9) | USA, Wyoming, Glenrock, soil, mine waste dump, 2010 | MAT1-1-1 |
| CCF 4413 = CMF ISB 2317 (F10) | USA, Wyoming, Glenrock, soil, mine waste dump, 2010 | MAT1-1-1 |
| CCF 4414 = CMF ISB 1974 = IFM 60856 (F13) | USA, Wyoming, Glenrock, soil, mine waste dump, 2010 | MAT1-1-1 (LT796762) |
| CCF 4415 = CMF ISB 2487 (F28) | USA, Wyoming, Glenrock, soil, mine waste dump, 2010 | MAT1-1-1 |
| CCF 4416 = CMF ISB 1976 = CBS 135455 (F29) | USA, Wyoming, Glenrock, soil, mine waste dump, 2010 | MAT1-2-1 (HF937388) |
| CCF 4418 = CMF ISB 2162 = IFM 60855 (F31) | USA, Wyoming, Glenrock, soil, mine waste dump, 2010 | MAT1-2-1 |
| CCF 4419 = CMF ISB 2495 (F53) | USA, Wyoming, Glenrock, soil, mine waste dump, 2010 | MAT1-2-1 |
| CCF 4420 = CMF ISB 2491 (F60) | USA, Wyoming, Glenrock, soil, mine waste dump, 2010 | MAT1-1-1 |
| IMI 133982 = CCF 4383 | Russia, Moscow, soil, < 1968 | MAT1-1-1 (LT796763) |
| IFM 59681 = CCF 4563 | China, Urumqi, soil, 2008 | MAT1-2-1 (LT796764) |
| DTO 155-G2 = CCF 5640 (Yogurt R.) | Australia, Melbourne, retrobulbar mass in a 1.5-year-old BSH cat, MN, 2010 | MAT1-2-1 |
| outgroup | ||
| A. lentulus NRRL 35552T = CBS 117885T = IBT 27201T = KACC 41940T | USA, human, clinical material | MAT1-2-1 |
1Culture collection acronyms: CBM-FA = Natural History Museum & Institute, Chiba, Japan; CBS = CBS culture collection housed at the Westerdijk Institute, Utrecht, The Netherlands; CCF = Culture Collection of Fungi, Prague, Czech Republic; CM = Filamentous fungus collection of the Spanish National Center for Microbiology, Madrid, Spain; CMF ISB = Collection of Microscopic Fungi, Institute of Soil Biology, Academy of Sciences of the Czech Republic, České Budějovice, Czech Republic; DTO = working collection of the Applied and Industrial Mycology department housed at the Westerdijk Institute, Utrecht, The Netherlands; FRR = Food Fungal Culture Collection, North Ryde, Australia; IBT = culture collection of the DTU Systems Biology, Lyngby, Denmark; IFM = Collection at the Medical Mycology Research Centre, Chiba University, Japan; IHEM = Belgian Coordinated Collections of Micro-organisms (BCCM/IHEM), Brussels, Belgium; IMI = CABI's collection of fungi and bacteria, Egham, UK; JCM = Japan Collection of Microorganisms, Tsukuba, Japan; KACC = Korean Agricultural Culture Collection, Wanju, South Korea; KUFC = Kasetsart University Fungal Collection, Bangkok, Thailand; NBRC (IFO) = Biological Resource Center, National Institute of Technology and Evaluation, Chiba, Japan; NRRL = Agricultural Research Service Culture Collection, Peoria, Illinois, USA.
2Original numbers of strains and personal strain designations are given in parentheses.
3BSH = British shorthair; DLH = domestic longhair; DSH = domestic shorthair; FN = female neutered (desexed); MN = male neutered; ND = not determined.
4When available, sequence number in public database is given in parentheses; in the remaining cases, the MAT idiomorph was confirmed only on the electrophoretogram (specific PCR and length of amplicons).
5Sequences generated in this study are in bold.
Phenotypic studies
The strains were grown on malt extract agar (MEA), Czapek Yeast Autolysate Agar (CYA), Czapek-Dox agar (CZA), yeast extract sucrose agar (YES), CYA supplemented with 20 % sucrose (CY20S), and creatine sucrose agar (CREA), and incubated at 25 °C. Agar media composition was based on that described by Samson et al. (2014). Malt extract and yeast extract were obtained from Oxoid (Basingstoke, UK) and Fluka Chemie GmbH (Switzerland), respectively. Growth at 42, 45 and 47 °C was tested on MEA plates sealed with Parafilm. Colour determination was performed according to the ISCC-NBS (Inter-Society Color Council – National Bureau of Standards) Centroid Colour Charts (Kelly 1964).
Micromorphology was observed on MEA. Lactic acid with cotton blue was used as a mounting medium. Photographs were taken on an Olympus BX-51 microscope (Olympus DP72 camera) using Nomarski contrast. Macromorphology of the colonies was documented using a stereomicroscope Olympus SZ61 (with Olympus Camedia C-5050 Zoom camera) or Canon EOS 500D.
Scanning electron microscopy (SEM) was performed using a JEOL-6380 LV scanning electron microscope (JEOL Ltd. Tokyo, Japan) as described by Hubka et al. (2013b). Briefly, pieces of colony or mature ascomata were fixed in osmium tetroxide vapours for one wk at 5–10 °C and gold coated using a Bal-Tec SCD 050 sputter coater. The specimens were observed using 40 μm spot size and 15–25 kV accelerating voltage.
Molecular studies
ArchivePure DNA yeast and Gram2+ kit (5 PRIME Inc., Gaithersburg, MD) was used for DNA isolation from 7-d-old cultures according to the manufacturer's instructions as updated by Hubka et al. (2015b). The purity and concentration of extracted DNA was evaluated by NanoDrop 1000 Spectrophotometer. ITS rDNA region was amplified using forward primers ITS1 or ITS5 (White et al. 1990) and reverse primers ITS4S (Kretzer et al. 1996) or NL4 (O’Donnell 1993); partial β-tubulin gene (benA) using forward primers Bt2a (Glass & Donaldson 1995) or Ben2f (Hubka & Kolařík 2012) and reverse primer Bt2b (Glass & Donaldson 1995); partial calmodulin gene (CaM) using forward primers CF1M or CF1L and reverse primer CF4 (Peterson 2008); partial actin gene (act) using primers ACT-512F and ACT-783R (Carbone & Kohn 1999); partial RNA polymerase II second largest subunit (RPB2) using forward primers fRPB2-5F (Liu et al. 1999) or RPB2-F50-CanAre (Jurjević et al. 2015) and reverse primer fRPB2-7cR (Liu et al. 1999); partial mcm7 gene encoding minichromosome maintenance factor 7 with primers Mcm7-709for and Mcm7-1348rev (Schmitt et al. 2009); and partial tsr1 gene encoding ribosome biogenesis protein with primers Tsr1-1453for and Tsr1-2308rev (Schmitt et al. 2009). Terminal primers were used for sequencing.
The PCR reaction volume of 20 μL contained 1 μL (50 ng) of DNA, 0.3 μL of both primers (25 pM/mL), 0.2 μL of MyTaqTM DNA Polymerase (Bioline, GmbH, Germany) and 4 μL of 5× MyTaq PCR buffer. The ITS rDNA, benA and CaM fragments were amplified using the following thermal cycle profile: 93 °C/2 min; 30 cycles of 93 °C/30 s; 55 °C/30 s; 72 °C/60 s; 72 °C/10 min. The annealing temperature for amplification of act gene was 60 °C (30 cycles); and that for tsr1 gene 50 °C (37 cycles). Partial RPB2 gene fragments were amplified using the above-mentioned cycle or touchdown thermal-cycling: 93 °C/2 min; 5 cycles of 93 °C/30 s, 65–60 °C/30 s, 72 °C/60 s; 38 cycles of 93 °C/30 s, 55 °C/30 s, 72 °C/60 s; 72 °C/10 min. The partial mcm7 gene was amplified using modified touchdown thermal-cycling: 93 °C/2 min; 5 cycles of 93 °C/30 s, 65–60 °C/30 s, 72 °C/60 s; 38 cycles of 93 °C/30 s, 60 °C/30 s, 72 °C/60 s; 72 °C/10 min. PCR product purification followed the protocol of Réblová et al. (2016). Automated sequencing was performed at Macrogen Sequencing Service (Amsterdam, The Netherlands) using both terminal primers. Sequences were deposited into the ENA (European Nucleotide Archive) database under the accession numbers listed in Table 2.
Table 2.
List of Aspergillus strains and sequences used in phylogenetic analysis; accession numbers in bold were generated for this study.
1Culture collection acronyms: CBM-FA = Natural History Museum & Institute, Chiba, Japan; CBS = CBS culture collection housed at the Westerdijk Institute, Utrecht, The Netherlands; CCF = Culture Collection of Fungi, Prague, Czech Republic; CM = Filamentous fungus collection of the Spanish National Center for Microbiology, Madrid, Spain; CMF ISB = Collection of Microscopic Fungi, Institute of Soil Biology, Academy of Sciences of the Czech Republic, České Budějovice, Czech Republic; DTO = working collection of the Applied and Industrial Mycology department housed at the Westerdijk Institute, Utrecht, The Netherlands; FRR = Food Fungal Culture Collection, North Ride, Australia; IBT = culture collection of the DTU Systems Biology, Lyngby, Denmark; IFM = Collection at the Medical Mycology Research Centre, Chiba University, Japan; IHEM = Belgian Coordinated Collections of Micro-organisms (BCCM/IHEM), Brussels, Belgium; IMI = CABI's collection of fungi and bacteria, Egham, UK; JCM = Japan Collection of Microorganisms, Tsukuba, Japan; KACC = Korean Agricultural Culture Collection, Wanju, South Korea; KUFC = Kasetsart University Fungal Collection, Bangkok, Thailand; NBRC (IFO) = Biological Resource Center, National Institute of Technology and Evaluation, Chiba, Japan; NRRL = Agricultural Research Service Culture Collection, Peoria, Illinois, USA.
Phylogenetic analysis
Sequences were inspected and assembled using Bioedit v. 7.2.5 (www.mbio.ncsu.edu/BioEdit/bioedit.html). Alignments of the benA, CaM, act and RPB2 regions were performed using the G-INS-i option implemented in MAFFT v. 7 (Katoh & Standley 2013). Alignments were trimmed, concatenated and then analysed using Maximum likelihood (ML) and Bayesian inference (BI) analyses. Suitable partitioning scheme and substitution models (Bayesian information criterion) for analyses were selected using the greedy algorithm implemented in PartitionFinder v. 1.1.1 (Lanfear et al. 2017) with settings allowing introns, exons and codon positions to be independent partitions. Proposed partitioning schemes and substitution models for each dataset are listed in Table 3. The alignment characteristics are listed in Table 4.
Table 3.
Partition-merging results and best substitution model for each partition according to Bayesian information criterion (BIC) as proposed by PartitionFinder v. 1.1.0. for combined dataset of benA, CaM, act and RPB2 genes.
| Dataset | Phylogenetic method | Partitioning scheme (substitution model) |
|---|---|---|
| Section Fumigati (Fig. 1) | Maximum likelihood | benA + CaM + act introns (TrNef+G); 3rd codon positions of benA (GTR+G); 1st codon positions of benA + CaM + act + RPB2 + 2nd codon positions of act + 3rd codon positions of act (TIM+I); 2nd codon positions of benA + CaM + RPB2 (HKY); 3rd codon positions of CaM + RPB2 (HKY+G) |
| Bayesian inference | benA + CaM + act introns (K80+G); 3rd codon positions of benA (GTR+G); 1st codon positions of benA + CaM + act + RPB2 + 2nd codon positions of act + 3rd codon positions of act (GTR+I); 2nd codon positions of benA + CaM + RPB2 (HKY); 3rd codon positions of CaM + RPB2 (HKY+G) | |
| A. viridinutans clade (Fig. 5) | Maximum likelihood | benA + CaM + act introns (K80+G); 3rd codon positions of benA + CaM + RPB2 (TrN+G); 1st codon positions of benA + CaM + act + RPB2 + 3rd codon positions of act (TrN); 2nd codon positions of benA + CaM + act + RPB2 (F81) |
| Bayesian inference | benA + CaM + act introns (K80+G); 3rd codon positions of benA + CaM + RPB2 (HKY+G); 1st codon positions of benA + CaM + act + RPB2 + 3rd codon positions of act (HKY); 2nd codon positions of benA + CaM + act + RPB2 (F81) |
Table 4.
Overview of alignments characteristics used for phylogenetic analyses.
| Alignment characteristic | benA | CaM | act | RPB2 | mcm7 | tsr1 | Combined dataset |
|---|---|---|---|---|---|---|---|
| Section Fumigati (Fig. 1) | |||||||
| Length (bp) | 534 | 697 | 431 | 999 | – | – | 2661 |
| Variable position | 268 | 322 | 234 | 280 | – | – | 1104 |
| Parsimony informative sites | 184 | 226 | 148 | 186 | – | – | 744 |
| A. viridinutans complex (Fig. 5) | |||||||
| Length (bp) | 475 | 697 | 344 | 967 | – | – | 2483 |
| Variable position | 115 | 168 | 102 | 135 | – | – | 520 |
| Parsimony informative sites | 84 | 114 | 70 | 81 | – | – | 349 |
| A. felis clade (Fig. 3) | |||||||
| Length (bp) | 474 | 681 | 329 | 967 | 623 | 761 | 3835 |
| Variable position | 72 | 73 | 35 | 59 | 38 | 103 | 380 |
| Parsimony informative sites | 50 | 49 | 18 | 32 | 24 | 58 | 231 |
The ML tree was constructed with IQ-TREE v. 1.4.4 (Nguyen et al. 2015) with nodal support determined by non-parametric bootstrapping (BS) with 1 000 replicates. Bayesian posterior probabilities (PP) were calculated using MrBayes v. 3.2.6 (Ronquist et al. 2012). The analyses ran for 107 generations, two parallel runs with four chains each were used, every 1 000th tree was retained, and the first 25 % of trees were discarded as burn-in. The trees were rooted with Aspergillus clavatus NRRL 1 and A. lentulus NRRL 35552, respectively. All alignments are available from the Dryad Digital Repository (https://doi.org/10.5061/dryad.38889).
Species delimitation and species tree inference
Several species delimitation methods were applied to elucidate the species boundaries within the AVSC. We followed the recommendation of Carstens et al. (2013) and compared the results of several different methods. The analysis was divided into two parts. Four genetic loci were examined in the first analysis which comprised all species from the AVSC while six genetic loci were examined in the second analysis focused on the clade comprising Aspergillus felis, A. pseudofelis, A. parafelis and A. pseudoviridinutans (A. aureolus was used as an outgroup). The alignment characteristics are listed in Table 4.
Only unique nucleotide sequences, selected with DAMBE v. 6.4.11 (Xia 2017) were used in the analyses. Nucleotide substitution models for particular loci were determined using jModeltest v. 2.1.7 (Posada 2008) based on Bayesian information criterion (BIC) and were as follows: 1st analysis - K80+G (benA), K80+I (CaM), K80+G (act), K80+G (RPB2); 2nd analysis - K80+I (benA), K80+G (CaM), K80 (act), K80 (RPB2), HKY+I+G (tsr1), K80 (mcm7).
In the first analysis, only unique sequences of four loci were used, i.e., benA, CaM, act and RPB2. The number of isolates of A. felis and A. pseudoviridinutans was reduced to two, because this clade was examined in detail in the second analysis based on six loci. Three single-locus species delimitation methods, i.e., bGMYC (Reid & Carstens 2012), GMYC (Fujisawa & Barraclough 2013) and PTP (Zhang et al. 2013), and one multilocus species delimitation method STACEY (Jones 2017) were used to find putative species boundaries. The bGMYC and GMYC methods require ultrametric trees as an input, while PTP does not. Therefore, single locus ultrametric trees were constructed using a Bayesian approach in BEAST v. 2.4.5 (Bouckaert et al. 2014) with both Yule and coalescent tree models. We also looked at possible differences between strict and relaxed clock models, but since these parameters had no effect on the number of delimited species, only the results with strict clock model are presented here. Chain length for each tree was 1 × 107 generations with 25 % burn-in. The highest credibility tree was used for the GMYC method and 100 trees randomly sampled throughout the analysis were used for the bGMYC method. Both methods were performed in R v. 3.3.4 (R Core Team 2015) using bgmyc (Reid & Carstens 2012) and splits (SPecies’ LImits by Threshold Statistics) (Fujisawa & Barraclough 2013) packages. The non-ultrametric trees for the PTP method were constructed using the ML approach in RAxML v. 7.7.1 (Stamatakis et al. 2008) and IQ-TREE v. 1.5.3 (Nguyen et al. 2015) with 1 000 bootstrap replicates. The PTP method was performed on the web server http://mptp.h-its.org/ (Kapli et al. 2017) with p-value set to 0.001. The multilocus species delimitation was performed in BEAST v. 2.4.5 with add-on STACEY v. 1.2.2 (Jones 2017). The chain length was set to 5 × 108 generations, priors were set as follows: the species tree prior was set to the Yule model, growth rate prior was set to lognormal distribution (M = 5, S = 2), clock rate priors for all loci were set to lognormal distribution (M = 0, S = 1), PopPriorScale prior was set to lognormal distribution (M = -7, S = 2) and relativeDeathRate prior was set to beta distribution (α = 1, β = 1 000). The output was processed with SpeciesDelimitationAnalyzer (Jones 2017).
The species tree was inferred using *BEAST (Heled & Drummond 2010) implemented in BEAST v. 2.4.5. The isolates were assigned to a putative species according to the results of the above-mentioned species delimitation methods. The MCMC analysis ran for 1 × 108 generations, 25 % of trees were discarded as a burn-in. The strict molecular clock was chosen for all loci and population function was set as constant. Convergence was assessed by examining the likelihood plots in Tracer v. 1.6 (http://tree.bio.ed.ac.uk/software/tracer). We also constructed the phylogenetic tree based on concatenated alignment of all four loci in IQ-TREE v. 1.5.3 with 1 000 bootstrap replicates and the optimal partitioning scheme determined by PartitionFinder v. 2.1.1 (Lanfear et al. 2017).
The validation of the species hypotheses was performed in BP&P v. 3.3 (Bayesian phylogenetics and phylogeography) (Yang & Rannala 2010). The isolates were assigned to the species based on the results of species delimitation methods and the species tree inferred with *BEAST was used as a guide tree. Three different combinations of the prior distributions of the parameters θ (ancestral population size) and τ0 (root age) were tested as proposed by Leaché & Fujita (2010), i.e., large ancestral population sizes and deep divergence: θ ~ G (1, 10) and τ0 ~ G (1, 10); small ancestral population sizes and shallow divergences among species: θ ~ G (2, 2000) and τ0 ~ G (2, 2000); large ancestral populations sizes and shallow divergences among species: θ ~ G (1, 10) and τ0 ~ G (2, 2000).
The second analysis with six protein-coding loci, i.e., benA, CaM, act, RPB2, mcm7 and tsr1, consisted of the same steps as described above. Instead of PTP, we used the programme mPTP (Kapli et al. 2017) with IQ-TREE and RAxML trees as an input. Within the mPTP programme we used the following settings: Maximum likelihood species delimitation inference (option ML) and a different coalescent rate for each delimited species (option multi). R package ggtree (Yu et al. 2017) and the programme densitree (Bouckaert 2010) were used for visualization of the phylogenetic trees.
Mating experiments
The MAT idiomorph was determined using the primer pairs alpha1 and alpha2 located in MAT1-1-1 locus (alpha box domain), and HMG1 and HMG2 primers located in MAT1-2-1 locus (high-mobility-group domain) as described by Sugui et al. (2010). The MAT idiomorphs were differentiated based on the different lengths of PCR products visualized by gel electrophoresis; absence of opposite MAT idiomorph was also verified in all isolates. The identity of PCR products was proved by DNA sequencing in several isolates (accession numbers in Table 1); product purification and sequencing were performed at Macrogen Europe (Amsterdam, The Netherlands) using terminal primers. Selected opposite mating type strains were paired within and between major phylogenetic clades on MEA and oatmeal agar (OA; Difco, La Ponte de Claix, France) plates and incubated at 25, 30 and 37 °C in the dark. The plates were sealed with Parafilm and examined weekly from the third wk of cultivation for two months under a stereomicroscope for the production of ascomata. The presence of ascospores was determined using light microscopy. Width and height of ascospores were recorded at least 35 times for each successful mating pair.
Statistical analysis
Statistical differences in the width and height of the ascospores of particular species and interspecific hybrids were tested with one-way ANOVA followed by Tukey's HSD (honest significant difference) post hoc test in R v. 3.3.4 (R Core Team 2015). R package multcomp (Hothorn et al. 2008) was used for the calculation and package ggplot2 (Wickham 2009) for visualization of the results.
Exometabolite analysis
The extracts were prepared according to Houbraken et al. (2012). High-performance liquid chromatography with diode-array detection was performed according to Frisvad & Thrane (1987, 1993) as updated by Nielsen et al. (Nielsen et al. 2011). Fungi were incubated for 1 wk at 25 °C in darkness on CYA and yeast extract sucrose (YES) agars for exometabolite analysis.
RESULTS
Phylogenetic definition of AVSC
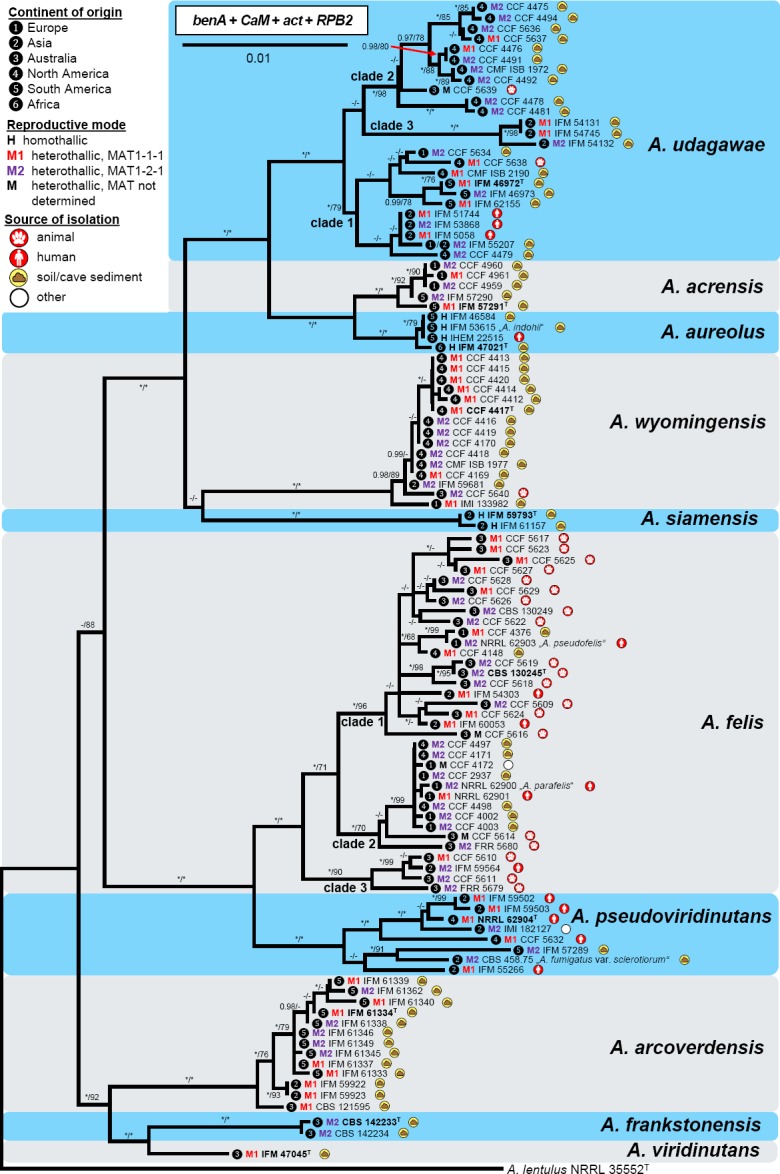
In the phylogenetic analysis, 76 combined benA, CaM, act and RPB2 sequences were assessed for members of sect. Fumigati. The analysis was based on the modified alignment previously used by Hubka et al. (2017) and enriched by taxa from AVSC. In the Bayesian tree shown in Fig. 1, members of sect. Fumigati are resolved in several monophyletic clades. The analysis showed that AVSC is a phylogenetically well-defined group and the clade gained full support. Similarly, some other clades are well-supported by both BI and ML analyses including A. spinosus clade, A. brevipes clade, A. tatenoi clade, A. thermomutatus clade and A. fennelliae clade; A. spathulatus forms a single-species lineage distantly related to other clades. Other clades have moderate or low support and the species represented therein may differ based on genetic loci used for phylogenetic reconstruction and taxa included in the analysis. Heterothallic species are dispersed across sect. Fumigati (Fig. 1) but the majority of them cluster in AVSC and A. fumigatus clades. These two clades also encompass the highest number of human and animal pathogens in sect. Fumigati not only in terms of their number but also their clinical relevance.
Fig. 1.
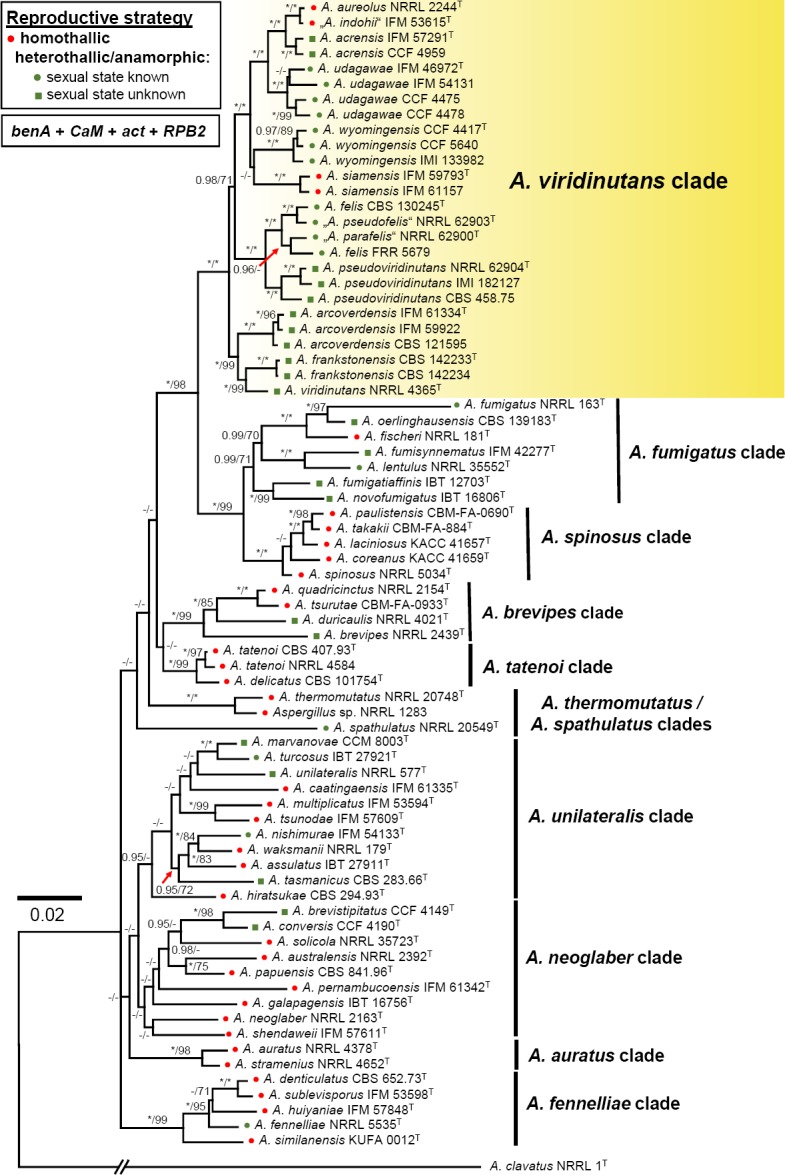
Phylogenetic relationships of the sect. Fumigati members inferred from Bayesian analysis of the combined, 4-gene dataset of β-tubulin (benA), calmodulin (CaM), actin (act) and RNA polymerase II second largest subunit (RPB2) genes. Bayesian posterior probabilities (PP) and Maximum likelihood bootstrap supports (BS) are appended to nodes; only PP ≥ 95 % and BS ≥ 70 % are shown; lower supports are indicated with a hyphen, whereas asterisks indicate full support (1.00 PP or 100 % BS); ex-type strains are designated by a superscript T; species names in quotes are considered synonyms; the bar indicates the number of substitutions per site. The tree is rooted with Aspergillus clavatus NRRL 1T. The reproductive mode of each species is designated by icons before the species name (see legend).
Species delimitation and validation in AVSC
In the first analysis, four genetic loci were examined across species of AVSC, isolates of A. felis and its close relatives were reduced to two individuals, because a separate analysis based on six loci was performed for this clade. Eleven tentative species were delimited in AVSC using STACEY. The results are summarised in Fig. 2, the differences in the colour of the tree branches reflect species delimited by the analysis. The analysis supported recognition of three putative species in A. udagawae lineage, delimitation of A. acrensis (described below) from A. aureolus was not supported, other AVSC species were supported by STACEY without differences from their current concept.
Fig. 2.
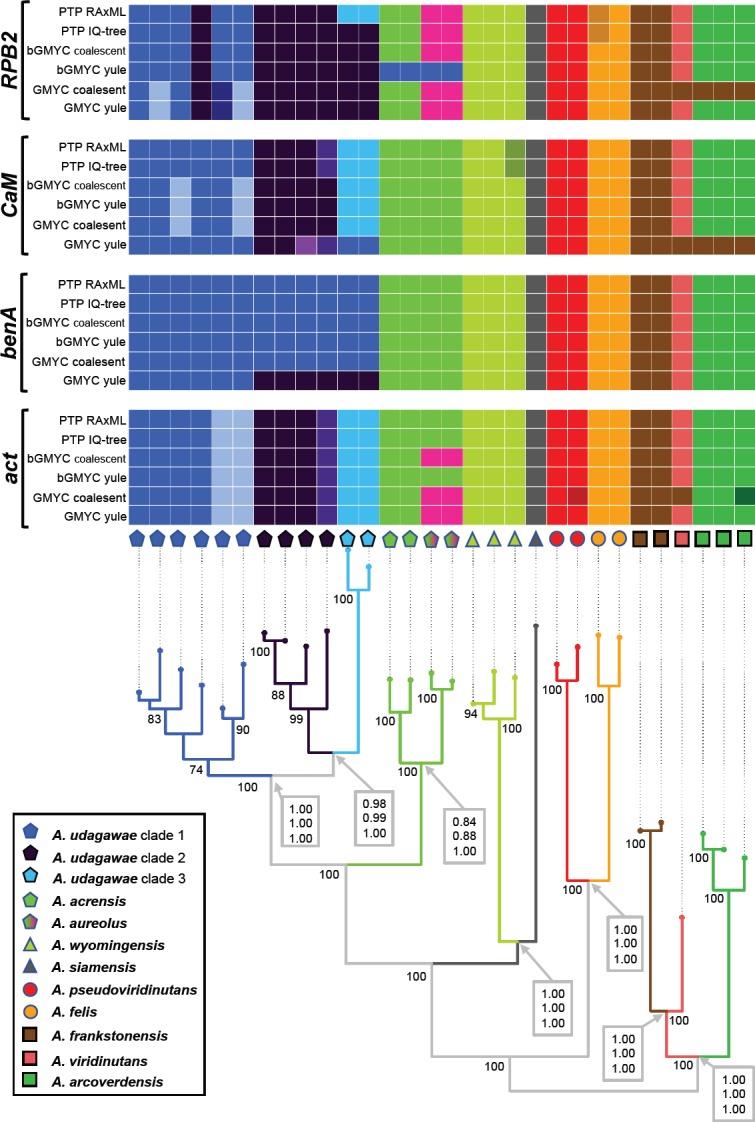
Schematic representation of results of species delimitation methods in Aspergillus viridinutans species complex based on four genetic loci. The results of multilocus method (STACEY) are compared to results of single-locus methods (PTP, bGMYC, GMYC). The results of STACEY are shown as tree branches with different colours, while the results of single-locus methods are depicted with coloured bars highlighting congruence across methods. The displayed tree is derived from IQ-TREE analysis based on a concatenated dataset and is used solely for the comprehensive presentation of the results from different methods. The species validation analysis results (BP&P) are appended to nodes and shown in grey bordered boxes; the values represent posterior probabilities calculated in three scenarios having different prior distributions of parameters θ (ancestral population size) and τ0 (root age). The top value represents the results of analysis with large ancestral population sizes and deep divergence: θ ~ G (1, 10) and τ0 ~ G (1, 10); the middle value represents the results of analysis with large ancestral populations sizes and shallow divergences among species: θ ~ G (1, 10) and τ0 ~ G (2, 2000); and the bottom value small ancestral population sizes and shallow divergences among species: θ ~ G (2, 2000) and τ0 ~ G (2, 2000).
The results derived from STACEY were compared to those from three single-locus species delimitation methods. The consensual results from single-locus species delimitation methods are generally in agreement with the results of STACEY for the majority of species but vary greatly for A. udagawae, A. aureolus and A. acrensis lineages (Fig. 2). Recognition of three putative species in A. udagawae lineage was supported only based on the CaM locus, while based on benA locus, none of these three sublineages gained support. Various delimitation schemes were proposed by different single-locus species delimitation methods in the A. udagawae lineage based on the RPB2 gene (results even varied between the analyses based on different input trees for the PTP and GMYC methods), while five putative species were identically delimited based on the act locus. The methods relatively consistently supported delimitation of the A. acrensis lineage based on the RPB2 locus and similarly, bGMYC and GMYC methods supported this species based on the act locus. In contrast, lineages of A. acrensis and A. aureolus were not split by any method when analyzing benA and CaM loci.
The species validation analysis results are appended to nodes of the tree in Fig. 2. A reasonable support is defined by posterior probabilities ≥ 0.95 under all three scenarios simulated by different prior distributions of parameters θ (ancestral population size) and τ0 (root age). Delimitation of all putative species (those delimited by STACEY, A. acrensis and A. aureolus) were supported by the posterior probability 0.98 or higher based on the analysis in BP&P v. 3.1 (Yang & Rannala 2010) under all three scenarios. The only exception was lower support for splitting of A. acrensis and A. aureolus; this scenario was supported by the posterior probabilities 0.84, 0.88, 1.00, respectively.
Species delimitation and validation in A. felis clade and its relatives
In the second analysis, six genetic loci were examined across isolates of A. felis, A. parafelis, A. pseudofelis and A. pseudoviridinutans. Only two tentative species, A. felis and A. pseudoviridinutans, were delimited in this clade using STACEY. The results are shown as branches designated by different colours in Fig. 3. The analysis did not support separation of A. pseudofelis and A. parafelis from A. felis; A. fumigatus var. sclerotiorum is included in the lineage of A. pseudoviridinutans.
Fig. 3.
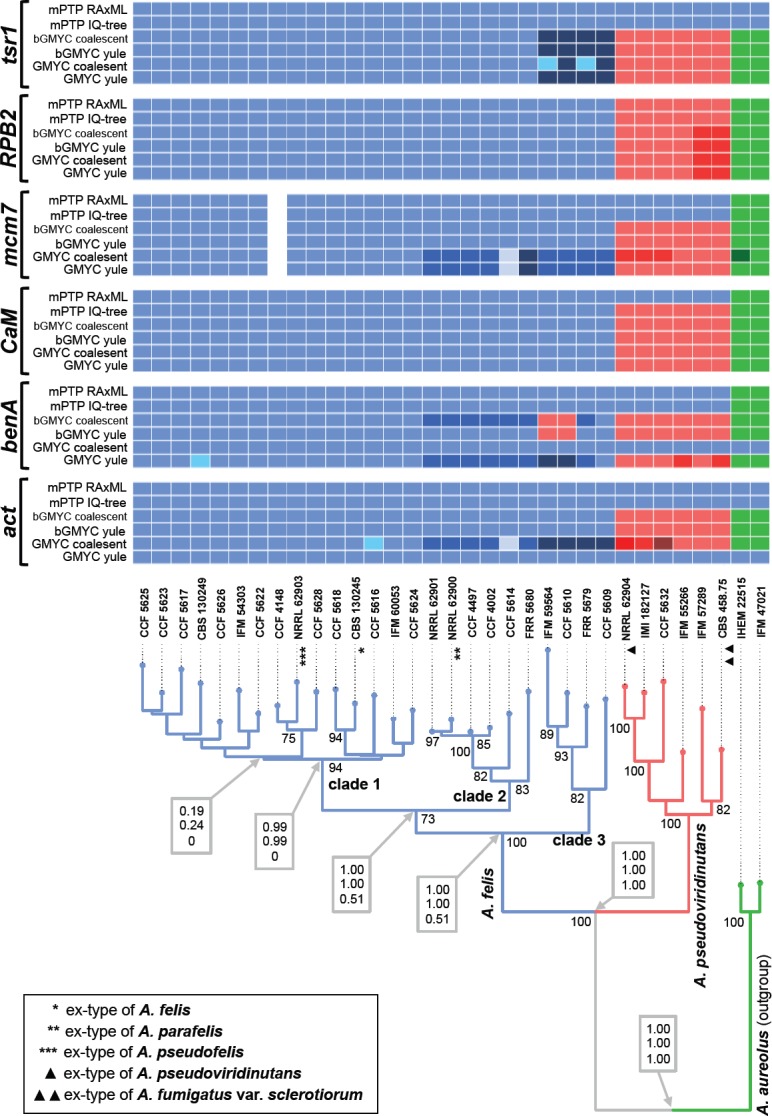
Schematic representation of results of species delimitation methods in Aspergillus felis clade based on six genetic loci. The results of multilocus method (STACEY) are compared to results of single-locus methods (mPTP, bGMYC, GMYC). The results of STACEY are shown as tree branches with different colours, while the results of single-locus methods are depicted with coloured bars highlighting congruence across methods. The displayed tree is derived from IQ-TREE analysis based on a concatenated dataset and is used solely for the comprehensive presentation of the results from different methods. The species validation analysis results (BP&P) are appended to nodes and shown in grey bordered boxes; the values represent posterior probabilities calculated in three scenarios having different prior distributions of parameters θ (ancestral population size) and τ0 (root age). The top value represents the results of analysis with large ancestral population sizes and deep divergence: θ ~ G (1, 10) and τ0 ~ G (1, 10); the middle value represents the results of analysis with large ancestral populations sizes and shallow divergences among species: θ ~ G (1, 10) and τ0 ~ G (2, 2000); and the bottom value small ancestral population sizes and shallow divergences among species: θ ~ G (2, 2000) and τ0 ~ G (2, 2000).
The results of three single-locus species delimitation methods were compared to those from STACEY, and the consensual results showed a general agreement (Fig. 3). Delimitation of A. pseudofelis from A. felis was not supported by any of the used methods. Only a negligible number of analyses supported delimitation of basal clades in A. felis as tentative species (designated as clade 2 and 3 in Fig. 3). But even in these minority scenarios, there were no clear consensual delimitation patterns that would support delimitation of A. parafelis. Interestingly, mPTP analysis based on act, benA, CaM (with RAxML trees as an input only), mcm7 and tsr1 loci together with GMYC analysis based on benA (only input tree based on coalescent tree model) and act (only input tree based on Yule tree model) loci did not support delimitation of A. pseudoviridinutans from a robust clade of A. felis. An incomplete lineage sorting was observed between A. felis and A. pseudoviridinutans (Fig. 3) evidencing that there was probably an ancestral gene flow between these lineages. Two isolates from A. felis lineage (IFM 59564 and CCF 5610) have benA sequences that cluster with A. pseudoviridinutans while sequences of the remaining 5 loci placed them in the A. felis lineage (single-gene trees not shown).
The species validation analysis results are appended to nodes of the tree in Fig. 3. Delimitation of A. felis and A. pseudoviridinutans gained absolute support in BP&P analysis (Yang & Rannala 2010) under all three scenarios simulated by different prior distributions of parameters θ (ancestral population size) and τ0 (root age). Delimitation of three putative species within A. felis lineage gained no support (posterior probability 0.51) under the scenario with small ancestral population sizes and shallow divergences among species: θ ~ G (2, 2000) and τ0 ~ G (2, 2000).
Species tree
The species tree topology was inferred with *BEAST (Heled & Drummond 2010) and is shown in Fig. 4. It was used as a guide tree during species validation using BP&P but it also represents the most probable evolutionary relationships between species in the AVSC. The analysis confirmed recombination between three subclades of A. felis (Fig. 4) which include also recently proposed species A. parafelis and A. pseudofelis thus representing the synonyms of A. felis. Similarly, the recombination between three subclades of A. udagawae rejected the hypothesis that they could be considered separate species (Fig. 4). The remaining species delimited in previous steps (Fig. 4), including A. pseudoviridinutans and A. acrensis (introduced in this study), were supported by *BEAST analysis. The species tree had identical topology with the trees inferred by ML and BI analyses of the concatenated and partitioned dataset (Fig. 5), and all species supported by *BEAST had 100 % ML bootstrap support (ML BS) and 1.00 BI posterior probabilities (BI PP). Several deep nodes in the species tree had only limited support similarly to ML and BI analyses. Thus clear positions of A. wyomingensis and A. siamensis within the clade also containing A. udagawae, A. acrensis and A. aureolus remains unresolved, while A. acrensis with A. aureolus form a sister clade to A. udagawae (this topology gained absolute support in all further analyses – see below). Another robust clade contained sister species A. felis and A. pseudoviridinutans. The remaining species, i.e., A. viridinutans, A. frankstonensis and A. arcoverdensis, formed a basal clade in the AVSC and their positions within the clade are fully resolved (Fig. 4).
Fig. 4.
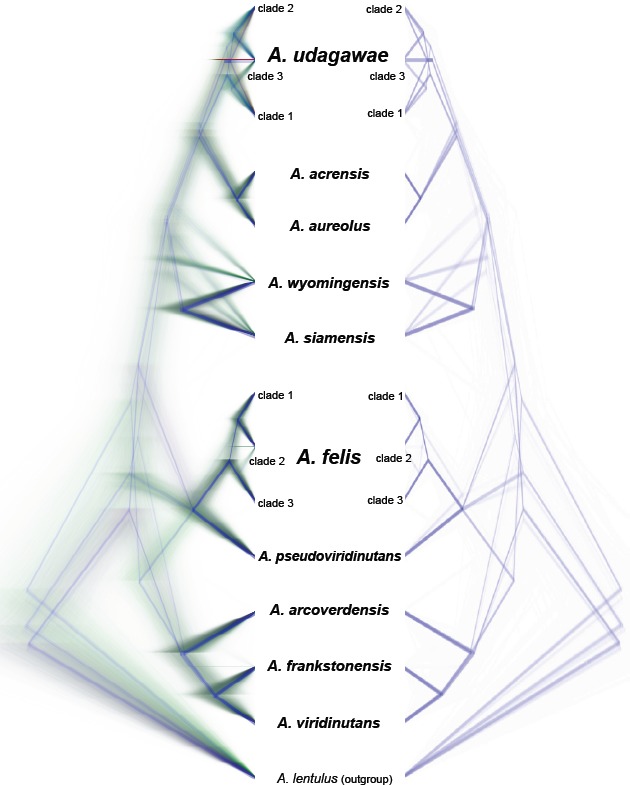
Species tree inferred with *BEAST visualized by using DensiTree (Bouckaert 2010). All trees created in the analysis (except 25 % burn-in phase) are displayed on the left side. Trees with the most common topology are highlighted by blue, trees with the second most common topology by red, trees with the third most common topology by pale green and all other trees by dark green. On the right side, the consensus trees of the three most common topologies are displayed.
Fig. 5.
Phylogenetic relationships of the Aspergillus viridinutans species complex members inferred from Bayesian analysis of the combined, 4-gene dataset of β-tubulin (benA), calmodulin (CaM), actin (act) and RNA polymerase II second largest subunit (RPB2) genes. Bayesian posterior probabilities (PP) and Maximum likelihood bootstrap supports (BS) are appended to nodes; only PP ≥ 90 % and BS ≥ 70 % are shown; lower supports are indicated with a hyphen, whereas asterisks indicate full support (1.00 PP or 100 % BS); ex-type strains are designated by a superscript T; species names in quotes are considered synonyms; the bar indicates the number of substitutions per site. The tree is rooted with Aspergillus lentulus NRRL 35552T. The geographic origin and reproductive mode with MAT idiomorph (if known) is designated by icons before the isolate number while substrate of origin is designated by icons after isolate number (see legend).
Clustering of isolates by origin and mating-type idiomorph
In the phylogenetic analysis, 111 combined benA, CaM, act and RPB2 sequences were assessed for members of AVSC. All species delimited by methods based on the coalescent model were fully supported by BI and ML analyses (Fig. 5).
The A. udagawae lineage included 25 isolates that clustered in three main clades. Mating type gene idiomorph MAT1-1-1 was detected in 10 isolates while 14 strains had MAT1-2-1 idiomorph (MAT idiomorph was not determined in one strain). The majority of North American isolates (10/14) clustered in clade 1 together with one strain from Australia; clade 2 comprised only three strains originating from Asia; isolates from four different continents were present in clade 3. There was no apparent clustering based on clinical or environmental origin of strains, or their MAT idiomorph. All three clinical isolates from Asia had an identical haplotype based on four studied protein-coding loci (Fig. 5) but one strain had MAT1-2-1 idiomorph in contrast to MAT1-1-1 idiomorph detected in the remaining two strains.
The A. acrensis lineage included five strains isolated from soil (Brazil) or cave sediment (Romania), two of which had MAT1-1-1 idiomorph and three had MAT1-2-1 idiomorph. This lineage is very closely related to a homothallic species A. aureolus represented by four strains in our analysis. The only known clinical isolate of A. aureolus (IHEM 22515) was isolated from the cornea of a patient in Peru. We were unable to source further information about this case and thus the clinical relevance of this isolate cannot be confirmed.
The mutual phylogenetic position of homothallic A. siamensis and heterothallic A. wyomingensis remains unresolved. Aspergillus siamensis was represented in our analysis by only two isolates from soil in Thailand, which were included in the original description (Eamvijarn et al. 2013). The A. wyomingensis lineage included 15 isolates; 12 of them came from Wyoming (USA) and were closely related to each other and to one isolate from China, while two isolates from Australia and Europe displayed a higher number of unique positions. The ratio of MAT1-1-1 isolates to MAT1-2-1 isolates was 8 : 7, and the majority of MAT1-1-1 isolates from the USA (6/7) clustered in a separate subclade that was only supported in the BI analysis.
The A. felis lineage comprised 35 isolates that clustered in three main clades. Mating type gene idiomorph MAT1-1-1 was detected in 12 isolates, while 20 strains had MAT1-2-1 idiomorph (MAT idiomorph was not determined in three strains). There was no clustering based on geographic origin as all three clades included isolates from two to four continents. Clade 3 contained only clinical isolates (n = 4). Clinical strains were predominant in clade 1 (18 : 2) whereas environmental strains dominated in clade 2 (7 : 4). The ratio of MAT1-1-1 isolates to MAT1-2-1 isolates in clade 1 was balanced (10 : 9) but was biased toward MAT1-2-1 idiomorph in clades 2 (1 : 7) and 3 (1 : 3). Eight isolates of A. pseudoviridinutans, a sister species of A. felis, were examined in this study; MAT1-1-1 idiomorph was determined in five of them and MAT1-2-1 idiomorph in three of them. There was no apparent clustering based on clinical or environmental origin of strains, or their MAT idiomorph (Fig. 5).
A basal clade of AVSC comprises three soil-borne species. Whilst A. viridinutans and A. frankstonensis are known only from one locality in Australia, 13 A. arcoverdensis strains included in the analysis were isolated on three continents, i.e., South America, Asia and Australia. Both, A. viridinutans and A. frankstonensis were represented only by one and two isolates, respectively, included in the original descriptions (McLennan et al. 1954, Talbot et al. 2017), and only isolates of one mating type are known for each of these species. Isolates of both mating types were present in A. arcoverdensis (MAT1-1-1 : MAT1-2-1 ratio, 8 : 5). A geographical clustering was apparent in A. arcoverdensis strains; two strains from China and one strain from Australia formed sublineages separate from the Brazilian strains (Fig. 5).
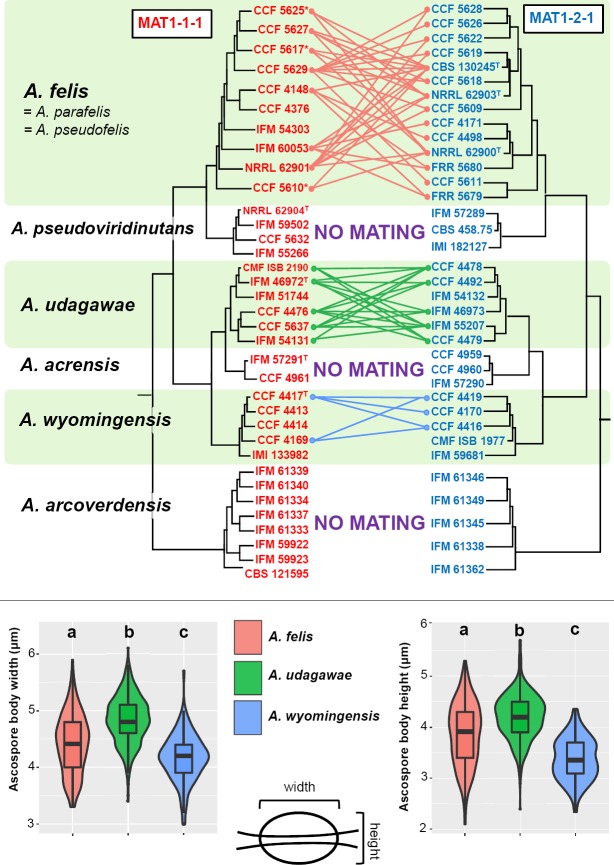
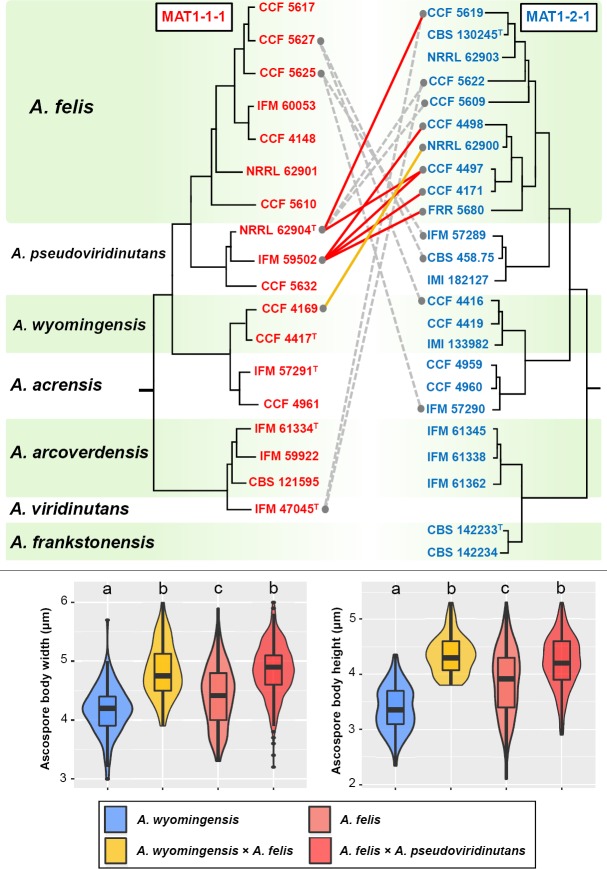
Mating experiments and morphology of spores
The MAT1-1-1 and MAT1-2-1 idiomorphs were determined for 100 of 104 isolates representing heterothallic species in AVSC (Table 1). Systematic mating experiments were first performed within major phylogenetic clades of the AVSC. Opposite mating type strains representing genetic and geographic diversity for each heterothallic species were selected for mating experiments and crossed in all possible combinations if not otherwise indicated (Fig. 6). Successful mating was observed in lineages of A. felis, A. udagawae and A. wyomingensis. At least some individuals representing all three phylogenetic subclades of A. felis (Fig. 3, 5) and A. udagawae (Fig. 2, 5) crossed successfully with individuals from the other subclades. The mating capacity of individual isolates was unequal. Whilst some isolates of a particular species were able to mate with a broad spectrum of opposite mating type strains of the same species, others produced fertile ascomata with only a limited set of strains or did not mate at all. The morphology of ascospores among different crosses in these three species was consistent (Fig. 7). The exception was great variability in the convex surface ornamentation of A. wyomingensis ascospores among and as well as within pairings of different isolates ranging from almost smooth, tuberculate to echinulate (Fig. 7). Although both the width and height of ascospores of A. felis, A. udagawae and A. wyomingensis overlapped significantly, their dimensions were statistically significantly different (Tukey's HSD test, p value < 0.05) (Fig. 6). No fertile cleistothecia were produced by crossing opposite mating type isolates of A. pseudoviridinutans, A. acrensis and A. arcoverdensis. Mating experiments were not performed in A. viridinutans and A. frankstonensis due to the absence of opposite mating type strains.
Fig. 6.
Schematic depiction of results of intraspecific mating experiments between opposite mating type isolates of heterothallic members of the Aspergillus viridinutans species complex. Only successful mating experiments are displayed by connecting lines between opposite mating type isolates; remaining mating experiments were negative. Isolates marked by asterisk were only crossed with ex-type strains of A. felis (CBS 130245T), A. parafelis (NRRL 62900T) and A. pseudofelis (NRRL 62903T). Boxplot and violin graphs were created in R 3.3.4 (R Core Team 2015) with package ggplot2 (Wickham 2009) and show the differences between the width and height of ascospores of A. udagawae, A. wyomingensis and A. felis. Different letters above the plot indicate significant difference (P < 0.05) in the size of the ascospores between different species based on Tukey's HSD test. Boxplots show median, interquartile range, values within ± 1.5 of interquartile range (whiskers) and outliers.
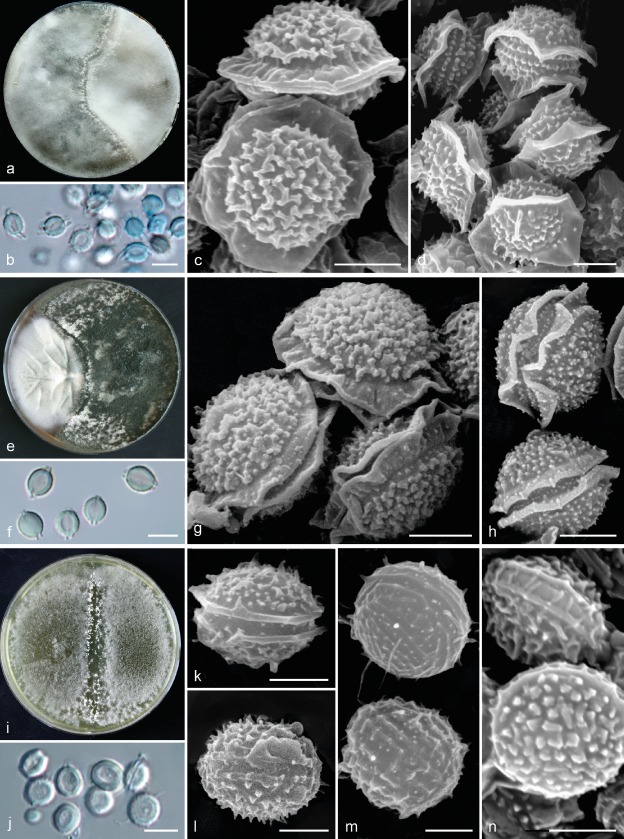
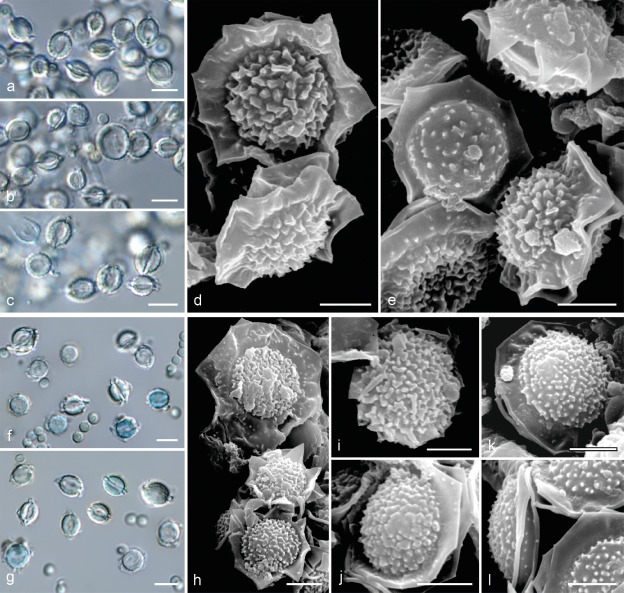
Fig. 7.
Comparison of morphology of sexual morphs of A. felis, A. udagawae and A. wyomingensis. a. Fertile cleistothecia of A. felis as a result of crossing of isolates IFM 60053 × FRR 5680; b. ascospores in light microscopy; c–d. ascospores in scanning electron microscopy: CBS 130245T × CCF 5627 (c), CBS 130245T × IFM 60053 (d); e. fertile cleistothecia of A. udagawae as a result of crossing of isolates IFM 46972T × IFM 46973; f. ascospores in light microscopy; g–h. ascospores in scanning electron microscopy; i. fertile cleistothecia of A. wyomingensis as a result of crossing of isolates CCF 4416 × CCF 4417T; j. ascospores in light microscopy (CCF 4416 × CCF 4169); k–n. ascospores in scanning electron microscopy: CCF 4416 × CCF 4417T (k–l), CCF 4417T × CCF 4419 (m–n). — Scale bars: b, f, j = 5 μm; c–d, g–h, k–n = 2 μm.
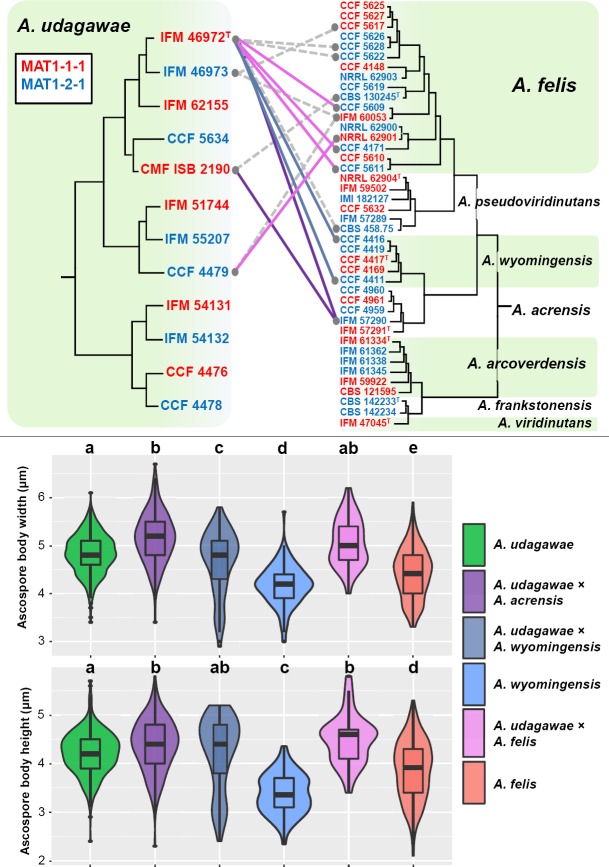
Opposite mating type isolates of each heterothallic species were also selected for interspecific mating assays and crossed in all possible combinations. Morphological characteristics of AVSC ascospores and induced hybrids are summarised in Table 5. Only three of 12 selected A. udagawae isolates produced fertile ascomata with some isolates of A. felis, A. wyomingensis or A. acrensis (Fig. 8). The highest mating capacity was observed in the ex-type strain of A. udagawae IFM 46972 that produced fertile ascomata when crossed with A. felis (CCF 5609, CCF 4171 and CCF 5611), A. wyomingensis (CCF 4416 and CCF 4411) and A. acrensis (IFM 57290). The width and height of ascospores of interspecific hybrids between A. udagawae and A. acrensis were significantly different (Tukey's HSD test, p value < 0.05) from A. udagawae (Fig. 8). Approximately 50 % of hybrid ascospores from mating CMF ISB 2190 with IFM 57290 lacked visible equatorial crests and if present, they were frequently interrupted or stellate (Fig. 9) in contrast to A. udagawae (visible crests present in > 90 % of ascospores, crests continuous). The ascomata from mating IFM 46972 with IFM 57290 contained only low numbers of ascospores that were globose or subglobose and glabrous (without crests and ornamentation on convex surface). This observation supported the hypothesis that A. acrensis is a separate species despite its close phylogenetic relationships to A. udagawae. The ascospore dimensions of hybrids between A. udagawae and A. wyomingensis were similar to those of A. udagawae and both width and height were significantly different (Tukey's HSD test, p value < 0.05) from A. wyomingensis (Fig. 8). These hybrid ascospores had well-defined equatorial crests that were most commonly 0.5–1 μm broad and similar to those of A. udagawae (Fig. 9). The hybrids of A. udagawae and A. felis had ascospores with similar equatorial crest length and body width to A. udagawae but were significantly different from A. felis, and their height was significantly different from both A. felis and A. udagawae (Fig. 8). The ascomata of hybrids between A. udagawae with A. wyomingensis and A. felis, respectively, usually contained only low numbers of ascospores. No mating or production of infertile ascomata only was observed between crosses of A. udagawae and the remaining heterothallic AVSC members (Fig. 8). Interestingly, the majority of interspecific hybrids produced approximately 1–10 % of globose or subglobose asco-spores with abnormally large dimensions, approximately 6.5–10.5 μm diam (their dimensions were not included for calculations of statistical measures in Fig. 8 and 10, and in Table 5). These cells had thick walls similar to normal ascospores, but lacked equatorial crests and had a glabrous or echinulate surface. Their dimensions were intermediate between normal ascospores and asci but their walls were dissimilar to those of thin-walled asci. These cells were not observed among progeny of the intraspecific crosses (intraspecific mating assay) and their presence probably indicates a defect in meiosis and ascospore development.
Table 5.
Ascospores characteristics of Aspergillus viridinutans complex species and interspecific hybrids.
| Species / interspecific hybrid (×) | Ascospore body (mean ± standard deviation; μm)) |
Ornamentation of ascospores |
||
|---|---|---|---|---|
| width | height | length of crests (μm)1 | surface ornamentation2 | |
| Aspergillus aureolus | 4.8 ± 0.5 | 4.4 ± 0.4 | (0.5–)1–1.5 | crests present3; CS tuberculate to echinulate (SEM) |
| A. felis | 4.4 ± 0.5 | 3.9 ± 0.6 | 0.5–1.5(–2) | crests present3; CS tuberculate to echinulate (SEM) |
| A. siamensis | 4.5 ± 0.5 | 3.7 ± 0.4 | (0.5–)1–1.5 | crests present3; CS tuberculate, echinulate to reticulate (SEM) |
| A. udagawae | 4.8 ± 0.4 | 4.2 ± 0.4 | (0–)0.5(–1) | visible crests absent in < 10 % of ascospores (LM); CS tuberculate to reticulate (SEM) |
| A. wyomingensis | 4.2 ± 0.4 | 3.4 ± 0.4 | 0–0.5 | visible crests absent in > 50 % of ascospores (LM); CS almost smooth, tuberculate, echinulate (SEM) |
| A. felis ཌ A. pseudoviridinutans | 4.9 ± 0.4 | 4.2 ± 0.5 | (0–)0.5–1 | visible crests absent in 5–20 % of ascospores (LM) depending on parental isolates; CS tuberculate to echinulate (SEM) |
| A. felis ཌ A. wyomingensis | 4.8 ± 0.5 | 4.3 ± 0.3 | (0–)0.5–1 | visible crests absent in ~ 10 % of ascospores (LM); CS tuberculate (SEM) |
| A. felis ཌ A. udagawae | 5.1 ± 0.5 | 4.5 ± 0.5 | 0–0.5(–1) | visible crests absent in ~ 20 % of ascospores (LM); CS echinulate, tuberculate to reticulate (SEM) |
| A. udagawae × A. wyomingensis | 5.0 ± 0.4 | 4.6 ± 0.3 | 0–1 | visible crests absent in ~ 15 % of ascospores (LM); CS tuberculate (SEM) |
| A. udagawae ཌ A. acrensis | 5.2 ± 0.5 | 4.4 ± 0.5 | 0–0.5 | visible crests absent in ~ 50 % of ascospores (LM) in CMF ISB 2190 ཌ IFM 57290 and in 100 % of ascospores in IFM 46972 ཌ IFM 57290; CS tuberculate to echinulate in CMF ISB 2190 ཌ IFM 57290 (SEM) and glabrous in IFM 46972 ཌ IFM 57290 (LM) |
1 Values in parentheses are less common (less than 10 % of measurements).
2 LM = light microscopy; SEM = scanning electron microscopy; CS = convex surface.
3 Crests may absent in < 1 % of ascospores in some isolates / crosses.
Fig. 8.
Schematic depiction of results of interspecific mating experiments between opposite mating type isolates of A. udagawae and other heterothallic members of Aspergillus viridinutans species complex. Only successful mating experiments are displayed by coloured connecting lines between opposite mating type isolates (different colours correspond to hybrids between different species); grey dashed lines indicate production of infertile ascomata; remaining mating experiments were negative. Boxplot and violin graphs were created in R 3.3.4 (R Core Team 2015) with package ggplot2 (Wickham 2009) and show the differences between the width and height of ascospores of particular species and their hybrids. Different letters above the plot indicate significant difference (P < 0.05) in the size of the ascospores based on Tukey's HSD test. Boxplots show median, interquartile range, values within ± 1.5 of interquartile range (whiskers) and outliers.
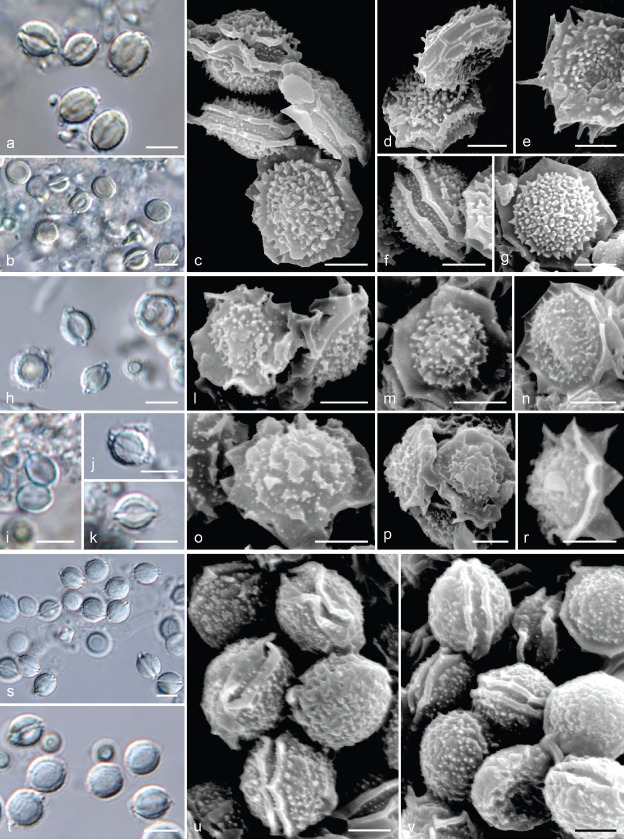
Fig. 9.
Ascospore morphology of interspecific hybrids between A. udagawae and other species. a–g. Hybrid of A. udagawae CMF ISB 2190 × A. acrensis IFM 57290; a–b. ascospores in light microscopy; c–g. ascospores in scanning electron microscopy; h–r. hybrid of A. udagawae CCF 4479 × A. felis NRRL 62901; h–k. ascospores in light microscopy; l–r. ascospores in scanning electron microscopy; s–v. hybrid of A. udagawae IFM 46972T × A. wyomingensis CCF 4411; s–t. ascospores in light microscopy; u–v. ascospores in scanning electron microscopy. — Scale bars: a–b, h–k, s–t = 5 μm; c–g, l–r, u–v = 2 μm.
Fig. 10.
Schematic depiction of results of interspecific mating experiments between opposite mating type isolates of heterothallic members of Aspergillus viridinutans species complex except of A. udagawae. Only successful mating experiments are displayed by coloured connecting lines between opposite mating type isolates (different colours correspond to hybrids between different species); grey dashed lines indicate production of infertile ascomata; remaining mating experiments were negative. Boxplot and violin graphs were created in R 3.3.4 (R Core Team 2015) with package ggplot2 (Wickham 2009) and show the differences between the width and height of ascospores of particular species and their hybrids. Different letters above the plot indicate significant difference (P < 0.05) in the size of the ascospores based on Tukey's HSD test. Boxplots show median, interquartile range, values within ± 1.5 of interquartile range (whiskers) and outliers.
Two MAT1-1-1 isolates of A. pseudoviridinutans selected for interspecific mating assays, namely the ex-type strain NRRL 62904 and strain IFM 59502, were able to mate with a relatively high number of MAT1-2-1 isolates of A. felis (Fig. 10). The ascospores of these hybrids were statistically significantly different in their width and height from A. felis. Equatorial crests were absent in approximately 5–20 % hybrid ascospores and, if present, they were shorter than those of A. felis (Table 5). These observations suggest that A. pseudoviridinutans should be treated as a separate species as proposed by species delimitation methods despite the close phylogenetic relationships of both species and incomplete lineage sorting detected between these two species (Fig. 3). Only one interspecific hybrid was induced in our assay between A. wyomingensis CCF 4169 and A. felis NRRL 62900. The ascospore body width and height of this hybrid was significantly different from both parental species (Fig. 10). In contrast to A. wyomingensis, equatorial crests were present in the majority of hybrids and they were occasionally interrupted and stellate (Fig. 11). Infertile ascomata were observed in some crosses between A. felis and following species: A. acrensis, A. wyomingensis and A. viridinutans.
Fig. 11.
Ascospore morphology of interspecific hybrids between A. felis, A. pseudoviridinutans and A. wyomingensis. a–e. Hybrid of A. felis × A. pseudoviridinutans; a–c. ascospores of hybrid CCF 4497 × IFM 59502 in light microscopy; d–e. ascospores in scanning electron microscopy: CCF 4497 × IFM 59502 (d), CCF 4171 × IFM 59502 (e); f–l. hybrid of A. felis NRRL 62900 × A. wyomingensis CCF 4169; f–g. ascospores in light microscopy; h–l. ascospores in scanning electron microscopy. — Scale bars: a–c, f–g = 5 μm; d–e, h–l = 2 μm.
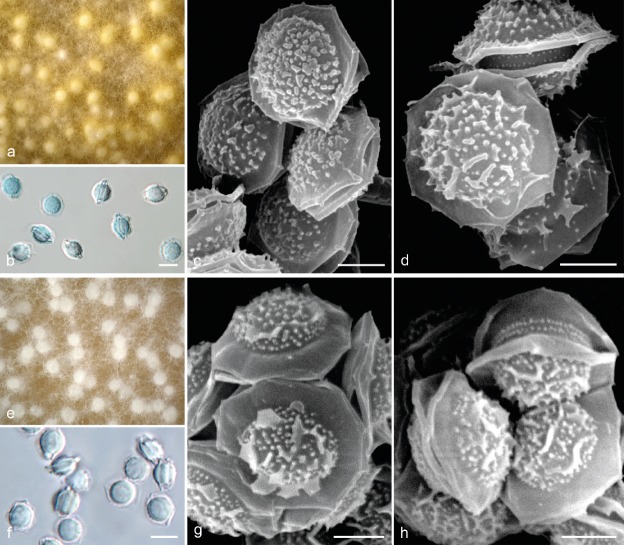
Aspergillus aureolus and A. siamensis are the only two homothallic species in the AVSC and readily produce ascomata on a broad spectrum of media and growth temperatures and are easily distinguishable from the eight heterothallic AVSC members. Most A. aureolus isolates produce distinctive yellow colonies in contrast to the whitish colonies of A. siamensis (Fig. 12). The ascospores of both species have similar dimensions, convex surface ornamentation and equatorial crest length (Table 5, Fig. 12) and most closely resemble those of A. felis from among heterothallic species.
Fig. 12.
Sexual morph morphology of homothallic species from Aspergillus viridinutans complex. a–d. Aspergillus aureolus isolates IFM 47021T (a–b, d) and IFM 46584 (c); a. Macromorphology of ascomata after 3 wk of incubation on MEA at 37 °C; b. ascospores in light microscopy; c–d. ascospores in scanning electron microscopy; e–h. Aspergillus siamensis isolate IFM 59793T; e. macromorphology of ascomata after 3 wk of incubation on MEA at 37 °C; f. ascospores in light microscopy; g–h. ascospores in scanning electron microscopy. — Scale bars: b, f = 5 μm; c–d, g–h = 2 μm.
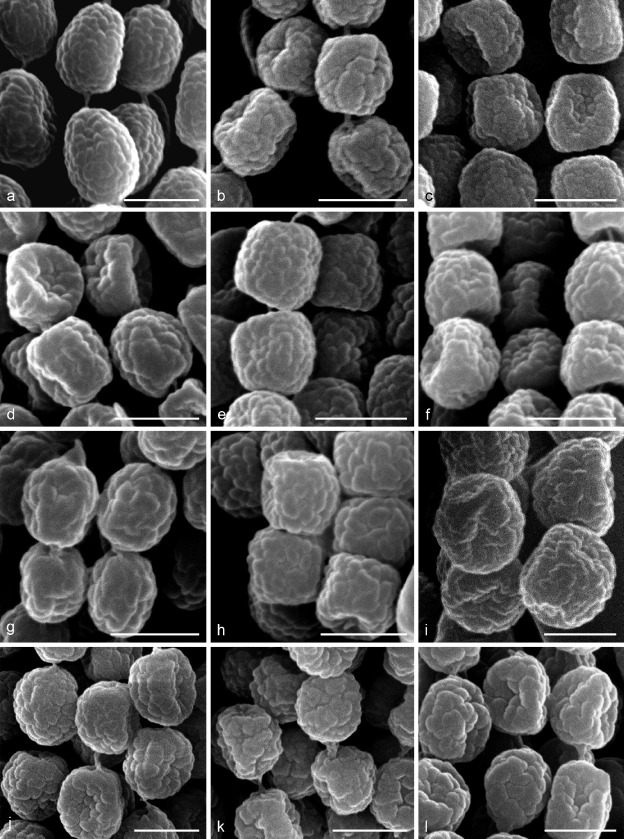
The macromorphology of colonies, micromorphology of asexual morphs and physiology have only limited discriminatory power in AVSC members, as recognized in previous studies (Nováková et al. 2014, Matsuzawa et al. 2015). We compared surface ornamentation of conidia in all currently recognized species using SEM. The ornamentation showed a micro-tuberculate pattern and was broadly identical across all species (Fig. 13).
Fig. 13.
Conidia with micro-tuberculate surface ornamentation pattern observed by scanning electron microscopy. a. Aspergillus acrensis IFM 57290; b. A. arcoverdensis IFM 61334T; c. A. aureolus IFM 46584; d. A. felis CBS 130245T; e. A. felis NRRL 62900 (ex-type of A. parafelis); f. A. felis NRRL 62903 (ex-type of A. pseudofelis); g. A. frankstonensis CBS 142234; h. A. pseudoviridinutans CBS 458.75; i. A. siamensis IFM 59793T; j. A. udagawae IFM 46972T; k. A. viridinutans IFM 47045T; l. A. wyomingensis CCF 4414. — Scale bars = 2 μm.
TAXONOMY
Aspergillus acrensis Hubka, A. Nováková, Yaguchi, Matsuz. & Y. Horie, sp. nov. — MycoBank MB822542; Fig. 14
Fig. 14.
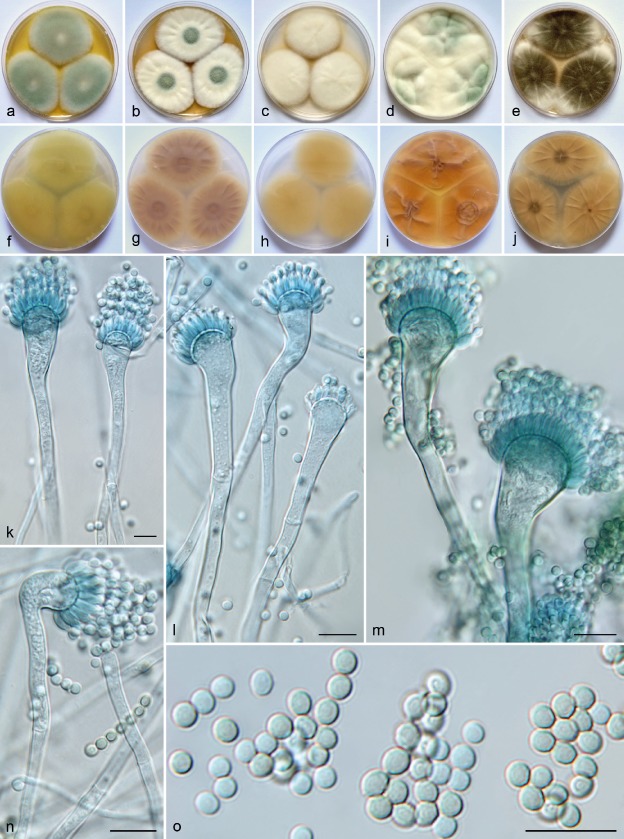
Micromorphology and macromorphology of Aspergillus acrensis. a–e. Colonies of IFM 57291T incubated 7 d at 25 °C on MEA, CYA, CZA, YES, and on CYA at 37 °C (from left to right); f–j. reverse of colonies of IFM 57291T incubated 7 d at 25 °C on MEA, CYA, CZA, YES, and on CYA at 37 °C (from left to right); k–n. conidiophores; o. conidia. — Scale bars = 10 μm.
Etymology. Named after the region of origin of the ex-type strain – state Acre located in the northern Brazil.
Mycelium composed of hyaline, branched, septate, smooth-walled hyphae. Conidial heads greyish green, loosely columnar, up to 140 μm long, 15–25 μm diam. Conidiophores uniseriate, arising from aerial hyphae or the basal mycelium, hyaline to pale yellowish brown, frequently nodding, smooth, 150–600 μm long; stipes 3–5.5(–8) μm wide in the middle; vesicles hyaline to greyish green, pyriform, subclavate to clavate, (6–)9–16(–20) μm diam; phialides ampulliform, hyaline to greyish green, 4.5–6(–7.5) × 1.5–2.5(–3) μm, covering approximately the apical half of the vesicle. Conidia hyaline to greyish green, globose, subglobose to broadly ellipsoidal, smooth-walled to delicately roughened, microtuberculate in SEM, 2.5–3 × 2–2.5 μm (mean ± standard deviation, 2.8 ± 0.2 × 2.4 ± 0.2; length/width ratio 1.1–1.3, 1.2 ± 0.1). Heterothallic, sexual morph unknown.
Culture characteristics (7 d at 25 °C, unless otherwise stated) — Colonies on MEA attained 51–62 mm diam, sparsely lanose, slightly raised, flat, yellowish white (ISCC–NBS No. 92) to pale green (No. 149), no exudate, soluble pigment light greyish yellow (No. 101), reverse light greenish yellow (No. 101) to brilliant greenish yellow (No. 98). Colonies on CYA attained 33–48 mm diam, floccose, slightly raised, flat to slightly radially furrowed, yellowish white (No. 92) to greenish white (No. 153), sporulation in the colony centre pale green (No. 149) to greyish green (No. 150), no exudate, soluble pigment dark greyish yellow (No. 91), reverse deep yellow (No. 85), light olive brown (No. 94) to moderate olive brown (No. 95) with light yellow (No. 86) margin. Colonies on CYA at 37 °C grow more rapidly compared to 25 °C and attained 60–70 mm diam, lanose, slightly raised, flat to radially furrowed, white mycelium in margins, sporulation light olive grey (No. 112) to olive grey (No. 113), no exudate, no soluble pigment, reverse colourless, moderate yellow (No. 87) to greyish yellow (No. 90). Colonies on CZA attained 36–48 mm diam, lanose, slightly raised, flat, yellowish white (No. 92), no exudate, no or light greyish yellow (No. 101) soluble pigment, reverse light yellow (No. 86), light greenish yellow (101) to brilliant greenish yellow (No. 98). Colonies on YES lanose, yellowish white (No. 92), irregularly furrowed, no exudate, soluble pigment brilliant yellow (No. 83), reverse brilliant yellow (No. 83). Colonies on CY20S attained 58–65 mm diam, lanose, slightly raised, flat, yellowish white (No. 92), no exudate, no soluble pigment, reverse moderate brown (No. 58) to moderate reddish brown (No. 43). Colonies on CREA attained 32–35 mm diam, sparsely lanose, plane, mycelium yellowish white, no visible sporulation, reverse strong brown (No. 55), no acid production. Growth on MEA at 45 °C, no growth on MEA at 47 °C.
Exometabolites — Isolate IFM 57291 produced an aszonapyrone, a fumigatin, tryptoquivalines, tryptoquivalones; isolate IFM 57290 an aszonapyrone, fumagillin, fumigatins, helvolic acid, pseurotin A, tryptoquivalines, and a trytoquivalone; isolate CCF 4959 pseurotin A, viriditoxin and several potential naphtho-gamma-pyrones; CCF 4960 antafumicins, fumagillin, a fumigatin, helvolic acid, pseurotin A, and a tryptoquivalone; and CCF 4961 an aszonapyrone, fumagillin, fumigatins, pseurotin A, tryptoquivalines and tryptoquivalones. In general, similar metabolites are also produced by the two most closely related species, i.e., A. aureolus and A. udagawae. Aspergillus aureolus produces fumagillin, helvolic acid, pseurotin A, tryptoquivalines, tryptoquivalones and viriditoxin as well as several unique yellow secondary metabolites. Aspergillus udagawae produces fumagillin, fumigatins, tryptoquivalines and tryptoquivalones (Frisvad & Larsen 2015a).
Specimens examined. Brazil, State of Acre, Xapuri, grassland soil in a cattle farm, 6 Nov. 2001, Y. Horie (holotype IFM 57291H, isotypes PRM 935088 and PRM 935089, culture ex-type IFM 57291T = CCF 4670T); State of Amazonas, Manaus, forest soil in tropical rain forest, 11 Nov. 2001, Y. Horie, culture IFM 57290 (= CCF 4666). –Romania, Movile cave, above the Lake Room, cave sediment, 8 June 2014, A. Nováková, culture CCF 4959; Movile cave, cave corridor, cave sediment, 8 June 2014, A. Nováková, culture CCF 4960; Movile cave, Lake Room, cave sediment, 8 June 2014, A. Nováková, culture CCF 4961.
Notes — The morphology of A. acrensis strongly resembles that of several other A. viridinutans complex members. The closely related taxa A. aureolus and A. siamensis are readily distinguished from A. acrensis by the production of ascomata under standard cultivation conditions (both are homothallic). Aspergillus viridinutans and A. frankstonensis grow more slowly at 25 °C and have smaller vesicles. The macromorphology of colonies and micromorphology of the asexual morph does not distinguish A. acrensis reliably from A. arcoverdensis, A. felis, A. pseudoviridinutans, A. udagawae and A. wyomingensis. Some of these species can be differentiated each from the other by their characteristic sexual morph, but the production of ascomata was not induced in A. acrensis despite our attempts, similarly to A. arcoverdensis and A. pseudoviridinutans. Although isolate IFM 57290 was successfully crossed with isolates of A. udagawae IFM 46972 and CMF ISB 2190 in vitro, both the width and height of ascospores were statistically different from A. udagawae. Also, abnormalities in the shape and superficial ornamentation (Fig. 9) were present in a significant number of spores (equatorial crests were absent in ~ 50 % of ascospores). Reliable identification of A. acrensis can currently only be achieved by molecular methods.
Aspergillus udagawae Horie et al., Mycoscience 36: 199. 1995.
Epitypification. Brazil, São Paulo State, Botucatú, Lagoa Seka Avea, soil in a plantation, 23 Aug. 1993, M. Takada (holotype CBM-FA-0711, designated by Horie et al. (1995), epitype designated here PRM 945579, isoepitypes PRM 945580 and 945581, MycoBank MBT378451, culture ex-epitype IFM 46972 = CBS 114217 = DTO 157-D7 = CBM-FA 0702 = KACC 41155 = CCF 4558).
Notes — Horie et al. (1995) designated the specimen CBM-FA-0711 as a holotype of A. udagawae, a dried culture with ascomata created by crossing the isolates CBM-FA-0702 (MAT1-1-1) × CBM-FA-0703 (MAT1-2-1). Although this specimen demonstrates the sexual and asexual morph of the life cycle, it is not suitable for the purposes of the recent taxonomy for several reasons. First of all, it is not clear which of the two cultures contained within the type should be considered the ex-holotype culture. Additionally, interspecific hybrids can be induced by crossing opposite mating type strains of unrelated species in vitro as shown in this study and some previous studies (see Discussion), and deposition of a resultant ‘hybrid’ type could lead to ambiguities. Although this second argument does not apply to A. udagawae as both isolates included in the holotype are closely related phylogenetically, we believe that a more clearly defined type of this species will facilitate future taxonomic work. Because it is not possible to recognize which part of the holotype belongs to particular isolate, lectotype designation (in this case part of holotype specimen) is difficult. For this reason we decided to select an epitype PRM 945579 derived from the IFM 46972 (= CBM-FA 0702) culture.
dISCUSSION
Changing species concepts in the AVSC
The AVSC members show considerable phenotypic variability but usually share production of nodding heads (some vesicles borne at an angle to the stipe) and relatively poor sporulation with abundant aerial mycelium. All species have a maximum growth temperature of 42 or 45 °C and the macromorphology and diameter of their colonies are similar, except for A. viridinutans and A. frankstonensis, which grow more slowly than remaining species. In addition, the morphology of conidiophores and conidia is relatively uniform across species, including the superficial ornamentation of conidia as shown here (Fig. 13). For these reasons heterothallic AVSC members have resisted taxonomic classification and were only identified to a species complex level, until recently.
Due to the absence of taxonomically informative characters, most recently described species in the AVSC were delimited using the GCPSR rules. Using this approach, the species are recognized based on concordance between single-gene phylogenies and the absence of tree incongruities. The GCPSR has found wide application in the taxonomy of fungi (Dettman et al. 2006, Hubka et al. 2013a, Peterson et al. 2015, Visagie et al. 2017). Huge progress has been made recently in the development of statistical methods for multilocus species delimitation, driven by advances in the multispecies coalescent model (Bouckaert et al. 2014, Flot 2015, Fontaneto et al. 2015, Schwarzfeld & Sperling 2015, Jones 2017). Although the ideology of MSC delimitation methods is relatively similar to GCPSR, these methods are more robust because the species are delimited in three steps, i.e., species discovery, species tree construction and species validation (Carstens et al. 2013). The determination of species boundaries is more objective in contrast to GCPSR rules that are based on relatively subjective evaluation and comparison of single-gene trees. In addition, MSC methods are able to deal better with phenomena such as incomplete lineage sorting, recombination or non-reciprocal monophyly that lead to incongruences between single-gene trees. Compared to the phylogenetic analysis of concatenated gene datasets (including partitioned datasets) and in part also GCPSR, the MSC methods are less prone to over-delimitation of species (Degnan & Rosenberg 2006, Kubatko & Degnan 2007, Heled & Drummond 2010, Rosenberg 2013), especially when the results of multiple delimitation methods are compared in one analysis.
The GCPSR rules together with evaluation of limited phenotypic data were recently used for description of A. felis, A. arcoverdensis and A. frankstonensis in the AVSC (Barrs et al. 2013, Matsuzawa et al. 2015, Talbot et al. 2017). Genealogical analysis using five genetic loci was carried out for delimitation of A. parafelis, A. pseudofelis and A. pseudoviridinutans, three close relatives of A. felis (Sugui et al. 2014). Although the authors found no conflict between single-gene phylogenies, only two isolates of each of these four species were used in analysis, and sequences of A. felis, A. parafelis and A. pseudofelis strains included were almost invariable. These isolates did not cover sufficiently the genetic diversity of these species as shown here. Species delimitation results based on MSC in this study showed that A. parafelis and A. pseudofelis are included in the genetically diverse lineage of A. felis (Fig. 3). The intraspecific pairwise genetic distances in A. felis (Table 6) range from 0.6 % (RPB2) to 4.2 % (benA). Similarly, pairwise genetic distances in A. udagawae (Table 6) are 1.1 % (benA) to 4.9 % (act). Such high intraspecific diversity in these genetic loci is unusual in Aspergillus and it reflects the intense recombination. Thus, when only limited number of strains from such species are selected for phylogenetic analysis, the results of species delimitation techniques may be biased and prone to overestimate the number of species. As we have shown here, this was clearly the case in the study of Sugui et al. (2014). This problem is probably widespread in current fungal taxonomy and limits possibilities of correct species boundaries delimitation. Also in this study, the number of strains of some closely related and phenotypically similar species is underrepresented, e.g., A. viridinutans and A. frankstonensis. In these cases, the species boundaries cannot be reliably defined using neither GCPSR rules nor MSC-based methods used in this study.
Table 6.
Highest intraspecific pairwise genetic distances in members of Aspergillus viridinutans complex (%).
| Species (no. of isolates) | Highest genetic distances between two isolates according to different genetic loci |
|||||
|---|---|---|---|---|---|---|
| benA | CaM | RPB2 | act | mcm7 | tsr1 | |
| A. acrensis (5) | 0.2 | 0.9 | 0.2 | 0 | ND | ND |
| A. arcoverdensis (13) | 0 | 0.9 | 0.5 | 1.4 | ND | ND |
| A. aureolus (4) | 0.4 | 0 | 0.1 | 0 | ND | ND |
| A. felis (35) | 4.2 | 2.4 | 0.6 | 2.5 | 1.3 | 3.3 |
| A. frankstonensis (2) | 0 | 0.2 | 0 | 0 | 0 | ND |
| A. pseudoviridinutans (8) | 2.6 | 2.2 | 1.9 | 2.1 | 0.7 | 1.4 |
| A. siamensis (2) | 0 | 0.1 | 0.1 | 0 | ND | ND |
| A. udagawae (25) | 1.1 | 2.8 | 1.2 | 4.9 | ND | ND |
| A. wyomingensis (15) | 0.4 | 0.9 | 0.4 | 0.9 | ND | ND |
ND, not determined.
Clinically relevant species and their identification in clinical setting
Although sect. Fumigati harbours many important pathogenic species, members of the AVSC have been overlooked by both clinicians and mycologists until recently. The presence of these soil-borne species in clinical material was first reported by Katz et al. (2005) who examined phylogenetic positions of several ‘atypical’ (poorly sporulating) clinical isolates of A. fumigatus. The majority of these strains grouped with, but were not identical to, A. viridinutans and A. aureolus from the AVSC. Since then many similar epidemiological and clinical studies have reported the pathogenic role of AVSC members in humans and animals, as reviewed by Talbot & Barrs (2018). In humans the most common manifestation of disease is chronic invasive pulmonary aspergillosis in immunocompromised patients. AVSC species are also frequently reported as a cause of sino-orbital aspergillosis (SOA) in cats that is chronic, but frequently fatal. In contrast to humans and dogs, the disease usually affects ostensibly immunocompetent cats. This increasingly recognised clinical entity is most frequently caused by the AVSC species and less frequently by other cryptic species in sect. Fumigati (Barrs et al. 2012, 2013, 2014).
Based on the species boundaries redefined in this study, the AVSC encompasses four species that are confirmed opportunistic pathogens. According to a number of reported cases, in humans A. udagawae is the most important opportunistic pathogenic from the AVSC followed by A. felis and A. pseudoviridinutans. In contrast, SOA in cats is most commonly caused by A. felis and much less frequently by A. udagawae and A. wyomingensis (Barrs et al. 2013, 2014).
Medically important species from the AVSC demonstrate elevated minimum inhibitory concentrations (MICs) of itraconazole and voriconazole in vitro, and a variable susceptibility to amphotericin B, while posaconazole and echinocandins have potent in vitro activities (Lyskova et al. 2018). Since the intraspecific variation in MICs of particular antifungals is usually high, the use of reliable methods for MIC determinations takes precedence over correct identification to a species level. The latter may be challenging or even impossible in the clinical setting. However, identification to the level of species complex and differentiation from A. fumigatus is important due to strikingly different antifungal susceptibility patterns.
In contrast to A. fumigatus, the AVSC species do not grow at 47 and 50 °C, usually sporulate less and a proportion of their vesicles are borne at an angle to the stipe. In addition, some isolates produce acidic compounds detectable on CREA (Barrs et al. 2013, Nováková et al. 2014, Talbot & Barrs 2018). Despite the fact that ITS rDNA region sequences are not available for all AVSC members, this universal marker for fungal species identification and barcoding can be used to achieve identification to a species complex level. The sequences from all six protein-coding genes included in this study (Table 2) have sufficient discriminatory power for species level identification of all clinically relevant species. Among these genes, sequences of β-tubulin and calmodulin belong to the most commonly used in the clinical practice when correct identification is required (epidemiological studies, outbreak investigations or when dealing with infections refractory to antifungal therapy). However, the discrimination between A. felis and A. pseudoviridinutans can be limited when using the β-tubulin gene due to the incomplete lineage sorting phenomenon detected in this study (Fig. 3).
Additionally, the increasingly used method of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) gives promising results for rapid and accurate discrimination between A. fumigatus and other clinically relevant aspergilli from sect. Fumigati (Alanio et al. 2011, Nakamura et al. 2017). The development of more robust, curated and accessible MALDI-TOF spectrum databases should enable the implementation of MALDI-TOF MS for routine identification of less common aspergilli in future. Several PCR assays targeting protein-coding or microsatellite loci have also been developed and show good efficiency in discrimination of less common pathogenic species in sect. Fumigati (Araujo et al. 2012, Fernandez-Molina et al. 2014, Chong et al. 2017).
Mating behaviour in the AVSC – heterothallic species
The increasing availability of PCR-based tools for identification of fungal genes responsible for sexual and somatic incompatibility has facilitated the ability to induce the sexual morph in fungi (Dyer & O’Gorman 2011). The characterisation of mating type (MAT) genes became routine when inducing the sexual morph of heterothallic species in vitro. Using this approach, the sexual morph has been induced recently in at least five members of sect. Fumigati (O’Gorman et al. 2009, Barrs et al. 2013, Swilaiman et al. 2013, Nováková et al. 2014, Hubka et al. 2017). The discovery of a sexual cycle in pathogenic and mycotoxigenic fungi has many important consequences, because fungi with a functional sexual cycle have greater potential to increase their virulence and to develop resistance to antifungals, fungicides, etc. (Kwon-Chung & Sugui 2009, Lee et al. 2010, Swilaiman et al. 2013).
Here, we induced the sexual morph with a relatively high rate of success in A. felis, A. udagawae and A. wyomingensis (Fig. 6). We demonstrated that ascospores of these three species have relatively stable morphology (Fig. 7) and that the size of their ascospores is significantly different from one another (Fig. 6) and can be differentiated by equatorial crest length (Table 5). However, not all opposite mating type strains of the same species are able to produce ascomata in vitro as demonstrated in all three mentioned species (Fig. 6). A similar decline in mating capacity was also demonstrated in previous studies on the AVSC (Sugui et al. 2010, Nováková et al. 2014), but also in A. lentulus (Swilaiman et al. 2013) and A. fumigatus (O’Gorman et al. 2009). These species require relatively rigid conditions to complete their sexual cycle and some crosses produce low numbers of or infertile ascomata or do not mate at all (Balajee et al. 2006, Yaguchi et al. 2007, Kwon-Chung & Sugui 2009, Sugui et al. 2010, Nováková et al. 2014). For instance, fertility between two opposite mating-type isolates may be influenced by the vegetative incompatibility genes (Olarte et al. 2015), regulators of cleistothecium development and hyphal fusion (Szewczyk & Krappmann 2010).
We were not able to induce the sexual morph in three heterothallic members of the AVSC, i.e., A. acrensis, A. arcoverdensis and A. pseudoviridinutans, despite the relatively high number of opposite mating-type strains that was available for the mating assays (Fig. 6). It is not clear if these species require different conditions for successful mating, if there are other unidentified pre-zygotic mating barriers between opposite mating type strains, or if they have lost the ability to complete their sexual cycle. The evidence that two of these species were able to mate with different species from AVSC makes the last possibility improbable (Fig. 8, 10). These hybrids can be differentiated from A. udagawae and A. felis, respectively, by their dimensions (Fig. 8, 10) and surface ornamentation (Fig. 9, 11; Table 5). It demonstrates that both A. acrensis and A. pseudoviridinutans should be treated as separate taxonomic entities from their related species. Similar deviations in size and surface ornamentation of ascospores were demonstrated in other interspecific hybrids (Fig. 8–11) when they were compared to parental species.
Mating behaviour in the AVSC – homothallic species
Although homothallic species prevail over heterothallic in sect. Fumigati (Fig. 1), only two homothallic species are present in the AVSC. It is supposed that heterothallism is ancestral to homothallism in fungi (Nauta & Hoekstra 1992), including Aspergillus (Rydholm et al. 2007, Lee et al. 2010). It is obvious from phylogenetic studies across different subgenera of Aspergillus, that reproductive strategy is evolutionary conservative and homothallic as well as heterothallic (or asexual) species are typically clustered in clades with a uniform reproductive strategy. For instance in subg. Aspergillus, the 31 currently accepted species of sect. Aspergillus are all homothallic (Chen et al. 2016a) while sister sect. Restricti encompasses 20 asexual and only one distantly related homothallic species, A. halophilicus (Sklenář et al. 2017). Similarly, subg. Polypaecilum harbours only asexual species (Martinelli et al. 2017, Tanney et al. 2017). Asexual species also predominate in subg. Circumdati (Jurjević et al. 2015) although most, if not all, probably have a cryptic sexual cycle as highlighted by sexual morph induction in A. flavus, A. nomius, A. parasiticus, A. terreus and A. tubingensis (Horn et al. 2009a, b, 2011, 2013, Arabatzis & Velegraki 2013). A strikingly different situation is present in subgenera Nidulantes (Chen et al. 2016a, Hubka et al. 2016a), Fumigati (Fig. 1) and Cremei (Hubka et al. 2016b) where heterothallic and homothallic species interchange like a mosaic along the phylogenetic tree.
Common genetic distances between closely related sister species across aspergilli usually range between 2–4 % in benA and CaM loci and 1–2 % in RPB2 locus; the situation in AVSC is very similar (Table 7). Interestingly, there are only few examples of closely related homothallic and heterothallic/asexual species in Aspergillus despite their common origin. Genetic similarities between related couples of homothallic and heterothallic/asexual exceeding 95 % are rare, with only two examples in subg. Circumdati and one in subg. Cremei (Table 8). Section Fumigati is exceptional because it contains at least five pairs of highly related homothallic and heterothallic species (Table 8; Fig. 1). Aspergillus acrensis, described here, and A. aureolus represent the most closely related pair across genus Aspergillus (Table 8) and thus could be an ideal model for studying the evolution of reproductive modes. If we accept the hypothesis about the derived origin of homothallic species, it is probable that A. aureolus evolved in the lineage of A. acrensis relatively recently, due to the extremely low genetic distances of both species. This is also likely the reason why the multilocus species delimitation method STACEY and also some single-locus methods failed to segregate A. acrensis from A. aureolus (Fig. 2) in this study.
Table 7.
Genetic similarities between the ex-type isolates of Aspergillus viridinutans complex members based on identities from BLAST similarity search1.
| Species | Genetic similarities between species: benA
/ CaM / RPB2 (%) 1 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1. | 2. | 3. | 4. | 5. | 6. | 7. | 8. | 9. | 10. | |
| 1.A. acrensis | – | |||||||||
| 2.A. arcoverdensis | 94.5/95.2/98.0 | – | ||||||||
| 3. A. aureolus | 99.6/98.8/99.0 | 94.5/95.6/98.1 | – | |||||||
| 4. A. felis | 92.0/95.6/97.7 | 93.4/96.8/97.6 | 92.4/95.9/97.8 | – | ||||||
| 5.A. frankstonensis | 95.3/94.7/98.0 | 95.6/97.1/98.3 | 95.3/94.9/98.2 | 92.6/96.2/97.7 | – | |||||
| 6.A. pseudoviridinutans | 94.7/95.2/97.6 | 95.7/96.0/97.4 | 94.9/95.5/97.8 | 95.5/97.6/98.1 | 96.0/95.3/97.5 | – | ||||
| 7.A. siamensis | 96.6/95.8/98.9 | 95.5/95.6/98.5 | 96.7/96.0/98.9 | 93.0/95.7/98.2 | 95.6/94.7/98.0 | 95.5/95.4/97.9 | – | |||
| 8.A. udagawae | 97.4/96.8/99.0 | 94.7/95.6/98.2 | 97.4/97.1/99.1 | 92.0/95.9/97.9 | 95.3/95.1/98.1 | 94.5/95.6/97.7 | 96.2/96.3/99.1 | – | ||
| 9.A. viridinutans | 95.3/94.8/98.6 | 96.5/97.3/99.1 | 95.5/95.1/98.6 | 93.8/95.4/98.2 | 97.5/97.8/98.8 | 96.5/94.7/97.9 | 96.3/95.3/98.8 | 95.6/95.3/98.8 | – | |
| 10. A. wyomingensis | 95.8/96.5/98.6 | 94.5/96.0/97.8 | 96.0/96.5/98.3 | 92.1/96.3/97.5 | 95.4/95.8/97.6 | 94.9/95.9/97.3 | 96.9/96.5/98.9 | 95.8/96.9/98.6 | 95.6/95.7/98.3 | – |
1 nucleotide BLAST with default setting (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Table 8.
Genetic similarities between selected homothallic species and their most closely related heterothallic / anamorphic relatives across diversity of the genus Aspergillus.
| Homothallic species (section) – closest heterothallic / anamorphic species | Genetic similarities (%): benA / CaM / RPB21 |
|---|---|
| subg. Aspergillus | |
| A. halophilicus (Restricti) – any species | ≤89 |
| A. montevidensis (Aspergillus) – any species | ≤88 |
| subg. Circumdati | |
| A. alliaceus (Flavi) – A. lanosus | 96.4 / 95.7 / 99.1 |
| A. muricatus (Circumdati) – A. ochraceus | ≤91 |
| A. neoflavipes (Flavipedes) – A. micronesiensis | 94.8 / 91.9 / 97.5 |
| A. neoniveus (Terrei) – any species | ≤90 |
| subg. Cremei | |
| A. chrysellus (Cremei) – A. wentii | 97.1 / 97.2 / 97.7 |
| A. cremeus (Cremei) – any species | ≤91 |
| A. stromatoides (Cremei) – any species | ≤93 |
| subg. Fumigati | |
| A. acanthosporus (Clavati) – A. clavatus | ≤93 |
| A. aureolus (Fumigati) – A. acrensis | 99.6 / 98.8 / 99.0 |
| A. cejpii (Clavati) – any species | ≤88 |
| A. fischeri (Fumigati) – A. fumigatus | 94.3 / 94.5 / 97.9 |
| A. posadasensis (Clavati) – A. clavatus | 95.1 / 92.6 / 93.5 |
| A. quadricinctus (Fumigati) – A. duricaulis | 92.6 / 95.0 / 99.1 |
| A. siamensis (Fumigati) – A. wyomingensis | 97.1 / 96.5 / 98.9 |
| A. waksmanii (Fumigati) – A. nishimurae | 97.8 / 98.4 / 96.6 |
| subg. Nidulantes | |
| A. discophorus (Nidulantes, A. aeneus clade) – A. karnatakaensis | ≤92 |
| A. falconensis (Nidulantes, A. nidulans clade) – A. recurvatus | ≤93 |
| A. monodii (Usti) – any species | ≤90 |
| A. nidulans (Nidulantes, A. nidulans clade) – any species | ≤92 |
| A. pluriseminatus (Nidulantes, A. multicolor clade) – any species | ≤92 |
| A. purpureus (Nidulantes, A. spelunceus clade) – any species | ≤90 |
| A. undulatus (Nidulantes, A. stellatus clade) – any species | ≤89 |
1If none of three genetic similarities exceed 95 %, the values are replaced by only one highest value (usually RPB2 locus).
Interspecific hybridization in fungi and its consequences
Interspecific hybridization is an important process affecting speciation and adaptation of micro- and macroorganisms, however, relatively little is still known about the frequency of hybridization in fungi and its role in evolution of fungal species. Fungal hybrids may form either by a partial or complete sexual cycle or by a parasexual process. Mating between two species may be prevented by pre-zygotic barriers (e.g., gamete recognition) and various post-zygotic barriers (developmental problems, hybrid viability and ability to reproduce, etc.). The disagreement between phylogenetic/morphological species concepts and biological species compatibilities has been repeatedly described in fungi. Phylogenetic divergence in some fungal groups might have preceded development of reproductive barriers as shown by interspecific hybrids induced in vitro between primary human and animal pathogenic Trichophyton species (Kawasaki et al. 2009, 2010, Anzawa et al. 2010, Kawasaki 2011), opportunistic pathogenic Candida albicans and C. dubliniensis (Pujol et al. 2004), members of Aspergillus sect. Fumigati (Sugui et al. 2014, Talbot et al. 2017), mycotoxigenic A. flavus and A. parasiticus (Olarte et al. 2015), A. flavus and A. minisclerotigenes (Damann & DeRobertis 2013), phytopathogenic species from the Fusarium graminearum complex (Bowden & Leslie 1999) and species of Neurospora (Dettman et al. 2003). Natural interspecific hybrids resulting from recombination between species or parasexual reproduction are most commonly reported and have been extensively studied in saprophytic yeasts (González et al. 2008, Sipiczki 2008, Nakao et al. 2009, Louis et al. 2012), the plant endophyte Epichloë (Cox et al. 2014, Charlton et al. 2014, Shymanovich et al. 2017) and in various plant pathogenic fungi including species of Fusarium graminearum complex (O’Donnell et al. 2004, Starkey et al. 2007), Ophiostoma (Brasier et al. 1998, Solla et al. 2008), Microbotryum (Gladieux et al. 2010), Melampsora (Spiers & Hopcroft 1994, Newcombe et al. 2000), Botrytis (Staats et al. 2005), Verticillium (Inderbitzin et al. 2011) and Heterobasidion (Gonthier et al. 2007, Lockman et al. 2014).
Considering that in vitro induction of hybrids is relatively successful, it is surprising that reports on the isolation of naturally occurring hybrids are infrequent in human and animal pathogenic fungi. It suggests that post-zygotic mating barriers play a fundamental role in the maintenance of species boundaries. Naturally occurring hybrids have been detected in yeasts and dimorphic fungi, including between Candida spp. (Schröder et al. 2016), Malassezia spp. (Wu et al. 2015), Cryptococcus neoformans and C. gattii (Bovers et al. 2006, 2008, Kwon-Chung & Varma 2006, Aminnejad et al. 2012) and Coccidioides immitis and C. posadasii (Johnson et al. 2015). However, to date, reports on these hybrids in filamentous fungi are restricted to the Neocosmospora solani complex (Short et al. 2013, 2014). Species definition has become a controversial issue in some of these species complexes with naturally occurring hybrids because of differing opinions on species concepts among taxonomists (Kwon-Chung & Varma 2006, Kawasaki 2011, Kwon-Chung et al. 2017).
Even in cases where interspecific hybrids with high fitness and fertility can be demonstrated, the intensity of gene flow between natural populations must be sufficient to oppose genetic drift in order to have a significant impact on genetic isolation of species. In fungi, these processes cannot be evaluated rigorously by in vitro mating assays, as these cannot be extrapolated fully to a natural setting (Starkey et al. 2007, Sugui et al. 2014, Hubka et al. 2015a). Indeed, natural interspecific hybrids have never been reported for the majority of species that readily produce hybrids in vitro, including Aspergillus, dermatophytes and Neurospora. The MSC and GCPSR approaches provide practical tools for evaluating the significance of gene flow between natural populations and for assessing species limits. The interpretation of in vitro mating assays without a robust phylogeny is thus controversial, because a number of clearly phylogenetically, morphologically and ecologically distinct species lack effective reproductive barriers. In addition, the evaluation of biological species limits using mating assays requires determination of the fitness and fertility of progeny, which is demanding in both time and cost.
In general, mating success between different species under laboratory conditions is much lower compared to intraspecific mating, suggesting strong reproductive isolation between species and adherence to the biological species concept. In agreement with this, only a limited number of strains with exceptional mating capacity are usually capable of interspecific hybridization with strains of different species, e.g., A. udagawae strain IFM 46972 (Fig. 8) or A. pseudoviridinutans strain IFM 59502 (Fig. 10).
Several studies demonstrated that interspecific hybrids express genetic abnormalities or have decreased fertility and viability. Genetic analysis of the progeny of a cross between F. asiaticum × F. graminearum detected multiple abnormalities that were absent in intraspecific crosses of F. graminearum, i.e., pronounced segregation distortion, chromosomal inversions, and recombination in several studied linkage groups (Jurgenson et al. 2002, Gale et al. 2005). Matings between C. neoformans × C. gattii produced only a low percentage of viable progeny. It has been suggested that C. neoformans and C. gattii produce only stable diploid hybrids, but not true recombinants (Kwon-Chung & Varma 2006). Although Olarte et al. (2015) obtained hybrid progeny of A. flavus and A. parasiticus, fertile crosses were rare and involved only one parental strain of A. flavus. Viable ascospores were extremely rare, suggesting extensive genetic incompatibility and post-zygotic incompatibility mechanisms. Morphologically, the progeny differed from parental strains in growth rate, sclerotium production, stipe length, conidial head seriation and conidial features (Olarte et al. 2015). Decreased viability of hybrid ascospores was also detected among Neurospora spp. (Dettman et al. 2003) and in Aspergillus sect. Fumigati, in addition to abnormalities in their surface ornamentation visualised by SEM (Sugui et al. 2014), which is in agreement with the present study (Fig. 9, 11). Apart from ascospore ornamentation, we also found significant differences in hybrid ascospore dimensions from parental species (Fig. 8, 10).
The relatively recent globalization of trade in horticultural and agricultural plants, and introduction of non-native plant species has resulted in the inadvertent introduction of alien plant pathogens into non-endemic areas, contributing to the emergence of some devastating plant diseases (Brasier 2001, Mehrabi et al. 2011, Dickie et al. 2017). Anthropogenic activities or changes in the distribution of fungi (e.g., in response to climate changes) may bring together related, previously allopatric pathogenic species. Subsequent interspecific hybridization could give rise to pathogens with new features, including adaptation to new niches and host species, and varying degrees of virulence, as evidenced in Verticillium longisporum, Zymoseptoria pseudotritici, Blumeria graminis f. sp. triticale, and hybrids between Ophiostoma novo-ulmi and O. ulmi (Brasier 2001, Schardl & Craven 2003, Depotter et al. 2016).
As far as we know, the occurrence of Aspergillus interspecific hybrids in nature has not been proven despite successful hybridization of some species in vitro. However, there is no reason to assume that this phenomenon does not occur occasionally. Genetic recombination similar to that found in intraspecific mating occurred in half of the progeny produced by mating A. fumigatus with A. felis, while the other half were probably diploids or aneuploids (Sugui et al. 2014). Progeny resulting from mating between A. flavus and A. minisclerotigenes was fertile when crossed with parental strains and the frequency of successful matings was similar to that within pairs of A. flavus and A. minisclerotigenes strains, respectively (Damann & DeRobertis 2013). Ultimately, the viable hybrid must present some characteristics that promotes its survival (Turner et al. 2010, Mixão & Gabaldón 2018). For instance Olarte at al. (2015) showed that some F1 progeny of A. flavus × A. parasiticus produced higher aflatoxin concentrations compared to midpoint parent aflatoxin levels, and some hybrids synthesized G aflatoxins that were not produced by the parents. This suggested that hybridization is an important diversifying force generating novel toxin profiles (Olarte et al. 2015). Although interspecific hybridization in aspergilli is a relatively newly discovered phenomenon, it is likely to have played an important role in the evolution of the genus.
The relationship between hybridization and changes in virulence potential is not well understood in human and animal fungal pathogens but its role in the emergence of novel plant fungal pathogens is well documented, as discussed. The evidence of biological compatibility between major pathogens in Aspergillus sect. Fumigati sheds new light on possible interspecific transfer of virulence genes, genes responsible for antifungal resistance, and other genes influencing adaptation of these fungi to a changing environment. Further studies should elucidate to what extent interspecific hybridization shaped the evolution of these opportunistic pathogens.
CONCLUSIONS
Based on consensus results of species delimitation methods and after evaluation of mating assay results and phenotypic data, we now recognise 10 species within the AVSC. This number comprises nine previously recognised and one new species proposed here. Aspergillus pseudofelis and A. parafelis are placed in synonymy with A. felis. All four genetic loci used for phylogenetic analysis across the AVSC have sufficient variability for reliable species identification and can be used as DNA barcodes. Though more laborious, the MSC are a suitable tool for delimitation of genetically diverse cryptic species in cases where classical phylogenetic, morphological and mating compatibility data do not yield satisfactory results.
Acknowledgments
This research was supported by the project of the Charles University Grant Agency (GAUK 1434217), Czech Science Foundation (No. 17-20286S), Charles University Research Centre program No. 20406, the project BIOCEV (CZ.1.05/1.1.00/02.0109) provided by the Ministry of Education, Youth and Sports of CR and ERDF, and by a Thompson Research Fellowship from the University of Sydney. We thank Milada Chudíčková and Alena Gabrielová for their invaluable assistance in the laboratory, CCF collection staff (Ivana Kelnarová and Adéla Kovaříčková) for lyophilization of the cultures, Miroslav Hyliš for assistance with scanning electron microscopy, Stephen W. Peterson, Kyung J. Kwon-Chung, Adrian M. Zelazny, Maria Dolores Pinheiro and Dirk Stubbe for providing important cultures for this study. Vit Hubka is grateful for support from the Czechoslovak Microscopy Society (CSMS scholarship 2016).
REFERENCES
- Alanio A, Beretti J-L, Dauphin B, et al. 2011. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry for fast and accurate identification of clinically relevant Aspergillus species. Clinical Microbiology and Infection 17:750–755. [DOI] [PubMed] [Google Scholar]
- Alastruey-Izquierdo A, Mellado E, Peláez T, et al. 2013. Population-based survey of filamentous fungi and antifungal resistance in Spain (FILPOP Study). Antimicrobial Agents and Chemotherapy 57:3380–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcazar-Fuoli L, Mellado E, Alastruey-Izquierdo A, et al. 2008. Aspergillus section Fumigati: Antifungal susceptibility patterns and sequence-based identification. Antimicrobial Agents and Chemotherapy 52:1244–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aminnejad M, Diaz M, Arabatzis M, et al. 2012. Identification of novel hybrids between Cryptococcus neoformans var. grubii VNI and Cryptococcus gattii VGII. Mycopathologia 173:337–346. [DOI] [PubMed] [Google Scholar]
- Anzawa K, Kawasaki M, Mochizuki T, et al. 2010. Successful mating of Trichophyton rubrum with Arthroderma simii. Medical Mycology 48:629–634. [DOI] [PubMed] [Google Scholar]
- Arabatzis M, Velegraki A. 2013. Sexual reproduction in the opportunistic human pathogen Aspergillus terreus. Mycologia 105:71–79. [DOI] [PubMed] [Google Scholar]
- Araujo R, Amorim A, Gusmão L. 2012. Diversity and specificity of microsatellites within Aspergillus section Fumigati. BMC Microbiology 12: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balajee SA, Gribskov J, Brandt M, et al. 2005a. Mistaken identity: Neosartorya pseudofischeri and its anamorph masquerading as Aspergillus fumigatus. Journal of Clinical Microbiology 43:5996–5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balajee SA, Gribskov JL, Hanley E, et al. 2005b. Aspergillus lentulus sp. nov., a new sibling species of A. fumigatus. Eukaryotic Cell 4:625–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balajee SA, Kano R, Baddley JW, et al. 2009. Molecular identification of Aspergillus species collected for the transplant-associated infection surveillance network. Journal of Clinical Microbiology 47:3138–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balajee SA, Nickle D, Varga J, et al. 2006. Molecular studies reveal frequent misidentification of Aspergillus fumigatus by morphotyping. Eukaryotic Cell 5:1705–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrs V, Beatty J, Dhand NK, et al. 2014. Computed tomographic features of feline sino-nasal and sino-orbital aspergillosis. The Veterinary Journal 201:215–222. [DOI] [PubMed] [Google Scholar]
- Barrs VR, Halliday C, Martin P, et al. 2012. Sinonasal and sino-orbital aspergillosis in 23 cats: Aetiology, clinicopathological features and treatment outcomes. The Veterinary Journal 191:58–64. [DOI] [PubMed] [Google Scholar]
- Barrs VR, Van Doorn TM, Houbraken J, et al. 2013. Aspergillus felis sp. nov., an emerging agent of invasive aspergillosis in humans, cats and dogs. PLoS One 8: e64871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert RR. 2010. DensiTree: making sense of sets of phylogenetic trees. Bioinformatics 26:1372–1373. [DOI] [PubMed] [Google Scholar]
- Bouckaert R[R], Heled J, Kühnert D, et al. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Computational Biology 10: e1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovers M, Hagen F, Kuramae EE, et al. 2006. Unique hybrids between the fungal pathogens Cryptococcus neoformans and Cryptococcus gattii. FEMS Yeast Research 6:599–607. [DOI] [PubMed] [Google Scholar]
- Bovers M, Hagen F, Kuramae EE, et al. 2008. AIDS patient death caused by novel Cryptococcus neoformans × C. gattii hybrid. Emerging Infectious Diseases 14:1105–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden RL, Leslie JF. 1999. Sexual recombination in Gibberella zeae. Phytopathology 89:182–188. [DOI] [PubMed] [Google Scholar]
- Brasier CM. 2001. Rapid evolution of introduced plant pathogens via interspecific hybridization. Bioscience 51:123–133. [Google Scholar]
- Brasier CM, Kirk SA, Pipe ND, et al. 1998. Rare interspecific hybrids in natural populations of the Dutch elm disease pathogens Ophiostoma ulmi and O. novo-ulmi. Mycological Research 102:45–57. [Google Scholar]
- Carbone I, Kohn LM. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91:553–556. [Google Scholar]
- Carstens BC, Pelletier TA, Reid NM, et al. 2013. How to fail at species delimitation. Molecular Ecology 22:4369–4383. [DOI] [PubMed] [Google Scholar]
- Charlton ND, Craven KD, Afkhami ME, et al. 2014. Interspecific hybridization and bioactive alkaloid variation increases diversity in endophytic Epichloë species of Bromus laevipes. FEMS Microbiology Ecology 90:276–289. [DOI] [PubMed] [Google Scholar]
- Chen A, Frisvad J, Sun B, et al. 2016a. Aspergillus section Nidulantes (formerly Emericella): polyphasic taxonomy, chemistry and biology. Studies in Mycology 84:1–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Varga J, Frisvad JC, et al. 2016b. Polyphasic taxonomy of Aspergillus section Cervini. Studies in Mycology 85:65–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AJ, Hubka V, Frisvad JC, et al. 2017. Polyphasic taxonomy of Aspergillus section Aspergillus (formerly Eurotium), and its occurrence in indoor environments and food. Studies in Mycology 88:37–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong GM, Vonk AG, Meis JF, et al. 2017. Interspecies discrimination of A. fumigatus and siblings A. lentulus and A. felis of the Aspergillus section Fumigati using the AsperGenius® assay. Diagnostic Microbiology and Infectious Disease 87:247–252. [DOI] [PubMed] [Google Scholar]
- Coelho D, Silva S, Vale-Silva L, et al. 2011. Aspergillus viridinutans: an agent of adult chronic invasive aspergillosis. Medical Mycology 49:755–759. [DOI] [PubMed] [Google Scholar]
- Cox MP, Dong T, Shen G, et al. 2014. An interspecific fungal hybrid reveals cross-kingdom rules for allopolyploid gene expression patterns. PLoS Genetics 10: e1004180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damann K, DeRobertis C. 2013. Mating of Aspergillus flavus × Aspergillus minisclerotigenes hybrids: are they functionally mules? Phytopathology 103: S2.32–S2.33. [Google Scholar]
- Degnan JH, Rosenberg NA. 2006. Discordance of species trees with their most likely gene trees. PLoS Genetics 2: e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depotter JR, Seidl MF, Wood TA, et al. 2016. Interspecific hybridization impacts host range and pathogenicity of filamentous microbes. Current Opinion in Microbiology 32:7–13. [DOI] [PubMed] [Google Scholar]
- Dettman JR, Jacobson DJ, Taylor JW. 2006. Multilocus sequence data reveal extensive phylogenetic species diversity within the Neurospora discreta complex. Mycologia 98:436–446. [DOI] [PubMed] [Google Scholar]
- Dettman JR, Jacobson DJ, Turner E, et al. 2003. Reproductive isolation and phylogenetic divergence in Neurospora: Comparing methods of species recognition in a model eukaryote. Evolution 57:2721–2741. [DOI] [PubMed] [Google Scholar]
- Dickie IA, Bufford JL, Cobb RC, et al. 2017. The emerging science of linked plant-fungal invasions. New Phytologist 215:1314–1332. [DOI] [PubMed] [Google Scholar]
- Domsch KH, Gams W, Anderson T-H. 2007. Compendium of soil fungi, 2nd taxonomically revised edition. IHW-Verlag, Eching. [Google Scholar]
- Dyer PS, O’Gorman CM. 2011. A fungal sexual revolution: Aspergillus and Penicillium show the way. Current Opinion in Microbiology 14:649–654. [DOI] [PubMed] [Google Scholar]
- Eamvijarn A, Manoch L, Chamswarng C, et al. 2013. Aspergillus siamensis sp. nov. from soil in Thailand. Mycoscience 54:401–405. [Google Scholar]
- Fernandez-Molina JV, Abad-Diaz-de-Cerio A, Sueiro-Olivares M, et al. 2014. Rapid and specific detection of section Fumigati and Aspergillus fumigatus in human samples using a new multiplex real-time PCR. Diagnostic Microbiology and Infectious Disease 80:111–118. [DOI] [PubMed] [Google Scholar]
- Flot J-F. 2015. Species delimitation's coming of age. Systematic Biology 64:897–899. [DOI] [PubMed] [Google Scholar]
- Fontaneto D, Flot J-F, Tang CQ. 2015. Guidelines for DNA taxonomy, with a focus on the meiofauna. Marine Biodiversity 45:433–451. [Google Scholar]
- Frisvad JC, Larsen TO. 2015a. Extrolites of Aspergillus fumigatus and other pathogenic species in Aspergillus section Fumigati. Frontiers in Microbiology 6: 1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisvad JC, Larsen TO. 2015b. Chemodiversity in the genus Aspergillus. Applied Microbiology and Biotechnology 99:7859–7877. [DOI] [PubMed] [Google Scholar]
- Frisvad JC, Thrane U. 1987. Standardized high-performance liquid chromatography of 182 mycotoxins and other fungal metabolites based on alkylphenone retention indices and UV-VIS spectra (diode array detection). Journal of Chromatography A 404:195–214. [DOI] [PubMed] [Google Scholar]
- Frisvad JC, Thrane U. 1993. Liquid column chromatography of mycotoxins. In: Betina V. (eds), Chromatography of mycotoxins: techniques and applications. Journal of Chromatography Library 54: 253–372. [Google Scholar]
- Fujisawa T, Barraclough TG. 2013. Delimiting species using single-locus data and the Generalized Mixed Yule Coalescent approach: a revised method and evaluation on simulated data sets. Systematic Biology 62:707–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale LR, Bryant J, Calvo S, et al. 2005. Chromosome complement of the fungal plant pathogen Fusarium graminearum based on genetic and physical mapping and cytological observations. Genetics 171:985–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier M, Normand A-C, Ranque S. 2016. Previously unknown species of Aspergillus. Clinical Microbiology and Infection 22:662–669. [DOI] [PubMed] [Google Scholar]
- Gladieux P, Vercken E, Fontaine MC, et al. 2010. Maintenance of fungal pathogen species that are specialized to different hosts: allopatric divergence and introgression through secondary contact. Molecular Biology and Evolution 28:459–471. [DOI] [PubMed] [Google Scholar]
- Glass NL, Donaldson GC. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61:1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonthier P, Nicolotti G, Linzer R, et al. 2007. Invasion of European pine stands by a North American forest pathogen and its hybridization with a native interfertile taxon. Molecular Ecology 16:1389–1400. [DOI] [PubMed] [Google Scholar]
- González SS, Barrio E, Querol A. 2008. Molecular characterization of new natural hybrids of Saccharomyces cerevisiae and S. kudriavzevii in brewing. Applied and Environmental Microbiology 74:2314–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heled J, Drummond AJ. 2010. Bayesian inference of species trees from multilocus data. Molecular Biology and Evolution 27:570–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie Y, Miyaji M, Nishimura K, et al. 1995. New and interesting species of Neosartorya from Brazilian soil. Mycoscience 36:199–204. [Google Scholar]
- Horn BW, Moore GG, Carbone I. 2009a. Sexual reproduction in Aspergillus flavus. Mycologia 101:423–429. [DOI] [PubMed] [Google Scholar]
- Horn BW, Moore GG, Carbone I. 2011. Sexual reproduction in aflatoxin-producing Aspergillus nomius. Mycologia 103:174–183. [DOI] [PubMed] [Google Scholar]
- Horn BW, Olarte RA, Peterson SW, et al. 2013. Sexual reproduction in Aspergillus tubingensis from section Nigri. Mycologia 105:1153–1163. [DOI] [PubMed] [Google Scholar]
- Horn BW, Ramirez-Prado JH, Carbone I. 2009b. The sexual state of Aspergillus parasiticus. Mycologia 101:275–280. [DOI] [PubMed] [Google Scholar]
- Hothorn T, Bretz F, Westfall P. 2008. Simultaneous inference in general parametric models. Biometrical Journal 50:346–363. [DOI] [PubMed] [Google Scholar]
- Houbraken J, Spierenburg H, Frisvad JC. 2012. Rasamsonia, a new genus comprising thermotolerant and thermophilic Talaromyces and Geosmithia species. Antonie van Leeuwenhoek 101:403–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubka V, Dudová Z, Kubátová A, et al. 2017. Taxonomic novelties in Aspergillus section Fumigati: A. tasmanicus sp. nov., induction of sexual state in A. turcosus and overview of related species. Plant Systematics and Evolution 303:787–806. [Google Scholar]
- Hubka V, Kolařík M. 2012. β-tubulin paralogue tubC is frequently misidentified as the benA gene in Aspergillus section Nigri taxonomy: primer specificity testing and taxonomic consequences. Persoonia 29:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubka V, Kolařík M, Kubátová A, et al. 2013a. Taxonomical revision of Eurotium and transfer of species to Aspergillus. Mycologia 105:912–937. [DOI] [PubMed] [Google Scholar]
- Hubka V, Kubatova A, Mallatova N, et al. 2012. Rare and new aetiological agents revealed among 178 clinical Aspergillus strains obtained from Czech patients and characterised by molecular sequencing. Medical Mycology 50:601–610. [DOI] [PubMed] [Google Scholar]
- Hubka V, Nissen C, Jensen R, et al. 2015a. Discovery of a sexual stage in Trichophyton onychocola, a presumed geophilic dermatophyte isolated from toenails of patients with a history of T. rubrum onychomycosis. Medical Mycology 53:798–809. [DOI] [PubMed] [Google Scholar]
- Hubka V, Nováková A, Jurjević Ž, et al. 2018. Polyphasic data support the splitting of Aspergillus candidus into two species; proposal of A. dobrogensis sp. nov. International Journal of Systematic and Evolutionary Microbiology 68: 995–1011. doi: 10.1099/ijsem.0.002583. [DOI] [PubMed] [Google Scholar]
- Hubka V, Nováková A, Kolařík M, et al. 2015b. Revision of Aspergillus section Flavipedes: seven new species and proposal of section Jani sect. nov. Mycologia 107:169–208. [DOI] [PubMed] [Google Scholar]
- Hubka V, Nováková A, Peterson SW, et al. 2016a. A reappraisal of Aspergillus section Nidulantes with descriptions of two new sterigmatocystin producing species. Plant Systematics and Evolution 302:1267–1299. [Google Scholar]
- Hubka V, Nováková A, Samson R, et al. 2016b. Aspergillus europaeus sp. nov., a widely distributed soil-borne species related to A. wentii (section Cremei). Plant Systematics and Evolution 302:641–650. [Google Scholar]
- Hubka V, Peterson SW, Frisvad JC, et al. 2013b. Aspergillus waksmanii sp. nov. and Aspergillus marvanovae sp. nov., two closely related species in section Fumigati. International Journal of Systematic and Evolutionary Microbiology 63:783–789. [DOI] [PubMed] [Google Scholar]
- Inderbitzin P, Davis RM, Bostock RM, et al. 2011. The ascomycete Verticillium longisporum is a hybrid and a plant pathogen with an expanded host range. PLoS One 6: e18260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SM, Carlson EL, Pappagianis D. 2015. Coccidioides species determination: does sequence analysis agree with restriction fragment length polymorphism? Mycopathologia 179: 373–379. [DOI] [PubMed] [Google Scholar]
- Jones G. 2017. Algorithmic improvements to species delimitation and phylogeny estimation under the multispecies coalescent. Journal of Mathematical Biology 74:447–467. [DOI] [PubMed] [Google Scholar]
- Jurgenson J, Bowden R, Zeller K, et al. 2002. A genetic map of Gibberella zeae (Fusarium graminearum). Genetics 160:1451–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurjević Ž, Kubátová A, Kolařík M, et al. 2015. Taxonomy of Aspergillus section Petersonii sect. nov. encompassing indoor and soil-borne species with predominant tropical distribution. Plant Systematics and Evolution 301:2441–2462. [Google Scholar]
- Kapli P, Lutteropp S, Zhang J, et al. 2017. Multi-rate Poisson tree processes for single-locus species delimitation under maximum likelihood and Markov chain Monte Carlo. Bioinformatics 33:1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz ME, Dougall AM, Weeks K, et al. 2005. Multiple genetically distinct groups revealed among clinical isolates identified as atypical Aspergillus fumigatus. Journal of Clinical Microbiology 43:551–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki M. 2011. Verification of a taxonomy of dermatophytes based on mating results and phylogenetic analyses. Medical Mycology Journal 52:291–295. [DOI] [PubMed] [Google Scholar]
- Kawasaki M, Anzawa K, Mochizuki T, et al. 2009. Successful mating of a human isolate of Arthroderma simii with a tester strain of A. vanbreuseghemii. Medical Mycology Journal 50:15–18. [DOI] [PubMed] [Google Scholar]
- Kawasaki M, Anzawa K, Wakasa A, et al. 2010. Matings among three teleomorphs of Trichophyton mentagrophytes. Japanese Journal of Medical Mycology 51:143–152. [DOI] [PubMed] [Google Scholar]
- Kelly KL. 1964. Inter-Society Color Council – National Bureau of Standards color name charts illustrated with centroid colors. US Government Printing Office, Washington DC. [Google Scholar]
- Klich MA. 2002. Biogeography of Aspergillus species in soil and litter. Mycologia 94: 21–27. [PubMed] [Google Scholar]
- Kocsubé S, Perrone G, Magistà D, et al. 2016. Aspergillus is monophyletic: evidence from multiple gene phylogenies and extrolites profiles. Studies in Mycology 85:199–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzer A, Li Y, Szaro T, Bruns TD. 1996. Internal transcribed spacer sequences from 38 recognized species of Suillus sensu lato: phylogenetic and taxonomic implications. Mycologia 88:776–785. [Google Scholar]
- Kubatko LS, Degnan JH. 2007. Inconsistency of phylogenetic estimates from concatenated data under coalescence. Systematic Biology 56:17–24. [DOI] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Bennett JE, Wickes BL, et al. 2017. The case for adopting the “species complex” nomenclature for the etiologic agents of cryptococcosis. mSphere 2: e00357-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Sugui JA. 2009. Sexual reproduction in Aspergillus species of medical or economical importance: why so fastidious? Trends in Microbiology 17: 481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Varma A. 2006. Do major species concepts support one, two or more species within Cryptococcus neoformans? FEMS Yeast Research 6: 574–587. [DOI] [PubMed] [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM, et al. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution 34:772–773. [DOI] [PubMed] [Google Scholar]
- Leaché AD, Fujita MK. 2010. Bayesian species delimitation in West African forest geckos (Hemidactylus fasciatus). Proceedings of the Royal Society of London B: Biological Sciences 277:3071–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Ni M, Li W, et al. 2010. The evolution of sex: a perspective from the fungal kingdom. Microbiology and Molecular Biology Reviews 74:298–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Wang M, Damm U, et al. 2016. Species boundaries in plant pathogenic fungi: a Colletotrichum case study. BMC Evolutionary Biology 16: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ, Whelen S, Hall BD. 1999. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution 16:1799–1808. [DOI] [PubMed] [Google Scholar]
- Lockman B, Mascheretti S, Schechter S, et al. 2014. A first generation Heterobasidion hybrid discovered in Larix lyalli in Montana. Plant Disease 98: 1003. [DOI] [PubMed] [Google Scholar]
- Louis VL, Despons L, Friedrich A, et al. 2012. Pichia sorbitophila, an interspecies yeast hybrid, reveals early steps of genome resolution after polyploidization. G3: Genes, Genomes, Genetics 2:299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyskova P, Hubka V, Svobodova L, et al. 2018. Antifungal susceptibility of the Aspergillus viridinutans complex: comparison of two in vitro methods. Antimicrobial Agents and Chemotherapy 62: e01927-17. doi: 10.1128/AAC.01927-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli L, Zalar P, Gunde-Cimerman N, et al. 2017. Aspergillus atacamensis and A. salisburgensis: two new halophilic species from hypersaline/arid habitats with a phialosimplex-like morphology. Extremophiles 21:755–773. [DOI] [PubMed] [Google Scholar]
- Matsuzawa T, Takaki GMC, Yaguchi T, et al. 2015. Aspergillus arcoverdensis, a new species of Aspergillus section Fumigati isolated from caatinga soil in State of Pernambuco, Brazil. Mycoscience 56:123–131. [Google Scholar]
- Mayr A, Lass-Flörl C. 2011. Epidemiology and antifungal resistance in invasive aspergillosis according to primary disease-review of the literature. European Journal of Medical Research 16: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLennan EI, Tucker S, Thrower L. 1954. New soil fungi from Australian heathland: Aspergillus, Penicillium, Spegazzinia. Australian Journal of Botany 2:355–364. [Google Scholar]
- Mehrabi R, Bahkali AH, Abd-Elsalam KA, et al. 2011. Horizontal gene and chromosome transfer in plant pathogenic fungi affecting host range. FEMS Microbiology Reviews 35:542–554. [DOI] [PubMed] [Google Scholar]
- Meyer V, Wu B, Ram AF. 2011. Aspergillus as a multi-purpose cell factory: current status and perspectives. Biotechnology Letters 33:469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mixão V, Gabaldón T. 2018. Hybridization and emergence of virulence in opportunistic human yeast pathogens. Yeast 35:5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Sato H, Tanaka R, et al. 2017. Ribosomal subunit protein typing using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDITOF MS) for the identification and discrimination of Aspergillus species. BMC Microbiology 17: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao Y, Kanamori T, Itoh T, et al. 2009. Genome sequence of the lager brewing yeast, an interspecies hybrid. DNA Research 16:115–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nauta M, Hoekstra R. 1992. Evolution of reproductive systems in filamentous ascomycetes. I. Evolution of mating types. Heredity 68:405–410. [DOI] [PubMed] [Google Scholar]
- Negri C, Gonçalves S, Xafranski H, et al. 2014. Cryptic and rare Aspergillus species in Brazil: prevalence in clinical samples and in vitro susceptibility to triazoles. Journal of Clinical Microbiology 52:3633–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcombe G, Stirling B, McDonald S, et al. 2000. Melampsora × columbiana, a natural hybrid of M. medusae and M. occidentalis. Mycological Research 104:261–274. [Google Scholar]
- Nguyen L-T, Schmidt HA, Von Haeseler A, et al. 2015. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen KF, Månsson M, Rank C, et al. 2011. Dereplication of microbial natural products by LC-DAD-TOFMS. Journal of Natural Products 74:2338–2348. [DOI] [PubMed] [Google Scholar]
- Nováková A, Hubka V, Dudová Z, et al. 2014. New species in Aspergillus section Fumigati from reclamation sites in Wyoming (USA) and revision of A. viridinutans complex. Fungal Diversity 64:253–274. [Google Scholar]
- O’Donnell K. 1993. Fusarium and its near relatives. In: Reynolds DR, Taylor JW. (eds), The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics: 225–233. CAB International, Wallingford. [Google Scholar]
- O’Donnell K, Ward TJ, Geiser DM, et al. 2004. Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genetics and Biology 41:600–623. [DOI] [PubMed] [Google Scholar]
- O’Gorman CM, Fuller HT, Dyer PS. 2009. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature 457:471–474. [DOI] [PubMed] [Google Scholar]
- Olarte RA, Worthington CJ, Horn BW, et al. 2015. Enhanced diversity and aflatoxigenicity in interspecific hybrids of Aspergillus flavus and Aspergillus parasiticus. Molecular Ecology 24:1889–1909. [DOI] [PubMed] [Google Scholar]
- Peterson SW. 2008. Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia 100:205–226. [DOI] [PubMed] [Google Scholar]
- Peterson SW, Jurjević Ž, Frisvad JC. 2015. Expanding the species and chemical diversity of Penicillium section Cinnamopurpurea. PLoS One 10: e0121987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt JI, Hocking AD. 2009. Spoilage of stored, processed and preserved foods. In: Pitt JI, Hocking AD. (eds), Fungi and food spoilage. Springer, London: 401–421. [Google Scholar]
- Posada D. 2008. jModelTest: phylogenetic model averaging. Molecular Biology and Evolution 25:1253–1256. [DOI] [PubMed] [Google Scholar]
- Pujol C, Daniels KJ, Lockhart SR, et al. 2004. The closely related species Candida albicans and Candida dubliniensis can mate. Eukaryotic Cell 3:1015–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2015. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. [Google Scholar]
- Réblová M, Hubka V, Thureborn O, et al. 2016. From the tunnels into the treetops: new lineages of black yeasts from biofilm in the Stockholm metro system and their relatives among ant-associated fungi in the Chaetothyriales. PLoS One 11: e0163396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid NM, Carstens BC. 2012. Phylogenetic estimation error can decrease the accuracy of species delimitation: a Bayesian implementation of the general mixed Yule-coalescent model. BMC Evolutionary Biology 12: 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Van der Mark P, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA. 2013. Discordance of species trees with their most likely gene trees: a unifying principle. Molecular Biology and Evolution 30:2709–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydholm C, Dyer P, Lutzoni F. 2007. DNA sequence characterization and molecular evolution of MAT1 and MAT2 mating-type loci of the self-compatible ascomycete mold Neosartorya fischeri. Eukaryotic Cell 6:868–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabino R, Veríssimo C, Parada H, et al. 2014. Molecular screening of 246 Portuguese Aspergillus isolates among different clinical and environmental sources. Medical Mycology 52:519–529. [DOI] [PubMed] [Google Scholar]
- Samson RA, Visagie CM, Houbraken J, et al. 2014. Phylogeny, identification and nomenclature of the genus Aspergillus. Studies in Mycology 78:141–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardl C, Craven K. 2003. Interspecific hybridization in plant-associated fungi and oomycetes: a review. Molecular Ecology 12:2861–2873. [DOI] [PubMed] [Google Scholar]
- Schmitt I, Crespo A, Divakar PK, et al. 2009. New primers for promising single-copy genes in fungal phylogenetics and systematics. Persoonia 23:35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder MS, De San Vicente KM, Prandini TH, et al. 2016. Multiple origins of the pathogenic yeast Candida orthopsilosis by separate hybridizations between two parental species. PLoS Genetics 12: e1006404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzfeld MD, Sperling FA. 2015. Comparison of five methods for delimitating species in Ophion Fabricius, a diverse genus of parasitoid wasps (Hymenoptera, Ichneumonidae). Molecular Phylogenetics and Evolution 93:234–248. [DOI] [PubMed] [Google Scholar]
- Shigeyasu C, Yamada M, Nakamura N, et al. 2012. Keratomycosis caused by Aspergillus viridinutans: an Aspergillus fumigatus-resembling mold presenting distinct clinical and antifungal susceptibility patterns. Medical Mycology 50:525–528. [DOI] [PubMed] [Google Scholar]
- Short DP, O’Donnell K, Geiser DM. 2014. Clonality, recombination, and hybridization in the plumbing-inhabiting human pathogen Fusarium keratoplasticum inferred from multilocus sequence typing. BMC Evolutionary Biology 14: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short DP, O’Donnell K, Thrane U, et al. 2013. Phylogenetic relationships among members of the Fusarium solani species complex in human infections and the descriptions of F. keratoplasticum sp. nov. and F. petroliphilum stat. nov. Fungal Genetics and Biology 53:59–70. [DOI] [PubMed] [Google Scholar]
- Shymanovich T, Charlton ND, Musso AM, et al. 2017. Interspecific and intraspecific hybrid Epichloë species symbiotic with the North American native grass Poa alsodes. Mycologia 109:459–474. [DOI] [PubMed] [Google Scholar]
- Singh G, Dal Grande F, Divakar PK, et al. 2015. Coalescent-based species delimitation approach uncovers high cryptic diversity in the cosmopolitan lichen-forming fungal genus Protoparmelia (Lecanorales, Ascomycota). PLoS One 10: e0124625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipiczki M. 2008. Interspecies hybridization and recombination in Saccharomyces wine yeasts. FEMS Yeast Research 8:996–1007. [DOI] [PubMed] [Google Scholar]
- Sklenář F, Jurjević Ž, Zalar P, et al. 2017. Phylogeny of xerophilic aspergilli (subgenus Aspergillus) and taxonomic revision of section Restricti. Studies in Mycology 88:161–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solla A, Dacasa M, Nasmith C, et al. 2008. Analysis of Spanish populations of Ophiostoma ulmi and O. novo-ulmi using phenotypic characteristics and RAPD markers. Plant Pathology 57:33–44. [Google Scholar]
- Spiers A, Hopcroft D. 1994. Comparative studies of the poplar rusts Melampsora medusae, M. larici-populina and their interspecific hybrid M. medusae-populina. Mycological Research 98:889–903. [Google Scholar]
- Staats M, Van Baarlen P, Van Kan JA. 2005. Molecular phylogeny of the plant pathogenic genus Botrytis and the evolution of host specificity. Molecular Biology and Evolution 22:333–346. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology 57:758–771. [DOI] [PubMed] [Google Scholar]
- Starkey DE, Ward TJ, Aoki T, et al. 2007. Global molecular surveillance reveals novel Fusarium head blight species and trichothecene toxin diversity. Fungal Genetics and Biology 44:1191–1204. [DOI] [PubMed] [Google Scholar]
- Stewart JE, Timmer LW, Lawrence CB, et al. 2014. Discord between morphological and phylogenetic species boundaries: incomplete lineage sorting and recombination results in fuzzy species boundaries in an asexual fungal pathogen. BMC Evolutionary Biology 14: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugui JA, Kwon-Chung KJ, Juvvadi PR, et al. 2015. Aspergillus fumigatus and related species. Cold Spring Harbor Perspectives in Medicine 5: a019786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugui JA, Peterson SW, Figat A, et al. 2014. Genetic relatedness versus biological compatibility between Aspergillus fumigatus and related species. Journal of Clinical Microbiology 52:3707–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugui JA, Vinh DC, Nardone G, et al. 2010. Neosartorya udagawae (Aspergillus udagawae), an emerging agent of aspergillosis: How different is it from Aspergillus fumigatus? Journal of Clinical Microbiology 48: 220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swilaiman SS, O’Gorman CM, Balajee SA, et al. 2013. Discovery of a sexual cycle in Aspergillus lentulus, a close relative of A. fumigatus. Eukaryotic Cell 12:962–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk E, Krappmann S. 2010. Conserved regulators of mating are essential for Aspergillus fumigatus cleistothecium formation. Eukaryotic Cell 9:774–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot JJ, Barrs VR. 2018. One-health pathogens in the Aspergillus viridinutans complex. Medical Mycology 56:1–12. [DOI] [PubMed] [Google Scholar]
- Talbot JJ, Houbraken J, Frisvad JC, et al. 2017. Discovery of Aspergillus frankstonensis sp. nov. during environmental sampling for animal and human fungal pathogens. PLoS One 12: e0181660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanney JB, Visagie CM, Yilmaz N, et al. 2017. Aspergillus subgenus Polypaecilum from the built environment. Studies in Mycology 88:237–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner E, Jacobson D, Taylor JW. 2010. Reinforced postmating reproductive isolation barriers in Neurospora, an Ascomycete microfungus. Journal of Evolutionary Biology 23:1642–1656. [DOI] [PubMed] [Google Scholar]
- Vinh DC, Shea YR, Jones PA, et al. 2009. Chronic invasive aspergillosis caused by Aspergillus viridinutans. Emerging Infectious Diseases 15:1292–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Yilmaz N, Renaud JB, et al. 2017. A survey of xerophilic Aspergillus from indoor environment, including descriptions of two new section Aspergillus species producing eurotium-like sexual states. MycoKeys 19:1–30. [Google Scholar]
- White TJ, Bruns T, Lee S, et al. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, et al. (eds), PCR protocols: a guide to methods and applications: 315–322. Academic Press, San Diego. [Google Scholar]
- Wickham H. 2009. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag, New York. [Google Scholar]
- Wu G, Zhao H, Li C, et al. 2015. Genus-wide comparative genomics of Malassezia delineates its phylogeny, physiology, and niche adaptation on human skin. PLoS Genetics 11: e1005614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X. 2017. DAMBE6: new tools for microbial genomics, phylogenetics, and molecular evolution. Journal of Heredity 108:431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaguchi T, Horie Y, Tanaka R, et al. 2007. Molecular phylogenetics of multiple genes on Aspergillus section Fumigati isolated from clinical specimens in Japan. Japanese Journal of Medical Mycology 48:37–46. [DOI] [PubMed] [Google Scholar]
- Yang Z, Rannala B. 2010. Bayesian species delimitation using multilocus sequence data. Proceedings of the National Academy of Sciences 107:9264–9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Smith DK, Zhu H, et al. 2017. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods in Ecology and Evolution 8:28–36. [Google Scholar]
- Zhang J, Kapli P, Pavlidis P, et al. 2013. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29:2869–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]