Abstract
Malassezia is a genus of medically-important, lipid-dependent yeasts that live on the skin of warm-blooded animals. The 17 described species have been documented primarily on humans and domestic animals, but few studies have examined Malassezia species associated with more diverse host groups such as wildlife. While investigating the skin mycobiota of healthy bats, we isolated a Malassezia sp. that exhibited only up to 92% identity with other known species in the genus for the portion of the DNA sequence of the internal transcribed spacer region that could be confidently aligned. The Malassezia sp. was cultured from the skin of nine species of bats in the subfamily Myotinae; isolates originated from bats sampled in both the eastern and western United States. Physiological features and molecular characterisation at seven additional loci (D1/D2 region of 26S rDNA, 18S rDNA, chitin synthase, second largest subunit of RNA polymerase II, β-tubulin, translation elongation factor EF-1α, and minichromosome maintenance complex component 7) indicated that all of the bat Malassezia isolates likely represented a single species distinct from other named taxa. Of particular note was the ability of the Malassezia sp. to grow over a broad range of temperatures (7–40 °C), with optimal growth occurring at 24 °C. These thermal growth ranges, unique among the described Malassezia, may be an adaptation by the fungus to survive on bats during both the host's hibernation and active seasons. The combination of genetic and physiological differences provided compelling evidence that this lipid-dependent yeast represents a novel species described herein as Malassezia vespertilionis sp. nov. Whole genome sequencing placed the new species as a basal member of the clade containing the species M. furfur, M. japonica, M. obtusa, and M. yamatoensis. The genetic and physiological uniqueness of Malassezia vespertilionis among its closest relatives may make it important in future research to better understand the evolution, life history, and pathogenicity of the Malassezia yeasts.
Keywords: Chiroptera, evolution, hibernation, Malassezia, Myotis, new species, phylogeny
INTRODUCTION
Members of the genus Malassezia are lipid-dependent fungi specialised to live on the skin of humans and other euthermic animals. Malassezia is the sole genus in the class Malasseziomycetes, and the 17 described species appear to be part of the natural skin mycobiome of animals (Wang et al. 2014). At least five species have been regularly associated with dermatitis or other types of skin disorders in humans (reviewed by Gaitanis et al. 2012), and M. pachydermatis is described as the cause of otitis externa in domestic dogs (Gustafson 1955, Bond et al. 2004). However, these potentially pathogenic species of Malassezia are also found on areas of normal skin of afflicted patients and on asymptomatic individuals, making it unclear what role the fungi play in skin disease (reviewed by Gaitanis et al. 2012). Malassezia furfur, M. pachydermatis, and M. sympodialis have also occasionally been implicated as causes of sepsis in infants and immunocompromised patients (reviewed by Gaitanis et al. 2012, Aguirre et al. 2015, Patron 2016), and it has even been hypothesised that Malassezia could play a role in promoting certain forms of skin cancer (Gaitanis et al. 2011).
In addition to their medical ambiguity, relatively little is known about the diversity and ecology of Malassezia. Members of the genus were traditionally identified on the basis of phenotypic traits, and prior to 1996 there were only three recognised species of Malassezia. More recent application of molecular techniques to assist with species characterisation has facilitated the ability to distinguish cryptic species and has increased the known diversity of the genus (e.g., Sugita et al. 2002, Hirai et al. 2004, Cabañes et al. 2007, 2016, Honnavar et al. 2016). Sugita et al. (2010) present a list of the animal hosts from which various species of Malassezia have been recovered. However, many reports used to generate that list lacked molecular data to support the identification of the Malassezia species that were isolated, and potentially novel taxa may have been overlooked. Indeed, diversity of the genus is likely much higher than currently documented (Amend 2014, Cabañes 2014). Ten of the 17 Malassezia species are most closely associated with humans, and were discovered through culture-based surveys of diseased skin; the remaining seven species (M. brasiliensis, M. caprae, M. cuniculi, M. equina, M.nana, M.pachydermatis, and M. psittaci) have been isolated primarily from animals (reviewed by Sugita et al. 2010, Cabañes et al. 2016), with M. pachydermatis reported as a zoonotic pathogen (Chang et al. 1998). Some of the zoophilic members of the group appear to have a broad host range, while others are more host-specific (reviewed by Guého-Kellerman et al. 2010, Sugita et al. 2010). Little work has been done with broader taxonomic host groups, and the relatively small number of described species of Malassezia is likely the result of sampling bias, which is skewed toward humans and domestic animals. Given the host specificity of some species of Malassezia, many more taxa may be discovered when a broader range of host species (especially wildlife) are sampled. Such undiscovered species of Malassezia could be important in further elucidating the taxonomy, evolution, ecology, and pathogenicity of this group of medically important fungi.
While investigating the mycobiota on the skin of bats, we detected a putative Malassezia sp. that was genetically distinct from other known members of the genus. Here we describe the isolation, occurrence, and characterisation of this novel species.
MATERIALS AND METHODS
Isolation of Malassezia from bats
Samples were collected in 2014, 2016, and 2017 under the U.S. Geological Survey - National Wildlife Health Center (NWHC) Animal Care and Use Committee Protocols #EP140212 and #EP081124-A2, with all necessary permits and permissions for the sites and species sampled. Hibernating bats (Fig. 1) were captured by hand and active bats were captured in mist nets. Gloves were changed between animals to prevent cross-contamination. The animals were sampled non-lethally using sterile Pur-Wraps® polyester-tipped swabs (Puritan Medical Products Company LLC, Guilford, Maine, USA) pre-moistened with 150 μL of sterile nuclease-free water. Swabs were gently rolled back-and-forth three times across the skin of the forearm and wing membrane between the elbow and wrist joints. Samples were then placed in sterile microcentrifuge tubes, stored chilled for up to 48 h, and shipped on ice to the NWHC. A total of 264 samples were obtained from thirteen sites in seven states (one site in Alabama, USA; one site in California, USA; one site in Kentucky, USA; one site in Missouri, USA; one site in Pennsylvania, USA; two sites in New York, USA; and six sites in Wisconsin, USA), representing ten bat species (Lasionycteris noctivagans, Myotis californicus, Myotis grisescens, Myotis leibii, Myotis lucifugus, Myotis septentrionalis, Myotis sodalis, Myotis thysanodes, Myotis yumanensis, and Perimyotis subflavus). A list of all individual bats sampled for the project is provided in Table 1.
Fig. 1.

Hibernating bats, such as these Myotis sp., were sampled for this study by swabbing wing skin.
Table 1.
List of individual bats sampled for this study.
| Individual identifier | Location | Host species | Sampling date | Malassezia vespertilionis isolated |
|---|---|---|---|---|
| 24716-001 | Wisconsin, USA (site #1) | Myotis septentrionalis | 03 March 2014 | no |
| 24716-002 | Wisconsin, USA (site #1) | Perimyotis subflavus | 03 March 2014 | no |
| 24716-003 | Wisconsin, USA (site #1) | Myotis septentrionalis | 03 March 2014 | no |
| 24716-004 | Wisconsin, USA (site #1) | Perimyotis subflavus | 03 March 2014 | no |
| 24716-005 | Wisconsin, USA (site #1) | Perimyotis subflavus | 03 March 2014 | no |
| 24716-006 | Wisconsin, USA (site #1) | Perimyotis subflavus | 03 March 2014 | no |
| 24716-007 | Wisconsin, USA (site #1) | Myotis septentrionalis | 03 March 2014 | yes |
| 24716-008 | Wisconsin, USA (site #1) | Myotis septentrionalis | 03 March 2014 | yes |
| 24716-009 | Wisconsin, USA (site #1) | Perimyotis subflavus | 03 March 2014 | no |
| 24716-010 | Wisconsin, USA (site #1) | Perimyotis subflavus | 03 March 2014 | no |
| 24716-011 | Wisconsin, USA (site #1) | Perimyotis subflavus | 03 March 2014 | no |
| 24716-012 | Wisconsin, USA (site #1) | Perimyotis subflavus | 03 March 2014 | no |
| 24716-013 | Wisconsin, USA (site #1) | Perimyotis subflavus | 03 March 2014 | no |
| 24716-014 | Wisconsin, USA (site #1) | Myotis septentrionalis | 03 March 2014 | yes |
| 24716-015 | Wisconsin, USA (site #1) | Myotis lucifugus | 03 March 2014 | no |
| 24716-016 | Wisconsin, USA (site #1) | Perimyotis subflavus | 03 March 2014 | no |
| 24716-017 | Wisconsin, USA (site #1) | Myotis septentrionalis | 03 March 2014 | no |
| 24716-018 | Wisconsin, USA (site #1) | Myotis septentrionalis | 03 March 2014 | no |
| 24716-019 | Wisconsin, USA (site #1) | Myotis septentrionalis | 03 March 2014 | no |
| 24716-020 | Wisconsin, USA (site #1) | Myotis sp.* | 03 March 2014 | no |
| 24738-002 | Wisconsin, USA (site #2) | Myotis lucifugus | 10 March 2014 | yes |
| 24738-005 | Wisconsin, USA (site #2) | Myotis lucifugus | 10 March 2014 | no |
| 24738-006 | Wisconsin, USA (site #2) | Myotis lucifugus | 10 March 2014 | no |
| 24738-008 | Wisconsin, USA (site #2) | Myotis lucifugus | 10 March 2014 | no |
| 24738-011 | Wisconsin, USA (site #2) | Myotis lucifugus | 10 March 2014 | no |
| 24738-013 | Wisconsin, USA (site #2) | Myotis lucifugus | 10 March 2014 | yes |
| 24738-022 | Wisconsin, USA (site #2) | Myotis lucifugus | 10 March 2014 | no |
| 24738-025 | Wisconsin, USA (site #2) | Myotis sp.* | 10 March 2014 | yes |
| 44767-001 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-002 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-003 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-004 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-005 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-006 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-007 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-008 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-009 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-010 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-011 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-012 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-013 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-014 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-015 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-016 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-017 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-018 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-019 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-020 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-021 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-022 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-023 | Kentucky, USA | Myotis grisescens | 04 March 2014 | no |
| 44767-024 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-025 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-026 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-027 | Kentucky, USA | Myotis sodalis | 04 March 2014 | no |
| 44767-028 | Kentucky, USA | Myotis sodalis | 04 March 2014 | no |
| 44767-029 | Kentucky, USA | Myotis lucifugus | 04 March 2014 | no |
| 44767-030 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-031 | Kentucky, USA | Myotis grisescens | 04 March 2014 | yes |
| 44767-032 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-033 | Kentucky, USA | Myotis sodalis | 04 March 2014 | yes |
| 44767-034 | Kentucky, USA | Myotis sodalis | 04 March 2014 | yes |
| 44767-035 | Kentucky, USA | Myotis sodalis | 04 March 2014 | yes |
| 44767-036 | Kentucky, USA | Myotis sodalis | 04 March 2014 | yes |
| 44767-037 | Kentucky, USA | Myotis sodalis | 04 March 2014 | yes |
| 44767-038 | Kentucky, USA | Myotis sodalis | 04 March 2014 | yes |
| 44767-039 | Kentucky, USA | Myotis sodalis | 04 March 2014 | yes |
| 44767-040 | Kentucky, USA | Myotis sodalis | 04 March 2014 | yes |
| 44767-041 | Kentucky, USA | Myotis sodalis | 04 March 2014 | yes |
| 44767-042 | Kentucky, USA | Myotis sodalis | 04 March 2014 | yes |
| 44767-043 | Kentucky, USA | Myotis sodalis | 04 March 2014 | no |
| 44767-044 | Kentucky, USA | Myotis sodalis | 04 March 2014 | yes |
| 44767-045 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-046 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-047 | Kentucky, USA | Perimyotis subflavus | 04 March 2014 | no |
| 44767-048 | Kentucky, USA | Myotis sodalis | 04 March 2014 | no |
| 44768-001 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | no |
| 44768-002 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | no |
| 44768-003 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | no |
| 44768-004 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | no |
| 44768-005 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | no |
| 44768-006 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | no |
| 44768-007 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | no |
| 44768-008 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | no |
| 44768-009 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | no |
| 44768-010 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | no |
| 44768-011 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | no |
| 44768-012 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | yes |
| 44768-013 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | no |
| 44768-014 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | no |
| 44768-015 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | no |
| 44768-016 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | no |
| 44768-017 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | no |
| 44768-018 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | no |
| 44768-019 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | no |
| 44768-020 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | yes |
| 44768-021 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | yes |
| 44768-022 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | no |
| 44768-023 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | no |
| 44768-024 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | no |
| 44768-025 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | yes |
| 44768-026 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | no |
| 44768-027 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | no |
| 44768-028 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | no |
| 44768-029 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | no |
| 44768-030 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | no |
| 44768-031 | New York, USA (site #1) | Myotis lucifugus | 19 March 2014 | no |
| 44769-001 | Wisconsin, USA (site #3) | Myotis lucifugus | 28 March 2014 | yes |
| 44769-002 | Wisconsin, USA (site #3) | Myotis lucifugus | 28 March 2014 | no |
| 44769-003 | Wisconsin, USA (site #3) | Myotis lucifugus | 28 March 2014 | no |
| 44769-004 | Wisconsin, USA (site #3) | Myotis lucifugus | 28 March 2014 | yes |
| 44769-005 | Wisconsin, USA (site #3) | Myotis lucifugus | 28 March 2014 | no |
| 44769-006 | Wisconsin, USA (site #3) | Myotis lucifugus | 28 March 2014 | yes |
| 44769-007 | Wisconsin, USA (site #3) | Myotis lucifugus | 28 March 2014 | no |
| 44769-008 | Wisconsin, USA (site #3) | Myotis lucifugus | 28 March 2014 | yes |
| 44769-009 | Wisconsin, USA (site #3) | Myotis lucifugus | 28 March 2014 | no |
| 44769-010 | Wisconsin, USA (site #3) | Myotis lucifugus | 28 March 2014 | yes |
| 44769-011 | Wisconsin, USA (site #3) | Myotis lucifugus | 28 March 2014 | no |
| 44769-012 | Wisconsin, USA (site #3) | Myotis lucifugus | 28 March 2014 | no |
| 44769-013 | Wisconsin, USA (site #3) | Myotis lucifugus | 28 March 2014 | yes |
| 44769-014 | Wisconsin, USA (site #3) | Myotis lucifugus | 28 March 2014 | yes |
| 44769-015 | Wisconsin, USA (site #3) | Myotis lucifugus | 28 March 2014 | no |
| 44769-016 | Wisconsin, USA (site #3) | Myotis lucifugus | 28 March 2014 | yes |
| 44769-017 | Wisconsin, USA (site #3) | Myotis lucifugus | 28 March 2014 | yes |
| 44769-018 | Wisconsin, USA (site #3) | Myotis lucifugus | 28 March 2014 | yes |
| 44769-019 | Wisconsin, USA (site #3) | Myotis lucifugus | 28 March 2014 | yes |
| 44769-020 | Wisconsin, USA (site #3) | Myotis lucifugus | 28 March 2014 | yes |
| 44769-021 | Wisconsin, USA (site #3) | Myotis lucifugus | 28 March 2014 | yes |
| 44769-022 | Wisconsin, USA (site #3) | Myotis lucifugus | 28 March 2014 | yes |
| 44769-023 | Wisconsin, USA (site #3) | Myotis lucifugus | 28 March 2014 | yes |
| 44769-024 | Wisconsin, USA (site #3) | Myotis lucifugus | 28 March 2014 | yes |
| 44769-025 | Wisconsin, USA (site #3) | Myotis lucifugus | 28 March 2014 | yes |
| 44797-032 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | no |
| 44797-033 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | no |
| 44797-034 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | no |
| 44797-035 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | yes |
| 44797-036 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | no |
| 44797-037 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | no |
| 44797-038 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | no |
| 44797-039 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | no |
| 44797-040 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | no |
| 44797-041 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | no |
| 44797-042 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | no |
| 44797-043 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | no |
| 44797-044 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | no |
| 44797-045 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | no |
| 44797-046 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | no |
| 44797-047 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | no |
| 44797-048 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | no |
| 44797-049 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | no |
| 44797-050 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | no |
| 44797-051 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | no |
| 44797-052 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | no |
| 44797-053 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | no |
| 44797-054 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | no |
| 44797-055 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | no |
| 44797-056 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | no |
| 44797-057 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | no |
| 44797-058 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | no |
| 44797-059 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | no |
| 44797-060 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | no |
| 44797-061 | New York, USA (site #2) | Myotis lucifugus | 15 January 2015 | no |
| 44797-062 | Missouri, USA | Myotis sodalis | 24 February 2015 | no |
| 44797-063 | Missouri, USA | Perimyotis subflavus | 24 February 2015 | no |
| 44797-064 | Missouri, USA | Myotis sodalis | 24 February 2015 | yes |
| 44797-065 | Missouri, USA | Myotis sodalis | 24 February 2015 | no |
| 44797-066 | Missouri, USA | Myotis sodalis | 24 February 2015 | no |
| 44797-067 | Missouri, USA | Perimyotis subflavus | 24 February 2015 | no |
| 44797-068 | Missouri, USA | Myotis sodalis | 24 February 2015 | no |
| 44797-069 | Missouri, USA | Myotis sodalis | 24 February 2015 | no |
| 44797-070 | Missouri, USA | Perimyotis subflavus | 24 February 2015 | no |
| 44797-071 | Missouri, USA | Myotis sodalis | 24 February 2015 | no |
| 44797-072 | Missouri, USA | Myotis sodalis | 24 February 2015 | no |
| 44797-073 | Missouri, USA | Myotis sodalis | 24 February 2015 | no |
| 44797-074 | Missouri, USA | Myotis sodalis | 24 February 2015 | no |
| 44797-075 | Missouri, USA | Perimyotis subflavus | 24 February 2015 | no |
| 44797-076 | Missouri, USA | Myotis sodalis | 24 February 2015 | no |
| 44797-077 | Missouri, USA | Perimyotis subflavus | 24 February 2015 | no |
| 44797-078 | Missouri, USA | Perimyotis subflavus | 24 February 2015 | no |
| 44797-100 | Wisconin, USA (site #4) | Myotis septentrionalis | 27 January 2015 | yes |
| 44797-101 | Wisconin, USA (site #4) | Myotis septentrionalis | 27 January 2015 | no |
| 44797-102 | Wisconin, USA (site #4) | Myotis septentrionalis | 27 January 2015 | no |
| 44797-103 | Wisconsin, USA (site #1) | Myotis septentrionalis | 28 January 2015 | yes |
| 44797-104 | Wisconsin, USA (site #1) | Myotis septentrionalis | 28 January 2015 | yes |
| 44797-105 | Wisconsin, USA (site #1) | Myotis septentrionalis | 28 January 2015 | no |
| 44797-106 | Wisconsin, USA (site #1) | Myotis septentrionalis | 28 January 2015 | no |
| 44797-107 | Wisconsin, USA (site #1) | Myotis septentrionalis | 28 January 2015 | no |
| 44797-108 | Wisconsin, USA (site #1) | Myotis septentrionalis | 28 January 2015 | no |
| 44797-109 | Wisconsin, USA (site #1) | Myotis septentrionalis | 28 January 2015 | no |
| 44797-110 | Wisconsin, USA (site #1) | Myotis septentrionalis | 28 January 2015 | no |
| 44797-111 | Wisconsin, USA (site #5) | Myotis septentrionalis | 29 January 2015 | no |
| 44797-112 | Wisconsin, USA (site #5) | Myotis septentrionalis | 29 January 2015 | no |
| 44797-113 | Wisconsin, USA (site #5) | Myotis septentrionalis | 29 January 2015 | no |
| 44797-114 | Wisconsin, USA (site #5) | Myotis septentrionalis | 29 January 2015 | no |
| 44797-115 | Wisconsin, USA (site #5) | Myotis septentrionalis | 29 January 2015 | yes |
| 44797-116 | Wisconsin, USA (site #5) | Myotis septentrionalis | 29 January 2015 | no |
| 44797-123 | Wisconsin, USA (site #6) | Myotis septentrionalis | 02 March 2015 | no |
| 44797-124 | Wisconsin, USA (site #6) | Myotis septentrionalis | 02 March 2015 | yes |
| 44797-125 | Wisconsin, USA (site #6) | Myotis septentrionalis | 02 March 2015 | no |
| 44797-126 | Wisconsin, USA (site #6) | Myotis septentrionalis | 02 March 2015 | no |
| 44797-127 | Wisconsin, USA (site #6) | Myotis septentrionalis | 02 March 2015 | yes |
| 44797-128 | Wisconsin, USA (site #6) | Myotis septentrionalis | 02 March 2015 | no |
| 44797-129 | Wisconsin, USA (site #6) | Perimyotis subflavus | 02 March 2015 | no |
| 44797-130 | Wisconsin, USA (site #6) | Myotis septentrionalis | 02 March 2015 | no |
| 44797-131 | Wisconsin, USA (site #6) | Myotis septentrionalis | 02 March 2015 | yes |
| 44797-132 | Alabama, USA | Perimyotis subflavus | 11 February 2015 | no |
| 44797-133 | Alabama, USA | Myotis grisescens | 11 February 2015 | no |
| 44797-134 | Alabama, USA | Perimyotis subflavus | 11 February 2015 | no |
| 44797-135 | Alabama, USA | Myotis sodalis | 11 February 2015 | yes |
| 44797-136 | Alabama, USA | Myotis grisescens | 11 February 2015 | no |
| 44797-137 | Alabama, USA | Myotis grisescens | 11 February 2015 | yes |
| 44797-138 | Alabama, USA | Myotis grisescens | 11 February 2015 | yes |
| 44797-139 | Alabama, USA | Myotis grisescens | 11 February 2015 | yes |
| 44797-140 | Alabama, USA | Perimyotis subflavus | 11 February 2015 | no |
| 44797-141 | Alabama, USA | Myotis grisescens | 11 February 2015 | yes |
| 44797-142 | Alabama, USA | Myotis grisescens | 11 February 2015 | no |
| 44797-143 | Alabama, USA | Myotis grisescens | 11 February 2015 | yes |
| 44797-144 | Alabama, USA | Myotis grisescens | 11 February 2015 | no |
| 44797-145 | Alabama, USA | Perimyotis subflavus | 11 February 2015 | no |
| 44797-146 | Alabama, USA | Myotis grisescens | 11 February 2015 | no |
| 44797-147 | Alabama, USA | Myotis grisescens | 11 February 2015 | no |
| 44797-148 | Alabama, USA | Myotis grisescens | 11 February 2015 | yes |
| 44797-149 | Alabama, USA | Perimyotis subflavus | 11 February 2015 | no |
| 44797-150 | Alabama, USA | Myotis grisescens | 11 February 2015 | yes |
| 44797-151 | Alabama, USA | Myotis grisescens | 11 February 2015 | no |
| 44797-152 | Alabama, USA | Myotis grisescens | 11 February 2015 | yes |
| 44797-153 | Alabama, USA | Myotis sodalis | 11 February 2015 | yes |
| 44797-154 | Alabama, USA | Myotis grisescens | 11 February 2015 | no |
| 44797-155 | Alabama, USA | Myotis grisescens | 11 February 2015 | no |
| 45701-660 | California, USA | Myotis yumanensis | 07 May 2016 | no |
| 45701-661 | California, USA | Myotis sp.** | 02 May 2016 | no |
| 45701-663 | California, USA | Myotis yumanensis | 18 April 2016 | no |
| 45701-664 | California, USA | Myotis yumanensis | 02 May 2016 | yes |
| 45701-665 | California, USA | Myotis californicus | 07 May 2016 | yes |
| 45701-666 | California, USA | Myotis californicus | 02 May 2016 | no |
| 45701-668 | California, USA | Myotis yumanensis | 09 May 2016 | yes |
| 45701-669 | California, USA | Myotis yumanensis | 18 April 2016 | no |
| 45701-671 | California, USA | Myotis yumanensis | 09 May 2016 | no |
| 45701-672 | California, USA | Myotis californicus | 02 May 2016 | no |
| 45701-674 | California, USA | Myotis californicus | 26 April 2016 | no |
| 45701-675 | California, USA | Myotis yumanensis | 07 May 2016 | no |
| 45701-676 | California, USA | Myotis sp.** | 02 May 2016 | yes |
| 45701-677 | California, USA | Myotis yumanensis | 18 April 2016 | yes |
| 45701-678 | California, USA | Myotis californicus | 18 April 2016 | yes |
| 45701-680 | California, USA | Myotis sp.** | 18 April 2016 | no |
| 45701-681 | California, USA | Myotis sp.** | 09 May 2016 | no |
| 45701-682 | California, USA | Myotis californicus | 19 April 2016 | yes |
| 45701-683 | California, USA | Myotis yumanensis | 20 April 2016 | no |
| 45701-684 | California, USA | Myotis californicus | 09 May 2016 | no |
| 45701-685 | California, USA | Myotis yumanensis | 20 April 2016 | no |
| 45701-686 | California, USA | Myotis thysanodes | 26 April 2016 | yes |
| 45701-687 | California, USA | Myotis yumanensis | 20 April 2016 | no |
| 45701-688 | California, USA | Myotis yumanensis | 20 April 2016 | yes |
| 45701-689 | California, USA | Myotis sp.** | 02 May 2016 | no |
| 45701-691 | California, USA | Myotis californicus | 16 May 2016 | yes |
| 45701-696 | California, USA | Myotis californicus | 16 May 2016 | no |
| 45701-699 | California, USA | Lasionycteris noctivagans | 16 May 2016 | no |
| 45701-714 | California, USA | Lasionycteris noctivagans | 16 May 2016 | yes |
| 45701-719 | California, USA | Myotis californicus | 16 May 2016 | yes |
| 45704-161 | Pennsylvania, USA | Myotis leibii | 26 February 2016 | yes |
| 46375-001 | California, USA | Myotis californicus | 08 May 2017 | yes |
| 46375-002 | California, USA | Myotis californicus | 08 May 2017 | yes |
| 46375-003 | California, USA | Lasionycteris noctivagans | 08 May 2017 | yes |
| 46375-004 | California, USA | Lasionycteris noctivagans | 08 May 2017 | no |
*either Myotis lucifugus or Myotis septentrionalis.
**either Myotis lucifugus or Myotis yumanensis.
Upon arrival at the laboratory, swabs were streaked onto Leeming and Notman agar (LNA; 10 g bacteriological peptone, 0.1 g yeast extract, 5 g glucose, 8 g desiccated ox bile, 1 mL glycerol, 0.5 g glycerol monostearate, 0.5 g Tween 60, 10 mL whole fat cow's milk, 0.5 g chloramphenicol, 0.5 g cycloheximide, 15 g agarose per litre, pH 6.0; modified slightly from Leeming & Notman (1987)) and incubated at 7 °C. Plates were checked weekly for a total of 12 wk, and any colonies resembling Malassezia were transferred to fresh LNA. Isolates were identified by sequencing the ITS as described by Lorch et al. (2015).
Whole genome sequence analysis
Isolate CBS 15041 (NWHC 44797-103; UAMH 11924) was selected for whole genome sequencing to further resolve the taxonomy of the bat-associated Malassezia. Nucleic acid was obtained using a phenol-chloroform extraction. Library preparation and next-generation sequencing was performed by the University of Wisconsin Biotechnology Center DNA Sequencing Facility using the genomic Nextera XT DNA Library Prep Kit (Illumina Inc., San Diego, CA) and the Illumina MiSeq Next Generation Sequencer platform. Sequence data was processed and assembled using JAAWS (https://github.com/nextgenusfs/jaaws). Briefly, the paired-end 250-bp MiSeq sequence reads (2 × 250) were processed with trimmomatic v. 0.36 (Bolger et al. 2014) to remove adapter sequences and phiX spike-in was removed using bowtie2 v. 2.3.2 (Langmead & Salzberg 2012) alignment to the phiX genome (NC_001422). The data were then assembled into scaffolds with Spades v. 3.9.0 (Bankevich et al. 2012). The subsequent assembly was cleaned using Blobtools v. 0.9.19 (Laetsch & Blaxter 2017) and filtered for unexpected coverage, mitochondrial DNA, contamination, and scaffolds less than 1 kb in length. Finally, the cleaned assembly was error corrected using five iterations of Pilon v. 1.22 (Walker et al. 2014). The genome of the bat-associated Malassezia was annotated with funannotate v. 0.7.0 while the 28 genomes of Malassezia species previously sequenced (Wu et al. 2015; Table 2) were annotated using funannotate v. 0.5.3 (https://github.com/nextgenusfs/funannotate). Conserved orthologues were identified using BUSCO2 (Simão et al. 2015) basidiomycete database using the busco wrapper script in Phyloma (https://github.com/nextgenusfs/phyloma). The concatenated protein sequences of 254 conserved BUSCO2 orthologues were aligned using MAFFT v. 7.305b (Katoh & Standley 2013) and trimmed using trimAl v. 1.4.rev15 (Capella-Gutiérrez et al. 2009). A maximum-likelihood phylogeny was estimated using RAxML v. 8.2.10 (Stamatakis 2014) (PROTGAMMALG, 100 bootstrap replicates). As a secondary method, a Bayesian phylogeny was estimated using MrBayes v. 3.2.6 (Ronquist & Huelsenbeck 2003) through the CIPRES Science Gateway (Miller et al. 2010). For the Bayesian analysis, an LG model with gamma-distributed rate variation across sites was used. To generate the 50 % majority rule consensus tree, two runs, each with 1 000 000 generations and four chains, were performed. The chains were sampled every 250 generations with the first 25 % of sampled values discarded as burn-in.
Table 2.
Fungal genomes and summary statistics used for whole genome phylogenic analysis.
| Species | Strain identifier | GenBank accession number | Locus_tag | Assembly size (Mb) | Number of scaffolds | Scaffold N50 (Kb) | Percent GC | Protein coding gene models |
|---|---|---|---|---|---|---|---|---|
| Malassezia caprae | CBS 10434* | GCA_001264625.1 | MCA1 | 7.58 | 229 | 110 | 59.78% | 3.553 |
| M. cuniculi | CBS 11721* | GCA_001264635.1 | MCU1 | 7.459 | 76 | 522 | 58.99% | 3.167 |
| M. dermatis | CBS 9169* | GCA_001264665.1 | MDM1 | 7.54 | 111 | 189 | 59.10% | 3.538 |
| JCM 11348 | GCA_001600775.1 | MDM2 | 7.551 | 18 | 1.325 | 58.98% | 3.544 | |
| M. equina | CBS 9969* | GCA_001264685.1 | MEQ1 | 7.658 | 117 | 372 | 58.00% | 3.232 |
| M. furfur | JPLK 23 | GCA_001265065.1 | MFU6 | 7.79 | 2 092 | 14 | 64.18% | 3.087 |
| CBS 1878* | GCA_001265055.1 | MFU1 | 13.865 | 3460 | 14 | 63.91% | 5.418 | |
| CBS 4172 | GCA_001264895.1 | MFU2 | 14.347 | 3453 | 15 | 64.23% | 5.672 | |
| CBS 7019* | GCA_001264875.1 | MFU3 | 13.707 | 3262 | 15 | 63.70% | 5.565 | |
| CBS 7710 | GCA_001264865.1 | MFU4 | 15.232 | 4053 | 14 | 63.89% | 5.835 | |
| CBS 7982 | GCA_001265045.1 | MFU5 | 7.877 | 1694 | 20 | 64.06% | 3.158 | |
| M. globosa | CBS 7966* | GCA_001264805.1 | MGL2 | 8.94 | 113 | 724 | 52.02% | 4.245 |
| CBS 7874 | GCA_001264815.1 | MGL1 | 8.938 | 138 | 398 | 51.87% | 4.191 | |
| CBS 7990 | GCA_001264795.1 | MGL3 | 8.884 | 108 | 414 | 52.07% | 3.703 | |
| M. japonica | CBS 9431* | GCA_001264785.1 | MJA1 | 8.341 | 295 | 66 | 62.38% | 4.215 |
| JCM 11963 | GCA_001600795.1 | MJA2 | 8.364 | 16 | 814 | 62.33% | 4.122 | |
| M. nana | CBS 9557* | GCA_001265015.1 | MNA1 | 7.607 | 95 | 492 | 57.93% | 3.785 |
| JCM 12085 | GCA_001600835.1 | MNA2 | 7.579 | 13 | 1.323 | 57.96% | 3.734 | |
| M. obtusa | CBS 7876* | GCA_001264985.1 | MOB1 | 7.842 | 1709 | 22 | 62.15% | 2.893 |
| M. pachydermatis | CBS 1879* | GCA_001264975.1 | MPA1 | 8.158 | 61 | 957 | 55.08% | 4.134 |
| M. restricta | CBS 7877* | GCA_001264765.1 | MRE1 | 7.249 | 90 | 402 | 55.83% | 3.556 |
| CBS 8742 | GCA_001264725.1 | MRE2 | 7.26 | 69 | 666 | 55.79% | 3.569 | |
| M. slooffiae | CBS 7956* | GCA_001264965.1 | MSL1 | 8.425 | 1641 | 15 | 65.82% | 3.422 |
| M. sympodialis | ATCC 42132 | GCA_001264925.1 | MSY1 | 7.546 | 824 | 54 | 58.77% | 3.055 |
| ATCC 44340 | GCA_001264715.1 | MSY2 | 7.562 | 769 | 59 | 58.88% | 3.080 | |
| ATCC 96806 | GCA_001264705.1 | MSY3 | 7.526 | 1030 | 44 | 58.80% | 3.946 | |
| M. vespertilionis | CBS 15041* | GCA_002818225.1 | MVES | 7.581 | 14 | 844 | 56.62% | 3.791 |
| M. yamatoensis | CBS 9725* | GCA_001264885.1 | MYA1 | 8.106 | 49 | 1.447 | 49.62% | 3.971 |
* type or neotype strain.
Multilocus sequence analysis
To determine whether the isolates of Malassezia from bats represented a single species, seven loci (in addition to ITS) from 12 isolates (Table 3) were also examined: the D1/D2 region of 26S rDNA (hereafter referred to as D1/D2), the 18S rDNA, chitin synthase CHS2, second largest subunit of RNA polymerase II (RPB2), β-tubulin (β-tub), translation elongation factor EF-1α (TEF1), and minichromosome maintenance complex component 7 (MCM7). DNA was extracted using the methods of Lorch et al. (2015). The D1/D2 region was amplified using primers NL4 (5’-GGT CCG TGT TTC AAG ACG G-3’) and NL1 (5’-GCA TAT CAA TAA GCG GAG GAA AAG-3’) (O’Donnell 1993); cycling conditions: 94 °C for 5 min; 30 cycles of 94 °C for 45 s, 51 °C for 1 min, and 72 °C for 3 min; and a final extension of 72 °C for 10 min. The 18S rDNA was amplified with forward (5’-ATC TGG TTG ATC CTG CCA GT-3’) and reverse (5’-TCC TCC GCT TAT TGA TAT GC-3’) primers described by Sugita & Nakase (1999); cycling conditions: 94 °C for 3 min; 30 cycles of 94 °C for 30 s, 55 °C for 1 min, and 72 °C for 2 min and 30 s; and a final extension of 72 °C for 10 min. A portion of CHS2 was amplified with primers ChiSyn2f (5’-CTG AAG CTT CAN ATG TAY AAY GAR GAY-3’) and ChiSyn2r (5’-GTT CTC GAG YTT RTA YTC RAA RTT YTG-3’) (Bowen et al. 1992, Cabañes et al. 2007); cycling conditions: 94 °C for 5 min; 45 cycles of 94 °C for 1 min, 50 °C for 2 min, and 72 °C for 3 min; and a final extension of 72 °C for 6 min. A fragment of RPB2 was amplified with primers fRPB2-5F (5’-GAY GAY MGW GAT CAY TTY GG-3’) and fRPB2-7cR (5’-CCC ATR GCT TGY TTR CCC AT-3’) (Liu et al. 1999); cycling conditions: 94 °C for 4 min; 40 cycles of 94 °C for 1 min, 50 °C for 1 min, and 72 °C for 1 min; and a final extension of 72 °C for 8 min. A portion of β-tub was amplified with primers F-βtub (5’-CAR GCY GGT CAR TGY GGT AAC CA-3’) and F- βtub4r (5’-GCC TCA GTR AAY TCC ATY TCR TCC AT-3’) (Einax & Voigt 2003); cycling conditions: 95 °C for 5 min; 30 cycles of 95 °C for 30 s, 50 °C for 1 min, and 72 °C for 1 min; and a final extension of 72 °C for 10 min. A portion of TEF1 was amplified with primers EF1-983F (5’-GCY CCY GGH CAY CGT GAY TTY AT-3’) and EF1-2218R (5’-ATG ACA CCR ACR GCR ACR GTY TG-3’) (Rehner & Buckley 2005); cycling conditions: 94 °C for 2 min; 47 cycles of 94 °C for 1 min, 53 °C for 1 min, and 72 °C for 1 min and 40 s; and a final extension of 72 °C for 10 min. A portion of MCM7 was amplified with primers MCM7-709 (5’-ACN MGN GTN TCV GAY GTH AAR CC-3’) and MCM7-1348 (5’-GAY TTD GCN ACN CCN GGR TCW CCC AT-3’) (modified slightly from Schmitt et al. 2009); cycling conditions as described for TEF1. Reactions were carried out using GoTaq® Flexi DNA polymerase (Promega Corporation, Madison, WI) according to the manufacturer's instructions (with final concentrations of 1.5 mM MgCl2 solution and 1 μM of each primer) except that twice the recommended amount of Taq polymerase (2.5 U) and 1–3 μL of template were added per 50 μL reaction. When necessary, PCR products were gel-purified prior to sequencing using the QIAquick Gel Extraction Kit (Qiagen Inc., Valencia, CA). All PCR products were sequenced in both directions using the same primers described for the initial amplifications. Additional internal sequencing primers were used for some loci: 18S rDNA, forward primers (5’-GCT ACC ACA TCC AAG GAA GG-3’, 5’-CTG CGA AAG CAT TTG CCA AGG-3’, 5’-TCT GGG CCG CAC GCG CGC TAC ACT G-3’) and reverse primers (5’-TGG AAT TAC CGC GGC TGC TGG CAC C-3’, 5’-TCC TTG GCA AAT GCT TTC GCA G-3’, 5’-CCG TCA ATT CCT TTA AGT TTC AGC C-3’, 5’-AAG GTC TCG TTC GTT ATC G-3’, 5’-GAC GGG CGG TGT GTA CAA AGG GCA G-3’) (Sugita & Nakase 1999); RPB2, RPB2-6f (5’-TGG GGH ATG GTV TGY CCB GC-3’) and RPB2-6r (5’-GCV GGR CAB ACC ATD CCC CA-3’) (modified slightly from Liu et al. 1999); and TEF1, EF1-1577F (5’-CAG GAY GTN TAC AAG ATY GGT GG-3’) and EF1-1567R (5’-ACH GTR CCR ATA CCA CCR ATC TT-3’) (Rehner & Buckley 2005).
Table 3.
List of GenBank accession numbers for individual locus Malassezia sequence data generated for this project. Existing sequence data used for the multilocus phylogenetic analysis are listed at the end of the table. GenBank accession numbers beginning with ‘L’ represent the whole genome contig from which sequence data for a given locus was obtained.
| Isolate identifier | Other identifier | Species | GenBank accession numbers |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ITS | D1/D2 | 18S rDNA | β-tub | TEF1 | RPB2 | MCM7 | CHS2 | |||
| NWHC 24716-007 | M. vespertilionis | MF669399 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 24716-008 | M. vespertilionis | MF669400 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 24716-014 | CBS 15042 | M. vespertilionis | MF669401 | MF669387 | MF669375 | MF669327 | MF669315 | MF669351 | MF669363 | MF669339 |
| NWHC 24738-002 | M. vespertilionis | MF669402 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 24738-013 | M. vespertilionis | MF669403 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 24738-025 | M. vespertilionis | MF669404 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44767-031 | M. vespertilionis | MF669405 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44767-033 | M. vespertilionis | MF669406 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44767-034 | CBS 15043 | M. vespertilionis | MF669407 | MF669388 | MF669376 | MF669328 | MF669316 | MF669352 | MF669364 | MF669340 |
| NWHC 44767-035 | M. vespertilionis | MF669408 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44767-036 | M. vespertilionis | MF669409 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44767-037 | UAMH 11925 | M. vespertilionis | MF669410 | MF669389 | MF669377 | MF669329 | MF669317 | MF669353 | MF669365 | MF669341 |
| NWHC 44767-038 | M. vespertilionis | MF669411 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44767-039 | M. vespertilionis | MF669412 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44767-040 | M. vespertilionis | MF669413 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44767-041 | M. vespertilionis | MF669414 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44767-042 | M. vespertilionis | MF669415 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44767-044 | M. vespertilionis | MF669416 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44768-012 | M. vespertilionis | MF669417 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44768-020 | M. vespertilionis | MF669418 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44768-021 | M. vespertilionis | MF669419 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44768-025 | CBS 15044 | M. vespertilionis | MF669420 | MF669390 | MF669378 | MF669330 | MF669318 | MF669354 | MF669366 | MF669342 |
| NWHC 44769-001 | M. vespertilionis | MF669421 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44769-004 | M. vespertilionis | MF669422 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44769-006 | M. vespertilionis | MF669423 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44769-008 | M. vespertilionis | MF669424 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44769-010 | M. vespertilionis | MF669425 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44769-013 | M. vespertilionis | MF669426 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44769-014 | M. vespertilionis | MF669427 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44769-016 | M. vespertilionis | MF669428 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44769-017 | M. vespertilionis | MF669429 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44769-018 | M. vespertilionis | MF669430 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44769-019 | M. vespertilionis | MF669431 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44769-020 | M. vespertilionis | MF669432 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44769-021 | M. vespertilionis | MF669433 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44769-022 | M. vespertilionis | MF669434 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44769-023 | M. vespertilionis | MF669435 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44769-024 | M. vespertilionis | MF669436 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44769-025 | M. vespertilionis | MF669437 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44797-035 | CBS 15045 | M. vespertilionis | MF669438 | MF669391 | MF669379 | MF669331 | MF669319 | MF669355 | MF669367 | MF669343 |
| NWHC 44797-064 | M. vespertilionis | MF669439 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44797-100 | M. vespertilionis | MF669440 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44797-103* | CBS 15041*, UAMH 11924* | M. vespertilionis | MF669441 | MF669392 | MF669380 | MF669332 | MF669320 | MF669356 | MF669368 | MF669344 |
| NWHC 44797-104 | M. vespertilionis | MF669442 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44797-115 | M. vespertilionis | MF669443 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44797-124 | M. vespertilionis | MF669444 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44797-127 | M. vespertilionis | MF669445 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44797-131 | M. vespertilionis | MF669446 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44797-135 | M. vespertilionis | MF669447 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44797-137 | CBS 15046 | M. vespertilionis | MF669448 | MF669393 | MF669381 | MF669333 | MF669321 | MF669357 | MF669369 | MF669345 |
| NWHC 44797-138 | M. vespertilionis | MF669449 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44797-139 | M. vespertilionis | MF669450 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44797-141 | M. vespertilionis | MF669451 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44797-143 | M. vespertilionis | MF669452 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44797-148 | M. vespertilionis | MF669453 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44797-150 | M. vespertilionis | MF669454 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44797-152 | M. vespertilionis | MF669455 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 44797-153 | M. vespertilionis | MF669456 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 45701-664 | M. vespertilionis | MF669457 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 45701-665 | CBS 15048 | M. vespertilionis | MF669458 | MF669394 | MF669382 | MF669334 | MF669322 | MF669358 | MF669370 | MF669346 |
| NWHC 45701-668 | M. vespertilionis | MF669459 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 45701-676 | M. vespertilionis | MF669460 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 45701-677 | M. vespertilionis | MF669461 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 45701-678 | M. vespertilionis | MF669462 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 45701-682 | CBS 15051 | M. vespertilionis | MF669463 | MF669395 | MF669383 | MF669335 | MF669323 | MF669359 | MF669371 | MF669347 |
| NWHC 45701-686 | CBS 15049 | M. vespertilionis | MF669464 | MF669396 | MF669384 | MF669336 | MF669324 | MF669360 | MF669372 | MF669348 |
| NWHC 45701-688 | M. vespertilionis | MF669465 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 45701-691 | CBS 15050 | M. vespertilionis | MF669466 | MF669397 | MF669385 | MF669337 | MF669325 | MF669361 | MF669373 | MF669349 |
| NWHC 45701-714 | M. vespertilionis | MF669467 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 45701-719 | M. vespertilionis | MF669468 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 45704-161 | M. vespertilionis | MF669469 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 46375-001 | M. vespertilionis | MF669470 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 46375-002 | M. vespertilionis | MF669471 | NA | NA | NA | NA | NA | NA | NA | |
| NWHC 46375-003 | CBS 15047 | M. vespertilionis | MF669472 | MF669398 | MF669386 | MF669338 | MF669326 | MF669362 | MF669374 | MF669350 |
| M9927* | CBS 9169* | M. dermatis | AB070356 | AB070361 | KF706452.1 | LFFX01000095.1 | LFFX01000013.1 | LFFX01000105.1 | LFFX01000023.1 | LFFX01000059.1 |
| CBS 1878* | M. furfur | AY743634 | AY743602 | EU192363.1 | LFGI01000839.1 | LFGI01000568.1 | LFGI01001900.1 | LFGI01002103.1 | LFGI01003290.1 | |
| CBS 7019* | M. furfur | AY743635 | AY743603 | NA | LFGG01001873.1 | LFGG01000118.1 | LFGG01002621.1 | LFGG01003117.1 | LFGG01000889.1 | |
| NCPF 3349 | M. furfur | NA | NA | AY083223.1 | NA | NA | NA | NA | NA | |
| CBS 9431* | M. japonica | EF140669 | EF140672 | KF706458.1 | LFDB01000216.1 | LFDB01000004.1 | LFDB01000183.1 | LFDB01000119.1 | LFDB01000111.1 | |
| CBS 7876* | M. obtusa | AY387137 | AY743629 | NA | LFGC01001328.1 | LFGC01000155.1 | LFGC01000795.1 | LFGC01001067.1 | LFGC01000917.1 | |
| CBS 7968 | M. obtusa | NA | NA | EU192365.1 | NA | NA | NA | NA | NA | |
| CBS 1879* | M. pachydermatis | AY387139 | AY743605 | LFGB01000057.1 | LFGB01000029.1 | LFGB01000018.1 | LFGB01000009.1 | LFGB01000036.1 | LFGB01000046.1 | |
* type or neotype strain.
NA = sequence data not available or not used for analyses.
A phylogenetic analysis was conducted using newly generated sequences from the bat-associated isolates of Malassezia and from existing sequence data in GenBank for type cultures of a subset of the recognised species of Malassezia for which sufficient sequence data were available (Table 3). Members of the genus residing in the same core clade as the type isolate (as determined by the whole-genome analysis described above) were included in this analysis as were representatives from the other two core clades described by Wu et al. (2015). Sequence data for protein-coding genes were obtained from whole genome sequences deposited in GenBank by Wu et al. (2015), while that for multicopy genes originated from various sources (see Table 3). For M. obtusa and one isolate of M.furfur, 18S sequence data were not available for the type isolates; instead, sequence data from non ex-type strains were substituted for that locus.
Nucleotide sequences were aligned independently for each locus using MUSCLE in MEGA v. 6 (Tamura et al. 2013), and all gaps were deleted. MEGA 6 was also used to determine the best substitution model for each locus. A multigene phylogenetic analysis was then conducted by concatenating the final alignments of all eight loci. A Bayesian analysis was run as described above, except that 5 000 000 generations were used for each run and the sampling frequency was set to 1 000. Data was partitioned by locus and (for coding genes) by nucleotide codon position. A Kimura 2-parameter model with gamma distribution was applied to the non-coding loci (ITS, 18S rDNA, and D1/D2); a Kimura 2-parameter model with gamma distribution and invariant sites was used for CHS2; a general time-reversible model with gamma distribution was used for β-tub, TEF1, and RPB2; and an HKY model with gamma distribution and invariant sites was applied to MCM7.
Physiological and morphological characterisation of isolates
Seven isolates of the bat-associated Malassezia sp. that were analysed genetically were also characterised physiologically and morphologically using criteria commonly employed to distinguish species in the genus (Guého-Kellerman et al. 2010). To determine the influence of temperature on growth, all isolates were incubated at various temperatures as described by Guého et al. (1996). Due to the fastidious nature of the isolates, growth temperature experiments were conducted on LNA instead of modified Dixon's agar (mDA; Guillot et al. 1996). Growth was assessed at the following temperatures: 7, 24, 30, 37, and 40 °C. Inclusion of growth characteristics at 7 °C and 24 °C are not standard for Malassezia, but were performed because the fungus was isolated from hibernating bats, which maintain low body temperatures. Plates were inoculated by transferring cells with an inoculating loop and streaking for isolated colonies. The diameter of isolated colonies was measured and colony morphology descriptions were recorded every 10 d for a total of 50 d. Cell morphology was assessed from wet mounts with lactophenol cotton blue stain conducted on 10-d-old cultures that were grown on LNA at 24 °C.
The ability of the bat-associated Malassezia isolates to grow on mDA and without lipid supplementation on Sabouraud dextrose agar (SDA) was tested by inoculating these media as described above. Utilisation of different types of Tween (i.e., 0.1 % Tween 80, 0.5 % Tween 40, 0.5 % Tween 60, and 10 % Tween 20) was tested according to the methodologies of Guého et al. (1996). The seven isolates were also characterised using the Tween Diffusion and Cremophor EL Assimilation Tests (Guillot et al. 1996). For all of these experiments, cultures were incubated at 32 °C and checked every seven days for a total of 50 d. Tests were also performed at 24 °C to ensure that lack of positive results was not due to incubation at temperatures outside the optimal growth range for the Malassezia sp.
Additional physiological tests included catalase reaction and β-glucosidase activity. The catalase reaction was performed by harvesting a loop full of cells from a 7-d-old culture grown on LNA at 24 °C, smearing the cells onto a glass slide, and adding one drop of 3 % hydrogen peroxide (Guého et al. 1996). The ability to hydrolyse esculin (i.e., β-glucosidase activity) was tested following the methods of Mayser et al. (1997) except that 7-d-old cultures grown on LNA were used to inoculate the medium due to the slower growth rate of the bat-associated isolates relative to other species of Malassezia. The esculin agar tubes were incubated at 32 °C and checked daily for 14 d, and thereafter they were checked every seven days for an additional 30 d. The test was considered positive if a black precipitate was produced. Mating experiments were not performed.
RESULTS
Isolation of Malassezia from bats
Fungal colonies that resembled Malassezia were visible on LNA medium after 40–50 d of incubation at 7 °C. These putative Malassezia colonies were observed growing in culture from samples collected from 75 of the 264 (28 %) bats examined. These included nine host species from seven U.S. states (Table 1; Fig. 2). The ITS sequences of 74 isolates shared 99.7–100 % sequence identity with one another across the approximately 760-bp fragment that was analysed. The remaining isolate had an ITS sequence divergent from the other 74 isolates and was not included in further analyses. When searched against the GenBank database, ITS sequences of the 74 bat-associated isolates most closely matched M. japonica (92 % sequence identity) within the portion of the sequences corresponding to the 5.8S rDNA; however, ITS1 and ITS2 were highly divergent from sequences that resided within GenBank. The ITS sequence data are available in GenBank (MF669451–MF669472).
Fig. 2.
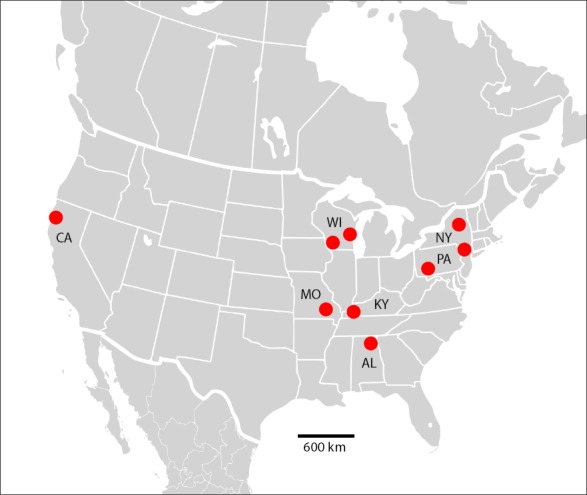
Map of the United States, showing the locations of sampling sites from which bats yielded isolates of Malassezia vespertilionis sp. nov. States from which isolates were obtained are labelled (AL = Alabama; CA = California; KY = Kentucky; MO = Missouri; NY = New York; PA = Pennsylvania; WI = Wisconsin).
Some isolates of the Malassezia sp. lost vigour and eventually failed to grow after several passes on LNA. Transfer of these isolates to mDA did not cause reinvigoration but rather seemed to facilitate further decline of the cultures. The reason for this was not determined. Despite losing some isolates, the majority grew well in the laboratory. Representative isolates were deposited in the Westerdijk Fungal Biodiversity Institute and the UAMH Centre for Global Microfungal Biodiversity culture collections (Table 3).
Whole genome sequence analysis
The Whole Genome Shotgun project was deposited at DDBJ/ENA/GenBank under the WGS Project PECA00000000 (accession GCA_002818225.1). The version described in this paper is version PECA01000000. Raw sequencing data is available from the NCBI SRA via the accession SRP121079. The genome of the Malassezia isolated from bats was 7.581 Mb, contained in 14 scaffolds, which is consistent with other Malassezia species (Boekhout et al. 1998, Wu et al. 2015). The annotated genome is estimated to contain 3 791 protein coding gene models (Table 2). Twenty-seven genomes of Malassezia species were obtained from NCBI GenBank, however most of these data contained only assemblies without annotated gene models. Thus, all genome assemblies were re-annotated using funannotate in order to generate comparable annotations between species. The number of predicted genes was similar to previously published reports (Table 2) (Wu et al. 2015). Using the annotated genomes and Ustilago maydis as an outgroup, we identified core conserved gene models in each genome using BUSCO v. 2 orthologous groups and generated a concatenated alignment of 254 gene models found in all 29 genomes studied. Using both maximum likelihood and Bayesian methods, the bat-associated Malassezia was placed as the most basal member of clade A (Fig. 3).
Fig. 3.
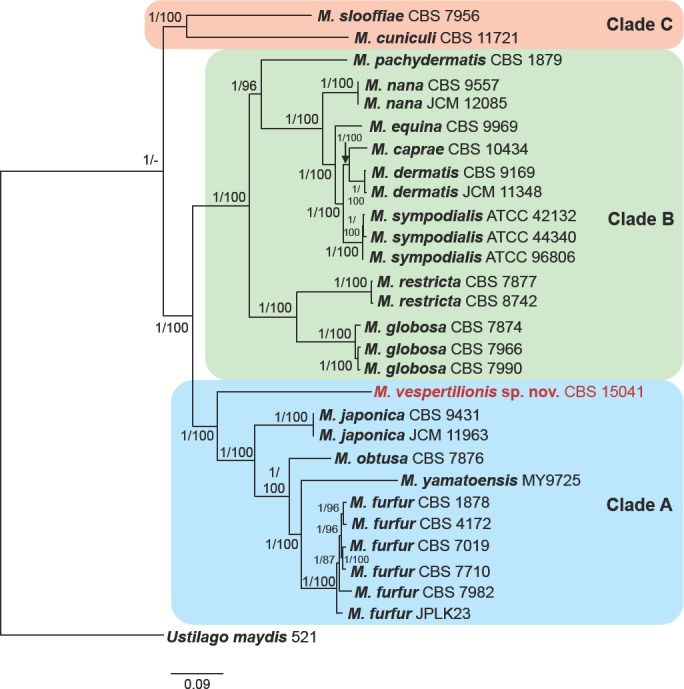
Phylogenetic tree of the genus Malassezia based on concatenated amino acid sequences of 254 conserved orthologues. The tree from the Bayesian analysis is shown, but the tree generated from the maximum likelihood analysis had an identical topology. Posterior probabilities (Bayesian)/bootstrap values (maximum likelihood), respectively, are shown at the nodes. Ustilago maydis was used to root the tree. Clades A, B, C as described by Wu et al. (2015) are illustrated. Based on the analyses, M. vespertilionis sp. nov. is a basal member of clade A.
Multilocus sequence analysis
To demonstrate that all of the Malassezia isolates from bats represented a single taxon, multilocus sequencing analysis was performed on 12 isolates (including the type isolate). The isolates for the analysis were selected to represent a broad range of host species, geographic locations, and strains with slight sequence variations in the ITS region. The portions sequenced of D1/D2 region, β-tub, TEF1, and RPB2 were 100 % identical among the 12 isolates, and the ITS region, 18S rDNA, MCM7, and CHS2 shared at least 99.7, 99.9, 99.8, and 99.8 % sequence identity, respectively, between the isolates examined. The sequences generated for the multilocus analysis are available in GenBank (D1/D2: MF669394–MF669398; 18S rDNA: MF669382–MF669386; β-tub: MF669334–MF669338; TEF1: MF669322–MF669326; RPB2: MF669358–MF669362; MCM7: MF669370–MF669374; CHS2: MF669346–MF669350).
The final alignments used for the phylogenetic analysis consisted of the following numbers of characters: ITS region, 402 characters; 18S rDNA, 1 065 characters; D1/D2 region, 541 characters; β-tub, 1 041 characters; TEF1, 987 characters; MCM7, 600 characters; RPB2, 1 086 characters; and CHS2, 534 characters. The small number of characters included in the ITS region alignment was due to high divergence between the different Malassezia species within the ITS1 and ITS2 regions, resulting in their subsequent removal from the alignment. Thus, the final ‘ITS’ alignment consisted almost entirely of sequence data representing the 5.8S rDNA which is more highly conserved among species of Malassezia. The Bayesian analysis based on the eight concatenated sequences produced a tree that showed the same relationships among the subset of species included as the phylogenetic analysis based on whole genome sequencing. The 12 Malassezia isolates from bats all grouped together into a well-supported clade that likely represented a single species (Fig. 4).
Fig. 4.
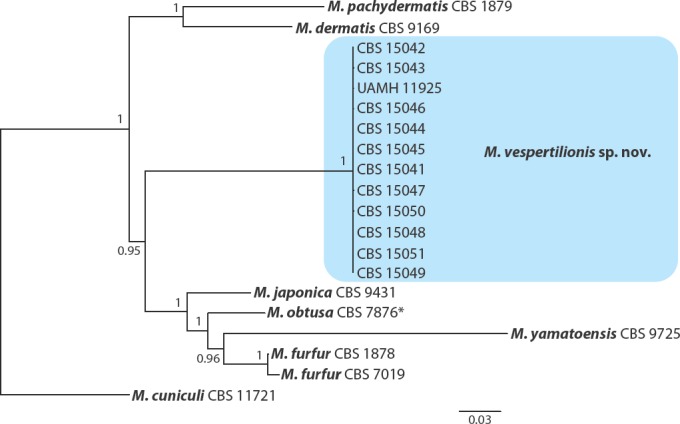
Phylogenetic tree resulting from a Bayesian analysis of concatenated nucleotide sequences from eight loci (ITS, 18S rDNA, D1/D2 region, and portions of the β-tub, TEF1, MCM7, RPB2, CHS2 genes) of 12 Malassezia isolates from bats, all Malassezia species from clade A for which sufficient genetic data was available, and representative members from clades B and C (Wu et al. 2015). Posterior probabilities are presented at each node. All examined isolates from bats formed a well-supported clade, suggesting that they represent a single taxon referred to herein as M. vespertilionis sp. nov.
Physiological and morphological characterisation of isolates
The Malassezia sp. isolated from bats grew at all temperatures tested (7, 24, 30, 37, and 40 °C), with best growth (i.e., largest colony diameters) occurring at 24 °C. At higher temperatures, growth was slower. This was problematic since standard physiological tests used to characterise species of Malassezia are typically conducted at 32 °C for incubation periods that were too brief to allow for sufficient growth of this novel taxon from bats (Guého et al. 1996). Thus, we conducted most tests at both 24 °C and 32 °C and allowed cultures to incubate for up to 50 d. Test results at the two temperatures were generally equivalent, although positive test results often required longer incubation times and produced weaker results at 32 °C compared to 24 °C. Detailed descriptions of growth at different temperatures, cell morphology, and colony morphology are presented in the species description and in Table 4. Colony and cell morphology are also shown in Fig. 5.
Table 4.
Physiological characteristics of the various species of Malassezia. Seven isolates of M. vespertilionis were characterized and the characteristics are summarized (‘Overall’) for the species based on those results. Information from other species of Malassezia were taken from previous summaries and original data by Cabañes et al. 2011, 2016, and Honnavar et al. 2016. Results are displayed as pos (positive reaction/test), neg (negative reaction/test), weak (weak positive reaction/test), variable (different isolates produce variable results), and ? (no information available for this specific test). When multiple results are listed for a given test/reaction, the one listed first is the most common result obtained from the isolates examined; results in parentheses are observed only rarely.
| Species | Cell morphology | Growth mDA | Lipid dependency | Utilization |
Tween Diffusion |
Activity |
Growth |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tween 20 | Tween 40 | Tween 60 | Tween 80 | Cremophor EL | Tween 20 | Tween 40 | Tween 60 | Tween 80 | Catalase | β-glucosidase | 37 °C | 40 °C | ||||
| M. arunalokei | ovoidal, globose | pos | pos | neg | neg | neg | variable | neg | ? | ? | ? | ? | neg | neg | pos | neg |
| M. brasiliensis | ovoidal, ellipsoidal | pos | pos | pos | pos | pos | pos | pos | ? | ? | ? | ? | pos | neg | pos | pos |
| M. caprae | globose, ellipsoidal | pos | pos | neg | pos | pos | pos, (neg) | neg | neg | pos | pos | pos, (neg) | pos | pos, (neg) | neg, (weak) | neg |
| M. cuniculi | globose | neg, weak | pos | neg | neg | neg | neg | neg | neg | neg | neg | neg | pos | pos | pos | pos |
| M. dermatis | ellipsoidal, globose | pos | pos | pos | pos | pos | pos | pos, weak | pos | pos | pos | pos | pos | ? | pos | pos |
| M. equina | ellipsoidal | pos | pos | weak | pos | pos | pos | neg | weak | pos | pos | pos | pos | neg | weak | neg |
| M. furfur | globose, ellipsoidal, cylindrical | pos | pos | pos | pos | pos | pos | pos | pos, (neg) | pos, (neg) | pos, (neg) | pos, (neg) | pos, (neg) | neg, (weak) | pos | pos |
| M. globosa | globose | pos | pos | neg | neg | neg | neg | neg | neg | neg | neg | neg | pos | neg | neg, (weak) | neg |
| M. japonica | globose, ellipsoidal | pos | pos | neg | weak | pos | neg | ? | neg | weak | pos | neg | pos | ? | pos | neg |
| M. nana | ellipsoidal | pos | pos | variable | pos | pos | weak | neg | variable | pos | pos | weak | pos | neg | pos | variable |
| M. obtusa | ellipsoidal, cylindrical | pos | pos | neg | neg | neg | neg | neg | neg | neg | neg | neg | pos | pos | neg, (weak) | neg |
| M. pachydermatis | ellipsoidal | pos | neg, weak | pos | pos | pos | pos | pos | pos | pos | pos | pos | pos, (weak) | pos, (neg) | pos | pos |
| M. psittaci | globose, ovoidal | pos | pos | pos | pos | pos | pos | pos | ? | ? | ? | ? | pos | neg | neg | neg |
| M. restricta | globose, ellipsoidal | pos | pos | neg | neg | neg | neg | neg | neg | neg | neg | neg | neg | neg | variable | neg |
| M. slooffiae | ellipsoidal, cylindrical | pos | pos | pos, weak | pos | pos | neg, weak | neg | pos, weak, (neg) | pos | pos | neg, (weak) | pos | neg | pos | pos |
| M. sympodialis | ellipsoidal | pos | pos | neg, weak | pos | pos | pos | neg, weak | neg, weak | pos | pos | pos | pos | pos | pos | pos |
| M. yamatoensis | ellipsoidal | pos | pos | pos | pos | pos | pos | ? | pos | pos | pos | pos | pos | ? | pos | neg |
| M. vespertilionis (Overall) | ellipsoidal/ovoid; rarely globose | pos | pos | weak* | pos | pos | weak* | neg | neg, (weak) | weak, pos | pos | variable | neg | neg | weak | weak |
| M. vespertilionis isolate CBS 15041 | ellipsoidal/ovoid; rarely globose | pos | pos | weak | pos | pos | weak | neg | neg | weak | pos | weak | neg | neg | weak | weak |
| M. vespertilionis isolate CBS 15042 | ellipsoidal/ovoid; rarely globose | pos | pos | weak | pos | pos | weak | neg | neg | weak | pos | neg | neg | neg | weak | weak |
| M. vespertilionis isolate CBS 15043 | ellipsoidal/ovoid; rarely globose | pos | pos | weak | pos | pos | weak | neg | weak | pos | pos | pos | neg | neg | weak | weak |
| M. vespertilionis isolate CBS 15044 | ellipsoidal/ovoid; rarely globose | pos | pos | weak | pos | pos | weak | neg | neg | pos | pos | weak | neg | neg | weak | weak |
| M. vespertilionis isolate CBS 15045 | ellipsoidal/ovoid; rarely globose | pos | pos | weak | pos | pos | weak | neg | neg | weak | pos | weak | neg | neg | weak | weak |
| M. vespertilionis isolate CBS 15046 | ellipsoidal/ovoid; rarely globose | pos | pos | weak | pos | pos | weak | neg | neg | weak | pos | weak | neg | neg | weak | weak |
| M. vespertilionis isolate UAMH 11925 | ellipsoidal/ovoid; rarely globose | pos | pos | weak | pos | pos | weak | neg | neg | weak | pos | weak | neg | neg | weak | weak |
* generally grew well when test was performed at 24 °C
Fig. 5.
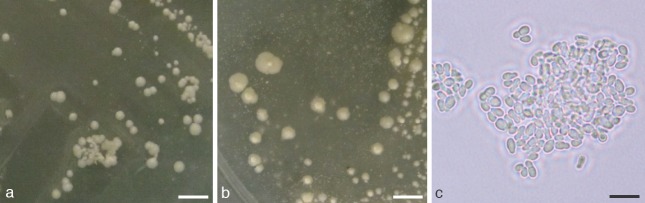
Colony and cell morphology of M. vespertilionis sp. nov. grown on Leeming and Notman Agar at 24 °C. a. Colony size and morphology after 10 d of growth; b. colony size and morphology after 40 d of growth; c. cell morphology of 10-d-old culture. — Scale bars: a, b = 4 mm; c = 5 μm.
The bat-associated Malassezia grew on mDA, but some isolates began to lose vigour (even more so than previously described on LNA) after several transfers on the medium. All isolates were lipid-dependent, failing to grow on SDA. Growth occurred on a variety of tween lipid sources, but not in the presence of Cremophor EL. The isolates were catalase and β-glucosidase negative. More detailed information is provided in the species description and in Table 4.
SPECIES DESCRIPTION
Malassezia vespertilionis J.M. Lorch & Vanderwolf, sp. nov.— MycoBank MB822382; Fig. 5
Etymology. The species epithet refers to the host from which the fungus was isolated (n. vespertilio, Latin for bat; gen. n. vespertilionis, of a bat).
Holotype. USA, Wisconsin, swab of wing skin of hibernating Myotis septentrionalis, 28 Jan. 2014, J.P. White (U.S. National Fungus Collections BPI 910536; culture ex-type CBS 15041 = UAMH 11924).
Colonies are approximately 0.5–1.0 mm diam after 10 d of growth at 24 °C on LNA; 2–5 mm diam after 40 d. At 10 d, colonies are cream-coloured, flat to slightly convex, somewhat glossy, have entire margins, and have a crumbly or waxy consistency. At 40 d, colonies have irregular margins, are slightly raised, and have a prominent papilla near the centre (Fig. 5). Cells are ellipsoid or ovoid to (rarely) globose, ranging in size from 2–3 × 2–4 μm (typically 2 × 3 μm) (Fig. 5). Buds are formed monopolarly, usually on a narrow base. Growth occurs (sometimes poorly) on mDA; no growth observed on SDA. Isolates are catalase and β-glucosidase negative. Variable in lipid utilisation: no growth observed for Cremophor EL, weak or no growth for Tween 20, usually weak growth for Tween 80, weak to good growth for Tween 40, and good growth for Tween 60. Growth occurs across a range of temperatures on LNA, but is slower at temperatures above and below 24 °C; specifically, individual pinpoint colonies are visible by day 30 at 7 °C and 30 °C, and on day 40 at 37 °C. Growth is evident at 40 d for cultures grown at 40 °C on areas of the medium where cells are placed at high densities; however, individual colonies are not grossly discernible. A sexual state was not observed; however, mating experiments were not explicitly performed.
Additional specimens examined for physiological characteristics and multilocus DNA sequencing. USA, Wisconsin, swab of wing skin of a hibernating Myotis septentrionalis, 3 Mar. 2014, J.P. White, CBS 15042; Kentucky, swab of wing skin of hibernating Myotis sodalis, 4 Mar. 2014, M.L. Verant, CBS 15043; Kentucky, swab of wing skin of hibernating Myotis sodalis, 4 Mar. 2014, M.L. Verant, UAMH 11925; New York, swab of wing skin of hibernating Myotis lucifugus, 19 Mar. 2014, M.L. Verant, CBS 15044; New York, swab of wing skin of hibernating Myotis lucifugus, 15 Jan. 2015, M.L. Verant, CBS 15045; Alabama, swab of wing skin of hibernating Myotis grisescens, 11 Feb. 2015, N. Sharp, CBS 15046.
Additional isolates for which multilocus DNA sequencing was conducted. USA, California, swab of wing skin of Lasionycteris noctivagans, 8 May 2017, T.J. Weller, CBS 15047; California, swab of wing skin of Myotis californicus, 7 May 2016, T.J. Weller, CBS 15048; California, swab of wing skin of Myotis thysanodes, 26 Apr. 2016, T.J. Weller, CBS 15049; California, swab of wing skin of Myotis californicus, 16 May 2016, T.J. Weller, CBS 15050; California, swab of wing skin of Myotis californicus, 19 Apr. 2016, T.J. Weller, CBS 15051.
DISCUSSION
The diversity of species within the genus Malassezia has been expanded in recent years due to increased sampling and use of genetic and molecular tools for distinguishing taxa. In the current study, we report on a species of Malassezia that is relatively common on skin of North American bats (i.e., cultured from 28 % of 264 individuals sampled). Isolates of this Malassezia sp. from bats were physiologically and genetically similar to one another. Specifically, sequences of four loci (D1/D2 region, β-tub, TEF1, and RPB2) were identical between isolates. Minimal variation in the ITS region (99.7 % sequence identity), 18S rDNA (99.9 % sequence identity), and portions of the MCM7 (99.8 % sequence identity) and CHS2 (99.8 % sequence identity) genes were within the intraspecific variation documented in other species of Malassezia (e.g., Makimura et al. 2000, Hirai et al. 2004, Cabañes et al. 2007). As a group, the bat-associated isolates were highly similar to one another, and they were sufficiently distinct from all other known taxa. Thus, we propose that these bat-associated isolates represent the novel species M. vespertilionis.
All isolates of M. vespertilionis subjected to physiological tests were catalase negative, a trait shared only with M. arunalokei, M. restricta, and some strains of M. furfur and M. pachydermatis (Guého et al. 1996, Guillot et al. 1998, reviewed by Batra et al. 2005, Honnavar et al. 2016). In contrast to M. arunalokei and M. restricta, M. vespertilionis is capable of growth (albeit slow) at 40 °C and can utilise multiple types of Tween. It can be distinguished from catalase negative strains of M. pachydermatis based on ability of the latter to grow on SDA without the supplementation of lipids. Differentiation of M. vespertilionis from M. furfur based on physiological tests alone may be problematic due to reported variation in M. furfur (Batra et al. 2005). We examined a number of isolates of M. vespertilionis and found some variability in results of the physiological tests for this species as well. Interpretation of physiological tests can be challenging (Gupta et al. 2004), and the inability of M.vespertilionis to grow sufficiently to produce positive results under the standard incubation procedures set forth by Guého et al. (1996) further complicates the use of these tests for identification. Thus, although physiological tests can often separate M. vespertilionis from other described taxa and may be helpful for some applications, we encourage the use of DNA sequencing (e.g., ITS region) to confirm identification.
To date, Malassezia species have been isolated primarily from euthermic animals that maintain constant core body temperature near 37 °C. This is consistent with the statement made by Guého-Kellerman et al. (2010) that members of the genus do not endure temperatures below 28 °C. Most of the bat species from which M. vespertilionis was isolated hibernate for up to seven months of the year at which time their body temperature is close to that of the surrounding environment. Specifically, Myotis lucifugus, Myotis sodalis, and Myotis septentrionalis were reported to prefer winter hibernacula with average air temperatures around 7.2, 8.5, and 9.1 °C, respectively (Brack 2007). In the active season, a bat's body temperature may exceed 40 °C; however, bats often use bouts of torpor even during the active season, such that their body temperatures frequently fluctuate, sometimes approaching ambient temperature (Hock 1951, Studier 1981, Willis & Cooper 2009). Thus, the skin temperature of bats is highly variable, and this may explain the wide range of temperatures under which M. vespertilionis is capable of growth (from at least 7 °C to 40 °C). The ability to grow at such cool temperatures is noteworthy and may be unique to M. vespertilionis among the Malassezia. However, the lower growth limits of most Malassezia species have not been expressly described in literature, making comparisons difficult. The detection of DNA of uncultured malassezia-like organisms on corals and in terrestrial and marine environments suggests that other undescribed species of Malassezia may be capable of growth at ambient temperatures or reside on poikilothermic hosts (reviewed by Amend 2014).
Previous phylogenetic studies of Malassezia have demonstrated disparities in the relationships between species when different loci were analysed (Cabañes et al. 2007, Castellá et al. 2014). This genealogical discordance has made deciphering taxonomic relationship among all members of the genus difficult. Wu et al. (2015) was able to better resolve Malassezia phylogeny through whole genome sequencing and concatenation of 164 genes. This phylogeny resulted in three main clades: clade A consists of M. furfur, M. obtusa, M. yamatoensis, and M. japonica; clade B contains M. sympodialis, M. dermatis, M.caprae, M. equina, M. nana, M. pachydermatis, M. globosa, and M. restricta; and clade C is comprised of M. cuniculi and M. slooffiae. Using the sequence data generated by Wu et al. (2015) and the whole genome sequence of M. vespertilionis produced in this study, we conducted a phylogenetic analysis using amino acid sequences of 254 core genes. The resulting phylogenetic tree was identical to that of Wu et al. (2015) except that our analysis suggested that M. obtusa is basal to the subclade consisting of M. furfur and M. yamatoensis (Fig. 3). The Malassezia tree of Wu et al. (2015) had placed M. yamatoensis basal to M. furfur and M. obtusa; however, in that study the node was less well-supported compared to other species-level relationships. The slight change in tree topology and greater support for relationships within clade A in our tree may be due to the inclusion of more genes and M. vespertilionis in the analysis. These findings suggest that while sequencing loci traditionally used to differentiate species of Malassezia (e.g., ITS, D1/D2, β-tub, and CHS2 (Cabañes et al. 2007, 2011, 2016, Castellá et al. 2014)) can be useful in identifying novel species, whole genome sequencing may be necessary to generate enough genetic data to fully resolve the relationship of those novel species with existing taxa.
Our phylogenetic analyses indicate that M. vespertilionis is the most basal member of clade A. Malassezia arunalokei, M.brasiliensis, and M. psittaci were not included in the analyses because genetic data is available for only three loci each for these newly-described species (Cabañes et al. 2016, Honnavar et al. 2016). Based on existing sequence data, M. arunalokei, M.brasiliensis, and M. psittaci share only about 75–87 %, 90–91 %, and 78–84 % DNA sequence identity, respectively, with M. vespertilionis in the ITS region, D1/D2 region, and portion of the β-tubulin gene. Furthermore, previous analyses indicate that M. arunalokei is a member of clade B (Honnavar et al. 2016). Malassezia brasiliensis and M. psittaci are sister taxa to M. furfur and M. yamatoensis, respectively, which are both divergent from M. vespertilionis (Cabañes et al. 2016).
The genus Malassezia may be more diverse than currently documented due to the difficulty of transporting and culturing many fragile and fastidious members of the genus, the historic use of morphological and physiological characteristics as the sole criteria to identify species (which can fail to distinguish cryptic species), and a lack of sampling of diverse taxonomic host groups (Amend 2014, Cabañes 2014). In the only other published study in which bats were specifically surveyed for Malassezia, Gandra et al. (2008) reportedly cultured M. furfur, M. globosa, M. pachydermatis, and M. sympodialis (based on physiological and morphological characteristics) from Pallas’ mastiff bats (Molossus molossus) in Brazil. No DNA sequence data were generated and the isolates were apparently not deposited in a public culture collection, making it difficult to ascertain their true species assignments. However, because all isolates from Gandra et al. (2008) were either catalase positive or lipid-independent (representing significant physiological deviations from M. vespertilionis) it may be that M. vespertilionis has not previously been isolated due to a lack of sampling effort of temperate bat species. Indeed, few studies have examined the fungal communities associated with bats, and those that have did not utilise fungal growth media suitable for the isolation of lipid-dependent species of Malassezia (Grose & Marinkelle 1966, Larcher et al. 2003, Voyron et al. 2011, Johnson et al. 2013, Vanderwolf et al. 2013). Njus (2014) detected Malassezia spp. on the skin of bats by conducting fungal community analyses with next generation sequencing. Based on reported sequence identities with other species of Malassezia, at least some of the Malassezia detected by Njus (2014) may represent M. vespertilionis. However, DNA sequence data from that project were not available in GenBank at the time of our study to confirm this.
Malassezia yeasts are best known for their association with certain skin ailments. For example, M. pachydermatis has been implicated as the cause of dermatitis in (among other species) rhinoceroses (Bauwens et al. 1996), dogs (Gustafson 1955, Bond et al. 2004), and sea lions (Guillot et al. 1998, Nakagaki et al. 2000). More often, the role of Malassezia in skin diseases of animals is unknown, and targeting skin lesions for culture and PCR analyses likely masks the frequency with which these fungi act as commensals. For example, Neves et al. (2017) isolated Malassezia from a relatively high percentage (32.8%) of free-ranging golden-headed lion tamarins (Leontopithecus chrysomelas) in Brazil; none of the animals had skin lesions. Similarly, none of the 74 bats from which isolates of M. vespertilionis were recovered in this study showed visible signs of dermatitis at the time they were sampled. Thus, there is no current evidence that M. vespertilionis acts as a pathogen; instead, the yeast is likely a normal component of the skin mycobiome of bats.
In this study, M. vespertilionis was isolated from nine species of bats in seven U.S. states. With the exception of Lasionycteris noctivagans, all of these bats were species of Myotis, a diverse and widely distributed genus in North America. This, coupled with the detection of the fungus in both the eastern and western United States (Fig. 2), may indicate that the yeast is more widespread than our survey indicates. Myotis also has representatives throughout much of South America, Eurasia, Africa, and Australia, and additional sampling is necessary to determine whether M. vespertilionis may have a more global distribution. If this is shown to be the case, M. vespertilionis could prove to be an important species in which to study Malassezia genetics and evolution. All other known species of Malassezia occur on humans and domestic animals, which have been transported across the world. This, combined with the possibility of interspecific recombination (Cabañes et al. 2007, Castellá et al. 2014) when different species come into contact with one another, makes it difficult to understand historic patterns of geographic distribution, variation, and genetic exchange between strains. Bat species, however, have not been subjected to such extensive and recent human-assisted global movements, nor do they frequently come into close contact with other animal hosts that might facilitate genetic transfer between different species of Malassezia. Thus, if M. vespertilionis is found on other continents, genetic distinctions between strains may still be intact and provide important opportunities for future research into the evolution of these fungi and the role that humans have played in shaping the genetic structure and pathogenicity of Malassezia species.
Data accessibility
All relevant metadata related to this manuscript can be found in the tables of this manuscript, in GenBank, or in TreeBASE (http://purl.org/phylo/treebase/phylows/study/TB2:S21466).
Acknowledgments
We thank Mike Armstrong, Elizabeth Bohuski, Shelly Colatskie, Carl Herzog, Heather Kaarakka, Julie Milbroda, Jennifer Redell, Mike Scafini, Nick Sharp, and J. Paul White for assisting with sample collection and Anne Ballmann for helping to coordinate submission for some of the samples. We acknowledge Carol Meteyer and Nancy Thomas for discussions regarding the presence of possible Malassezia observed on bat wings prior to the initiation of this project, Andrew Minnis for providing technical advice, and Teun Boekhout for providing constructive feedback on the manuscript. This work was funded by the U.S. Fish and Wildlife Service and the U.S. Geological Survey. We thank the University of Wisconsin Biotechnology Center DNA Sequencing Facility for providing next-generation sequencing services. The original map modified for Fig. 2 is attributed to Alan Rockefeller, is available on Wikimedia Commons, and was used under the terms of the GNU Free Documentation License. The use of trade, product, or firm names is for descriptive purposes only and does not imply endorsement by the US Government.
REFERENCES
- Aguirre C, Euliarte C, Finquelievich J, et al. 2015. Fungemia and interstitial lung compromise caused by Malassezia sympodialis in a pediatric patient. Revista Iberoamericana de Micología 32(2): 118–121. [DOI] [PubMed] [Google Scholar]
- Amend A. 2014. From dandruff to deep-sea vents: Malassezia-like fungi are ecologically hyper-diverse. PLoS Pathogens 10 (8): e1004277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology 19: 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batra R, Boekhout T, Guého E, et al. 2005. Malassezia Baillon, emerging clinical yeasts. FEMS Yeast Research 5: 1101–1113. [DOI] [PubMed] [Google Scholar]
- Bauwens L, De Vroey C, De Meurichy W. 1996. A case of exfoliative dermatitis in a captive Southern white rhinoceros (Ceratotherium simum simum). Journal of Zoo and Wildlife Medicine 27: 271–274. [Google Scholar]
- Boekhout T, Kamp M, Guého E. 1998. Molecular typing of Malassezia species with PFGE and RAPD. Medical Mycology 36: 365–372. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond R, Patterson-Kane JC, Lloyd DH. 2004. Clinical, histopathological and immunological effects of exposure of canine skin to Malassezia pachydermatis. Medical Mycology 42: 165–175. [DOI] [PubMed] [Google Scholar]
- Bowen AR, Chen-Wu JL, Momany M, et al. 1992. Classification of fungal chitin synthases. Proceedings of the National Academy of Sciences, USA 89: 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brack V., Jr 2007. Temperature and locations used by hibernating bats, including Myotis sodalis (Indiana bat), in a limestone mine: implications for conservation and management. Environmental Management 40: 739–746. [DOI] [PubMed] [Google Scholar]
- Cabañes FJ. 2014. Malassezia yeasts: how many species infect humans and animals? PLoS Pathogens 10: e1003892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabañes FJ, Coutinho SD, Puig L, et al. 2016. New lipid-dependent Malassezia species from parrots. Revista Iberoamericana de Micologia 33: 92–99. [DOI] [PubMed] [Google Scholar]
- Cabañes FJ, Theelen B, Castellá G, et al. 2007. Two new lipid-dependent Malassezia species from domestic animals. FEMS Yeast Research 7: 1064–1076. [DOI] [PubMed] [Google Scholar]
- Cabañes FJ, Vega S, Castellá G. 2011. Malassezia cuniculi sp. nov., a novel yeast species isolated from rabbit skin. Medical Mycology 49: 40–48. [DOI] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellá G, Coutinho SD, Cabañes FJ. 2014. Phylogenetic relationships of Malassezia species based on multilocus sequence analysis. Medical Mycology 52: 99–105. [DOI] [PubMed] [Google Scholar]
- Chang HJ, Miller HL, Watkins N, et al. 1998. An epidemic of Malassezia pachydermatis in an intensive care nursery associated with colonisation of health care workers' pet dogs. New England Journal of Medicine 338: 706–711. [DOI] [PubMed] [Google Scholar]
- Einax E, Voigt K. 2003. Oligonucleotide primers for the universal amplification of β-tubulin genes facilitates phylogenetic analyses in the regnum Fungi. Organisms Diversity and Evolution 3: 185–194. [Google Scholar]
- Gaitanis G, Magiatis P, Hantschke M, et al. 2012. The Malassezia genus in skin and systemic diseases. Clinical Microbiology Reviews 25: 106–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitanis G, Velegraki A, Magiatis P, et al. 2011. Could Malassezia yeasts be implicated in skin carcinogenesis through the production of aryl-hydrocarbon receptor ligands? Medical Hypotheses 77: 47–51. [DOI] [PubMed] [Google Scholar]
- Gandra RF, Gambale W, De Cássia Garcia Simão R, et al. 2008. Malassezia spp. in acoustic meatus of bats (Molossus molossus) of the Amazon Region, Brazil. Mycopathologia 165: 21–26. [DOI] [PubMed] [Google Scholar]
- Grose E, Marinkelle CJ. 1966. Species of Sporotricum, Trichophyton and Microsporum from Columbian bats. Tropical and Geographical Medicine 18: 260–263. [PubMed] [Google Scholar]
- Guého E, Midgley G, Guillot J. 1996. The genus Malassezia with description of four new species. Antonie van Leeuwenhoek 69: 337–355. [DOI] [PubMed] [Google Scholar]
- Guého-Kellerman E, Boekhout T, Begerow D. 2010. Biodiversity, phylogeny and ultrastructure. In: Boekhout T, Guého-Kellerman E, Mayser P, et al. (eds), Malassezia and the skin: science and clinical practice: 17–63. Springer, Germany. [Google Scholar]
- Guillot J, Guého E, Lesourd M, et al. 1996. Identification of Malassezia species: A practical approach. Journal de Mycologie Médicale 6: 103–110. [Google Scholar]
- Guillot J, Petit F, Degorce-Rubiales F, et al. 1998. Dermatitis caused by Malassezia pachydermatis in a California sea lion (Zalophus californianus). Veterinary Record 142: 311–312. [DOI] [PubMed] [Google Scholar]
- Gupta AK, Boekhout T, Theelen B, et al. 2004. Identification and typing of Malassezia species by amplified fragment length polymorphism and sequence analyses of the internal transcribed spacer and large-subunit regions of ribosomal DNA. Journal of Clinical Microbiology 42: 4253–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson BA. 1955. Otitis externa in the dog: a bacteriological and experimental study. PhD thesis, Royal Veterinary College, Sweden. [Google Scholar]
- Hirai A, Kano R, Makimura K, et al. 2004. Malassezia nana sp. nov., a novel lipid-dependent yeast species isolated from animals. International Journal of Systematic and Evolutionary Microbiology 54: 623–627. [DOI] [PubMed] [Google Scholar]
- Hock RJ. 1951. The metabolic rates and body temperatures of bats. The Biological Bulletin 101: 289–299. [Google Scholar]
- Honnavar P, Prasad GS, Ghosh A, et al. 2016. Malassezia arunalokei sp. nov., a novel yeast species isolated from seborrheic dermatitis patients and healthy individuals from India. Journal of Clinical Microbiology 54: 1826–1834. doi: 10.1128/JCM.00683-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LJAN, Miller AN, McCleery RA, et al. 2013. Psychrophilic and psychrotolerant fungi on bats and the presence of Geomyces spp. on bat wings prior to the arrival of white-nose syndrome. Applied and Environmental Microbiology 79: 5465–5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laetsch DR, Blaxter ML. 2017. BlobTools: Interrogation of genome assemblies [version 1; referees: awaiting peer review]. F1000Research 6: 1287 [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larcher G, Bouchara JP, Pailley P, et al. 2003. Fungal biota associated with bats in western France. Journal de Mycologie Médicale 13: 29–34. [Google Scholar]
- Leeming JP, Notman FH. 1987. Improved methods for isolation and enumeration of Malassezia furfur from human skin. Journal of Clinical Microbiology 25: 2017–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YL, Whelen S, Hall BD. 1999. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Molecular Biology and Evolution 16: 1799–1808. [DOI] [PubMed] [Google Scholar]
- Lorch JM, Minnis AM, Meteyer CU, et al. 2015. The fungus Trichophyton redellii sp. nov. causes skin infections that resemble white-nose syndrome of hibernating bats. Journal of Wildlife Diseases 51: 36–47. [DOI] [PubMed] [Google Scholar]
- Makimura K, Tamura Y, Kudo M, et al. 2000. Species identification and strain typing of Malassezia species stock strains and clinical isolates based on the DNA sequences of nuclear ribosomal internal transcribed spacer 1 regions. Journal of Medical Microbiology 49: 29–35. [DOI] [PubMed] [Google Scholar]
- Mayser P, Haze P, Papavassilis C, et al. 1997. Differentiation of Malassezia species: selectivity of Cremophor EL, castor oil and ricinoleic acid for M. furfur. British Journal of Dermatology 137: 208–213. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE), 14 Nov. 2010: 1–8. New Orleans, Louisiana. [Google Scholar]
- Nakagaki K, Hata K, Iwata E, et al. 2000. Malassezia pachydermatis isolated from a South American sea lion (Otaria byronia) with dermatitis. The Journal of Veterinary Medical Science 62: 901–903. [DOI] [PubMed] [Google Scholar]
- Neves JJ, Francelino M, Silva FG, et al. 2017. Survey of Malassezia sp. and dermatophytes in the cutaneous microbiome of free-ranging golden-headed lion tamarins (Leontopithecus chrysomelas – Kuhl, 1820). Journal of Medical Primatology 46: 65–69. [DOI] [PubMed] [Google Scholar]
- Njus KA. 2014. Molecular techniques for the identification of commensal fungal populations on cave roosting bats. Master's thesis. University of Akron, USA. http://rave.ohiolink.edu/etdc/view?acc_num=akron1403716687. [Google Scholar]
- O'Donnell K. 1993. Fusarium and its near relatives. In: Reynolds DR, Taylor JW. (eds), Fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics: 225–233. CAB International, UK. [Google Scholar]
- Patron RL. 2016. A 34-year-old man with cough, lung nodules, fever, and eosinophilia. Clinical Infectious Diseases 63 (11): 1525–1526. [DOI] [PubMed] [Google Scholar]
- Rehner SA, Buckley E. 2005. A Beauveria phylogeny inferred from nuclear ITS and EF1-alpha sequences: evidence for cryptic diversification and likes to Cordyceps teleomorphs. Mycologia 97: 84–98. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Schmitt L, Crespo A, Divakar PK, et al. 2009. New primers for promising single-copy genes in fungal phylogenetics and systematics. Persoonia 23: 35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, et al. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier EH. 1981. Energetic advantages of slight drops in body temperature in little brown bats, Myotis lucifugus. Comparative Biochemistry and Physiology Part A: Physiology 70: 537–540. [Google Scholar]
- Sugita T, Boekhout T, Velegraki A, et al. 2010. Epidemiology of Malassezia-related skin diseases. In: Boekhout T, Guého-Kellerman E, Mayser P, et al (eds), Malassezia and the skin: science and clinical practice: 65–119. Springer, Germany. [Google Scholar]
- Sugita T, Nakase T. 1999. Non-universal usage of the leucine CUG codon and the molecular phylogeny of the genus Candida. Systematic and Applied Microbiology 22: 79–86. [DOI] [PubMed] [Google Scholar]
- Sugita T, Takashima M, Shinoda T, et al. 2002. New yeast species, Malassezia dermatis, isolated from patients with atopic dermatitis. Journal of Clinical Microbiology 40: 1363–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, et al. 2013. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderwolf KJ, McAlpine DF, Malloch D, et al. 2013. Ectomycota associated with hibernating bats in eastern Canadian caves prior to the emergence of white-nose syndrome. Northeastern Naturalist 20: 115–130. [Google Scholar]
- Voyron S, Lazzari A, Riccucci M, et al. 2011. First mycological investigation on Italian bats. Hystrix 22: 189–197. [Google Scholar]
- Walker BJ, Abeel T, Shea T, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9: e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q-M, Theelen B, Groenewald M, et al. 2014. Moniliellomycetes and Malasseziomycetes, two new classes in the Ustilaginomycotina. Persoonia 33: 41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis CKR, Cooper CE. 2009. Techniques for studying thermoregulation and thermal biology in bats. In: Kunz TH, Parsons S. (eds), Ecological and behavioral methods for the study of bats: 646–658. The John Hopkins University Press, USA. [Google Scholar]
- Wu G, Zhao H, Li C, et al. 2015. Genus-wide comparative genomics of Malassezia delineates its phylogeny, physiology, and niche adaptation on human skin. PLoS Genetics 11: e1005614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant metadata related to this manuscript can be found in the tables of this manuscript, in GenBank, or in TreeBASE (http://purl.org/phylo/treebase/phylows/study/TB2:S21466).