Abstract
On 18 January 2016, the French National Reference Centre for Salmonella reported to Santé publique France an excess of Salmonella enterica serotype Dublin (S. Dublin) infections. We investigated to identify the source of infection and implement control measures. Whole genome sequencing (WGS) and multilocus variable-number tandem repeat analysis (MLVA) were performed to identify microbiological clusters and links among cases, animal and food sources. Clusters were defined as isolates with less than 15 single nucleotide polymorphisms determined by WGS and/or with identical MLVA pattern. We compared different clusters of cases with other cases (case–case study) and controls recruited from a web-based cohort (case–control study) in terms of food consumption. We interviewed 63/83 (76%) cases; 2,914 controls completed a questionnaire. Both studies’ findings indicated that successive S. Dublin outbreaks from different sources had occurred between November 2015 and March 2016. In the case–control study, cases of distinct WGS clusters were more likely to have consumed Morbier (adjusted odds ratio (aOR): 14; 95% confidence interval (CI): 4.8–42) or Vacherin Mont d’Or (aOR: 27; 95% CI: 6.8–105), two bovine raw-milk cheeses. Based on these results, the Ministry of Agriculture launched a reinforced control plan for processing plants of raw-milk cheeses in the production region, to prevent future outbreaks.
Keywords: Salmonella Dublin, outbreaks, raw-milk, cheese, France, WGS
Background
Nontyphoidal Salmonella is a main cause of bacterial food-borne infection in Europe [1,2]. The majority of human infections is caused by a limited number of Salmonella serotypes among the 2,600 described to date [3,4]. Salmonella enterica serotype Dublin (S. Dublin) is particularly invasive in humans and more often leads to severe disease and higher mortality rates compared with other serotypes [4-7]. S. Dublin is host-adapted to bovines and is frequently isolated from cattle, with raw milk or raw-milk cheeses as a typical vehicle for food-borne outbreaks [8,9]. In 2012, a major S. Dublin outbreak occurred in France, with 103 cases linked to Saint-Nectaire (bovine raw-milk cheese) consumption [10,11]. In 2015, 34 S. Dublin cases were reported linked to the consumption of Reblochon (bovine raw-milk cheese) (data not shown; Santé publique France).
In France, the National Reference Center for Salmonella (NRC) and the French Agency for Food, Environmental and Occupational Health and Safety (ANSES) routinely collect and serotype human and non-human Salmonella isolates, respectively [12-14], using the Kauffmann–White–Le Minor scheme [3]. The S. Dublin isolates collected are frequently susceptible to all antibiotics and show an indistinguishable pulsed-field gel electrophoresis (PFGE) pattern. To better distinguish S. Dublin isolates, multilocus variable-number tandem repeat analysis (MLVA) has recently been used for surveillance and outbreak investigations [11,15]. Moreover, whole genome sequencing (WGS) of Salmonella has been shown to discriminate between closely related isolates of S. Dublin [16,17].
Outbreak detection
On 18 January 2016, the French NRC reported to Santé publique France (SpFrance, the French national public health agency) an excess of S. Dublin infections across the country, with 37 S. Dublin isolates identified between mid-November 2015 and mid-January 2016, compared with 10 S. Dublin isolates during the same period in the two previous years. An outbreak investigation team with experts from SpFrance, NRC, ANSES and the French Directorate General for Food (DGAL) launched extensive epidemiological, microbiological and food investigations to confirm the outbreak, identify the vehicle of transmission and propose appropriate control measures.
Methods
We carried out both epidemiological and microbiological investigations on subsets of S. Dublin cases and isolates, respectively.
Microbiological investigations
Salmonella Dublin isolates
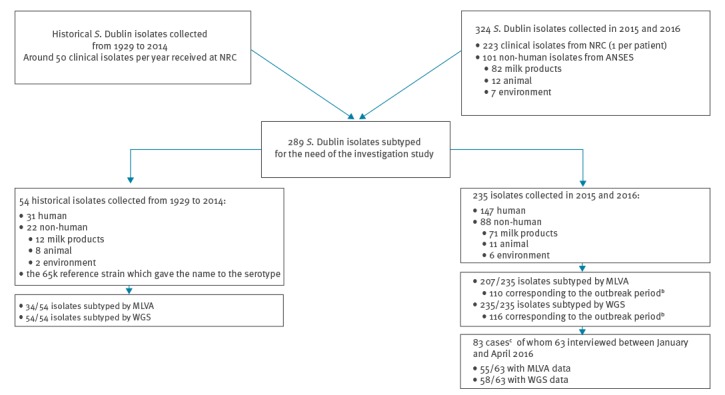
During the years 2015 and 2016, a total of 324 S. Dublin isolates were collected, 223 from clinical NRC isolates (108 in 2015 and 115 in 2016) and 101 from non-human ANSES isolates (62 and 39, in each year respectively). Of those, a total of 235, including 147 (83 in 2015 and 64 in 2016) clinical and 88 (62 and 26, respectively) non-human isolates, were extensively studied by WGS and/or MLVA. We also analysed 54 ‘historical’ subtyped S. Dublin isolates collected between 1929 and 2014, 31 from humans and 22 from non-human specimens, as well as the human isolate which gave the name to the serotype in 1929 (number 65k) [18]. In total, we included 289 isolates in this study (Figure 1).
Figure 1.
Flowchart of human and non-human Salmonella Dublin isolatesa subtyped and included in the study, France, 2015–2016 (n = 289)
ANSES: French Agency for Food, Environmental and Occupational Health and Safety; MLVA: multilocus variable-number tandem repeat analysis; NRC: national reference center; WGS: whole genome sequencing.
a While the study was conducted between 2015 and 2016, the subtyped isolates also included historical strains isolated between 1929 and 2014.
b The outbreak period is from 17 November 2015 to 11 March 2016.
c Case definition: residents in mainland France with a S. Dublin infection reported to the NRC between 17 November 2015 and 11 March 2016.
Whole genome sequencing
The NRC used WGS to subtype all the 289 isolates of which 116 corresponded to the outbreak period from November 2015 to March 2016. High-throughput genome sequencing was carried out at the ‘Plateforme de microbiologie mutualisée’ (P2M) of the Pasteur International Bioresources network (Institut Pasteur, Paris, France). After extraction with the MagNA Pure 96 System (Roche, Basel, Switzerland), DNA was further processed for sequencing with Illumina systems (libraries using the Nextera XT DNA Library Prep kit and the sequencing with the NextSeq 500 system) generating 100 to 146 bp paired-end reads. Reads were trimmed and assembled as previously described [19]. Genomic data as multilocus sequence typing (MLST) type and resistance genes were detected from assembled sequences using web-tools (http://www.genomicepidemiology.org/). For each isolate, the paired-end reads were aligned against the S. Dublin str. 3246 reference genome (GenBank accession number: CM001151.1) using Bowtie2 with default parameters [20]. A core-genome multi-alignment of assembled genomes was also done using Harvest v1.0.1 f ParSNP function [21]. For each approach, the resulting single nucleotide polymorphisms (SNPs) were concatenated to generate a filtered multiple alignment that was used as input for the construction of a phylogenetic tree using Molecular Evolutionary Genetics Analysis (MEGA)6 [22] with a maximum-likelihood (ML) approach. The final trees were visualised in the interactive Tree Of Life [23]. All reads generated in this study have been deposited in project PRJEB28817.
Multilocus variable-number tandem repeat analysis
ANSES used MLVA as described elsewhere [11], to analyse 241 isolates (including 148 human and 93 non-human), of which 110 corresponded to the outbreak period. The measured lengths for each fragment were obtained using an ABI3500 capillary electrophoresis system (Applied Biosystems, France). Data were imported into GeneMapper software (Applied Biosystems, France) where each fragment was identified according to colour and size. A normalisation of the results was done with the free access MLVA_Normalizer software [24].
Epidemiological investigation
Case definition
We defined cases as residents in mainland France with a S. Dublin infection reported to the NRC between 17 November 2015 and 11 March 2016, without travel history outside of France within 7 days prior symptom onset and without history of S. Dublin infection before 17 November 2015 (i.e. no S. Dublin strain ever isolated and received at the NRC before 17 November 2015).
Cluster definition
We defined a cluster as isolate sequences with < 15 SNP divergence obtained by core-genome comparison and/or with identical MLVA pattern. Among these clusters, we also defined subclusters as isolate sequences having < 5 intra SNPs.
Study design
We compared cases belonging to a specific cluster/subcluster with other cases belonging to all other clusters/subclusters in a case–case study and with controls in a case–control study. For the case–control study, controls were recruited from a cohort of individuals with children and adults registered on GrippeNet.fr (https://www.grippenet.fr), an online population-based surveillance system for influenza-like illness [25,26]. During winter 2015/16, 6,515 participants reported online the presence or absence of basic symptoms on a weekly basis using a list of 19 predefined symptoms commonly or rarely related to influenza. We excluded controls reporting travel abroad during the Salmonella outbreak period and those with digestive symptoms.
Data collection
Epidemiologists from the regional offices of SpFrance interviewed the cases by telephone using a trawling questionnaire on clinical symptoms, medical history, detailed food consumption, contact with other persons experiencing diarrhoea, travel history and contact with animals. Loyalty card information was collected to trace-back supermarket purchases.
On week 8 (22–28 February), 2016, controls from the GrippeNet.fr cohort received a web link to complete an online questionnaire about health, travel history and food consumption during 11–17 January 2016 (a 7-day ordinary non-festive period).
Statistical analyses
We calculated proportions, using the number of non-missing values as denominators. For the case–case study, we calculated crude odds ratios (OR). For the case–control study, we calculated adjusted odds ratios (aOR) for age and sex using multivariable logistic regression. The initial regression models included age, sex and food items consumed by at least 50% of the cases. We performed this analysis for WGS clusters/subclusters and MLVA clusters with at least 10 cases. We used STATA version 12.0 (Stata Corporation, Texas, United States) for this analysis.
Ethical considerations
The study was approved by the French Commission for Data Protection (Commission Nationale de l’Informatique et des Libertés). Interviewees or next of kin provided verbal consent. Only anonymised data were analysed and used for the purpose of the study.
Food production chain and animal trace-back investigations
DGAL conducted a trace-back investigation on potential contaminated products identified by the epidemiological investigations. Points of purchase, like supermarkets or cheese retailers, were reported by cases. Where possible, customer loyalty card numbers were used to identify the exact point of purchase and product batch numbers and specific production facilities. The trace-back investigation was conducted for a retrospective period of up to a month before symptom onset of given cases, if date of onset was available, or if not, up to two months before the date of isolation of S. Dublin in the patient specimen. Suspected food products were tested for Salmonella if food samples were available.
Results
Description of cases
Between 17 November 2015 and 11 March 2016, 83 cases were identified. Median age was 70 years (range: 1–94), 44 (53%) were female and respondents originated from 12/13 regions in mainland France, with 19 (23%) cases coming from the Bourgogne-Franche-Comté region (Table 1). S. Dublin was isolated from blood (n = 39; 47%) but also from stool (25; 30%), urine (11; 13%), and other samples (8; 10%). Ten (12%) deaths were reported with no information available on the cause of death. Questionnaires were not completed for 20 cases (9 deaths, 7 unreachable, 4 refusals), leading to 63 (76%) cases included in further analyses.
Table 1. Characteristics of Salmonella Dublin outbreak cases, France, November 2015–March 2016 (n = 83, including 63 interviewed cases).
| Characteristics of cases | Category | n | Na | % |
|---|---|---|---|---|
| All cases (n = 83) | ||||
| Sex | Female | 44 | 83 | 53 |
| Age | 1–17 | 6 | 83 | 7 |
| 18–44 | 10 | 83 | 12 | |
| 45–64 | 19 | 83 | 23 | |
| 65–84 | 31 | 83 | 37 | |
| 85–94 | 17 | 83 | 21 | |
| Region of residence | Auvergne-Rhône-Alpes | 8 | 83 | 10 |
| Bourgogne-Franche-Comté | 19 | 83 | 23 | |
| Bretagne | 6 | 83 | 7 | |
| Grand-Est | 6 | 83 | 7 | |
| Haut-de-France | 7 | 83 | 8 | |
| Île-de-France | 9 | 83 | 11 | |
| Nouvelle-Aquitaine | 9 | 83 | 11 | |
| Pays-de-la-Loire | 7 | 83 | 8 | |
| Otherb | 12 | 83 | 15 | |
| Type of human sample | Blood | 39 | 83 | 47 |
| Stool | 25 | 83 | 30 | |
| Urine | 11 | 83 | 13 | |
| Articular fluid | 3 | 83 | 4 | |
| Pus | 2 | 83 | 2 | |
| Other | 3 | 83 | 4 | |
| Deceased | Yesc | 10 | 83 | 12 |
| Interviewed cases (n = 63) | ||||
| Comorbidities | Yesd | 39 | 59 | NA |
| Clinical symptomse | Feverf | 38 | 55 | NA |
| Nausea | 15 | 45 | NA | |
| Vomiting | 19 | 51 | NA | |
| Abdominal pain | 27 | 47 | NA | |
| Diarrhoea | 30 | 56 | NA | |
| Blood in faeces | 5 | 38 | NA | |
| Hospitalisation | Yes | 41 | 60 | 68 |
NA: Not applicable due to low figure in the denominator (< 60).
a Number of cases for which the information was available.
b Centre Val-de-Loire, Normandie, Occitanie and Provence-Alpes-Côte d’Azur.
c Information provided by the National Reference Center for Salmonella (NRC), without confirmation that cause of death was attributable to S. Dublin infection.
d Asthma, cancer, cardiac problems, diabetes, renal failure.
e Cases could have more than one clinical symptom.
f Fever was defined as body temperature > 38°C or perception of having fever.
Among these cases, 39 of 59 with available information reported having pre-existing chronic medical conditions (asthma, cancer, cardiac problems, diabetes, renal failure). Most frequently reported symptoms included fever (either reported as a measurement of body temperature > 38°C or reported as a perception; 38/55), abdominal pain (27/47) and diarrhoea (30/56). Among cases with data on hospitalisation, 68% (41/60) were hospitalised.
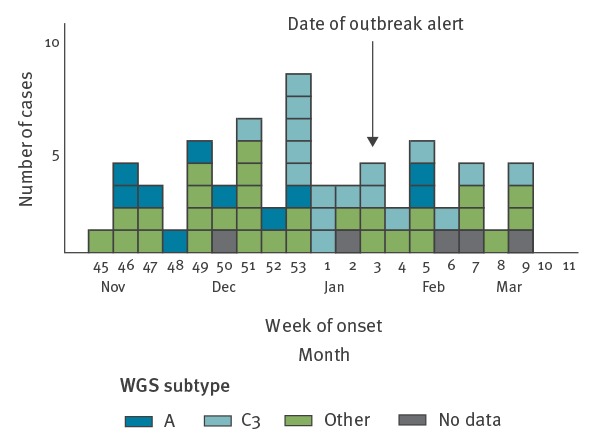
The number of cases peaked during week 53 (28 December–3 January) (Figure 2).
Figure 2.
Number of cases by reporteda date of onset of symptomsb, Salmonella Dublin outbreak, France, November 2015–March 2016 (n = 83b)
WGS: whole genome sequencing.
a 55 cases reported a precise day or week of onset and six cases reported a vague period of symptom onset time (within several weeks); we then recorded the last week of that period as the week of onset of symptoms.
b For 22 cases, no date of onset was recorded; of these, we did not have a completed questionnaire for 20.
Description of controls
Among the 6,200 GrippeNet.fr participants (i.e. controls) who could be reached, 2,914 (47%) completed the questionnaire. Of those, 2,690 (92%) did this within 2 days; 1,916 (66%) were female; median age was 56 years (range: 2–90). The whole process, i.e. from the beginning of the recruitment of the controls to the end of data collection, took 12 days.
There were significant differences between cases and controls in terms of proportion of female (53% in cases vs 66% in controls; p value = 0.01) and age (58% of cases vs 29% of controls were > 65 years-old; p value = 0.00).
Microbiological findings
Among 241 MLVA performed for S. Dublin isolates, 49 different MLVA patterns were obtained and among these, seven gathered 10 or more isolates (Table 2).
Table 2. Number of subtyped human and non-human Salmonella Dublin isolates by MLVA patterns (rows) and whole genome sequencing clusters (columns) included in the study, France, 2015–2016a (n = 289).
| MLVA | WGS | Total | ||||||
|---|---|---|---|---|---|---|---|---|
| A | B | C3 | Cother | F | K | Otherb | ||
| 15–8-10–7-5–3 | 0 | 0 | 0 | 1 | 19c | 0 | 0 | 20 |
| 17–8-10–7-5–4 | 0 | 9d | 0 | 6 | 0 | 0 | 0 | 15 |
| 18–8-10–7-5–4 | 0 | 14e | 28 | 3 | 0 | 0 | 2 | 47 |
| 19–8-10–7-5–3 | 0 | 0 | 0 | 22f | 0 | 0 | 0 | 22 |
| 19–8-10–7-5–4 | 1 | 4 | 1 | 18g | 0 | 0 | 3 | 27 |
| 20–8-10–7-5–3 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 10 |
| 20–8-10–7-5–4 | 15 | 1 | 0 | 0 | 0 | 0 | 1 | 17 |
| Otherh | 0 | 5 | 1 | 11 | 15 | 11 | 40 | 83 |
| Missing | 2 | 2 | 5 | 17 | 4 | 0 | 18 | 48 |
| Total | 18 | 35 | 35 | 88 | 38 | 11 | 64 | 289 |
MLVA: multilocus variable-number tandem repeat analysis; WGS: whole genome sequencing.
a While the study was conducted between 2015 and 2016, the subtyped isolates included historical strains isolated between 1929 and 2014.
b There were 23 other WGS subtypes.
c Among these 19, 13 were associated with WGS subcluster F4.
d All nine isolates with MLVA 17–8-10–7-5–4 pattern were associated with WGS subcluster B3.
e Eight isolates with MLVA 18–8-10–7-5–4 pattern were associated with WGS subcluster B4.
f Fourteen isolates with MLVA 19–8-10–7-5–3 pattern were associated with WGS subcluster C8.
g Eight isolates with MLVA 19–8-10–7-5–4 pattern were associated with WGS subcluster C6.
h There were 42 other MLVA patterns.
The WGS analysis of all the 289 S. Dublin isolates, including historical ones, indicated a unique MLST, ST10 that had previously been related to serotype Dublin in both the literature and public databases (https://enterobase.warwick.ac.uk/) [27]. As the mapping against the S. Dublin str. 3246 reference genome (GenBank accession number: CM001151.1) showed high divergence with 19,213 SNPs, the core-genome multi-alignment of assembled genomes using ParSNP function was preferred. A generated matrix file revealed that maximum divergence for all isolates sequenced during the outbreak including the reference strain was 791 SNPs suggesting a relative homogeneous population of S. Dublin isolates that were circulating in France. We identified 28 different clusters with < 15 SNPs, including five clusters with > 10 isolates (clusters A, B, C, F and K). Three of those, A, B and C, accounted for the majority of human (70%, 125/179) and non-human isolates (46%, 51/110) (Figure 3).
Figure 3.
Phylogenetic analysis of all the subtyped human and non-human Salmonella Dublin isolates included in the study that were whole genome sequenced, France, 2015–2016a (n = 289)
a While the study was conducted between 2015 and 2016, the subtyped isolates included historical strains isolated between 1929 and 2014.
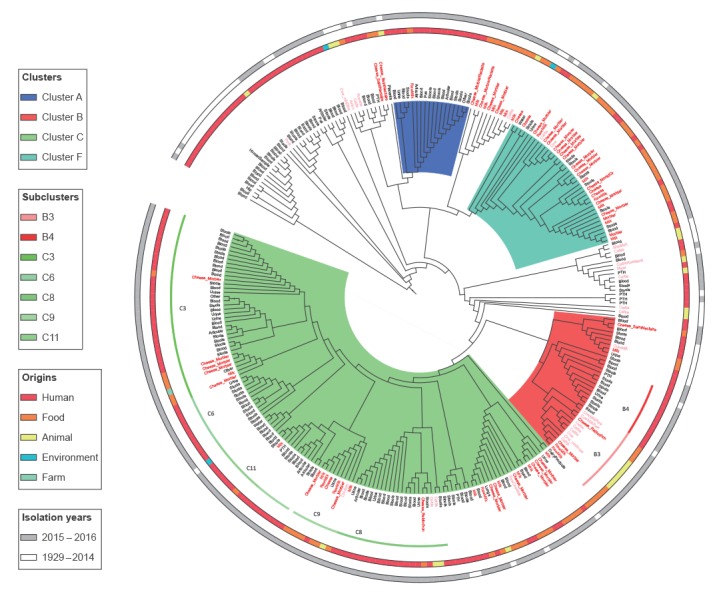
A total of 18 isolates (17 human and 1 non-human) with date of isolation between October 2015 and March 2016 were identified as belonging to the WGS cluster A and were mainly associated to the MLVA cluster 20–8-10–7-5–4 (Table 2, Figure 3). In terms of food items, only one WGS cluster A isolate from October 2015 was found in raw milk.
The WGS cluster B comprised a total of 35 isolates (20 human and 15 non-human). Two main subclusters were identified; one B3 (mainly associated to the MLVA cluster 17–8-10–7-5–4) was found in relation to cattle, milk and raw-milk cheeses (Reblochon and Morbier) with date of isolation mainly in January 2016 and the other B4 (mainly associated to MLVA cluster 18–8-10–7-5–4) was found in cases with date of isolation mainly in December 2015 and January 2016.
The WGS cluster C was the most prevalent with 123 isolates (88 human and 35 non-human). It was subdivided into nine subclusters (< 5 SNPs), C3 and Cother (grouping eight smaller subclusters including C8). All the 35 isolates belonging to the C3 subcluster presented a very limited intra SNP difference (< 2) and a sufficient divergence (> 5 SNPs) to the other eight C subclusters (Cother), indicating high level of genetic relationship due to a putative common source of contamination. The WGS subcluster C3 was mainly associated with MLVA cluster 18–8-10–7-5–4 and was found in clinical isolates from patients between January and April 2016. This molecular signature was also found in one isolate from Morbier cheese in February 2016 during this investigation. It was found as well in several isolates in Morbier cheeses tested in 2015 through company internal microbiological monitoring system. The WGS subcluster C8 was mainly associated with the MLVA cluster 19–8-10–7-5–3 and harboured isolates from patients at the beginning of 2015. In that period, another S. Dublin outbreak had been investigated between February and April 2015 in seven French regions, and was found to be possibly associated with consumption of Reblochon cheese (data not shown; Santé publique France).
Among the non-human isolates, we revealed several other WGS cluster groups (in particular F and K) but no or few linked human isolates (data not shown). The review of the historical human and non-human specimens suggested that WGS B and C cluster isolates have been circulating in France for decades, while WGS cluster A isolates seem to be more recent.
Among the 63 cases for whom a completed questionnaire was available, 58 had had an isolate subtyped by WGS, 55 had had an isolate subtyped by MLVA, and 55 both. Of the 58 cases with WGS subtype, 17 belonged to subcluster C3, 11 to cluster A, and 30 to 14 other subclusters. Of the 55 cases with MLVA pattern, 23 belonged to cluster 18–8-10–7-5–4, 10 to cluster 20–8-10–7-5–4 and 22 to six other clusters.
Epidemiological investigations
Case–case study
Compared with cases belonging to the other WGS clusters, WGS cluster A cases seemed to have consumed Vacherin Mont d’Or cheese more frequently, although this result was not statistically significant (odds ratio (OR): 5.1; 95% confidence interval (CI): 0.9–34). Similarly WGS subcluster C3 cases appeared to be related to Morbier cheese consumption (OR: 3.3; 95% CI: 0.8–15) (Table 3). Compared with cases belonging to other MLVA patterns, MLVA cluster 18–8-10–7-5–4 cases (which frequently coincided with WGS cluster A) seemed to have more often consumed Morbier cheese, however this was not significant (OR: 2.1; 95% CI: 0.5–8.3) and MLVA cluster 20–8-10–7-5–4 cases (which frequently coincided with WGS cluster C3) appeared as consuming more often Vacherin Mont d’Or cheese (OR: 5.1; 95% CI: 0.9–35). No associations were found with other raw-milk cheeses, nor with any other food items.
Table 3. Frequency of reported cheese consumptiona according to MLVA and whole genome sequencing clusters for the case–case study and the case–control study, Salmonella Dublin outbreak, France, November 2015–March 2016 (n cases = 58; n controls = 2,914).
| Type of cheese | Cluster cases | Other casesb | Controls | Case–case studyc | Case–control studyd | Cluster cases | Other casesb | Controls | Case–case studyc | Case–control studyd | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | OR | 95% CI | aORe | 95% CI | n | % | n | % | n | % | OR | 95% CI | aORe | 95% CI | |
| Type of cheese | MLVA 20–8-10–7-5–4 (n = 10 cases) | WGS A (n = 11 cases) | ||||||||||||||||||
| Vacherin Mont d'Or | 7 | 70 | 11 | 31 | 233 | 8 | 5.1 | 0.9–35 | 27 | 6.8–104 | 7 | 70 | 12 | 32 | 233 | 8 | 5.1 | 0.9–34 | 27 | 6.8–105 |
| Comté | 6 | 67 | 21 | 58 | 1,386 | 48 | 1.4 | 0.3–10.2 | NS | NA | 6 | 67 | 23 | 59 | 1,386 | 48 | 1.4 | 0.2–9.8 | NS | NA |
| Gruyère | 7 | 78 | 26 | 76 | 1,841 | 63 | 1.1 | 0.2–13 | NS | NA | 7 | 78 | 28 | 78 | 1,841 | 63 | 1.0 | 0.1–12 | NS | NA |
| Camembert | 6 | 67 | 23 | 66 | 756 | 26 | 1.0 | 0.2–7.6 | NS | NA | 6 | 67 | 25 | 66 | 756 | 26 | 1.0 | 0.2–7.5 | NS | NA |
| Type of cheese | MLVA 18–8-10–7-5–4 (n = 23 cases) | WGS C3 (n = 17 cases) | ||||||||||||||||||
| Morbier | 12 | 60 | 10 | 42 | 361 | 12 | 2.1 | 0.5–8.3 | 11 | 4.2–29 | 10 | 67 | 12 | 37 | 361 | 12 | 3.3 | 0.8–15 | 14 | 4.8–42 |
| Goat cheese | 9 | 53 | 13 | 54 | 1,384 | 47 | 1.0 | 0.2–4.0 | NS | NA | Not consumed by 50% of the cases | NA | NA | NA | NA | |||||
| Camembert | 12 | 63 | 17 | 68 | 756 | 26 | 0.8 | 0.2–3.4 | NS | NA | 11 | 73 | 20 | 62 | 756 | 26 | 1.6 | 0.4–8.7 | NS | NA |
| Gruyère | 13 | 68 | 20 | 83 | 184 | 63 | 0.4 | 0.1–2.3 | NS | NA | 10 | 71 | 25 | 81 | 184 | 63 | 0.6 | 0.1–3.6 | NS | NA |
| Comté | Not consumed by 50% of the cases | NA | NA | NA | NA | 7 | 50 | 22 | 65 | 1,386 | 48 | 0.5 | 0.1–2.3 | NS | NA | |||||
aOR: adjusted odds ratio; CI: confidence interval; MLVA: Multilocus variable-number tandem repeat analysis; NA: not applicable; NS: did not remain significant in the final logistic regression model; OR: odds ratio; WGS: whole genome sequencing.
a Only cheeses consumed by at least 50% of the cases are included in the analysis; cases and controls could report more than one cheese consumed.
b Cases who belong to the other known subtypes.
c Compares cluster cases with other cases with other known subtypes.
d Compares cluster cases with controls.
e Adjusted for age and sex.
Case–control study
After adjustment for age and sex, compared with controls, cases belonging to the WGS cluster A were more likely to have consumed Vacherin Mont d’Or cheese (aOR: 27; 95% CI: 6.8–105) and cases belonging to the WGS subcluster C3 were more likely to have consumed Morbier cheese (aOR: 14; 95% CI: 4.8–42) (Table 3). Compared with controls, cases belonging to the MLVA cluster 18–8-10–7-5–4 were more likely to have consumed Morbier cheese (aOR: 11; 95% CI: 4.2–29) and cases belonging to the MLVA cluster 20–8-10–7-5–4 were more likely to have consumed Vacherin Mont d’Or cheese (aOR: 27; 95% CI: 6.8–104). No other significant associations were found with other raw-milk cheeses, nor with any other food items.
Food trace-back investigations
We collected 39 (62%) loyalty card numbers from 63 cases. Based on the available information, trace-back investigations were conducted among 10 supermarket brands. Twelve cheese producers were identified as potential origin of the cheeses consumed by the cases. The trace-back investigations linked one Morbier producer and three different Vacherin Mont d’Or producers, to 11, five, four and three cases, respectively. All those producers were located in the same region, i.e. Bourgogne-Franche-Comté (Eastern part of France).
Food and veterinary investigations
From the 101 non-humans isolates collected in 2015 and 2016, 82 (81%) were collected from milk products (54 from cheese, 27 from milk and one from other dairy), 12 (12%) from animal samples (cattle, meat, faeces) and seven (7%) from environmental samples (milk filter, trough). For the 54 cheese samples, S. Dublin was detected in Morbier (n = 37), in Saint-Nectaire (n = 6), in Reblochon (n = 5), in Vacherin Mont d’Or (n = 1) and in other or unknown cheeses (n = 5).
Discussion
We reported one of the largest S. Dublin outbreaks in France in the past few years. Two different bovine raw-milk cheeses, Morbier and Vacherin Mont d’Or, were the most likely vehicles of transmission for this food-borne outbreak. For the present outbreak investigation we used two different typing methods on a large panel of strains (both historical and obtained during the 2015–16 outbreak period, as well as from human and non-human origins). The first method was MLVA, which had already been used in previous investigations of other outbreaks in France [11]. The second, WGS, was used for the first time in the current S. Dublin investigation and demonstrated increased capacity to discriminate clusters. We also used two different epidemiological methods: a case–case study that allowed a rapid analysis and identification of suspected sources and a case–control study that was more statistically powerful to confirm the suspected associations.
In this investigation, MLVA was deemed sufficient to identify a link between human cases, food and animal sources. However, MLVA could not distinguish some of the clusters identified by WGS. Our results suggested that at least two outbreaks of S. Dublin occurred during the same period, and potentially originated from two different sources. WGS cluster A and subcluster C3 occurred in different periods indicating that they might belong to distinct outbreaks. The retrospective use of WGS also confirmed the occurrence of different S. Dublin outbreaks in 2012 [10,11]. High resolution molecular tools like WGS may facilitate linkage of human cases to sources, especially in serotypes with limited intrinsic genetic variation, and may also provide a more detailed picture of the extent and context of the outbreak.
Recruiting controls from an online health cohort survey for an ongoing outbreak investigation was novel in France and served as a pilot to evaluate the suitability of this method in future food-borne outbreak investigations. This method allowed conducting the case–control study in a timely manner with minimum resources, achieving a high response rate [28].
Our investigation pointed towards several cheese producers from the same region as sources of the outbreaks. In this region, an increase in salmonellosis incidence was observed in cattle at the end of summer 2015 (data not shown; Santé publique France). This could explain the increase of contaminated cheese batches in autumn and winter 2015. Veterinary and food investigations were challenging due to (i) high number of possibly implicated processing plants of raw-milk cheeses, and (ii) the high frequency of cheese consumption by cases and the variety of cheeses and places of purchase. It was difficult to identify the exact batches that cases consumed because some cheeses were sold at the deli counter, sliced on demand. Furthermore, the probable low levels of contamination of the implicated cheeses may have led to false negative test results, possibly allowing some contaminated batches to enter the market.
Previous studies indicated that S. Dublin is frequently isolated in live cattle, and that cheeses made with unpasteurised milk may be contaminated with S. Dublin [8,9]. S. Dublin infection is also responsible for substantial losses in the dairy industry [29]. A modelling exercise in Denmark [29] estimated the gross margin losses due to S. Dublin infection in a 200-cow stall-herd to be up to EUR 188 per stall annually averaged over the 10-year period following introduction of infection. In that study, relative simple and cheap control measures such as improving calving and colostrum management could lead to significant decreases in prevalence of S. Dublin in some herds. In other herds, it was reported that these measures might have to be supplemented by changes in hygiene and feeding practices. It might be worthwhile conducting studies in France to evaluate the impact of recent or future measures on S. Dublin prevalence at herd level. In addition, infected cattle might carry chronic and possibly asymptomatic infections while still contributing to onwards transmission by excreting pathogens in faeces [30,31]. SNP-typing based on WGS is a promising tool to monitor the routes and the spread of S. Dublin between herds in traditional regions of cheese production, as already reported in previous studies [16,17]. The combination of epidemiological studies in human and non-human sectors and the use of WGS may improve the cost effectiveness of control measures for S. Dublin in France, by targeting contaminated herds.
Following the investigations currently reported here, the group of producers of Morbier and Vacherin Mont d’Or cheeses implemented an action plan, including (i) systematic testing for Salmonella of batches of Morbier and Vacherin Mont d’Or sold since 1 February 2016, (ii) more regular farm visits by veterinarians, (iii) detection and containment of infected cattle, (iv) expert mission from the Ministry of Agriculture and Food to support milk industry professionals in Bourgogne-Franche-Comté region to identify and recommend better practices for detection and management of contaminated raw-milk products and (v) creation of a working group with experts on Salmonella issues from different organisations. The Morbier processing plants union reinforced their sanitary protocols, including more frequent testing of milk.
Limitations
Our investigations suffered from several limitations. First, cases were interviewed by phone, while controls completed a shorter online questionnaire, which could have led to obtaining exposure data with different degrees of accuracy. To minimise this bias, we used the same questions for cases and controls. Second, age and sex distribution of controls differed from that of the cases. We thus included age and sex in the multivariable analysis to adjust for those characteristics. Third, it was difficult to identify the exact sources of contamination due to probably low levels of contamination by S. Dublin of the cheese batches. As the cattle contamination was diffuse, it was difficult to incriminate specific cheese producers in the Bourgogne-Franche-Comté region as sources of contamination. Furthermore, suspected batches of cheese identified through trace-back investigations were no longer available for testing. Then, even if epidemiological investigations were carried out within a very constrained period of time to allow the ad hoc microbiological analyses to be launched to support the generated hypotheses, we had to deal with the impossibility to get through the whole process due to the lack of products.
Conclusions and recommendations
Microbiological, epidemiological and environmental evidence pointed towards two raw-milk cheeses, Morbier and Vacherin Mont d’Or, as vehicles of the S. Dublin infections. The use of MLVA and WGS subtyping methods allowed the identification of different clusters and of the potential vehicles of infection, highlighting the importance of adequate subtyping methods during Salmonella outbreaks and the relevance of company internal microbiological monitoring system. As a result, WGS has now been routinely implemented at the French NRC and findings of this multi-disciplinary investigation led to a reinforced control plan for processing plants of raw-milk cheeses to prevent future outbreaks.
Acknowledgements
We are grateful to all the microbiological laboratories processing human and non-human samples, who participate in the Salmonella network for isolates processing. We acknowledge Santé publique France epidemiologists from the regional offices who conducted the interviews, Edith Laurent who performed the data entry and Marie-José Letort who helped contacting laboratories. We thank all the persons involved in the microbiological investigations in Institut Pasteur (in particular Vincent Enouf, Estelle Serre and François Gravey) and in the French Agency for Food, Environmental and Occupational Health and Safety. We also thank the persons who conducted the food trace-back investigations in the Ministry of Agriculture and Food. We are grateful for the persons in the French National Institute of Health and Medical Research in the GrippeNet.fr project who worked with us in implementing the control recruitment method. Finally, we thank all the cases, their relatives and the nursing home staff and the controls who participated in the studies.
Conflict of interest: None declared.
Authors’ contributions: AU, KD and NJ coordinated the epidemiological part of the investigation. EL implemented the electronic version of the questionnaire, AU was in charge of the case–case study, DVC was in charge of the case–control study. DVC, LR and CG coordinated the part of the investigation regarding the GrippeNet.fr study. MLV, SCS, RL and SLH coordinated the microbiological part of the investigation regarding MLVA: MLV, SCS and RL collected food and animal samples and planned the MLVA analysis of human and non-human isolates, AB, NF, LF and SLH analysed the molecular data, AB, NF, LF, SL and SLH merged and analysed microbiological data, MLV, SCS performed MLVA analyses. AB, NF, LF and SLH coordinated the microbiological part of the investigation regarding WGS. AM and MPD coordinated the food trace-back part of the investigation. AU and AB took lead in manuscript writing, NJ, KD and SLH the general coordination. All authors read, commented and approved the final manuscript.
References
- 1.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ, et al. International Collaboration on Enteric Disease ‘Burden of Illness’ Studies The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis. 2010;50(6):882-9. 10.1086/650733 [DOI] [PubMed] [Google Scholar]
- 2.Langridge GC, Fookes M, Connor TR, Feltwell T, Feasey N, Parsons BN, et al. Patterns of genome evolution that have accompanied host adaptation in Salmonella. Proc Natl Acad Sci USA. 2015;112(3):863-8. 10.1073/pnas.1416707112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grimont PAD, Weill FX. Antigenic formulae of the Salmonella serovars, 9th ed. Paris: World Health Organization Collaborating Center for Reference and Research on Salmonella, Institut Pasteur; 2007. Available from: https://www.pasteur.fr/sites/default/files/veng_0.pdf
- 4.Jones TF, Ingram LA, Cieslak PR, Vugia DJ, Tobin-D’Angelo M, Hurd S, et al. Salmonellosis outcomes differ substantially by serotype. J Infect Dis. 2008;198(1):109-14. 10.1086/588823 [DOI] [PubMed] [Google Scholar]
- 5.Helms M, Vastrup P, Gerner-Smidt P, Mølbak K. Short and long term mortality associated with foodborne bacterial gastrointestinal infections: registry based study. BMJ. 2003;326(7385):357. 10.1136/bmj.326.7385.357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohammed M, Cormican M. Whole genome sequencing provides insights into the genetic determinants of invasiveness in Salmonella Dublin. Epidemiol Infect. 2016;144(11):2430-9. 10.1017/S0950268816000492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El Sayed F, Sapriel G, Fawal N, Gruber A, Bauer T, Heym B, et al. In-Host Adaptation of Salmonella enterica Serotype Dublin during Prosthetic Hip Joint Infection. Emerg Infect Dis. 2018;24(12):2360-3. 10.3201/eid2412.180214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selander RK, Smith NH, Li J, Beltran P, Ferris KE, Kopecko DJ, et al. Molecular evolutionary genetics of the cattle-adapted serovar Salmonella dublin. J Bacteriol. 1992;174(11):3587-92. 10.1128/jb.174.11.3587-3592.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Werner SB, Humphrey GL, Kamei I. Association between raw milk and human Salmonella dublin infection. BMJ. 1979;2(6184):238-41. 10.1136/bmj.2.6184.238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giron S, Le Hello S, Salah S, Mazur M, Lailler R, Barataud D, et al. Salmonella Dublin: what is its potential severity? Investigating a national foodborne outbreak of Salmonellosis associated with the consumption of raw milk cheese, France, August-September 2012. Poster session presented at: 6th International Symposium Salmonella and Salmonellosis (I3S); 2013 May 27-29; Saint-Malo, France. [Google Scholar]
- 11.Vignaud M-L, Cherchame E, Marault M, Chaing E, Le Hello S, Michel V, et al. MLVA for Salmonella enterica subsp. enterica Serovar Dublin: Development of a Method Suitable for Inter-Laboratory Surveillance and Application in the Context of a Raw Milk Cheese Outbreak in France in 2012. Front Microbiol. 2017;8:295. 10.3389/fmicb.2017.00295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.David JM, Sanders P, Bemrah N, Granier SA, Denis M, Weill FX, et al. Attribution of the French human Salmonellosis cases to the main food-sources according to the type of surveillance data. Prev Vet Med. 2013;110(1):12-27. 10.1016/j.prevetmed.2013.02.002 [DOI] [PubMed] [Google Scholar]
- 13.Jones G, Le Hello S, Jourdan-da Silva N, Vaillant V, de Valk H, Weill F, et al. The French human Salmonella surveillance system: evaluation of timeliness of laboratory reporting and factors associated with delays, 2007 to 2011. Euro Surveill. 2014;19(1):20664. 10.2807/1560-7917.ES2014.19.1.20664 [DOI] [PubMed] [Google Scholar]
- 14.Jourdan-Da Silva N, Le Hello S. [Salmonellosis in 2002-2010 in France: trends in human epidemiology, monophasic serotype emergence, main food implicated in the latest outbreaks]. Bulletin Epidémiologique Hebdomadaire Hors-série. 2012;25-33. French. [Google Scholar]
- 15.Kjeldsen MK, Torpdahl M, Campos J, Pedersen K, Nielsen EM. Multiple-locus variable-number tandem repeat analysis of Salmonella enterica subsp. enterica serovar Dublin. J Appl Microbiol. 2014;116(4):1044-54. 10.1111/jam.12441 [DOI] [PubMed] [Google Scholar]
- 16.Mohammed M, Delappe N, O’Connor J, McKeown P, Garvey P, Cormican M. Whole genome sequencing provides an unambiguous link between Salmonella Dublin outbreak strain and a historical isolate. Epidemiol Infect. 2016;144(3):576-81. 10.1017/S0950268815001636 [DOI] [PubMed] [Google Scholar]
- 17.Ågren EC, Wahlström H, Vesterlund-Carlson C, Lahti E, Melin L, Söderlund R. Comparison of whole genome sequencing typing results and epidemiological contact information from outbreaks of Salmonella Dublin in Swedish cattle herds. Infect Ecol Epidemiol. 2016;6(1):31782. 10.3402/iee.v6.31782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gossner C, Le Hello S, De Jong B, Rolfhamre P, Faensen D, Weill FX, et al. Around the World in 1,475 Salmonella Geo-serotypes. Emerg Infect Dis. 2016;22(7):1298-302. 10.3201/eid2207.141678 [DOI] [Google Scholar]
- 19.Fonteneau L, Jourdan Da Silva N, Fabre L, Ashton P, Torpdahl M, Müller L, et al. Multinational outbreak of travel-related Salmonella Chester infections in Europe, summers 2014 and 2015. Euro Surveill. 2017;22(7):30463. 10.2807/1560-7917.ES.2017.22.7.30463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357-9. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15(11):524. 10.1186/s13059-014-0524-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725-9. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44(W1):W242-5. 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bachelerie P, Felten A, Vignaud ML, Glasset B, Feurer C, Lailler R, et al. MLVA_Normalizer: Workflow for Normalization of MLVA Profiles and Data Exchange between Laboratories. J Proteomics Bioinform. 2016;9(2):25-7. 10.4172/jpb.1000385 [DOI] [Google Scholar]
- 25.Debin M, Turbelin C, Blanchon T, Bonmarin I, Falchi A, Hanslik T, et al. Evaluating the feasibility and participants’ representativeness of an online nationwide surveillance system for influenza in France. PLoS One. 2013;8(9):e73675. 10.1371/journal.pone.0073675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guerrisi C, Turbelin C, Blanchon T, Hanslik T, Bonmarin I, Levy-Bruhl D, et al. Participatory syndromic surveillance of influenza in Europe. J Infect Dis. 2016;214(suppl_4):S386-92. 10.1093/infdis/jiw280 [DOI] [PubMed] [Google Scholar]
- 27.Achtman M, Wain J, Weill FX, Nair S, Zhou Z, Sangal V, et al. S. Enterica MLST Study Group Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog. 2012;8(6):e1002776. 10.1371/journal.ppat.1002776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mook P, Kanagarajah S, Maguire H, Adak GK, Dabrera G, Waldram A, et al. Selection of population controls for a Salmonella case-control study in the UK using a market research panel and web-survey provides time and resource savings. Epidemiol Infect. 2016;144(6):1220-30. 10.1017/S0950268815002290 [DOI] [PubMed] [Google Scholar]
- 29.Nielsen TD, Kudahl AB, Østergaard S, Nielsen LR. Gross margin losses due to Salmonella Dublin infection in Danish dairy cattle herds estimated by simulation modelling. Prev Vet Med. 2013;111(1-2):51-62. 10.1016/j.prevetmed.2013.03.011 [DOI] [PubMed] [Google Scholar]
- 30.Spier SJ, Smith BP, Cullor JS, Olander HJ, Roden LD, Dilling GW. Persistent experimental Salmonella dublin intramammary infection in dairy cows. J Vet Intern Med. 1991;5(6):341-50. 10.1111/j.1939-1676.1991.tb03148.x [DOI] [PubMed] [Google Scholar]
- 31.Uzzau S, Brown DJ, Wallis T, Rubino S, Leori G, Bernard S, et al. Host adapted serotypes of Salmonella enterica. Epidemiol Infect. 2000;125(2):229-55. 10.1017/S0950268899004379 [DOI] [PMC free article] [PubMed] [Google Scholar]


