Abstract
Tissues used in pathology laboratories are typically stored in the form of formalin-fixed, paraffin-embedded (FFPE) samples. One important consideration in repurposing FFPE material for next generation sequencing (NGS) analysis is the sequencing artifacts that can arise from the significant damage to nucleic acids due to treatment with formalin, storage at room temperature and extraction. One such class of artifacts consists of chimeric reads that appear to be derived from non-contiguous portions of the genome. Here, we show that a major proportion of such chimeric reads align to both the ‘Watson’ and ‘Crick’ strands of the reference genome. We refer to these as strand-split artifact reads (SSARs). This study provides a conceptual framework for the mechanistic basis of the genesis of SSARs and other chimeric artifacts along with supporting experimental evidence, which have led to approaches to reduce the levels of such artifacts. We demonstrate that one of these approaches, involving S1 nuclease-mediated removal of single-stranded fragments and overhangs, also reduces sequence bias, base error rates, and false positive detection of copy number and single nucleotide variants. Finally, we describe an analytical approach for quantifying SSARs from NGS data.
INTRODUCTION
Formalin-fixed paraffin-embedded (FFPE) tissue is the most common form of tissue preparation used in pathology laboratories. Characterization of FFPE-derived nucleic acids is frequently employed in both retrospective and prospective analysis of clinical samples, in lieu of corresponding fresh or fresh-frozen (FF) tissue. Recent progress in the automation of nucleic acid extraction from FFPE tissues for NGS studies has added further impetus for sequencing library construction protocols optimized for use with this invaluable clinical material (1). An overarching challenge is that treatment of samples with formalin and paraffin, along with sample storage and extraction procedures, can result in significant fragmentation, denaturation and chemical modifications of nucleic acids (2). These changes result in sequencing artifacts. DNA damage can, in part, be mitigated by enzymatic treatment to repair nicks and deaminated or oxidized bases (1–4). However, artifacts still persist at much higher prevalence compared to data from NGS libraries that are prepared from fresh or FF tissues (1).
In this study, we used a cohort of matched FF and FFPE samples (n = 38) to identify differences that led to the investigation of the mechanistic basis for FFPE-associated chimeric read artifacts. We then explored approaches to reduce the levels of such artifacts, and other noise and bias in FFPE data, including GC-bias, base ‘error’ rate and aberrant detection of copy number variants (CNVs) and single nucleotide variants (SNVs). Of note, a fraction of base error rates may include true SNVs and, to a degree, comparisons with matched FF samples allow us to discern true SNVs from artifact SNVs that arise due to FFPE-associated base errors.
MATERIALS AND METHODS
Samples and extraction
Mouse FFPE nucleic acid samples were prepared from one block of C57BL/6 mouse liver tissue. The tissue was fixed in 10% neutral buffered formalin and the FFPE block was prepared using Sakura Tissue-Tek VIP Tissue Processor (GMI) according to manufacturer's instructions. The FFPE block was stored at room temperature for 4 years prior to extraction. 10 μm tissue sections were prepared using a Leica RM2255 rotary microtome (Leica Biosystems). All experiments that involved this mouse sample were performed in three technical replicates. Three of the human FFPE tissue blocks, used in the Nextera versus ligation-based library construction comparison, were previously described (1) while the rest (n = 38; used for the comparisons between FF and FFPE tumor samples) were obtained from the Burkitt Lymphoma Genome Sequencing Project (BLGSP) (5). The FFPE blocks from these human samples were stored for 1–4 years.
Nucleic acids were extracted from the 2–5 FFPE scrolls using a modified version of Agencourt's FormaPure protocol as described previously (1) or a combination of Qiagen's AllPrep FFPE and Roche's High Pure protocols (Zmuda et al., submitted). Nucleic acids were extracted from the FF samples using the DNA/RNA AllPrep kit (Qiagen). For experiments evaluating the effects of various reverse cross-linking durations using the mouse FFPE sample, lysates were pooled and re-aliquoted after the deparaffinization/lysis step in order to avoid the otherwise confounding variability in cellular composition between FFPE tissue sections.
Genomic DNA libraries and sequencing
Libraries from FF tissue were generated from 500 to 1000 ng DNA using a PCR-free library construction protocol (details of the protocol are described in the Supplementary Methods and Materials). FFPE libraries were generated from 100 to 200 ng DNA/total nucleic acid (TNA) using eight cycles of PCR. The former are enriched for 400–500 bp insert sizes as opposed to 200–300 bp for the latter. The FFPE genome library construction protocol and sequencing steps were as described previously (1) with some modifications. S1 Nuclease was purchased from Thermo Fisher Scientific (Catalog# MAN0013722) and the treatment of 100–300 ng gDNA or TNA was as described in the manufacturer's instructions. Following treatment, DNA was purified using magnetic beads (PCR Clean-DX magnetic beads from Aline Biosciences; Catalog# C-1003–450) at a ratio of 1.8 (beads):1 (reaction). Other purification conditions and subsequent shearing, size selection and library construction steps were as described previously (1). The matched FF and FFPE libraries from the 38 human sources were sequenced to >30× coverage. The libraries that were generated as part of the optimization process to improve FFPE data quality were sequenced to obtain at least 10–20 million reads and the libraries that were deep sequenced for evaluation of S1 nuclease treatment were sequenced to >30x coverage.
Sequence analysis
Sequence analysis was performed as described previously (1). Briefly, libraries were sequenced on an Illumina HiSeq 2500 lane (paired-end 125 bp) or HiSeq X lane (paired-end 150 bp). Sequencing reads were aligned to the human reference (hg19) using BWA-MEM version 0.7.6.a. Sequencing data from human tissue samples was deposited in the database of Genotypes and Phenotypes (dbGAP; accession no. phs000527) and data from mouse tissues was deposited in the National Center for Biotechnology Information's Sequence Read Archive (identifier SRP150031). The Integrative Genomics Viewer (IGV) (6) was used for manual evaluation of read alignments. GC bias and error rates were estimated using Picard (http://broadinstitute.github.io/picard) and Qualimap (7). Data were visualized using MultiQC (8).
Quantification of strand-split artifact reads (SSAR) levels
SSARs were identified in reads whose alignments to the reference genome were broken into two or more disparate segments of which at least two segments aligned (i) on opposite strands and (ii) within a pre-defined maximum distance of each other. As above, the alignments were created using BWA-MEM 0.7.6a, which is capable of splitting a single chimeric read across a non-continuous reference sequence in a strand-agnostic manner. In libraries that were prepared from FFPE samples, >80% such artifacts exhibited non-contiguous alignments within 500 bp windows (Supplementary Figure S1). In contrast, <25% of such chimeric reads were within this distance in libraries that were derived from matching FF tissue samples. Thus, we used 500 bp as a cut off for defining SSARs.
Detection of single nucleotide and copy number variants
Each of the three tumor samples (FF, S1nuclease- treated FFPE, and untreated FFPE) were compared to a FF germline-derived tissue sample from the same individual to create three sets of somatic variant calls.
Somatic SNVs were called using Strelka (9) Version 1.0.6. We found that a minimum Strelka somatic variant quality score (QSS) greater than 34 was found to be the threshold above which the majority of called variants in the FFPE data were also in the matched FF data. Implementation of this threshold allowed for analysis of FFPE-called somatic variants that had significantly reduced numbers of false positives.
CNVs were identified by comparing the depth of coverage in the matched tumor and normal (T:N) BAM files as described previously (10–11). Briefly, after GC correction was applied to the T:N sequence read depth ratio, the genome was partitioned into regions of consistent copy number using a Hidden Markov Model. The Jaccard index, which describes similarity between sets, was used to compare CNV results from FF and FFPE in the presence and absence of S1 nuclease. After determining copy number status of the 57773 genes in Ensembl 75, intersections were calculated by comparing genes that were copy number gained or lost in each of the samples. The Jaccard index was calculated independently for the gains or losses by dividing the intersection of the gene sets by the union in each of the comparisons. For example, the Jaccard's index for copy number gains between FF and FFPE-S1 was calculated by dividing the number of genes that were copy number gained in both sets by the number of genes that were copy number gained in either set.
RESULTS AND DISCUSSION
The aims of the current study were to (1) characterize the nature of artifact reads associated with FFPE samples and (2) improve the quality of sequence data derived from FFPE genome libraries.
Chimeric reads are significantly more abundant in sequencing data from FFPE genome libraries
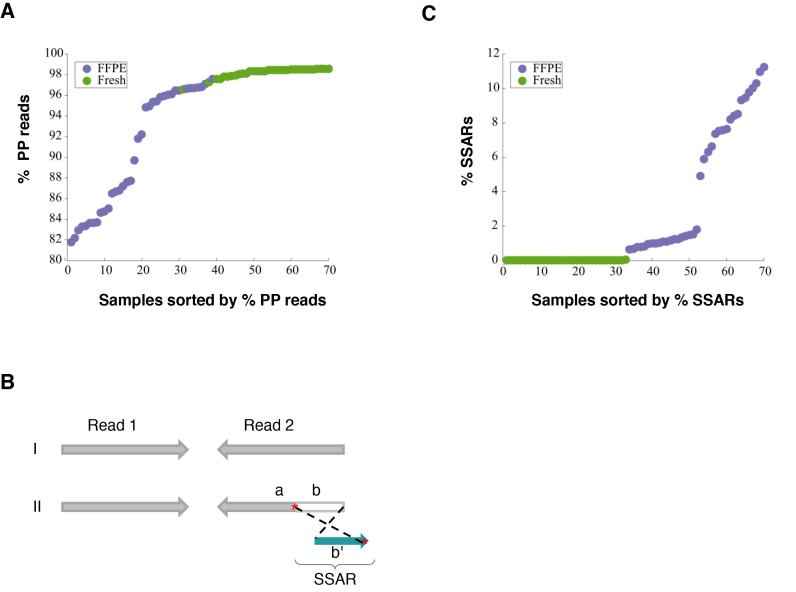
Sequencing from both ends of a library insert, referred to as paired-end sequencing (PE-seq), is often the preferred sequencing approach in NGS analyses. Considered alongside a defined range of insert sizes, specified during sequencing template size selection in library preparation, PE-seq in genome analysis allows the characterization of structural rearrangements including insertions, deletions, and translocations. Such rearrangements may represent the underlying genome or alternatively may be artifacts that arise during sample preparation and/or library construction. The latter include chimeric reads that apparently are derived from non-contiguous genome sequences. Thus, the proportion of properly paired read alignments (%PP) is an important quality metric in NGS analysis. Aligned reads whose insert size distribution fit within the distribution estimated by the majority of the aligned fragments were defined as PP reads. Systematic comparison of such artifact levels and other genome library sequencing metrics using data from a large cohort of matched FFPE and fresh or FF tissue is currently limited. In this study, we used such data for 38 samples that were generated as part of the BLGSP project (5). Genome libraries from both FFPE and FF materials were prepared and sequenced. The average % PP for the majority of the libraries that were generated from FF tissue was >98% versus a wide range of PP reads (81.7–97.6%) for those that were derived from FFPE samples (Figure 1A).
Figure 1.
Differences in library quality between matched FFPE and FF tissue samples and possible underlying mechanism. (A) Comparison of the frequency of properly paired (PP) reads between matched FFPE (n = 38) and FF (n = 38) tissue samples. (B) Diagrammatic depiction of Strand-split artifact reads (SSARs). Typical PP reads (I) and an example of PP reads with SSAR in Read 2 (II) are shown. For III, Read 1 is contiguously aligned to the reference genome while Read 2 is split into two parts. Part ‘a’ of Read 2 aligns in the expected paired-end orientation while the distal end of Read 2 (part ‘b’) does not match the reference sequence at that position. Part ‘b’ of Read 2 does match the reference genome near Part ‘a’, but aligns in the opposite orientation (b’). Sequence homologies in the reference genome between the regions are marked with red asterisks ‘*’. (C) Comparison of SSAR levels between matched FFPE (n = 38) and FF (n = 38) tissue samples.
Strand-split read artifacts are predominant among FFPE-associated chimeric read artifacts
In the process of using IGV (6) to review the alignments of reads obtained from sequencing FFPE genome libraries, we noted the existence of anomalous reads in which one portion of the read aligned to the ‘Watson’ strand and another portion of the read aligned to the ‘Crick’ strand (Figure 1B). These artifact reads are hereafter referred to as SSARs. We established a bioinformatics approach (see Materials and Methods section) to quantify SSARs. SSAR levels were barely detectable in libraries that were generated from FF tissue (<0.01%), compared to prevalence ranging from 0.67–12.26% for those libraries that were derived from FFPE samples (Figure 1C). A strong negative correlation between %PP and SSAR levels was observed (Pearson's correlation of −0.8778), which indicated the possibility of a common mechanism underlying the high levels of improperly paired reads and SSAR levels in FFPE libraries (see below). For the FFPE libraries, ∼86% of SSARs were in PP reads (Supplementary Figure S2), suggesting that, although they correlate, rates of improperly paired reads and SSARs are largely independent measures of FFPE library quality.
Variability in artifact levels in FFPE data is in part associated with extraction protocols
Of the 38 BLGSP FFPE samples, 19 were extracted using our automated FormaPure protocol (F) (1) and the rest (n = 19) were extracted using a low-throughput manual column-based AllPrep/HighPure protocol (A–H) developed by The Cancer Genome Atlas (TCGA) project. The proportion of PP reads in libraries that were generated from gDNA that was extracted using the FormaPure protocol (mean = 92.5%) was notably lower than in libraries prepared from nucleic acid extracted using the A–H protocol (mean = 99.2%) (Supplementary Figure S3A). In contrast, SSAR proportions were notably higher in the libraries from the FormaPure extracted material (average 9.4%) than in the libraries from the A–H protocol (average 1.1%) (Supplementary Figure S3B).
Other quality differences between sample types and extraction protocols
In addition to chimeric artifact levels, the quality of libraries was also evaluated on base error rate, GC bias, and sensitivity and specificity of SNV/CNV detection. Based on these quality metrics, the following general trend in overall quality was observed: FF tissue > A–H FFPE > FormaPure FFPE (Supplementary Figures S4–S7). There were several differences between the two FFPE extraction protocols (Supplementary Figure S8) that may account for the observed trend. In particular, the FormaPure protocol includes longer incubations at higher temperature and is magnetic bead-based (as opposed to column-based).
Origin of strand-split read artifacts
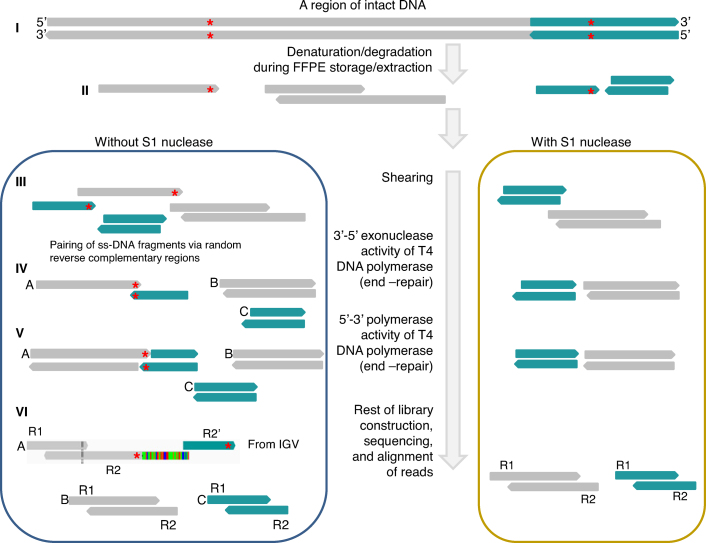
We hypothesized that SSAR artifacts originated from single-stranded DNA (ss-DNA) fragments, which are plentiful in FFPE DNA (12). One premise supporting this hypothesis is that standard ligation-based NGS library construction may produce chimeric fragments. T4 DNA ligase, which is used in NGS library construction, acts on double-stranded DNA (ds-DNA) substrates (13). The 3′-5′ exonuclease and 5′-3′ polymerase activities of T4 DNA polymerase are used to repair ‘ragged’ overhangs of bona fide ds-DNA fragments, but may also act on spurious ds-DNA fragments that result from opportunistic annealing of ss-DNA fragments, which may be mediated by repetitive DNA sequences (Figure 2). Indeed, 100% of the 24 SSAR cases we queried were found to contain short reverse complementary regions that could mediate SSAR formation. Interestingly, a similar phenomenon was described previously for a strand-specific RNA-seq protocol, in which the first strand cDNA was used to generate libraries via T4 DNA ligase-mediated library construction (14).
Figure 2.
SSAR mapping and diagrammatic depiction of the proposed mechanism. The SSAR example shown is a screen shot of an actual IGV image. At the top (I) we depict a ds-DNA region of intact gDNA. In the process of FFPE preparation, storage and extraction (II), gDNA is fragmented and denatured. In the absence of S1 nuclease (III left), ss-DNA fragments from non-contiguous regions of the genome anneal via short complementary repetitive sequences (red asterisks). In contrast, ss-DNA fragments and overhangs are removed upon treatment with S1 nuclease (III right). During the end-repair step of library construction, T4 DNA polymerase removes overhangs (IV) and fills ends (V), resulting in the formation of double-stranded chimeric fragments (‘A’ in V). One class of such chimeric fragments yield SSARs (‘A’ in VI). R1 = read; R2 = read 2. For SSARs, part of Read 2 aligns in the expected paired-end orientation while the distal end of Read 2 does not match the reference at that position and instead aligns to a nearby region of the reference genome in the opposite orientation (denoted as R2′).
The effect of high temperature incubation during extraction on FFPE library quality
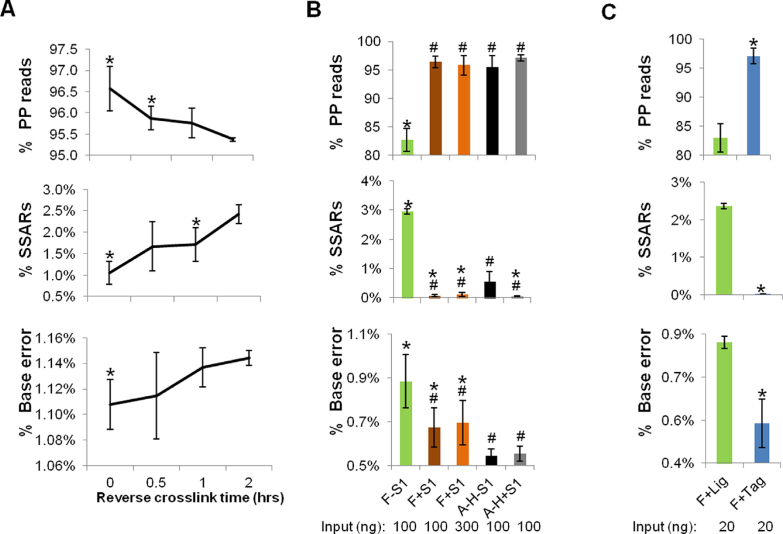
The elevated temperature for deparaffinization (70°C), lysis (55°C) and reverse cross-linking (90°C) steps during DNA purification from FFPE tissue extraction contribute to denatured DNA, thus increasing the frequency of ss-DNA. We therefore sought to measure the effect of the duration of the reverse cross-linking incubation on FFPE-associated artifact levels. For this, we used a 4-year-old FFPE mouse liver FFPE block. Following deparaffinization and lysis steps, lysates were pooled and aliquoted for increasing durations of reverse cross-linking (0–2 h). As expected, extraction/library yield was positively correlated with the duration of reverse cross-linking (data not shown). Sequencing of the resulting libraries revealed that the proportion of PP reads was inversely correlated to the duration of reverse cross-linking (Figure 3A; upper panel). Conversely, SSAR levels and error rates were positively correlated with the duration of reverse cross-linking (Figure 3A; middle and lower panels). We further noted that the quality of the FormaPure libraries was not as high as those prepared from DNA that was extracted using the A–H protocol (Supplementary Figure S9). For example, SSAR levels were 1.05% for the former (reverse cross-linking time = 0 min) as opposed to 0.3% for the latter. Consistent with our findings, Robbe et al. reported recently that temperature reductions from 90°C to 65°C along with an increased salt concentration during reverse cross-linking, in an extraction protocol using the QIAamp DNA FFPE Tissue kit (Qiagen), led to improved CNV detection (15).
Figure 3.
Measures to improve FFPE library quality and their effects on the levels of properly paired (PP) reads, chimeric reads and base error rates. (A) Effects of reverse cross-link timing. A time course experiment was performed using mouse liver FFPE tissue as the starting material. 100 ng of FormaPure extracted total nucleic acid (TNA) was used. N = 3 (technical replicates). Error bars = Standard deviations. P<0.05 (relative to 2 h). (B) Effects of S1 nuclease treatment. 100 and/or 300 ng TNA extracted using the FormaPure protocol was used with (F+S1) or without (F-S1) S1 nuclease treatment. 100 ng DNA extracted using the Qiagen/HiPure protocol was used with (A–H+S1) or without (A–H–S1) S1 nuclease treatment. N = 5 (FFPE samples from five patients). Of note, these samples were not patient-matched between the extraction protocols (A–H and F). Error bars = Standard deviations. *P< 0.05 (relative to A–H-S1); #P< 0.05 (relative to F-S1). (C) Comparisons of ligation-based and tagmentation-based library construction protocols. 20 ng TNA from FormaPure extracted TNA was used for library construction using the ligation-based protocol (F+Lig) or the tagmentation-based protocol (F+Tag). N = 3 (FFPE samples from 3 patients). Error bars = Standard deviations. *P< 0.05 (relative to F+Lig).
A modified library construction protocol for improved FFPE library quality
Using the mouse FFPE sample, we also tested post-extraction conditions. According to our proposed mechanism of SSAR formation (Figure 2), the removal of ss-DNA fragments should reduce SSAR levels and increase the proportion of PP reads. To remove ss-DNA, we experimented with S1 nuclease, which digests ss-DNA and RNA (16).Total nucleic acid was treated with S1 nuclease before shearing to ensure that ss-DNA fragments were removed prior to end repair. Sequencing of the resulting libraries revealed that the proportion of PP reads was increased from 96.1% to 98.4% with S1 nuclease treatment (Supplementary Figure S9A). As predicted, SSAR levels were reduced from 1.8% to 0.04%, which was even lower than what we obtained (0.3%) for libraries that were prepared from DNA that was extracted using the A–H protocol (Supplementary Figure S9B). Likewise, there was a significant (P<0.05), albeit modest, decrease in base error rates, from 1.09% to 1.07%, compared to 0.99% for the A–H protocol (Supplementary Figure S9C).
To address whether these improvements could be extended to human FFPE samples, we next tested the effect of S1 nuclease on nucleic acids that were extracted from 5 human FFPE samples, using both the FormaPure and A–H protocols. Our results were consistent with those obtained for the mouse sample (Figure 3B), except that the improvements seen in the human samples were notably higher than in mouse, perhaps due to the generally poorer quality of the human FFPE samples. For the FormaPure extracted samples, the percentage of PP reads increased from 82.7% to 96.4% upon treatment with S1 nuclease, compared to an increase from 95.5% to 97.1% for samples that were extracted using the A–H samples (Figure 3B; upper panel). SSAR levels were reduced from 2.95% to 0.07% for the FormaPure samples, compared to a reduction from 0.56% to 0.054% for the A–H protocol (Figure 3B; middle panel). Base error rates were reduced from 0.885% to 0.675% for the FormaPure extracted samples, compared to a change from 0.545% to 0.553% for the A–H protocol (Figure 3B; lower panel). These data show that S1 nuclease treated libraries from FormaPure extracted samples were of comparable quality to those prepared using the A–H protocol. However, the duplicate rates of the libraries produced using the FormaPure protocol were higher compared to those produced using the A–H protocol regardless of S1 nuclease treatment (Supplementary Figure S10). This may in part be explained by the use of total nucleic acid (TNA) in the FormaPure protocol used here, which includes RNA along with DNA that can result in overestimation of the amount of DNA present in the sample (1). The addition of S1 nuclease to TNA further reduced apparent diversity (Supplementary Figure S10). This loss was partially ameliorated by increasing the TNA input amount from 100 ng to 300 ng (Supplementary Figure S10).
Motivated by these observations, we next hypothesized that library construction protocols that did not employ end-repair would result in relatively low levels of SSARs. One such method is the tagmentation-based Nextera™ protocol (17). We applied this protocol to TNA that was purified from three human FFPE samples using the FormaPure protocol. In parallel, we also generated libraries from the same samples and equivalent input amounts using a library construction protocol involving end-repair and ligation steps, but without prior S1 nuclease treatment. Consistent with our hypothesis, the tagmentation-based libraries displayed a higher percentage of PP reads (97%) as compared to those that were generated using the ligation-based approach (83%) (Figure 3C; upper panel). Conversely, the tagmentation-based libraries had reduced SSAR levels (0.02% versus 2.4%) and base error rates (0.54% versus 0.82%) (Figure 3C; middle and lower panels).
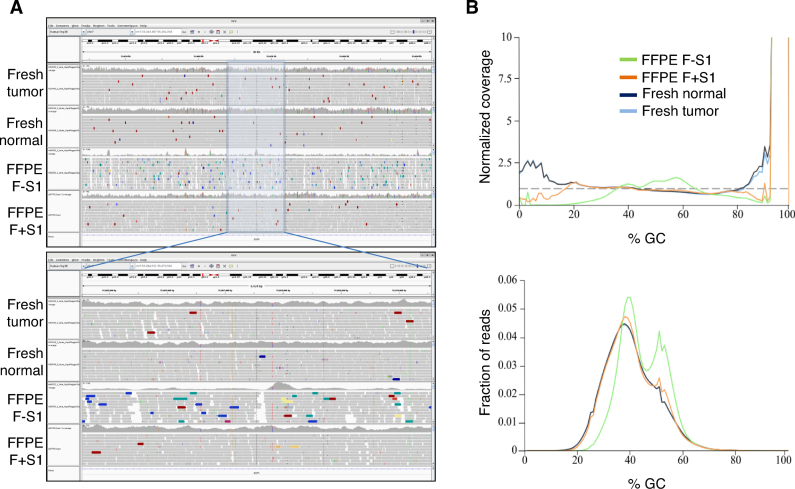
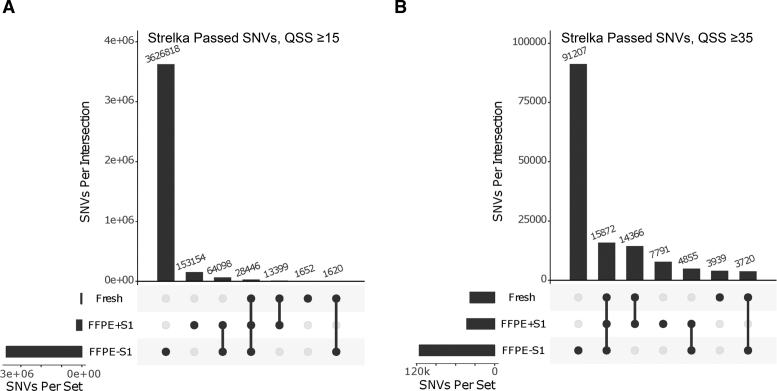
The above comparisons, evaluating the effect of S1 nuclease, were computed using relatively shallow (10–20 million reads) genome sequence of the genome. To validate our observations using deeper redundancy of sequence coverage, we sequenced one of the libraries, generated from 300 ng FormaPure TNA + S1 nuclease treatment, to more than 30× using a HiSeq X instrument. The alignment metrics from this library were compared to those of the libraries that were generated from untreated 100 ng TNA as well as patient-matched fresh normal and tumor tissues (Supplementary Figure S11). The trends we observed analyzing shallow sequencing data were confirmed using deeper sequencing data, including an improved % PP, lower SSAR levels, and lower base error rates from S1 nuclease-treated FFPE nucleic acids. In addition, reads were distributed more evenly across the genome (Figure 4A) and appeared to exhibit less GC-content bias (Figure 4B), approaching the results obtained for libraries from matched FF tissues. Somatic SNV false positives were also reduced (Figure 5A). For example, analysis of the FF tumor tissue data revealed 45,117 SNVs, compared to 259,097 SNVs detected in the library from S1 nuclease-treated FFPE TNA, and 3.7 million SNVs detected in the library from untreated FFPE TNA. 16.2% of the SNVs in the S1-treated FFPE library were also detected in the library from the FF tumor tissue. In contrast, only 0.81% of the SNVs in the library from the untreated FFPE nucleic acids were also detected in the library from FF tumor tissue. These numbers are based on the default quality threshold of the SNV detection pipeline (QSS score≥15; Materials and Methods). In another study (manuscript in preparation), our analysis of a cohort of patient-matched FF and FFPE established that a higher threshold (QSS score≥35) is required to filter out false positives in FFPE data. When this stringent filter is applied, the number of SNVs detected is reduced by 97% and 83% for the untreated and S1 –treated FFPE libraries, respectively (Figure 5B). 71% of the SNVs in the S1-treated FFPE library were also detected in the library from the FF tumor tissue, compared to an overlap of only 17% between FF tumor tissue and the untreated FFPE library. The more stringent filter also reduced SNVs with low variant allelic frequencies (VAFs) (<20%) with the following descending order of the degree of reduction of low VAFs: untreated FFPE > S1-treated FFPE > FF (Supplementary Figure S12). The frequency of somatic single nucleotide deletion/insertion artifacts was also reduced with S1 treatment (Supplementary Figure S13).
Figure 4.
Effects of S1 nuclease treatment on genome coverage and sequence bias. (A) Genome coverage. A screen shot of an IGV image of a representative chromosomal region is shown for libraries that were prepared from fresh normal and tumor samples, and matching FormaPure FFPE samples with (F+S1) or without (F-S1) S1 nuclease treatment. The lower panel is an enlarged portion of the region shown in the upper panel. Colored vertical lines within coverage histograms designate consensus SNVs that are also shown as colored vertical lines within reads. Colored arrow boxes within reads represent SSARs and improperly paired artifacts (Red = insert size too large relative to consensus insert size range; Blue = insert too small; Green = SSARs. Other colors depict paired reads that aligned to regions from different chromosomes). (B) Effects of S1 nuclease treatment on GC-bias. Samples are the same as in (A). Upper panel shows normalized coverage data at various levels of GC-content and lower panel shows read distribution as a function of GC-content.
Figure 5.
Effects of S1 nuclease treatment on FFPE-associated somatic SNV noise. Libraries were prepared from fresh normal and tumor samples from the same patient, and matching FormaPure FFPE samples with (F+S1) and or without (F-S1) S1 nuclease treatment. For each of the three latter libraries, SNVs were identified relative to the library from the normal blood sample. Upset plots indicating data overlaps are shown. In (A) are data obtained using a QSS score cutoff ≥15 as a cut-off. In (B) are data generated using a QSS score cutoff ≥35.
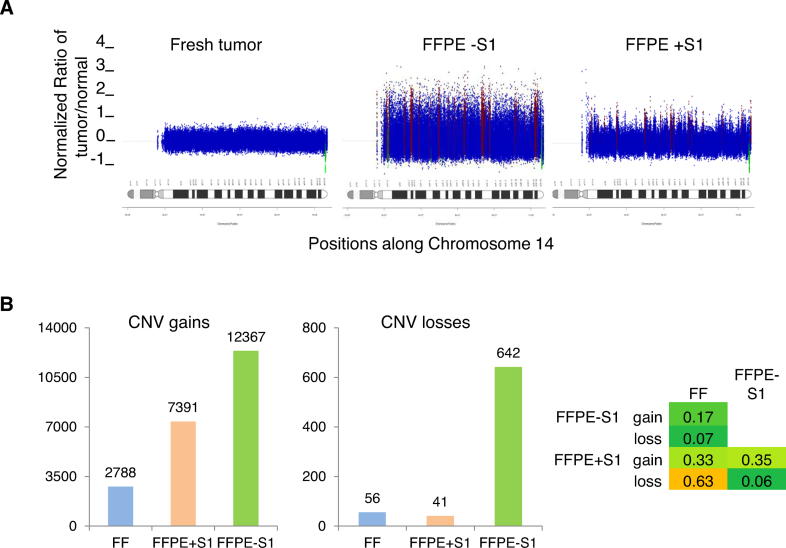
We next evaluated the impact of S1 treatment on CNV detection. When we compared data from tumor samples to normal tissue using a bin size of 200 reads (Materials and Methods), it was evident that the FFPE libraries yielded ‘noisy’ CNV data compared to the FF library (Figure 6A). Importantly, data from the S1 nuclease treated FFPE TNA appeared less noisy than data from the untreated comparator (Figure 6A). We quantified the noise reduction achieved with S1 nuclease treatment, detecting 50 CNV segments in the FF tumor library, 1790 in the library from S1 nuclease treated FFPE TNA and 2588 from the untreated FFPE sample. The 50 segments in the FF library results included seven regions of copy number loss and nine regions of copy number gain. Each of these CNVs was detected in both sets of FFPE data. There were 141 regions of loss and 1027 regions of gain that were unique to the untreated FFPE library, consistent with a relatively high apparent false positive rate. The S1-treated library had fewer apparent false positives, with only three regions of loss and 798 regions of gain that were not detected in the FF tissue. CNV profiles for all chromosomes are shown in Supplementary Figures S14–S16. Consistent with these data, an increased overlap of gene-level copy number changes between FFPE and FF was observed in the presence compared to the absence of S1 treatment (Figure 6B). Of note, while the majority of the differences between the FFPE and FF tumor libraries may be associated with FFPE sample processing, some aspects could be due to the differences in library construction strategies. In addition, some of those differences could also be related to differential sampling of the tumor specimens as previously reported (15).
Figure 6.
Effects of S1 nuclease treatment on FFPE-associated CNV noise. (A) Example illustrating CNV noise. Samples are the same as in Figure 5. Using a bin size of 200 reads, CNV segments were calculated in the tumor samples relative to the normal blood sample and the resulting profile is shown for chromosome 14. (B) CNV counts at the gene level. Gains are shown on the left and losses are shown in the middle panel. Jaccard's intersection index (Materials and Methods) is shown in the right panel as a measure of overlap of gene-level CNVs between the three samples.
Our observations are consistent with the notion that S1 nuclease treatment significantly improves the quality of libraries, particularly from FormaPure extracted samples. As shown in Supplementary Figure S17 and using several quality metrics, the FormaPure + S1 nuclease library was shown to be within the range of the quality reported for the 15 FFPE libraries that were prepared from gDNA that was extracted using the A–H protocol.
Overall, we only observed improvements in quality or no significant differences for S1 nuclease treated libraries versus untreated libraries for all the sequencing quality metrics we evaluated. These improvements come at the cost of increased amounts of nucleic acid for library construction.
In summary, based on our systematic analysis of a relatively large cohort (n = 38) of human cancer samples, FFPE genome libraries displayed generally lower quality compared to libraries from matched FF tissue, including reductions in the proportion of PP reads and increased frequencies of chimeric artifacts. Characterization of these FFPE–associated chimeric read artifacts led to the observation that many of them were non-contiguous sequences that mapped to the Watson and Crick strands. Our proposed mechanism for the genesis of these artifacts identified ss-DNA from denatured DNA fragments as their probable source. Short stretches of sequence complementarity in the ss-DNA regions can link fragments together, yielding the chimeras after end-repair, which then become templates for T4 DNA ligase. We present lines of evidence supporting this proposed mechanism, which include (i) verification of the existence of short complementary regions in 100% of the SSARs we studied; (ii) the relationship between reductions in nucleic acid heat exposure (which presumably also reduces denaturation and therefore ss-DNA) and increased quality of FFPE libraries; (iii) the impact of removing ss-DNA fragments via S1 nuclease treatment and (iv) the use of a tagmentation-based library construction protocol that lacks an end-repair step. Some of our protocol improvements, most notably S1 nuclease treatment, yield reduced base error rates and reduced GC-bias, resulting in more uniform sequence coverage of the genome and decreased rates of false positive SNV and CNV.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful for the contribution from members of the various groups at Canada's Michael Smith Genome Sciences Centre including those from the Biospecimen, Quality Assurance, Library Construction, Instrumentation, Sequencing, LIMS, Purchasing, Project Management and Bioinformatics teams. We also thank Dr Daniela Gerhard (Office of Cancer Genomics, National Cancer Institute, NIH, Bethesda, MD, USA) for her help in procuring the BLGSP FFPE samples.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
U.S. Federal funds from the National Cancer Institute, National Institutes of Health [Contract No. HHSN261200800001E] (in part). The contents of this publication do not necessarily reflect the views of policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. BC Cancer Foundation and grants from Genome Canada/Genome British Columbia [212SEQ/202SEQ]; Canadian Institutes of Health Research [FDN-143288 to M.A.M.]. Funding for open access charge: Genome Canada.
Conflict of interest statement. None declared.
REFERENCES
- 1. Haile S., Pandoh P., McDonald H., Corbett R.D., Tsao P., Kirk H., MacLeod T., Jones M., Bilobram S., Brooks D. et al. . Automated high throughput nucleic acid purification from formalin-fixed paraffin-embedded tissue samples for next generation sequence analysis. PLoS One. 2017; 12:e0178706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Do H., Dobrovic A.. Sequence artifacts in DNA from Formalin-Fixed Tissues: Causes and strategies for minimization. Clin. Chem. 2015; 61:64–71. [DOI] [PubMed] [Google Scholar]
- 3. Hosein A.N., Song S., McCart Reed A.E., Jayanthan J., Reid L.E., Kutasovic J.R., Cummings M.C., Waddell N., Lakhani S.R., Chenevix-Trench G. et al. . Evaluating the repair of DNA derived from formalin-fixed paraffin-embedded tissues prior to genomic profiling by SNP-CGH analysis. Lab Invest. 2014; 93:701–710. [DOI] [PubMed] [Google Scholar]
- 4. Mouttham N., Klunk J., Kuch M., Fourney R., Poinar H.. Surveying the repair of ancient DNA from bones via high-throughput sequencing. BioTechniques. 2015; 59:19–25. [DOI] [PubMed] [Google Scholar]
- 5. Gerhard D.S., Grande B., Griner N., Casper C., Gerdts S.E., Omoding A., Orem J., Mbulaiteye S.M., Ogwang M.D., Reynolds S.J. et al. . Burkitt Lymphoma Genome Sequencing Project (BLGSP): Iintroduction. Blood. 2016; 128:1760. [Google Scholar]
- 6. Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P.. Integrative genomics viewer. Nat. Biotechnol. 2011; 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. García-Alcalde F., Okonechnikov K., Carbonell J., Cruz L.M., Götz S., Tarazona S., Dopazo J., Meyer T.F., Conesa A.. Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics. 2012; 28:2678–2679. [DOI] [PubMed] [Google Scholar]
- 8. Ewels P., Magnusson M., Lundin S., Käller M.. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016; 32:3047–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Saunders C.T., Wong W.S., Swamy S., Becq J., Murray L.J., Cheetham R.K.. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012; 28:1811–1817. [DOI] [PubMed] [Google Scholar]
- 10. Jones S.J, Laskin J., Li Y.Y., Griffith O.L., An J., Bilenky M., Butterfield Y.S., Cezard T., Chuah E., Corbett R. et al. . Evolution of an adenocarcinoma in response to selection by targeted kinase inhibitors. Genome Biol. 2010; 11:R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morin R.D., Mendez-Lago M., Mungall A.J., Goya R., Mungall K.L., Corbett R.D, Johnson N.A., Severson T.M., Chiu R., Field M. et al. . Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2010; 476:298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bass B.P., Engel K.B., Greytak S.R., Moore H.M.. A review of preanalytical factors affecting molecular, protein, and morphological analysis of formalin fixed, paraffin-embedded (FFPE) tissue: how well do you know your FFPE specimen?. Arch. Pathol. Lab. Med. 2014; 138:1520–1530. [DOI] [PubMed] [Google Scholar]
- 13. Engler M.J., Richardson C.C.. Boyer PD. DNA ligases. The Enzymes. 1982; 15B:San Diego: Academic Press Inc; 3–30. [Google Scholar]
- 14. Croucher N.J., Fookes M.C., Perkins T.T., Turner D.J., Marguerat S.B., Keane T., Quail M.A., He M., Assefa S., Bähler J. et al. . A simple method for directional transcriptome sequencing using Illumina technology. Nucleic Acids Res. 2009; 37:e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Robbe P., Popitsch N., Knight S.J.L, Antoniou P., Becq J., He M., Kanapin A., Samsonova A., Vavoulis D.V., Ross M.T. et al. . Clinical whole-genome sequencing from routine formalin-fixed, paraffin-embedded specimens: pilot study for the 100,000 Genomes Project. Genet Med. 2018; 20:1196–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lehman R.I. Boyer PD. Endonucleases specific for single-stranded polynucleotides. The Enzymes. 1981; 4:3rd edn193–201. [Google Scholar]
- 17. Adey A., Morrison H.G., Asan, Xun X., Kitzman J.O., Turner E.H., Stackhouse B., MacKenzie A.P., Caruccio N.C., Zhang X. et al. . Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010; 11:R119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.