Abstract
RNA-binding proteins (RBPs) control and coordinate each stage in the life cycle of RNAs. Although in vivo binding sites of RBPs can now be determined genome-wide, most studies typically focused on individual RBPs. Here, we examined a large compendium of 114 high-quality transcriptome-wide in vivo RBP–RNA cross-linking interaction datasets generated by the same protocol in the same cell line and representing 64 distinct RBPs. Comparative analysis of categories of target RNA binding preference, sequence preference, and transcript region specificity was performed, and identified potential posttranscriptional regulatory modules, i.e. specific combinations of RBPs that bind to specific sets of RNAs and targeted regions. These regulatory modules represented functionally related proteins and exhibited distinct differences in RNA metabolism, expression variance, as well as subcellular localization. This integrative investigation of experimental RBP–RNA interaction evidence and RBP regulatory function in a human cell line will be a valuable resource for understanding the complexity of post-transcriptional regulation.
INTRODUCTION
Of the 20,345 annotated protein-coding genes in human, at least 1542 are RNA-binding proteins (RBPs) (1). RBPs interact with RNA regulatory elements within RNA targets to control splicing, nuclear export, localization, stability and translation (2). RBPs have specificity to bind one or multiple RNA categories, including messenger RNA (mRNA) and diverse categories of non-coding RNA such as ribosomal RNA (rRNA), transfer RNA (tRNA), small nuclear and nucleolar RNA (snRNA/snoRNA), microRNA (miRNA) and long non-coding RNA (lncRNA). Mutations in RBPs or RNA regulatory elements can result in defects in RNA metabolism that cause human disease (3,4).
A standard technique for in vivo global identification of RBP–RNA interaction sites consists of immunoprecipitating the ribonucleoprotein (RNP) complex, isolating the bound RNA, and quantifying the RNA targets by microarrays or deep sequencing (5,6). The introduction of cross-linking prior to immunoprecipitation (CLIP) as well as RNase digestion enabled the biochemical mapping of individual interaction sites (7). Subsequent modifications to CLIP increased the resolution of the interaction sites (8,9). One of these methods, photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation (PAR-CLIP), utilizes 4-thiouridine or 6-thioguanosine combined with 365 nm UV crosslinking to produce single-nucleotide RBP–RNA interaction evidence that is utilized to define binding sites (9–11).
Experimentally-derived RBP binding sites provide valuable functional insights. First, they can reveal the rules for regulatory site recognition by the RBP, whether due to sequence and/or structural characteristics. Second, the region and position of the interaction sites of an RBP within transcripts provides insights into its role in RNA metabolism and its subcellular localization. For example, if most of the mapped interaction sites are intronic and adjacent to splice sites, the RBP is highly likely to be a nuclear splicing factor rather than a cytoplasmic translation factor. Finally, these data reveal the target transcripts and therefore the potential biological role for the RBP.
Throughout the life of an RNA, interactions with many different RBPs determine the ultimate fate of the transcript. Even though profiling of the interaction sites of a single RBP is clearly powerful, it does not provide information on other RBPs potentially targeting the same RNA or on other regulatory elements within the RNA. Small comparative efforts focusing on the regulation of splicing, 3′ end processing, RNA stability by AU-rich elements, and miRNA-mediated silencing have demonstrated the value of integrating interaction sites from multiple RBPs (12–15). Therefore, a large-scale comparative examination of interaction sites for many RBPs will yield valuable knowledge regarding the architecture and determinants of RNA regulatory networks.
At least 173 PAR-CLIP experiments have been performed in HEK293 cells to date, laying the groundwork for a large-scale integrative analysis and complementing efforts of ENCODE, which focused on other cell types and utilized other CLIP protocols (16). We describe a concerted effort to identify and uniformly process all high-quality PAR-CLIP data sets by evaluating the characteristic T-to-C transitions induced by photocrosslinking. Using the resulting compendium of high-quality in vivo RBP interaction maps from the same cell line enabled us to determine the relationship between RBPs with respect to their preferred category of target RNA and any underlying sequence specificity. We uncovered regulatory modules reflected by combinatorial binding events and assessed their role and functional implications on RNA metabolism. Finally, our results support the role of RBPs in buffering gene expression variance.
MATERIALS AND METHODS
Processing, filtering, and quality control of PAR-CLIP libraries
Each PAR-CLIP library was subject to two rounds of quality control. First, all PAR-CLIP libraries generated in HEK293 cells were subject to the quality control pipeline PAR-CLIP Suite v1.0 (https://rnaworld.rockefeller.edu/PARCLIP_suite/). Using raw Illumina sequencing data, this pipeline identified the predominant target RNA category or categories for each RBP and provided the T-to-C conversion frequency resolved by read length and RNA category (Supplementary Figure S1). The mapped reads of each RNA category were resolved by error distance 0 (d0), error distance 1 (d1; split in T-to-C and d1 other than T-to-C), and error distance 2 (d2). This process discriminated for each library true target RNA categories from non-crosslinked background RNA categories populated by fragments of abundant cellular RNAs. In order to disqualify experiments comprising too many non-crosslinked RBP-specifically bound RNAs or co-purified non-crosslinked background RNAs, we pursued only datasets which collect at least 10 000 redundant d1 reads ≥20 nt in at least one of major RNA annotation categories with d1(T-to-C)/(d0 + d1) ≥ 30%, and d1(T-to-C)/(d1-total) ≥65%.
For the libraries passing the first threshold, we defined and annotated binding sites using PARpipe (https://github.com/ohlerlab/PARpipe), which is a pipeline wrapper for PARalyzer (10,14). The threshold for additional filtering were determined by comparisons with the reference library (17). This reference library was generated using a modified PAR-CLIP protocol in which there was no immunoprecipitation and the addition of an rRNA depletion step after proteinase K digestion, followed by a partial digestion using RNase T1. We required libraries had to have an average fraction T-to-C over remaining reads >0.32 (the average fraction T-to-C over remaining reads greater of the reference library), an average conversion specificity >0, >20 000 aligned reads, not be digested only with micrococcal nuclease, a redundant read copy fraction <0.98 (Supplementary Figure S1B, C and Supplementary Table S1). For RBPs with three or more libraries, we removed outlier based on correlation of 6-mer frequency calculated from PARalyzer-utilized reads.
Annotation category preference and positional analysis of binding density
For calculating the annotation category preference, we calculated the difference in the fraction of T-to-C reads per annotation category between each RBP library and the reference library. For example, if the fraction of miRNA annotated reads with T-to-C transitions in a specific RBP library was 0.20 compared to 0.05 in the reference library, the miRNA preference value for this specific RBP is 0.15. For the positional binding analysis, we selected genes (n = 15 120) using GENCODE v19 as annotation based on our earlier work on HEK293 RNA processing and turnover dynamics (18). Isoform expression was calculated using RSEM (19). For each gene, we selected the transcript isoform with the highest isoform percentage or chose one randomly in case of ties (n = 8298). The list of selected transcript isoforms was used to calculate the median 5′ UTR, CDS and 3′ UTR length proportions (5′ UTR = 0.06, CDS = 0.53, 3′ UTR = 0.41) using R Bioconductor packages GenomicFeatures and GenomicRanges. For regions downstream annotated transcription ends (TES) and adjacent to splice sites, we chose windows of fixed sizes (TES 500nt, 5′ and 3′ splice sites 250nt each). We generated coverage tracks from the PARalyzer output alignment files and intersected those with the filtered transcripts. Each annotation category was binned according to its relative coverage averaged according to each bin. For intronic coverage, we averaged across all introns per gene, given a minimal intron length of 500nt. All bins were stitched to one continuous track per transcript. Altogether 6632 intron containing transcripts showed coverage in at least one PARCLIP library. For each library, we required transcripts to have a minimal coverage maximum of >2. For each transcript, we scaled the binned coverage dividing by its maximal coverage (min-to-1 scaling) to emphasize spatial patterns independent from transcript expression levels. Replicate RBP PARCLIP libraries were combined at this point. Transcripts targeted in more than one replicate library were aggregated using the average of their binned coverage. RBPs with <50 filtered target transcripts (after aggregation) were not considered. Next, we split transcript coverage in two parts, separating 5′ UTR to TES regions and intronic regions. To generate the scaled meta coverage across all targeted transcripts per RBP, we used the heatMeta function from the Genomation package. For the 5′UTR to TES, we scaled each RBP meta-coverage track independent of other RBPs. For each RBP, we subtracted the scaled meta coverage of PARCLIP reference library (17). For intronic sequences, we scaled each RBP relative to all other RBPs to highlight RBPs with more substantial intronic binding patterns. Finally, we visualized the density using pheatmap.
Sequence analysis
We performed sequence analysis on the sequences of binding sites (clusters) called by PARalyzer each library. These clusters represent regions of overlapping reads that exhibited a T-to-C conversion density above background conversion density. For the 6-mer analysis, we used Jellyfish to count 6-mer frequencies from each unique read that overlapped a PARalyzer called binding site irrespective of annotation category. For each RBP, we selected the library with the lowest percent of duplicated sequences (see Supplementary Table S1) to serve as a representative library for the sequence analysis and factor analysis. We counted the number of 6-mers with a frequency of x or higher, where x was from 1/4096 to 1/4. To evaluate the 6-mers enriched by a given RBP relative to the reference library, we regressed the RBP 6-mer frequency against the the reference library 6-mer frequency and collected the residuals (the unexplained variance). Next, identified all 6-mers that were found as the top 5 enriched over the reference library for any of the analyzed RBPs. We clustered the enrichment scores for the 6-mers across all RBPs and generated a heatmap using the ‘aheatmap’ function in the R package NMF v0.21.0. We ran SSMART using all binding sites found in mRNA-derived annotation categories ranked by the library size normalized enrichment over the reference library.
Factor analysis
For each site identified annotated as mRNA and lncRNA, we calculated a library size normalized enrichment compared to the reference library using DESeq2. Next, we calculated the sum of all enrichment scores for per annotated mRNA or lncRNA gene. Finally, we normalized this enrichment score for expression levels by performing a regression against log-transformed HEK293 expression levels and collected the residuals to create the final input matrix for the factor analysis. Factor analysis was performed using the ‘factanal’ function from the R package ‘stats’ v3.5.1. The number of factors, 10, was determined using multiple approaches to estimate the number of factors using the R package ‘nFactors’ v2.3.3. Two out of the four approaches estimated the number of factors to be 10, while the others reported 1 and >14 (Supplementary Figure S3A). Clustering of the score matrix to associate genes with specific factors was performed using the most stable results from multiple iterations of k-means clustering.
Gene ontology analysis
Multiple-test corrected gene ontology enrichment values were calculated using the PantherDB (http://www.pantherdb.org/). For each set of genes, we used all 13 299 genes in the factor analysis as the background or gene universe.
Precursor and mature RNA quantification
Mature- and premature-transcript expression, transcripts per million (TPM), was quantified with RSEMv1.2.11 (http://deweylab.biostat.wisc.edu/rsem/src/rsem-1.2.11.tar.gz) as described previously (18). Briefly, for each gene we included an additional isoform corresponding to the sequence of the full gene locus. Specifically, we modified the GENCODEv19 gtf and used this as the input for the ‘rsem-prepare-reference’ function to generate a modified index used for quantification. For each gene, we calculated the expression of ‘mature’ RNA as the sum of all isoforms for that gene excluding the ‘primary’ transcript. For intronless genes, premature and mature expression values were summed. We performed this analysis on the ELAVL1 knockdown RNA-seq experiments (20).
Cell-to-cell expression variability
RNA-seq gene expression data for individual HEK293 cells were downloaded from (21). We calculated the mean, standard deviation, and coefficient of variation (100 × standard deviation/mean) for each gene across all 25 cells.
Data access and visualization
We provide an overview analysis of each independent PAR-CLIP library that can be accessed at https://github.com/BIMSBbioinfo/RCAS_meta-analysis. We have also added all RBP binding sites to DoRiNA (https://dorina.mdc-berlin.de/), which enables the user to query all binding sites and perform additional analyses. Sequencing reads for the 7 new PAR-CLIP libraries are available at SRP154398.
RESULTS
A high-quality map of in vivo RBP–RNA interactions across 64 proteins
In order to generate a comprehensive quantitative resource of RBP–RNA interactions within a human cell line, we identified 166 published PAR-CLIP data sets performed predominantly in HEK293 cells and added seven new libraries generated in our laboratories (Supplementary Table S1). Typically, these datasets were generated using transgenic HEK293 cell lines in which each individual RBP was FLAG-tagged and recombined into the same chromosomal locus containing a strong promoter. In this way, the expression of each RBP as well as the strength of its immunoprecipitation were generally comparable. Furthermore, the availability of orthogonal transcriptome-wide datasets quantifying individual steps of RNA metabolism made HEK293 cells ideal for examining the functional characteristics of RNA targets (18).
Each of the 173 PAR-CLIP libraries generated in HEK293 were subject to a stringent analysis strategy to retain high-quality datasets (Supplementary Table S1). First, each library was analyzed using the PAR-CLIP Suite v1.0 (https://rnaworld.rockefeller.edu/PARCLIP_suite) (11) to discriminate significant target RNA categories from non-crosslinked background RNA categories populated by fragments of abundant cellular RNAs (see Methods, Supplementary Figure S1A). Next, we defined binding sites based on the local density of T-to-C transitions using PARpipe (https://github.com/ohlerlab/PARpipe) (10) and only retained those libraries with sufficiently high read counts and T-to-C transition specificity compared to a deeply sequenced background reference library (Supplementary Figure S1B) (17). Since the immunoprecipitation step was omitted in this reference library it served as an effective comparison point to score read count and T-to-C transition for all RBPs. Finally, for RBPs with more than three libraries available, outlier libraries exhibiting poor correlation of 6-mer frequencies were excluded (Supplementary Figure S1D, E). This resulted in 114 libraries corresponding to 64 RBPs that were the basis for downstream analysis. There were eight RBP families represented by two or more RBPs.
Grouping RBPs by annotation category and positional binding site preferences
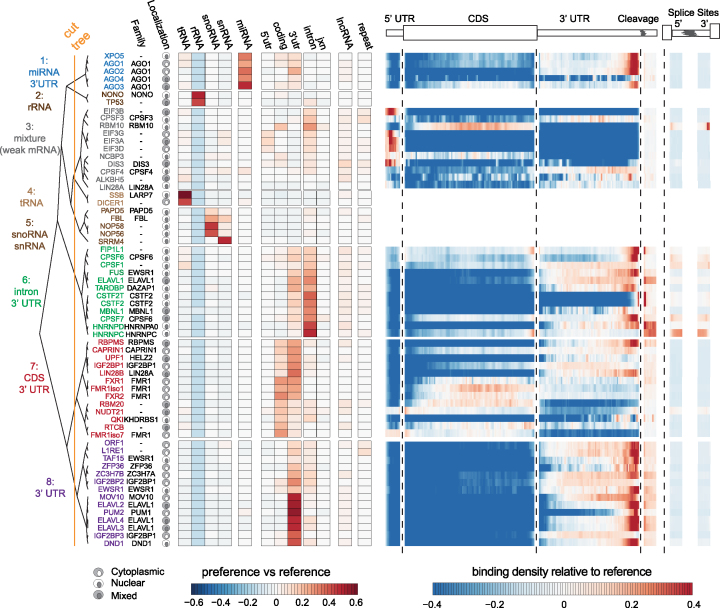
As first step to describe RBP–RNA regulatory networks, we determined the relative binding preference of each RBP for specific target RNA annotation categories (Supplementary Table S2). For each library, we calculated an RNA annotation category preference value, defined as the difference in the fraction of T-to-C reads per annotation category between each RBP library and the reference library. We performed hierarchical clustering of RBPs by annotation category preference, using Ward's method and Euclidean distances. This yielded eight clusters of binding preference (Figure 1A, orange line demarcates cluster definitions) with varying enrichment or depletion for individual or combinations of specific annotation categories. For each of these clusters, we compiled a detailed table summarizing the reported functions for each of the RBPs (Supplementary Table S3). Taken together, clustering by RNA annotation category separated RBPs into groups according to their known subcellular localization and functions.
Figure 1.
RBP analyzed and binding preferences by RNA category. Heatmap of reference normalized annotation category preference for each RBP clustered into 8 branches by color (left) and subcellular localization (Supplementary Table S5). The heatmap represents the difference in the proportion of sites for a given annotation category in the RBP library versus the reference library. Heatmap of the reference library normalized relative positional binding preference of the 55 RBPs with enriched binding in at least one mRNA-relevant annotation category per branch (right). RBP-specific binding preferences were averaged across selected transcripts (see methods). The relative spatial proportion of 5′UTR, coding regions and 3′UTR were averaged across all selected transcript isoforms. For TES (regions beyond transcription end site), 5′ splice site, and 3′ splice site, we chose fixed windows (250nt for TES and 500nt for splice sites). For each RBP, meta-coverage was scaled between 5′UTR to TES. The 5′ and 3′ intronic splice site coverage was scaled separately from other regions but relative to each other.
Three of the eight clusters (clusters 2, 4 and 5) contained nine RBPs that exhibited preference for categories of non-coding RNA (rRNA, snRNA, snoRNA and tRNA), but not mRNA, precursor mRNA (pre-mRNA), or lncRNA. The remaining five clusters contained 55 RBPs exhibiting preference for binding to mRNA, pre-mRNA and long-noncoding RNA (lncRNA) annotation categories. The RBPs in clusters 1, 6, 7 and 8 exhibited strong preferences for various mRNA annotation categories. The RBPs in cluster 3 did not exhibiting strong preference for specific mRNA annotation categories. Additionally, for each of the RBPs in the cluster, we performed a positional meta-analysis of binding sites with respect to major transcript landmarks within target mRNAs. Many of the RBPs also showed strong preferences for binding to specific positions within mRNAs relating to their role in specific steps of mRNA processing (Supplementary Table S3).
We hypothesized that target annotation category preferences and positional binding preferences should reflect subcellular localization of the RBP and its role(s) in mRNA processing. Cluster 6 contained twelve RBPs and exhibited strong preference for intronic regions and to a lesser degree 3′ UTRs of mRNAs and lncRNAs. The intronic preference was consistent with the predominantly nuclear localization of these RBPs and the pre-mRNA splicing process. ELAVL1, which is the sole member of the ELAVL1 family of RBPs that is predominantly localized in the nucleus but capable of shuttling to the cytoplasm, exhibited positional binding flanking the end of the 3′ UTR and for 5′ and 3′ splice sites. Cluster 8 contained fourteen RBPs and exhibited distinct preference for 3′ UTR regions. This included the unpublished and predominantly cytoplasmic ELAVL1 family members, ELAVL2, ELAVL3, and ELAVL4, which exhibited a strong positional preference for binding in the distal region of the 3′ UTR and acting predominantly on mature mRNA (22). In summary, the annotation category preferences and positional binding preferences implicated the specific steps of mRNA processing the RBPs potentially regulate.
The spectrum of RNA sequence specificity
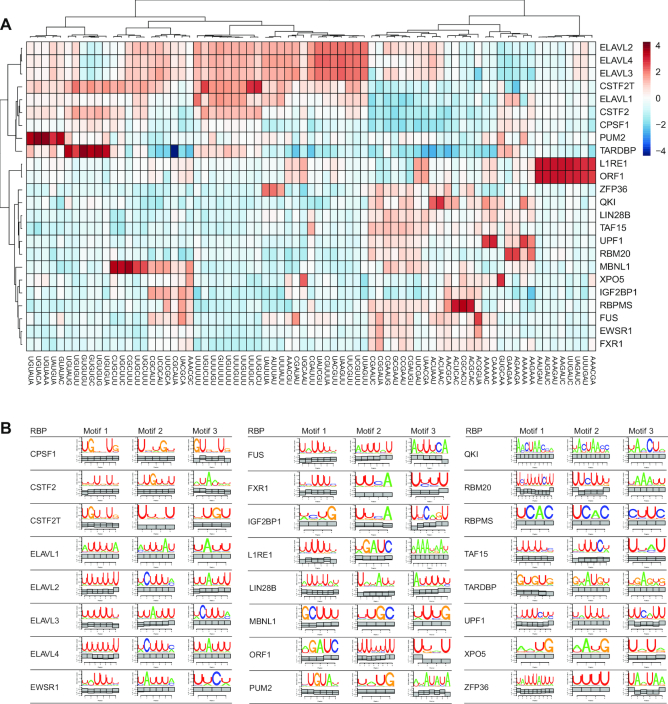
RBPs exist on a spectrum of specificity depending on a variety of primary and secondary structure features (23). Here, our goal was to identify the RBPs with substantial primary sequence specificity and then examine their sequence preference. For each of the 55 RBPs, we counted all possible 6-mers using Jellyfish (24) for the reads contributing to PARalyzer-defined binding sites. We observed 6-mer frequencies ranging as high as 512-fold to as low as 5-fold over a uniform distribution of 6-mers (Supplementary Figure S2). In contrast, our reference background library exhibited 16-fold enrichment of at least one 6-mer compared to uniform. AGO1-4 libraries were excluded from 6-mer analysis due to the overwhelming sequence contribution from crosslinked miRNAs. Twenty-seven RBPs did not have a single 6-mer found at higher frequency than present in the reference sample. Amongst these RBPs established or expected to display low sequence-specificity were the RNA helicase MOV10, the nuclear exosome component DIS3, and the EIF3 complex translation initiation factors.
For each of the 24 RBPs with stronger sequence enrichment than the reference library, we clustered the top 5 sequences enriched over the reference library (Figure 2A). Our results recapitulated the sequence preference for the RBPs in this group with well-characterized sequence motifs (detailed in Supplementary Table S4). The ELAVL1 family proteins, which bound to different regions and positions of mRNA, showed similar preference for U- and AU-rich 6-mers, while ZFP36 only enriched a subset of the AU-rich 6-mers (14). Complementing the 6-mer enrichment analysis, we performed motif analysis for each RBP library with the motif finding algorithm SSMART (sequence-structure motif identification for RNA-binding proteins, (25)) (Figure 2B). For most RBPs, we observed strong concordance between the two analyses. RBM20 was a clear exception, for which we observed the established UCUU-containing motifs (26) with SSMART, but a GA-rich sequence in the 6-mer enrichment analysis. However, we do observe UCUU-containing motifs in the top fifteen, but not the top five 6-mers for RBM20. Altogether, our analysis was remarkably consistent with previously reported motifs in spite of differences in data processing and analysis (detailed Supplementary Table S4).
Figure 2.
RBP binding sequence specificity and elements. (A) Heatmap of reference normalized 6-mer enrichment for top 5 enriched 6-mers for each RBP in the set of RBPs exhibiting more sequence specificity than the reference. (B) Top 3 SSMART motif results using all binding sites found in mRNA-derived annotation categories ranked by the library size normalized enrichment over reference library.
Identification of RNA regulatory modules
To understand the functional impact of co-regulation by multiple RBPs, we analyzed the co-variation in binding patterns of all 55 RBPs across 13 299 target RNA encoding genes to probe for the existence of regulatory modules, i.e. specific subsets of RNAs implicated in similar function bound by subsets of RBPs. To this end, we employed Factor Analysis (FA), which reduces a large number of observed variables to a smaller number of latent factors. Here, our observed variables represented the normalized RBP binding (see methods) for each of the 55 RBPs across all target RNA encoding genes (n = 13 299). The latent factors represented similar binding patterns to RNA targets by one or more of the 55 RBPs. RBPs exhibiting high loadings for the same factor would have very similar binding patterns to RNA targets. Importantly in this framework, a single RBP could be assigned to multiple factors, just as a single RBP can participate in multiple RNPs and regulate different aspects of RNA metabolism.
The FA model decomposed the 55 × 13 299 normalized RBP binding matrix into a 55 × 10 factor loading matrix (representing the strength of the dependence of each of the 55 RBP target RNA binding pattern on each of the 10 factorsSupplementary Table S6), a 13 299 × 10 factor score coefficient matrix (representing the dependence between the binding of the 13 299 target RNA encoding gene and each of the 10 factors), and residual error (Supplementary Figure S3A and methods).
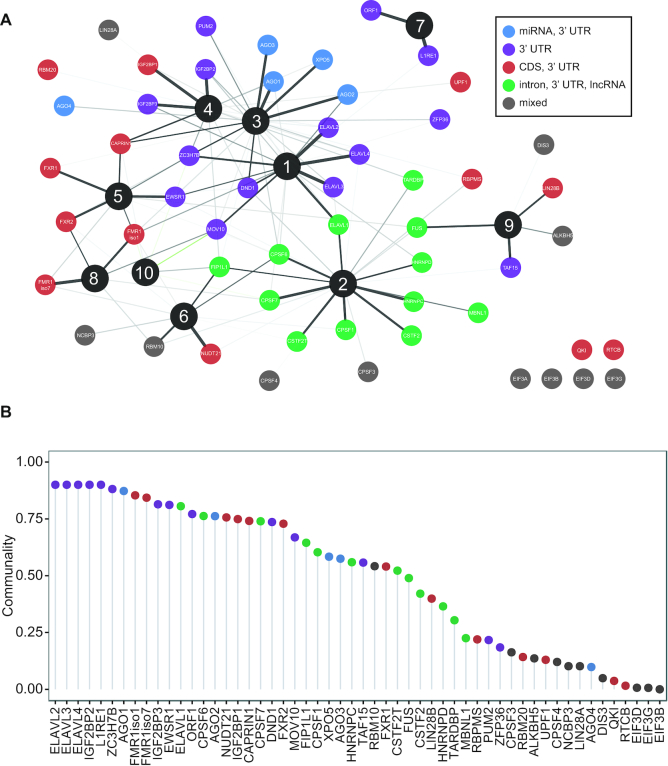
The FA model uncovered interesting parallels between the similarity in the binding of target RNA encoding genes and the target annotation category preferences (from Figure 1A). We observed that individual factors contained RBPs that preferred binding to either mature (Factors 1, 3, 4, 5, 8) or precursor transcripts (Factors 2, 6), reflecting involvement in different stages of RNA metabolism (Figure 3A). Furthermore, individual factors contained RBPs exhibiting similar patterns of binding to specific regions of the mRNA (i.e. intron, coding, 3′ UTR). Indeed, RBPs from the same family, or known to regulate a specific aspect of RNA processing, had high loadings for the same factors. For example, the ELAVL1 family members were associated with Factor 1; the AGO1 family were associated with Factor 3; the IGF2BP1 family were associated with Factor 4; the FMR1 family had were associated with Factor 5 and Factor 8; LINE-1 encoded proteins were associated with Factor 7. One of the interesting associations was that of HNRNPC with Factor 2, which contained many cleavage and polyadenylation factors. HNRNPC was shown to interact with U-rich sequences downstream of a viral poly-adenylation signal nearly three decades ago (27), and more recently, to repress cleavage and poly-adenylation in humans (28). These examples highlight the specific hypotheses generated by an integrative analysis that are not obvious when examining a single RBP in isolation.
Figure 3.
RNA regulatory modules. (A) Factor analysis of target RNA encoding genes binding normalized by the reference library and expression for the 55 RBPs binding to mRNAs and lncRNAs for 13 299 genes (see ‘factor analysis’ section in methods for details). Spring-embedded graph of the factor loading matrix, indicating the association between each of the 55 RBPs and one of the 10 factors. Nodes color-coded by RNA annotation category preference cluster membership from Figure 1. Edge width scales with factor loadings (thicker edge = higher factor loading = stronger association). Only edges with a factor loading > 0.2 (positive values in black) or <−0.2 (negative values in green) depicted. (B) Dot plot of the communality, or the variance in a given RBP cumulatively explained by the all factors color-coded by RNA annotation category preference.
Cumulatively, the FA model explained ∼60% of the variance in the observed data. The remaining unexplained variance was expected due to the challenges of integrating data sets of varying depth and quality, in spite of our efforts to control these aspects. The communality, which is the amount of variance explained by the model for each RBP-binding variable, varied drastically for all 55 RBPs; the model explained at least 80% of the variance in enrichment scores for 12 RBPs, and at least 50% of the variance in enrichment scores for 30 RBPs (Figure 3B). RBPs with lower communality often coincided with shallow depth of their PAR-CLIP libraries.
By clustering the factor score coefficients, i.e. the specific linear combination of RBP binding for that target RNA, we identified target RNA encoding genes constituting putative regulatory modules associated with a given factor. Therefore, each regulatory module was associated with an RBP component (the subset of RBPs exhibiting similar binding pattern) and an RNA component (the subsets of target RNA encoding genes bound by those RBPs). These regulatory modules did not imply physical interactions between RBPs; rather, it identified RBPs that may cooperate in controlling RNA metabolism for specific subsets of RNA targets. Almost 1/4 of the target RNA encoding genes (3,180/13,299) were assigned to regulatory modules by exhibiting high factor score coefficients for a single factor (Supplementary Figure S3B). We did not identify target RNA encoding genes with high factor score coefficients for Factor 9 or 10. The remaining target RNA encoding genes did not exhibit high factor score coefficients for any specific factor in our analysis, suggesting that the targets were either not bound by specific combinations of these RBPs, bound broadly by all RBPs, or not bound by the subset of RBPs in the analysis. As such, we labeled this target RNA encoding gene category as ‘non-specific’. The RNA regulatory modules encoding genes were enriched for different GO categories. Factor 1 RNA regulatory modules were enriched for ‘AU-rich element binding’ and Factor 3 RNA regulatory modules were enriched for ‘gene silencing by miRNA’; AU-rich RBPs and AGO proteins were strongly associated with Factor 1 and Factor 3, respectively. This was consistent with the recurrent observation that RBPs target the mRNAs encoding themselves (5,29). In turn, the RNAs encoding ‘non-specific’ genes contained ribosomal proteins and mitochondrial electron-transport proteins.
RNA regulatory modules underlie distinct patterns of RNA metabolism
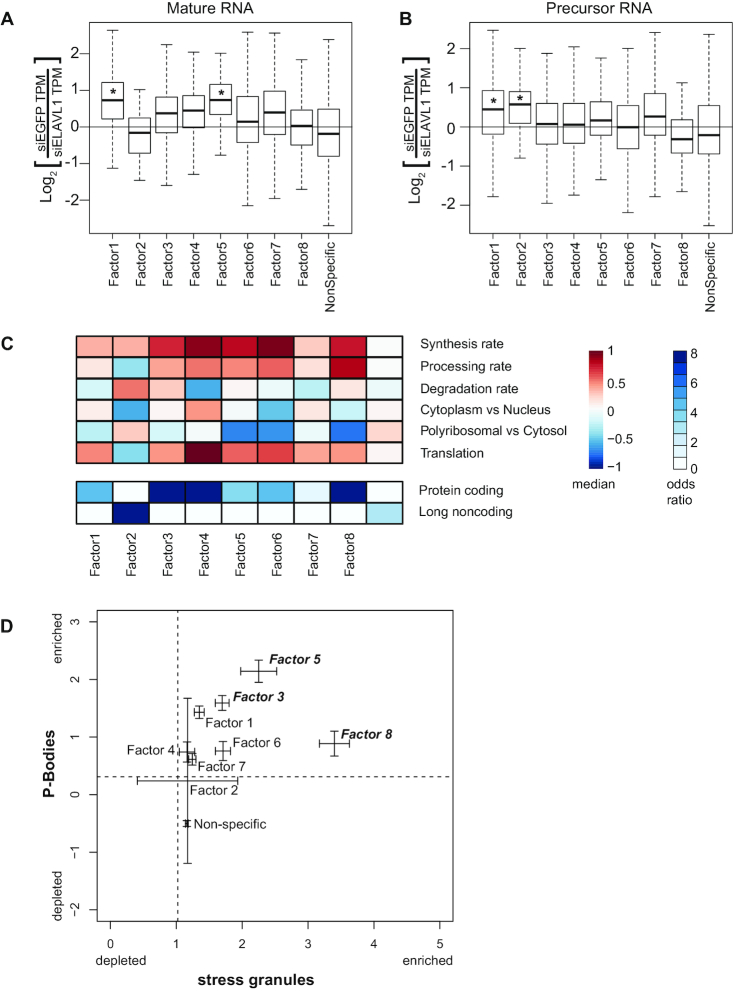
In order to test the functional importance of these RNA regulatory modules, we reasoned that depletion of an RBP will lead to pronounced effects only for the RNA regulatory modules assigned to the specific factor(s) that RBP is associated with. We examined mature and precursor RNA expression changes induced by siRNA knockdown of ELAVL1, which was strongly associated with Factor 1 and 2 (20). ELAVL1 stabilizes mature (Figure 4A) and precursor (Figure 4B) RNAs associated with Factor 1 RNA regulatory modules. However, ELAVL1-dependent stabilization was restricted to precursor RNAs associated with Factor 2 RNA regulatory modules. Indeed, the RNAs associated with Factor 2 RNA regulatory modules exhibit lower processing rates and more nuclear localization. Mature RNAs associated with Factor 5 RNA regulatory modules also exhibited ELAVL1-dependent stabilization, which is consistent with positive loadings for Factor 1 and therefore ELAVL1-binding (Supplementary Figure S3B, column 1, row 5). Taken together, the model was able to identify and distinguish ELAVL1-dependent stabilization of both precursor and mature RNA for different populations of target RNAs (30,31).
Figure 4.
Functional characterization of RNA regulatory modules. (A) The difference in either (A) primary or (B) mature RNA expression TPM (transcripts per million) upon ELAVL1 knockdown by siRNA treatment (y-axis), for each set of factor-associated RNAs. *P < 0.05 Mann–Whitney U-test. (C) Heatmap of the median value of synthesis rate, processing rates, degradation rates, cytoplasmic versus nuclear localization, polyribosomal versus cytoplasmic localization, and translational status from ribosome profiling data for each gene set (top). Heatmap of the odds-ratio of the overlap between factor associated gene sets with annotation (bottom). (D) Plot of the confidence intervals for P-body localization (y-axis from (35)) and stress granule localization (x-axis from (36)) for each set of factor-associated RNAs. Dashed lines represent median enrichment of RNA in P-bodies (y-axis) and stress granules (x-axis).
We hypothesized that the subsets of RNAs assigned to the different regulatory module would exhibit differences in RNA metabolism driven by the RBPs in the factor associated with the regulatory module. Therefore, we compared six aspects of RNA metabolism previously quantified in HEK293 cells (18), for each of the RNA regulatory modules associated with each of the factors. The factor-associated RNA regulatory modules exhibited very distinct RNA metabolic profiles compared to each other and to non-specific category (Figure 4C, Supplementary Figure S4A). Factor 2 RNA regulatory modules, which was the only factor associated with RBPs binding to precursor mRNA and lncRNA, had low processing rates, high degradation rates and their encoded RNAs were preferentially localized in the nucleus versus the cytoplasm. Factor 2 RNA regulatory modules were strongly enriched for lncRNAs (Figure 4C). Indeed, these genes strongly overlapped with a set of lncRNAs likely to be functional (Supplementary Figure S4B) (18).
We also examined regulatory differences in RNA metabolism for genes associated with cytoplasm-enriched factors. For example, factor 1 RNA regulatory modules were more stable than Factor 3 RNA regulatory modules (Figure 4C). Factor 1 was strongly associated with ELAVL1 family proteins, which stabilize target mRNAs. Factor 3 was strongly associated with AGO1 family proteins, which execute miRNA-mediated degradation of target mRNAs. Additionally, Factor 4 RNA regulatory modules, which are bound by IGF2BP1 family proteins, were highly synthesized, processed, stabilized, and translated (Figure 4C). The RNA targets of IGF2BP1 family RBPs were strongly localized to the ER (Supplementary Figure S4C) (32), which is also consistent with the proposed role of IGF2BP1 family proteins for RNA localization and translation (33,34). Although correlative, these results indicate that different RBP binding patterns beget different consequences for RNA metabolism.
Specific RNA regulatory modules also exhibited preferential localization to processing bodies (P-bodies), which are cytoplasmic granules associated with translational repression (35) and stress-granules (36) (Figure 4D). Factor 3 and Factor 5 RNA regulatory modules were most strongly enriched for localizing to both P-bodies and stress granules and were associated with AGO1 family proteins as well as CAPRIN and FMR1 family proteins, respectively. Consistent with this the AGO2 protein and FMR1 protein were 90-fold and 16-fold enriched in P-bodies, respectively (37). Interestingly, Factor 8 RNA regulatory modules, which contained FMR1 family proteins, exhibited preferential localization to stress granules. FMR1 preferentially bound CDS regions that would be exposed during stress-induced translational repression. Indeed, these RBPs may be more stably localized to stress granule cores rather than the periphery.
Fine-tuning of gene expression has been postulated to be an important function of post-transcriptional regulation by RBP and miRNAs. Therefore, we examined the cell-to-cell variability in gene expression across 25 individual HEK293 cells with respect to the RNA regulatory modules. The single-cell RNA-seq data was very deeply sequenced and generated using the massively parallel single-cell RNA-sequencing (MARS-Seq) protocol (21). Most RNA regulatory modules exhibited lower expression variability than the non-specific category (Supplementary Figure S4). In particular, Factor 4 RNA regulatory modules exhibited the lowest variation and highest median expression across the 25 cells (Supplementary Figure S4e). These results supported the notion that post-transcriptional gene regulation confers robustness and fine-tuning of RNA expression levels.
DISCUSSION
Our study presents a curation of existing datasets, followed by systematic analysis of high-quality and high-resolution RBP–RNA interaction data. We focused on the RBPs that preferentially bound to mRNA and lncRNA and examined their sequence specificity and sequence motif preferences. Our survey of the RBP regulatory landscape identified the most prevalent subsets of RNAs targeted by a specific subset of RBPs, which we refer to as RNA regulatory modules.
We only utilized high quality PAR-CLIP datasets for which the immunoprecipitation efficiency was comparable since most RBPs were FLAG-tagged. Nevertheless, several caveats need to be pointed out. Despite several measures of quality control to decide which datasets to include in our analysis, the libraries varied in depth, quality, and digestion biases. Furthermore, a small number of RBPs in our analysis are not endogenously expressed in HEK293, and their natural expression is tissue-specific and/or context-dependent. The FA model quantitatively assessed the degree to which we could explain the full complement of RBP–RNA target binding patterns, which is critical in the face of such confounders that contributed to the ∼40% of variance not explained by the FA model. In comparison, the ENCODE eCLIP datasets (16) would suffer from varying IP efficiency since they were generated antibodies against endogenous proteins expressed. (38). This represents the trade-offs in experimental design between endogenous and epitope-tagged protein.
We utilized two complementary methods to examine sequence specificity. SSMART additionally assesses local secondary structure and could explain some individual cases in which the two approaches did not concur with each other or previous studies. Many issues complicate comparative motif analyses, including crosslinking bias, RNase digestion, immunoprecipitation purity, the effect of 4SU on binding, library depth, use of adapters to minimize ligation bias, and complexity. For example, RNaseI was exclusively used in the HNRNPC IP and may explain the lower than expected observe 6mer sequence specificity for an RBP that binds with high affinity to U-rich sequences. We observed a wide spectrum of sequence specificity, which was quite extensive and specific, as in the case of Pumilio, or minimal and non-sequence specific, as in the case the translation initiation factor EIF3. In vitro approaches avoiding many of the concerns listed above are better suited for making quantitative comparison of the sequence/structural-specificity between individual RBPs (39) though lack physiological context of in vivo binding data. Assuming the RBPs investigated here are a representative sample of the ∼1542 RBPs encoded in the human genome, there may be a considerable fraction of RBPs with substantial primary sequence preferences or specificity.
In our FA model, we integrated RBP binding data at the scale of all RNAs encoded by a given gene rather than individual binding sites. Hence, our model does not test position-specific binding effects, which can influence RNA fate decisions most prominently alternative splicing. There are numerous challenges in assessing the impact of individual binding sites on the full transcript, which has been partially investigated in a complementary study (40). At least 25% of target RNA encoding genes were assigned to RNA regulatory modules, which is probably an underestimation due to noisy data, as well as, a biased and incomplete sampling of all RBPs. This approach can scale to binding data for the ∼700 proteins bound to poly-adenylated RNA in HEK293 cells or the ∼1542 known RBPs (41).
The RNA regulatory modules exhibited different patterns of RNA processing, degradation, localization, and translation driven by individual or the combinations of RBPs associated with that regulatory module. We provide an example of this for the targetes of the ELAVL1 family of RBPs. Each human ELAV1 family protein contains three RRM domains (>90% sequence identity), but the hinge region between the second and third RRM of ELAVL1 contains a shuttling sequence responsible for its nuclear localization (42). Due to the lack of this shuttling sequence, ELAVL2/3/4 are mostly cytoplasmic and strongly associated with Factor 1, but not Factor 2. Furthermore, we observed ELAVL1-dependent stabilization is more prominent for different stages of RNA processing for different populations of target RNAs consistent with the known localization pattern of ELAVL1 protein and RNA targets. We show that RNA regulatory modules may specify both RNA and protein localization. Furthermore, the RNA regulatory modules encoded proteins with similar molecular functions or multi-component complexes though not for members of signaling pathways (Supplementary Figure S3B). These lines of evidence provide support for the coordinate regulation of ‘functionally coherent’ RNA regulatory modules as proposed by the post-transcriptional operon/regulon model (43). Our results and data provide a rationale for experimentally testing the RNA operon/regulon model by manipulating specific combinations of RNA regulatory elements (binding sites) and RBPs.
Our results have important implications for the buffering transcriptional noise (44,45) by RBP–RNA regulatory networks. The mRNA targets within specific regulatory modules encoded the RBP themselves, a generalization of a frequently made observation that RBPs bind to the mRNAs encoding them (46). These potential auto-regulatory feedback loops may buffer the expression range of the targeted mRNAs and partially explain the observation that most RNA regulatory modules exhibited low cell-to-cell RNA expression variance. Systematic and minimally-disrupting perturbation of individual and combinations of RBPs will aid the quantitative modeling of these emergent properties of RBP–RNA regulatory networks.
The binding preference and targets of the vast majority of human RBPs remain unknown. The insights gained from this study demonstrate the value of large-scale efforts by ENCODE and others in the research community to globally map RBP binding sites. Of the 64 RBPs in this study, 44 were not represented in the ENCODE cell lines. Cumulatively these efforts interrogate ∼10% of human RBPs with known RNA-binding domains. Thus, these two large scale efforts offer the potential to complement one another in our continuing attempts to crack RBP–RNA regulatory networks.
DATA AVAILABILITY
We provide an overview analysis of each independent PAR-CLIP library that can be accessed at https://github.com/BIMSBbioinfo/RCAS_meta-analysis. We have also added all RBP binding sites to DoRiNA (https://dorina.mdc-berlin.de/), which enables the user to query all binding sites and perform additional analyses. Sequencing reads for the 7 new PAR-CLIP libraries are available at SRP154398.
Supplementary Material
ACKNOWLEDGEMENTS
Author Contributions: Conceptualization, N.M., T.T. and U.O.; Methodology, N.M., H.W., M.S., M.G., A.G., A.M., T.T. and U.O.; Investigation, N.M., H.W., M.G., A.G., A.M., T.F., J.I.H. and K.A; Formal Analysis, N.M., H.W., M.S., M.G., A.G. and A.M.; Writing – Original Draft, N.M., T.T. and U.O.; Writing – Review & Editing, N.M., S.V., A.G., M.S., T.T. and U.O.; Funding Acquisition, N.M., T.T. and U.O.; Resources, N.M., T.T. and U.O. Supervision, N.M., T.T. and U.O.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
US National Institutes of Health [R01-GM104962 to U.O. and T.T.]; ETIUDA scholarship [2014/12/T/NZ1/00497 to M.S.] from National Science Center, Poland. N.M. acknowledges support from EU Marie Curie IIF; RNA Bioscience Initiative for startup funds. S.L. and U.O. acknowledge support by Deutsche Forschungsgemeinschaft under Priority Programme “Deciphering the mRNP code: RNA-bound Determinants of Post-transcriptional Gene Regulation” (SPP 1935). Funding for open access charge: MDC Berlin.
Conflict of interest statement. T.T. is cofounder and advisor to Alnylam Pharmaceuticals.
REFERENCES
- 1. Gerstberger S., Hafner M., Tuschl T.. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014; 15:829–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moore M.J. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005; 309:1514–1518. [DOI] [PubMed] [Google Scholar]
- 3. Cooper T.A., Wan L., Dreyfuss G.. RNA and disease. Cell. 2009; 136:777–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fredericks A.M., Cygan K.J., Brown B.A., Fairbrother W.G.. RNA-binding proteins: splicing factors and disease. Biomolecules. 2015; 5:893–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tenenbaum S.A., Carson C.C., Lager P.J., Keene J.D.. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:14085–14090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhao J., Ohsumi T.K., Kung J.T., Ogawa Y., Grau D.J., Sarma K., Song J.J., Kingston R.E., Borowsky M., Lee J.T.. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell. 2010; 40:939–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ule J., Jensen K.B., Ruggiu M., Mele A., Ule A., Darnell R.B.. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003; 302:1212–1215. [DOI] [PubMed] [Google Scholar]
- 8. König J., Zarnack K., Rot G., Curk T., Kayikci M., Zupan B., Turner D.J., Luscombe N.M., Ule J.. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 2010; 17:909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M., Jungkamp A.-C., Munschauer M. et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010; 141:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Corcoran D.L., Georgiev S., Mukherjee N., Gottwein E., Skalsky R.L., Keene J.D., Ohler U.. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 2011; 12:R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Garzia A., Meyer C., Morozov P., Sajek M., Tuschl T.. Optimization of PAR-CLIP for transcriptome-wide identification of binding sites of RNA-binding proteins. Methods. 2017; 118–119:24–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang C., Frias M.A., Mele A., Ruggiu M., Eom T., Marney C.B., Wang H., Licatalosi D.D., Fak J.J., Darnell R.B.. Integrative modeling defines the Nova splicing-regulatory network and its combinatorial controls. Science. 2010; 329:439–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pandit S., Zhou Y., Shiue L., Coutinho-Mansfield G., Li H., Qiu J., Huang J., Yeo G.W., Ares M., Fu X.-D.. Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Mol. Cell. 2013; 50:223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mukherjee N., Jacobs N.C., Hafner M., Kennington E.A., Nusbaum J.D., Tuschl T., Blackshear P.J., Ohler U.. Global target mRNA specification and regulation by the RNA-binding protein ZFP36. Genome Biol. 2014; 15:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martin G., Gruber A.R., Keller W., Zavolan M.. Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′ UTR length. Cell Rep. 2012; 1:753–763. [DOI] [PubMed] [Google Scholar]
- 16. Van Nostrand E.L., Pratt G.A., Shishkin A.A., Gelboin-Burkhart C., Fang M.Y., Sundararaman B., Blue S.M., Nguyen T.B., Surka C., Elkins K. et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods. 2016; 13:508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Friedersdorf M.B., Keene J.D.. Advancing the functional utility of PAR-CLIP by quantifying background binding to mRNAs and lncRNAs. Genome Biol. 2014; 15:R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mukherjee N., Calviello L., Hirsekorn A., de Pretis S., Pelizzola M., Ohler U.. Integrative classification of human coding and noncoding genes through RNA metabolism profiles. Nat. Struct. Mol. Biol. 2017; 24:86–96. [DOI] [PubMed] [Google Scholar]
- 19. Li B., Dewey C.N.. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011; 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kishore S., Jaskiewicz L., Burger L., Hausser J., Khorshid M., Zavolan M.. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat. Methods. 2011; 8:559–564. [DOI] [PubMed] [Google Scholar]
- 21. Guillaumet-Adkins A., Rodríguez-Esteban G., Mereu E., Mendez-Lago M., Jaitin D.A., Villanueva A., Vidal A., Martinez-Marti A., Felip E., Vivancos A. et al. Single-cell transcriptome conservation in cryopreserved cells and tissues. Genome Biol. 2017; 18:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mansfield K.D., Keene J.D.. Neuron-specific ELAV/Hu proteins suppress HuR mRNA during neuronal differentiation by alternative polyadenylation. Nucleic Acids Res. 2012; 40:2734–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jankowsky E., Harris M.E.. Specificity and nonspecificity in RNA-protein interactions. Nat. Rev. Mol. Cell Biol. 2015; 16:533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marçais G., Kingsford C.. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011; 27:764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Munteanu A., Mukherjee N., Ohler U.. SSMART: sequence-structure motif identification for RNA-binding proteins. Bioinformatics. 2018; 492:382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maatz H., Jens M., Liss M., Schafer S., Heinig M., Kirchner M., Adami E., Rintisch C., Dauksaite V., Radke M.H. et al. RNA-binding protein RBM20 represses splicing to orchestrate cardiac pre-mRNA processing. J. Clin. Invest. 2014; 124:3419–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wilusz J., Feig D.I., Shenk T.. The C proteins of heterogeneous nuclear ribonucleoprotein complexes interact with RNA sequences downstream of polyadenylation cleavage sites. Mol. Cell. Biol. 1988; 8:4477–4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gruber A.J., Schmidt R., Gruber A.R., Martin G., Ghosh S., Belmadani M., Keller W., Zavolan M.. A comprehensive analysis of 3′ end sequencing data sets reveals novel polyadenylation signals and the repressive role of heterogeneous ribonucleoprotein C on cleavage and polyadenylation. Genome Res. 2016; 26:1145–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pullmann R., Kim H.H., Abdelmohsen K., Lal A., Martindale J.L., Yang X., Gorospe M.. Analysis of turnover and translation regulatory RNA-binding protein expression through binding to cognate mRNAs. Mol. Cell. Biol. 2007; 27:6265–6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lebedeva S., Jens M., Theil K., Schwanhäusser B., Selbach M., Landthaler M., Rajewsky N.. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol. Cell. 2011; 43:340–352. [DOI] [PubMed] [Google Scholar]
- 31. Mukherjee N., Corcoran D.L., Nusbaum J.D., Reid D.W., Georgiev S., Hafner M., Ascano M., Tuschl T., Ohler U., Keene J.D.. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol. Cell. 2011; 43:327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jønson L., Vikesaa J., Krogh A., Nielsen L.K., Hansen T.V., Borup R., Johnsen A.H., Christiansen J., Nielsen F.C.. Molecular composition of IMP1 ribonucleoprotein granules. Mol. Cell Proteomics. 2007; 6:798–811. [DOI] [PubMed] [Google Scholar]
- 33. Farina K.L., Hüttelmaier S., Musunuru K., Darnell R., Singer R.H.. Two ZBP1 KH domains facilitate β-actin mRNA localization, granule formation, and cytoskeletal attachment. J. Cell Biol. 2003; 160:77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nielsen F.C., Nielsen J., Christiansen J.. A family of IGF-II mRNA binding proteins (IMP) involved in RNA trafficking. Scand. J. Clin. Lab. Invest. Suppl. 2001; 234:93–99. [PubMed] [Google Scholar]
- 35. Sheth U., Parker R.. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003; 300:805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Khong A., Matheny T., Jain S., Mitchell S.F., Wheeler J.R., Parker R.. The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol. Cell. 2017; 68:808–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hubstenberger A., Courel M., Bénard M., Souquere S., Ernoult-Lange M., Chouaib R., Yi Z., Morlot J.-B., Munier A., Fradet M. et al. P-Body purification reveals the condensation of repressed mRNA regulons. Mol. Cell. 2017; 68:144–157. [DOI] [PubMed] [Google Scholar]
- 38. Sundararaman B., Zhan L., Blue S.M., Stanton R., Elkins K., Olson S., Wei X., Van Nostrand E.L., Pratt G.A., Huelga S.C. et al. Resources for the comprehensive discovery of functional RNA elements. Mol. Cell. 2016; 61:903–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dominguez D., Freese P., Alexis M.S., Su A., Hochman M., Palden T., Bazile C., Lambert N.J., Van Nostrand E.L., Pratt G.A. et al. Sequence, structure, and context preferences of human RNA binding proteins. Mol. Cell. 2018; 70:854–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li Y.E., Xiao M., Shi B., Yang Y.-C.T., Wang D., Wang F., Marcia M., Lu Z.J.. Identification of high-confidence RNA regulatory elements by combinatorial classification of RNA-protein binding sites. Genome Biol. 2017; 18:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Baltz A.G., Munschauer M., Schwanhäusser B., Vasile A., Murakawa Y., Schueler M., Youngs N., Penfold-Brown D., Drew K., Milek M. et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell. 2012; 46:674–690. [DOI] [PubMed] [Google Scholar]
- 42. Fan X.C., Steitz J.A.. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:15293–15298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Keene J.D. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 2007; 8:533–543. [DOI] [PubMed] [Google Scholar]
- 44. Bahar Halpern K., Caspi I., Lemze D., Levy M., Landen S., Elinav E., Ulitsky I., Itzkovitz S.. Nuclear retention of mRNA in Mammalian tissues. Cell Rep. 2015; 13:2653–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Battich N., Stoeger T., Pelkmans L.. Control of transcript variability in single Mammalian cells. Cell. 2015; 163:1596–1610. [DOI] [PubMed] [Google Scholar]
- 46. Mesarovic M.D., Sreenath S.N., Keene J.D.. Search for organising principles: understanding in systems biology. Syst. Biol. (Stevenage). 2004; 1:19–27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We provide an overview analysis of each independent PAR-CLIP library that can be accessed at https://github.com/BIMSBbioinfo/RCAS_meta-analysis. We have also added all RBP binding sites to DoRiNA (https://dorina.mdc-berlin.de/), which enables the user to query all binding sites and perform additional analyses. Sequencing reads for the 7 new PAR-CLIP libraries are available at SRP154398.