Abstract
In Trypanosoma brucei, genes are arranged in Polycistronic Transcription Units (PTUs), which are demarcated by transcription start and stop sites. Transcription start sites are also binding sites of Origin Recognition Complex 1 (ORC1). This spatial coincidence implies that transcription and replication in trypanosomes must occur in a highly ordered and cooperative manner. Interestingly, a previously published genetic screen identified the T. brucei MCM-BP, which interacts with subunits of MCM helicase, as a protein whose downregulation results in the loss of transcriptional silencing at subtelomeric loci. Here, I show that TbMCM-BP is required for DNA replication and transcription. TbMCM-BP depletion causes a significant reduction of replicating cells in S phase and genome-wide impairments of replication origin activation. Moreover, levels of sense and antisense transcripts increase at boundaries of PTUs in the absence of TbMCM-BP. TbMCM-BP is also important for transcriptional repression of the specialized subtelomeric PTUs, the Bloodstream-form Expression-Sites (BESs), which house the major antigenic determinant (the Variant Surface Glycoprotein, VSG gene) as well as TbORC1 binding sites. Overall, this study reveals that TbMCM-BP, a replication initiation protein, also guides the initiation, termination and directionality of transcription.
INTRODUCTION
DNA replication initiates by the binding of Origin Recognition Complex (ORC) to replication origins. Origins must be activated by a mechanism termed ‘replication licensing’, which occurs via a cascade of protein recruitment to, and protein phosphorylation on, the origin (reviewed (1–3)). The hexameric ORC complex recruits the replication licensing factors CDC6 and CDT1 and the replicative helicase MCM (MCM2–7, a hexamer). CDC45-SLD3 and GINS-DPB11-SLD2 are also recruited to the origin, bringing with them the leading and lagging strand polymerases. Phosphorylation of SLD2 and SLD3 by the S-phase kinase DDK (DBF4-Dependent Kinase, CDC7-DBF4) initiates DNA replication, and cells enter S-phase.
While many of these processes have been elucidated in model organisms (like yeast), in the protozoan parasite Trypanosoma brucei, nuclear DNA replication is relatively a new field: replication origins were only recently identified and although many replication proteins, especially at initiation and licensing steps, have been identified in the past several years through homology with their yeast and mammalian counterparts (4–7), there are still much to be elucidated to understand the mechanism of nuclear DNA replication in T. brucei and its relationship with other cellular processes, such as with chromosome segregation, cell-cycle progression and DNA damage response as well as with transcription. For example, the TbORC complex appears to be distinct from other ORCs particularly in subunit primary sequence and in complex composition (4,6,8). So far, five subunits of TbORC complex have been found, including TbORC1, TbORC4 and three ORC-like proteins (TbORC1B, Tb7980 and Tb3120). But a CDC6 homologue has not been found in T. brucei and TbORC1 shows sequence similarity to both the ORC1 and CDC6 of other organisms. In contrast, all 6 subunits of T. brucei MCM complex (MCM2–7) have been identified and the helicase activity of the complex has been characterized in vitro (5). Furthermore, in yeast and human, MCM subunits interact with MCM-Binding Protein (MCM-BP) to form an alternative MCM complex (MCM-BP complex) (9,10). The work described herein centers upon some surprising functions of T. brucei MCM-BP, which, despite the fact that it is part of the replicative complex, was initially identified in a genetic screen designed to isolate genes important for transcriptional repression at subtelomeres (11).
MATERIALS AND METHODS
Trypanosoma brucei Strain
Trypanosoma brucei bloodstream form (the Lister 427 antigenic type MITat1.2 clone 221a) was cultured in HMI-9 at 37°C (12). TbMCM-BP conditional knock out (TbMCM-BP cKO) strains were generated previously (11) and two independent clones, HSTB-660 and 661, were used in this study. Briefly, they are of ‘single marker (SM)’ background expressing T7 RNA polymerase and Tet repressor (TetR). CRE recombinase gene was integrated at rDNA spacer and is under the control of TetR. In the absence of tetracycline, TetR binds the Tet Operator (TetO) sequences upstream of the CRE gene (transcription off). With tetracycline addition, TetR is removed from TetO and the CRE gene is expressed (on). One allele of TbMCM-BP was knocked out and the remaining allele was epitope-tagged at the C-terminus with 3xmyc followed by Hygromycin-Resistance Gene (HYG) conjugated with Herpes Simplex Virus Thymidine Kinase gene (HSVTK or TK). This cassette is flanked by loxP sites that can be recombined by Cre recombinase. Tetracycline addition induces Cre expression and Cre removes TbMCM-BP-myc allele, resulting in TbMCM-BP depletion. HSTB-904, the isogenic wild type strain, was used as a control for some experiments.
Trypanosome cells were maintained in HMI-9 media containing necessary antibiotics at the following concentrations: 2.5 μg/ml of G418 (Sigma); 5 μg/ml blasticidin (Invivogen); 1 μg/ml phleomycin (Invivogen); 5 μg/ml hygromycin (Invivogen); 1–2.5 μg/ml tetracycline (Sigma)
Western blot
Equal numbers of cells were collected and suspended in the Laemmli buffer and separated on an SDS-PAGE gel. Following transfer to nitrocellulose membrane, the proteins were analyzed using mouse anti-myc, mouse anti-tubulin, rabbit anti-VSG3, or mouse anti-HA antibodies.
Flow cytometry
WT and TbMCM-BP cKO strains were collected at days 0, 1 and 2 after tetracycline addition. Cells were fixed with ice-cold 70% ethanol and DNA was stained with 50 μg/ml propidium iodide (PI) in PBS at 37°C for 30 min. Cell-cycle progression was analyzed using the LSR II, FACSdiva and flowjo.
BrdU pulse experiment
BrdU pulse experiments were performed as described previously (13) with some modifications. BrdU (Sigma) was added to a final concentration of 500 μM to cell cultures and incubated for 40 min. Cells were then fixed with ice-cold 70% ethanol, incubated in 0.1N HCl with 0.5% Triton X-100 and washed with PBS containing 0.5% Tween 20 and 1% BSA. To detect BrdU incorporation, fixed cells were incubated with 1 μg/ml mouse anti-BrdU antibody at room temperature for 2 h and incubated with donkey anti-mouse-Alexa488 at 4 μg/ml for 45 min at room temperature in the dark. Cells were then resuspended in PBS containing 5 μg/ml PI and analyzed by flow cytometry (LSR II, FACSdiva, and flowjo).
MFA-seq
MFA-seq was performed as reported previously (7) with some modifications. The TbMCM-BP cKO cell line was treated with tetracycline for 0 and 1 day. About 50 million cells at ∼1 × 106/ml density were fixed with ice-cold 70% ethanol and DNA was stained with 50 μg/ml PI in PBS. Cells in G1, early S, late S and G2 phases were sorted by flow cytometry (BD Aria3). Genomic DNA was isolated from these sorted cells using the QIAamp DNA Blood mini kit (Qiagen). To prepare high-throughput sequencing libraries, 5–10 ng of genomic DNA was fragmented using the NEBNext dsDNA fragmentase (NEB) and sequencing libraries were generated using the NEBNext Ultra DNA Library prep kit and NEB multiplex oligos for Illumina according to the manufacturer's protocol. Sequencing was performed on an Illumina HiSeq 2000 sequencer (50-bp single end read). Read quality was analyzed using the FastQC program and reads were trimmed using the TrimGalore program from Babraham Bioinformatics (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) (parameters: trim_galore -phred33 -q 20 -a GATCGGAAGAGCACACGTCTGAACTCCAGTCAC -stringency 5 -length 20 -e 0.1) and aligned with Bowtie (14) to the reference genome (Tb927v5 was retrieved from http://tryitrypdb.org/tritrypdb/), allowing only for uniquely aligning reads with a maximum of 2 mismatches (parameters: -a -best -M 1 -v 2). About 2.2 ∼ 11 million reads were analyzed using the SeqMonk algorithm (http://www.bioinformatics.babraham.ac.uk/projects/seqmonk/) from Babraham Bioinformatics. To examine replication initiation profiles, chromosomes were binned at 10 kb resolution with 2.5 kb step and Reads Per Million mapped reads (RPM) values were generated with SeqMonk. RPM values from the S phase samples were normalized to values from G1 phase and plotted using Excel.
Stranded RNA-seq
TbMCM-BP cKO cells were grown in the absence or presence of tetracycline (for 1 or 2 days) and ∼50 million cells were collected. RNA was extracted using the RNA Stat-60 (Tel-Test) according to the manufacturer's protocol and quantified on a NanoDrop2000c and further cleaned using RNeasy kit (Qiagen). RNA-seq libraries were prepared from 1μg of RNA samples using Sera-Mag oligo-dT beads, the NEBNext Ultra Directional RNA Library Prep Kit for Illumina, and Illumina Tru-Seq adapters. Single-end, 50-cycle sequencing was then performed for the 12 libraries (TbMCM-BP cKO strain treated with tetracycline for 0, 1 and 2 days, in four replicates) on a single lane using the Illumina HiSeq 2000. Sequence reads were quality checked, trimmed, and aligned to the reference genome, as described above, and ∼9.8–19 million reads were analyzed.
For transcriptomic analysis, raw reads mapping to ∼9300 genes were analyzed by the DEseq program from Bioconductor (https://bioconductor.org/packages/release/bioc/html/DESeq.html) with default parameters. To examine transcription profiles at chromosome level, aligned reads were analyzed using SeqMonk. Each chromosome was binned at 5 kb resolution with 1 kb step (sliding window analysis: 5 kb bin, 1kb step). RPM values from forward or reverse reads only were generated and plotted over the length of chromosome 7. Fold changes between Tet-treated day 2 sample (or day 1) and uninduced (day 0) were plotted for all 11 megabase chromosomes. The correlation coefficient between replicates was >0.99.
To get a snapshot of transcription defects at PTU boundaries, reads mapping to TTSs, TSSs or HTs were counted and read per kilobase per million mapped reads (RPKM) values were generated. I used new lists of TSSs, TTSs and HTs modified from a previous study (15) by removing sites of ambiguity. For example, several of TSSs and TTSs located at subtelomeres were removed, due to ambiguity associated with disorderly organization of genes (constant change of transcription directions). Scatter and box plots were generated using the ggplot2 package in the R program supported by the R Foundation for Statistical Computing (https://www.r-project.org). Differences between TbMCM-BP cKO uninduced (day 0) and induced (day 1 or 2) were summarized using descriptive statistics performed in R, as described previously (15). A Kruskal-Wallis one-way analysis of variance test (command kruskal.test()) was first applied to determine whether a significant difference exists across median log2(RPKM) counts in uninduced and induced samples. Once significance was established across groups, a Mann-Whitney U test (command wilcox.test()) was performed between median log2(RPKM) counts for induced and uninduced samples. The same analysis was performed to compare derepression phenotypes at all PTU borders and borders that replicate early. The 30 kb region encompassing TSSs, TTSs or HTs were analyzed with sliding windows (5 kb bin, 0.2 kb step).
To examine VSG expression, trimmed reads were bowtie-aligned to the VSGnome (retrieved from http://tryps.rockefeller.edu) (16) and differential expression was analyzed using the DEseq. Scatter and box plots were generated using the ggplot2 in R program. To examine expression profiles within BESs, reads were aligned with Bowtie to the BES sequences (14 BES sequences were obtained from L. Figueiredo, IMM, Lisbon) and analyzed with sliding windows (1kb bin and 0.2kb step). RPM values were generated and fold changes relative to the uninduced sample were plotted.
RPA1 IF
Cells were fixed with 0.5% paraformaldehyde for 10 min and permeabilized with 0.2% NP-40 in PBS. Cells were incubated with rat anti-HA antibody and then with secondary antibody conjugated with Alexa 488. DNA was stained with 0.5 mg/ml DAPI. Images were captured using a Zeiss Axioplan 2 fluorescence microscope and edited with Adobe Photoshop.
S9.6 dot blot
Genomic DNA was isolated from WT and TbMCM-BP cKO uninduced and induced with tetracycline for 0, 1 and 2 days, using the QIAamp DNA Blood mini prep kit. Genomic DNA was treated with or without RNase H and spotted directly on a positively charged Nylon membrane (Amersham) using a Dot blot apparatus, then UV cross-linked (0.12 J/m2). To detect DNA:RNA hybrid (R loop), membrane was probed with mouse S9.6 antibody in PBS containing 1% milk and 0.1% Tween 20 (17).
RESULTS
TbMCM-BP is required for DNA synthesis
We previously showed that TbMCM-BP is required for proper cell-cycle progression, as the absence of TbMCM-BP resulted in the accumulation of cells in G2 and then within a sub-G1 population (11). Additionally, we found that TbMCM-BP strongly interacts with other MCM subunits (11), suggesting that TbMCM-BP has roles in DNA replication. These initial findings implied that the mutant cells are experiencing DNA damage but were unable to properly block nuclear and cell divisions. To determine how TbMCM-BP functioned, I needed a finer analysis of how DNA replication was impacted. To do this, I used the previously established TbMCM-BP conditional knockout (TbMCM-BP cKO) cell line in bloodstream form (11) (Supplementary Figure S1, removal of TbMCM-BP-myc can be detected by western blot using anti-myc antibody).
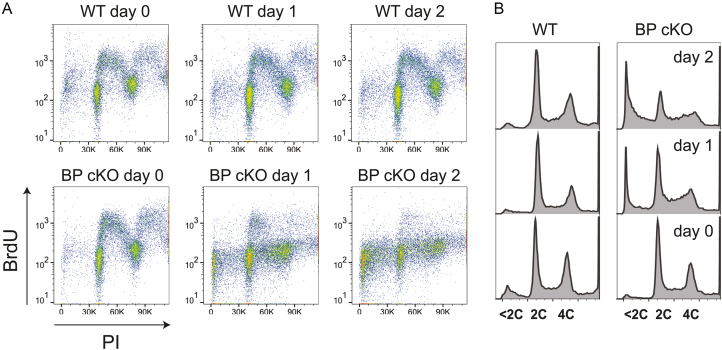
To take a closer look at DNA replication, I labeled wild type and TbMCM-BP cKO cells (treated with tetracycline for 0, 1 and 2 days) with 500 μM 5-bromo-2′deoxyuridine (BrdU, an analog of dT) for 40 min. BrdU substituted DNA was then detected using anti-BrdU antibodies and bulk DNA was stained with Propidium Iodide (PI). Cells were then analyzed by flow cytometry (Figure 1A). TbMCM-BP cKO at day 0 showed the same staining pattern as the wild type samples. However, at day 1 upon Cre induction, BrdU-incorporated S phase population was greatly reduced and only a small portion of cells in early S phase were positive for BrdU staining. DNA synthesis was further reduced at day 2 in the mutant. Consistent with a previous study (11), the sub-G1 population increased greatly in the absence of TbMCM-BP (Figure 1B) confirming previous findings that TbMCM-BP is required for DNA synthesis.
Figure 1.
TbMCM-BP is required for DNA replication. (A) BrdU pulse to label replicating cells. WT and TbMCM-BP cKO cells treated with tetracycline for 0, 1 and 2 days were pulse labeled with 500 μM BrdU for 40 min. BrdU incorporation was detected using anti-BrdU antibody. BrdU pulse labeled cells were stained with Propidium Iodide (PI) and analyzed by flow cytometry. (B) Cell cycle profile with PI channel only.
TbMCM-BP is required for replication initiation at early replicating regions
BrdU staining can be used to delineate cell cycle progression by FACS. To gain a finer understanding of a defect associated with replication, I performed MFA-seq (or Sort-seq) experiment as described previously (7) with some modifications. This technique relies on the fact that replicated regions (2n) have double the ‘read’ depth compared to regions that are not yet replicated which would have the 1n genomic complement. Therefore, early replicating regions (and locations of origins) have a 2n genomic complement in early S phase cells. By plotting the ratio between sequence reads from S and G1 phase, one can identify the location, strength and timing of replication origins within the cell population.
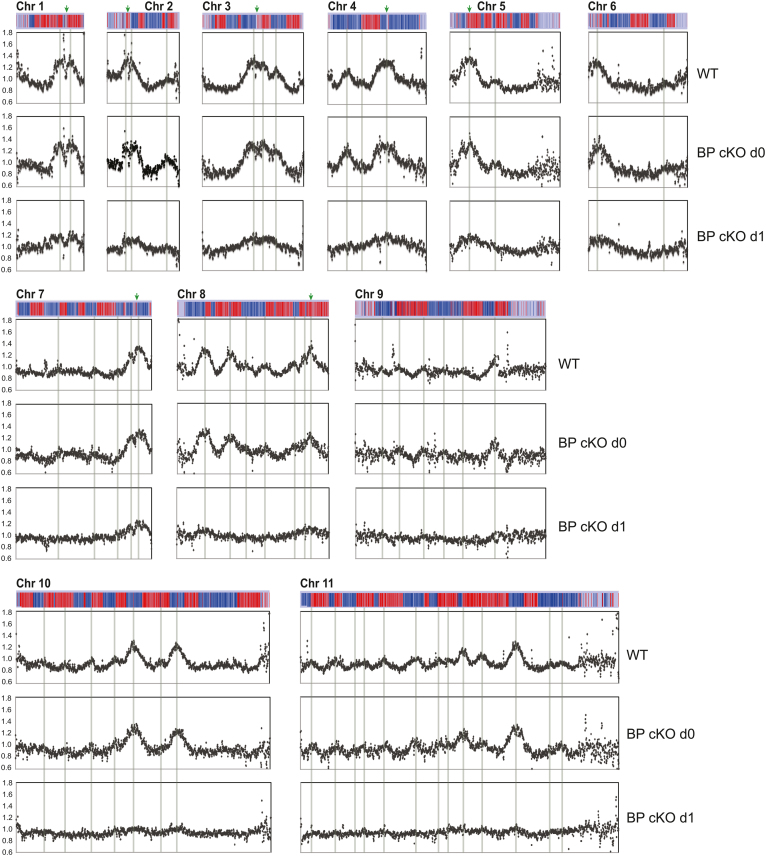
I sorted PI-stained cells with G1, early S, late S, or G2 DNA content using flow cytometry (Supplementary Figure S2), isolated genomic DNA and prepared high-throughput sequencing libraries. These were sequenced by Illumina, and sequence reads were aligned to the Tb927v5 reference genome (with 11 megabase chromosomes). Due to the severity of cell-cycle defects at day 2, I analysed the TbMCM-BP cKO only at day 1 after Cre induction. To examine replication initiation profiles, chromosomes were binned at 10 kb resolution with 2.5 kb step (sliding window analysis) and reads mapping to each bin was counted. Ratios between the early S and G1 samples were plotted along each chromosome (Figure 2). Replication profiles of the wild type strain were comparable to those reported previously from the McCulloch lab (7,18); overall peak patterns were almost identical to the previous profiles, including the number of peaks, location and height (strength of origin) of peaks, and also replication origins near centromere that become activated early with strong activity (shown in green arrows). Replication profiles at the late S phase also agreed well with the previous study (18), losing resolution and displaying broadened peaks compared to the early S phase (Supplementary Figure S3). Replication profiles of TbMCM-BP cKO day 0 cells were comparable to those of wild type. However, at day 1 after TbMCM-BP removal, no major peaks were observed, indicating that none of early replicating origins are activated in the absence of TbMCM-BP and thus, that TbMCM-BP is required for replication initiation.
Figure 2.
TbMCM-BP is required for DNA replication initiation at early firing origins. Replication was examined by MFA-seq. Cells were collected at day 0 and 1 after tetracycline addition and stained with PI. Cells in G1, early S, late S and G2 phase were sorted by flow cytometry (Supplementary Figure S2), genomic DNA was isolated and high-throughput sequenced. To examine replication profiles, chromosomes were binned at 10 kb resolution with 2.5 kb step. Read count ratio between early S to G1 was plotted for 11 megabase chromosomes. Chromosomes are depicted as bars above each graph. Blue genes are transcribed reverse and red are forward direction. Grey lines indicate the early replicating origins previously identified by the McCulloch lab [7]. Green arrows indicate locations of centromere.
In the absence of TbMCM-BP, replication may be initiated promiscuously at random places and therefore no clear major peaks are detected. However, considering that very little of BrdU incorporation was observed in the absence of TbMCM-BP (Figure 1A), even if replication initiates promiscuously at random places, DNA synthesis must not occur vigorously. With lack of DNA synthesis and no clear replication origin firing detected, my data indicate that TbMCM-BP is required for replication initiation.
TbMCM-BP depletion increases expression levels of genes near PTU boundaries and at subtelomeres
Unlike in other eukaryotes, in T. brucei, genes are assembled in Polycistronic Transcription Units (PTUs) with ∼100 genes on average, transcribed unidirectionally by RNA Polymerase II (Pol II). Boundaries of PTUs (termed Transcription Start Sites (TSSs) and Transcription Termination Sites (TTSs)) define where transcription starts and terminates. Surprisingly, McCulloch and colleagues recently showed that TbORC1 associates with most of TSSs and some TTSs, and that at least about 41 of those TbORC1 binding sites are early replicating origins (7). This raised the possibility that some of replication factors may have dual roles in controlling DNA replication and transcription. This, together with the identification TbMCM-BP in a loss of transcriptional silencing screen, prompted me to ask whether TbMCM-BP has a general role in transcriptional control of T. brucei. Toward that, I performed stranded RNA-seq analysis. RNA samples were collected from four independent cultures (two independent cultures from two independent cKO strains, HSTB-660 and HSTB-661) at day 0, 1 and 2 after Cre induction (Supplementary Figure S4B) and high-throughput sequencing libraries were generated and sequenced on an Illumina sequencer.
Raw reads mapping to about 9,300 genes were analyzed by DEseq and gene expression levels were compared between day 0 and day 1 (or day 2) after TbMCM-BP depletion (Supplementary Figure S4C and D). Genes with significant changes (Padj < 0.01) are shown as red dots. Genes that are >2-fold up or down (50% reduction) with Padj < 0.01 are also listed in Supplementary Table S1. TbMCM-BP did not significantly influence overall transcription—only 71 were at least 2-fold (>50%) downregulated and 301 were at least 2-fold upregulated at day 2 compared to day 0. The list of genes also includes some of rRNA, tRNA, and snoRNA (not included in Supplementary Table S1). Of 71 of 2-fold downregulated genes, 38 were rRNA and 13 were H2B, and 17 were known or hypothetical genes and one has no information. Of 301 of 2-fold upregulated ones, 113 were in fact snoRNA (108), tRNA (1), small nuclear RNA (2), and 5S rRNA (2) (not included in Supplementary Table S1), which are usually clustered near TTSs. Among the rest of the 188 genes, 51 were developmentally regulated genes including EPs, a GPEET, and PAGs that are expressed in procyclic form but repressed in BF T. brucei (19), and genes including VSGs and ESAGs that are repressed in BF T. brucei. These genes are normally clustered either in subtelomeres or at the end of RNA pol II transcribed PTUs. Genes that encode Retrotransposon Hot Spot proteins (RHS) were also found and 73 were hypothetical conserved or hypothetical genes. Locations of these 188 genes with >2-fold upregulation were mapped (Supplementary Figure S5). Interestingly, most of these genes were closely located at borders of PTUs or subtelomeric regions, suggesting that aside from its role in replication, TbMCM-BP is also required for genome-wide transcription regulation, specifically at transcription start and stop sites as well as subtelomeres.
TbMCM-BP deficiency accumulates antisense transcripts at transcription start and termination sites throughout the genome
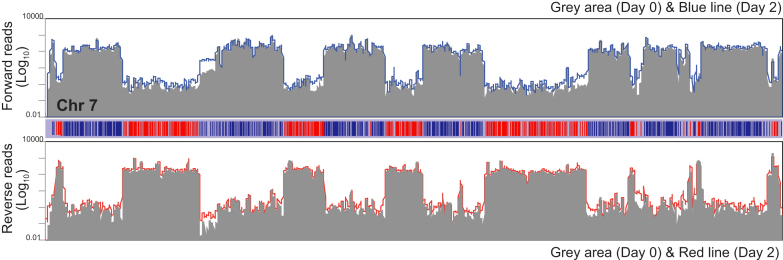
To assess whether TbMCM-BP was required for transcription initiation, termination, or directionality, I analyzed forward and reverse reads separately. Chromosome 7 is shown as an example in Figure 3. Forward reads correspond to transcripts produced from reverse strand (blue PTUs, transcribing right to left) and reverse reads from forward strand (red PTUs, left to right). Grey areas indicate the number of forward or reverse reads count from day 0, blue and red lines indicate the forward and reverse reads count from day 2 after TbMCM-BP depletion. Sense transcription appears to remain unaffected by the TbMCM-BP absence, while antisense transcripts (defined as polyadenylated transcripts that do not terminate properly but read-through to the next PTU—which they enter in the ‘wrong’ direction- or transcripts that do not initiate properly), increased in the absence of TbMCM-BP, especially at the borders of PTUs.
Figure 3.
TbMCM-BP is required for transcription regulation. Transcription was examined by stranded RNA-seq. Forward and reverse reads were analyzed separately. Chromosomes were analyzed with sliding window (5 kb bin, 1 kb step). Log10(RPM) was plotted over the chromosome length. Chromosome 7 is shown as an example. The graph on the top represents forward read map: grey area is day 0 and blue line is day 2 after TbMCM-BP depletion. The graph on the bottom represents reverse read map: grey area is day 0 and red line is day 2 after TbMCM-BP deletion. The bar diagram in the middle depicts PTUs. Blue genes are transcribed in reverse PTUs and red are forward PTUs.
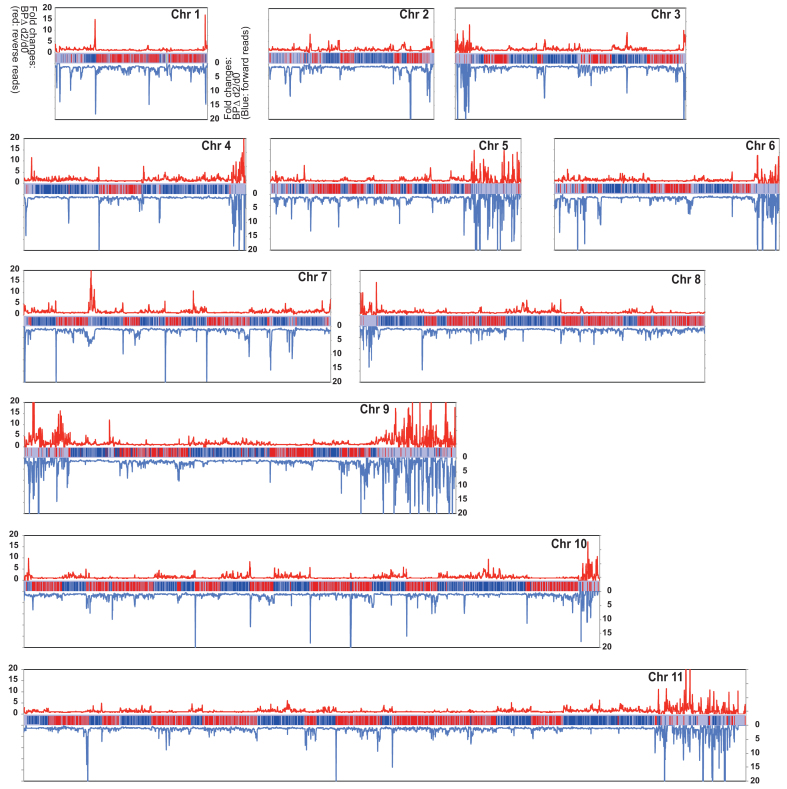
To see the difference between WT and TbMCM-BP mutant more clearly, fold changes between day 2 and day 0 were plotted. Figure 4 shows all 11 megabase chromosomes and has the same color usage as in Figure 3—i.e. red lines indicate the fold changes from the reverse reads (mapping to the forward strands, red PTUs) and blue lines indicate the fold changes from the forward reads (mapping to the reverse strands, blue PTUs). Throughout all chromosomes, sense transcription remained unaffected, while antisense transcription increased. Significant fold increases of antisense transcription were observed near transcription borders, generally, upstream of normal start sites and downstream of normal stop sites. This indicates that transcription initiates upstream of the start sites and terminates past the stop sites. This is observed throughout all chromosomes. In addition, levels of transcripts from subtelomeres greatly increased from both directions in the absence of TbMCM-BP. Transcription profiles comparing day 0 and day 1 after TbMCM-BP removal are shown in Supplementary Figure S6.
Figure 4.
TbMCM-BP deficiency accumulates antisense transcripts at PTU borders. Fold changes comparing transcription levels between day 0 and day 2 after TbMCM-BP deletion were calculated and plotted over the chromosome length. Reverse and forward reads were analyzed separately. Red lines indicate fold changes from reverse reads (mapping to forward strand, red PTUs) and blue lines indicate fold changes from forward reads (mapping to reverse strands, blue PTUs). Chromosome diagrams are shown in the middle of each graph.
TbMCM-BP affects the direction of transcription
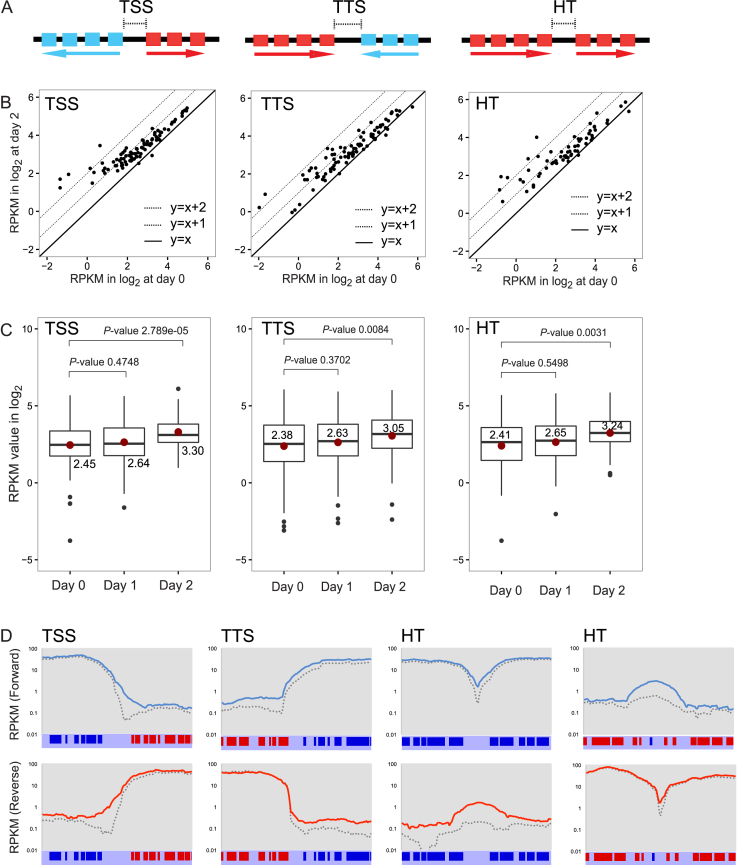
I then examined transcription more closely at boundaries of PTUs—TSS, TTS and HT. Levels of transcripts at these sites were quantified. As we reported previously (15) and shown in Figure 5A, TSS and TTS are computationally defined regions where coding strands switch. Head-to-Tail (HT) sites are defined as TbBDF3 bound regions (20), where one PTU stops and another starts in the same direction. Reads mapping to either TSS, TTS, or HT were counted and log2(RPKM) values were generated. I then compared day 0 and day 2 log2(RPKM) values for TSSs (91 sites), TTSs (89) or HTs (64) in scatter plots (Figure 5B). Many of TSSs, TTSs, and HT sites were upregulated in the absence of TbMCM-BP. Boxplots comparing median log2(RPKM) values also indicated that all three types of PTU boundaries were upregulated in the absence of TbMCM-BP (Figure 5C). Fold increases at day 2 were 1.8-fold for TSS, 1.6 for TTS and 1.8 for HT. The differences were statistically significant (Supplementary Table S2).
Figure 5.
TbMCM-BP affects transcription initiation and termination, and direction of transcription. Levels of transcripts at TSSs, TTSs, and HTs increased in TbMCM-BP mutant. (A) Diagrams of a TSS, a TTS, and a HT: boxes are genes in the PTUs and arrow indicates PTUs and transcription direction. (B) Scatter plots comparing levels of transcripts between day 0 and day 2 after TbMCM-BP depletion. Each dot represents a transcription boundary. Dotted lines are changes that are 2-fold or 4-fold up. (C) Box plots compare levels of transcripts from TSSs, TTSs, and HTs at day 0, 1, and 2 after TbMCM-BP deletion. Median values are shown and indicated as red dots. Black dots are outliers. Differences between day 0 and day 1 (day 0 and day 2) were measured by a Mann-Whitney U statistical test (Supplementary Table S2). (D) The region encompassing 15 kb upstream and downstream from a midpoint of a representative TSS (or a TTS or two HTs) was binned at 5 kb resolution with 0.2 kb step. RPKM values of forward or reverse reads mapping to each bin were calculated and plotted over the 30 kb length. Blue lines are forward reads and red are reverse reads from day 2 samples and dotted lines are from day 0. PTUs and genes are shown.
To see whether increased levels of transcripts at these sites are due to transcription readthrough or improper transcription initiation, I examined forward and reverse reads separately at regions of 15 kb up and downstream from a midpoint of each site (Figure 5D). I examined sites that have at least 2-fold increase in transcript levels (62 sites were examined, and differences at only 2 sites were statistically insignificant when analyzed by DEseq). Examples for each boundary type are shown in Figure 5D. At TSS, the region upstream of the start site produced more transcripts in the TbMCM-BP mutant (solid lines) than in the wild type (dotted lines) when forward or reverse reads were analyzed separately, indicating that transcription initiates upstream of its start site in the absence of TbMCM-BP. This causes the elevation of antisense transcripts from genes that are located in the beginning of the neighboring PTU. At a TTS, more transcripts were generated past the stop site, accumulating antisense transcripts on the converging PTU. At HT sites, where both PTUs transcribe in the same direction, more sense transcripts were generated at the HT sites in the absence of TbMCM-BP, indicating that transcription may initiate upstream of normal start sites and/or readthrough stop sites. More interestingly, in the absence of TbMCM-BP, antisense transcripts increased around HT sites, indicating that TbMCM-BP is required for determining the direction of the transcription machinery.
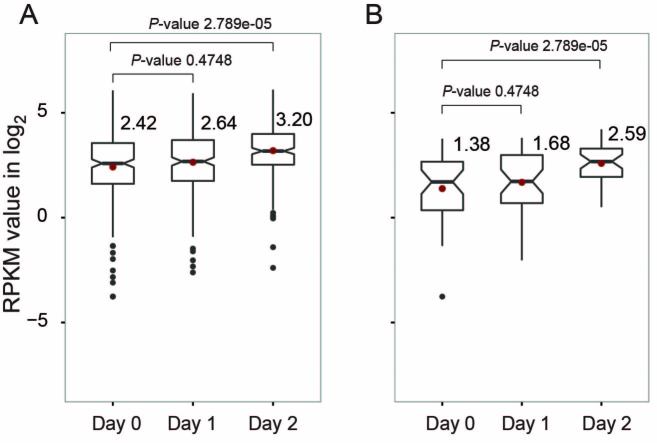
Replication origin activity correlates with transcription levels in human (21). Therefore, blocking origin activation in the absence of TbMCM-BP may also cause transcription defects. To see whether there is correlation between replication and transcription defects, I compared transcript levels from all PTU borders (91 TSSs, 89 TTSs, and 64 HT sites) and PTU borders that are fired early (40 early origins (7); 19 TSSs, 4 TTSs and 17 HT sites) (Figure 6, right). The median difference between day 0 and 2 was bigger in early replicating PTU borders (2.3-fold up) than that in all borders (1.7-fold up). The difference between day 0 and day 2 was statistically significant (Supplementary Table S3). These data suggest that transcription boundaries with stronger origin activity may be more prone to transcriptional dysregulation when replication is compromised, leading to elevation of antisense transcription.
Figure 6.
PTU borders that are early replicating are more strongly influenced by the TbMCM-BP absence. Levels of transcripts produced from all PTU borders and borders that are replicating early were compared. Box plots compare between day 0, day 1, and day 2 after TbMCM-BP deletion for all PTU borders (A) and for borders that are replicated early in S phase (B). Median values from each sample are shown and indicated as red dots. Black dots are outliers. Differences between day 0 and day 1 (or day 2) were measured by a Mann–Whitney U statistical test (Supplementary Table S3).
Finally, it is unlikely that transcription and replication defects observed in the absence of TbMCM-BP are indirect effects caused by downregulation of replication genes, as expression levels of known replication licensing and initiation factors were not changed significantly (Supplementary Table S4).
TbMCM-BP regulates transcriptional repression of the subtelomeric PTUs, BESs
Besides TSSs and TTSs, replication proteins have a major influence on transcription at subtelomeric regions in T. brucei (7,11,22). Both TbORC1 and TbMCM-BP are required for repressing the expression of Variant Surface Glycoprotein (VSG), the surface coat gene that is monoallelically expressed from one of ∼15–20 telomeric expression sites (BES) in the mammalian-infectious Bloodstream Form (BF) (11,22). VSG silencing is part of the strategy that T. brucei uses to evade the host immune response—by expressing one species of VSG gene at a time and while repressing the rest (23). A Bloodstream form Expression Site (BES), is the last PTU of the majority of T. brucei chromosomes. It houses a VSG gene immediate upstream of telomere repeats and several Expression Site Associated Genes (ESAGs) upstream of the VSG (24). The size of the BES PTUs ranges between 10 and 60 kb in length all BES PTUs are transcribed by RNA Pol I (25). Only one BES promoter is transcriptionally active at any given time in a cell, while the rest are transcriptionally repressed, allowing monoallelic VSG expression. Moreover, TbORC1 associates with telomeres and the active BES origin appears to be activated early in S phase (7,18).
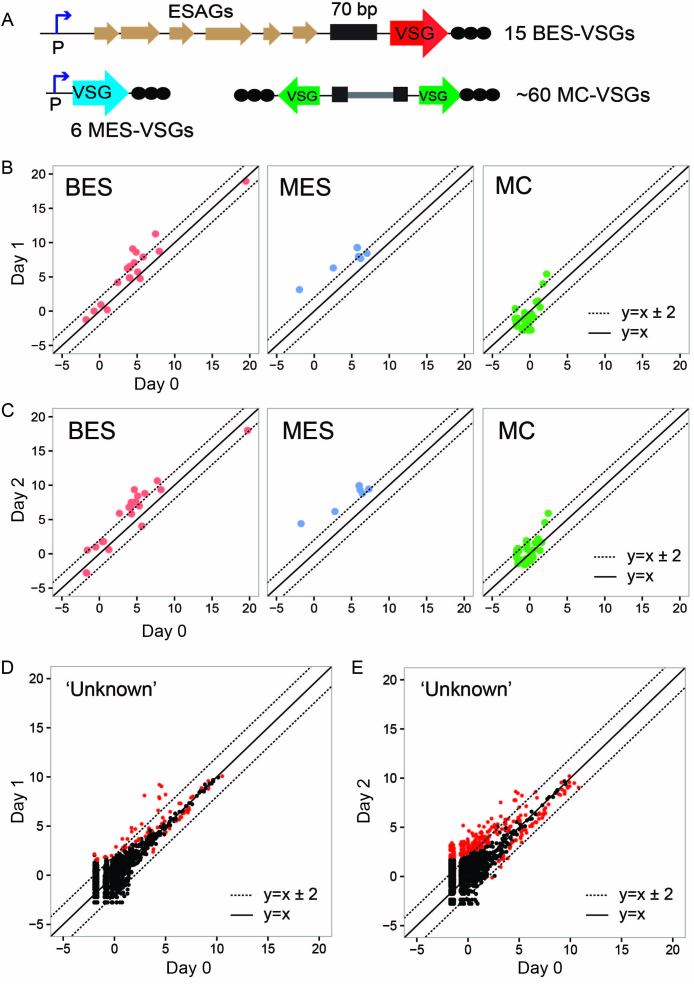
I already knew that subtelomeric gene expression was greatly influenced by TbMCM-BP (11). To study the global function of TbMCM-BP in VSG expression control, RNA-seq reads mapping to the VSGnome (16) were analyzed using DEseq. Figure 7 shows expression levels of four different types of VSGs (BES; minichromosomal (MC) which are also subtelomeric (26); MES or metacyclic—which are subtelomeric and transcribed monocitronically by Pol I (16,27); and chromosome internal VSGs which are mostly unknown (16)). Upon TbMCM-BP depletion, BES- and MES-VSGs transcribed by RNA Pol I promoter (polycistronic and monocistronic transcription respectively) were greatly derepressed (Figure 7B and C, Supplementary Table S5 and 6). On the other hand, MC VSGs lacking a promoter were not significantly affected; only 2 and 6 of 41 were upregulated at day 1 and day 2 (Padj < 0.1). Many chromosome-internal VSGs showed increased level of transcription after TbMCM-BP removal (Figure 7D and E). These VSGs are thought to be located at subtelomeric regions in arrays, however their exact locations are not known. Overall, these data indicate that TbMCM-BP regulates transcriptional silencing of special PTUs such as BES and MES PTUs, that are RNA Pol I and telomere associated, and subtelomeric ‘unknown’ VSGs.
Figure 7.
TbMCM-BP is required for silencing of subsets of VSG genes. (A) Schematic diagrams of VSG types; BES VSG (Bloodstream-form Expression Site VSG), MES VSG (metacyclic VSG), and MC VSG, (minichromosomal VSG). Reads from the RNA-seq experiment were aligned to the VSGnome database [16] and raw reads mapping to each VSG was analyzed with the DEseq. Log2(normalized count) values were obtained and graphed in scatter plots. (B) Scatter plots showing comparison between day 0 and day 1 for BES, MES or MC VSGs. Dotted diagonal lines indicate changes that are 4-fold up or down. (C) Scatter plots showing comparison between day 0 and day 2 for BES, MES or MC VSGs. Dotted diagonal lines indicate changes that are 4-fold up or down. (D, E) The rest of VSGs (‘Unknown’) excluding BES, MC, and MES were graphed in scatter plots. Plot D compares day 0 and day 1 and plot E compares day 0 and day 2. Red dots indicate VSG genes whose expression level changed significantly (Padj < 0.01). Dotted diagonal lines indicate changes that are 4-fold up or down.
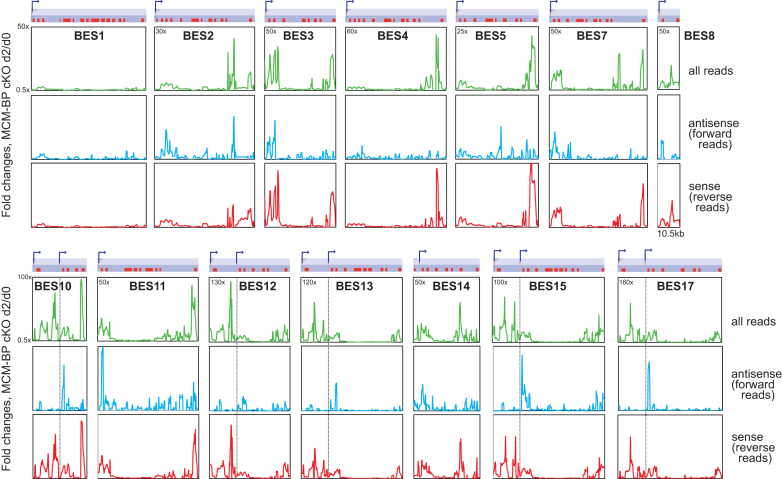
TbMCM-BP was initially isolated as a regulator of promoter silencing from procyclic form in which all BESs and VSGs are repressed (11). Loss of VSG silencing in the absence of TbMCM-BP showed strong dependence on the presence of Pol I promoter (Figure 7), suggesting that TbMCM-BP function may be needed at both ends of subtelomeric PTU, similar to its role in chromosome internal PTU transcription. To examine this and to see how far the derepression phenotype can spread from promoter or telomere within BES PTUs, I analyzed RNA-seq reads mapping to the 14 BESs. Reads were aligned to the T. brucei Lister 427 BES sequences (24) (one active BES (BES1) and 13 silent BESs) with Bowtie. The BESs were binned at 1 kb resolution with 200 bp step and all reads mapping to each bin were counted and RPM values were generated. All reads (green), forward only (antisense, blue), and reverse (sense, red) reads only were separately analyzed, and fold changes of read counts between day 0 and day 2 are plotted in Figure 8. The locations of BES promoters are indicated as bent arrows and also as dotted black lines in the BES10, 12, 13, 15 and 17. The transcription of the active BES1 was not affected by the TbMCM-BP absence, when analyzed all reads, forward or reverse reads only. When all or reverse reads (sense) were analyzed, all 13 silent BESs showed similar trend; effect of TbMCM-BP absence was polarized to the ends, promoter and VSG regions. The derepression spread to a certain distance from the promoter or the VSG, about 10 kb, not significantly affecting genes in the middle of silent BESs. Interestingly, changes in antisense transcription (forward reads) were stronger near the promoter regions. Levels of antisense transcripts increased downstream of their BES promoters. This phenotype is more clearly depicted in those BESs with two promoters. The highest peak was observed downstream of the second promoter, but not upstream. These data suggest that RNA Pol I machinery may access to the silent promoter region better in the absence of TbMCM-BP, increasing sense transcripts near the promoter, and it may also bind downstream of the silent promoters and travel opposite direction, increasing antisense transcripts. TbMCM-BP may control VSG silencing by blocking the access of RNA Pol I to silent promoter regions.
Figure 8.
TbMCM-BP inhibits transcription, both sense and antisense, near the silent promoters. RNA-seq reads were aligned to the T. brucei Lister 427 BES sequences (14 BESs) (24) with Bowtie. All reads, forward (antisense) or reverse (sense) reads were analyzed with sliding window (1 kb bin and 0.2 kb step). Fold changes comparing day 0 and day 2 were plotted over each BES length. Diagram indicates each BES PTUs. Red boxes are genes and a VSG is situated furthest to the right. Promoters are indicated as bent arrows and five of BESs have two promoters. Location of BES Pol I promoter is also indicated with dotted lines across the graphs in the BES 10, 12, 13, 15 and 17. The length of BES8 is ∼10.5 kb. Maximum y-axis values for each BES are indicated and minimum y-axis value is 0.5 for all plots.
TbMCM-BP absence induces DNA damage but does not significantly affect overall R loop level
Replication problems can cause replication fork stalling and collapsing, which could result in DNA damage. To see whether the genome is damaged in the TbMCM-BP mutant, I examined T. brucei Replication Protein A 1 (TbRPA1). Replication Protein A is well characterized in other organisms and consists of three subunits, RPA1–3. RPA plays essential roles in DNA replication, repair, recombination and DNA damage sensing (28,29). In the absence of exogenous DNA damaging agents, replication stress is the major source of DNA damage (30). Single stranded DNA generated during damage repair invariably recruits RPA, which coats the exposed ssDNA, becomes phosphorylated, forms repair foci, and activates cell-cycle checkpoint to arrest cells at S or G2/M phase (28).
To assess DNA damage in the absence of TbMCM-BP, I tagged one copy of TbRPA1 allele at the C-terminus with 3xHA and examined its localization by immunofluorescence (IF) (Figure 9A). TbRPA1 was nuclear and exhibited punctated signals inside the nucleus in uninduced cKO cells. But at day 2 after TbMCM-BP depletion, TbRPA1 formed bright nuclear foci in some cells, suggesting that TbMCM-BP depletion accumulates DNA lesions with extended ssDNA regions. Although the TbRPA1–3xHA protein was localized in the nucleus, this tagged version is not fully functional, as I was not able to generate a tagged version in the TbRPA1Δ/+ heterozygote strain. A second tagged version, TbRPA1-GFP was also tested for localization and TbRPA1-GFP also showed nuclear localization and formed a bright nuclear focus when a DSB was induced (data not shown). Although both tagged versions were not able to fully complement the lethality of TbRPA1 null mutant (TbRPA1 is an essential protein (31) they are located in the nucleus and appear to respond to DNA damage.
Figure 9.
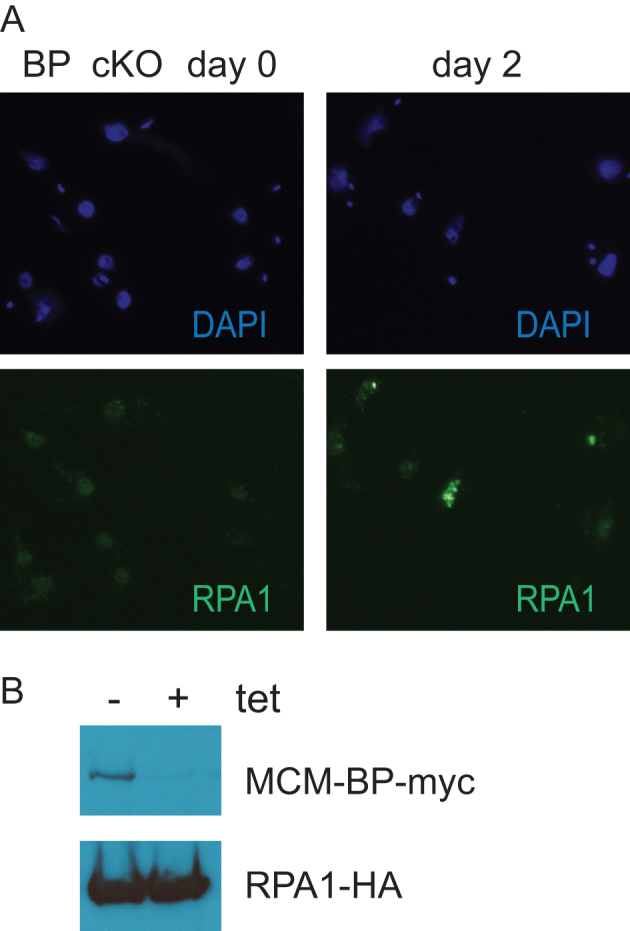
TbMCM-BP absence leads to DNA damage. (A) TbRPA1 forms nuclear foci in the absence of TbMCM-BP. TbMCM-BP cKO cells treated with tetracycline for 0 and 2 days were analyzed by immunofluorescence. Bulk DNA was stained with DAPI (blue) and TbRPA1 with rat anti-HA followed by secondary antibodies conjugated with A488 (green). (B) Level of RPA1-HA protein does not change upon TbMCM-BP removal. Western blot shows expression of TbMCM-BP-myc and TbRPA1-HA.
Transcription-replication fork collisions can also generate R-loops (DNA:RNA hybrids created as the transcriptional machinery backtracks) (32–34). These can be sensed by RPA1 (35), so I asked whether the effects I observe (deregulation at the RNA level, DNA damage) were indirect measurements of increased R-loop formation in the mutant. I therefore examined global R loop levels using serial-dilution dot-blot analysis based on the R-loop specific antibody S9.6 (Supplementary Figure S7). Using this analysis, I did not find significant changes between wildtype and TbMCM-BP cKO cells, suggesting either that R loop maintenance was not impaired in the absence of TbMCM-BP, or, that if it was impaired, it was a localized (site-specific) effect.
DISCUSSION
Formation of a pre-Replication Complex (pre-RC) occurs via stepwise recruitment of proteins to the replication origins (reviewed in (2,3)). The hexameric MCM complex (MCM2–7) is an essential replicative helicase in the pre-RC. The MCM unwinds the double strand DNA so that two strands can be separated and DNA synthesis can begin by DNA polymerases and accessary replication proteins (reviewed in (36,37)). MCM-BP is a conserved replication factor that has been found in yeasts, plants, frogs and humans as well as trypanosomes (9–11,38–41).
The ways the MCM-BP binds the MCM complex and functions are, to some extent, dissimilar between organisms. MCM-BP binds the whole complex in plant (39) but in human and yeast it replaces the MCM2 subunit and forms a complex with the rest of the subunits, MCM3–7 (9,10). TbMCM-BP is yet different: I had previously shown that in T. brucei, two MCM subunits, MCM2 and MCM3, are replaced by MCM-BP and MCM8 (11). Experiments with Xenopus egg extracts showed that at the end of S phase, MCM-BP removed the MCM complex from the DNA, which terminates replication and also blocks re-replication (40). In human (41), MCM-BP interacts with DBF4, a regulatory subunit of an early S-phase specific kinase complex DDK (DBF4-Dependent Kinase) consisting of DBF4 and CDC7 kinase (42) and partially inhibits the MCM phosphorylation by the DDK. In fission yeast (10), similar to TbMCM-BP mutant, accumulation of cells less than 1N (2C) content and fragmented nuclei was observed when MCM-BP (mcb1 in Schizosaccharomyces pombe) was overexpressed, suggesting that cells entered into mitosis and cytokinesis with damage DNA and/or incompletely replicated DNA. Collectively, given the specific function of MCM-BP in DNA replication in these model organisms, T. brucei MCM-BP may also have a specific mode of replication control mechanism. I showed here that TbMCM-BP is required for DNA synthesis likely at the replication initiation step, and especially at replication origins that are activated early in S phase. But because MFA-seq is inadequate in examining late replicating origins due to low resolution (because the peaks of early replicating regions broaden as S phase progresses), one cannot exclude the possibility that TbMCM-BP may have roles at late origins as well.
At the same time, it is quite clear that replication and transcription are intimately related in T. brucei. Previous MFA-seq experiments estimated a total of ∼100 replication origins, of which, 41 appear to be early replicating (7,18). A study using the Single Molecule Analysis of Replicated DNA (SMARD) identified a dormant origin that was activated in response to replication stress, indicating more TbORC1 sites are expected to have origin activity (43). A second, independent SMARD experiment using trypanosome DNA estimated about 170 active origins (44) and interestingly, about 14.4% of the DNA fiber analyzed had unidirectional replication forks (substantially higher origin numbers than Leishmania major (∼1.6%) and mouse embryonic fibroblast (∼4%). Head-on collisions between replication and transcription are inevitable if replication occurs bidirectionally at HT sites and collisions are possible at TTSs, therefore some of TbORC1 bound HT sites might initiate undirectional replication. So in addition to controlling DNA replication, TbMCM-BP may guide transcription machineries to properly position for start and stop, and to move in the right direction, so as to avoid conflicts between replication and transcription,
Overall the data I present, together with prior work, suggest that TbMCM-BP is required for replication and for transcription. The question remaining is how TbMCM-BP coordinates replication and transcription. Are replication problems in the TbMCM-BP mutant transformed into transcription problems? Or does TbMCM-BP control replication and transcription independently? As replication and transcription initiate in the same chromatin environment, replication defects caused by TbMCM-BP deficiency, such as replication fork stalling and/or fork collapse and the generation of DNA lesions (evidenced as RPA-bound foci, and eventually as abnormal DNA content), may change both DNA and chromatin structure, misleading transcription machineries to start or stop at wrong places. More generally, emerging literature suggests that MCM-BP is important for DNA replication, repair, and the DNA damage response in other organisms as well, and it is also associated with cancer predisposition through genome instability (45,46). It is necessary to determine whether DNA damage triggered by TbMCM-BP deficiency accumulates at specific genomic loci and whether there is any correlation between sites of DNA damage and sites with transcription problems.
The genetic analysis indicates that TbMCM-BP may bind the replication initiation sites or transcription boundaries. To examine whether TbMCM-BP binds these loci, ChIP-seq experiment were performed. However, genomic DNA was not detected in the TbMCM-BP pulled down fraction. It could be due to that TbMCM-BP complex may bind the DNA via other subunits, e.g., MCM3–8 instead of TbMCM-BP, or via some other factors. Alternatiely, TbMCM-BP association with DNA may be transient and occur only at a certain cell-cycle stage during S phase. Although it is important to determine TbMCM-BP interaction with DNA, additional methodologies with ChIP-seq need to be developed to study whether and where TbMCM-BP binds the genome of T. brucei.
Alternatively,TbMCM-BP may have direct roles in transcription control, independent from replication. We and the Sabatini lab showed previously that T. brucei H3vΔ JΔ cells derepressed the PTU borders, especially at TTSs and HTs, accumulating antisense transcripts at these sites, and also derepressed silent VSGs (15,47). These chromatin related phenotypes are highly similar to what I report for the TbMCM-BP mutant. This similarity suggests that chromatin factors might play important roles in TbMCM-BP placement or function. In a sense, the strongest evidence that TbMCM-BP has replication-independent roles in transcription comes from the effects of the mutant at the specialized PTUs termed ‘BESs’. Specifically, strong derepression was observed at BESs either near the promoter or distal from the promoter (at telomeric VSG regions). However, the VSG derepression phenotype of TbMCM-BP mutant was dependent on the presence of promoter, indicating that Pol I access to a silent promoter is required for VSG expression. However, the genes located in the middle of the BES PTU are not derepressed very much at all—suggesting either that they are not entirely unique (and there is non-unique read-loss at the bioinformatic level) or that promoters and VSG-linked telomeres are proximal in 3D space (through the folding back of telomeres (48,49)) and therefore co-regulated. Overall, these observations and the association of specific chromatin marks at each type of PTU borders suggest that chromatin structure may play key roles in the control of replication and transcription. Specific regulatory mechanisms may operate at each of these borders in addition to a central one. Further studies are necessary to understand how replication comes in to play in the TbMCM-BP-mediated transcription control at this special location. Identification and characterization of non-lethal separation-of-function TbMCM-BP alleles will be necessary to understand molecular detail of how TbMCM-BP functions in replication and transcription, and how it might coordinate these processes in the context of specific chromatin structure.
DATA AVAILABILITY
MFA-seq and stranded RNA-seq raw sequence files and processed files have been deposited to the GEO with accession number GSE118232.
Supplementary Material
ACKNOWLEDGEMENTS
I would like to thank Svetlana Mazel and Songyan Han (Rockefeller University Flow Cytometry Core) for help with cell sorting; Chingwen Yang (Rockefeller University Genomics Core) for help with HT-sequencing; Dewi Harjanto (Rockefeller University and Neon Labs) for discussions with regard to bioinformatics analysis tools; Eric Fritz for generating the stranded RNA-seq NGS library; Luisa Figueiredo (Instituto de Medicina Molecular) for the BES sequences.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Allergy and Infectious Diseases of the National Institutes of Health (NIH) [R01AI127562 to H.K.]. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1. Li Z. Regulation of the cell division cycle in Trypanosoma brucei. Eukaryot. Cell. 2012; 11:1180–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pospiech H., Grosse F., Pisani F.M.. The initiation step of eukaryotic DNA replication. Subcell. Biochem. 2010; 50:79–104. [DOI] [PubMed] [Google Scholar]
- 3. Kang S., Kang M.S., Ryu E., Myung K.. Eukaryotic DNA replication: Orchestrated action of multi-subunit protein complexes. Mutat. Res. 2018; 809:58–69. [DOI] [PubMed] [Google Scholar]
- 4. Godoy P.D., Nogueira-Junior L.A., Paes L.S., Cornejo A., Martins R.M., Silber A.M., Schenkman S., Elias M.C.. Trypanosome prereplication machinery contains a single functional orc1/cdc6 protein, which is typical of archaea. Eukaryot. Cell. 2009; 8:1592–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dang H.Q., Li Z.. The Cdc45.Mcm2-7.GINS protein complex in trypanosomes regulates DNA replication and interacts with two Orc1-like proteins in the origin recognition complex. J. Biol. Chem. 2011; 286:32424–32435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tiengwe C., Marcello L., Farr H., Gadelha C., Burchmore R., Barry J.D., Bell S.D., McCulloch R.. Identification of ORC1/CDC6-Interacting Factors in Trypanosoma brucei Reveals Critical Features of Origin Recognition Complex Architecture. PLoS One. 2012; 7:e32674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tiengwe C., Marcello L., Farr H., Dickens N., Kelly S., Swiderski M., Vaughan D., Gull K., Barry J.D., Bell S.D. et al. Genome-wide analysis reveals extensive functional interaction between DNA replication initiation and transcription in the genome of Trypanosoma brucei. Cell Rep. 2012; 2:185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tiengwe C., Marques C.A., McCulloch R.. Nuclear DNA replication initiation in kinetoplastid parasites: new insights into an ancient process. Trends Parasitol. 2014; 30:27–36. [DOI] [PubMed] [Google Scholar]
- 9. Sakwe A.M., Nguyen T., Athanasopoulos V., Shire K., Frappier L.. Identification and characterization of a novel component of the human minichromosome maintenance complex. Mol. Cell. Biol. 2007; 27:3044–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ding L., Forsburg S.L.. Schizosaccharomyces pombe Minichromosome Maintenance-binding Protein (MCM-BP) antagonizes MCM helicase. J. Biol. Chem. 2011; 286:32918–32930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim H.S., Park S.H., Gunzl A., Cross G.A.. MCM-BP is required for repression of life-cycle specific genes transcribed by RNA polymerase I in the mammalian infectious form of Trypanosoma brucei. PLoS One. 2013; 8:e57001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hirumi H., Hirumi K.. Continuous cultivation of Trypanosoma brucei bloodstream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 1989; 75:985–989. [PubMed] [Google Scholar]
- 13. Ryba T., Battaglia D., Pope B.D., Hiratani I., Gilbert D.M.. Genome-scale analysis of replication timing: from bench to bioinformatics. Nat. Protoc. 2011; 6:870–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schulz D., Zaringhalam M., Papavasiliou F.N., Kim H.-S.. Base J and H3.V regulate transcriptional termination in trypanosoma brucei. PLoS Genet. 2016; 2:e1005762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cross G.A., Kim H.S., Wickstead B.. Capturing the variant surface glycoprotein repertoire (the VSGnome) of Trypanosoma brucei Lister 427. Mol. Biochem. Parasitol. 2014; 195:59–73. [DOI] [PubMed] [Google Scholar]
- 17. Sollier J., Stork C.T., Garcia-Rubio M.L., Paulsen R.D., Aguilera A., Cimprich K.A.. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol. Cell. 2014; 56:777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Devlin R., Marques C.A., Paape D., Prorocic M., Zurita-Leal A.C., Campbell S.J., Lapsley C., Dickens N., McCulloch R.. Mapping replication dynamics in Trypanosoma brucei reveals a link with telomere transcription and antigenic variation. Elife. 2016; 5:e12765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Biebinger S., Rettenmaier S., Flaspohler J., Hartmann C., Penadiaz J., Wirtz L.E., Hotz H.R., Barry J.D., Clayton C.. The PARP promoter of Trypanosoma brucei is developmentally regulated in a chromosomal context. Nucleic Acids Res. 1996; 24:1202–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schulz D., Mugnier M.R., Paulsen E.M., Kim H.S., Chung C.W., Tough D.F., Rioja I., Prinjha R.K., Papavasiliou F.N., Debler E.W.. Bromodomain proteins contribute to maintenance of bloodstream form stage identity in the african trypanosome. PLoS Biol. 2015; 13:e1002316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dellino G.I., Cittaro D., Piccioni R., Luzi L., Banfi S., Segalla S., Cesaroni M., Mendoza-Maldonado R., Giacca M., Pelicci P.G.. Genome-wide mapping of human DNA-replication origins: levels of transcription at ORC1 sites regulate origin selection and replication timing. Genome Res. 2013; 23:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Benmerzouga I., Concepcion-Acevedo J., Kim H.S., Vandoros A.V., Cross G.A., Klingbeil M.M., Li B.. Trypanosoma brucei Orc1 is essential for nuclear DNA replication and affects both VSG silencing and VSG switching. Mol. Microbiol. 2013; 87:196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Horn D., McCulloch R.. Molecular mechanisms underlying the control of antigenic variation in African trypanosomes. Curr. Opin. Microbiol. 2010; 13:700–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hertz-Fowler C., Figueiredo L.M., Quail M.A., Becker M., Jackson A., Bason N., Brooks K., Churcher C., Fahkro S., Goodhead I. et al. Telomeric expression sites are highly conserved in Trypanosoma brucei. PLoS One. 2008; 3:e3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gunzl A., Bruderer T., Laufer G., Schimanski B., Tu L.C., Chung H.M., Lee P.T., Lee M.G.. RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryot. Cell. 2003; 2:542–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wickstead B., Ersfeld K., Gull K.. The small chromosomes of Trypanosoma brucei involved in antigenic variation are constructed around repetitive palindromes. Genome Res. 2004; 14:1014–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kolev N.G., Ramey-Butler K., Cross G.A., Ullu E., Tschudi C.. Developmental progression to infectivity in Trypanosoma brucei triggered by an RNA-binding protein. Science. 2012; 338:1352–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Allen C., Ashley A.K., Hromas R., Nickoloff J.A.. More forks on the road to replication stress recovery. J. Mol. Cell. Biol. 2011; 3:4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Broderick S., Rehmet K., Concannon C., Nasheuer H.P.. Eukaryotic single-stranded DNA binding proteins: central factors in genome stability. Subcell. Biochem. 2010; 50:143–163. [DOI] [PubMed] [Google Scholar]
- 30. Mirkin E.V., Mirkin S.M.. Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev. 2007; 71:13–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alsford S., Turner D.J., Obado S.O., Sanchez-Flores A., Glover L., Berriman M., Hertz-Fowler C., Horn D.. High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res. 2011; 21:915–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Santos-Pereira J.M., Aguilera A.. R loops: new modulators of genome dynamics and function. Nat. Rev. Genet. 2015; 16:583–597. [DOI] [PubMed] [Google Scholar]
- 33. Chen P.B., Chen H.V., Acharya D., Rando O.J., Fazzio T.G.. R loops regulate promoter-proximal chromatin architecture and cellular differentiation. Nat. Struct. Mol. Biol. 2015; 22:999–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Aguilera A., Garcia-Muse T.. R loops: from transcription byproducts to threats to genome stability. Mol. Cell. 2012; 46:115–124. [DOI] [PubMed] [Google Scholar]
- 35. Nguyen H.D., Yadav T., Giri S., Saez B., Graubert T.A., Zou L.. Functions of replication protein A as a sensor of R loops and a regulator of RNaseH1. Mol. Cell. 2017; 65:832–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Forsburg S.L. Eukaryotic MCM proteins: beyond replication initiation. Microbiol. Mol. Biol. Rev. 2004; 68:109–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bochman M.L., Schwacha A.. The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiol. Mol. Biol. Rev. 2009; 73:652–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Takahashi N., Quimbaya M., Schubert V., Lammens T., Vandepoele K., Schubert I., Matsui M., Inze D., Berx G., De Veylder L.. The MCM-binding protein ETG1 aids sister chromatid cohesion required for postreplicative homologous recombination repair. PLoS Genet. 2010; 6:e1000817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Takahashi N., Lammens T., Boudolf V., Maes S., Yoshizumi T., De Jaeger G., Witters E., Inze D., De Veylder L.. The DNA replication checkpoint aids survival of plants deficient in the novel replisome factor ETG1. EMBO J. 2008; 27:1840–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nishiyama A., Frappier L., Mechali M.. MCM-BP regulates unloading of the MCM2-7 helicase in late S phase. Genes Dev. 2011; 25:165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nguyen T., Jagannathan M., Shire K., Frappier L.. Interactions of the human MCM-BP protein with MCM complex components and Dbf4. PLoS One. 2012; 7:e35931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Larasati. Duncker B.P. Mechanisms governing DDK regulation of the initiation of DNA replication. Genes (Basel). 2016; 8:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Calderano S.G., Drosopoulos W.C., Quaresma M.M., Marques C.A., Kosiyatrakul S., McCulloch R., Schildkraut C.L., Elias M.C.. Single molecule analysis of Trypanosoma brucei DNA replication dynamics. Nucleic Acids Res. 2015; 43:2655–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stanojcic S., Sollelis L., Kuk N., Crobu L., Balard Y., Schwob E., Bastien P., Pagès M., Sterkers Y.. Single-molecule analysis of DNA replication reveals novel features in the divergent eukaryotes Leishmania and Trypanosoma brucei versus mammalian cells. Sci. Rep. 2016; 6:23142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Quimbaya M., Raspé E., Denecker G., De Craene B., Roelandt R., Declercq W., Sagaert X., De Veylder L., Berx G.. Deregulation of the replisome factor MCMBP prompts oncogenesis in colorectal carcinomas through chromosomal instability. Neoplasia. 2014; 16:694–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jagannathan M., Sakwe A.M., Nguyen T., Frappier L.. The MCM-associated protein MCM-BP is important for human nuclear morphology. J. Cell Sci. 2012; 125:133–143. [DOI] [PubMed] [Google Scholar]
- 47. Reynolds D., Hofmeister B.T., Cliffe L., Alabady M., Siegel T.N., Schmitz R.J., Sabatini R.. Histone H3 Variant Regulates RNA Polymerase II Transcription Termination and Dual Strand Transcription of siRNA Loci in Trypanosoma brucei. PLoS Genet. 2016; 12:e1005758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. de Bruin D., Zaman Z., Liberatore R.A., Ptashne M.. Telomere looping permits gene activation by a downstream UAS in yeast. Nature. 2001; 409:109–113. [DOI] [PubMed] [Google Scholar]
- 49. Poschke H., Dees M., Chang M., Amberkar S., Kaderali L., Rothstein R., Luke B.. Rif2 promotes a telomere fold-back structure through Rpd3L recruitment in budding yeast. PLoS Genet. 2012; 8:e1002960. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MFA-seq and stranded RNA-seq raw sequence files and processed files have been deposited to the GEO with accession number GSE118232.