Abstract
Dmc1 catalyzes homology search and strand exchange during meiotic recombination in budding yeast and many other organisms including humans. Here we reconstitute Dmc1 recombination in vitro using six purified proteins from budding yeast including Dmc1 and its accessory proteins RPA, Rad51, Rdh54/Tid1, Mei5-Sae3 and Hop2-Mnd1 to promote D-loop formation between ssDNA and dsDNA substrates. Each accessory protein contributed to Dmc1’s activity, with the combination of all six proteins yielding optimal activity. The ssDNA binding protein RPA plays multiple roles in stimulating Dmc1’s activity including by overcoming inhibitory effects of ssDNA secondary structure on D-loop reactions, and by elongating D-loops. In addition, we demonstrate that RPA limits inhibitory interactions of Hop2-Mnd1 and Rdh54/Tid1 that otherwise occur during assembly of Dmc1-ssDNA nucleoprotein filaments. Finally, we report interactions between the proteins employed in the biochemical reconstitution including a direct interaction between Rad51 and Dmc1 that is enhanced by Mei5-Sae3.
INTRODUCTION
During meiosis, chromosomes undergo high levels of homologous recombination. Meiotic recombination generates genetic diversity as well as the reciprocal crossover chromosomes required to promote high fidelity reductional chromosome segregation. Meiotic recombination is initiated by the transesterase Spo11 which generates double-stranded DNA breaks (DSBs) (1,2). DNA ends created by DSBs are processed by nucleases to form 3′ overhanging tracts of ssDNA. The strand exchange protein Dmc1, assembles de-novo on these ssDNA ends, via its high affinity binding site (Site I), with the help of a subset of its accessory proteins. Dmc1 then searches for homologous duplex DNA sequences in a process involving its low affinity binding site (Site II) and carries out strand exchange to form tracts of hybrid dsDNA in which the incoming Dmc1-bound ssDNA and the complementary strand of the original duplex are basepaired. The non-complementary strand is displaced as ssDNA, forming a displacement loop (D-loop).
Replication protein A (RPA) is an essential and abundant protein with no enzymatic activity. It is composed of three subunits: RPA1 (70 kDa), RPA2 (30 kDa) and RPA3 (14 kDa). RPA has four well defined DNA binding domains that cooperate to bind ssDNA tightly and selectively (relative to dsDNA) with affinity of ∼1010 M−1 (3,4). In vivo, binding of RPA to ssDNA is known to play key roles in a wide array of DNA metabolic pathways including DNA replication, nucleotide excision repair, base excision repair, mismatch repair, mitotic repair by homologous recombination, and meiotic recombination. However, the mechanisms through which RPA contributes to these pathways is not fully understood, particularly in the case of meiotic recombination (5–8).
In addition to RPA, genetic and biochemical studies have provided evidence for key accessory proteins that stimulate the activity of Dmc1 (9–18). The main accessory proteins include Rdh54 (a.k.a. Tid1), Rad51, and two heterodimers Mei5-Sae3 and Hop2-Mnd1. All these proteins bind both ssDNA and dsDNA. Rdh54/Tid1 is a dsDNA-specific, ATP hydrolysis-dependent, DNA translocase (19). Rdh54/Tid1and its paralogue Rad54 prevent sequestration of Rad51 and Dmc1 by displacing them from toxic non-recombinogenic complexes formed by direct binding to dsDNA (20–22); and the two translocases show a high degree of functional redundancy in vivo (23–25). Rad54 is thought to serve as a ‘heteroduplex pump’ that stabilizes nascent D-loops while displacing the cognate strand exchange protein from strand exchange products (26). Mei5 and Sae3 form a heterodimer that stimulates Dmc1 filament assembly on RPA-coated ssDNA (13,15). In vivo, Mei5-Sae3 is strongly required for DSB-dependent immunostaining foci formed by Dmc1 and for Dmc1-dependent D-loop formation, as assayed by 2-D gels and Southern blotting at a meiotic recombination hotspot (27). Hop2 and Mnd1 also functions as a heterodimer with a significant preference for binding dsDNA over ssDNA (10). Although Hop2-Mnd1’s in vivo function is related to that of Mei5-Sae3 in that it is strongly required for D-loop formation in vivo, Hop2-Mnd1 is unique among recombination proteins in that it binds chromosomes to form visible immunostaining foci that are completely independent of DSBs and display low levels of colocalization to the foci formed by Dmc1 (17,18). Finally, Rad51 is a structural and functional relative of Dmc1 that plays the direct catalytic role in promoting strand exchange during mitotic recombination (25,28). In addition, and particularly important for this study, Rad51 also serves as an accessory protein to Dmc1 during meiosis (11,29–31). Rad51’s strand exchange activity is inhibited during meiosis and is not required for normal levels of meiotic recombination in budding yeast (24). Rad51 is required for normal recruitment of Dmc1 to DSBs in vivo; the Dmc1 foci formed in rad51 mutants are faint compared to those in wild type nuclei (32,33). Rad51 has also been shown to be capable of enhancing Dmc1’s D-loop activity in conjunction with Mei5-Sae3 (11).
When assayed in vitro, individual accessory proteins stimulate Dmc1’s D-loop activity (10,11,15,16,34–37). However, no reconstitution of the activities of all Dmc1 accessory proteins has been reported. Here we describe a 6 protein (10 polypeptide) in vitro reconstitution system involving Dmc1 and 5 accessory proteins that stimulate its activity, RPA, Rad51, Mei5-Sae3, Hop2-Mnd1 and Rdh54/Tid1. Our findings show that RPA is particularly important for the high level of Dmc1 activity in the reconstituted system, with three distinct mechanisms contributing to RPA’s overall activity. Finally, we examine protein-protein interactions associated with D-loop activity and present evidence for a novel interaction between Dmc1 and Rad51 that is enhanced by interactions involving Mei5-Sae3.
MATERIALS AND METHODS
Protein and DNA preparation
All proteins used in this study are Saccharomyces cerevisiae proteins. RPA was expressed from p11d-sctRPA in E. coli and purified as described by Binz et al. (38). His6-tagged Dmc1, Mei5-Sae3, Rad51-II3A, Rdh54/Tid1 and Hop2-Mnd1 were expressed and purified as described previously (11,16,39) (Supplementary Figure S1). All these His6-tagged proteins have been previously shown to be functional in vivo (16,40–44). Supercoiled pRS306 and its variant plasmids used for D-loop assays were purified without DNA denaturation using triton lysis buffer and fractionated by cesium chloride density gradient banding (40). The 90C ssDNA is a 90 nt ssDNA and its sequence is homologous to the DNA sequence at position 764 - 853 in pRS306. The 90NF is a 90 nt ssDNA (sequence 5′GGCACCAACACAAAACACATCTACACTCAACAATCACTTTTTATACAACACTTCTTCTCTCACATACAACACTTCTGGCACCAACACAAA) and it was transposed from an RNA sequence that was experimentally determined to have no secondary structure above 25°C (45). The 90NF ssDNA is also predicted to have no secondary structure at 37°C by the computer programs mFold and OligoAnalyzer tool (IDT). Both 90 nt ssDNA were synthesized by IDT and purified on denaturing gels. The dsDNA version of 90NF was inserted into pRS306 replacing the sequence of 90C at exactly the same location (position 764 – 853 in pRS306) forming pNRB722 which was used as the homologous dsDNA template in D-loop assays with 90NF ssDNA. The 563 nt ssDNA used is identical to the nt's 306–868 of the Crick strand of pRS306. In order to obtain the 563 nt ssDNA, a dsDNA fragment tagged with biotin at the 5′ end of one strand was generated by PCR, then bound to Dynabeads MyOne Streptavidin (Invitrogen) following vendor's instructions. Subsequently, the strand of 563 nt ssDNA without biotin was released from the bound dsDNA using 0.4 N NaOH with incubation on ice for 2 min. The released 563 nt ssDNA was immediately neutralized upon strand separation and recovered by ethanol precipitation. The ssDNAs were 5′-32P labeled by a standard protocol and the unreacted γ-32P-ATP in the labeling reactions was removed by G25 microspin column (GE Healthcare), and residual T4 polynucleotide kinase was heat-inactivated. The 200 nt, 300 nt, and 400 nt ssDNA have sequences identical to the 3′ segments of the 563 nt ssDNA. The 200 nt ssDNA was synthesized and purified by IDT. The 300 nt and 400 nt ssDNA were isolated by the same method used for the 563 nt ssDNA. Desthiobiotin-90C ssDNA is a variant of the 90C oligo that carries a desthiobiotin moiety with a TEG spacer arm at its 5′ end, and was synthesized and purified by IDT.
Antibodies
Antibodies against purified Mei5-Sae3, Rad51-II3A, Hop2 and Rdh54/Tid1 were raised in rabbits, antibodies against purified Dmc1 were raised in a goat at the Pacific Immunology Corp. Antibodies against RPA2 (the 30 kilodaltons (kDa) subunit of RPA), raised in a rabbit, were a gift from A. Shinohara (Osaka University, Japan). Antibodies were used at 1:1000 to 1:3000 dilution in Western blotting. Arp7 anti-goat antibody was purchased from Santa Cruz Biotechnology. The Hop2 antibody cross-reacts with Mnd1, and Dmc1 with Rad51 albeit weakly. The IgG fractions from antisera were affinity purified by protein A or protein G agarose (GE Healthcare) using standard methods.
D-loop assay
Reactions (10 μl) were carried out at 37°C in D-loop buffer (25 mM Tris–HCl (pH 7.5), 1 mM DTT, 5 mM MgCl2, 3 mM ATP, 0.25 mM CaCl2 and 100 μg/ml BSA). Dmc1 functions best with both MgCl2 and CaCl2 in the reaction (16). Concentrations of proteins and their abbreviations are D = Dmc1 (3 μM), A = RPA (0.2 μM), M = Mei5-Sae3 (0.5 μM), R = Rad51-II3A (0.25 μM), H = Hop2-Mnd1 (0.2 μM), T = Rdh54/Tid1 (0.1 μM). ssDNA in the reactions for 90C and 90NF were 30 nM (2.7 μM nt); for 563 nt was 4 nM (2.2 μM nt). dsDNA was supercoiled plasmid pRS306 or pNRB722 at 5 nM (22 μM bp) for reactions using 90-mer oligos, or 2.5 nM (11 μM bp) for reactions using the 563nt oligo. Proteins in various combinations were added to reactions using either a pre-mixed or a pre-staged regimen. For the pre-mixed regimen, proteins were first mixed on ice before adding ssDNA. After DNA addition, samples were at 37°C for 5 min to allow formation of nucleoprotein filaments; dsDNA was then added and the reactions incubated for an additional 30 min to form D-loops. For the pre-staged regimen, protein mixtures were added to ssDNA and incubated for 5 min. Hop2-Mnd1 and/or Rdh54/Tid1 were then added as a mixture with dsDNA, followed by a final incubation for 30 min. Hop2-Mnd1 and/or Rdh54/Tid1 were pre-incubated with dsDNA at room temperature for 3 min before adding to nucleoprotein filaments. For both regimens, D-loop formation was terminated by deproteination with the addition of 0.5% SDS and 0.5 mg/ml proteinase K (final concentrations) and incubation was for 5 min at 37°C. 0.2 volumes of loading buffer (25 mM Tris–HCl (pH 7.5), 50% glycerol, 0.1% bromphenol blue, and 0.1% xylene cyanol) was added, and reaction products separated by electrophoresis in 0.9% agarose gel (10 cm × 14.5 cm) in TAE buffer (40 mM Tris–HCl, pH 7.5, 5 mM sodium acetate, 1 mM EDTA). Electrophoresis was at room temperature at 8 V/cm for 1 h 45 min. The gel was dried onto positively charged nylon membranes (Roche), exposed to imaging plate, analyzed using the Typhoon 9200 Imager, and quantified using the computer software Quantity One (BioRad). The intensity of both the D-loop band and the unreacted ssDNA band were imaged at the linear range. The D-loop yield was expressed as a percentage of input plasmid DNA. The mean values from three independent trials were plotted for each reaction and error bars show s.e.m.
Bead ‘catch and release’ assay for DNA binding
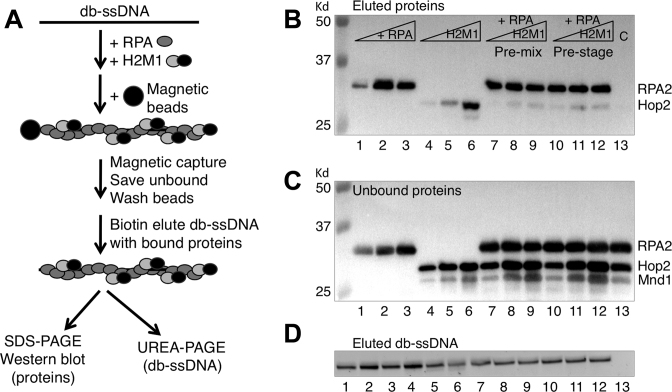
RPA and/or Hop2-Mnd1, at concentrations indicated in the legend of Figure 5, was first incubated with desthiobiotin-90C ssDNA (db-ssDNA, 70 nM) in 30 μl D-loop buffer at 37°C for 8 min to form db-ssDNA–protein complexes (Figure 5A). To capture the db-ssDNA–protein complexes, 2 μl of streptavidin magnetic beads (Roche Diagnostics) was added to the reaction. The bead mixture was incubated for 10 min each at 37°C and at room temperature on a rotator. The bead bound and unbound fractions were separated using a magnet. The beads were washed two times with 40 μl D-loop buffer containing 0.05% NP40. Protein–ssDNA complexes were then eluted from the beads with 30 μl of biotin buffer (4 mM biotin in 50 mM Tris–HCl, pH 7.5 with 50 mM NaCl) at 37°C for 13 min. The advantage of the catch and release method compared to standard bead capture assays is that db-ssDNA–protein complexes are eluted from beads to eliminate the fraction of protein bound to beads rather than DNA. Half of the eluted db-ssDNA–protein fraction (15 μl) was analyzed for protein content by 12% SDS-PAGE and western blotting. The other half of each eluted fraction was analyzed to determine the yield of eluted DNA by adding an equal volume of urea buffer (8 M urea in 50 mM Tris–HCl, pH 7.5, 2 mM EDTA), boiling for 2 min, and running on an 8% urea–PAGE, and staining the gel with SYBR-gold (Figure 5D).
Figure 5.
RPA outcompetes Hop2-Mnd1 for binding to ssDNA. (A) Scheme for bead ‘catch and release’ of protein complexes bound to ssDNA. db-ssDNA stands for desthiobiotin modified ssDNA. (B) The captured protein complexes on ssDNA were analyzed by Western blotting. RPA were 0.125, 0.25 and 0.5 μM in lanes 1–3; 0.5 μM in lanes 7–13. Hop2-Mnd1 were 0.2, 0.4 and 0.6 μM in lanes 4–6, lanes 7–9, and lanes 10–12. Lane 13 is a control that lacked db-ssDNA but contained 0.5 μM RPA and 0.4 μM Hop2-Mnd1. (C) The unbound protein complexes analyzed by western blotting. (D) The eluted db-ssDNA–protein complexes were denatured and analyzed for DNA content on 8% urea-PAGE via staining with SYBR-gold.
Ni affinity pulldown
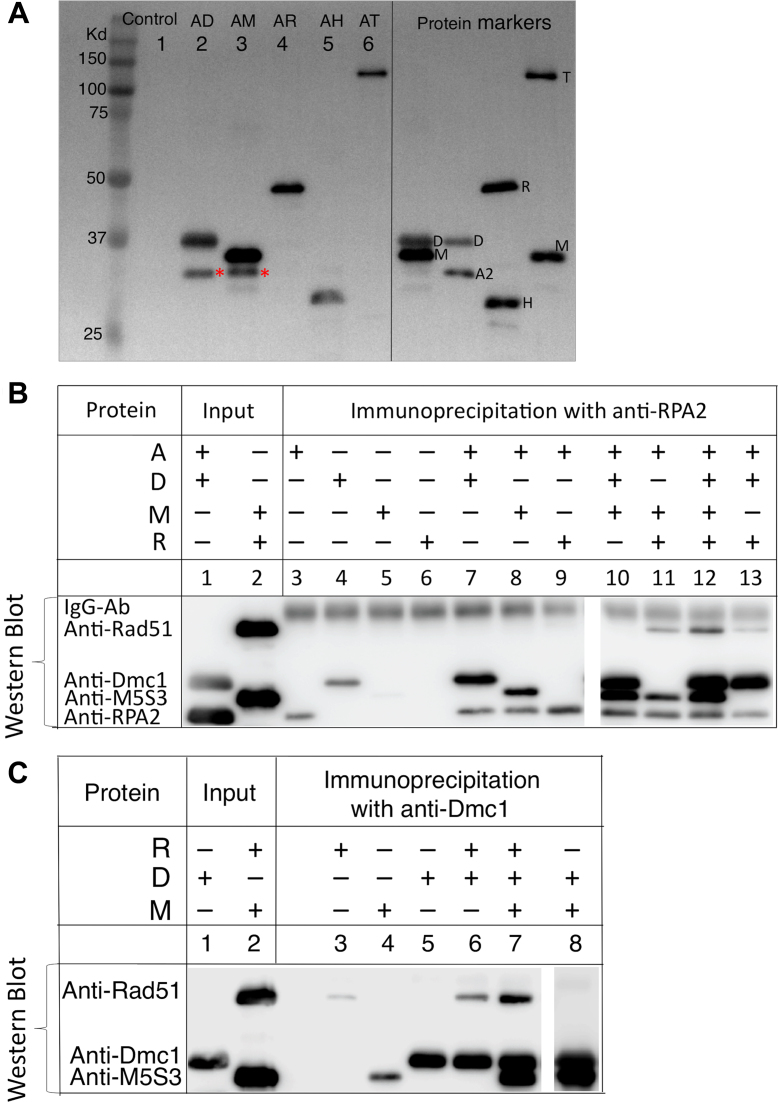
Purified Dmc1 (D, 0.5 μM), Mei5-Sae3 (M, 0.5 μM), Rad51-II3A (R, 0.25 μM), Hop2-Mnd1 (H, 0.2 μM) and Rdh54/Tid1 (T, 0.2 μM) were His6-tagged. Each of these proteins was first incubated individually with untagged RPA (A, 0.2 μM) in 20 μl D-loop buffer at 37°C for 10 min, followed by the addition of 1.7 μl Ni sepharose beads (30% slurry of Ni high performance sepharose, GE Healthcare). The mixture was then incubated at 4°C for 30 min on a rotator to capture the interacting protein complexes through the His6 tag. Beads were pelleted by centrifugation for 0.5 min at 3000 rpm and washed one time with 100 μl of wash buffer A (25 mM Tris–HCl (pH 7.5), 1 mM DTT, 1 mM MgCl2, 1 mM ATP, 50 μM CaCl2, 0.5 M NaCl, 50 mM imidazole, 100 μg/ml BSA and 0.05% NP40), and three times with 100 μl wash buffer B (wash buffer A with 0.1 M NaCl). The Ni captured proteins were eluted by boiling in 20 μl 1.7%SDS and 0.1 M DTT solution, separated on 12% SDS-PAGE, and analyzed by western blotting.
Co-immunoprecipitation
Physical interaction between RPA (0.2 μM) and Dmc1 (0.5 μM), RPA and Mei5-Sae3 (0.5 μM), or RPA and Rad51-II3A (0.25 μM) was tested by co-immunoprecipitation assay. Proteins were mixed (as indicated in Figure 6B, C) in a 20 μl reaction containing D-loop buffer and incubated at 37°C for 10 min. Purified antibodies (0.5 μl) against either the RPA2 or Dmc1 were added as indicated and the mixtures were incubated at 4°C for 1 h on a rotator. Equal volumes of 0.5 μl magnetic-conjugated protein G (Dynabeads protein G, Invitrogen) and magnetic-conjugated protein A (NEB) were added to the mixture, and further incubated at 4°C for 1 h. The immunoprecipited complexes immobilized on magnetic beads were then washed with 100 μl of wash buffer (25 mM Tris–HCl (pH 7.5), 1 mM DTT, 1 mM MgCl2, 1 mM ATP, 50 μM CaCl2 and 100 μg/ml BSA, 0.1 M NaCl and 0.05% NP40) four times. The bead-bound protein was eluted by boiling in 40 μl 1.7% SDS and 0.1 M DTT denaturing buffer, and a 20 μl eluate was loaded onto 12% SDS-PAGE for analysis. Proteins were detected by Western blot using antibodies against all proteins in the reactions. Three independent trials were done for each reaction.
Figure 6.
Protein-protein interactions. (A) Detection of direct protein-protein interactions by His6-affinity pulldowns: purified Dmc1 (D), Mei5-Sae3 (M), Rad51-II3A (R), Hop2-Mnd1 (H), and Rdh54/Tid1 (T) were His6-tagged. Each of these proteins was first incubated individually with untagged RPA, followed by the addition of Ni sepharose beads to capture the interacting protein complex through the His6-tag. The Ni captured complexes were eluted with a solution containing 1.7% SDS and 0.1 M DTT and separated on 12% SDS-PAGE, further analyzed by Western blotting using antibodies against all the proteins involved in the analysis for detection. An asterisk indicates the position of RPA2 (A2) pulled down by Dmc1 and Mei5-Sae3. (B) Detection of direct protein-protein interactions by in vitro co-immunoprecipitation of anti-RPA2 antibody with purified proteins as indicated. (C) Detection of direct protein-protein interactions by in vitro co-immunoprecipitation with anti-Dmc1 antibody. A mixture of antibodies against all proteins was used for protein detection.
DNA binding assay by fluorescence polarization
The fluorescence polarization assays were in D-loop buffer and performed essentially as described previously (46).The 84 mer ssDNA with 5′ Alexa Fluor-488 (5′GGTAGCGGTTGGGTGAGTGGTGGGGAGGGTCGGGAGGTGGCGTAGAAACATGATAGGAATGTGAATGAATGAAGTACAAGTAAA) was synthesized by IDT. The Alexa Fluor-488 labeled 162 bp double-hairpin linear duplex DNA was prepared as detailed previously (47). The protein concentrations were as indicated in the legends to the figures.
Quantitative Western analysis of protein levels in vivo
200 ml cultures of SK-1 strain DKB1772 (ho::hisG/’, HIS4::LEU2-(NBam)/his4X::LEU2-(NBam)-URA3, leu2::hisG/’, ura3 (DPst-Sma)/’) were prepared for liquid sporulation and sporulation/meiosis was induced using a previously published method (32). Yeast whole cell protein extracts prepared by the TCA method from three independent meiotic cultures by taking culture aliquots at 0, 2, 3, 4, 5 and 7 h following induction of meiosis. Antibodies used were as described above. Because steady state levels of Arp7 do not change during meiosis, it was used as an internal loading control to normalize band intensity across all time points. A dilution series of purified target protein of known amount was used to construct a standard curve, and the amount of yeast proteins present at different meiotic time points calculated by linear regression. For calculations, we used the median budding yeast diploid cell volume of 86 × 10−15 l, and nuclear volume at 7% of the cell volume (48,49).
RESULTS
In vitro reconstitution of Dmc1 dependent recombination with five accessory proteins
In order to reconstitute Dmc1 mediated recombination in vitro, we developed a system in which the influence of 5 accessory proteins on Dmc1’s activity was determined. These proteins included RPA (abbreviated ‘A’ in all figures), Rad51-II3A (‘R’), Rdh54/Tid1 (‘T’) and two heterodimers Mei5-Sae3 (‘M’) and Hop2-Mnd1 (‘H’). Rad51-II3A was used in place of the wild type Rad51 protein to ensure that all D-loop activity observed under the various reaction conditions was contributed by Dmc1 and not Rad51. Rad51-II3A retains DNA binding but not strand exchange activity (11). Rad51 and Rad51-II3A stimulate Dmc1’s D-loop activity in cooperation with Mei5-Sae3. The final concentrations of proteins in our reconstitution experiments were arrived at by titration by varying the concentration of one protein, but keeping all other five proteins constant (Supplementary Figure S1). The protein concentration range explored for each protein in the titrations was informed by results of previous studies (11,16,35). The concentration titration range for RPA and Dmc1 was above its theoretical saturation value on ssDNA which was estimated using RPA’s binding site size of 25 nucleotides and Dmc1’s binding site size of three nucleotides per protomer. The optimal concentrations of proteins used in the experiments described below are listed in MATERIALS AND METHODS and in Table 1 below.
Table 1.
Concentration of proteins in biochemical reconstitution and in vivo
| Protein | Concentration in reconstitution (μM) | In vitro ratio relative to Dmc1 | Meiosis peak hour | Molecules x 104 Per cell | Concentration per cell (μM) | Concentration per nucleus (μM) | In vivo nuclear ratio relative to Dmc1 |
|---|---|---|---|---|---|---|---|
| Dmc1 | 3 | = 1.0 | 4 | 8.9 ± 1.4 | 1.74 | 24.9 | = 1.0 |
| Rad51 | 0.25 | 0.08 | 4 | 1.8 ± 0.9 | 0.36 | 5.1 | 0.2 |
| Mei5 | 0.5 | 0.17 | 4 | 5.5 ± 2.2 | 1.08 | 15.4 | 0.6 |
| Rdh54/Tid1 | 0.1 | 0.03 | 5 | 0.92 ± 0.4 | 0.18 | 2.6 | 0.1 |
| RPA | 0.2 | 0.07 | 4 | 39.2 ± 3.0 | 7.70 | 110.0 | 4.4 |
| Hop2 | 0.2 | 0.07 | 4 | 24.7 ± 2.0 | 4.84 | 69.1 | 2.8 |
For the heterodimers, only one protein concentration in the pair was determined in vivo. For the values of RPA, it is assumed that all proteins are completely localized to the nucleus.
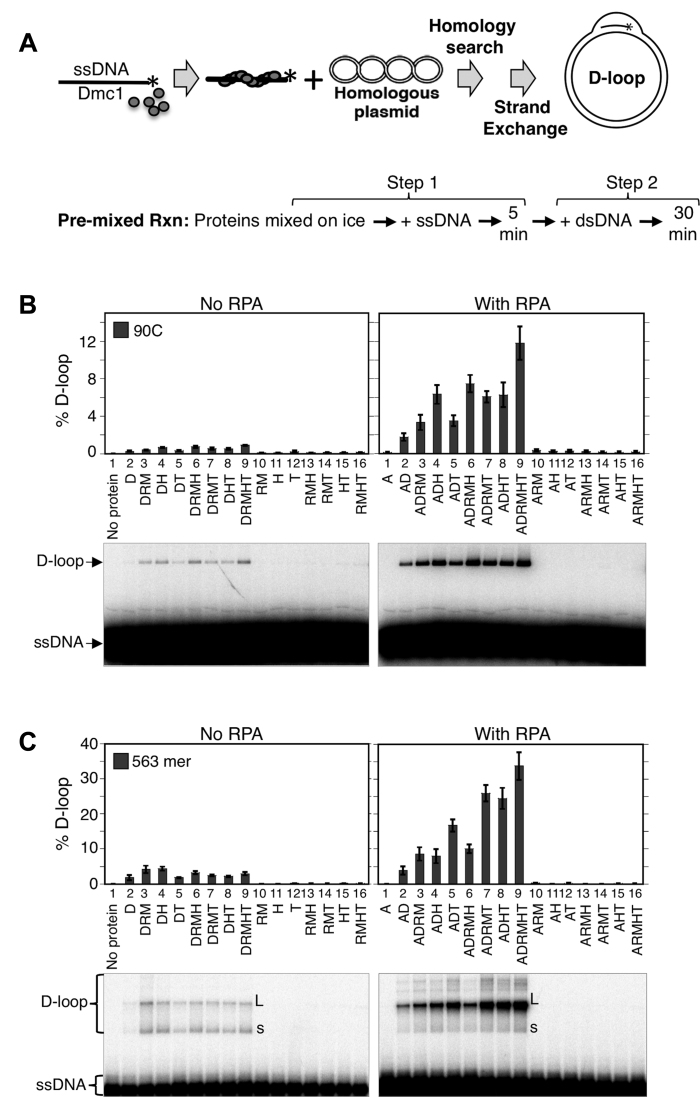
After determining the optimum concentration at which each of the five accessory proteins stimulated Dmc1, we carried out D-loop reactions in which different combinations of the proteins were mixed on ice prior to addition of the ssDNA substrate (Figure 1A). D-loop reactions were carried out using a variant of the standard two step method in which ssDNA is added to the strand exchange protein, incubated to allow nucleoprotein filament formation in Step 1, and the supercoiled dsDNA substrate then added to initiate D-loop formation in Step 2. In this case, Dmc1 was added to the ssDNA either alone or as a mixture with accessory proteins. Proteins were premixed rather than added sequentially to ssDNA, as was done in our previous studies (11,16). This regimen was used to better mimic in vivo conditions in which formation of ssDNA initiates assembly of recombination complexes (see legend of Figure 1). Using the ‘pre-mixed’ version of the 2-step protocol, we determined D-loop formation using a 90-mer ssDNA oligonucleotide (designated 90C) as ssDNA substrate and a 4.4 kb supercoiled plasmid carrying a sequence identical to 90C (Figure 1B). During the initial analysis of the data it became clear that RPA had a particularly strong stimulatory effect. We therefore present the data from these experiments as a comparison between a set of mixtures that do not contain RPA to a corresponding set of mixtures that do. We found that (1) D-loop formation is completely Dmc1-dependent regardless of whether or not RPA was present (lanes 10 - 16), confirming that none of the other proteins, including Rad51-II3A, contribute significant D-loop activity under any of the conditions analyzed. (2) In the absence of RPA, the yield of D-loops was low (<0.9%) even in the presence of all the other accessory proteins (left panel, lanes 2–9). When RPA was included in the same set of reactions, D-loop yields were ∼10-fold higher than the corresponding reactions lacking RPA (right panel, lanes 2–9). Importantly, omitting any one of the proteins reduced D-loop yield indicating that each protein in the reaction contributes to the efficiency of D-loop formation.
Figure 1.
In vitro reconstitution of Dmc1-dependent recombination with five accessory proteins. (A) Scheme for the 2-step D-loop assay with the pre-mixed reaction regimen. For the first step, reaction components, including pre-mixed proteins, were incubated with ssDNA at 37°C for 5 min to make Dmc1 filaments. The second step is initiated by addition of target dsDNA followed by incubation for 30 min to form D-loops. (B) D-loop assay reactions using the 90C ssDNA (30 nM or 2.7 μM nt) and various mixtures of proteins. Abbreviations for proteins and their concentrations in the reactions: D = Dmc1 (3 μM), A = RPA (0.2 μM), M = Mei5-Sae3 (0.5 μM), R = Rad51-II3A (0.25 μM), H = Hop2-Mnd1 (0.2 μM), T = Rdh54/Tid1 (0.1 μM); dsDNA was supercoiled plasmid pRS306 (5 nM or 22 μM bp). (C) D-loop assay reactions using the 563 nt ssDNA (4 nM or 2.2 μM nt) and various combinations of accessory proteins at the same concentrations as in B, dsDNA was at 2.5 nM (11 μM bp). ‘L’ stands for position of the largest D-loops detected, and ‘s’ for position of the smallest D-loops. The D-loop yield was expressed as a percentage of input plasmid DNA. The gel images from D-loop assays were shown and data were quantified and graphed (n = 3, ± s.e.m.). [NB: Pre-mixing results in lower levels of D-loop activity than observed when proteins are added sequentially to ssDNA, accounting for the difference in D-loop yields seen here as compared to our previously published papers (11,16)].
To determine if the results obtained with the 90-mer ssDNA substrate would hold for a longer substrate, one similar in length to that of ssDNA tracts that form in the cell (50), we repeated analysis of the activity of protein mixtures using a 563-mer ssDNA (Figure 1C). The results we obtained with the 563-mer were similar to those obtained with the 90C. RPA greatly stimulated D-loop yields for the 563-mer regardless of the combination of other accessory proteins present; additive effects of Rad51+Mei5-Sae3, Hop2-Mnd1, or Rdh54/Tid1 were observed in reactions containing RPA (compare Figure 1C to Figure 1B). As for 90C, the 563-mer substrate yielded the most D-loops when all proteins were present in the reaction with a yield of about 34% (Figure 1C, right panel, lane 9). Interestingly, reactions that employed the 563-mer without RPA differed from those that employed the 90-mer without RPA in that, D-loop yields with the 563-mer were not enhanced by combining accessory proteins. When RPA was omitted from reactions, the highest yields from the 563-mer were obtained by adding either Rad51+Mei5-Sae3 alone or Hop2-Mnd1 alone. This indicates that in the absence of RPA, the activities of the other accessory proteins conflict with one another. Addition of RPA to the reactions with the 563-mer not only increased yields of all reactions, but also created a situation in which each accessory protein was able to contribute to the overall yield of the reaction, such that reactions containing all 6 proteins generated the highest yield. Note that we standardly added Rad51 and Mei5-Sae3 together because our previous work showed they function cooperatively (11). However, additional experiments were carried out to show that Rad51-II3A and Mei5-Sae3 are individually required for optimal yield of the ‘complete’ reaction (Supplementary Figure S2).
We note that the yield of D-loops from reactions containing all 6 proteins was roughly 3-fold higher for the 563-mer compared to the 90-mer. The relatively high yield of D-loops observed with the 563-mer was largely dependent on Rdh54/Tid1; reactions containing Rdh54/Tid1 yielded 3- to 4-fold more D-loops with the 563-mer as compared to the 90-mer (compare Figure 1C to Figure 1B, right panels, lanes 5 to 2, 7 to 3, 8 to 4 and 9 to 6). We note that the mechanism underlying this difference remains to be determined. One possibility, based on previous observations with Rdh54’s paralogue Rad54 (26), is that Rdh54 may have both D-loop forming and D-loop dissociation activities with the D-loop dissociation activity being more predominant for shorter substrates.
Another notable observation obtained for the 563-mer was that, when RPA was present, the mobility of the predominant D-loop species was lower than that observed when RPA was absent; with RPA most D-loop migrated at the position of open circles (L), while without RPA a wider range of D-loop sizes, including a band corresponding to the rapidly migrating super-coiled pRS306 (s) was observed (Figure 1C, lanes 2–9, compare both panels). These results imply that RPA increases the average size of D-loops. This interpretation is supported by the known impact of D-loop formation on negatively supercoiled plasmids; one negative supercoil is expected to be lost per 10.5 bp hybrid DNA formed in a D-loop. Given that negative supercoiling is required to drive D-loop formation in this system (40), it is expected that the longest D-loops that formed will be those in which all energy stored in negative supercoils is expended and the open-circle configuration is achieved. Again, consistent with this interpretation, L migrates at the position of open circles of pRS306. The average number of negative supercoils for a 4.4 kb plasmid grown in Escherichia coli is 29/plasmid (supercoil density σ = –0.07 sc/turn) and that 10.5 bp of hybrid DNA is expected to form for each supercoil unwound, the longest stable D-loop length following protein removal is expected to be 305 bp, i.e. it is expected that just over half of the 563-mer is incorporated into D-loops in the deproteinized samples. To confirm this expectation, we examined the mobility of D-loop species formed by 90, 200, 300, 400 and 563 nt substrates (Supplementary Figure S3). As predicted from the size and expected super-helicity of the 4.4 kb plasmid, the 300 and 400 nt substrates yield D-loops with the same mobility as that obtained with the 563-mer substrate: a single predominate species with a mobility equivalent to that of open circles was observed for both the 300 and 400 bp species. In contrast, a 200 nt ssDNA substrate yielded a variety of D-loop species of greater mobility than that of the open circle form. The 90C ssDNA yielded D-loops which migrated at the same position as the pure negative supercoiled pRS306 plasmid suggesting that the reduction in supercoiling associated with formation of D-loops by the 90-mer substrate is not detected in this gel system. These observations provide support for our conclusion that the slow running D-loop species observed with the 563-mer (L) represents the maximum length D-loop that can be observed in this system. The results also confirm that RPA increases the averages size of D-loops observed in this system.
To further characterize the mechanism of stimulation of Dmc1 by the accessory proteins, we carried out kinetic analysis on the 563-mer ssDNA substrate to obtain apparent endpoints and rates of approach to those endpoints for D-loops formation in the presence of Dmc1 and RPA. We find that Rad51+Mei5-Sae3, Rdh54-Tid1 and Hop2-Mnd1 increase both the rate and extent of reaction (Supplementary Figure S4). The results also show that all of the reactions examined are nearing their endpoints by 30 min, the time used to report yields in all other D-loop experiments reported in this paper (Supplementary Figure S5). The results of this kinetic analysis are consistent with the possibility that addition of each protein increases the fraction of ssDNA–protein complexes that is capable of forming stable D-loops. However, the results do not exclude alternative, or more complex, kinetic models.
We also examined the species specificity of RPA in reconstitution reactions under two D-loop assay regimens (Supplementary Figure S6). We found that human RPA (hRPA) could not fully substitute for budding yeast RPA (ScRPA). The hRPA supported formation of only about 10% the level of D-loops seen for ScRPA in the pre-mixed regimen, and only about 30% of the level in the pre-staged regimen. Note that we showed the hRPA we used in our assay binds ssDNA efficiently (the calculated saturation of 2.7 μM nt 90C ssDNA by hRPA is 100 nM). This result suggests that species-specific protein–protein interactions between ScRPA and one or more other yeast proteins is/are required for optimal stimulation of D-loop formation by ScRPA.
Removal of secondary structures from ssDNA by RPA enhances the D-loop forming activity of Dmc1
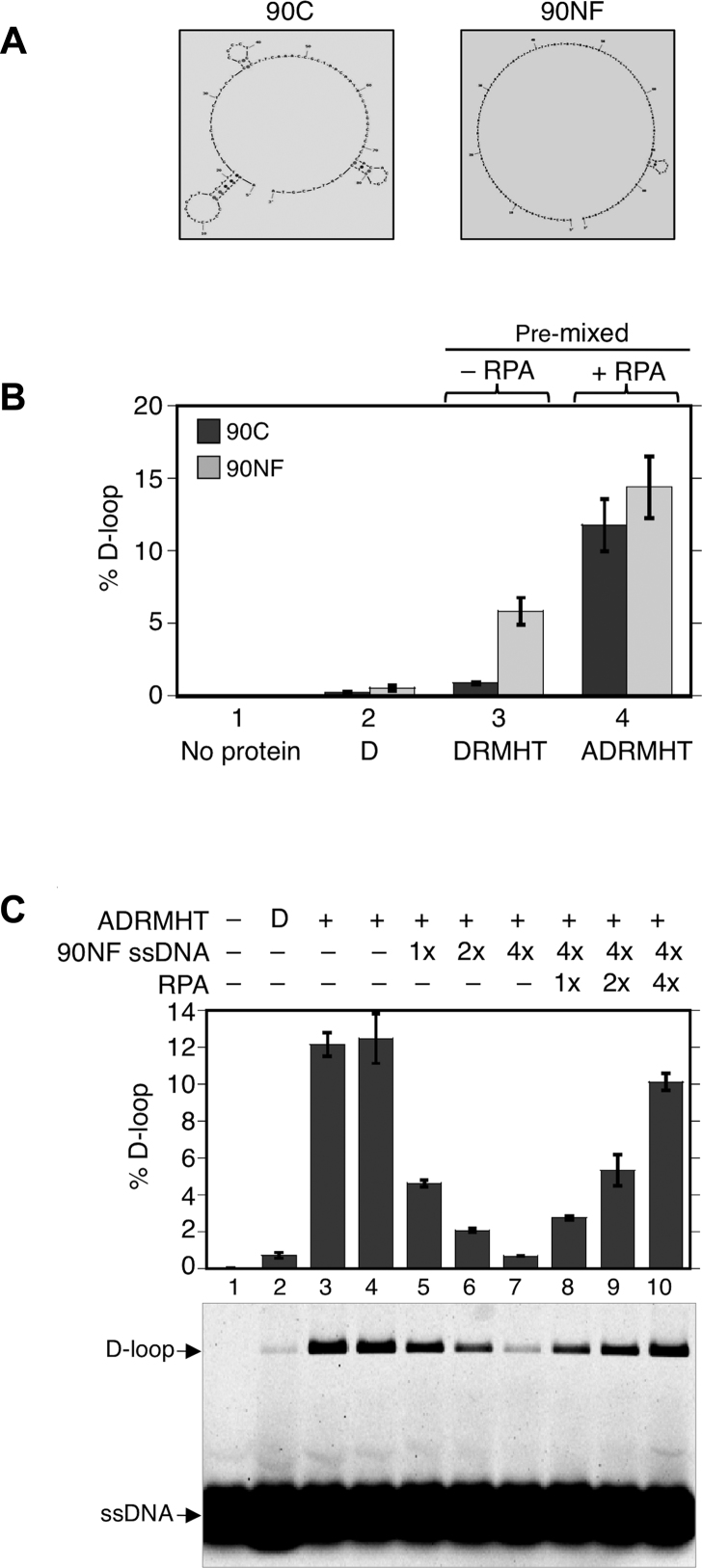
To determine if the overall stimulation of reactions by RPA involved previously described mechanisms through which RPA, or its prokaryotic functional homolog SSB, stimulates strand exchange reactions. One well known stimulatory activity of RPA, and bacterial SSB, is to remove the secondary structure from ssDNA substrates that otherwise limits the activity of strand exchange proteins (51,52). We therefore asked if secondary structure was limiting D-loop yield in the absence of RPA, and if the stimulatory activity of RPA could be fully accounted for as an effect of eliminating secondary structure. To answer these questions, we compared the results obtained using the 90C oligonucleotide, which is predicted to have substantial secondary structure at 37°C, to the results obtained using another 90-mer, designated 90NF (for ‘not folded’), which is predicted to lack secondary structure under the same conditions (Figure 2A). To confirm that 90C has more secondary structure than 90NF, we ran the two oligos under both denaturing and non-denaturing conditions. We found that while the two oligos have near identical mobilities on denaturing gel, 90C runs faster on native gel than 90NF (Supplementary Figure S7). In addition, only 90C was stained by ethidium bromide which intercalates at helical folds. These results show that 90C contains regions of secondary structure that 90NF lacks. Using these two oligos, we found that, in the absence of RPA, the D-loop yield was 6-fold higher for 90NF as compared to 90C when all proteins except RPA were added to reactions (Figure 2B, lane 3). These findings suggest that secondary structure of 90C limits the yield of D-loops in the absence of RPA. Importantly, although 90NF is less dependent on RPA than 90C for generation of D-loops; addition of RPA enhances the level of D-loops of 90NF by 2.5-fold from 6 to 15% (Figure 2B, compare lanes 3 and 4), indicating that RPA’s stimulatory activity is not limited to overcoming inhibitory effects of secondary structure. These results indicate that some, but not all, of RPA’s stimulatory activity on 90C results from eliminating the inhibitory effects of secondary structure.
Figure 2.
Effects of RPA on secondary structure and inhibitory binding of non-homologous ssDNA to Dmc1-Site II. (A) Cartoons of predicted secondary structures of 90C ssDNA (significant secondary structure) and 90NF ssDNA (no secondary structure at 37°C). [NB: the cartoon for 90NF was produced by a computer program using 25°C rather then 37°C as the input temperature because the program does not produce an output image if no secondary structure is detected. The small stem loop shown in the cartoon is not predicted to form at 37°C.]. (B) D-loop formation by Dmc1 was examined using 90C ssDNA (30 nM or 2.7 μM nt) and 90NF ssDNA (30 nM or 2.7 μM nt). The pre-mixed regimen was used. Abbreviations and concentrations were as in Figure 1. (C) Competition of RPA with Dmc1-Site II for ssDNA binding. In the pre-mixed regimen, all six proteins (ADRMHT) were first added to 90C ssDNA (1× = 30 nM or 2.7 μM nt) to make Dmc1-ssDNA filaments. After filament formation, increasing amounts of non-homologous ssDNA (90NF, 1x = 30 nM or 2.7 μM nt) were added to reactions 5–10 as indicated to allow binding to the secondary Site II in Dmc1-ssDNA filaments. RPA (1× = 0.2 μM) was then added in increasing amounts to reactions 8–10 before the addition of plasmid pRS306 to each sample to initiate D-loop formation (n = 3, ± s.e.m.).
RPA suppresses inhibition of D-loop formation by heterologous DNA
In addition to stimulating recombination reactions by removing secondary ssDNA structures, RPA was previously implicated in enhancing strand exchange reactions by binding to the displaced strand of D-loops thereby stabilizing them (53,54). Although it is technically difficult to directly demonstrate that RPA binds the displaced strand, an indirect approach was used previously to provide evidence for this activity. The secondary low affinity DNA binding site (Site II) of E. coli RecA is known to bind the displaced (a.k.a. ‘outgoing’) ssDNA strand briefly before it is released from the interior of the filament following strand exchange. The indirect approach used a non-homologous ssDNA to inhibit strand exchange by binding to Site II. Subsequent addition of single strand binding protein to the inhibited reaction competes for the heterologous ssDNA and restores D-loop activity (53). In our reconstitution system, 90C ssDNA produced ∼12% D-loops at 0.2 μM RPA (Figure 2C, lanes 3 and 4). Addition of a 4-fold excess of a non-homologous 90NF ssDNA almost completely inhibited D-loop formation (lane 7). This inhibition was efficiently relieved when additional RPA was added to reaction mixtures; D-loops yields were restored to up to 80% of those observed without heterologous ssDNA addition (lanes 8–10). Our preferred interpretation of these results is as follows. First, the efficient inhibition of D-loop formation by heterologous ssDNA is very likely to occur by the binding of the heterologous ssDNA to the low affinity binding sites (Site II) within Dmc1-ssDNA filaments. If this is the correct interpretation, the ability of RPA to suppress inhibition is easily explained; RPA simply outcompetes the lower affinity Dmc1 Site II for binding to the heterologous ssDNA. We note that we cannot strictly eliminate a possible alternative explanation; it is possible that addition of heterologous ssDNA causes the accessory protein complexes required for efficient D-loop formation to be redistributed to heterologous ssDNA molecules, thereby reducing the yield of D-loops. However, this alternative explanation for inhibition does not explain why addition of RPA reverses the inhibition, because here is no obvious mechanism through which RPA would restore the normal level of accessory protein complexes on the minority population of homologous ssDNA molecules. Nonetheless, it could still be argued that one or more accessory protein may bind the additional ssDNA while also remaining bound to the original Dmc1 filament. Thus, the notion that RPA enhances D-loop length by binding the displaced D-loop strand should be considered a model rather than a proven mechanism. If RPA does reverse inhibition by heterologous ssDNA as a consequence of competing with Dmc1-Site II, this suggests a mechanism through which RPA increases D-loop size; RPA may bind the displaced strand during D-loop formation forming a complex that drives D-loop elongation via polymerization.
Reconstituted Dmc1 reactions lacking RPA are more efficient when Hop2-Mnd1 and Rdh54/Tid1 are added late to reactions
Although the results presented above provide evidence for two mechanisms through which RPA enhances Dmc1’s D-loop activity in reconstituted reactions, neither of these mechanisms explains how RPA enables the cooperation of accessory proteins seen in the pre-mixed protocol described above. Previous in vivo studies suggest that the Dmc1 stimulatory activities of Rdh54/Tid1 and Hop2-Mnd1 are likely to involve mechanisms acting after the assembly of Dmc1 filaments; Dmc1 focus formation does not depend on Hop2-Mnd1 or Rdh54/Tid1 in vivo. Furthermore, previous studies showed that Hop2-Mnd1’s stimulation of Dmc1 activity is optimal only when the protein is added to reactions late, along with the dsDNA template (Figure 3A) (10,16,55), suggesting that its presence during Dmc1 filament assembly may have an inhibitory effect. To determine if Rdh54/Tid1 displays similar sensitivity to order of addition, we carried out a set of reactions in which Rdh54/Tid1 was added at different stages (Figure 3B). As for Hop2-Mnd1, Rdh54/Tid1 stimulatory activity was 16-fold greater if the protein was added at the second step of the 2-step D-loop protocol, along with the dsDNA target plasmid, as compared to when it was added at the first step, in a mixture with Dmc1, to ssDNA (Figure 3B; compare reactions 1 and 2). The relatively low yields of D-loops formed when Hop2-Mnd1 or Rdh54/Tid1 was added at the first step are likely to reflect inhibitory interactions of Hop2-Mnd1 or Rdh54/Tid1 with ssDNA, and/or binding to secondary structures formed by folding of ssDNA. If this were the case, such an inhibitory interaction might limit the ability of Hop2-Mnd1, or Rdh54/Tid1, to cooperate with each other, or with the other accessory factors, for stimulation of Dmc1’s activity. To test this, we delayed the time of addition of Hop2-Mnd1, or Rdh54/Tid1, to protein mixtures containing Dmc1 and Rad51+Mei5-Sae3, but lacking RPA. Using this ‘pre-staged’ regimen, we found that delayed addition of Hop2-Mnd1 or Rdh54/Tid1 increased the yield of D-loops 4.4-fold from about 1.4% to ∼6% (Figure 3C). This result indicates that, in the absence of RPA, early addition of Hop2-Mnd1 or Tid1 results in less efficient stimulation than occurs when proteins are added with the dsDNA substrate at the second step of the two-step D-loop protocol.
Figure 3.
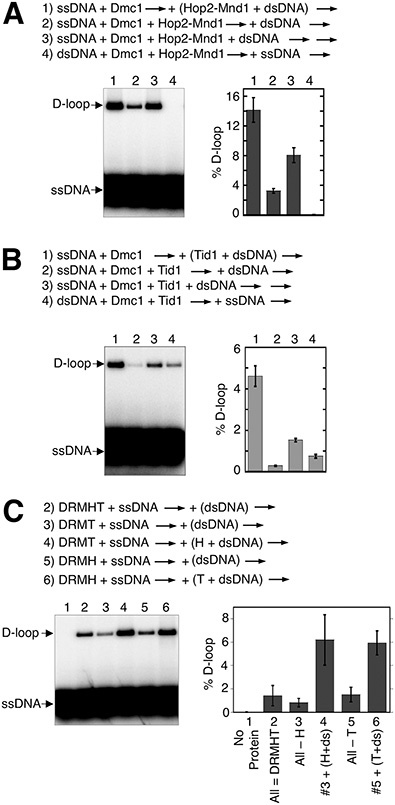
Dmc1 reactions lacking RPA are more efficient when Hop2-Mnd1 and Rdh54/Tid1 are added late to reactions. (A) The order of addition of reaction components is as indicated. The concentrations of reactants are as detailed in Figure 1 and in Materials and Methods. The first arrow indicates incubation was at 37°C for 10 min followed by the second arrow, which indicates an additional incubation at 37°C for 15 min. Hop2-Mnd1 and dsDNA in reaction 1 were incubated at room temperature for 3 min prior to adding to Dmc1-ssDNA filaments. The gel images from D-loop assays are shown and results graphed (n = 3, ± s.e.m.). Data shown here was published previously (16) and are presented for comparison to B with permission [NB: permission pending]. (B) The same experimental procedure as in A except Rdh54/Tid1 was used in place of Hop2-Mnd1. (C) Deletion and add-back reactions in the pre-mixed regimen of Hop2-Mnd1 or Rdh54/Tid1 in the absence of RPA. The order of addition of components were as indicated. Abbreviations for proteins and protein and DNA concentration are as in Figure 1.
Adding Hop2-Mnd1 and Rdh54/Tid1 at the second step of the D-loop assay reduces the impact of RPA on D-loop yield
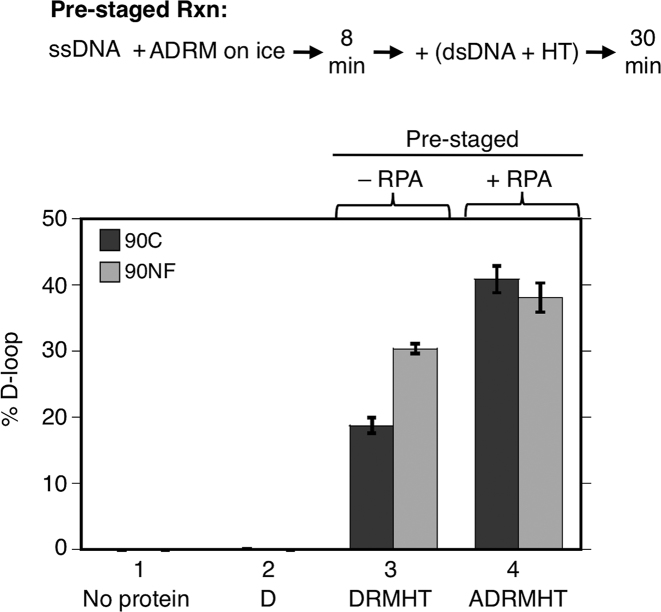
If pre-staging the order of protein addition in reconstitution reactions avoids inhibitory interactions between Hop2-Mnd1 and ssDNA and/or between Rdh54/Tid1 and ssDNA, during Dmc1 filament formation, and if RPA reduces or eliminates such inhibitory interactions in the pre-mixed protocol, then RPA’s stimulatory activity is predicted to be less pronounced in the pre-staged protocol as compared to the pre-mixed protocol. To test this prediction, we compared the relative impact of adding RPA to pre-mixed vs. pre-staged D-loop reactions using both 90C and 90NF. As described above (Figure 2B), RPA improved yields in the pre-mix protocol by 13-fold stimulation for 90C and 2.5-fold for 90NF. The amount of stimulation by RPA in the pre-staged reaction was less dramatic than that in the pre-mixed reaction; addition of RPA increased D-loop yield 2-fold for 90C and about 1.3-fold for 90NF (Figure 4, lanes 3 and 4). The results support the conclusion that a substantial amount of RPA’s stimulatory activity in the pre-mixed regimen involves eliminating inhibitory interactions of Hop2-Mnd1 and Rdh54/Tid1 during pre-synaptic filament formation. On the other hand, our results clearly show that addition of RPA increased the yield of D-loops in the pre-staged regimen even for 90NF; therefore, reducing inhibitory interactions during Step 1 and eliminating inhibitory effects of secondary structure, does not fully account for the stimulatory activity of RPA. The residual stimulatory activity seen in the pre-staged reactions for 90NF may be accounted for by binding of RPA to the displaced strand of nascent D-loops as described above.
Figure 4.
Effect of RPA in pre-staged reactions. D-loop formation by Dmc1 and its accessory proteins was examined using 90C ssDNA and 90NF ssDNA (30 nM or 2.7 μM nt). Reactions were with or without RPA as indicated. Abbreviations and reaction component concentrations were as in Figure 1 (n = 3, ±s.e.m.).
We also considered an alternative explanation for the difference in the efficiency of the pre-mixed versus the pre-staged regimens. Rdh54/Tid1 is unstable at 37°C as is its paralogue Rad54 (56). If Rdh54/Tid1’s activity contributes to D-loop yield during Step 2, and the protein's activity is lost during Step 1, this could account for why the pre-staged protocol is relatively efficient. To test this, we set up parallel reactions in which the only difference was the time of incubation of Rdh54/Tid1 prior to addition of the dsDNA substrate. We found that adding Rdh54/Tid1 5 min prior to addition of the dsDNA plasmid reduced yields ∼10% compared to yields obtained when the translocase was added at the same time as the dsDNA (Supplementary Figure S8). A longer, 40 min incubation of Rdh54/Tid1 prior to dsDNA addition reduced yields about 50%. These results indicate that, although Rdh54/Tid1 is unstable as expected, the loss of activity associated with 5 min incubations, such as those used in Step 1 in our standard pre-mixed protocol, does not account for degree of difference between the efficiency of the pre-mix and pre-stage regimens. The data are more consistent with a model in which conflicts between accessory proteins are the predominant cause of the relative inefficiency of the pre-mixed protocol.
RPA outcompetes Hop2-Mnd1 binding to ssDNA
The finding that early addition of Hop2-Mnd1, or Rdh54/Tid1, dramatically limits the yield of D-loops in the absence of RPA, raised the obvious possibility that the mechanism through which RPA promotes cooperation of Hop2-Mnd1 and Rdh54/Tid1 involves competition of RPA with the accessory proteins for binding to ssDNA. RPA is well known to bind ssDNA with sub-nanomolar affinity, while Hop2-Mnd1 binds less tightly (57,58). To confirm this, we used a bead ‘catch and release’ method to measure protein binding to DNA (Figure 5A). This method allowed us to determine: (i) if Hop2-Mnd1 binds the ssDNA substrate under our reaction conditions and (ii) if addition of RPA blocks Hop2-Mnd1 binding to ssDNA under the same conditions. Proteins were allowed to bind a variant of the 90C ssDNA oligo that carries a desthiobiotin moiety at its 5′ end (designated as db-ssDNA). Following binding reactions, protein bound oligos were mixed with streptavidin magnetic beads. Unbound protein and oligo were then removed by washing the beads with buffer, and biotin added to release DNA from beads, along with bound protein. The amount of DNA-bound protein was determined by Western blotting, and the amount of DNA by urea-PAGE. As shown in Figure 5B, Hop2-Mnd1 bound ssDNA when added alone, and addition of RPA to binding reactions blocked Hop2-Mnd1 binding. Analysis of unbound proteins (Figure 5C) confirmed this conclusion. No protein was detected in a no DNA control (lane 13), highlighting the advantage of our catch and release method; it avoids background caused by non-specific binding of proteins to beads. These results support our model that RPA enhances reconstituted reactions by blocking inhibitory interaction of Hop2-Mnd1 with ssDNA. We were unable to carry out equivalent catch and release experiments for Rdh54/Tid1 presumably because binding of Rdh54/Tid1 was too weak to withstand the washing steps required for the method, but we were able to demonstrate by fluorescence polarization assay that the apparent binding affinity of Rdh54/Tid1 for an 84-mer ssDNA is about 230 ± 17 nM (Supplementary Figure S9) which is ∼4600-fold lower than that of RPA at high- affinity mode (Kd ∼0.05 nM) with a binding site size of ∼30 nucleotides (59). Therefore, RPA is very likely to improve D-loop yields in the pre-mixed protocol by blocking inhibitory interactions of Rdh54/Tid1 with ssDNA, as for Hop2-Mnd1
The ability of RPA to outcompete Hop2-Mnd1 for binding to ssDNA led us to consider an alternative mechanism for the role of RPA in enhancing the efficiency of the pre-mixed reaction. RPA’s ability to outcompete Hop2-Mnd1 might make more of these accessory proteins available for binding to dsDNA, thereby improving their D-loop activity. We were able to test this alternative model for Hop2-Mnd1 using fluorescence polarization to measure the amount of Hop2-Mnd1 bound to dsDNA in the presence or absence of ssDNA. We found that preincubation of Hop2-Mnd1 with ssDNA under the conditions used for D-loop formation resulted in only a 20% decrease in the amount of Hop2-Mnd1 bound to dsDNA (Supplementary Figure S10). This finding argues against the idea that RPA allows Hop2-Mnd1 binding to dsDNA by preventing its sequestration on ssDNA. Instead the results support the idea that RPA blocks an inhibitory interaction of Hop2-Mnd1 with ssDNA.
Multiple protein-proteins interactions involving RPA, Dmc1, Rad51 and Mei5-Sae3
A ‘hand-off’ model was proposed previously to account for RPA function; RPA binds other DNA processing proteins to order and guide the activities of the proteins that trade places on ssDNA (7). However, ‘hand-off’ mechanisms remain poorly understood. To gain a better understanding of how RPA exerts its functions during our in vitro reconstitution reactions, we analyzed the interactions of RPA with Dmc1 and its accessory proteins. Pairwise interactions with RPA were examined using Dmc1, Rad51-II3A, Rdh54/Tid1, Mei5-Sae3 and Hop2-Mnd1. The approach took advantage of the fact that all proteins used in the study, with the exception of RPA, carry His6 tags allowing use of His6-Ni affinity pulldowns. Each of these proteins was incubated individually with RPA; then Ni resin was added to the mixture to capture protein or protein complexes through the His6 tag. We found Dmc1 and Mei5-Sae3 each pulled down RPA (Figure 6A, lanes 2 and 3), but Rad51-II3A, Hop2-Mnd1, and Rdh54/Tid1 did not (Figure 6A, lanes 4–6). To verify these findings, we used a reciprocal method of in vitro co-immunoprecipitation by antibodies against RPA2 to pulldown proteins that interact with RPA. We confirmed the above physical interactions between RPA and Dmc1, and between RPA and Mei5-Sae3 (Figure 6B, lanes 7, 8, 10). Although we did not detect interaction between RPA and Rad51 (Figure 6B, lane 9), Rad51 was pulled down with RPA if Mei5-Sae3 and/or Dmc1 were also present, with the highest level of Rad51 recovered when both were present (Figure 6B, lanes 11–13). To further verify these protein-protein interactions, we carried out co-immunoprecipitation using antibodies against Dmc1 (Figure 6C). Importantly, we detected direct interaction between Dmc1 and Rad51 (Figure 6C, lanes 6), and a previously described interaction between Dmc1 and Mei5-Sae3 (13,15) (Figure 6C, lanes 8). Moreover, we showed Mei5-Sae3 enhanced the interaction between Dmc1 and Rad51 (Figure 6C, lanes 7). The results provide evidence for a network of interactions amongst these yeast proteins. RPA physically interacts with Dmc1 and Mei5-Sae3, and a complex of RPA-Dmc1-(Mei5-Sae3) contacts Rad51 through interactions involving Dmc1 and Mei5-Sae3.
Physiological concentrations of meiotic recombination proteins
To compare our reconstituted conditions with relative protein levels in vivo we determined in vivo protein concentrations by quantitative Western blotting. Yeast whole cell extracts were prepared from synchronous meiotic cultures at various times following meiotic induction (Supplementary Figure S11A and S11B). Peak protein concentrations were calculated, under the assumption that all proteins are localized to the nucleus (Table 1, Supplementary Figure S11C). The peak protein concentrations showed that RPA and Hop2 are the most abundant, and Rdh54/Tid1 the least. After normalizing the protein levels to that of Dmc1, we found that the relativein vivo levels of Rdh54/Tid1, Rad51 and Mei5-Sae3 were within 3-fold of the relative levels of the same proteins under our optimized D-loop reconstitution conditions. The relative concentrations of RPA and Hop2-Mnd1 were much higher than those in the reconstituted system (see Discussion).
DISCUSSION
Reconstitution of Dmc1-dependent D-loop formation
We have reconstituted meiotic D-loop formation using six purified Saccharomyces cerevisiae proteins including the strand exchange protein Dmc1 and its accessory proteins RPA, Rad51-II3A, Mei5-Sae3, Hop2-Mnd1 and Rdh54/Tid1. We found that, when added as a protein mixture to DNA substrates, each accessory protein contributes to the yield of D-loop products. The reconstitution conditions we identified support a much higher level of D-loop activity than previously reported (11,16), with up to 34% of duplex substrate converted to products for the pre-mixed regimen and 41% for the pre-staged regimen. In vivo measurements of the steady state levels of these proteins indicate that, although the absolute concentrations at which the activities of the proteins are optimal in our biochemical experiments are much lower than their steady state concentrations in vivo, the relative optimum concentrations in vitro display significant similarity to the relative concentrations of the same proteins in wild type cells. This similarity suggests that the reconstitution conditions are biologically relevant. Two exceptions to the correspondence of relative protein levels in vivo and in vitro were Hop2-Mnd1 and RPA, which are present at much higher relative concentrations in vivo. We speculate that the effective concentration of Hop2-Mnd1 in the vicinity of a particular DNA recombination event is much lower than the overall nuclear concentration as a consequence of sequestration of the majority of protein at distant, non-recombining chromosomal sites. Our speculation is supported by previous cytological studies, as described above. We further note that our assumption that the majority of each recombination protein is localized to the nucleus is invalid for RPA as a significant fraction of total cellular protein localizes to the cytoplasm (60,61). If we assume that RPA is uniformly distributed in the cell, there is good agreement between the relative levels of RPA in the reconstituted system and in the nucleus. Further studies are required to determine the true nuclear concentration of budding yeast RPA.
Our progress in biochemical reconstitution of meiotic recombination builds a strong foundation for future work. This work will be directed at improving the similarity between properties of the reconstituted system and those of Dmc1-mediated recombination in vivo. One significant way in which the current biochemical system differs from the in vivo situation is that mutant cells lacking either Hop2-Mnd1 or Mei5-Sae3 have almost no residual Dmc1-dependent D-loop activity, while omitting either protein from the reconstituted system causes only a modest reduction of Dmc1 activity. This suggests that in vivo conditions are more restrictive to D-loop formation than our in vitro conditions. An obvious possibility to account for the relative permissiveness of the in vitro system is that duplex DNA substrates are packaged in chromatin in vivo, but not in vitro. Therefore, future studies will employ chromatinized plasmids to determine if they impose more strict requirements for accessory proteins than those observed with naked DNA. It is also worth pointing out that our optimized conditions for D-loop yield employ 250 μM Ca2+ which is higher than the estimated in vivo concentration of 70 μM (16). Given that 1 mM Ca2+ is known to activate Dmc1 in the absence of co-factors (35,62), it is possible that the intermediate level of Ca2+ used in our experiments partially suppressed the dependency of the reaction on one or more accessory proteins. Thus, future efforts will include further efforts to determine the impact of metal cofactor concentration on the dependency of D-loop formation on each accessory protein in the reconstituted system. Finally, we note that not all proteins which have been implicated as playing a role in the efficiency of Dmc1-mediated D-loop formation in vivo were included in this first effort to establish a reconstituted system. For example, Rdh54/Tid1’s semi-redundant paralogue Rad54, which is required for normal spore viability (23), remains to be tested for its ability to enhance D-loop yields in this system.
RPA plays multiple roles in stimulating Dmc1
A particularly important observation that arose from our effort to optimize the efficiency of Dmc1-dependent D-loop formation is that RPA is critical for the efficiency of the process. Our experiments provide evidence for three distinct mechanisms that contribute to the RPA’s stimulatory activity. RPA stimulates D-loop formation by: (1) eliminating secondary structure (51,52); (2) elongating D-loops by a mechanism likely to involve binding the displaced strand of the D-loop (53,54); (3) a novel mechanism that prevents inhibitory interactions of Hop2-Mnd1 and Rdh54/Tid1, that can occur during Dmc1 binding to ssDNA. At least one cause of accessory protein conflict is binding to ssDNA, which is prevented by RPA’s ability to outcompete the other proteins for binding to ssDNA.
Protein-protein interactions associated with Dmc1 filament assembly
Protein-protein interactions have been found to induce conformational rearrangement of RPA’s modular domains in a manner that alters the mode of RPA-ssDNA binding resulting in hand-offs of ssDNA from RPA to other ssDNA binding proteins (7). As a first step towards examining this potential mechanism for RPA’s activity, we showed that RPA binds directly to Dmc1, demonstrating evolutionary conservation of an interaction previously reported for the corresponding mammalian orthologues (63). We also showed that human RPA cannot fully substitute for budding yeast RPA in our budding yeast system, suggesting that at least one protein-protein interaction involving RPA is species-specific and functionally important.
Previous in vivo studies suggested that both Rad51 and Mei5-Sae3 act to recruit or stabilize Dmc1 at sites of recombination in vivo (13,14) and that Rad51 and Mei5-Sae3 can cooperate to stimulate Dmc1’s activity in vitro (11). However, the biochemical mechanism underlying the influence of Rad51 and Mei5-Sae3 on Dmc1’s activity has yet to be determined. Previous studies showed that Dmc1 binds Mei5-Sae3, Mei5-Sae3 binds Rad51, and RPA binds Mei5-Sae3 (13,15,41). Here we report detection of protein-protein interaction between Rad51 and Dmc1. A conventional two-hybrid system failed to detect this interaction (our unpublished results), and a recent biochemical study showed that Rad51-Rad51 and Dmc1-Dmc1 homotypic interactions are strong enough to result in predominance of homofilaments when the two are allowed to bind DNA as a mixture (64). Cytological observations also argue for a predominance of homofilaments (12,24,65). Therefore, detection of direct Rad51-Dmc1 binding is significant because it provides a molecular explanation for the role of Rad51 in stimulating Dmc1 activity. We also show that Mei5-Sae3 can enhance interaction between Rad51 and Dmc1, consistent with our previous work that showed Rad51 and Mei5-Sae3 cooperate to stimulate Dmc1’s D-loop activity (11).
Model
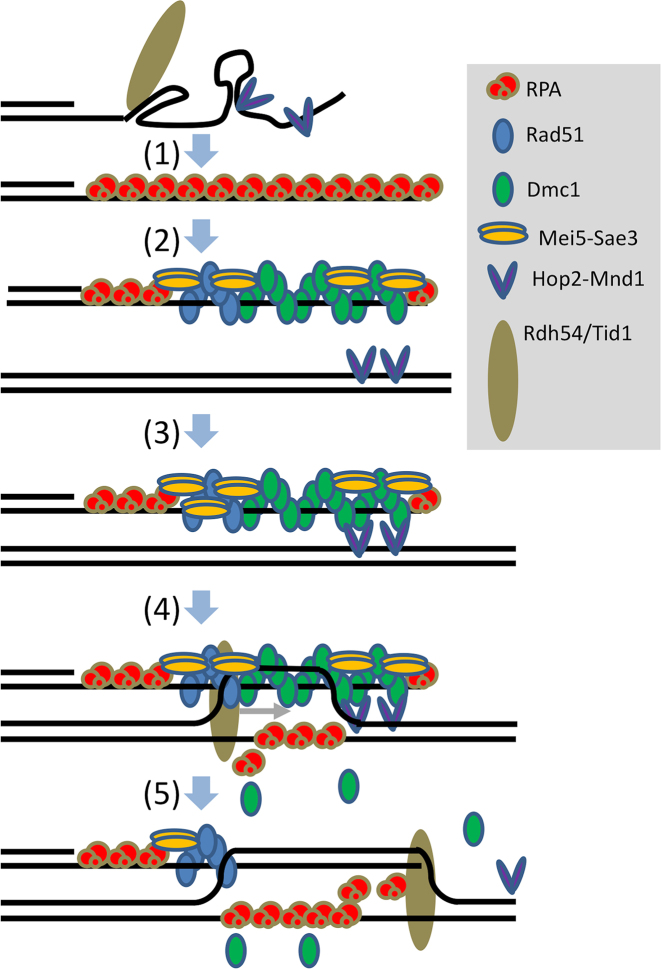
Our results may be incorporated into a model for the functions of Dmc1 accessory proteins that involves roles of RPA during both pre-synaptic filament formation and during strand exchange (Figure 7). The model incorporates many previous findings with those presented here. The steps in the model are as follows. (i) RPA binds ssDNA with high affinity, coating ssDNA. RPA removes secondary structures from ssDNA and blocks Hop2-Mnd1 and Rdh54/Tid1 from accessing ssDNA. (ii) Protein–protein interactions involving Dmc1, Rad51, Mei5-Sae3 and RPA contribute to the assembly of functional Dmc1 filaments, with interaction of Rad51 and Dmc1 stabilized by protein-protein interactions with Mei5-Sae3 (66). (iii) Hop2-Mnd1 stabilizes non-homology dependent interactions between Dmc1-ssDNA filaments and dsDNA (25,58,67). (iv) Rdh54/Tid1 stabilizes nascent loops by virtue of its ability to simultaneously bind both a ssDNA and a dsDNA molecule and/or its translocase activity (26 and references therein). (v) As D-loops form, RPA binds the displaced strand, blocking D-loop dissociation and extending D-loop length.
Figure 7.
Model. (1) RPA binds ssDNA thereby removing secondary structure and preventing binding of Hop2-Mnd1 and Rdh54/Tid1. (2) RPA carries out hand-off reactions allowing Rad51+Mei5-Sae3 to assist Dmc1 filament formation. The protein configuration shown is speculative but depicts potentially relevant protein-protein interactions between RPA and Mei5-Sae3, RPA and Dmc1, Rad51 and Mei5-Sae3, Dmc1 and Mei5-Sae3, and, particularly importantly, between Dmc1 and Rad51. (3) dsDNA bound Hop2-Mnd1 captures filaments via homology-independent interactions with Dmc1. (4) Homology recognition occurs to form a homology-dependent nascent D-loop which is stabilized by RPA binding to the displaced ssDNA strand. (5) Rdh54/Tid1 binds the D-loop, via contacts with both ssDNA and dsDNA and then translocates across the D-loop stabilizing it by displacing Dmc1 and elongating the D-loop. RPA can elongate the D-loop, even in the absence of Rdh54/Tid1, but does not afford the stability associated with Dmc1 displacement and/or simultaneous binding by Rdh54/Tid1 to both ssDNA and dsDNA.
Conclusion
We report major progress in reconstitution of meiotic recombination reactions by identifying conditions under which a set of five accessory proteins cooperate to enhance the efficiency of Dmc1-mediated D-loop formation. Our results demonstrate the critical importance of RPA to the system, with evidence for three mechanisms of RPA-mediated stimulation of Dmc1-dependent D-loop reactions. The mechanisms of RPA stimulation include one that is particularly important and previously undescribed; RPA resolves inhibitory and conflicting activities of Hop2-Mnd1 and Rdh54/Tid1. We also detect novel protein-protein interactions including direct interaction between Rad51 and Dmc1, a result that further supports the view that meiotic recombination events require cooperation of Rad51 and Dmc1, with Dmc1 serving as enzyme, and Rad51 as accessory protein.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Diedre Reitz and Brian Budke for comments on the manuscript, and Joseph Piccirilli for advice on analysis of kinetic data. We thank A. Shinohara (Osaka University, Japan) for providing antibodies against RPA2, and Marc Wold (University of Iowa) for providing human RPA. We are also grateful to Dr. Wold for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [GM050936 to D.K.B.]. Funding for open access charge: NIGMS [R01-GM50936].
Conflict of interest statement. None declared.
REFERENCES
- 1. Keeney S., Giroux C.N., Kleckner N.. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997; 88:375–384. [DOI] [PubMed] [Google Scholar]
- 2. Neale M.J., Keeney S.. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006; 442:153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim C., Snyder R.O., Wold M.S.. Binding properties of replication protein A from human and yeast cells. Mol. Cell. Biol. 1992; 12:3050–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim C., Paulus B.F., Wold M.S.. Interactions of human replication protein A with oligonucleotides. Biochemistry. 1994; 33:14197–14206. [DOI] [PubMed] [Google Scholar]
- 5. Chen R., Wold M.S.. Replication protein A: single-stranded DNA’s first responder: dynamic DNA-interactions allow replication protein A to direct single-strand DNA intermediates into different pathways for synthesis or repair. Bioessays. 2014; 36:1156–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Prakash A., Borgstahl G.E.O.. The structure and function of replication protein A in DNA replication. Subcell. Biochem. 2012; 62:171–196. [DOI] [PubMed] [Google Scholar]
- 7. Fanning E., Klimovich V., Nager A.R.. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006; 34:4126–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Oakley G.G., Patrick S.M.. Replication protein A: directing traffic at the intersection of replication and repair. Front. Biosci. (Landmark Ed.). 2010; 15:883–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gerton J.L., DeRisi J.L.. Mnd1p: an evolutionarily conserved protein required for meiotic recombination. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:6895–6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen Y.-K., Leng C.-H., Olivares H., Lee M.-H., Chang Y.-C., Kung W.-M., Ti S.-C., Lo Y.-H., Wang A.H.-J., Chang C.-S. et al. Heterodimeric complexes of Hop2 and Mnd1 function with Dmc1 to promote meiotic homolog juxtaposition and strand assimilation. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:10572–10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cloud V., Chan Y.-L., Grubb J., Budke B., Bishop D.K.. Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science. 2012; 337:1222–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shinohara M., Gasior S.L., Bishop D.K., Shinohara A.. Tid1/Rdh54 promotes colocalization of Rad51 and Dmc1 during meiotic recombination. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:10814–10819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hayase A., Takagi M., Miyazaki T., Oshiumi H., Shinohara M., Shinohara A.. A protein complex containing Mei5 and Sae3 promotes the assembly of the meiosis-specific RecA homolog Dmc1. Cell. 2004; 119:927–940. [DOI] [PubMed] [Google Scholar]
- 14. Tsubouchi H., Roeder G.S.. The budding yeast mei5 and sae3 proteins act together with dmc1 during meiotic recombination. Genetics. 2004; 168:1219–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ferrari S.R., Grubb J., Bishop D.K.. The Mei5-Sae3 protein complex mediates Dmc1 activity in Saccharomyces cerevisiae. J. Biol. Chem. 2009; 284:11766–11770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chan Y.-L., Brown M.S., Qin D., Handa N., Bishop D.K.. The third exon of the budding yeast meiotic recombination gene HOP2 is required for calcium-dependent and recombinase Dmc1-specific stimulation of homologous strand assimilation. J. Biol. Chem. 2014; 289:18076–18086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zierhut C., Berlinger M., Rupp C., Shinohara A., Klein F.. Mnd1 is required for meiotic interhomolog repair. Curr. Biol. 2004; 14:752–762. [DOI] [PubMed] [Google Scholar]
- 18. Tsubouchi H., Roeder G.S.. The Mnd1 protein forms a complex with hop2 to promote homologous chromosome pairing and meiotic double-strand break repair. Mol. Cell. Biol. 2002; 22:3078–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Daley J.M., Gaines W.A., Kwon Y., Sung P.. Regulation of DNA pairing in homologous recombination. Cold Spring Harb. Perspect Biol. 2014; 6:a017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Holzen T.M., Shah P.P., Olivares H.A., Bishop D.K.. Tid1/Rdh54 promotes dissociation of Dmc1 from nonrecombinogenic sites on meiotic chromatin. Genes Dev. 2006; 20:2593–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shah P.P., Zheng X., Epshtein A., Carey J.N., Bishop D.K., Klein H.L.. Swi2/Snf2-related translocases prevent accumulation of toxic Rad51 complexes during mitotic growth. Mol. Cell. 2010; 39:862–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mason J.M., Dusad K., Wright W.D., Grubb J., Budke B., Heyer W.-D., Connell P.P., Weichselbaum R.R., Bishop D.K.. RAD54 family translocases counter genotoxic effects of RAD51 in human tumor cells. Nucleic Acids Res. 2015; 43:3180–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shinohara M., Shita-Yamaguchi E., Buerstedde J.M., Shinagawa H., Ogawa H., Shinohara A.. Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics. 1997; 147:1545–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brown M.S., Bishop D.K.. DNA strand exchange and RecA homologs in meiosis. Cold Spring Harb. Perspect. Biol. 2014; 7:a016659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. San Filippo J., Sung P., Klein H.. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008; 77:229–257. [DOI] [PubMed] [Google Scholar]
- 26. Wright W.D., Heyer W.-D.. Rad54 functions as a heteroduplex DNA pump modulated by its DNA substrates and Rad51 during D loop formation. Mol. Cell. 2014; 53:420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lao J.P., Cloud V., Huang C.-C., Grubb J., Thacker D., Lee C.-Y., Dresser M.E., Hunter N., Bishop D.K.. Meiotic crossover control by concerted action of Rad51-Dmc1 in homolog template bias and robust homeostatic regulation. PLoS Genet. 2013; 9:e1003978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haber J.E. DNA repair: the search for homology. Bioessays. 2018; 40:e1700229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schwacha A., Kleckner N.. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997; 90:1123–1135. [DOI] [PubMed] [Google Scholar]
- 30. Bishop D.K., Park D., Xu L., Kleckner N.. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992; 69:439–456. [DOI] [PubMed] [Google Scholar]
- 31. Tsubouchi H., Roeder G.S.. Budding yeast Hed1 down-regulates the mitotic recombination machinery when meiotic recombination is impaired. Genes Dev. 2006; 20:1766–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bishop D.K. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell. 1994; 79:1081–1092. [DOI] [PubMed] [Google Scholar]
- 33. Gasior S.L., Olivares H., Ear U., Hari D.M., Weichselbaum R., Bishop D.K.. Assembly of RecA-like recombinases: distinct roles for mediator proteins in mitosis and meiosis. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:8411–8418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Petukhova G.V., Pezza R.J., Vanevski F., Ploquin M., Masson J.-Y., Camerini-Otero R.D.. The Hop2 and Mnd1 proteins act in concert with Rad51 and Dmc1 in meiotic recombination. Nat. Struct. Mol. Biol. 2005; 12:449–453. [DOI] [PubMed] [Google Scholar]
- 35. Busygina V., Gaines W.A., Xu Y., Kwon Y., Williams G.J., Lin S.-W., Chang H.-Y., Chi P., Wang H.-W., Sung P.. Functional attributes of the Saccharomyces cerevisiae meiotic recombinase Dmc1. DNA Repair (Amst.). 2013; 12:707–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chi P., Kwon Y., Moses D.N., Seong C., Sehorn M.G., Singh A.K., Tsubouchi H., Greene E.C., Klein H.L., Sung P.. Functional interactions of meiotic recombination factors Rdh54 and Dmc1. DNA Repair (Amst.). 2009; 8:279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nimonkar A.V., Dombrowski C.C., Siino J.S., Stasiak A.Z., Stasiak A., Kowalczykowski S.C.. Saccharomyces cerevisiae Dmc1 and Rad51 proteins preferentially function with Tid1 and Rad54 proteins, respectively, to promote DNA strand invasion during genetic recombination. J. Biol. Chem. 2012; 287:28727–28737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Binz S.K., Dickson A.M., Haring S.J., Wold M.S.. Functional assays for replication protein A (RPA). Meth. Enzymol. 2006; 409:11–38. [DOI] [PubMed] [Google Scholar]
- 39. Chan Y.-L., Bishop D.K.. Purification of saccharomyces cerevisiae homologous recombination proteins Dmc1 and Rdh54/Tid1 and a fluorescent D-Loop assay. Meth. Enzymol. 2018; 600:307–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hong E.L., Shinohara A., Bishop D.K.. Saccharomyces cerevisiae Dmc1 protein promotes renaturation of single-strand DNA (ssDNA) and assimilation of ssDNA into homologous super-coiled duplex DNA. J. Biol. Chem. 2001; 276:41906–41912. [DOI] [PubMed] [Google Scholar]
- 41. Say A.F., Ledford L.L., Sharma D., Singh A.K., Leung W.-K., Sehorn H.A., Tsubouchi H., Sung P., Sehorn M.G.. The budding yeast Mei5-Sae3 complex interacts with Rad51 and preferentially binds a DNA fork structure. DNA Repair (Amst.). 2011; 10:586–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Namsaraev E., Berg P.. Characterization of strand exchange activity of yeast Rad51 protein. Mol. Cell. Biol. 1997; 17:5359–5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ferrari M., Nachimuthu B.T., Donnianni R.A., Klein H., Pellicioli A.. Tid1/Rdh54 translocase is phosphorylated through a Mec1- and Rad53-dependent manner in the presence of DSB lesions in budding yeast. DNA Repair (Amst). 2013; 12:347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chi P., Kwon Y., Seong C., Epshtein A., Lam I., Sung P., Klein H.L.. Yeast recombination factor Rdh54 functionally interacts with the Rad51 recombinase and catalyzes Rad51 removal from DNA. J. Biol. Chem. 2006; 281:26268–26279. [DOI] [PubMed] [Google Scholar]
- 45. Nallagatla S.R., Hwang J., Toroney R., Zheng X., Cameron C.E., Bevilacqua P.C.. 5′-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science. 2007; 318:1455–1458. [DOI] [PubMed] [Google Scholar]
- 46. Jayathilaka K., Sheridan S.D., Bold T.D., Bochenska K., Logan H.L., Weichselbaum R.R., Bishop D.K., Connell P.P.. A chemical compound that stimulates the human homologous recombination protein RAD51. Proc. Natl. Acad. Sci. U/S.A. 2008; 105:15848–15853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Budke B., Chan Y.-L., Bishop D.K., Connell P.P.. Real-time solution measurement of RAD51- and RecA-mediated strand assimilation without background annealing. Nucleic Acids Res. 2013; 41:e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jorgensen P., Nishikawa J.L., Breitkreutz B.-J., Tyers M.. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002; 297:395–400. [DOI] [PubMed] [Google Scholar]
- 49. Jorgensen P., Edgington N.P., Schneider B.L., Rupes I., Tyers M., Futcher B.. The size of the nucleus increases as yeast cells grow. Mol. Biol. Cell. 2007; 18:3523–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mimitou E.P., Yamada S., Keeney S.. A global view of meiotic double-strand break end resection. Science. 2017; 355:40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sugiyama T., Zaitseva E.M., Kowalczykowski S.C.. A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem. 1997; 272:7940–7945. [DOI] [PubMed] [Google Scholar]
- 52. Wold M.S. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997; 66:61–92. [DOI] [PubMed] [Google Scholar]
- 53. Mazin A.V., Kowalczykowski S.C.. The function of the secondary DNA-binding site of RecA protein during DNA strand exchange. EMBO J. 1998; 17:1161–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Eggler A.L., Inman R.B., Cox M.M.. The Rad51-dependent pairing of long DNA substrates is stabilized by replication protein A. J. Biol. Chem. 2002; 277:39280–39288. [DOI] [PubMed] [Google Scholar]
- 55. Ploquin M., Petukhova G.V., Morneau D., Déry U., Bransi A., Stasiak A., Camerini-Otero R.D., Masson J.-Y.. Stimulation of fission yeast and mouse Hop2-Mnd1 of the Dmc1 and Rad51 recombinases. Nucleic Acids Res. 2007; 35:2719–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Swagemakers S.M., Essers J., de Wit J., Hoeijmakers J.H., Kanaar R.. The human RAD54 recombinational DNA repair protein is a double-stranded DNA-dependent ATPase. J. Biol. Chem. 1998; 273:28292–28297. [DOI] [PubMed] [Google Scholar]
- 57. Pezza R.J., Petukhova G.V., Ghirlando R., Camerini-Otero R.D.. Molecular activities of meiosis-specific proteins Hop2, Mnd1, and the Hop2-Mnd1 complex. J. Biol. Chem. 2006; 281:18426–18434. [DOI] [PubMed] [Google Scholar]
- 58. Chi P., Filippo San, Sehorn J., Petukhova M.G., Sung P.. Bipartite stimulatory action of the Hop2-Mnd1 complex on the Rad51 recombinase. Genes Dev. 2007; 21:1747–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wyka I.M., Dhar K., Binz S.K., Wold M.S.. Replication protein A interactions with DNA: differential binding of the core domains and analysis of the DNA interaction surface. Biochemistry. 2003; 42:12909–12918. [DOI] [PubMed] [Google Scholar]
- 60. Sundin B.A., Chiu C.-H., Riffle M., Davis T.N., Muller E.G.D.. Localization of proteins that are coordinately expressed with Cln2 during the cell cycle. Yeast. 2004; 21:793–800. [DOI] [PubMed] [Google Scholar]
- 61. Murti K.G., He D.C., Brinkley B.R., Scott R., Lee S.H.. Dynamics of human replication protein A subunit distribution and partitioning in the cell cycle. Exp. Cell Res. 1996; 223:279–289. [DOI] [PubMed] [Google Scholar]
- 62. Lee M.-H., Chang Y.-C., Hong E.L., Grubb J., Chang C.-S., Bishop D.K., Wang T.-F.. Calcium ion promotes yeast Dmc1 activity via formation of long and fine helical filaments with single-stranded DNA. J. Biol. Chem. 2005; 280:40980–40984. [DOI] [PubMed] [Google Scholar]
- 63. Golub E.I., Gupta R.C., Haaf T., Wold M.S., Radding C.M.. Interaction of human rad51 recombination protein with single-stranded DNA binding protein, RPA. Nucleic Acids Res. 1998; 26:5388–5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Crickard J.B., Kaniecki K., Kwon Y., Sung P., Greene E.C.. Spontaneous self-segregation of Rad51 and Dmc1 DNA recombinases within mixed recombinase filaments. J. Biol. Chem. 2018; 293:4191–4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dresser M.E., Ewing D.J., Conrad M.N., Dominguez A.M., Barstead R., Jiang H., Kodadek T.. DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 pathway. Genetics. 1997; 147:533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bishop D.K. Rad51, the lead in mitotic recombinational DNA repair, plays a supporting role in budding yeast meiosis. Cell Cycle. 2012; 11:4105–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pezza R.J., Voloshin O.N., Vanevski F., Camerini-Otero R.D.. Hop2/Mnd1 acts on two critical steps in Dmc1-promoted homologous pairing. Genes Dev. 2007; 21:1758–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.