Abstract
Recoding of stop codons as amino acid-specifying codons is a co-translational event that enables C-terminal extension of a protein. Synthesis of selenoproteins requires recoding of internal UGA stop codons to the 21st non-standard amino acid selenocysteine (Sec) and plays a vital role in human health and disease. Separately, canonical stop codons can be recoded to specify standard amino acids in a process known as stop codon readthrough (SCR), producing extended protein isoforms with potential novel functions. Conventional computational tools cannot distinguish between the dual functionality of stop codons as stop signals and sense codons, resulting in misannotation of selenoprotein gene products and failure to predict SCR. Manual curation is therefore required to correctly represent recoded gene products and their functions. Our goal was to provide accurately curated and annotated datasets of selenoprotein and SCR transcript and protein records to serve as annotation standards and to promote basic and biomedical research. Gene annotations were curated in nine vertebrate model organisms and integrated into NCBI’s Reference Sequence (RefSeq) dataset, resulting in 247 selenoprotein genes encoding 322 selenoproteins, and 93 genes exhibiting SCR encoding 94 SCR isoforms.
INTRODUCTION
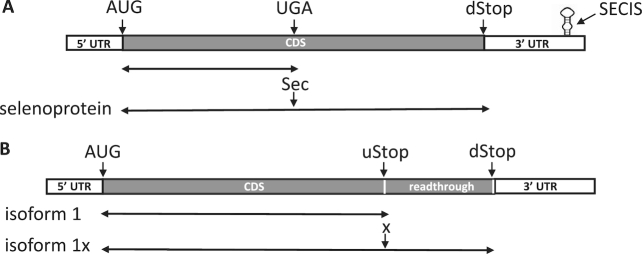
Recoding is an mRNA-specific, non-standard decoding of the genetic code that is stimulated by cis-acting signals. The genetic information transmitted to mRNA via transcription can be redefined during translation by several types of recoding events, including ribosomal frameshifting and translational or stop codon readthrough. During translational readthrough, a stop codon is co-translationally recoded to specify an amino acid, which results in translation termination at an in-frame downstream stop codon and extension of the protein. Two types of stop codon recoding events are known to occur in eukaryotes (Table 1): selenocysteine (Sec) insertion, which involves recoding a UGA stop codon to specify Sec for the synthesis of full-length Sec-containing proteins, selenoproteins (Figure 1A); and stop codon readthrough (SCR), which involves recoding any of the three stop codons (UAA, UAG or UGA) to specify a standard amino acid to generate C-terminally extended protein isoforms (Figure 1B).
Table 1.
Comparison of Sec insertion and SCR
| Features | Sec insertion | SCR |
|---|---|---|
| Upstream stop codon | UGA | UGA>UAG>UAAa |
| Downstream stop codon | UAA, UAG, UGA | UAA, UAG, UGA |
| Amino acid specified | Sec | Several standard amino acids |
| Stimulatory signal | SECIS element | CUAG, stem loop, other |
| End product | Selenoprotein | C-terminally extended isoform |
aFrequency of occurrence as readthrough codon.
Figure 1.
Schematic representing Sec insertion (A) and SCR (B). The coding regions are shown as filled black bars, and the 5′ and 3′ UTRs as open bars. (A) Selenoprotein mRNA containing an AUG start codon, in-frame UGA and downstream stop (dStop) codons, and SECIS element is shown. Double arrowed lines indicate proteins resulting from translation termination at UGA (top) or dStop following recoding of UGA as Sec (bottom). (B) SCR candidate mRNA containing an AUG start codon, canonical upstream stop (uStop) and in-frame downstream stop (dStop) codons, and inter-stop readthrough region encoding C-terminal extension is shown. Double arrowed lines indicate isoforms resulting from translation termination at uStop (top) or dStop following recoding of uStop to specify a standard amino acid (denoted as ‘x’) (bottom).
Selenoproteins are found in all three domains of life: bacteria, archaea and eukaryota. They contain the essential trace element selenium (Se), which is of fundamental importance in humans and animals (1). Most of the biological effects of dietary Se are mediated by Sec, a Se-containing amino acid, which is co-translationally inserted into nascent polypeptides by recoding UGA codon via a sophisticated machinery involving cis- and trans- acting factors. A conserved stem-loop structure, the Sec insertion sequence (SECIS) element, present in the 3′ UTRs of selenoprotein mRNAs in eukaryotes (Figure 1A), is essential for the recognition of UGA as a Sec codon rather than a stop signal. The requirement of the SECIS element for Sec insertion and its conservation have been exploited to identify selenoprotein genes in sequenced vertebrate genomes. The first complete set of selenoproteins (i.e. selenoproteome) in vertebrates was determined for human and consists of 25 selenoproteins (2). Subsequently, selenoprotein genes in 44 vertebrate species were identified and analyzed to study the composition and evolution of selenoproteomes (3). The size of the selenoproteome varies greatly among eukaryotes; larger selenoproteomes have been observed in aquatic organisms, such as fish, which contain several selenoproteins not found in mammals. Except for the characteristic presence of Sec in all selenoproteins, they are a diverse group with varied sequences and involved in a variety of processes, ranging from antioxidant defense, fertility, immune function, muscle development, thyroid hormone metabolism, and in cancer prevention and promotion (1,4,5). The specific functions of many selenoproteins are not known. Many of the best characterized selenoproteins, such as the members of the DIO, GPX and TXNRD gene families, function as enzymes containing a Sec residue at the catalytic site, which is thought to participate in redox reactions. GPX4 is unusual in that it has a dual function in male reproduction: as a peroxidase in spermatids, and as a structural protein in mature spermatozoa (6). SELENOP, another well studied selenoprotein with multiple Sec residues, is implicated in selenium transport (7). SELENON has a role in myogenesis; mutations in this gene have been associated with several muscle disorders, collectively known as SEPN1-related myopathies (8).
SCR is used extensively in viruses to expand the proteomes encoded by their small genomes (9). However, SCR in eukaryotes was known only for a few cellular genes (10–12), until recently when two complementary approaches, a phylogenetic analysis using protein-coding evolutionary signatures (13) and ribosome profiling (14), identified abundant SCR in Drosophila, and to a lesser degree in yeast and mammals. Together, these and subsequent studies (15–19) predicted the presence of over 50 SCR candidate genes in mammals and showed that the C-terminal extensions in several mammalian candidate genes were also conserved in other vertebrates. Experimental evidence for readthrough, however, exists for just a few genes. It is believed that unlike selenoproteins for which the property of Sec is functionally important, for SCR it is the C-terminal extension and not the identity of the specified amino acid that is functionally relevant in the majority of cases. Beier and Grimm (20) showed that the misreading of stop codons in eukaryotes is achieved by a variety of naturally occurring tRNAs that recognize near-cognate codons. Thus, each stop codon has been reported to specify more than one amino acid: Arg, Cys, Trp, Ser for UGA; Gln, Tyr, Leu, Lys for UAG; and Gln and Tyr for UAA (10,13,20). Studies have shown that the stop codon context, including the stop codon itself, plays a significant role in influencing SCR efficiency. The hierarchy of the leakiest SCR-prone codon observed is UGA>UAG>UAA, and the leakiest downstream base is C>U>G>A, suggesting that the UGA-C context is most effective in enabling SCR in eukaryotes (13). Other elements, including adjacent short sequence motifs (16,21), 3′ cis-acting elements (15), and stem loop structures (22,23), have also been implicated in stimulating SCR. A trans-acting factor has also been reported to promote SCR (15). In experimentally verified genes, those with the highest readthrough efficiency had a UGA stop codon immediately followed by a short motif, CUAG (16). Little is known about the functional importance of the readthrough isoforms; however, recent studies have shown that the extended C-terminus may expose additional signals or domains, which may modulate their localization, activity, and/or stability. For instance, the extended isoforms of LDHB and MDH1 were targeted to the peroxisomes compared to the cytosolic canonical isoforms (18,19), the extended isoform of VEGFA was reported to have antiangiogenic activity (15) but this has been disputed recently (24), and the extended isoforms of MPZ (12), AQP4 (25) and VDR (17) exhibit different properties that may modulate the original gene function.
Recoding of stop codons to sense codons bends the rules of standard decoding of the genetic code. Conventional computational tools cannot distinguish between the dual functionality of UGA codon as a stop signal and as Sec, resulting in misannotation of selenoprotein coding sequences (CDSes) on transcript and genomic sequences, and loss of crucial biological information. Predicting SCR systematically is even more challenging, given three potential readthrough stop codons and the diversity of stimulatory signals promoting readthrough. Manual curation is thus essential for accurate representation of these recoding events and their gene products. Our goal was to provide an accurately curated and annotated set of selenoprotein transcript and protein records and represent experimentally validated cases of SCR in several vertebrate model organisms in NCBI’s Reference Sequence (RefSeq) database (26). The RefSeq database is a comprehensive collection of genomic, transcript, and protein sequences spanning the tree of life. The RefSeq collection is based on both automated and manual curation approaches, and the highly curated subset is vital for additional resources at NCBI, including Gene and Genomes, and helps guide NCBI’s evidence-based eukaryotic genome annotation pipeline. RefSeq records are also widely used by the scientific community for basic and biomedical research, as well as for large scale genome annotation and comparative analyses.
MATERIALS AND METHODS
Several different approaches were used to identify selenoprotein genes in vertebrates similar to the methods described before for antizyme genes (27). Briefly, the earliest selenoprotein records in the RefSeq database were generated by automatic processes: provisional RefSeq records were created by automatic processing based on primary sequence data submitted to the International Nucleotide Sequence Database Collaboration (INSDC) (28); and predicted RefSeq records were generated by the NCBI’s eukaryotic genome annotation pipeline. Many of the provisional and predicted RefSeq records contained misannotated selenoprotein CDS propagated from the primary sequence data or from computational prediction. These records were subjected to manual review as guided by our in-house QA analyses, which for instance would flag candidates for nonsense-mediated mRNA decay (NMD) (29). The manually curated records from well-studied organisms, such as human and mouse, facilitated prediction of orthologous models by the NCBI’s eukaryotic genome annotation pipeline, which then became targets for manual review. Selenoprotein genes were also identified by literature search, search of specialized databases, such as SelenoDB 2.0 (30) and Recode-2 (31), and BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) search of publicly available databases at NCBI with related sequences. The SECISearch3 tool (32) was used to search for SECIS elements in the RefSeq transcript sequences. Selenoprotein gene nomenclature was updated where necessary as per recommendation by Gladyshev et al. (33), in collaboration with organism-specific nomenclature authorities (34–39). A brief RefSeq summary describing the salient features of a gene and its function (if known), a biological attribute (‘protein contains selenocysteine’) with supporting evidence for the recoding event, and an INSDC-approved regulatory feature annotation with a ‘recoding_stimulatory_region’ qualifier for the SECIS element were added to the curated RefSeq records. Additional phylogenetically conserved elements shown to modulate Sec insertion, such as the stop-codon redefinition element (SRE) in SELENON mRNA (40) and the two stem-loop structures (stem–loop 1 and 2) in SELENOS mRNA (41), were also annotated on the respective RefSeq transcript records. Separate RefSeq records were created for alternatively spliced transcript variants supported by transcript, EST, RNA-seq data or publications. Each transcript variant record is accompanied by a brief text highlighting its uniqueness and difference from the predominant variant.
SCR genes were selected for inclusion in the RefSeq database based on experimental support demonstrating readthrough and conservation of the C-terminal extension; readthrough candidate genes identified only by systematic bioinformatic or computational analysis were not included in this review. The RefSeq data model for eukaryotes allows only one transcript explicitly linked to one protein; therefore, to represent SCR, a second RefSeq transcript identical to the one encoding the canonical isoform was created and manually annotated with a CDS terminating at an alternative in-frame, downstream stop codon. As the identity of the amino acid specified by a readthrough stop codon is often not known, the amino acid inserted at the readthrough site was generally denoted as an undefined amino acid (‘X’). In rare instances, when the specified amino acid was identified as a single species, as for VEGFA (15), then that specific amino acid (serine, S) was annotated at the readthrough site. The names of C-terminally extended isoforms were generally appended with an ‘x’ (for extended, as per convention in this field) to distinguish them from their standard counterparts (e.g. isoform 1 and isoform 1x), except when an alternate name was reported in literature (e.g. isoform MPZ and isoform L-MPZ) (12). The reviewed SCR records also included a RefSeq summary, transcript variant text, attribute (‘stop codon readthrough’) and a regulatory feature for SCR signal (if known) as described above for selenoprotein records.
The analysis, assembly and annotation of RefSeq records was performed using NCBI’s Genome Workbench application (Gbench; http://www.ncbi.nlm.nih.gov/tools/gbench) as described before (27). RefSeqs are generally created from a combination of conventional transcript sequences (INSDC transcripts and ESTs), transcriptome shotgun assemblies (TSAs) and genomic sequence— the latter two are used when a gene or region(s) of a gene lacks conventional transcript support. Assembling a RefSeq transcript for the chicken SEPHS2 gene posed a challenge (see Results) as none of the sources of sequence data mentioned above was available for a portion of the 3′ UTR of the SEPHS2 transcript; therefore, a full-length RefSeq transcript (NM_001366334.2) was assembled using ESTs and two (18 and 52 bp) inserts based on RNA-seq reads available in SRA. The details of the assembly components are shown on the public NM_001366334.2 record under the COMMENT section.
RESULTS
Curation and annotation of selenoprotein genes in vertebrates
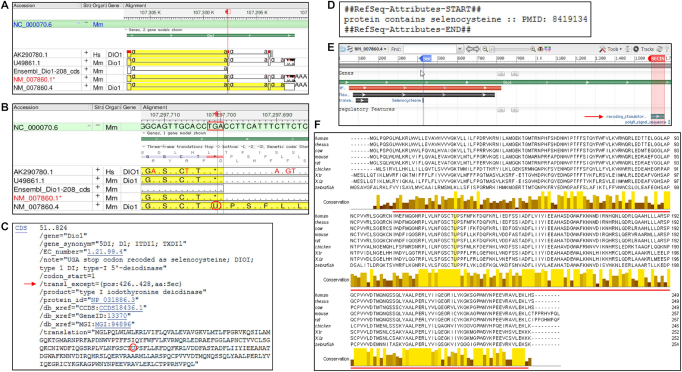
The dual role of the UGA codon as a stop signal and as encoding Sec poses a problem for automatic annotation of selenoprotein genes by standard computational tools, which often results in misannotation of selenoprotein CDSes on transcripts and genomic sequences, as illustrated by the mouse Dio1 gene (Figure 2A and B). INSDC sequences AK290780.1 (human) and U49861.1 (mouse) were misannotated with a CDS terminating at TGA and encoding a C-terminally truncated protein of 125 amino acid residues (aa), rendering these transcripts candidates for NMD. A similar misannotated CDS was found on an Ensembl transcript (ENSMUST00000150974.1/Dio1-208). The misannotation was also automatically propagated to NM_007860.1, a provisional RefSeq transcript based on U49861.1. Subsequent manual review led to the replacement of the erroneous record with an updated version (NM_007860.4) containing the accurately annotated CDS encoding the full-length selenoprotein of 257 aa (NP_031886.3). A translation exception (/transl_except) qualifier was manually added to the CDS feature indicating the position of the TGA-specifying Sec using feature annotation details approved by the INSDC (Figure 2C). This allowed annotation of the full-length CDS and inclusion of Sec as ‘U’ in the selenoprotein sequence. The ‘protein contains selenocysteine’ attribute (Figure 2D) and feature annotations, including the locations of Sec and SECIS element (Figure 2E), were also added to the RefSeq transcript record to highlight the functionally relevant features of the selenoprotein transcript. Although UGA recoding to Sec has been reported to be inefficient (42) and modulated by Se availability (43), which can result in the production of both the full-length and truncated isoforms of selenoproteins, we have opted to only represent CDSes for the full-length gene products in accordance with RefSeq's emphasis on representing full-length, functional products.
Figure 2.
Manual curation, annotation and analysis of selenoprotein. (A) Genome Workbench display of alignment of a few select transcripts to the mouse genomic sequence (chr4, NC_000070.6) at the location of Dio1gene (GeneID:13370). The annotated CDS is highlighted in yellow; the red flag marks the location of TGA in exon 2. (B) Expansion of the region around the red marker shows translation of TGA (boxed in red) as a stop signal (*) or Sec (U, circled in red) in the -2 frame of the 3-frame translation track. (C) GenBank format display of manually added /transl_except qualifier (red arrow) and the incorporated Sec residue (U circled in red) on the public RefSeq record, NM_007860.4. (D) Display of a portion of the COMMENT section on NM_007860.4 showing manually added selenoprotein-specific attribute with supporting publication. (E) Display of the graphical view of NM_007860.4 configured to add two feature annotations highlighting the location of Sec (nt 426–428) and SECIS element (nt 1582–1651). (F) Alignment of type I iodothyronine deiodinase (DIO1) amino acid sequence from nine model organisms. Rows 1–9 represent human (NP_000783.2), rhesus (NP_001116124.1), cow (NP_001116065.1), mouse (NP_031886.3), rat (NP_067685.5), chicken (NP_001091083.1), X. tropicalis (NP_001243226.1), X. laevis (NP_001089136.1) and zebrafish (NP_001007284.3) RefSeq protein sequences. The position of Sec (U) is highlighted in yellow and the extent of the thioredoxin_like super family domain (aa 8–247, mouse NP_031886.3) is indicated by the red line. The color gradation from yellow to darkest brown in the conservation histogram indicates highest (identical amino acids) to lowest (least conserved) scoring columns. The multiple protein sequence alignment was performed using Clustal Omega, version 1.2.4 (https://www.ebi.ac.uk/Tools/msa/clustalo/) and the alignment was viewed with Jalview, version 2.10.4b1 (http://www.jalview.org). The thioredoxin_like super family domain was identified by searching NCBI’s conserved domain database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi).
Alignment of manually curated DIO1 orthologs across phylogenetically distant model organisms showed conservation in selenoprotein length, location of Sec, and sequence, especially around the Sec residue (found at the catalytic site) and the thioredoxin_like super family domain (Figure 2F). The high sequence conservation of selenoproteins is used to predict orthologous models in other vertebrate species by the NCBI’s eukaryotic genome annotation pipeline.
Selenoprotein genes and selenoproteomes of vertebrate model organisms
Curation of 247 selenoprotein genes in nine model organisms resulted in 361 curated RefSeq records, including 115 alternatively spliced transcript variants consisting of 76 protein coding variants with Sec, 10 without Sec, and 29 non-protein coding variants (designated by NR_ accessions). The Sec-containing protein coding variants retained the conserved Sec-UGA codon and SECIS element. The non-Sec containing protein coding variants included those that lacked the exon containing the conserved Sec-UGA codon (e.g. NM_001039715.2 for DIO1) or the SECIS-containing 3′ exon (e.g. NM_203472.2 for SELENOS (41); thus, they were not expected to incorporate Sec. Most of the non-protein coding variants were likely candidates for NMD (e.g. NR_136692.1 for DIO1). The GeneID, RefSeq identifiers, transcript variant and isoform names, and other related information are provided in Supplementary Table 1. Comparative analysis of selenoproteins in nine model organisms is shown in Table 2.
Table 2.
Seleoprotein genes in vertebrate model organisms
| Froga | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Human | Rhesus macaque | Cow | Mouse | Rat | Chicken | X.tr | X.la | Zebrafishb |
| DIO1 | + | + | + | + | + | + | + | + (L) | + |
| DIO2 | + | + | + | + | + | + | + | + (L&S) | + |
| DIO3 | + | + | + | + | + | + | + | + (L&S) | + (a&b) |
| GPX1 | + | + | + | + | + | + | + | + (L&S) | + (a&b) |
| GPX2 | + | + | + | + | + | + | + | + (L&S) | + |
| GPX3 | + | + | + | + | + | + | + | + (L&S) | + |
| GPX4 | + | + | + | + | + | + | + | + (L&S) | + (a&b) |
| GPX6 | + | + | + | − | − | − | − | − | − |
| MSRB1 | + | + | + | + | + | + | + | + (L) | + (a&b) |
| SELENOF | + | + | + | + | + | + | + | + (L&S) | + |
| SELENOH | + | + | + | + | + | + | − | − | + |
| SELENOI | + | + | + | + | + | + | + | + (L&S) | + |
| SELENOK | + | + | + | + | + | + | + | + (L&S) | + |
| SELENOM | + | + | + | + | + | + | + | + (L&S) | + |
| SELENON | + | + | + | + | +c | + | + | + (L&S) | + |
| SELENOO | + | + | + | + | + | + | + | + (L) | + |
| SELENOP | + | + | + | + | + | + | + | + (L) | + |
| SELENOS | + | + | + | + | + | + | + | + (L&S) | + |
| SELENOT | + | + | + | + | + | + | + | + (L&S) | + (a&b) |
| SELENOV | + | + | + | + | + | − | − | − | − |
| SELENOW | + | + | + | + | + | + | + | + (L) | + |
| SEPHS2 | + | + | + | + | + | +c | + | + (L) | + |
| TXNRD1 | + | + | + | + | + | + | + | + (L&S) | − |
| TXNRD2 | + | + | + | + | + | + | + | + (S) | + |
| TXNRD3 | + | + | + | + | + | + | + | + (L&S) | + |
| Non-mammalian vertebrate selenoprotein genes | |||||||||
| SELENOE | − | − | − | − | − | − | − | − | + |
| SELENOJ | − | − | − | − | − | − | − | − | + |
| SELENOL | − | − | − | − | − | − | − | − | + |
| SELENOO2 | − | − | − | − | − | − | − | − | + |
| SELENOP2 | − | − | − | − | − | − | − | − | + |
| SELENOT2 | − | − | − | − | − | − | − | − | + |
| SELENOU | − | − | − | − | − | − | − | − | + (a&b) |
| SELENOW2 | − | − | − | − | − | − | + | + (L&S) | + (a&b) |
| Seleno-proteome | 25 | 25 | 25 | 24 | 24 | 25 | 23 | 23(39) | 30(37) |
aX.tr (Xenopus tropicalis); X.la (Xenopus laevis); L&S represent homeologs of X.la.
ba and b represent paralogs of zebrafish.
crat Selenon and chicken SEPHS2 genes have genome problems (see text).
The selenoproteomes of human, rhesus macaque and cow consist of the same 25 selenoproteins. A common core of 21 selenoproteins was also found in the remaining model organisms reviewed here, which all lack GPX6—the only selenoprotein missing in mouse and rat. All non-mammalian vertebrate models also lack SELENOV. Chicken additionally has a non-mammalian paralog (SELENOP2) and selenoprotein (SELENOU). X. tropicalis lacks SELENOH, and has a non-mammalian paralog, SELENOW2. X. laevis has the same set of 23 selenoproteins as X. tropicalis; however, because of its tetraploid genome (44) X. laevis has two homeologs (L and S) for 16 and a single homolog for seven selenoproteins. Zebrafish lacks TXNRD1 and has duplicates (a and b) of five selenoproteins, as well as additional non-mammalian paralogs (three singles, one duplicate) and selenoproteins (three singles, one duplicate), some of which were found only in zebrafish. The duplication of several selenoprotein genes in zebrafish was likely the result of whole genome duplication in the early evolution of bony fishes (45).
Our selenoprotein results in five mammalian model species and chicken (more below) are consistent with previous reports (2,3). However, our estimate of the number of selenoproteins in X. tropicalis and zebrafish of 23 and 37, respectively, was lesser by one than previously reported (3). We could not find SELENOH in X. tropicalis nor X. laevis. Together, blast and orthology (https://www.ncbi.nlm.nih.gov/gene/?Term=ortholog_gene_280636[group]) results showed the presence of SELENOH in many different vertebrate lineages, including birds, fishes, and two frog species (Nanorana parkeri and Rana catesbeiana), but not in Xenopus, suggesting that the lack of SELENOH is restricted to a subset of amphibians. SelenoDB 2.0 also does not list SELENOH for X. tropicalis. Zebrafish was previously reported to have 38 selenoprotein genes including two GPX3 paralogs, gpx3a and gpx3b (3); however, we found only a single copy of gpx3 gene (GeneID:798788) on chromosome 14. SelenoDB 2.0 has entries for both gpx3a and gpx3b genes. Blast results showed that the gpx3a sequence corresponded to gpx3 gene on chromosome 14; whereas, the gpx3b sequence aligned to a locus on chromosome 12, previously annotated as si:ch73-111m19.2 and very recently renamed gpx9 (GeneID:794084, ZDB id: ZDB-GENE-130603-13). The full extent of the gpx3b transcript and the predicted gpx3b protein was not clear, and the N- and C-termini of gpx3b lacked protein support; therefore, it was not certain if GeneID:794084 on chromosome 12 represents the gpx3b paralog or even encodes a selenoprotein.
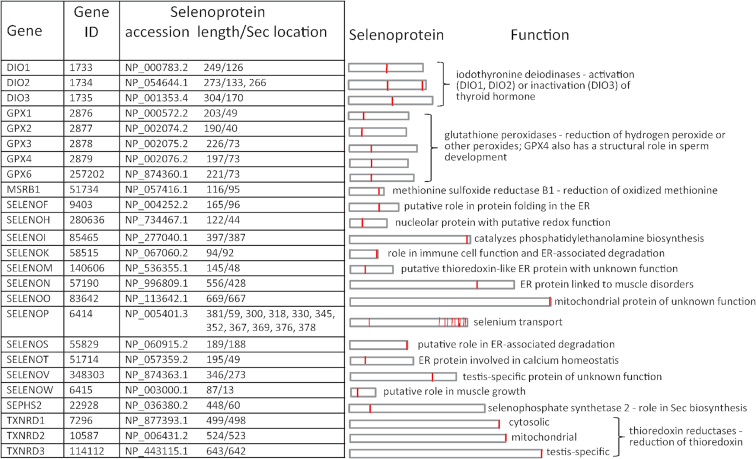
The human selenoproteome (Figure 3) exemplified some characteristics observed across all model organisms. Selenoproteins varied in length and in the location of their Sec residues. Most selenoproteins contained a single Sec residue; SELENOP was a major exception containing multiple Sec residues (10 in human and between 10–18 in other model organisms). SELENOP was also unique in that most of its mRNA contained two SECIS elements, while other selenoprotein mRNAs contained only one. The use of an alternative polyadenylation (polyA) site located between the two SECIS elements has been reported to result in 10–25% of SELENOP mRNA lacking the second SECIS element (46). Both polyA sites were annotated on the human (NM_005410.3) and rhesus macaque (NM_001159490.2) RefSeq transcripts (the upstream polyA site was not found in other model organisms). The same study showed that the two SECIS elements might have distinctive roles in recoding the various UGA codons, which could produce different SELENOP forms in plasma. Other forms of SELENOP have also been reported: under low Se conditions, cysteine was found to be inserted at various UGA sites (47) and C-terminally truncated isoforms were shown to result from termination at a subset of UGA sites (48). Given the complexity of SELENOP regulation, and the lack of information on the full-length nature of many isoforms, we elected to represent only the canonical SELENOP with all UGA-encoded Sec residues. DIO2 was also of interest because unlike the other two DIO family members (DIO1 and DIO3) its mRNA contained two in-frame UGA codons upstream of an in-frame universal stop codon. It has been reported (in human) that the second UGA codon can function either as a Sec or a stop codon resulting in two isoforms with one or two Sec residues; however, only the upstream Sec (conserved with the single Sec residue found at the active site in DIO1 and DIO3) was essential for enzyme activity (49). Additionally, the observation that the protein extension past the second UGA codon to the downstream stop codon is not conserved (3) suggests that the one-Sec form represents the canonical isoform. Therefore, both the one-Sec (NP_001353425.1) and two-Sec (NP_054644.1) forms were represented for the human DIO2 gene, and only the canonical one-Sec form for the DIO2 gene in other model organisms. Feature annotations with relevant comments for the potentially longer isoform were added to the latter RefSeq transcript records at the location of the second UGA and the downstream stop codon (e.g. NM_010050.4).
Figure 3.
Human selenoproteome. The table on the left shows details of 25 selenoprotein genes, RefSeq accessions for the encoded selenoproteins, their lengths (in aa) and the locations of Sec residue(s) within. The schematic on the right shows relative lengths of selenoproteins, with the vertical red lines indicating the positions of Sec residue(s). A brief description of selenoprotein function (known or putative) is also shown.
Several genome and annotation problems were encountered during the curation of this dataset. First, our initial genome blast analysis seemed to indicate that the SEPHS2 gene was missing in chicken (GRCg6a assembly) and many other avian genomes, corroborated by a similar conclusion in turkey (50). However, SEPHS2 is an essential gene for Sec synthesis (required for translation of all selenoproteins) whose function cannot be complemented by its paralog, SEPHS1 (51); therefore, a functional SEPHS2 gene was expected to be present in chicken and other birds. Tblastn analysis using human SEPHS2 protein (NP_036380.2) revealed the presence of many ESTs in chicken, which allowed the construction of a full-length RefSeq (NM_001366334.2, see Methods for details) for the chicken SEPHS2 gene (GeneID: 113219448). The encoded selenoprotein of 418 aa (NP_001353263.2) shared ∼70% and ∼80% identity with the mammalian and avian counterparts, respectively. NM_001366334.2 was overall very GC-rich and contained several tandem repeats in the 3′ region, which may contribute to the assembly problem seen with this gene. Similar problems observed in other avian species suggested a peculiarity of the genomic region containing the SEPHS2 gene that complicates proper genome assembly. However, there were a couple of exceptions: a complete 8-exon SEPHS2 gene could be identified in the genomes of Pseudopodoces humilis (Tibetan ground-tit, GeneID:102113712) and Parus major (Great Tit, GeneID:107198884). Curated RefSeqs for the SEPHS2 gene in both these avian species were created and included in Supplementary Table 1 to facilitate detection of this gene in other birds in the future. Also of note is that the multi-exon SEPHS2 gene found in non-mammalian vertebrates lacks synteny with the single-exon retrocopy found in placental mammals (3), suggesting that they are not true orthologs, though functionally alike. Second, the Selenon gene in rat (GeneID:362624) was found on chromosome 5, but because of gaps in the rat genome assembly (Rnor_6.0) in the 5′ region of this gene, and lack of suitable transcript data, it was not possible to provide a full-length Selenon RefSeq. The rat Selenon protein represented in SelenoDB 2.0 is N-terminally partial. Third, the mouse Smcp gene (GeneID: 17235) was previously thought to encode a selenoprotein due to the presence of three in-frame UGA codons in the 5′ region (52); however, this was subsequently shown to be incorrect (53). Based on the latter study, the previous mouse Smcp RefSeq (NM_008574.3) was updated to NM_008574.4 with a correct CDS encoding an N-terminally shorter protein of 143 aa (NP_032600.3), which is supported by the rat ortholog of similar length (NP_113724.1). Fourth, it was previously reported that fish have only two (txnrd1 and txnrd2) of the three txnrd family members found in mammals (3). We also detected only two txnrd genes in zebrafish; however, our analysis showed that they correspond to the mammalian TXNRD2 and TXNRD3 genes. Zebrafish txnrd2 (GeneID:798259) and txnrd3 (GeneID: 352924) genes are located on chromosomes 5 (3′ neighboring genes, tbx1 and gnb1l) and 6 (3′ neighboring genes, plxna1b, chchd6), respectively, in regions syntenic with their human counterparts. Zebrafish txnrd3 also had a better protein match to human TXNRD3 (GeneID: 114112) than to TXNRD1 (GeneID: 7296). Based on our analysis and consultation with ZFIN, the nomenclature on GeneID:352924 was updated to txnrd3.
Curation and annotation of SCR in vertebrates
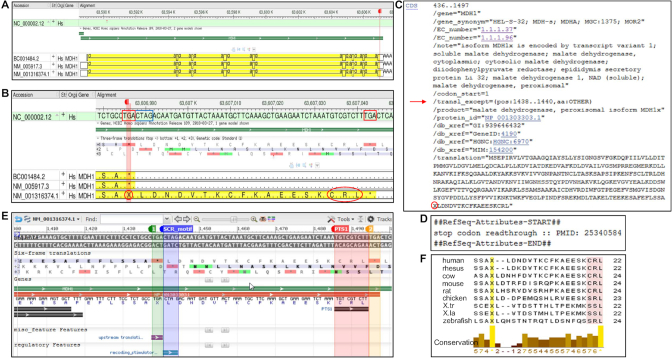
As with selenoprotein curation, representing recoding of stop codons as sense codons in cases of SCR required manual curation, illustrated by the human MDH1 gene (Figure 4). INSDC sequence BC001484.2 and NM_005917.3 were annotated with a conventional CDS terminating at the upstream stop codon (TGA) and encoding the canonical isoform of 334 aa (Figure 4A and B). A second RefSeq transcript (NM_001316374.1) identical to NM_005917.3 was created and manually annotated with a CDS terminating at an alternative in-frame stop codon (TGA) 19 codons downstream, resulting in a C-terminally extended isoform of 353 aa (NP_001303303.1). To ensure accurate annotation of the C-terminally extended CDS, a translation exception qualifier was manually added to the CDS as described for selenoprotein curation, except specifying the inserted amino acid as ‘OTHER’ (Figure 4C). This allowed annotation of the longer CDS, with the inclusion of ‘X’ in the protein sequence at the location of the upstream stop codon. A ‘stop codon readthrough’ attribute with published evidence (Figure 4D) and additional feature annotations (Figure 4E) were also manually added to the RefSeq record to highlight the functionally relevant features of this transcript. MDH1 contained a SCR-stimulatory motif (CTAG) adjacent to the upstream stop codon (TGA) and the encoded protein contained a tripeptide (CRL) at the C-terminus (Figure 4B and E), which has been shown to function as a peroxisomal targeting signal (PTS1) (19). The cryptic PTS1 signal in the C-terminal extension was conserved in mammals and other vertebrate model organisms (Figure 4F).
Figure 4.
Manual curation, annotation and analysis of SCR. (A) Genome Workbench display of alignment of few select transcripts to the human genomic sequence (chr2, NC_000002.12) at the location of the MDH1gene (GeneID:4190). The annotated CDS is highlighted in yellow; the red flag marks the location of the upstream stop codon in the 3′ terminal exon. (B) Expansion of the region around the red marker between the two in-frame TGA stop codons (boxed in red) shows translation of upstream TGA as a stop signal (*) or an amino acid (X, circled in red) in the +1 frame of the 3-frame translation track. The CTAG SCR_motif (boxed in blue) and the tri-peptide CRL (circled in red) at the C-terminus of NM_001316374.1 are shown. (C) GenBank format display of manually added /transl_except qualifier (red arrow) and the TGA-specified amino acid (X circled in red) on the public RefSeq record, NM_001316374.1. (D) Display of a portion of the COMMENT section on NM_001316374.1 showing manually added SCR-specific attribute with supporting publication. (E) Display of the graphical view of NM_001316374.1 configured to add four feature annotations highlighting the location of the two in-frame stop codons (1 and 2, nt 1438–1440 and 1495–1497, respectively), SCR_motif (nt 1441–1444) and PTS1 (CRL, nt 1486–1494). (F) Amino acid (aa) sequence alignment of MDH1 C-terminal extension from nine model organisms. Rows 1–9 represent human (NP_001303303.1, aa 332–353), rhesus (NP_001307034.1, aa 332–353), cow (NP_001307248.1, aa 332–355), mouse (NP_001303604.1, aa 332–355), rat (NP_001303806.1, aa 332–355), chicken (NP_001303820.1, aa 332–354), X. tropicalis (NP_001303830.1, 332–353), X. laevis (NP_001342383.1, aa 332–353) and zebrafish (NP_001303854.1, aa 332–354). The positions of TGA-specified amino acid (X) and the C-terminal tripeptide are highlighted. The multiple protein sequence alignment was performed using Clustal Omega and viewed with Jalview as described earlier.
SCR in vertebrate model organisms
Thirteen genes experimentally verified as authentic SCR cases and the corresponding orthologous model genes with conserved C-terminal extensions were selected for inclusion in the RefSeq database. Review of 93 genes exhibiting SCR in nine model organisms resulted in 94 curated SCR RefSeq records. The rabbit beta-hemoglobin gene (GeneID: 100009084) was also reviewed for historical reasons, being the first mammalian gene experimentally verified to undergo SCR (10,11), although this readthrough appears to be a species-specific event not conserved in other mammals. The GeneID, RefSeq identifiers, transcript and isoform names, and other related information are provided in Supplementary Table 2. Only the twin pair of RefSeqs encoding the canonical and C-terminally extended isoform (with or without scr attribute) are shown here. Other alternatively spliced transcript variants that differ only in the N-terminus may also undergo readthrough, resulting in additional C-terminally extended isoforms. However, we represented only one readthrough isoform per gene, unless there was published evidence for more readthrough forms, as described for AQP4 M1 and M23 isoforms (25). The summary of SCR curation in nine model organisms is shown in Table 3, and the alignment of C-terminal extensions in orthologs of various genes is shown in Supplementary Figure S1.
Table 3.
Summary of SCR curation in model organisms
| Froga | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Human | Rhesus macaque | Cow | Mouse | Rat | Chicken | X.tr | X.la | Zebrafishb | |
| Conservation of C-terminal extension | Rangec | |||||||||
| ACP2 | + | + | + | + | + | − | − | − | − | 11 |
| AGO1 | + | + | + | + | + | − | − | − | − | 34–38 |
| AQP4 | + | + | + | + | + | + | + | +(L&S) | + | 24–29 |
| LDHB | + | + | + | + | + | − | − | − | − | 7 |
| MAPK10 | + | + | + | + | + | + | + | +(L) | + | 14 |
| MDH1 | + | + | + | + | + | + | + | +(L&S) | +(a&b) | 19–21 |
| MPZ | + | + | + | + | + | + | + | +(L&S) | − | 64–68 |
| MTCH2 | + | + | + | + | + | − | − | − | − | 11 |
| OPRK1 | + | + | + | + | + | + | + | +(L&S) | + | 27–30 |
| OPRL1 | + | + | + | + | + | + | +d | +(L&S) | + | 29–32 |
| SACM1L | + | + | + | + | + | + | + | +(L) | − | 22–24 |
| VEGFA | + | − | + | + | + | − | − | − | − | 22–23 |
| VDR | + | + | − | − | − | − | − | − | − | 67 |
| Total # genes | 13 | 12 | 12 | 12 | 12 | 7 | 7 | 7(12) | 5(6) | |
aX.tr (Xenopus tropicalis); X.la (Xenopus laevis); L&S represent homeologs of X.la.
ba&b represent paralogs of zebrafish.
cRange of C-terminal extension (in aa) between orthologs, includes aa specified by the upstream stop codon.
dX.tr oprl1 gene has genome problem (see text).
SCR in many of the genes reviewed here was phylogenetically conserved. In seven (AQP4, MAPK10, MDH1, MPZ, OPRK1, OPRL1 and SACM1L) out of 13 genes, the C-terminal extension was conserved in vertebrates beyond mammals. For five genes (ACP2, AGO1, LDHB, MTCH2 and VEGFA), readthrough was conserved only in mammals. SCR in the VDR gene was represented only in human and rhesus macaque as it appears to be conserved only in the Old World monkey clade, including apes and human (17). Note, the X. tropicalis genome assembly (Xtropicalis_v7) has gaps in the 5′ region of the oprl1 gene (GeneID: 105945889) and in the absence of suitable transcript data, it was not possible to provide a full-length RefSeq for this gene. However, a predicted model (XM_012954875.1) had an intact 3′ coding region and 3′ UTR containing the conserved readthrough extension of 30 aa (see Supplementary Figure S1), so SCR in OPRL1 gene also seems to be conserved in this frog species. Interestingly, rhesus macaque VEGFA gene (GeneID:574209) contains a sequence variant in the downstream stop codon, changing from TGA in other primates to AGA. The AGA codon was confirmed to be valid and homozygous in reads from a DNA sample (SRR447492) from the same individual used for the Mmul_8.0 (GCA_000772875.3) assembly, and in an additional 15 samples of Indian-origin; however, 15 samples from Chinese-origin macaques showed a mix of homozygous AGA and heterozygous AGA/UGA genotypes (data not shown). The AGA codon would result in a predicted C-terminal extension twice the length found in other mammalian models (44 aa versus 22–23 aa). It is not clear if this sequence change would have an influence on SCR, so we elected to not represent the longer SCR form in rhesus macaque at this time.
The variation in the length of the C-terminal extension between species for a given gene ranged from 0 to 5 aa, mostly attributable to length differences between mammalian and non-mammalian species. Remarkably, the extension in MAPK10 (14 aa) and SACM1L (22 aa) was the same in all model organisms. Conservation of the sequence of the C-terminal extension, however, was quite variable. For instance, the extensions in a few genes (e.g. ACP2, AGO1, MAPK10, MTCH2 and VEGFA) were well conserved throughout, with the 11 aa of the MTCH2 extension being identical in all mammals. For some genes (e.g. OPRK1, OPRL1 and SACM1L), strongest conservation was seen near the C-terminus; and for LDHB and MDH1 (Figure 4E), it was mostly the conservation of the C-terminal tripeptide that functions as a peroxisomal targeting signal (PTS1). Thus, sequence conservation of the entire extension may not be important, depending on the role of the C-terminal extension.
The details of SCR curation in human are shown in Table 4 and exemplify some general observations. The upstream (readthrough) stop codon (uStop) in 12 out of 13 genes represented here is UGA, the leakiest of the three stop codons. MPZ is the only gene in this set that has UAG as uStop. The uStop is conserved across all model organisms for all genes. The downstream stop codon (dStop) was observed to be a mix of all three stop codons, and UGA was most prevalent even as a dStop, followed by UAG. The various signals that stimulate SCR in different genes are shown. There are a couple of genes (AGO1 and MTCH2) for which readthrough has been observed (15), but no stimulatory signal was identified. On the other hand, ACP2 and SACM1L have highly conserved stem-loop structures (thought to stimulate readthrough) and C-terminal extensions, but efficient readthrough has been difficult to measure in these genes (23). Genes with a UGACUAG motif have been best characterized (16–19) and constitute the majority of genes (7 out of 13) in this sampling. For MPZ with UAG as uStop, a quite different conserved six base pair sequence surrounding the UAG stop codon (AAA_UAG_CGG) was shown to be essential for readthrough (21). For VEGFA, a unique cis-acting element (Ax element) that binds a hnRNP A2/B1 trans-acting factor was shown to promote readthrough (15). The variety of stimulatory signals suggest the existence of different mechanisms for SCR in eukaryotes.
Table 4.
SCR curation in human
| Protein isoform | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Stop codonsa | Canonical | Readthrough | ||||||||
| Gene | Gene ID | uStop | dStop | SCR signal | Accession | Lengthb | Accession | Lengthb | C-terminal extensionb | Expt evidencec |
| ACP2 | 53 | UGA | UAG | 2° structure | NP_001601.1 | 423 | NP_001343945.1 | 434 | 11 | 23 |
| AGO1 | 26523 | UGA | UAG | ND | NP_036331.1 | 857 | NP_001304051.1 | 891 | 34 | 15 |
| AQP4 | 361 | UGA | UAA | UGACUAG | NP_001641.1 | 323 | NP_001304313.1 | 352 | 29 | 16 |
| LDHB | 3945 | UGA | UAG | UGACUAG | NP_002291.1 | 334 | NP_001302466.1 | 341 | 7 | 18, 19 |
| MAPK10 | 5602 | UGA | UGA | UGACUAG | NP_620448.1 | 464 | NP_001304998.1 | 478 | 14 | 16 |
| MDH1 | 4190 | UGA | UGA | UGACUAG | NP_005908.1 | 334 | NP_001303303.1 | 353 | 19 | 19 |
| MPZ | 4359 | UAG | UGA | AAAUAGCGG | NP_000521.2 | 248 | NP_001302420.1 | 312 | 64 | 12 |
| MTCH2 | 23788 | UGA | UAG | ND | NP_055157.1 | 303 | NP_001304160.1 | 314 | 11 | 15 |
| OPRK1 | 4986 | UGA | UGA | UGACUAG | NP_000903.2 | 380 | NP_001305426.1 | 409 | 29 | 16 |
| OPRL1 | 4987 | UGA | UAG | UGACUAG | NP_872588.1 | 370 | NP_001305782.1 | 399 | 29 | 16 |
| SACM1L | 22908 | UGA | UGA | 2° structure | NP_054735.3 | 587 | NP_001306000.1 | 609 | 22 | 23 |
| VEGFA | 7422 | UGA | UGA | Ax element | NP_001165097.1 | 191 | NP_001303939.1 | 213 | 22 | 15 |
| VDR | 7421 | UGA | UGA | UGACUAG | NP_000367.1 | 427 | NP_001351014.1 | 494 | 67 | 17 |
aUpstream (uStop) and downstream (dStop) stop codons, ND, not determined.
bThe numerical values indicate number of aa residues.
cReference for experimental evidence of readthrough.
DISCUSSION
The dual role of AUG in initiating translation and encoding an internal methionine was realized soon after the deciphering of the genetic code. However, the dual/multiple function of stop codon came to be known later with the discovery of UGA recoding to specify Sec (resulting in the expansion of the genetic code to include Sec as the 21st amino acid) and the recoding of all three stop codons as sense amino acids via SCR. The ambiguous nature of stop codons poses a problem for conventional computational tools to accurately predict selenoprotein genes or detect SCR, resulting in misannotation of selenoprotein CDSes on transcripts and genomic sequences, and failure to find novel readthrough isoforms. Several computational methods have been developed to detect selenoprotein genes: for example, one which combines the prediction of eukaryotic SECIS element using SECISearch3 with another tool (Seblastian) that predicts selenoprotein sequences encoded upstream of SECIS elements (32); and a homology-based tool (Selenoprofiles) designed to identify members of known selenoprotein families in sequenced genomes (54). However, their use, in conjunction with standard genome annotation pipelines, is not widespread. Our dataset of manually curated and annotated full-length selenoprotein transcript and protein products can serve as a valuable resource for individual research projects, large scale computational analyses, and genome annotation. It expands on the vertebrate selenoprotein data tracked by other databases, such as SelenoDB 2.0, and repositories of translational recoding events, such as Recode-2. Similarly, no single tool is available that can effectively predict SCR in genomes, so our curated set of RefSeqs representing readthrough in several model organisms can be a useful resource for SCR studies, and to populate Recode-2 database, which currently has no entry for readthrough in mammalian cellular genes.
The manually curated selenoprotein dataset described here is unique in its representation of full-length selenoprotein transcript and protein sequences, additional alternatively spliced transcript variants, and expanded functional annotation including gene summary, relevant publications, biological attributes, and feature annotations. The inclusion of nine model organisms in this curation effort allowed comparison of selenoproteomes across phylogenetically distant vertebrate species, and with the existing data on various selenoproteomes in publications and specialized databases, such as SelenoDB 2.0 and Recode-2. For instance, our analysis revealed the restricted loss of selenoh gene in Xenopus. The X. laevis selenoproteome reported here is novel. It has the same complement of selenoprotein genes as X. tropicalis; however, because of its tetraploid genome, X. laevis has additional selenoproteins derived from homeologous copies of some selenoprotein genes, making its selenoproteome larger (with 39 selenoproteins), comparable to that of zebrafish (with 37 selenoproteins). Manual curation was essential for correcting errors in the representation of selenoprotein transcripts, CDSes and proteins. Errors resulted from misannotation of UGA as a stop codon on primary transcripts or predicted models, or the opposite, i.e. misinterpretation of UGA codons as Sec codons, exemplified by the mouse Smcp gene mistaken to encode a selenoprotein. Manual curation also included standardizing nomenclature of selenoprotein genes across all model organisms. We encountered problems in representing full-length RefSeqs for rat Selenon and X. tropicalis oprl1 genes because of genome issues, which also affected correct model prediction in these regions. Besides the challenge of accurately predicting the dual meaning of stop codons, poor genome quality adds to the problem of correctly annotating full-length gene products by conventional annotation pipelines. This is no better illustrated than the curious problem we faced with the identification of SEPHS2 gene in chicken and other avian species. At first glance it appeared as if this very essential gene for Sec synthesis was missing in birds because it could not be detected in many of their genomes. However, the identification of ESTs for SEPHS2 gene in chicken and other avian species confirmed that this gene was indeed present in birds. It is thus important to realize the limitations of automated genome annotation of not only recoded genes, but other genes as well, and the impact of such limitations on evolutionary analyses. Therefore, manually curated data are an important complement to computational genome annotation by NCBI’s eukaryotic genome annotation pipeline.
Different mechanisms exist for proteins to acquire new domains or functional modules, such as alternative splicing, leaky AUG scanning, and SCR. SCR, which operates at the level of translation termination to generate diversity at the C-terminus, is complementary to leaky AUG scanning, a means for protein diversification at the N-terminus via alternative translation initiation. The different N- or C-termini can confer different subcellular localization or function; for example NP_001173.2 and NP_001188306.1 for human ALDH7A1 gene (GeneID:501) generated by alternative AUG usage are localized in the mitochondria and cytosol, respectively. Similarly, pairs of LDHB and MDH1 isoforms generated by alternative stop codon usage are localized in the cytosol and peroxisomes. The exact function of many C-terminally extended isoforms is not known; however, the conservation of C-terminal extension in vertebrates in many SCR cases described here suggests that readthrough is not simply due to chance, or an accidental misreading of the stop codon, and likely serves a function. Some readthrough isoforms may have normal physiological roles, for instance, peroxisomal LDHx in lactate/pyruvate shuttling and glyoxylate metabolism (18), while some may have pathological roles, such as L-MPZ in stimulating the production of anti L-MPZ antibodies in patients with neuropathies (12). Inducing stop codon readthrough with aminoglycoside antibiotics has gained prominence for treatment of inherited diseases, such as cystic fibrosis, Duchenne muscular dystrophy, and some cancers, caused by genes containing premature termination codon (PTC) mutations (55). Studies in animal models and cultured cells have shown that aminoglycosides can promote PTC readthrough to partially restore full-length protein synthesis and function. However, as for SCR, the efficiency of aminoglycoside-induced PTC readthrough is context dependent, requiring identification of pathogenic mutation context in each patient for effective treatment. While PTC readthrough mechanisms may not be the same as those for SCR, insights gained from SCR studies on factors influencing efficient readthrough, choice of amino acid inserted at the readthrough site, and the development of reliable assays to measure readthrough efficiency, will be valuable in designing therapies for treatment of PTC disorders.
The RefSeq database is unique, not only in its offering of annotated reference sequence dataset of transcripts, proteins and genomes across a broad taxonomic scope, but also in its active maintenance and provision of updates for a diverse set of species in a timely manner to incorporate new data types and current knowledge (26). The high quality of RefSeq products is achieved through a combinatorial approach leveraging computation, manual curation and collaboration, with a focus on representing only full-length and non-redundant data. Manual curation has a vital role in the annotation process in improving the quality of data, which in turn adds to the quality of genome annotation. This is particularly true for genes with exceptional biology that require manual curation for accurate annotation of their gene products. RefSeq is also notable as one of the major reference sequence databases to represent various recoding events, such as +1 ribosomal frameshifting in antizyme genes (27), –1 ribosomal frameshifting in mammalian PEG10 gene (e.g. NM_015068.3: NP_055883.2), and stop codon recoding (Sec insertion and SCR, this report). The RefSeq dataset is widely used, including by the recoding community (3,16,19,21,50), for a variety of research purposes. To cope with the growing number of vertebrate sequenced genomes, our goal going forward is to consider automatic annotation of selenoprotein genes. Recent improvements in NCBI’s eukaryotic genome annotation pipeline has enabled high quality selenoprotein model predictions, including automatic addition of /transl_except qualifier and annotation of Sec as ‘U’ (e.g. XM_009293291.2 for zebrafish gpx2). A similar approach was recently incorporated into the RefSeq prokaryotic genome annotation pipeline and used to successfully annotate several selenoproteins across many bacterial species (56). Manual curation, however, will continue to be needed to provide high quality annotations of important gene sets based on well-supported transcripts for key eukaryotic genomes, and to support improvements to the genome annotation pipeline to model both typical and atypical biology.
DATA AVAILABILITY
The RefSeq data presented here are publicly available from several resources at NCBI (https://www.ncbi.nlm.nih.gov). Information about selenoprotein and SCR genes, transcripts and proteins can be accessed interactively from the Gene, Nucleotide and Protein databases, respectively; from reciprocal links on the Gene and RefSeq records, and from BLAST databases. These data are also included in the bi-monthly comprehensive RefSeq release from: ftp://ftp.ncbi.nlm.nih.gov/refseq/release/. Shown below are examples of web queries relevant to recoding datasets, and useful for retrieving subsets of related records:
Query the Gene database with a gene symbol, for example, dio1[sym], to retrieve dio1 orthologs, which are displayed in a tabular format, and individual Gene records in the set can be retrieved via the link on the Name/GeneID column.
Query the Nucleotide or Protein database with a specific attribute, for example, ‘protein contains selenocysteine’[prop] or ‘stop codon readthrough’[prop] to retrieve all RefSeq records annotated with the respective attribute, not just the dataset represented here. For example, the ‘protein contains selenocysteine’[prop] search currently retrieves 552 RefSeq records for 472 selenoprotein genes from 37 taxa. The sidebar on the right shows per-taxon counts, which can be navigated to display taxon-specific data. Using ‘Find related data’ and selecting Gene database will show the number of selenoprotein genes. The datasets retrieved by both web queries can be directly downloaded using the available ‘Send to’ options.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to acknowledge RefSeq curators Catherine Farrell and David Webb, and developers Vamsi Kodali and Alexander Souvorov for helpful consults. We would also like to thank our NAR referees for their valuable insights and observations, which helped with annotation and improving this manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Research Program of the National Institutes of Health, National Library of Medicine. Funding for open access charge: Intramural Research Program of the National Institutes of Health, National Library of Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1. Hatfield D.L., Tsuji P.A., Carlson B.A., Gladyshev V.N.. Selenium and selenocysteine: Roles in cancer, health, and development. Trends Biochem. Sci. 2014; 39:112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kryukov G.V., Castellano S., Novoselov S.V., Lobanov A.V., Zehtab O., Guigo R., Gladyshev V.N.. Characterization of mammalian selenoproteomes. Science. 2003; 300:1439–1443. [DOI] [PubMed] [Google Scholar]
- 3. Mariotti M., Ridge P.G., Zhang Y., Lobanov A.V., Pringle T.H., Guigo R., Hatfield D.L., Gladyshev V.N.. Composition and evolution of the vertebrate and mammalian selenoproteomes. PLoS One. 2012; 7:e33066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Moghadaszadeh B., Beggs A.H.. Selenoproteins and their impact on human health through diverse physiological pathways. Physiology (Bethesda). 2006; 21:307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pitts M.W., Hoffmann P.R.. Endoplasmic reticulum-resident selenoproteins as regulators of calcium signaling and homeostasis. Cell Calc. 2018; 70:76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ursini F., Heim S., Kiess M., Maiorino M., Roveri A., Wissing J., Flohe L.. Dual function of the selenoprotein PHGPx during sperm maturation. Science. 1999; 285:1393–1396. [DOI] [PubMed] [Google Scholar]
- 7. Schomburg L., Schweizer U., Holtmann B., Flohe L., Sendtner M., Kohrle J.. Gene disruption discloses role of selenoprotein P in selenium delivery to target tissues. Biochem. J. 2003; 370:397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Castets P., Lescure A., Guicheney P., Allamand V.. Selenoprotein N in skeletal muscle: from diseases to function. J. Mol. Med. 2012; 90:1095–1107. [DOI] [PubMed] [Google Scholar]
- 9. Firth A.E., Brierley I.. Non-canonical translation in RNA viruses. J. Gen. Virol. 2012; 93:1385–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chittum H.S., Lane W.S., Carlson B.A., Roller P.P., Lung F.D., Lee B.J., Hatfield D.L.. Rabbit beta-globin is extended beyond its UGA stop codon by multiple suppressions and translational reading gaps. Biochemistry. 1998; 37:10866–10870. [DOI] [PubMed] [Google Scholar]
- 11. Geller A.I., Rich A.. A UGA termination suppression tRNATrp active in rabbit reticulocytes. Nature. 1980; 283:41–46. [DOI] [PubMed] [Google Scholar]
- 12. Yamaguchi Y., Hayashi A., Campagnoni C.W., Kimura A., Inuzuka T., Baba H.. L-MPZ, a novel isoform of myelin P0, is produced by stop codon readthrough. J. Biol. Chem. 2012; 287:17765–17776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jungreis I., Lin M.F., Spokony R., Chan C.S., Negre N., Victorsen A., White K.P., Kellis M.. Evidence of abundant stop codon readthrough in Drosophila and other metazoa. Genome Res. 2011; 21:2096–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dunn J.G., Foo C.K., Belletier N.G., Gavis E.R., Weissman J.S.. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. eLife. 2013; 2:e01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eswarappa S.M., Potdar A.A., Koch W.J., Fan Y., Vasu K., Lindner D., Willard B., Graham L.M., DiCorleto P.E., Fox P.L.. Programmed translational readthrough generates antiangiogenic VEGF-Ax. Cell. 2014; 157:1605–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Loughran G., Chou M.Y., Ivanov I.P., Jungreis I., Kellis M., Kiran A.M., Baranov P.V., Atkins J.F.. Evidence of efficient stop codon readthrough in four mammalian genes. Nucleic Acids Res. 2014; 42:8928–8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Loughran G., Jungreis I., Tzani I., Power M., Dmitriev R.I., Ivanov I.P., Kellis M., Atkins J.F.. Stop codon readthrough generates a C-terminally extended variant of the human vitamin D receptor with reduced calcitriol response. J. Biol. Chem. 2018; 293:4434–4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schueren F., Lingner T., George R., Hofhuis J., Dickel C., Gartner J., Thoms S.. Peroxisomal lactate dehydrogenase is generated by translational readthrough in mammals. eLife. 2014; 3:e03640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stiebler A.C., Freitag J., Schink K.O., Stehlik T., Tillmann B.A., Ast J., Bolker M.. Ribosomal readthrough at a short UGA stop codon context triggers dual localization of metabolic enzymes in Fungi and animals. PLos Genet. 2014; 10:e1004685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Beier H., Grimm M.. Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res. 2001; 29:4767–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yamaguchi Y., Baba H.. Phylogenetically conserved sequences around myelin P0 stop codon are essential for translational readthrough to produce L-MPZ. Neurochem. Res. 2018; 43:227–237. [DOI] [PubMed] [Google Scholar]
- 22. Firth A.E., Wills N.M., Gesteland R.F., Atkins J.F.. Stimulation of stop codon readthrough: frequent presence of an extended 3′ RNA structural element. Nucleic Acids Res. 2011; 39:6679–6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Loughran G., Howard M.T., Firth A.E., Atkins J.F.. Avoidance of reporter assay distortions from fused dual reporters. RNA. 2017; 23:1285–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xin H., Zhong C., Nudleman E., Ferrara N.. Evidence for Pro-angiogenic functions of VEGF-Ax. Cell. 2016; 167:275–284. [DOI] [PubMed] [Google Scholar]
- 25. De Bellis M., Pisani F., Mola M.G., Rosito S., Simone L., Buccoliero C., Trojano M., Nicchia G.P., Svelto M., Frigeri A.. Translational readthrough generates new astrocyte AQP4 isoforms that modulate supramolecular clustering, glial endfeet localization, and water transport. Glia. 2017; 65:790–803. [DOI] [PubMed] [Google Scholar]
- 26. O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D. et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016; 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rajput B., Murphy T.D., Pruitt K.D.. RefSeq curation and annotation of antizyme and antizyme inhibitor genes in vertebrates. Nucleic Acids Res. 2015; 43:7270–7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Karsch-Mizrachi I., Takagi T., Cochrane G.. The international nucleotide sequence database collaboration. Nucleic Acids Res. 2018; 46:D48–D51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kervestin S., Jacobson A.. NMD: A multifaceted response to premature translational termination. Nat. Rev. Mol. Cell Biol. 2012; 13:700–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Romagne F., Santesmasses D., White L., Sarangi G.K., Mariotti M., Hubler R., Weihmann A., Parra G., Gladyshev V.N., Guigo R. et al. SelenoDB 2.0: annotation of selenoprotein genes in animals and their genetic diversity in humans. Nucleic Acids Res. 2014; 42:D437–D443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bekaert M., Firth A.E., Zhang Y., Gladyshev V.N., Atkins J.F., Baranov P.V.. Recode-2: New design, new search tools, and many more genes. Nucleic Acids Res. 2010; 38:D69–D74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mariotti M., Lobanov A.V., Guigo R., Gladyshev V.N.. SECISearch3 and Seblastian: New tools for prediction of SECIS elements and selenoproteins. Nucleic Acids Res. 2013; 41:e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gladyshev V.N., Arner E.S., Berry M.J., Brigelius-Flohe R., Bruford E.A., Burk R.F., Carlson B.A., Castellano S., Chavatte L., Conrad M. et al. Selenoprotein gene nomenclature. J. Biol. Chem. 2016; 291:24036–24040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Burt D.W., Carre W., Fell M., Law A.S., Antin P.B., Maglott D.R., Weber J.A., Schmidt C.J., Burgess S.C., McCarthy F.M.. The chicken gene nomenclature committee report. BMC Genomics. 2009; 10:S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Eppig J.T. Mouse genome informatics (MGI) Resource: Genetic, genomic, and biological knowledgebase for the laboratory mouse. ILAR J. 2017; 58:17–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Karimi K., Fortriede J.D., Lotay V.S., Burns K.A., Wang D.Z., Fisher M.E., Pells T.J., James-Zorn C., Wang Y., Ponferrada V.G. et al. Xenbase: A genomic, epigenomic and transcriptomic model organism database. Nucleic Acids Res. 2018; 46:D861–D868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ruzicka L., Bradford Y.M., Frazer K., Howe D.G., Paddock H., Ramachandran S., Singer A., Toro S., Van Slyke C.E., Eagle A.E. et al. ZFIN, The zebrafish model organism database: Updates and new directions. Genesis. 2015; 53:498–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shimoyama M., De Pons J., Hayman G.T., Laulederkind S.J., Liu W., Nigam R., Petri V., Smith J.R., Tutaj M., Wang S.J. et al. The rat genome database 2015: Genomic, phenotypic and environmental variations and disease. Nucleic Acids Res. 2015; 43:D743–D750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yates B., Braschi B., Gray K.A., Seal R.L., Tweedie S., Bruford E.A.. Genenames.org: The HGNC and VGNC resources in 2017. Nucleic Acids Res. 2017; 45:D619–D625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Howard M.T., Aggarwal G., Anderson C.B., Khatri S., Flanigan K.M., Atkins J.F.. Recoding elements located adjacent to a subset of eukaryal selenocysteine-specifying UGA codons. EMBO J. 2005; 24:1596–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bubenik J.L., Miniard A.C., Driscoll D.M.. Alternative transcripts and 3′UTR elements govern the incorporation of selenocysteine into selenoprotein S. PLoS One. 2013; 8:e62102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hatfield D.L., Gladyshev V.N.. How selenium has altered our understanding of the genetic code. Mol. Cell. Biol. 2002; 22:3565–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Howard M.T., Carlson B.A., Anderson C.B., Hatfield D.L.. Translational redefinition of UGA codons is regulated by selenium availability. J. Biol. Chem. 2013; 288:19401–19413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Session A.M., Uno Y., Kwon T., Chapman J.A., Toyoda A., Takahashi S., Fukui A., Hikosaka A., Suzuki A., Kondo M. et al. Genome evolution in the allotetraploid frog Xenopus laevis. Nature. 2016; 538:336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Taylor J.S., Braasch I., Frickey T., Meyer A., Van de Peer Y.. Genome duplication, a trait shared by 22000 species of ray-finned fish. Genome Res. 2003; 13:382–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wu S., Mariotti M., Santesmasses D., Hill K.E., Baclaocos J., Aparicio-Prat E., Li S., Mackrill J., Wu Y., Howard M.T. et al. Human selenoprotein P and S variant mRNAs with different numbers of SECIS elements and inferences from mutant mice of the roles of multiple SECIS elements. Open Biol. 2016; 6:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Turanov A.A., Everley R.A., Hybsier S., Renko K., Schomburg L., Gygi S.P., Hatfield D.L., Gladyshev V.N.. Regulation of selenocysteine content of human selenoprotein P by dietary selenium and insertion of cysteine in place of selenocysteine. PLoS One. 2015; 10:e0140353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ma S., Hill K.E., Caprioli R.M., Burk R.F.. Mass spectrometric characterization of full-length rat selenoprotein P and three isoforms shortened at the C terminus. Evidence that three UGA codons in the mRNA open reading frame have alternative functions of specifying selenocysteine insertion or translation termination. J. Biol. Chem. 2002; 277:12749–12754. [DOI] [PubMed] [Google Scholar]
- 49. Salvatore D., Harney J.W., Larsen P.R.. Mutation of the Secys residue 266 in human type 2 selenodeiodinase alters 75Se incorporation without affecting its biochemical properties. Biochimie. 1999; 81:535–538. [DOI] [PubMed] [Google Scholar]
- 50. Sunde R.A., Sunde G.R., Sunde C.M., Sunde M.L., Evenson J.K.. Cloning, sequencing, and expression of selenoprotein transcripts in the turkey (Meleagris gallopavo). PLoS One. 2015; 10:e0129801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xu X.M., Carlson B.A., Irons R., Mix H., Zhong N., Gladyshev V.N., Hatfield D.L.. Selenophosphate synthetase 2 is essential for selenoprotein biosynthesis. Biochem. J. 2007; 404:115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Karimpour I., Cutler M., Shih D., Smith J., Kleene K.C.. Sequence of the gene encoding the mitochondrial capsule selenoprotein of mouse sperm: Identification of three in-phase TGA selenocysteine codons. DNA Cell Biol. 1992; 11:693–699. [DOI] [PubMed] [Google Scholar]
- 53. Cataldo L., Baig K., Oko R., Mastrangelo M.A., Kleene K.C.. Developmental expression, intracellular localization, and selenium content of the cysteine-rich protein associated with the mitochondrial capsules of mouse sperm. Mol. Reprod. Dev. 1996; 45:320–331. [DOI] [PubMed] [Google Scholar]
- 54. Mariotti M., Guigo R.. Selenoprofiles: profile-based scanning of eukaryotic genome sequences for selenoprotein genes. Bioinformatics. 2010; 26:2656–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bidou L., Allamand V., Rousset J.P., Namy O.. Sense from nonsense: therapies for premature stop codon diseases. Trends Mol. Med. 2012; 18:679–688. [DOI] [PubMed] [Google Scholar]
- 56. Haft D.H., DiCuccio M., Badretdin A., Brover V., Chetvernin V., O’Neill K., Li W., Chitsaz F., Derbyshire M.K., Gonzales N.R. et al. RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Res. 2018; 46:D851–D860. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RefSeq data presented here are publicly available from several resources at NCBI (https://www.ncbi.nlm.nih.gov). Information about selenoprotein and SCR genes, transcripts and proteins can be accessed interactively from the Gene, Nucleotide and Protein databases, respectively; from reciprocal links on the Gene and RefSeq records, and from BLAST databases. These data are also included in the bi-monthly comprehensive RefSeq release from: ftp://ftp.ncbi.nlm.nih.gov/refseq/release/. Shown below are examples of web queries relevant to recoding datasets, and useful for retrieving subsets of related records:
Query the Gene database with a gene symbol, for example, dio1[sym], to retrieve dio1 orthologs, which are displayed in a tabular format, and individual Gene records in the set can be retrieved via the link on the Name/GeneID column.
Query the Nucleotide or Protein database with a specific attribute, for example, ‘protein contains selenocysteine’[prop] or ‘stop codon readthrough’[prop] to retrieve all RefSeq records annotated with the respective attribute, not just the dataset represented here. For example, the ‘protein contains selenocysteine’[prop] search currently retrieves 552 RefSeq records for 472 selenoprotein genes from 37 taxa. The sidebar on the right shows per-taxon counts, which can be navigated to display taxon-specific data. Using ‘Find related data’ and selecting Gene database will show the number of selenoprotein genes. The datasets retrieved by both web queries can be directly downloaded using the available ‘Send to’ options.