Abstract
Timely stalling and resumption of RNA polymerases at damaged chromatin are actively regulated processes. Prior work showed an importance of FACT histone chaperone in such process. Here we provide a new role of OTUD5 deubiquitinase in the FACT-dependent process. Through a DUB RNAi screen, we found OTUD5 as a specific stabilizer of the UBR5 E3 ligase. OTUD5 localizes to DNA double strand breaks (DSBs), interacts with UBR5 and represses the RNA Pol II elongation and RNA synthesis. OTUD5 co-localizes and interacts with the FACT component SPT16 and antagonizes the histone H2A deposition at DSB lesions. OTUD5 interacts with UBR5 and SPT16 independently through two distinct regions, and both interactions are necessary for arresting the Pol II elongation at lesions. These analyses suggested that the catalytic (through UBR5 stabilization) as well as scaffolding (through FACT binding) activities of OTUD5 are involved in the FACT-dependent transcription. We found that a cancer-associated missense mutation within the OTUD5 Ubiquitin Interacting Motif (UIM) abrogates the FACT association and the Pol II arrest, providing a possible link between the transcriptional regulation and tumor suppression. Our work establishes OTUD5 as a new regulator of the DNA damage response, and provides an insight into the FACT-dependent transcription at damaged chromatin.
INTRODUCTION
Deubiquitinating enzymes (DUBs) are important regulators of many biological processes. DUBs process ubiquitin precursors to release free ubiquitins, cleave ubiquitin chains from substrates or edit chains to modify the functional outcome. DUBs are subject to various forms of regulations, such as phosphorylation and being bound to co-factors, which can regulate catalytic activity, stability or localization (1,2). DUBs are also often physically coupled to E3 ubiquitin ligases, with different functional consequences; a DUB may counteract E3 activity on substrate ubiquitination, or promote E3 activity by stabilizing the E3 itself (3,4). Among the several types of DUBs is the subfamily of OTU (Ovarian Tumor) DUBs, which are cysteine proteases that regulate various biological processes including the immune signaling responses (5). Of note, some of the OTU family members such as OTUB1, OTUB2, OTUD4 participate in the regulation of DNA repair or DNA damage responses (6–10).
In response to genotoxic stresses, various mechanisms operate to maintain the genome and transcriptome integrity. One such response is the rapid arrest of transcription at or nearby the DNA lesions. The transcriptional arrest may facilitate the access of DNA repair machineries to the lesions enabling the repair processes, which is followed by resumption of transcription upon recovery. Transcription obstacles, including DNA damage, can also lead to ubiquitination and degradation of elongating RNA polymerases as a last resort (11). DNA lesions such as UV-induced CPDs induce direct stalling of RNA polymerases in cis, triggering transcription-coupled nucleotide-excision repair, but transcription arrest can also occur distant from DNA breakage sites (12,13), through kinases or protein-modifying activities. For example, Ataxia Telangiectasia Mutated (ATM), Poly ADP-Ribose Polymerase (PARP) and DNA-dependent Protein Kinase (DNA-PK) repress transcription near or distant from double strand break (DSB) sites (13–15), Ataxia Telangiectasia and Rad3-related (ATR) induces transcription repression at stalled replication forks (16). A Polycomb gene repressor component BMI1 also regulates damage-associated transcriptional repression (both DSB and UV-induced lesions), and it may do so by inducing histone H2A ubiquitination, or at least in part in collaboration with UBR5 E3 ligase. For the latter, the proposed mechanism involves UBR5-induced repression of FACT histone chaperone complex and arrest of RNA Pol II elongation at UVC or nuclease-induced DSB lesions (17).
Here we identify that the OTUD5 DUB is a new regulator of the DNA damage-induced transcriptional repression. Through a DUB siRNA screen, we found that OTUD5 is a specific DUB that stabilizes and promotes foci formation of UBR5. Both UBR5 and OTUD5 localizes to DSB lesions, where they interact. Similar to UBR5 depletion, OTUD5-depleted cells show misregulation of the FACT-dependent Pol II elongation and histone H2A deposition at damaged chromatin. Through mapping analysis, we found that OTUD5 associates with UBR5 and the FACT component SPT16 through the disordered N- and C-termini, respectively. When the either interaction-deficient OTUD5 mutant is expressed in OTUD5 KO cells, the cells are defective in arresting Pol II elongation and nascent RNA synthesis, underscoring the importance of the engagement of the UBR5–OTUD5 complex with SPT16 in transcription repression. We have further identified that a cancer-associated missense mutation in the UIM of OTUD5 abrogates the association of OTUD5 with the FACT components SPT16 and SSRP1, and that this leads to loss of the transcriptional repression at the damaged sites. Our work provides a new insight into the FACT-mediated transcription during the DNA damage response.
MATERIALS AND METHODS
Cell lines, plasmids and chemicals
HeLa, 293T and U2OS cells were grown in Dulbecco’s-Modified Eagle’s Medium (DMEM) supplemented with 10% Fetal Bovine Serum (FBS) and L-glutamine. The HeLa CRISPR-Cas9 UBR5 knockout was described previously (17). OTUD5 knockout HeLa clones were generated using the CRISPR-Cas9 plasmid and Double Nickase (Cas9 D10A mutant) plasmid synthesized from Santa Cruz Biotechnology. In brief, HeLa cells were transfected with these plasmids, transiently selected with Puromycin (1 µg/ml) for 2 days, and surviving cells were isolated into single cells, amplified, then checked by anti-OTUD5 western blots. The targeted genomic regions of isolated clones were amplified and sequenced (by Washington University, GEiC) to confirm the knockouts (see Supplementary Figure S3). U2OS cells stably expressing mCherry-LacI-Fok1 fusion protein that induces DSBs at a single genomic locus is previously described (18). The coding sequence for histone H2A (isolated from T80 ovarian cells) was cloned into the pSNAPf Vector (NEB). The U2OS cells stably expressing SNAP-H2A were generated through G418 selection and clonal isolation. pDEST_Tet_CMV_FLAG-OTUD5 was a gift from Wade Harper (Addgene plasmid # 22610) (19) and was used for generating HeLa cells stably expressing FLAG-OTUD5. pDEST10-6xHis-UBR5 plasmid was a gift from Sally Kornbluth (20) and was used for recombinant protein production from SF9 cells. OTUD5 cDNA was cloned into p3xFlag-CMV, pBabe-puro, pGEX 6p-1 and pLEGFP-N vectors. Mutations were introduced using Quikchange Site-Directed Mutagenesis Kit (Stratagene). Etoposide (Selleck Chemical), Mitomycin C (Sigma Aldrich), Bleomycin (Selleck Chemical), 4-OHT (Sigma Aldrich), and Shield-1 (Chemipharma), SNAP-Cell® TMR-Star (NEB) were purchased.
RNAi
Cells were cultured in medium without antibiotics and transfected once with 20 nM siRNA (final concentrations) using the RNAiMAX (Invitrogen) reagent following the manufacturer’s protocol. The following siRNA sequences were synthesized by QIAGEN:
UBR5#1: CAGGUAUGCUUGAGAAAUAAU,
UBR5#2: GAAUGUAUUGGAACAGGCUACUAUU,
SPT16: ACCGGAGUAAUCCGAAACUGA,
OTUD5 #1: GGCCGGCUUGGACAAUGAATT
OTUD5 #2: GGGUGCCGAAGAIAGACAATT
The following siRNA sequences were synthesized by Bioneer:
OTUB1: CTCCGACTACCTTGTGGTCTA,
OTUB2: TTCCGTTTACCTGCTCTATAA
OTUD1: CTGGTGTACCTTCATCTATGA,
OTUD2: GAGUACUGUGACUGGAUCA
OTUD3: CAGAAGCGAAGCAGAGGCGAA,
OTUD4: CTGTATGAGAAGGTATTTAAA
OTUD6A: AAGAGTGAACAGCAGCGCATA,
OTUD6B: GGUCAUUGAUAGCAAGUAATT
OTUD7A: CACAAGUCGCAGACCUACA,
OTUD7B: ACCGAGTGGCTGATTCCTATA
TRABID: CACCUUAAAAGAUCCAUCU,
OTULIN: CGUAUGCCCUUGAUUGGUU
VCPIP1: CUGGUAACCCACACCUUGA,
A20: CUCAGUUUCGGGAGAUCAU
BRCA1: CUGAAACCAUACAGCUUCA
Western blots and antibodies
Cell extracts were run on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and then transferred to a PVDF membrane (Bio-Rad). Membranes were probed with primary antibodies overnight at 4°C, followed by incubation of HRP-conjugated secondary antibodies (Cell Signaling Technologies) for 1 hour. The bound antibodies were viewed via Pierce ECL Western Blotting Substrate (Thermo Scientific). The following primary antibodies were used: α-SPT16, α-SSRP1, α-UBR5, α-H2A, α-Rpb1, α-γH2AX, α-OTUD5, α-53BP1, α-SIRT3 rabbit polyclonal antibodies from Cell Signaling Technologies; α-NELF-E, α-RPB1 (CTD pSer2) rabbit polyclonal antibodies from Abcam; α-53BP1 goat polyclonal and α-FLAG mouse monoclonal antibodies from Sigma Aldrich; α-γH2AX and α-Tubulin mouse monoclonal antibodies from Millipore; α-SPT16 and α- GST mouse monoclonal antibodies from Santa Cruz Biotechnologies.
Immunoprecipitation and mass spectrometry analysis
293T or HeLa cells stably expressing the transgene (FLAG-OTUD5) or transiently transfected with plasmids grown to 70–80% confluency were harvested by scraping. The pellets were lysed with the Lysis buffer (25 mM Tris pH 7.4, 0.5% NP40, 100 mM NaCl, 0.1 mM ethylenediaminetetraacetic acid (EDTA) supplemented with protease inhibitor cocktail solution) for 10–15 minutes on ice. The lysates were cleared by centrifuging for 30 minutes at 14 000 RPM, and 10% of the supernatant was collected for ‘input’ samples while the remaining volume was incubated overnight with the anti-FLAG M2 agarose (Sigma Aldrich) at 4°C while rotating. The M2 beads were washed three times with the lysis buffer before elution by boiling at 95°C for 3 minutes in 1× Laemli buffer. For the mass spec sample, the bound proteins before eluted with the phosphate-buffered saline (PBS) containing 4% SDS. The eluate was processed using the FASP method, digested with trypsin-LysC and desalted using HYPERSEP C18 columns. Peptides were separated on an Acclaim PepMap C18 (75 μm × 50 cm) UPLC column (Thermo) using an EASY-nLC 1000 with gradient times of 60–90 minutes (2–40% acetonitrile in 0.1% formic acid). Mass spectrometry analysis was performed by a hybrid quadrupole-Orbitrap (Q Exactive Plus, Thermo) or hybrid linear ion trap-Orbitrap (Orbitrap XL) using a top 10 data-dependent acquisition method. For LC-MS/MS analysis using the Q Exactive, full scan and MS/MS resolution was 70 000 and 17 500, respectively. For LC-MS/MS analysis using the Orbitrap XL, full scan mass resolution was 60 000 (Orbitrap detection) with parallel MS/MS acquisition performed in the linear ion trap. Protein identifications were assigned through MaxQuant (version 1.5.0.30) using the UniProt Homo sapiens database. Carbamidomethyl (C) was set as a fixed modification and acetyl (protein N-terminus) and oxidation (M) were set as variable modifications. Trypsin/P was designated as the digestion enzyme with the possibility of two missed cleavages. A mass tolerance of 20 ppm (first search)/4.5 ppm (recalibrated second search) was used for precursor ions while fragment ion mass tolerance was 20 ppm and 0.6 Da for Q Exactive and Orbitrap XL data, respectively. All proteins were identified at a false discovery rate of <1% at the protein and peptide level.
Immunofluorescence and image quantification
Cells were seeded in 12-well plates onto coverslips and treated with indicated siRNA and damage treatments. For UV irradiation, cells were irradiated with 15–100 J/m2 UVC (UV stratalinker 2400), depending on the type of experiments. For inhibitor treatments, ATM inhibitor (KU60019, 1 µM), ATR inhibitor (AZ20, 100 nM), PARP inhibitor (ABT888, 5 µM), DNA-PK inhibitor (NU7441, 1 µM) were treated for 12 hours prior to fixing. The PTuner 263 cells (provided by Dr Roger Greenberg) were seeded on coverslips in 12-well plates at ∼30% confluence; indicated siRNAs were treated at 20 nM for 48 hours. Stabilization of the Fok1-mCherry fusion protein was induced with the addition of 4-hydroxytamoxifin (1 µM) and Shield-1 (1 µM) to the cell growth medium 3 hours prior to fixing. Transcription of YFP-MS2 was induced with the addition of tetracycline (1 µg/ml) to the growth medium 3 hours prior to fixing. Cells were fixed and stained for indicated antibodies following standard procedures. For rescue experiments, siRNA was treated 72 hours prior to fixation, and plasmids were transfected 24 hours prior to fixation. For fixation, coverslips were washed twice with ice-cold PBS and fixed for 10 minutes in the dark with cold 4% paraformaldehyde. Fixed cells were permeabilized for 5 minutes with 0.25% Trition, and incubated with primary antibodies (diluted in PBS for 1:300–1:500) for 1–2 hours in the dark, then with secondary antibodies (diluted in PBS for 1:1000) for 1 hour in the dark, followed by incubating with Vectashield mounting medium containing DAPI (Vector Laboratories Inc). Images were collected by a Zeiss Axiovert 200 microscope equipped with a Perkin Elmer ERS spinning disk confocal imager and a 63x/1.45NA oil objective using Volocity software (Perkin Elmer). All fluorescence quantification was performed using Image J. To measure relative fluorescence intensity (RFI), the fluorescence channel of interest was imported into Image J. The raw integrated density of the nucleus was measured and normalized to background in the image. The raw density measurements were normalized to a value of 10 (arbitrary) to the highest reading. Pearson’s overlap correlations were obtained with the use of the ‘Colocalization finder’ plugin for Image J. Full color images were imported into Image J and the channels were split into blue, red and green; the red and green channels were analyzed and the degree of colocalization was determined. All Pearson’s correlation graphs are representative of at least three independent experiments, error bars represent standard error. For measuring the SPT16/γH2AX foci area the pixel areas were automatically selected using the ‘wand tracing tool’ in the Image J. For vector quantification, single color images are opened in Image J, the straight line section tool is used to draw a transect of 250 pixels across the area of interest. The ‘plot profile’ function in the ‘analyze’ menu is used to measure the raw gray value across the transect. The raw gray values for each pixel is exported into Microsoft excel, the values are normalized to the highest raw value in the sample set using a 1–10 scale (RFI). These values are plotted for each pixel to obtain a fluorescence profile for the area of the selection.
Proximity ligation assay
Proximity ligation assays were preformed using the Duolink kit from Sigma Aldrich; cells were grown in a 12-well format on coverslips. Cells were fixed and permeabilized according to the standard immunofluorescence protocol (previously described), primary antibodies were added at a 1:500 dilution in PBS and incubated for 1 hour at room temperature. Proximity ligation assay (PLA) minus and plus probes were diluted 1:5 in the provided dilution buffer, 30 µl of the probe reaction was added to each coverslip and incubated for 1 hour at 37C; the coverslips were washed twice with buffer A. The provided ligation buffer was diluted 1:5 in water, the ligase was added at a 1:30 dilution; the ligation reaction was left at 37°C for 30 minutes before washing twice with wash buffer A. The provided amplification buffer was diluted 1:5 in water before adding the provided polymerase at a 1:80 ratio, the amplification reaction was left at 37°C for 100 minutes, the reaction was quenched by washing twice with buffer B. The coverslips were mounted on slides with DAPI containing mounting medium.
Cell fractionation assay
293T cells with or without Bleomycin (10 µM) treatment for ∼16 hours were harvested from 6 cm plates. Cells were lysed by 10 minutes incubation on ice with buffer containing 20 mM Tris (pH 7.4), 0.5% NP40 and 100 mM NaCl, then the lysate was subject to gentle centrifugation (5 minutes at 3000 RPM). The supernatant was collected and cleared by high-speed centrifugation (10 minutes at 14 000 RPM) and the cleared supernatant was preserved as ‘S100’ fraction. The remaining pellet was gently washed twice with the same buffer, then resuspended with Laemmli buffer (P fraction). P fraction protein loading amount was normalized proportional to the amount measured in S100 fractions.
Clonogenic survival assay
HeLa and U2OS cells were seeded into 24-well plates (100 cells per well) and treated with indicated siRNAs for 48 hours, then treated with Bleomycin with indicated concentrations, then allowed to grow for ∼10 days. The plating efficiencies (the number of cells that survive in the absence of drug treatment) were roughly equal between the groups. The cells were fixed with 10% methanol, 10% Acetic acid solution for 15 minutes at room temperature, followed by staining with crystal violet. Dried colonies were dissolved and resuspended with Sorensen buffer (0.1M sodium citrate, 50% ethanol), then the colorimetric intensity of each solution was quantified using Gen5 software on a Synergy 2 (BioTek, Winooksi, VT, USA) plate reader (OD at 595 nm). Error bars are representative of three independent experiments.
Protein purification and invitro binding assay
OTUD5 cDNA was cloned into pGEX-6p vector and transformed into BL21 Escherichia coli. strain. Protein expression was induced by addition of IPTG at a final concentration of 300 µM for 4 hours. Cells were harvested and lysed using the ice-cold Lysis buffer (150 mM NaCl, 1% Triton, 20 mM Tris pH 7.4, 0.1% EDTA, supplemented with PMSF and protease inhibitor cocktail) while rotating in 4°C. Resuspended pellets were sonicated three rounds (40 seconds pulse), and the lysate was cleared by centrifugation (20 000 RPM for 40 minutes). Glutathione beads (GE Healthcare) was added to the supernatant and incubated by rotating for 2 hours at 4°C. Beads were then washed three times with lysis buffer. Bound proteins were eluted with the Elution buffer (50 mM glutathione, 200 mM NaCl, 50 mM Tris) or treated overnight rotating at 4°C with Precision Protease to remove protein of interest from the beads. For UBR5 purification, pDEST10-6xHis-UBR5 plasmid was transformed into DH10bac for bacmid creation. Protein purification was performed as according to the Bac-to-Bac Baculovirus Expression System (Invitrogen). In brief, bacmid was transfected into SF9 cells and the virus in supernatant was harvested 72 hours later. Virus was amplified by infecting SF9 cells two more rounds. Infected (P3) cells were harvested and lysed using lysis buffer (25 mM Tris pH 7.4, 0.5% NP40, 100 mM NaCl plus protease inhibitor cocktail), and cleared by high speed centrifugation (20,000 RPM for 40 minutes). The supernatant was incubated with Ni2+ Sepharose (Qiagen) for 2 hours, washed three times, and eluted with the Elution buffer (20 mM Tris pH 7.4, 250 mM Imidazole). For the binding assays, purified proteins were mixed in 50 mM Tris buffer on a 1:1 stoichiometric ratio and allowed to incubate by rotation overnight at 4°C. The Ni2+ beads were added and allowed to incubate for additional 2 hours by rotating at 4°C. Beads were washed three times before eluting with 2× Laemli buffer and analyzed via gel electrophoresis and Coomasie stain.
SNAP technology
The U2OS cells stably expressing SNAP-H2A fusion proteins were cultured in DMEM medium on coverslips at 37°C and transfected with indicated siRNAs (20nM) for 72 hours. Cells were incubated with SNAP-Cell Block reagent (NEB) at 10 µM for 1 hour, then washed out twice with warm media to remove the blocking reagent. Subsequently, cells were irradiated with 100 J/m2 of UVC through 3 µm filters and allowed to recover for the indicated time points. Cells were labeled with SNAP-Cell TMR-Star (NEB) at 3 µM concentration immediately after UVC irradiation, to label the SNAP-H2A during the recovery time.
Limited proteolysis
Purified OTUD5 (5 μg) was mixed with Trypsin (Sigma Aldrich) at a 1:100 ratio of protease:protein and incubated in 100 mM Tris pH 8.5 for indicated times. The same amount of bovine serum albumin was used for control. Reaction was quenched by addition of 4× Laemli buffer and boiling for 5 minutes. Samples were run on SDS-PAGE and the gel was stained with Imperial Protein Stain (Thermo Fisher).
Cell cycle analysis
U2OS cells were treated with siRNAs for 72 hours. Sixteen hours prior to harvesting, indicated cells were irradiated with Bleomycin (5 μM). Cells were harvested with trypsin and fixed with 70% Ethanol for 1 hour in darkness, washed with PBS and incubated with Propidium Iodide and RNase (25 μg/ml) for 1 hour. Cell-cycle analysis was carried out in Accuri C6 Flow Cytometer and data was analyzed using BD Accuri C6 Software.
RESULTS
OTUD5 is a regulator of UBR5 stability and foci formation
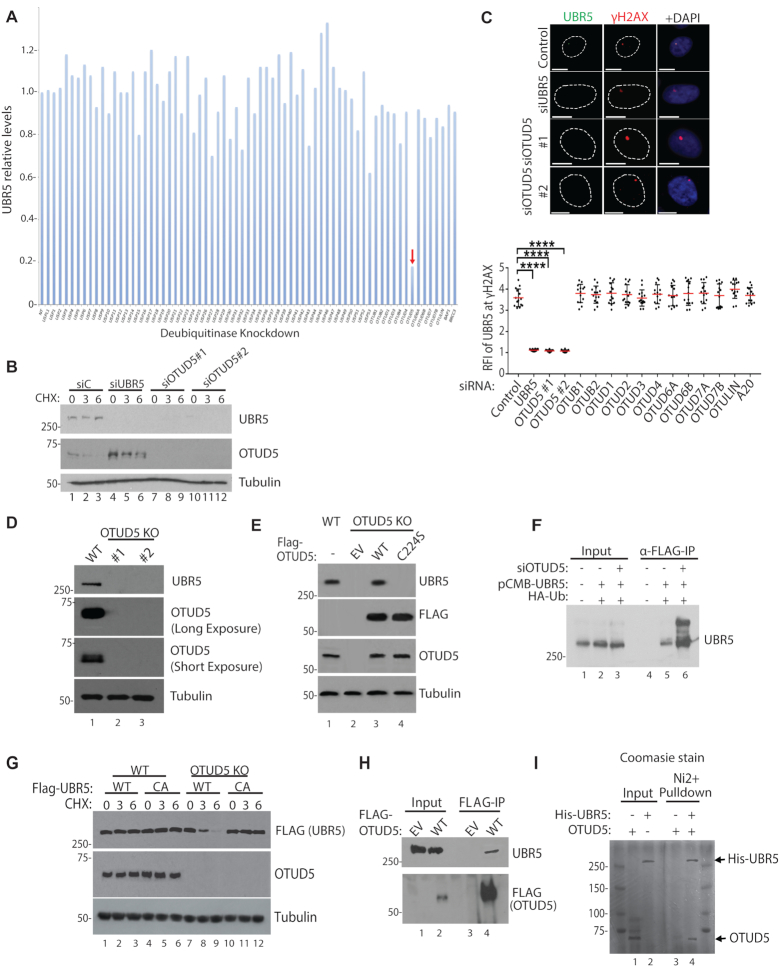
In an attempt to identify a putative DUB that regulates the UBR5 stability and damage-inducible foci formation, we screened siRNAs that target human DUBs, with the measurement of UBR5 protein amount by western blot as a readout. Among the siRNAs used for targeting ∼80 DUBs, siRNA targeting OTUD5 drastically reduced the level of UBR5 (Figure 1A; see Supplementary Figure S1 for representative western blots). The result was validated by treatment of two different siRNAs targeting OTUD5 (Figure 1B). OTUD5 protein levels slightly increased upon UBR5 knockdown, suggesting that a reverse regulation may also exist. We previously showed that UBR5 localizes to the UVC-induced γH2AX foci when cells were irradiated through micropore filters (17). The UVC-induced nuclear foci of UBR5 were also abrogated by OTUD5 siRNA treatment (Figure 1C). The UBR5 foci ablation was specific to OTUD5 knockdown, as siRNAs to other OTU DUB members had little effects (Figure 1C; the representative images are provided in Supplementary Figure S2). We generated two independent HeLa OTUD5 knockout clones (one generated with conventional CRISPR-Cas9 method and the other with the Double Nickase (Cas9 D10A mutant); genomic sequencing analysis for both clones confirmed that both alleles had out-of-frame mutations (Supplementary Figure S3). In both clones, UBR5 was depleted (Figure 1D). The depleted UBR5 proteins were effectively rescued by expressing FLAG-UBR5 wild-type (WT), but not by a catalytically inactive FLAG-OTUD5 mutant (C224S) (21) (Figure 1E), suggesting that the deubiquitinating activity of OTUD5 is necessary for preserving the UBR5 stability. Consistent with the notion of deubiquitination leading to protein stability, reduced UBR5 was recovered by treatment of proteasome inhibitor MG132, when OTUD5 was absent (Supplementary Figure S4). Ubiquitination of UBR5 was reported in proteomic studies (22,23), and we reasoned that OTUD5 may promote UBR5 stability by antagonizing the ubiquitination. Indeed, increased UBR5 polyubiquitination was observed in OTUD5 knockdown cells (Figure 1F). UBR5 polyubiquitination appears to be self-catalyzed, as FLAG-UBR5 WT was much less stable than the catalytically inactive CA mutant, in the absence of OTUD5 (Figure 1G). UBR5 co-precipitates with OTUD5 (Figure 1H), and the purified recombinant OTUD5 and UBR5 proteins interact in vitro (Figure 1I; Supplementary Figure S5 for western blot), suggesting that they interact directly. Altogether, these results suggest that OTUD5 is a specific stabilizer of UBR5.
Figure 1.
OTUD5 is a specific stabilizer of UBR5. (A) Indicated siRNAs (20 nM) were transfected to 293T cells, pellets were harvested after 72 hours and UBR5 levels were detected by western blots. Band intensities were internally normalized to tubulin and quantified using Image J. (B) siRNAs were transfected to HeLa, followed by Cycloheximide treatment (10 µM, for indicated hours) and western blotting. (C) UBR5 foci formation was induced by UVC through 3 µm micropore filter (100J/m2, 1 hour recovery) following the siRNA transfections (n = 20 each). See ‘Materials and Method’ section for RFI description. Bottom panel is for testing various (OTU DUB members) siRNAs for UBR5 foci formation (n = 20 each, **** indicates P-value < 0.0005). (D) Confirmation of OTUD5 CRISPR KO HeLa clones (#1 is CRISPR-Cas9 clone, #2 is Double-Nickase clone). (E) OTUD5 KO#1 cells were transfected with WT or C224S OTUD5 plasmids, and analyzed by western blotting. (F) 293T cells were transfected with indicated siRNAs and the plasmids, harvested pellets were lysed and anti-FLAG IP assay was performed. Bands above unmodified UBR5 are increased in OTUD5 Knockdown cells. (G) Cyclohexamide chase analysis (10 µM, for indicated hours) in OTUD5 KO cells complemented with either WT or catalytically inactive C2768A FLAG-UBR5 plasmids. (H) 293T cells were transfected with 3xFLAG-OTUD5 plasmid and anti-FLAG IP was performed. (I) Purified recombinant OTUD5 and 6xHis-UBR5 proteins were mixed, and the mixture was subject to Ni beads pulldown. In Ni2+ -only, OTUD5 proteins were added without UBR5. Shown is the coomassie stained gel.
OTUD5 localizes to UVC and nuclease-induced DSB lesions
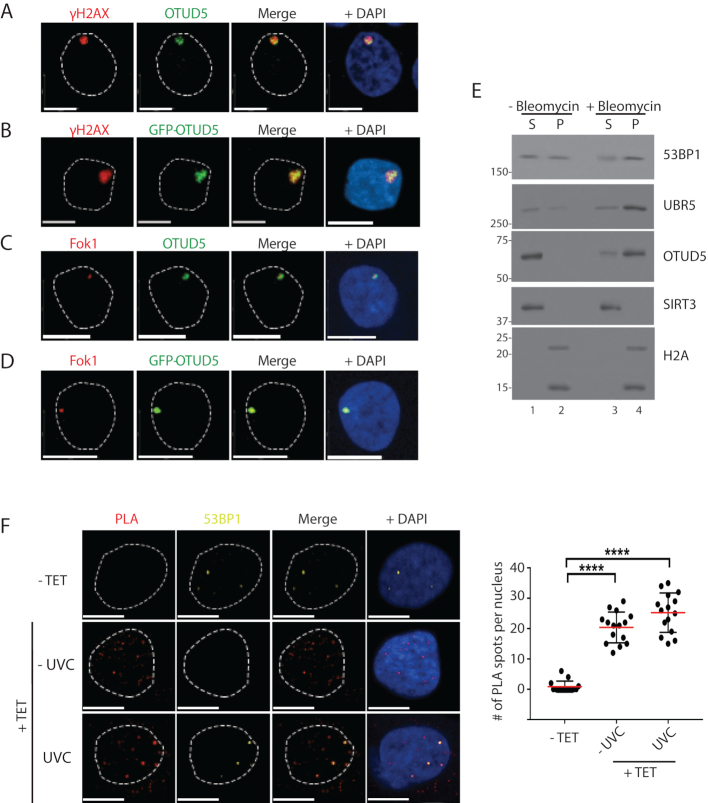
As UBR5 has been shown to localize to damaged chromatin sites and OTUD5 regulates the UBR5 foci, we reasoned that OTUD5 itself may localize to the damaged chromatin. Indeed, we have observed this in several ways; foci of endogenous OTUD5 and GFP-OTUD5 were detected at UVC-induced γH2AX spots (Figure 2A and B, respectively; FLAG-OTUD5 in Supplementary Figure S6). GFP-OTUD5 and endogenous OTUD5 were also observed at Fok1 nuclease-induced DSB sites (Figure 2C and D, respectively; mCherry-LacI-Fok1 nuclease fusion produces DSBs at LacO array inserted in a single chromosome locus in U2OS cells (18)). The endogenous OTUD5 signal was absent in the OTUD5 KO cells, validating the antibody for our assays (Supplementary Figure S7). In a cellular fractionation assay, both UBR5 and OTUD5 proteins were enriched in the chromatin-enriched fraction upon treatment of Bleomycin (Figure 2E) and Etoposide (Supplementary Figure S8).
Figure 2.
OTUD5 localizes to UV and DSB-induced chromatin lesions. (A) HeLa cells were irradiated with UVC (100J/m2) through 3 µm micropore filters, and 1 hour later cells were fixed and co-stained with anti-γH2AX and OTUD5 antibodies. (B) HeLa cells were transfected with GFP-OTUD5 plasmid, irradiated with UVC through micropore filter as in A, and the fixed cells were stained for γH2AX. (C) mCherry-LacI-Fok1 fusion proteins (expressed in PTuner 263 U2OS cells) were induced by Shield-1 and 4-OHT (see ‘Materials and Methods’ section), and the fixed cells were stained with endogenous OTUD5 antibody. (D) The PTuner U2OS reporter cells were transfected with GFP-OTUD5. (E) 293T cells treated with or without Bleomycin (5 µM, 16 hours) were subject to fractionation assay (S = NaCl 100 mM eluate, P = the rest of pellet containing chromatin fraction) and the eluates were analyzed by western blots. (F) HeLa cells stably expressing Dox-inducible FLAG-OTUD5 were treated with Tetracyclin (10 µg/ml), followed by UV (30 J/m2) irradiation through micropore (3 µm) filter. PLA was performed with anti-FLAG and anti-UBR5 antibodies (see ‘Materials and Methods’ section). The slides were also co-stained with anti-53BP1 antibody to mark the DSB lesions. On the right is quantification for relative number of interactions per nucleus (N = 17). Scale bars indicate 10 μm. (**** indicates P-value <0.0005). Experiments were done in triplicates.
Since both OTUD5 and UBR5 localize at DSB lesions, we wished to test whether the interaction occurs at the damaged chromatin spots. PLA detected the interaction between UBR5 and OTUD5 at damaged chromatin in situ, and that the interaction spots partially coincide with the DNA DSB marker 53BP1 (Figure 2F), suggesting that at least a fraction of the interaction indeed occurs at damage (DSB) sites. Altogether, these results show that both UBR5 and OTUD5 localizes to damaged chromatin sites, in particular DSB lesions, where they interact.
OTUD5 regulates transcription repression at damaged chromatin
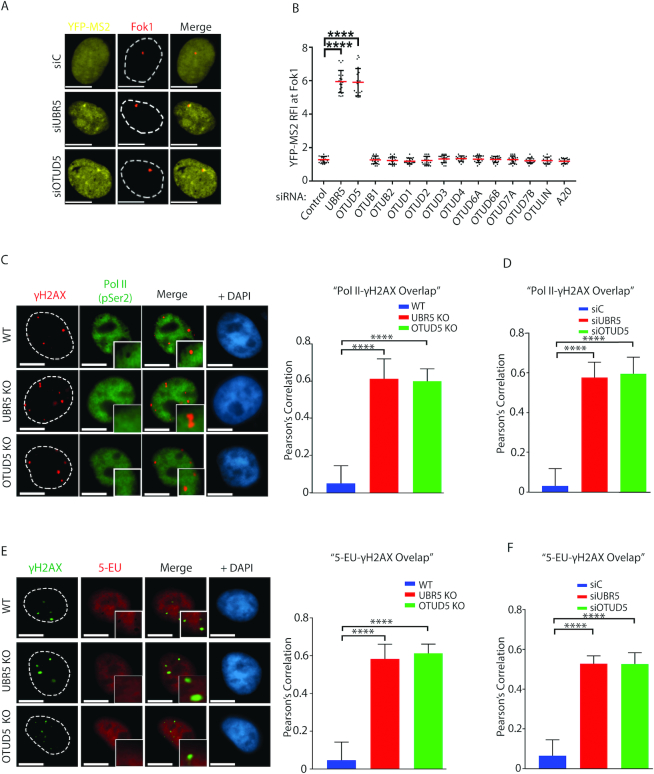
We previously showed that UBR5 regulates transcriptional repression at damaged chromatin (17); in this assay, we used the reporter cell line described in Tang et al. (18) in which DSBs are induced by mCherry-LacI-Fok1 fusion nuclease on the LacO arrays placed kilobases upstream of the transcription sites that produce YFP-MS2 reporter. Using this reporter, it was shown that inducible stabilization of Fok1 (by 4-OHT and Shield-1) suppress the transcription of YFP-MS2 reporter that become localized at the lesion. We found that when the cells were depleted of UBR5 or OTUD5 by siRNAs, the Fok1 (DSB)-mediated repression of YFP-MS2 transcription was no longer observed (Figure 3A). These results suggest that both OTUD5 and UBR5 repress transcription nearby DSB lesions. The transcriptional repression phenotype was unique to OTUD5 among OTU DUB members, as knockdown of other OTU DUBs did not induce such phenotype (Figure 3B; representative images are in Supplementary Figure S9). This phenomenon appears to be independent of ongoing DNA replication, as cells arrested at G0 also display such lack of regulation in the OTUD5 knockdown cells (Supplementary Figure S10). We have further confirmed the transcriptional repressive roles of OTUD5 by staining of the actively elongated form of RNA Pol II (RPB1 P-Ser2 CTD) and of nascent RNA by 5-ethynyl uridine (5-EU) at the γH2AX sites. When the normal WT cells are treated with Bleomycin, the staining of RNA Pol II (RPB1 P-Ser2 CTD) is excluded from the γH2AX sites, suggesting that access of Pol II is inhibited to the damaged area (Figure 3C). Under the same condition, this exclusion is no longer observed in OTUD5 KO and UBR5 KO cells. The results were quantified and presented as Pearson’s correlations that indicate the overlap coefficient. siRNA-mediated knockdown of the genes led to similar phenotypes (Figure 3D; representative images are shown in Supplementary Figure S11). Staining of 5-EU is also excluded at the γH2AX sites when Bleomycin is treated, but not in the OTUD5 KO or UBR5KO cells (Figures 3E) or in the siRNA-transfected cells (Figure 3F; representative images are shown in Supplementary Figure S12). These results altogether suggest that UBR5 and OTUD5 induce transcriptional silencing at DSB lesions by controlling the PolII access.
Figure 3.
OTUD5 regulates transcription repression at damaged chromatin. (A) Ptuner 263 cells transfected with indicated siRNAs were treated with Shield-1/4-OHT and Tetracyclin for inducing LacI-mCherry-Fok1 fusion nuclease and YFP-MS2 reporter, respectively, 3 hours prior to fixing. The representative images are shown. (B) As in A, the assays were performed with indicated siRNAs and the YFP-MS2 RFI at Fok1 sites were quantified and plotted. See ‘Materials and Methods’ section for RFI description. The assay was performed in triplicates (N = 35 each, **** indicates P-value < 0.0005). Scale bars indicate 10 μm. For C and E, the UBR5 and OTUD5 CRISPR KO HeLa cells were treated with Bleomycin (5 µM) for 16 hours, fixed, then co-stained with γH2AX and Pol II P-Ser2 (C), or γH2AX and 5-EU (E). On the right are the quantification of the γH2AX overlap with Pol II (P-Ser) or 5-EU in WT and KO cells. The Pearson’s correlation was measured using the ‘co-localization’ finder plug-in in Image J. The assay was performed in triplicates (N = 50 each). A score of ‘1’ indicates 100% correlation between red and green pixels; a score of ‘−1’ indicates inverse correlation. ‘0’ indicates no relationship. Similar experiments were done using siRNA transfected cells for the PolII-γH2AX overlap analysis (D) and the 5EU-γH2AX overlap analysis (F). Representative images for these assays are shown in Supplementary Figures S11 and 12, respectively). All analysis were done in triplicates. (**** indicates P-value < 0.0005).
OTUD5 interacts with SPT16 and regulates the SPT16-dependent Pol II elongation at damaged chromatin
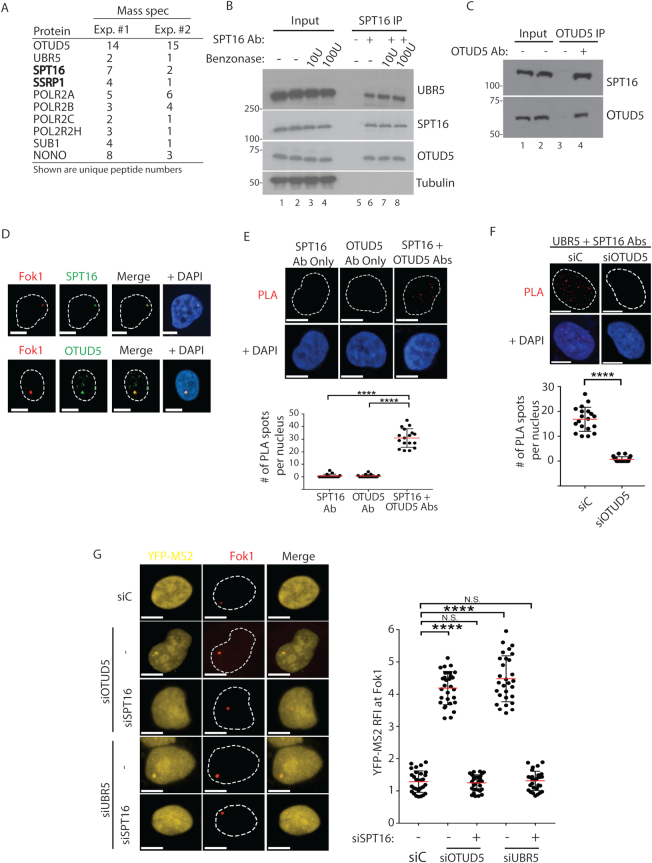
Our previous study showed that UBR5 interacts with SPT16 (17), a component of the FACT histone chaperon complex (24). To test whether OTUD5 also interacts with SPT16 in an unbiased method, we performed FLAG-OTUD5 IP and GST-OTUD5 pulldown experiments from HeLa S3 cells and subjected the eluate to mass spec analysis (Figure 4A). In both experiments, we found peptides derived from UBR5, a few RNA Pol II-associated factors, and the FACT components SPT16 and SSRP1. We confirmed the interaction with the FACT components by reverse immunoprecipitation (IP) of endogenous SPT16, which co-purified both UBR5 and OTUD5 (Figure 4B). The lysates were pre-treated with benzonase, which ruled out that the interactions are mediated by DNA (nuclease activity is confirmed in Supplementary Figure S13). IP of endogenous OTUD5 also co-precipitated SPT16 (Figure 4C), confirming the interaction. Supporting the interaction, we found that SPT16 and OTUD5 co-localize to Fok1-induced DSB lesions (Figure 4D), as well as to the UVC-irradiated lesions that contain a DSB marker 53BP1 (Supplementary Figure S14). Further, the interaction between OTUD5 and SPT16 can be detected in situ by the PLA (Figure 4E). The PLA results were further validated by the absence of PLA signal when each gene was depleted (Supplementary Figure S15). These altogether suggest that OTUD5 and UBR5 interact with SPT16. In addition, the PLA signal between UBR5 and SPT16 at the damaged spots was no longer observed in OTUD5 knockdown cells (Figure 4F), consistent with the observations that OTUD5 is necessary for UBR5 stabilization and thus the interaction with SPT16. Our previous work showed that UBR5 ubiquitinates SPT16, and suggested that this may repress the FACT activity (17). Consistent with this model, the aberrant expression of YFP-MS2 reporter at Fok1-induced DSB sites in OTUD5 or UBR5 knockdown cells was reversed by SPT16 knockdown (Figure 4G). These results suggest that the aberrant Pol II elongation near DSB sites caused by UBR5 or OTUD5 deficiency depend on the FACT activity.
Figure 4.
OTUD5 interacts with SPT16 and regulates the dynamics of SPT16 recruitment at damaged sites. (A) Mass spectrometry analysis of FLAG-OTUD5 IP in HeLa cells (experiment #1) or GST-OTUD5 pulldown with 293T lysate (experiment #2). Key reproducible peptides are shown. (B) 293T lysate were subject to anti-SPT16 IP, with or without pre-treatment of the lysate with benzonase for 1 hour at 37C (U = units), then the eluate was analyzed. (C). 293T cells were subject to anti-OTUD5 IP and the eluate was analyzed. (D) Ptuner 263 cells were treated with Shield1-4OHT to induce Fok1, and the cells were fixed and stained with SPT16 (top) or OTUD5 (bottom) antibodies. (E) SPT16-OTUD5 PLA was performed in HeLa cells (Top) and quantification is shown. The assay was performed in triplicates (N = 20). (F) PLA (anti-UBR5 + anti-SPT16 antibodies) was performed in HeLa cells transfected with either siControl or siOTUD5. Quantification of number of PLA signals per nucleus is shown. The assay was performed in triplicates (N = 20). (G) Ptuner cells transfected with indicated siRNAs were treated with Shield1/4OHT and Tetracyclin to induce Fok1 and YFP-MS2, respectively. The assay was performed in triplicates (N = 30 each) and the YFP-MS2 RFI was quantified on the right. All the scale bars indicate 10 μm. (**** indicates P-value < 0.0005, *** <0.005, N.S. = not significant).
OTUD5 regulates the SPT16 enrichment and deposition of new histone H2A at the damage sites
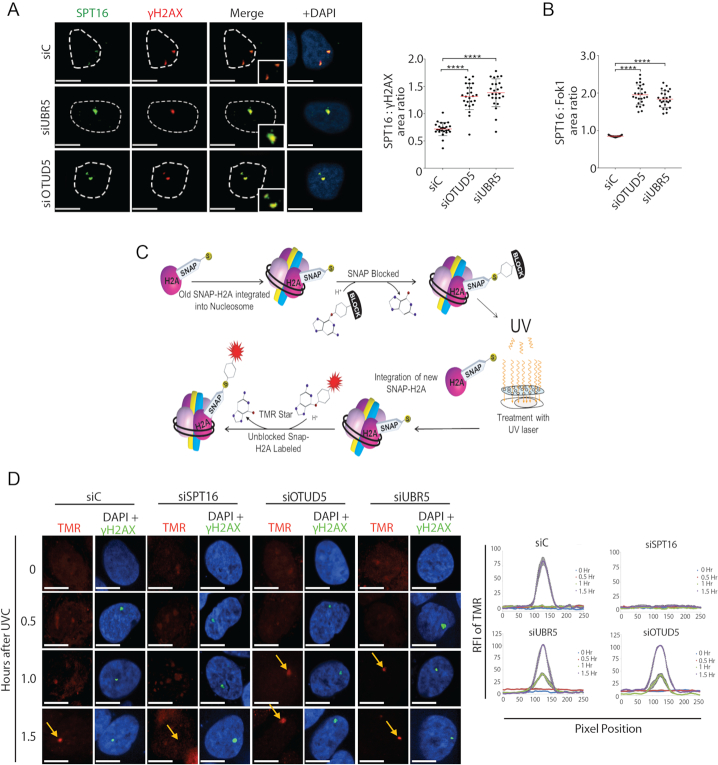
When we analyzed the effects of SPT16 foci at UVC lesions using micropore filter, we consistently noticed that knockdown of both UBR5 and OTUD5 led to the SPT16 foci enlargement at the sites of γH2AX (Figure 5A). These effects were reproducible when the DSBs were induced by the Fok1 nuclease (Figure 5B). These results suggested that UBR5 and OTUD5 negatively influence on FACT enrichment at the damage sites.
Figure 5.
OTUD5 and UBR5 regulates the deposition of new Histone H2A at the damage sites. (A) Enlargement of SPT16 foci in UBR5 and OTUD5 knockdown cells. HeLa cells were irradiated with UVC (100 J/m2) through 3 μm micropore filters and fixed 1 hour after. The number of pixels in each γH2AX and SPT16 co-localized foci was measured using Image J, with each pixel represents an area of 0.2 μm2. The ratio of SPT16 to γH2AX was calculated by dividing the SPT16 area by the γH2AX area. The assay was performed in triplicates (N = 35). Red bars indicate the median value for each set. (B) Ratio of SPT16 to Fok1 area in pTuner263 cells transfected with the siRNAs were quantified as in (A) The assay was performed in triplicates, N = 25. (C) Schematic for SNAP-H2A labeling and nucleosome integration. (D) U2OS cells stably expressing SNAP-H2A were first blocked with TMR Block and then irradiated with UVC (100 J/m2) through 3 μm micropore filters. Cells were labeled with TMR Star (red) and then fixed at indicated time points. Cells were counter-stained for γH2AX. The assay was performed in triplicates. Vector quantification of RFI of TMR signal (SNAP-H2A) (N = 20); see ‘Materials and Methods’ section for details. Scale bars indicate 10 μm. (**** indicates P-value < 0.0005).
A well characterization function of FACT is to facilitate the nucleosomal reorganization and transcriptional elongation of Pol II (25,26). Purified FACT directly binds histone H2A-H2B dimer (25,27,28), and deposit histones onto DNA (25). A study also showed that SPT16 induces accelerated exchange of H2A-H2B at UV-induced spots, which promotes transcriptional restart during recovery (29). Based on this and our results, we hypothesized that UBR5 and OTUD5 may temporarily antagonize the FACT activity to regulate the timely stalling and subsequent resumption of Pol II at the damage sites. We measured the incorporation dynamics of newly synthesized H2A molecules to chromatin using a cell line that stably expressing the SNAP-tagged H2A (Figure 5C: the schematic of assay). In this assay, knockdown of SPT16 effectively inhibited the incorporation of new H2A into the UVC-induced lesions that are marked by γH2AX (Figure 5D). This result is consistent with the previous finding (29), and perhaps reflects the role of FACT in depositing histones back into nucleosomes (30). Interestingly, under the same condition, the incorporation of new H2A was accelerated at the γH2AX-marked lesions when either UBR5 or OTUD5 is knocked down (Figure 5D; see 1 h time point). These results suggest that FACT-mediated histone deposition at the lesions are facilitated when UBR5 or OTUD5 is depleted. A caveat in this assay is the usage of UVC, which induces other types of damage (e.g. UV-photo-adducts) in addition to DSBs (as marked by γH2AX). However this finding, albeit in limited insight due to the mixed damage types induced, support our conclusion that increased SPT16 in UBR5 or OTUD5-depleted cells (Figure 5A) has a functional consequence, in prematurely facilitating the histone H2A exchange at the lesions.
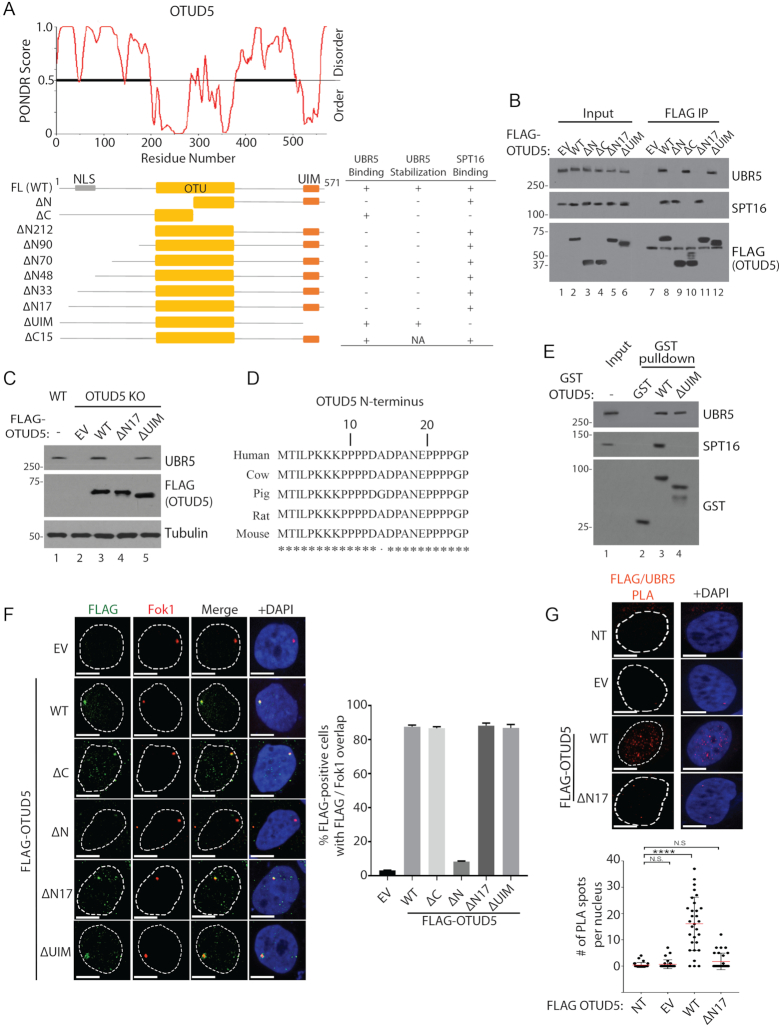
OTUD5 interacts with UBR5 and SPT16 independently through two distinct regions
Next we sought to identify the regions of OTUD5 that interacts with UBR5 and SPT16. OTUD5 has an OTU domain that possesses core catalytic residues in the central region of the polypeptide, and a UIM (Ubiquitin Interacting Motif) at the C-terminus (21). Protein secondary structure prediction analysis (PONDR) suggested that the area flanking both sides of the OTU domain is disordered (Figure 6A; scores greater than 0.5 is considered to be disordered). The N-terminal extension of ∼200 amino acids is predominantly disordered, with few secondary structures predicted. The IUPRED algorithm also generated similar disorder prediction pattern (Supplementary Figure S16). Consistent with the secondary structure predictions, limited proteolysis assay using the recombinant OTUD5 showed that exposure to trypsin led to rapid degradation and produced a fragment with the approximate size of the central structured domain (∼25 kDa; Supplementary Figure S17). Through co-IP analysis using a series of OTUD5 truncation constructs, we found that deletion of the N-terminal 17 amino acids disrupts the UBR5 interaction (Figure 6B; ΔN17). When the interaction deficient ΔN17 mutant is expressed in OTUD5 KO cells, it failed to rescue the UBR5 stability (Figure 6C), underscoring that the interaction is necessary for UBR5 stabilization. Based on the cross-species analysis of the N-terminus (Figure 6D), we deleted a few sequences in the region; deletion of a few selected residues in the N-terminus did not disrupt the interaction with or stability of UBR5 (Supplementary Figure S18), suggesting that the span of disordered residues is necessary for the UBR5 binding. As for the SPT16 binding, the interaction is retained in the ΔN17 IP, but interestingly, it is lost by deleting the C-terminus containing the UIM motif (ΔUIM; Figure 6B). Deleting the C-terminal tail of 15 amino acids flanking the UIM domain did not disrupt the SPT16 binding (Supplementary Figure S19), suggesting that it is the UIM domain that interacts with SPT16. Of note, the ΔUIM mutant retains the ability to stabilize UBR5 (Figure 6C), suggesting that the UIM domain or the SPT16 interaction is not necessary for UBR5 binding or stabilization. Recombinant GST-OTUD5 WT, but not GST-OTUD5 ΔUIM interacted with SPT16, further confirming that the UIM is necessary for the interaction (Figure 6E). The ΔN17 and ΔUIM mutants still localized to the Fok1-induced DSB lesions as efficiently as WT, suggesting that these interactions are not required for the localization (Figure 6F). Through the localization studies, we also found that OTUD5 is primarily localized in the nucleus, and disrupting the residues within 33–70 a.a. inhibited its nuclear localization (Supplementary Figure S20). These analyses also indicated that the nuclear localization and UBR5 interaction is separable. At last, the PLA assay showed that FLAG-OTUD5 WT interacted with endogenous UBR5, measured by increased PLA spots, but the FLAG-OTUD5 ΔN17 mutant did not (Figure 6G), further supporting the conclusions. Altogether, these results demonstrate that OTUD5 interacts with UBR5 and SPT16 through two distinct regions, and that the UBR5-SPT16 interaction occurs indirectly through OTUD5.
Figure 6.
OTUD5 interacts with UBR5 and SPT16 through distinct regions. (A) PONDR software predicts the disordered regions within OTUD5 protein. Below is a schematic for the OTUD5 truncation plasmids used for interaction and other functional analyses. The summary of the assay results are shown on the right: ‘Binding’ was measured by co-IP assays as shown in (B), and ‘Stabilization’ was measured by western blots as shown in (C). ‘NLS’ is indicated on the schematics based on our analysis in Supplementary Figure S20. (B) 293T cells transfected with the indicated plasmids, then anti-FLAG IP was performed and the eluates were analyzed by western blotting (C). HeLa OTUD5 KO (clone#2) cells were transfected with indicated plasmids and UBR5 protein was analyzed. (D) Cross-species alignment on the N-terminus of OTUD5. (E) Purified WT or ΔUIM GST-OTUD5 recombinant proteins was mixed with 293T cell lysate, pulled down with glutathione beads, then the eluates were analyzed by western blots. (F) Ptuner263 cells were transfected with indicated OTUD5 truncates (EV = empty vector) then the Fok1 nucleases were induced by Shield1 and 4-OHT. Cells were fixed then stained with anti-FLAG antibody. The experiments were done in triplicates and on the right is the statistical information (N = 30 each). (G) HeLa cells were transfected with indicated pBABE-FLAG-OTUD5 plasmids (NT = non transfected, EV = empty vector) and the PLA assay with anti-UBR5 and anti-FLAG (OTUD5) was performed (N = 30). Scale bars indicate 10 μm. (**** indicates P-value < 0.0005, N.S. = not significant).
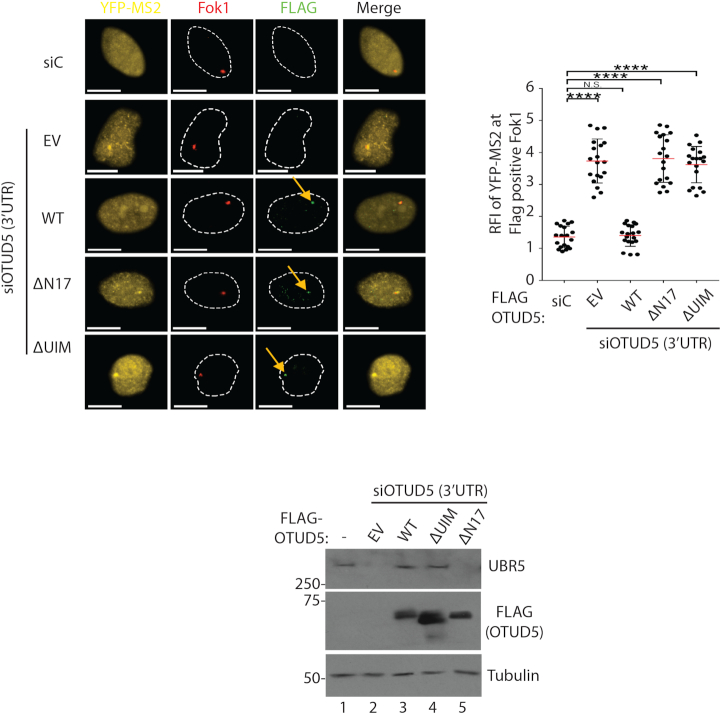
Interaction-deficient OTUD5 mutants cannot rescue the transcription phenotypes
In order to assess whether the above-described interactions are necessary for transcriptional repression at damaged chromatin, we performed a rescue analysis. First, we found that expressing the FLAG-OTUD5 WT reversed (repressed) the YFP-MS2 reporter expression at the Fok1-induced DSB sites in OTUD5 knockdown cells (Figure 7). However, neither ΔN17 nor ΔUIM mutant were able to rescue the phenotype (Figure 7; presence of FLAG-OTUD5 mutants at the spots indicate the normal expression and localization). These results unequivocally suggest that both the UBR5–OTUD5 interaction and OTUD5–SPT16 interaction are necessary for transcriptional repression at the damaged chromatin. Below panel shows the expression confirmation of the FLAG-OTUD5 truncates. These results suggest that the catalytic (deubiquitinating UBR5) as well as the scaffolding (binding to SPT16) roles of OTUD5 are important in the process.
Figure 7.
Interaction-deficient OTUD5 mutants cannot rescue the transcription repressive phenotypes. The PTuner 263 cells expressing indicated OTUD5 constructs (EV = empty vector) were transfected with siRNAs targeting the 3’UTR region of OTUD5, treated with 4-OHT and Shield-1 to stabilize Fok1, then the presence of YFP-MS2 expression at the Fok1 spots was measured. YFP-MS2 reporter was induced by tetracycline 3 hours prior to fixing. Only the cells with positive FLAG spots were counted. The assay was performed in triplicates (N = 50). On the center is the quantification of the MS2 RFI at Fok1 nuclease using Image J (N = 50). Below is the expression confirmation of each cell. Scale bars indicate 10 μm. (**** indicates P-value < 0.0005, ** <0.05.)
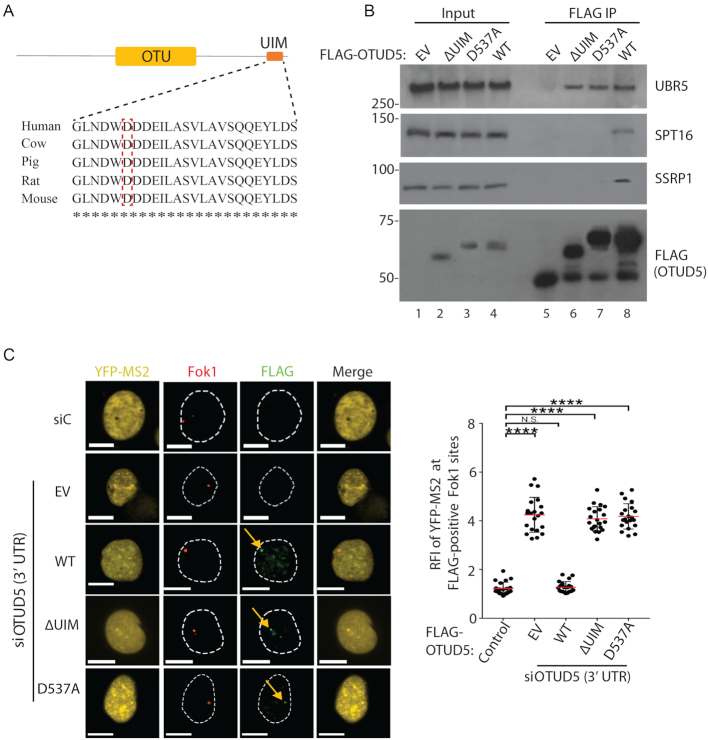
Cancer-associated OTUD5 missense mutation disrupts the FACT association
Numerous missense mutations are found in the OTUD5 locus in cancers (Cbioportal), however the functional implications of these mutations are not known. We noted that the D537 residue within the UIM motif is mutated to Ala in leukemia (Figure 8A; red-boxed). Interestingly, we found that the D537A mutation abrogates the interaction with the FACT components SPT16 and SSRP1 in the IP analysis (Figure 8B). The point mutant still interacted with UBR5, further supporting that the OTUD5-FACT association is independent of UBR5. Similar to the ΔUIM mutant, the D537A mutant failed to restore the transcriptional regulation in the YFP-MS2 reporter assay (Figure 8C). These results confirm that the interaction between OTUD5 and FACT is important for the transcriptional regulation, and provide a new possible link between the FACT-dependent transcriptional process and tumor suppression.
Figure 8.
Cancer-associated OTUD5 missense mutation disrupts the FACT association. (A) Sequence alignment of the UIM and indication of a cancer-associated missense mutation (D537A; Cbioportal). (B) anti-FLAG IPs were performed from 293T cells transfected with indicated OTUD5 constructs (EV = empty vector) and the eluates were analyzed by western blotting. (C) The PTuner 263 cells transfected with the indicated FLAG-OTUD5 constructs were treated with siRNA targeting the 3’UTR region of OTUD5, treated with 4-OHT and Shield-1 to stabilize Fok1, then stained with FLAG antibody. YFP-MS2 reporter transcription was induced by tetracycline 3 hour prior to fixing. The intensities (RFI) of YFP-MS2 expression at the FLAG-positive Fok1 sites was measured using Image J, then plotted on the right. The assay was performed in triplicates (N = 50). Scale bars indicate 10 μm. (**** indicates P-value < 0.0005, ** <0.05).
OTUD5 depletion causes genome instability
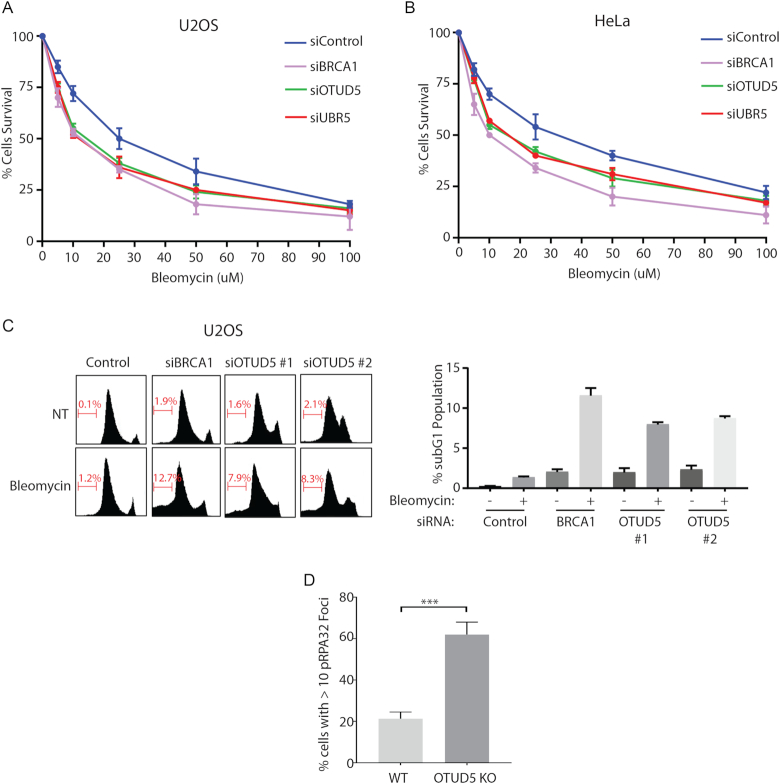
Our work establishes OTUD5 as a new regulator of transcriptional activity at damaged chromatin. Deregulation of transcription at the damaged chromatin can impair DNA repair and possibly DNA replication. We found that the cells depleted of OTUD5 or UBR5 are more sensitive to Bleomycin treatment compared to control cells, albeit to slightly milder degrees compared to BRCA1 knockdown. The phenotypes were consistent between U2OS (Figure 9A) and HeLa cells (Figure 9B). Furthermore, OTUD5 knockdown cells show increased apoptosis in response to Bleomycin treatment (Figure 9C; see sub-G1 populations). It is also noticeable that knockdown of OTUD5 itself caused increases in the populations in S and G2/M phases, possibly suggesting a DNA replication problem along with increased DNA damage. While we cannot exclude the possibility that OTUD5 has other functions in response to DSBs, the lack of transcriptional regulation may, at least partly, account for the Bleomycin sensitivity. In addition, we found an increased number of RPA foci in OTUD5 KO cells (Figure 9D). These data altogether suggest that OTUD5 is a bona fide regulator of the DNA damage response and is necessary for genomic integrity maintenance.
Figure 9.
OTUD5 depletion causes genome instability. U2OS (A) and HeLa (B) cells were transfected with indicated siRNAs, then 48 hours later treated with indicated concentrations of Bleomycin. Cells were incubated for 10 additional days, fixed, then stained using crystal violet. The staining intensities measured by colorimetric analysis as described in ‘Materials and Methods’ section. Assays were done in triplicates. (C) U2OS cells were transfected with siRNAs, then treated with Bleomycin (5 µM) for 16 hours before fixed for flow cytometer analysis. On the right is the quantification of the subG1 populations. The assays were done in triplicates. (D) HeLa WT or OTUD5 KO cells were fixed and pRPA32 foci were counted using Image J (N = 50). (*** indicates P-value < 0.005).
DISCUSSION
Herein, we describe a new role of OTUD5 as a regulator of transcriptional repression at damaged chromatin. Through a DUB siRNA screen, we found OTUD5 is a specific stabilizer of the UBR5 E3 ubiquitin ligase. We showed that OTUD5 localization to damaged chromatin sites is induced by UVC or nuclease-induced DSBs, where it interacts with UBR5. Mapping analysis demonstrated that OTUD5 interacts with UBR5 through the N-terminal disordered tail region, and deleting the tail (17 amino acids) abrogated its ability to stabilize UBR5. Independently of UBR5, OTUD5 also interacts with SPT16, through the UIM domain at the C-terminal tail. We previously reported that UBR5 ubiquitinates SPT16, and this is associated with suppression of SPT16 enrichment at UV and DSB-induced lesions (17). We now show that OTUD5 depletion phenocopies UBR5 depletion in all aspects of the transcriptional regulation we tested, such as misregulation of: (i) Pol II elongation through Bleomycin and nuclease-induced DSB lesions, (ii) nascent RNA synthesis at the lesions and (iii) increased cellular sensitivity to Bleomycin. We further support these findings by demonstrating that the kinetics of new H2A incorporation into damaged chromatin during the recovery is altered in UBR5 and OTUD5-depleted cells. We demonstrated that transcriptional deregulation in the OTUD5-depleted cells were rescued by exogenous OTUD5 WT, but not by either OTUD5 ΔN17 or ΔUIM mutant, in which the interaction sites for UBR5 or SPT16 are independently disrupted. These analyses suggest that the catalytic (through UBR5 stabilization) as well as scaffolding (through FACT binding) activities of OTUD5 are necessary in regulating the transcription at lesions. These data support a model, in which the OTUD5–UBR5 complex interacts with SPT16 and antagonizes its activity in facilitating the exchanges of new H2A molecules at the damaged chromatin. We propose that repression of FACT activity by the OTUD5–UBR5 complex is crucial for generating a barrier for the Pol II from gaining access to DNA breakages, and relieving this barrier is necessary for the timely elongation of Pol II during recovery from damage.
It is important to note that the mechanism of transcriptional arrest we describe is distinct from the direct stalling of Pol II by the lesions (e.g. UV-induced CPD). The Shanbhag et al. study suggested that DSBs do not themselves create a physical roadblock to transcription, but rather initiate a signal that represses transcription distant from damaged sites (13). This was demonstrated by inducing DSBs on LacO arrays placed kilobases upstream of the transcription sites (the system is also used throughout this study). Thus, our work propose that FACT is an important factor in this in cis regulation, and that FACT is subject to regulation by OTUD5–UBR5 complex for the non-lesion stalling of Pol II.
Our work leaves several open-ended questions; our data collectively suggest that OTUD5 and UBR5 repress FACT histone exchange activity, but how does this complex exactly achieve this? Models suggest that FACT can either facilitate the eviction of H2A/H2B dimer by directly binding to them, or engages in dynamic contacts with the histone core that weakens the DNA-core contacts (25–27,31–33). It is possible that OTUD5 prevents the FACT engagement on the histones or DNA, by directly binding to FACT. Alternatively, ubiquitination of SPT16 by UBR5 (17), which would be still supported by OTUD5, might have an effect in limiting the FACT access to nucleosomes. FACT is suggested to induce the ‘breathing’ of nucleosome off DNA template as well as the nucleosomal recovery, in order to provide access for Pol II (26,32,34–36). By inhibiting the FACT-mediated nucleosomal rearrangement and subsequent dimer exchange, which may be possibly reflected in the H2A deposition assay (Figure 5D), OTUD5 may prevent the efficient Pol II progression nearby the damaged chromatin. Further work is needed to understand the precise regulatory mechanism. How is OTUD5 recruited to the damage sites? Our data suggested that the recruitment is not dependent on ATM, ATR, PARP, SPT16 or UBR5 (Supplementary Figures S21 and 22). We do however show that the nuclear localization signal that we roughly mapped (between 33 and 70; Supplementary Figure S20) plays an important role in the foci formation. Further work is necessary to better understand the activating signal for nuclear localization and foci formation.
Our work suggests that the OTUD5–FACT interaction through the UIM motif may be an important tumor suppressive mechanism, as shown for the case of D537A mutation (Figure 8A and Cbioportal). FACT overexpression is observed in certain cancers and undifferentiated cells (33,37,38), and aberrant FACT activity may accelerate the oncogenicity through affecting various gene expression programs (39). It is possible that some tumors that bearing the D537A mutation in OTUD5 may have evolved through the loss of FACT-restraining regulation, which could mimic the effects of FACT overexpression. That being said, we cannot exclude a possibility that OTUD5 exerts a tumor suppressive function through interacting with other proteins in a UIM-dependent manner.
Our results also support an increasingly prevalent concept that the intrinsically disordered regions (IDRs) within enzymatic components play critical roles in protein interaction or substrate recognition (40,41). Our analysis predicts that the N-terminus of OTUD5 is highly disordered, containing no discernible domain. Several examples demonstrate critical roles of IDR in DUBs, including UBP10 (40), OTULIN (42), to name a few. As is the case for the interaction between OTULIN and the HOIP E3 ligase (42), our results highlight an importance of IDR in the E3–DUB interaction.
OTUD5, also known as DUBA, has been well described as a regulator of cytokine production in immune cells (21,43). OTUD5 downregulates the stability of the transcription factor RORγt, which induces IL-17 production in TH cells, and interestingly, it does so indirectly by stabilizing UBR5 (43); the model suggested that UBR5 ubiquitinates and degrades RORγt. Therefore, the role of OTUD5 in cytokine production has a similarity with the role described herein, in that it negatively impacts transcription, at least in part through UBR5 stabilization. In the study showing that OTUD5 suppresses the type I interferon (IFN-1) production, the proposed mechanism is that OTUD5 antagonizes the TRAF3 signaling (21). Our result that the UBR5-interaction region of OTUD5 is critical in the damage-associated transcription suggests that these two mechanisms are discrete. Interestingly, the inhibitory effect on the IFN-1 production was unique to OTUD5 knockdown among other OTU members (21), and so was in the reporter assay measuring the DSB-mediated transcription repression as shown in this study (Figure 3B). Thus, it is intriguing to speculate that the transcriptional repressive function of OTUD5 may have been co-opted during the DNA damage response, in non-immune cells. We speculate that the OTUD5–SPT16 interaction, which occurs through the UIM domain, may pose an additional role in the repression of cytokine production described in the earlier studies. To support this notion, the UIM-deficient OTUD5 mutant was unable to repress the IFN-1 production (21).
Our results further reaffirm the notion that transcription repression at DSBs is an actively regulated process. Multiple mechanisms have been proposed that regulate the process under different experimental contexts (reviewed in (44)). Known regulators of the DSB-associated repression of Pol II-mediated transcriptional include ATM (13), DNA-PK (14), PBAF (45), ENL (46), ZMYND8-NuRD (47), BMI1-UBR5 (17), NELF-E (48). Some degree of convergence have been reported among these factors (e.g. ATM phosphorylates ENL as well as a subunit in the PBAF complex). We have not yet been able to found evidence of crosstalk between the ATM or DNA-PK signaling pathway and the UBR5–OTUD5-FACT components in the transcriptional repression. We tested a possibility that OTUD5 or UBR5 may regulate the stability or DSB recruitment of Pol II negative regulator NEFL-E, but no supporting evidence was found (Supplementary Figure S23). However, we do believe that a crosstalk exist between the UBR5–OTUD5-FACT axis and the Polycomb factors BMI1 and RNF2 (17). Further studies are needed to better understand how these distinct pathways collaborate in the transcriptional repression processes. The role of Cockayne’s syndrome proteins (CSA and CSB) in UV-induced transcription regulation is also well documented (49,50), and it will be also important to investigate whether a collaboration exist between these pathways.
One of the known functions of UBR5 is to restrict the DSB-induced ubiquitin signaling by repressing RNF168-induced histone ubiquitination (51). In this study, UBR5 and TRIP12 knockdown leads to expanded 53BP1 markers at damaged chromatin, which is associated with reduced transcriptional activity at the lesions. This seemingly contradictory result can possibly be explained by the notion described in (52,53); formation of the 53BP1-containing nuclear body in G1 cells is a result of mitotic errors transmitted to next generation. We speculate that in UBR5 (and TRIP12) knockdown cells, 53BP1 accumulation is increased at the common fragile lesions as a result of incomplete DNA replication. Under the acute damage-inducible transcriptional arrest condition as we employed, we do not see evidence of 53BP1 foci enlargement upon UBR5 or OTUD5 knockdown (at the Fok1-induced DSB spots; Supplementary Figures S24 and 25). However, we did notice that the number of 53BP1 foci are increased upon UBR5 or OTUD5 depletion, suggesting that replication stress is incurred. We speculate that the sites of incomplete DNA replication can be, in part, caused by aberrant transcription activities.
In summary, our results establish that OTUD5 is a new regulator of the DNA damage response. Our work reveals a new layer of regulation for the FACT-dependent transcription at the damaged chromatin. Further investigation into the nature of the interaction between OTUD5 and FACT will provide more insight into the regulation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs Roger Greenberg for the PTuner263 cell line, Sally Kornbluth for the pDEST10-6xHis-UBR5 plasmid and Wade Harper for the pDEST_Tet_CMV_FLAG-OTUD5 plasmid (through Addgene). We thank the USF CDDI proteomics facility for mass spectrometry analysis. We also thank Robert Hill for technical support in using confocal microscopy, and former Kee lab members Li Ji and Maha Ahmad for technical assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [R01GM117062-01A1 to Y.K]. Funding for open access charge: NIH [R01GM117062-01A1].
Conflict of interest statement. None declared.
REFERENCES
- 1. Komander D., Clague M.J., Urbe S.. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009; 10:550–563. [DOI] [PubMed] [Google Scholar]
- 2. Reyes-Turcu F.E., Ventii K.H., Wilkinson K.D.. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 2009; 78:363–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kee Y., Huibregtse J.M.. Regulation of catalytic activities of HECT ubiquitin ligases. Biochem. Biophys. Res. Commun. 2007; 354:329–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nijman S.M., Luna-Vargas M.P., Velds A., Brummelkamp T.R., Dirac A.M., Sixma T.K., Bernards R.. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005; 123:773–786. [DOI] [PubMed] [Google Scholar]
- 5. Mevissen T.E., Hospenthal M.K., Geurink P.P., Elliott P.R., Akutsu M., Arnaudo N., Ekkebus R., Kulathu Y., Wauer T., El Oualid F. et al. . OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell. 2013; 154:169–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kee Y., Huang T.T.. Role of deubiquitinating enzymes in DNA repair. Mol. Cell. Biol. 2015; 36:524–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nakada S., Tai I., Panier S., Al-Hakim A., Iemura S., Juang Y.C., O’Donnell L., Kumakubo A., Munro M., Sicheri F. et al. . Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature. 2010; 466:941–946. [DOI] [PubMed] [Google Scholar]
- 8. Sun X.X., Challagundla K.B., Dai M.S.. Positive regulation of p53 stability and activity by the deubiquitinating enzyme Otubain 1. EMBO J. 2012; 31:576–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao Y., Majid M.C., Soll J.M., Brickner J.R., Dango S., Mosammaparast N.. Noncanonical regulation of alkylation damage resistance by the OTUD4 deubiquitinase. EMBO J. 2015; 34:1687–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kato K., Nakajima K., Ui A., Muto-Terao Y., Ogiwara H., Nakada S.. Fine-tuning of DNA damage-dependent ubiquitination by OTUB2 supports the DNA repair pathway choice. Mol. Cell. 2014; 53:617–630. [DOI] [PubMed] [Google Scholar]
- 11. Somesh B.P., Reid J., Liu W.F., Sogaard T.M., Erdjument-Bromage H., Tempst P., Svejstrup J.Q.. Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell. 2005; 121:913–923. [DOI] [PubMed] [Google Scholar]
- 12. Geijer M.E., Marteijn J.A.. What happens at the lesion does not stay at the lesion: transcription-coupled nucleotide excision repair and the effects of DNA damage on transcription in cis and trans. DNA Repair (Amst.). 2018; doi:10.1016/j.dnarep.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 13. Shanbhag N.M., Rafalska-Metcalf I.U., Balane-Bolivar C., Janicki S.M., Greenberg R.A.. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010; 141:970–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pankotai T., Bonhomme C., Chen D., Soutoglou E.. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat. Struct. Mol. Biol. 2012; 19:276–282. [DOI] [PubMed] [Google Scholar]
- 15. Chou D.M., Adamson B., Dephoure N.E., Tan X., Nottke A.C., Hurov K.E., Gygi S.P., Colaiacovo M.P., Elledge S.J.. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. PNAS. 2010; 107:18475–18480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Im J.S., Keaton M., Lee K.Y., Kumar P., Park J., Dutta A.. ATR checkpoint kinase and CRL1betaTRCP collaborate to degrade ASF1a and thus repress genes overlapping with clusters of stalled replication forks. Genes Dev. 2014; 28:875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sanchez A., De Vivo A., Uprety N., Kim J., Stevens S.M. Jr., Kee Y.. BMI1-UBR5 axis regulates transcriptional repression at damaged chromatin. PNAS. 2016; 113:11243–11248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tang J., Cho N.W., Cui G., Manion E.M., Shanbhag N.M., Botuyan M.V., Mer G., Greenberg R.A.. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat. Struct. Mol. Biol. 2013; 20:317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sowa M.E., Bennett E.J., Gygi S.P., Harper J.W.. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009; 138:389–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Matsuura K., Huang N.J., Cocce K., Zhang L., Kornbluth S.. Downregulation of the proapoptotic protein MOAP-1 by the UBR5 ubiquitin ligase and its role in ovarian cancer resistance to cisplatin. Oncogene. 2017; 36:1698–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kayagaki N., Phung Q., Chan S., Chaudhari R., Quan C., O’Rourke K.M., Eby M., Pietras E., Cheng G., Bazan J.F. et al. . DUBA: a deubiquitinase that regulates type I interferon production. Science. 2007; 318:1628–1632. [DOI] [PubMed] [Google Scholar]
- 22. Elia A.E., Boardman A.P., Wang D.C., Huttlin E.L., Everley R.A., Dephoure N., Zhou C., Koren I., Gygi S.P., Elledge S.J.. Quantitative proteomic atlas of ubiquitination and acetylation in the DNA damage response. Mol. Cell. 2015; 59:867–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim W., Bennett E.J., Huttlin E.L., Guo A., Li J., Possemato A., Sowa M.E., Rad R., Rush J., Comb M.J. et al. . Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 2011; 44:325–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Orphanides G., Wu W.H., Lane W.S., Hampsey M., Reinberg D.. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature. 1999; 400:284–288. [DOI] [PubMed] [Google Scholar]
- 25. Belotserkovskaya R., Oh S., Bondarenko V.A., Orphanides G., Studitsky V.M., Reinberg D.. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003; 301:1090–1093. [DOI] [PubMed] [Google Scholar]
- 26. Hsieh F.K., Kulaeva O.I., Patel S.S., Dyer P.N., Luger K., Reinberg D., Studitsky V.M.. Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. PNAS. 2013; 110:7654–7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hondele M., Stuwe T., Hassler M., Halbach F., Bowman A., Zhang E.T., Nijmeijer B., Kotthoff C., Rybin V., Amlacher S. et al. . Structural basis of histone H2A-H2B recognition by the essential chaperone FACT. Nature. 2013; 499:111–114. [DOI] [PubMed] [Google Scholar]
- 28. Kemble D.J., McCullough L.L., Whitby F.G., Formosa T., Hill C.P.. FACT disrupts nucleosome structure by binding H2A-H2B with conserved peptide motifs. Mol. Cell. 2015; 60:294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dinant C., Ampatziadis-Michailidis G., Lans H., Tresini M., Lagarou A., Grosbart M., Theil A.F., van Cappellen W.A., Kimura H., Bartek J. et al. . Enhanced chromatin dynamics by FACT promotes transcriptional restart after UV-induced DNA damage. Mol. Cell. 2013; 51:469–479. [DOI] [PubMed] [Google Scholar]
- 30. Chen P., Dong L., Hu M., Wang Y.Z., Xiao X., Zhao Z., Yan J., Wang P.Y., Reinberg D., Li M. et al. . Functions of FACT in breaking the nucleosome and maintaining its integrity at the single-nucleosome level. Mol. Cell. 2018; 71:284–293. [DOI] [PubMed] [Google Scholar]
- 31. Winkler D.D., Muthurajan U.M., Hieb A.R., Luger K.. Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J. Biol. Chem. 2011; 286:41883–41892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xin H., Takahata S., Blanksma M., McCullough L., Stillman D.J., Formosa T.. yFACT induces global accessibility of nucleosomal DNA without H2A-H2B displacement. Mol. Cell. 2009; 35:365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gurova K., Chang H.W., Valieva M.E., Sandlesh P., Studitsky V.M.. Structure and function of the histone chaperone FACT—resolving FACTual issues. Biochim. Biophys. Acta. 2018; doi:10.1016/j.bbagrm.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hondele M., Ladurner A.G.. Catch me if you can: how the histone chaperone FACT capitalizes on nucleosome breathing. Nucleus. 2013; 4:443–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Winkler D.D., Luger K.. The histone chaperone FACT: structural insights and mechanisms for nucleosome reorganization. J. Biol. Chem. 2011; 286:18369–18374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Valieva M.E., Armeev G.A., Kudryashova K.S., Gerasimova N.S., Shaytan A.K., Kulaeva O.I., McCullough L.L., Formosa T., Georgiev P.G., Kirpichnikov M.P. et al. . Large-scale ATP-independent nucleosome unfolding by a histone chaperone. Nat. Struct. Mol. Biol. 2016; 23:1111–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fleyshman D., Prendergast L., Safina A., Paszkiewicz G., Commane M., Morgan K., Attwood K., Gurova K.. Level of FACT defines the transcriptional landscape and aggressive phenotype of breast cancer cells. Oncotarget. 2017; 8:20525–20542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Garcia H., Fleyshman D., Kolesnikova K., Safina A., Commane M., Paszkiewicz G., Omelian A., Morrison C., Gurova K.. Expression of FACT in mammalian tissues suggests its role in maintaining of undifferentiated state of cells. Oncotarget. 2011; 2:783–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Garcia H., Miecznikowski J.C., Safina A., Commane M., Ruusulehto A., Kilpinen S., Leach R.W., Attwood K., Li Y., Degan S. et al. . Facilitates chromatin transcription complex is an “accelerator” of tumor transformation and potential marker and target of aggressive cancers. Cell Rep. 2013; 4:159–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reed B.J., Locke M.N., Gardner R.G.. A conserved deubiquitinating enzyme uses intrinsically disordered regions to scaffold multiple protein interaction sites. J. Biol. Chem. 2015; 290:20601–20612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van der Lee R., Buljan M., Lang B., Weatheritt R.J., Daughdrill G.W., Dunker A.K., Fuxreiter M., Gough J., Gsponer J., Jones D.T. et al. . Classification of intrinsically disordered regions and proteins. Chem. Rev. 2014; 114:6589–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Elliott P.R., Nielsen S.V., Marco-Casanova P., Fiil B.K., Keusekotten K., Mailand N., Freund S.M., Gyrd-Hansen M., Komander D.. Molecular basis and regulation of OTULIN-LUBAC interaction. Mol. Cell. 2014; 54:335–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rutz S., Kayagaki N., Phung Q.T., Eidenschenk C., Noubade R., Wang X., Lesch J., Lu R., Newton K., Huang O.W. et al. . Deubiquitinase DUBA is a post-translational brake on interleukin-17 production in T cells. Nature. 2015; 518:417–421. [DOI] [PubMed] [Google Scholar]
- 44. Capozzo I., Iannelli F., Francia S., d’Adda di Fagagna F.. Express or repress? The transcriptional dilemma of damaged chromatin. FEBS J. 2017; 284:2133–2147. [DOI] [PubMed] [Google Scholar]
- 45. Kakarougkas A., Ismail A., Chambers A.L., Riballo E., Herbert A.D., Kunzel J., Lobrich M., Jeggo P.A., Downs J.A.. Requirement for PBAF in transcriptional repression and repair at DNA breaks in actively transcribed regions of chromatin. Mol. Cell. 2014; 55:723–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ui A., Nagaura Y., Yasui A.. Transcriptional elongation factor ENL phosphorylated by ATM recruits polycomb and switches off transcription for DSB repair. Mol. Cell. 2015; 58:468–482. [DOI] [PubMed] [Google Scholar]
- 47. Gong F., Chiu L.Y., Cox B., Aymard F., Clouaire T., Leung J.W., Cammarata M., Perez M., Agarwal P., Brodbelt J.S. et al. . Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription-associated DNA damage that promotes homologous recombination. Genes Dev. 2015; 29:197–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Awwad S.W., Abu-Zhayia E.R., Guttmann-Raviv N., Ayoub N.. NELF-E is recruited to DNA double-strand break sites to promote transcriptional repression and repair. EMBO Rep. 2017; 18:745–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gregersen L.H., Svejstrup J.Q.. The cellular response to transcription-blocking DNA damage. Trends Biochem. Sci. 2018; 43:327–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Epanchintsev A., Costanzo F., Rauschendorf M.A., Caputo M., Ye T., Donnio L.M., Proietti-de-Santis L., Coin F., Laugel V., Egly J.M.. Cockayne's Syndrome A and B proteins regulate transcription arrest after genotoxic stress by promoting ATF3 degradation. Mol. Cell. 2017; 68:1054–1066. [DOI] [PubMed] [Google Scholar]
- 51. Gudjonsson T., Altmeyer M., Savic V., Toledo L., Dinant C., Grofte M., Bartkova J., Poulsen M., Oka Y., Bekker-Jensen S. et al. . TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell. 2012; 150:697–709. [DOI] [PubMed] [Google Scholar]
- 52. Lukas C., Savic V., Bekker-Jensen S., Doil C., Neumann B., Pedersen R.S., Grofte M., Chan K.L., Hickson I.D., Bartek J. et al. . 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat. Cell Biol. 2011; 13:243–253. [DOI] [PubMed] [Google Scholar]
- 53. Harrigan J.A., Belotserkovskaya R., Coates J., Dimitrova D.S., Polo S.E., Bradshaw C.R., Fraser P., Jackson S.P.. Replication stress induces 53BP1-containing OPT domains in G1 cells. J. Cell Biol. 2011; 193:97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.