Abstract
Retrograde Bone Morphogenetic Protein (BMP) signaling in neurons is essential for the differentiation and synaptic function of many neuronal subtypes. BMP signaling regulates these processes via Smad transcription factor activity, yet the scope and nature of Smad-dependent gene regulation in neurons are mostly unknown. Here, we applied a computational approach to predict Smad-binding cis-regulatory BMP-Activating Elements (BMP-AEs) in Drosophila, followed by transgenic in vivo reporter analysis to test their neuronal subtype enhancer activity in the larval central nervous system (CNS). We identified 34 BMP-AE-containing genomic fragments that are responsive to BMP signaling in neurons, and showed that the embedded BMP-AEs are required for this activity. RNA-seq analysis identified BMP-responsive genes in the CNS and revealed that BMP-AEs selectively enrich near BMP-activated genes. These data suggest that functional BMP-AEs control nearby BMP-activated genes, which we validated experimentally. Finally, we demonstrated that the BMP-AE motif mediates a conserved Smad-responsive function in the Drosophila and vertebrate CNS. Our results provide evidence that BMP signaling controls neuronal function by directly coordinating the expression of a battery of genes through widespread deployment of a conserved Smad-responsive cis-regulatory motif.
INTRODUCTION
With its extraordinarily high cellular diversity that underpins its multiple functions, the central nervous system (CNS) is the most complex organ in most animals. The generation of such a high diversity of neuronal subtypes requires that they undergo unique programs of differentiation as young maturing neurons. Extensive work has shown that these programs are primarily determined by the activities of subtype-specific combinatorial codes of transcription factors acting at cis-regulatory regions of target genes (1–4). Considerable evidence also supports a critical contribution of retrograde signaling from target cells to neuronal differentiation and function, in mammals and Drosophila (5–7). However, even though such a role is long established, the networks of genes they control and the genomic enhancers they operate through remain largely undiscovered. Thus, the molecular mechanisms underlying the contribution of these retrograde signals remains undefined for the most part.
In the larval Drosophila ventral nerve cord (VNC), retrograde Bone Morphogenetic Protein (BMP) signaling occurs in efferent neurons (8–13). It is required by neuropeptidergic efferents for neuropeptide gene specification (10–13) and by motor neurons for synaptic growth, stability, neurotransmission and homeostasis of the neuromuscular junction (NMJ) (9,14,15). The BMP ligand Glass bottom boat (Gbb) is secreted from postsynaptic cells and from motor neurons (12,16), to bind a presynaptic BMP receptor complex of type II BMP receptor, Wishful thinking (Wit), and the type I BMP receptor kinases, Thickveins (Tkv) and Saxophone (Sax) (9,14,17). Accumulating evidence indicates that after BMP activation, these receptors are endocytosed for retrograde transport to the soma (18–20). BMP receptor kinase activity phosphorylates the cytoplasmic R-Smad, Mother against decapentaplegic (Mad), which binds the co-Smad, Medea, to form a pMad/Medea (pSmad) complex that accumulates in the nucleus, where it regulates gene expression (10,12,17,21). Motor neuron and neuropeptidergic cell phenotypes characterized for gbb, wit, tkv and sax mutants are largely phenocopied in Mad and Med mutants and upon overexpression of DNA-binding defective Mad (9,17,21,22). Thus, BMP signaling via pMad/Medea-responsive gene regulation appears to play a critical role in motor neuron and neuropeptidergic cell function.
Numerous genes are regulated by retrograde BMP signaling in Drosophila efferent neurons. In neuropeptidergic efferents, this includes the activation of subtype-specific neuropeptides in FMRFa-Tv4, CCAP and Ilp7 neuropeptidergic cells (10,11,13). The GPI-anchored Ly6 gene, target of wit (twit) (23), and the guanine nucleotide exchange factor, trio (24), are upregulated in motor neurons by BMP signaling, and act as effectors of BMP function at the NMJ. However, neither twit nor trio mutants fully phenocopied wit mutants, and restoration of these genes in wit mutants only partially rescued the wit phenotype; implicating additional undefined genes as BMP effectors. A microarray study reported the differential expression of 101 genes in the late larval CNS of controls and wit mutants, which included twit as well as two other confirmed wit-responsive genes, Pburs and FMRFa (25). These additional genes represent candidate BMP effector genes, although they remain unverified as direct targets of pMad/Medea complexes; only in the cases of FMRFamide (FMRFa) and trio has such evidence been provided (24,26). Thus, we still know very little about the direct coordinated gene regulatory processes controlled by BMP signaling and pMad/Medea transcriptional activity that underlies neuronal differentiation, plasticity and synaptic function.
Numerous cis-regulatory motifs that are bound by pMad/Medea complexes have been described in Drosophila that mostly converge around GC-rich binding motifs (27–36). From these studies, two motifs have had their precise sequence requirements rigorously characterized in vitro and in vivo (33–35) and mediate cis-regulatory activity in a number of developmental contexts. These include a silencer element termed the BMP-SE, which mediates repression of brinker and other genes (35,37–42), and also an activating element (BMP-AE) that mediates activation of daughters against decapentaplegic (dad) and the enhancer activity for numerous genomic regions throughout fly development (34). The 15 bp consensus sequence of the BMP-AE comprises two distinct binding sites optimally separated by a 5-nucleotides linker, a GGCGCC site bound by two pMad, and a Medea-bound GNCV site (V = any nucleotide except T) (34) (Figure 1A). Although a function for the BMP-AE has not been demonstrated in the nervous system, we postulated that it may serve as a platform for retrograde BMP-activated gene transcription in neurons.
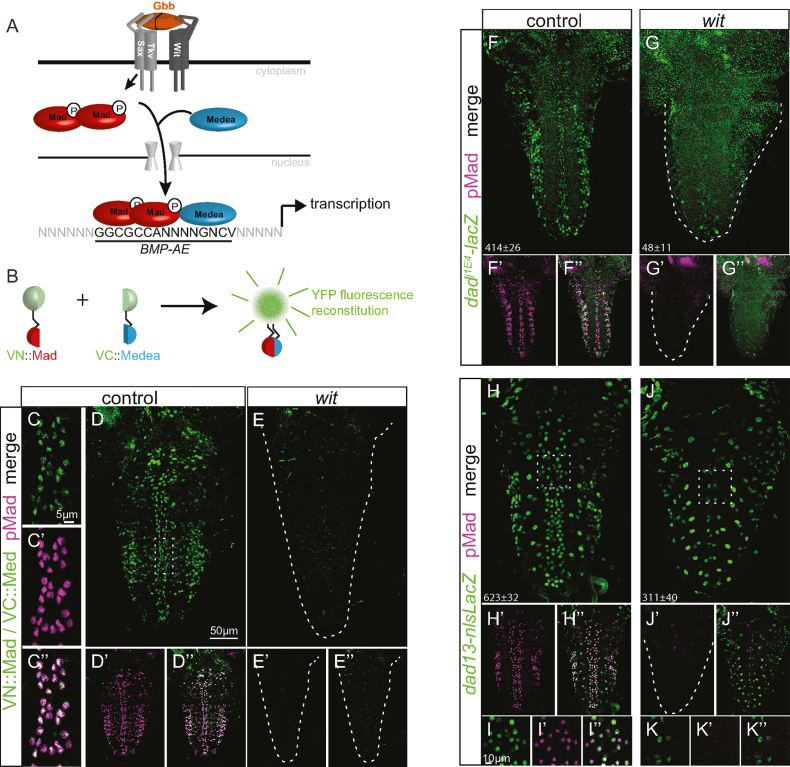
Figure 1.
A canonical BMP signaling pathway acts in neurons. (A) Schematic representation of the canonical fly BMP signaling pathway and binding of pMad and Medea to cis-regulatory BMP-AE sequences. (B) Schematic of the BiFC method. Mad and Medea are fused to two split non-fluorescent fragments of Venus YFP. Upon co-expression of both constructs, direct interaction between VN::Mad and VC::Medea allows for reconstitution of fluorescence. (C–E”) Bimolecular Venus YFP fluorescence is specific to nuclei with BMP activity in L3 VNCs, as revealed by overlap with immunoreactivity to pMad in control genotypes (C–D”), and its absence in wit mutants (E–E”). (F–K”) β-galactosidase expression of the dad enhancer trap dadj1E4 (F–G”) and the dad13-nlsLacZ reporter construct (H–K”), shown with pMad co-immunostaining in controls (F,H-I”) and wit mutants (G,J–K”). (I–I”, K–K”) Close up of immunoreactivity at the dorsal midline taken from the region of the square dotted box in H and J, respectively. The mean±SEM number of nuclei per VNC that express the reporter is indicated at the bottom of each images. Reporter activity of both dadj1E4and dad13-nlsLacZ was significantly reduced in wit mutants as dadj1E4: P = 0.0022 and dad13-nlsLacZ: P = 0.0012 (two-sided Wilcoxon rank-sum test). At least 5 VNCs were analyzed for each genotype. Genotypes: (C, D): pUbi-VC-Medea/+; pUbi-VN-Mad, witA12/+. (E): pUbi-VC-Medea/+; pUbi-VN-Mad, witA12/witB11. (F) dadj1E4, witA12/+. (G) dadj1E4, witA12/witB11. (H, I) dad13-nlsLacZ/+; witB11/+. (J, K) dad13-nlsLacZ/+; witA12/witB11.
The high cellular diversity of the larval Drosophila nervous system and the neuron subtype-specific expression of most defined BMP-responsive genes makes genomic approaches to identifying their cis-regulatory elements challenging. However, the sequence complexity of the BMP-AE allows for a computational approach to predict BMP-responsive enhancers and genes in neurons. Identification of functional cis-regulatory elements has been successfully performed using bioinformatics algorithms that scan the genome for conserved instances of transcription factor binding site sequences (43–47). Here, we combined the identification of highly conserved BMP-AE sequences near neuronally expressed genes with transgenic enhancer activity analysis in vivo to discover a co-regulated battery of 34 wit and pMad/Medea-responsive genomic fragments that are active in efferent (motor neurons and/or neuropeptide expressing) neurons. RNA-seq expression profiling of wit mutant VNCs revealed that functionally-validated BMP-AEs are enriched to BMP-activated genes and that most of them locate within 20 kb of the transcription start site of a BMP-activated gene. We further demonstrated a direct regulatory relationship between BMP-AEs and BMP-activated genes by functional testing of the three BMP-AEs within the twit gene locus. Finally, we showed that the BMP-AE motif has a conserved function in the nervous system by chick neural tube electroporation, and by the demonstration that a BMP-AE taken from the Xenopus bambi BMP-responsive enhancer functionally replaced a fly BMP-AE in vivo.
Our results show that the BMP-AE motif is a widely used, conserved pMad/Medea-responsive element that mediates co-regulation of a battery of BMP-activated genes in Drosophila efferent neurons. We further demonstrate the utility of such sequences in computational searches for BMP-responsive genes and enhancers in the nervous system, and provide a framework for analysis of the transcriptional mechanisms underlying synaptic growth and neurotransmission.
MATERIALS AND METHODS
BMP-AE identification and prioritization
Using merMER (http://www.insilicolabs.com/experiment/index.php), we identified 775 occurrences matching the BMP-AE sequence (GGCGCCANNNNGNCV) in the D. melanogaster genome (48). With the assistance of the Stark group, these were filtered by the controlled Branch Length Score (BLS) method to evaluate phylogenetic conservation over 12 Drosophila species and assigned a motif confidence score based on the relative conservation of the BMP-AE and control motifs (43,44) (Supplementary Figure S2, Supplementary Table S1). Of these, we selected the following BMP-AEs only if they were located in noncoding DNA within 12kb of the locus of a gene expressed in the late larval central nervous system: (A) All BMP-AEs with a motif confidence of 1 or 0.9 (where 0 is lowest confidence and 1 is highest confidence) were analyzed. (B) All BMP-AEs with a motif confidence of 0.8 were analyzed, if ‘paired’ with another BMP-AE of motif confidence ≥0.9 within 12 kb of the same annotated gene locus. (C) All BMP-AEs with a motif confidence of 0.8 were analyzed, if more than two were found within 12kb of the same annotated gene locus. (D) An additional two unpaired BMP-AEs with a motif confidence of 0.8 were also analyzed. This filtering led to the prediction of 62 BMP-AE candidates (43 intronic, 3 UTR, 16 intergenic) located near ∼46 neuronally-expressed genes. Release 3 of the Drosophila melanogaster reference genome assembly (Dm3) was used for this initial computational analysis (49). Coordinates for BMP-AE were later converted to Release 6 (Dm6) coordinates for matching their proximity to BMP-regulated genes, as determined by RNA-seq (see below). Determination of neuronally-expressed genes was taken from late third instar CNS gene expression data available, according to the FlyAtlas microarray database (50) or RNA-seq data available from the modENCODE consortium (51) or the Knoblich group (52).
Fly strains and chick embryos
Flies were reared on standard medium at 25°C, 70% humidity. VN::Mad and VC::Medea (53), dad13-lacZ (34), UAS-dad and dadj1E4-lacZ (54), UAS-tkvDN (55), witA12 and witB11 (9), elav-GeneSwitch-GAL4 (56), twit160 (23), Mi{MIC}twitMI06552 (57) were provided by the Bloomington Drosophila Stock Center. w1118 was used as control if not otherwise stated. Eggs from White-Leghorn chickens (Gallus gallus) were obtained from University of Alberta, Canada, incubated at 39°C and staged according to Hamburger and Hamilton (HH) (58).
Drosophila DNA constructs, transgenic flies and neuronal BMP signaling knock down
A list of all primers used in this study can be found in Supplementary Table S1. To generate reporter constructs, approximately 2 kb genomic DNA fragments were amplified by PCR and cloned into the pCR8/GW/TOPO entry vector by TA cloning (Invitrogen) or the pJet1.2/blunt vector (Thermo Fisher). The 2 kb genomic fragment size is in line with other studies performing large scale enhancer identification in Drosophila (34,59,60), and is a good compromise between cloning efficiency of large DNA fragments and reducing the chance of excluding important enhancer elements on either side of a putative BMP-AE. Genomic fragments were cloned into the pattBGWhZn destination vector (61) by Gateway cloning (Invitrogen) or into the EcoRI and BamHI sites of the pStingerattBDsRednls. For the twit genomic locus, a 6.1 kb DNA fragment was amplified from the BAC CH322-97H12 (P[acman] Resources) by PCR and cloned into the KpnI and HpaI sites of the pStdTomatoattB. Mutagenesis was performed by SOE-PCR, using primers designed to introduce the specific point mutations. All constructs were verified by sequencing.
For fly transgenesis, constructs were inserted via PhiC31 mediated site-specific integration (62) at either insertion site attP40 (25C6) or VK37 (22A3) on chromosome 2 or at attP2 (68A4) on chromosome 3 by Genetics Services Inc. (MA, USA) and Rainbow Transgenics Flies Inc. (CA, USA). Transgenic knock-down of BMP signaling in neurons was carried out using elav-GeneSwitch-GAL4 (56). To activate GeneSwitch-GAL4 from embryonic stages onwards, mothers were fed with yeast paste containing 10–12 μg/ml of RU486 (Mifepristone) for 2 days before being crossed with males. Larvae were then fed with standard food containing 8–10 μg/ml of RU486 until dissection.
Immunofluorescence, in situ hybridization and microscopy
Standard procedures were used for immunostaining of Drosophila VNCs and vibratome sections of chicken embryos. Primary antibodies were chicken anti-βgal (1:1000; ab9361) and chicken anti-GFP (1:1000, ab13970) (Abcam); mouse anti-βgal (40-1a, 1:5), mouse anti-Elav (9F8A9, 1:10) and mouse anti-Pax7 (1:5–10) (Developmental Studies Hybridoma Bank, University of Iowa, USA); guinea pig anti-Dimmed (1:500; a gift from Dr. S. Thor); rabbit anti-pSmad1/5 (1:100; 41D10; Cell Signaling Technology). Secondary antibodies were donkey anti-Mouse, anti-Chicken, anti-Rabbit, anti-Guinea Pig conjugated to FITC, DyLight 488, Cy3, Cy5 and Alexa 647 (1:500–1:5000; Jackson ImmunoResearch). Images of experimental groups were captured the same day using the same settings with an Olympus FV1000 confocal microscope.
For in situ hybridization of twit, the cDNA clone LD40063 was obtained from Drosophila Genomics Resource Center (DGRC, University of Indiana, USA) and was used to synthesize a 1141 bases Dig-labeled RNA probe by in vitro transcription using the DIG RNA labeling kit (Roche). Whole third instar larvae carcasses flipped inside out were fixed in 4.2% formaldehyde for 40 min (10 min. on ice and 30 min. at RT) and washed with RNase-free PBS. After methanol storage for at least 12 h at −20°C, they were rehydrated in PBS Tween 20 (0.1%) (PBSTw) and pre-hybridized in pre-hybridization buffer (50% formamide, 4× SSC, 0.01% Tween 20, pH 6) for 1 h at 55°C. The twit Dig-labeled RNA probe was diluted (1 ng/μl) in hybridization buffer (50% formamide, 4× SSC, 0.01% Tween-20, 5% dextran sulfate, pH 6), heated at 80°C for 3 min and chilled on ice cold water for 5 min. The riboprobe was added to the samples and hybridization was carried out for 24 h at 55°C in a Bambino Hybridization Oven (Boekel Scientific). After washing the samples five times in hybridization washing buffer (50% formamide, 2× SSC, 0.01% Tween 20, pH 6) with the last wash overnight at 55°C, they were washed four times in PBSTw and blocked for 1 h at RT in PBSTw with 5% normal donkey serum. Following overnight incubation of the samples with anti-Digoxygenin antibodies (1:2000, Roche) diluted in the blocking solution at 4°C, they were washed six times with PBSTw and two times with the alkaline phosphatase buffer (100 mM Tris–HCl pH 9.5, 50 mM MgCl2, 100 mM NaCl, 0.2% Tween 20) before incubation in NBT/BCIP staining solution (Roche).
Quantification of reporter expression in the ventral nerve cord
To compare reporter expression in control and wit mutant VNCs, we examined tissues fluorescent for nlsDsRed or immunofluorescent for nuclear localized βgal, anti-Elav (neuronal nuclei) and anti-pMad (nuclei of BMP signaling activated cells). In all cases, five or more VNC were dissected and imaged for each genotype. All tissues to be compared were processed in the same tube with the same reagents, mounted on the same slide and also imaged and analyzed in identical ways. To quantitate reporter activity, we used Bitplane:Imaris v9.2 software (in Spots Mode) to identify reporter-positive nuclei in the VNC (excluding the brain lobes), and in controls we additionally filtered these for pMad co-immunoreactivity (by median intensity thresholding). As there was no pMad immunoreactivity in wit mutants, we counted the number of reporter-positive nuclei in this genotype, which was compared to control numbers. We also compared the percentage loss of reporter expression in wit mutants, to the percentage of reporter-positive nuclei that were pMad-co-immunoreactive in controls. Imaris settings were established independently for each set of reporters, in order to provide optimal ‘spot’ marking of verifiable reporter and pMad co-immunoreactive nuclei, with minimal background fluorescence spot marking. Each image was further subtracted, manually, for spots that erroneously labelled background fluorescence.
For intensity measurements of reporter fluorescence in dorsal midline motor neurons, fluorescent intensity of individual motor neuron nuclei (Elav-positive) was measured in ImageJ (US National Institutes of Health). Mean pixel intensity for each motor neuron was measured from summed Z-projections to measure the total mean fluorescence intensity for each VNC. For each VNC, we subtracted the mean background fluorescence intensity, which was measured from 10 adjacent Elav-negative locations, using the same circular area as for Elav-positive nuclei. At least 5 VNCs with at least 40 motor neurons per VNC were analyzed. Each data point representing the mean of total reporter fluorescence intensity for a genotype was then expressed as a percentage of the mean of the control group.
Statistical analysis and graphing were performed using the online tool at https://ccb-compute2.cs.uni-saarland.de/wtest/ (63) and MS Excel, respectively. All multiple comparisons were carried out using the two-sided Wilcoxon rank-sum test. Differences between genotypes were considered significant when P < 0.05. Data are presented as mean ± standard error of the mean (SEM).
Electrophoretic mobility shift assay (EMSA)
FLAG::Mad, Myc::Medea, and/or TkvQD cDNA sequences were derived from previously described vectors (33,37) (a gift from Dr A. Laughon) and subcloned into a pAc5.1/V5-His vector backbone (ThermoFisher). For EMSA, 3 × 106Drosophila S2 cells in 2 ml medium were co-transfected with 2.4 μg of total expression plasmids consisting of 800 ng TkvQD, 800 ng FLAG::Mad and 800 ng Myc::Medea and/or empty pAc5.1 using the XtremeGENE HD transfection kit (Roche). Forty eight hours following transfection, cells were harvested, washed with PBS and lysed for 15 min in 90 μl ice-cold lysis buffer containing 100 mM Tris–HCl pH 7.6, 0.5% Tween-20, 1 mM DTT and 1× Roche cOmplete ULTRA EDTA-free Protease inhibitor cocktail. The lysate was cleared by centrifugation and stored at −80°C until use. Oligonucleotides synthesized and labelled with IRDye700 by Integrated DNA Technologies (IDT) were annealed to generate probes. Binding reactions were carried out in 20 μl of reaction buffer containing 25 mM Tris pH 7.5, 35 mM KCl, 80 mM NaCl, 3.5 mM DTT, 5 mM MgCl2, 0.25% Tween 20, 1 μg poly dIdC, 10% glycerol and 1× cOmplete ULTRA EDTA-free Protease inhibitor cocktail (Roche). DNA and protein binding was performed by incubating 20 μg of lysate protein with 1 μl of 50nM IRDye700-labeled probe for 30 min at room temperature. The reactions were resolved by non-denaturing 4% polyacrylamide gel electrophoresis and analysed using a Licor Odyssey Imager system (Lincoln, NE).
Chicken spinal cord electroporation
Eight repeat concatemers of the wildtype and mad site mutated dad13 BMP-AE were obtained using ultramers (IDT) that were cloned into the Acc65I and BglII sites of the ptk-EGFP (64) (a gift from Dr A. Kania) to generate BMP-AE8x-EGFP and BMP-AE8xΔmad-EGFP plasmids. Sequences can be found in Supplementary Table S1.
The BMP-AE8x-EGFP and BMP-AE8xΔmad-EGFP plasmids (390–500 ng/μl) were co-electroporated with a control pCAGGS-IRES-H2B-RFP plasmid (130–500 ng/μl) (a gift from Dr E. Marti) to verify electroporation efficiency or a pCAGGS-SmadSD-IRES-H2B-RFP plasmid (a gift from Dr E. Marti) to increase BMP signaling. Plasmid DNA diluted in water and 0.05% Fast Green (Sigma) were microinjected into the lumen of neural tubes of Hamburger Hamilton (HH) stage 14 embryos. Following microinjection of plasmid DNA using a Picospritzer® 2 (General Valve Corporation) microinjector, tungsten electrodes (2 mm) were placed at the lumbar spinal cord level with a spacing of 4 mm between the anode and the cathode and a current was applied using an Electro Square Porator ECM 830 (BTX) electroporator (settings: 30 V, 5 pulses of 50 ms wide in a 1 s interval). Following electroporation, Tyrode's saline solution supplemented with 10% Penicillin and Streptomycin and 1% Fungizone was added on top of the embryos. After sealing the egg shell, embryos were further incubated at 39°C for 24 or 48 h before harvesting.
RNA isolation, sequencing, analysis and enrichment of BMP-AE motifs near BMP-regulated genes
To minimize variation between experimental groups, first instar larvae were transferred to fresh vials at a density of 80 larvae/vial until the desired stage. Whole CNS from w1118;;witA12/+ and w1118;;witA12/witB11 wandering third instar female larvae were dissected free of all other tissues in ice cold PBS and brain lobes were removed to isolate VNCs. VNCs were placed in RNAlater (Ambion, ThermoFisher) and stored at −20°C until the day of RNA extraction. Four independent biological samples of 25 VNCs each were prepared for both genotypes. Total RNA was extracted using RNAzol® RT (Millipore, Sigma) following the manufacturer's protocol, except that the RNA pellet was washed four times with 75% ethanol before solubilization in RNase-free water. Total RNA quality and concentration was assessed using an Agilent 2100 Bioanalyzer (Agilent). Libraries for RNA-seq were generated using the Illumina Neoprep System and sequenced using the Illumina Nextseq500 (Illumina), collecting a total of 24–35 million read pairs (2 × 80 nucleotides long).
In a preliminary analysis, multiple pipelines have been applied. Briefly, for the alignment stage we used STAR (65) and HISAT2 (66) and the pseudo aligners kallisto (67) and Salmon (68). Quantification was performed directly with STAR, kallisto and Salmon or with RSEM (69) and StringTie (66) for the pipelines producing real alignments. In all cases the reference transcriptome was from release 6 of the Drosophila melanogaster reference genome assembly (Dm6) (70). The alignment rate was consistent at 93–94% for all the samples. In-house Perl scripts were used to sum the read counts at the transcript level for each gene and create matrices comprising the read counts for all of the genes for all of the samples. Differential expression analysis was then performed on the data from those matrices using the R package DESeq2 (71), edgeR (72), sleuth (73) and Ballgown (66). Each sample was assessed using the quality-control software RSeQC and the PtR script from the trinity suite (74). No potential outliers were detected when clustering the samples and therefore all the samples were kept for the differential expression analysis. The output for each pipeline is a list of genes ranked by the P-value for differential expression after correction for multiple testing with a significance cutoff set at P<0.05. A combined list was obtained by ranking the genes according to their median rank from the various analysis pipelines and only genes achieving the significance cutoff in at least half the pipelines were considered as significantly differentially expressed. Genes with differential expression not going in a consistent direction between pipelines were eliminated from that combined list of significant genes. The kallisto-DESeq2 pipeline gave results close to the combined list and for sake of simplicity only the numerical output from that pipeline is reported in the current work. Differential expression was determined by log2 fold change between control and wit mutant gene values. The display panel for differential expression shown in Figure 4A and Supplementary Figure S11 was made in R using the ggplot2 package (75).
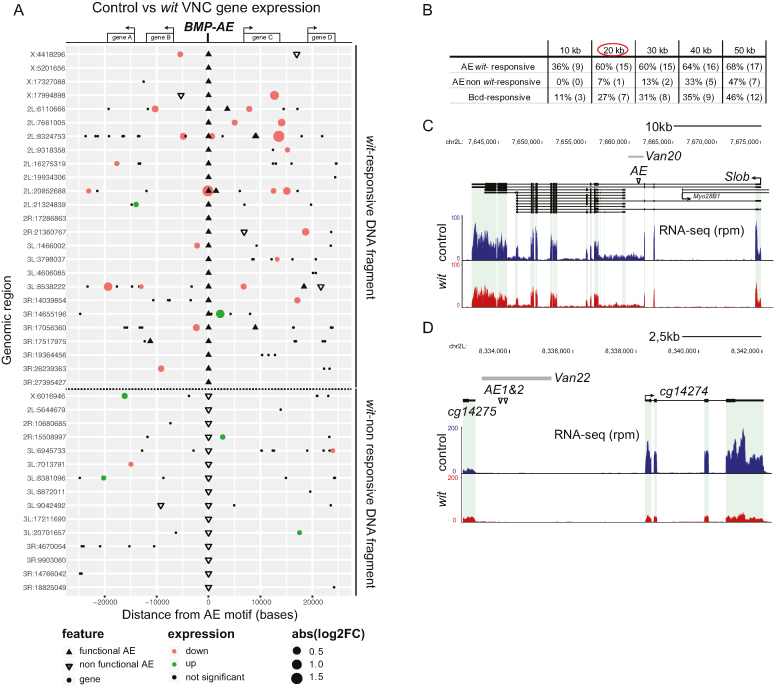
Figure 4.
Proximity of BMP-AEs to BMP-regulated genes. (A) Diagram showing genomic regions (version Dm6) containing BMP-AEs analyzed for wit-responsiveness (see Supplementary Table S1 and Table S2 for identity of all BMP-AEs, genomic fragments and wit-responsive genes). We do not show 5 of the BMP-AEs tested, for the sake of clarity. Black triangles denote BMP-AEs within wit-responsive fragments; a subset of these represent two closely spaced BMP-AEs (on lines 2L:8324753, 2L:20852688 and 3R:19364456). Open inverted triangles denote BMP-AEs within wit-non responsive genomic fragments. Circles denote the 5′-most transcription start site (TSS) of genes expressed in the VNC, within a span of 25 kb from the central BMP-AE shown; small black circles are genes that are wit-non responsive, red and green circles are genes that are significantly reduced or upregulated, respectively, in wit mutants; as determined by RNA-seq analysis of control (witA12/+) and wit mutant (witA12/witB11) VNCs. The size of red/green circles denotes absolute log2-fold change from control to wit mutants expression level. (B) Proximity to the TSS of BMP-activated genes of BMP-AEs within wit-responsive (AE wit-responsive) and non-responsive genomic fragments (AE non wit-responsive), and also Bicoid-responsive fragments (Bcd-responsive). The percentage (to total number of genomic regions) and the actual number of genomic regions containing each motif type that are within a specific distance to the TSS of at least one BMP-activated gene is shown. The enrichment for functional BMP-AEs over non-functional BMP-AEs or Bcd-responsive fragments are shown at 10–50 kb increments. (C, D) The genomic locus of two genes, Slob (C) and CG14274 (D) that were reduced in wit mutants, as well as the span of the genomic fragments tested (Van20 in C, and Van22 in D), and also the specific location of the BMP-AEs therein is shown. RNA-seq rpm denotes the number of reads per million reads.
To test for enrichment of wit-responsive genes to BMP-AEs or bicoid-responsive fragments (Figure 4B), we mapped the location of all wit-responsive genes identified by RNA-seq analysis, with the 26 bcd-responsive fragments as controls (8 of the 34 bicoid-responsive fragments listed in (76) had to be removed due to close proximity to one of the BMP-AE tested herein) and the 59 BMP-AE motifs contained within the 56 genomic fragments for which we could determine wit-responsiveness. Note that three BMP-AEs embedded into two genomic fragments, Van10 and Van19, were excluded because we could not determine whether these were wit-responsive. We then counted the number of genomic loci that contain at least one wit-responsive gene located within a given distance of a Bcd-responsive fragment or BMP-AE motif within wit-responsive and wit-non responsive fragments. This approach allowed us to perform statistical analysis that avoids re-counting of the same gene(s) multiple times.
To more selectively test whether BMP-AEs in wit-responsive fragments were more likely to be directly adjacent to a wit-responsive gene, with no intervening wit-non responsive gene, we repeated the above enrichment analysis, but only considered adjacent motifs. For this analysis, we were able to separate the genomic locus 2L:8324753 into two distinct loci, due to the presence of an intervening wit-non-responsive gene between a pair of BMP-AEs and their adjacent wit-responsive gene. This increased the total number of loci tested to 26 for BMP-AE in wit-responsive fragments.
Statistical significance of the enrichment of BMP-AE and Bcd-responsive genomic fragments near BMP-dependent genes was calculated using Pearson's Chi square test for count data in R (function chisq.test) with continuity correction without correcting for multiple testing (77).
BMP-AE motif sequence enrichment calculation
The Two Sample Logo software (78) available at http://www.twosamplelogo.org/ was used to calculate and visualize the sequence difference between the 37 BMP-AE motifs embedded into the wit-responsive fragments and the 22 BMP-AE motifs within the wit-non responsive fragments as well as the 10 BMP-AE motifs within tkv-responsive fragments (34). Statistical enrichment was calculated using Student's t-test with a significance cutoff of P < 0.05.
RESULTS
Canonical BMP signaling acts through BMP-activating elements (BMP-AE) in neurons
Efferent neurons of the VNC accumulate nuclear phosphorylated Mad (pMad) in response to synaptic Gbb (Figure 1A). While pMad is assumed to associate with Medea to regulate transcription in neurons, this had yet to be explicitly demonstrated. To test this, we examined pMad and Medea physical interaction by bimolecular fluorescent complementation (BiFC) of split Venus yellow fluorescent protein fragments, VN and VC (53,79–82) (Figure 1B). We ubiquitously expressed VN::Mad and VC::Medea fusion proteins and examined BiFC in the VNC. Expression of either VN::Mad or VC::Medea alone did not report any fluorescence (not shown), but co-expression of VN::Mad and VC::Medea led to strong fluorescence within the nuclei of BMP-activated neurons (Figure 1C and D). We examined BiFC in wit mutants that have a loss of neuronal BMP signaling (9,14), and observed an absence of fluorescence in the VNC (Figure 1E). Thus, BMP activation in neurons results in the formation of pMad/Medea complexes in the cytoplasm, followed by their accumulation in nuclei.
In vivo and in vitro studies have shown that pMad/Medea complexes act at cis-regulatory sequences, termed BMP-activating elements (BMP-AEs), to activate the expression of dad, an inhibitory Smad, and a number of BMP-responsive genomic fragments that are active in the embryonic dorsal epidermis and larval wing imaginal disc (Figure 1A) (34). To obtain evidence for BMP-AE activity in neurons, we examined whether BMP-AE-dependent dad expression occurs in the VNC. First, using a faithful lacZ enhancer trap reporter for dad (dadj1E4) (54), we found that dad was expressed in 414 ± 26 nuclei of the VNC (n = 6), and that 321 ± 19 (or 78%) of those nuclei were co-immunoreactive for pMad. Our counts indicate that there are approximately 742±20 pMad-positive nuclei in the VNC (n = 14), therefore the dad reporter is expressed in approximately 43% of pMad-positive nuclei. In wit mutants, reporter expression fell to 48 ± 11 nuclei (n = 6; P = 0.0022) (Figure 1F and G). This 88% loss of reporter expression correlates with the approximate 78% expression of the reporter in pMad co-immunoreactive nuclei, suggesting that reporter expression is largely lost from nuclei that are pMad-positive in controls. Next, to test a role for BMP-AEs in neuronal dad expression, we examined expression of the dad13-nlsLacZ transgenic reporter, which reports the activity of a 520 bp enhancer of dad mediated by a single BMP-AE (34). In the VNC, dad13-nlsLacZ was expressed more broadly than the dadj1E4 reporter, in 623±32 nuclei of which 486±13 (79%) nuclei were pMad-positive (n = 7). This represents approximately 65% of all pMad-positive nuclei. Expression in pMad-negative nuclei included glia, as evidenced by their lack of Elav co-immunoreactivity (Figure 1H–K, Supplementary Figure S1). In wit mutants, dad13-nlsLacZ reporter activity was reduced by 50% to 311 ± 40 nuclei (n = 6; P = 0.0012). This 50% loss of reporter expression was substantially less than the approximate 79% expression of the reporter in pMad co-immunoreactive nuclei, suggesting that in wit mutants the reporter is lost in most but not all nuclei that are pMad-positive in controls. Comparison of reporter expression in Elav-immunoreactive nuclei in the dorsal VNC of controls and mutants indicated that reporter expression was commonly retained in most glia but also in a subset of neurons (Supplementary Figure S1). Thus, we find that these dad reporters exhibit wit-responsive activity in neurons that are pMad-positive.
Identification of 34 wit-responsive genomic fragments in neurons
We wished to test whether the BMP-AE motif plays a widespread role as a cis-regulatory element that can be used in silico to efficiently predict pMad/Medea-responsive enhancers near BMP-responsive genes in neurons. Such a method offers the advantage that it can be performed regardless of neuronal subtype gene expression, whereas genomics approaches to identify pMad-bound genomic enhancers in the Drosophila CNS are challenging due to the relative low number of pMad-positive nuclei, the high cell subtype diversity of pMad-positive motor and neuropeptidergic efferent neuronal populations (83,84), and the neuronal subtype diversity of known BMP-dependent genes (10,11,13,23,24).
We identified all BMP-AE motifs in the D. melanogaster genome using merMer (48). These were filtered for high conservation using the Branch Length Score to motif confidence method (Supplementary Figure S2) (43,44) and for proximity to genes expressed in the larval nervous system, from available databases (50–52). This led to our prioritization of 62 BMP-AEs (see Materials and Methods and Supplementary Table S1 for details). To test the in vivo activity of these BMP-AEs, 58 genomic DNA fragments (of ∼2kb) containing one or more of the 62 BMP-AEs were cloned in front of lacZ or DsRed reporters, both with a nuclear location signal (nlsLacZ or nlsDsRed). These genomic fragment reporters were placed into attB transgenic vectors for integration into the genome using phiC31-integrase (62) (see Materials and Methods and Supplementary Table S1).
We examined reporter activity driven from these genomic fragments in the VNC of wandering third instar larvae. Out of the 58 reporters, 13 showed no expression in the VNC, while 45 exhibited robust reporter activity in the VNC (Supplementary Table S1). Of these active reporters, 42 exhibited expression in subsets of pMad-positive cells in the VNC (which at this developmental stage comprises efferent neurons, e.g. motor and/or neuropeptidergic neurons) (8–11,13), and in certain cases also pMad-negative glia and neurons (Figure 1H–K, Supplementary Figures S1, S3, S4, S5). We tested the BMP-responsiveness of these reporters by placing all 42 into a wit mutant background. Out of those 42 reporters, 32 showed a partial to total loss of reporter expression, and 8 reporters showed no change (Figures 1H–K, 2, Supplementary Figure S6, Supplementary Table S1). For 2 reporters (Van10 and Van19), we were unable to unambiguously determine if there was any wit-responsive activity, because they were expressed in dense clusters of pMad-positive and pMad-negative cells which we could not discriminate in wit mutants (due to the loss of pMad as a marker to distinguish these cells) (Supplementary Table S1).
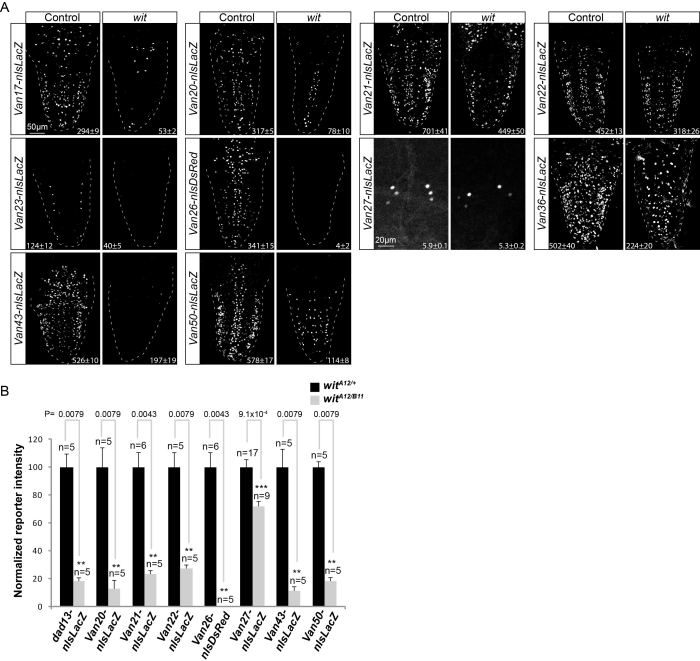
Figure 2.
Identification of wit-responsive genomic fragments in neurons. (A) Nuclear β-galactosidase and nlsDsRed expression patterns driven from 10 genomic fragments containing conserved BMP-AEs that are down-regulated in wit mutants (witA12/witB11) as compared to controls (witA12/+), in dissected late third instar larval VNCs. The observed down-regulation ranged from a total loss of all expression to loss of expression in a subset of neurons. The mean ± SEM number of nuclei per VNC that express the reporter is indicated at the bottom of each panel. Reporter activity of all fragments was significantly reduced in wit mutants; Van17, P = 5.83 × 10−4; Van20, P = 0.0079; Van21, P = 0.0087; Van22, P = 0.0159; Van23, P = 0.0022; Van26, P = 0.0022; Van27, P = 0.0154; Van36, P = 0.0022; Van43, P = 0.0012; Van50, P = 0.0079 (two-sided Wilcoxon rank-sum test). At least five VNCs were analyzed for each genotype. (B) Quantification of relative nuclear β-galactosidase and nlsDsRed intensities for 8 of the 11 wit-responsive genomic fragments shown in Figures 1H–K and 2A that show expression in motor neurons of the dorsal midline and 6 ventral neuropeptidergic cells. Bar plots illustrate mean ± SEM; n indicates the number of VNCs analyzed. ***P< 0.001; **P< 0.01, *P< 0.05 (two-sided Wilcoxon rank-sum test).
For wit-responsive reporters, we wished to test if the reporter was preferentially lost in cells that are pMad-positive in controls. To this end, we examined co-expression of the reporter with anti-pMad and anti-Elav in controls and wit mutants. As predicted, we observed that reporter activity was primarily lost in neurons that are pMad-positive in controls (Supplementary Figure S1 and Table S4). We further validated that these reporters respond cell-autonomously to the canonical BMP pathway, by finding that reporter expression was reduced upon co-expression of UAS-dad and a dominant negative form of tkv (UAS-tkvDN) in neurons, using the elav-GeneSwitch-GAL4 driver (Supplementary Figures S7 and S8).
For the 13 reporters that failed to express in the VNC, we postulated that these genomic fragments may be missing critical regulatory elements required to drive neuronal reporter activity. To test this, we searched the Janelia GAL4 collection (60) for reporters of larger DNA fragment size that contain any of the genomic fragments tested. We found three ∼3 kb genomic fragments (GMR49D01 GMR89G06 and GMR89H02) that encompass the BMP-AEs of Van16, Van37 and Van38, respectively. Upon testing, all of these genomic fragments drove reporter expression in subsets of pMad-positive neurons, and two of these, GMR89G06 and GMR89H02, exhibited reduced reporter expression in wit mutants (Supplementary Figure S6). We also considered the possibility that BMP signaling may directly repress the activity of these reporters. Therefore, we tested for de-repression of these remaining 10 reporters in wit mutants, but found that none of them increased expression in the absence of BMP signaling, suggesting that the BMP-AEs within these genomic fragments do not act as BMP-dependent silencer elements (Supplementary Figure S9).
Overall, we identified 34 genomic fragments responsive to the loss of neuronal BMP signaling in wit mutants, out of the 58 genomic fragments tested; giving our approach a discovery rate of 59%. These data provide experimental validation for a computational approach to efficiently identify genomic regions that contain wit-responsive neuronal enhancers, and further suggests that BMP-AEs serve as a common platform for BMP-responsive gene regulatory activity in VNC efferent neurons.
BMP-AEs recruit pMad/Medea complexes in vitro and are necessary for reporter expression in vivo
We tested if BMP-AEs embedded in wit-responsive fragments interact with pMad/Medea complexes. To this end, we analyzed the recruitment of an activated pMad/Medea complex to three BMP-AEs, by Electrophoretic Mobility Shift Assay (EMSA). These BMP-AEs were selected because they represent different sequences within the Medea GNCV consensus sequence, and because their corresponding genomic fragments reported broad activity that was strongly wit-responsive. Drosophila Schneider 2 cells (S2) were transfected with plasmids encoding Mad and Medea as well as constitutively activated tkv, tkvQD. Lysates from these cells were tested for their ability to band shift IRDye700 tagged probes containing these BMP-AE sequences in vitro (Figure 3A, B). In all three cases, the BMP-AE probes recruited a pMad/Medea complex, as shown by a strong band shift of labelled probe. The sequence specificity of this recruitment was shown by competition assays, in which an excess of untagged wildtype probe fully competed for the labelled band shift, while an untagged probe with substitution mutations through the pMad binding site abolished competition, and a strong band shift was retained (Figure 3B).
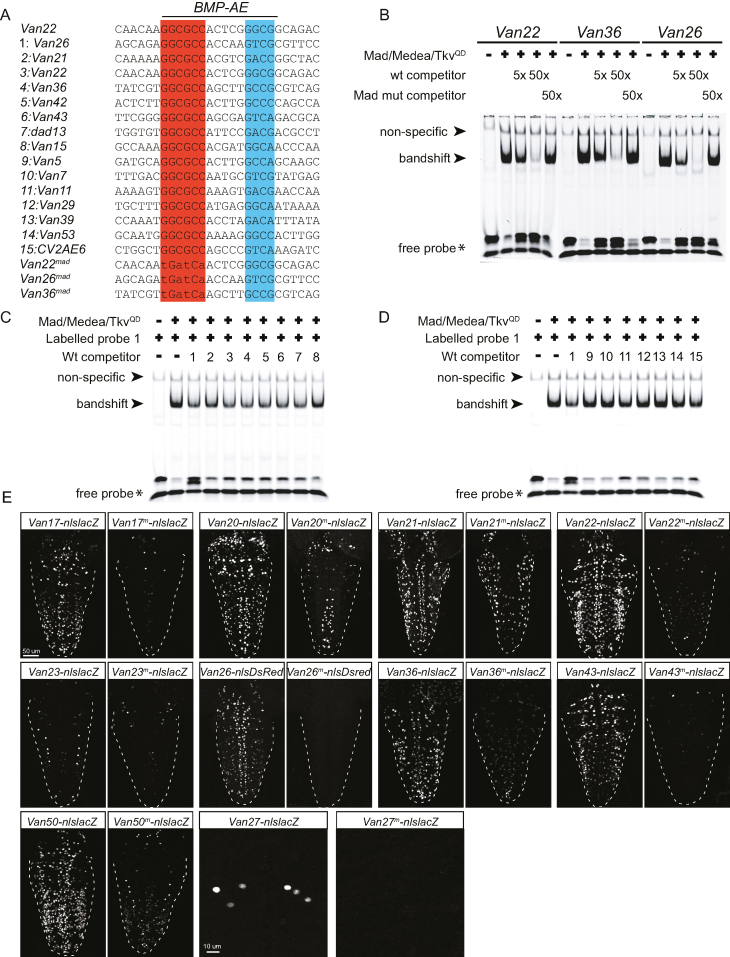
Figure 3.
The BMP-AE of wit-responsive genomic fragments is bound by pMad/Medea complexes with high affinity in vitro and is required in vivo. (A–D) Electrophoretic mobility shift assays (EMSA) for three selected BMP-AEs with S2 cell lysates transfected with Mad, Medea and TkvQD to activate BMP signaling. (A) DNA sequences used for EMSA analysis, highlighting the presumptive pMad (red) and Medea (blue) binding sites of the BMP-AE. The DNA sequences shown were taken from the predicted BMP-AE within the indicated genomic fragments. The BMP-AEs within Van22, Van36 and Van26 were used as IRDye700-labeled probes, as well as for unlabeled wildtype and mutated (Van36mad, Van26mad, Van22mad) competitors. All other sequences were used as unlabeled wildtype competitors. Mutant competitors were all mutated at the putative pMad-binding site. (B) EMSA with lysates from S2 cells transfected with the indicated plasmids (Mad/Medea/TkvQD) were incubated with the indicated IRDye700-labeled wildtype probes and run on non-denaturing gels. A band shift of each labeled probe was generated only in lanes loaded with S2 cell lysate transfected with Mad/Medea/TkvQD. Labeled probes were mixed with 5x or 50x stoichiometric excess of wildtype competitor (wt) or mutated (Mad mut) competitor. Five and 50x excess of wildtype competitor progressively diminished the band shift of the labeled probe, indicative of pMad/Medea complexes binding to the unlabeled competitor. In contrast, addition of the mutated competitor at a 50x excess resulted in retention of the labeled band shift, indicating that mutated competitors did not bind pMad/Medea complexes. (C, D) We tested the ability of fourteen BMP-AEs from wit-responsive (1–8 in A) and wit-non responsive (9–15 in A) genomic fragments to compete for pMad/Medea binding, as unlabeled DNA probes, with an IRDye700-labeled DNA probe for the BMP-AE of Van26. EMSA with lysates from S2 cells transfected with the indicated plasmids (Mad/Medea/TkvQD) were incubated with the IRDye700-labeled Van26 probe and the fourteen BMP-AE DNA unlabeled competitors. The labeled probe was mixed with a 5× stoichiometric excess of unlabeled competitors. The 5× excess of Van26 competitor diminished the band shift of the labeled probe, indicative of pMad/Medea complexes binding to the unlabeled competitor. The 5× excess of functional BMP-AEs within Van21, Van22, Van36, Van42, Van43, dad13 and Van15 competitors also resulted in a diminution of the band shift of the labeled probe (C). In contrast, the 5x excess of non-functional BMP-AEs within Van5, Van7, Van29, Van39 and Van53 did not result in a diminution of the band shift of the labeled probe (D), indicating that their affinity for the pMad/Medea complexes is lower than for the functional BMP-AEs within wit-responsive fragments. (E) Nuclear β-galactosidase and nlsDsred expression driven from 10 select wit-responsive fragments in which the BMP-AE motif was either wildtype or mutated at the Mad site. The point mutations that were introduced into the Mad site of the BMP-AEs are similar to the ones that abolished competition in vitro. In all cases, there was a reduction in the number of nuclei that expressed the nlsLacZ and nlsDsred reporters.
The 62 BMP-AEs tested above display a high degree of diversity within the consensus BMP-AE motif. Differences within this consensus motif may contribute to differential affinity for pMad/Medea recruitment, and potentially underlie differences observed between wit-responsive and wit-non responsive genomic fragments. To test this, we selected 7 BMP-AEs from each set, based on their divergent Medea-binding GNCV consensus sequence, and used these as untagged probes (in a 5:1 stoichiometry) to compete for the band shift generated from interaction of pMad/Medea with the BMP-AE from the wit-responsive Van26 fragment (as above) (Figure 3C, D). Notably, five out of the seven BMP-AEs from wit-responsive fragments strongly competed for the labelled band shift, while two weakly competed. In contrast, five out of the seven BMP-AEs from wit-non responsive fragments failed to compete for the labelled band shift, while two weakly competed. Thus, BMP-AEs within wit-responsive fragments typically have a higher affinity for pMad/Medea complexes than those from wit-non responsive genomic fragments, indicating that pMad/Medea affinity may underlie observed differences in enhancer activity for the majority of BMP-AE containing genomic fragments.
These results prompted us to compare the sequences of all 59 BMP-AE motifs (excluding the three BMP-AEs of Van10 and Van19, as above), embedded within the 56 genomic fragments for which we could determine responsiveness to wit. Interestingly, in comparing BMP-AEs from wit-responsive and wit-non responsive fragments, we found that BMP-AEs from wit-responsive fragments were significantly enriched for thymine at position 11 and depleted for guanine and adenine at position 11 and position 15, respectively (Supplementary Figure S10A, C). Although we could not detect any other sequence bias that could explain a difference of affinity between both sets of motifs, we noticed that a GCCA sequence within the Medea binding sequence is absent for BMP-AEs in wit-responsive fragments (0%, 0/37), whereas it is well represented for BMP-AE in wit non-responsive fragments (23%, 5/22) (Supplementary Figure S10C). These data led us to examine whether BMP-AE motifs that are active in the CNS differ from those that are active in other tissues. To do this, we compared the sequence of BMP-AEs within neuronal wit-responsive fragments to those that are embedded in genomic fragments shown to be tkv-responsive in the late embryo and wing imaginal disc (34) (Supplementary Figure S10B, C). Interestingly, we noticed that neuronally-active BMP-AEs were enriched for cytosine and depleted for guanine at position 8 in the linker region. Overall, these results demonstrate that that there are sequence biases that correlate with, and perhaps even confer pMad/Medea recruitment or even tissue-specific function, in positions previously found to not be critical in the BMP-AE motif. It will be interesting to analyze a larger set of BMP-AE motifs to demonstrate whether these biases are indeed functionally relevant.
We next tested whether the activity of the identified wit-responsive fragments was dependent on the embedded BMP-AEs. For ten selected wit-responsive fragments, we introduced specific mutations into the pMad site of the BMP-AE, and placed mutant genomic fragment reporters into the same insertion site as the corresponding wildtype reporter. In all cases tested, reporter expression driven from the mutant genomic fragments was lost in pMad-positive neurons, in a pattern that was strikingly similar to that observed for wildtype wit-responsive fragments in wit mutants (Figure 3E). These data strongly suggest that the BMP-AE motif serves as the platform for pMad/Medea-responsive reporter activity of these genomic fragments. We hereafter refer to these as functional BMP-AEs.
Identification of neuronal genes directly regulated by the BMP signaling pathway
The large collection of wit-responsive fragments identified suggested that a battery of genes may be directly regulated by BMP-activated pMad/Medea transcriptional regulation in efferent neurons. We next sought to resolve if the identified wit-responsive fragments are near BMP-responsive genes. Therefore, we dissected VNCs from control and wit mutant third instar larvae and compared transcript expression by RNA-sequencing (RNA-seq) of total poly-A transcripts. To evaluate the sensitivity of the technique and validate our approach, we examined the relative transcript level of five BMP-responsive neuronal genes with highly restricted expression; FMRFamide (FMRFa), Partner of bursicon (Pburs), Crustacean cardioactive peptide (CCAP), Myoinhibiting peptide precursor (Mip) and Insulin-like peptide 7 (Ilp7) (10,11,13). In the VNC, these genes all exhibit wit-dependent expression in a small subset of the neurons in which the gene is expressed. In spite of this, our RNA-seq analysis successfully identified all genes as being significantly downregulated in wit mutants, indicating that our RNA-seq analysis is sufficiently sensitive to identify neuronal subtype-specific wit-responsive gene regulation (Supplementary Table S2).
We next examined whether BMP-AEs in wit-responsive fragments were preferentially enriched over BMP-AEs within wit-non responsive fragments around wit-responsive genes. To avoid over-counting genomic regions where multiple BMP-AE motifs clustered around multiple wit-responsive genes, we instead quantified the number of genomic regions in which any number of BMP-AEs were up to 50 kb from the TSS (transcription start site) of any number of wit-responsive genes (Figure 4A, B, Supplementary Table S2). As an additional control, we examined enrichment of Bicoid (Bcd)-responsive genomic fragments to wit-responsive genes (Supplementary Figure S11A). These fragments are active during anterior-posterior patterning of the early Drosophila embryo (76) and are not predicted to enrich around wit-responsive neuronal genes. Our analysis was performed separately for BMP-activated genes (significantly downregulated in wit) versus BMP-repressed genes (significantly upregulated in wit) (see Materials and Methods for details of analysis).
Out of the 25 genomic loci that contain all of the 34 wit-responsive fragments (Figure 4A), we found that 36% of these loci (9/25) contain BMP-AEs within 10 kb of a BMP-activated gene's TSS, which increases to 60% (15/25) at 20 kb and then plateaus to reach 68% (17/25) at 50 kb (Figure 4B). For enrichment analysis of BMP-AEs within the 22 wit-non responsive fragments, we wished to eliminate the possible confound that the presence of a nearby wit-responsive fragments may explain the wit-responsiveness of the gene. Therefore, we only considered those regions that did not also include a wit-responsive fragment, which eliminated BMP-AEs within 4 wit-non responsive fragments. Thus, we examined 15 genomic loci containing the remaining 18 wit-non responsive fragments, and found that 0% of these loci (0/15) contain BMP-AEs within 10 kb of a BMP-activated gene's TSS, with only 7% (1/15) at 20 kb and steadily increasing to 47% (7/15) at 50 kb (Figure 4B). Similarly, only 11% of the loci (3/26) that contain Bcd-responsive fragments were within 10 kb of a BMP-activated gene's TSS, with 27% (7/26) by 20 kb and rising to 46% (12/26) by 50 kb (Figure 4B and Supplementary Figure S11A). These data indicate that functional BMP-AEs are selectively enriched within 20 kb of at least one BMP-activated gene (circled in red in Figure 4B). Indeed, at 20 kb this difference was found to be significant by chi2 analysis; P = 2.7 × 10−3 comparing wit-responsive versus wit-non responsive fragments, and P = 3.6 × 10−2 comparing wit-responsive versus Bcd-responsive fragments. We next tested for any enrichment of BMP-repressed genes at the same distance, and found no significant difference between BMP-AEs within wit-responsive fragments (12%: 3/25) and wit non-responsive fragments (20%: 3/15; P = 0.82), nor Bcd-responsive fragments (19%: 5/26; P = 0.75) (Supplementary Figure S11B). These results indicate that functional BMP-AEs are selectively enriched near BMP-activated genes, supporting the hypothesis of a positive regulatory relationship.
Since Drosophila embryonic enhancers most often regulate the adjacent gene (59), we addressed whether functional BMP-AEs enrich to adjacent wit-responsive genes, i.e. with no intervening wit-non responsive gene. We calculated the proportion of genomic loci containing a BMP-AE whose adjacent gene was a BMP-activated gene. For this analysis, we were able to examine 26 loci for BMP-AEs in wit-responsive fragments (as opposed to the 25 shown above) (see Methods and Materials for details). Notably, 61% (16/26) of genomic loci containing functional BMP-AEs harbor at least one adjacent BMP-activated gene. In contrast, only 7% (1/15) and 11% (3/26) of genomic loci containing non-functional BMP-AE (P = 1.9 × 10−3 compared to wit-responsive fragments) and Bcd-responsive fragments (P = 5.5 × 10−4 compared to wit-responsive fragments), respectively harbor at least one adjacent BMP-activated gene. These data suggest that a large proportion of functional BMP-AEs regulate the adjacent gene.
We observed numerous instances of multiple, reiterated BMP-AEs within 12 kb of a single annotated gene locus; therefore, we compared the proximity of individual versus reiterated BMP-AEs to BMP-activated genes. We found that 77% of reiterated BMP-AEs (10/13) were within or adjacent to BMP-activated genes, and that 46% (6/13) of single BMP-AEs were within or adjacent to BMP-activated genes. This corroborates our finding that 72% of reporters (21/29) from a locus with reiterated BMP-AE were wit-responsive, whereas 48% of reporters (13/27) containing a single BMP-AE were wit-responsive. These data indicate that reiteration of conserved BMP-AEs in a locus is a strong predictor of a wit-responsive genomic region and nearby wit-responsive genes in the Drosophila nervous system, but that the presence of multiple BMP-AEs within a gene locus is not a requirement for the wit-responsiveness of a genomic region.
Many genes have numerous alternate promoters that generate different transcript isoforms; therefore, we considered the possibility that BMP-AEs may selectively regulate specific isoforms of a gene, and not others. To test this hypothesis, we analyzed our RNA-seq dataset for differentially expressed transcripts within 100 kb of functional BMP-AEs. Interestingly, we found six instances that confirmed our hypothesis (Supplementary Table S2). For example, the sickie gene (sick) locus contains eight predicted promoters that generate twelve putative isoforms. We found that only the two shortest isoforms were significantly down-regulated in wit mutants, whereas the expression of other isoforms remained unchanged (Supplementary Figure S12). Interestingly, a functional BMP-AE (Van25) was located approximately 950bp upstream of the TSS of each differentially expressed isoform, suggesting specific isoform regulation by this BMP-AE.
In summary, these data suggest that BMP-AEs identified here are pMad/Medea-responsive cis-elements for the BMP-activation of a battery of genes and selective gene isoforms. Moreover, a subset of these genes are known to be regulators of synaptic growth and neurotransmission, suggesting that they are putative effectors of the BMP signaling pathway in neurons (see Discussion).
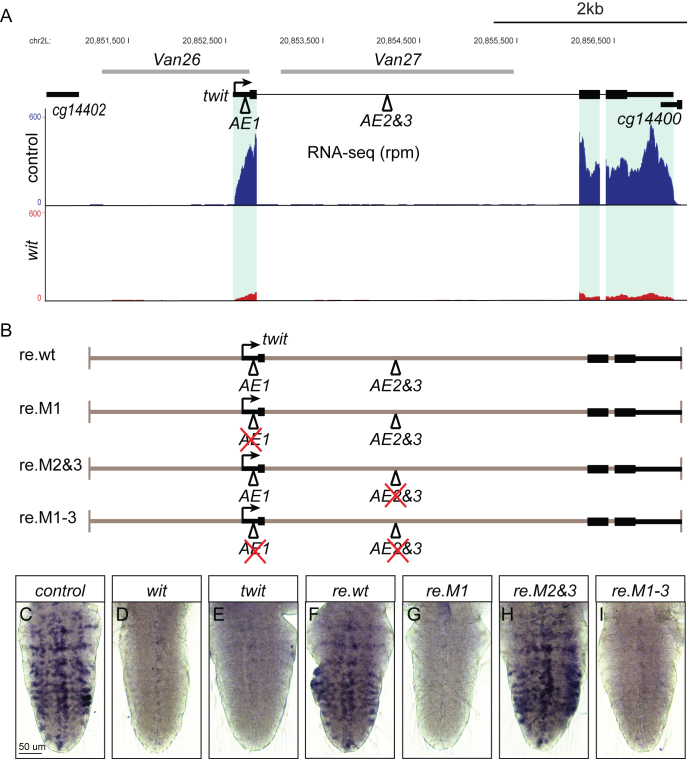
BMP-AEs are required for BMP-activated twit expression in vivo
To obtain additional evidence that identified BMP-AEs represent genuine cis-regulatory sites for BMP-responsive gene expression, we focused on the BMP-activated gene, twit, which plays an effector role in BMP-dependent neurotransmission at the larval NMJ (23,25). The twit locus harbors three conserved BMP-AEs; one in the 5′UTR (AE1 within the Van26 fragment) and two BMP-AEs (sharing a single GGCGCC palindromic Mad motif) are located in the first intron of twit (AE2&3 within the Van27 fragment) (Figure 5A). Our reporter analysis in Figures 2 and 3 suggested that the AE1 is required for widespread wit-responsive twit expression in motor neurons, whereas the inverted AE2&3 motifs are only required for wit-responsive twit expression in six ventral neurons (Supplementary Table S4). To examine whether either BMP-AEs mediate the wit-responsiveness of twit, we first compared reporter expression driven from Van26 and Van27 with that of twit-GFP, a Mi{MIC} transposon gene trap insertion in the twit intron (57). Notably, we observed that Van26 mostly recapitulated the extensive expression of twit-GFP expression in motor neurons, while Van27 recapitulated the expression of twit-GFP expression in six ventral neuropeptidergic cells in the VNC (Supplementary Figures S5, S13A, B). Interestingly, expression driven by Van26 and Van27 did not overlap (Supplementary Figure S13C). This suggests that the AE1 within Van26 is required for widespread wit-responsive expression of twit in motor neurons, whereas AE2&3 within Van27 are only required for twit expression in six ventral neuropeptidergic cells of the VNC.
Figure 5.
An exonic BMP-AE is required for twit transcription in motor neurons. (A) Genome browser showing the twit locus with three BMP-AEs and the two genomic fragments used for the reporter analysis, together with the tracks from RNA-seq (in rpm; reads per million reads). The expression of twit is strongly downregulated in witA12/witB11 mutants as compared to controls (witA12/+). (B) Schematic of four twit locus DNA constructs used for in vivo rescue of twit mutants shown in F–I. The BMP-AEs within this locus as well the BMP-AE motifs mutated in each construct are shown. (C–I) In situ hybridization using a Digoxigenin-labeled RNA twit probe reveals expression of twit in VNCs of controls (C), wit mutants (D), twit mutants (E) and twit mutants rescued with the constructs shown in B (F-I). The AE1 within the twit 5′UTR is critical for twit motor neuron expression. Genotypes: (C) w1118. (D) w1118;; witA12/witB11. (E) w1118; twit160 / twit160. (F) w1118; twit160 / twit160; re.wt/+. (G) w1118; twit160 / twit160; re.M1/+. (H) w1118; twit160 / twit160; re.M2&3/+. (I) w1118; twit160 / twit160; re.M1-3/+.
To test whether these BMP-AEs are required for twit expression, we generated a series of rescue transgenes containing ∼6 kb of genomic DNA encompassing the twit locus, and targeted these into the attP2 integrase site in twit deletion mutant flies. Rescue transgenes included the wildtype sequence (re.wt) and also a series of BMP-AE mutants, to eliminate pMad/Medea recruitment, including single mutants of AE1 (re.M1) and AE2&3 (re.M2&3), and also a double mutant of AE1 and AE2&3 (re.M1-3) (Figure 5B). Flies transgenic for these constructs were analyzed for neuronal twit transcript expression by in situ hybridization. We confirmed that re.wt rescued twit expression of twit mutants throughout the VNC (Figure 5C–F). In contrast, both re.M1 and re.M1-3 failed to rescue detectable VNC twit expression in twit mutants (Figure 5G, I), whereas re.M2&3 rescued broad twit transcript levels in twit mutants (Figure 5H). Thus, the single BMP-AE in the twit 5′ UTR is required for native wit-responsive motor neuron expression of twit in vivo. We could not detect any twit expression in the six ventral neuropeptidergic cells using re.M1, suggesting that the BMP-AE1 found in the twit 5′UTR also contributes to twit expression in these cells, or reflects the insufficient resolution of the in situ hybridization protocol to efficiently detect expression in these six cells. Regardless, we identified the BMP-AE motif that acts as the primary functional pMad/Medea-responsive element for wit-responsive twit expression in motor neurons.
The neuronal function of the BMP-AE is evolutionarily conserved
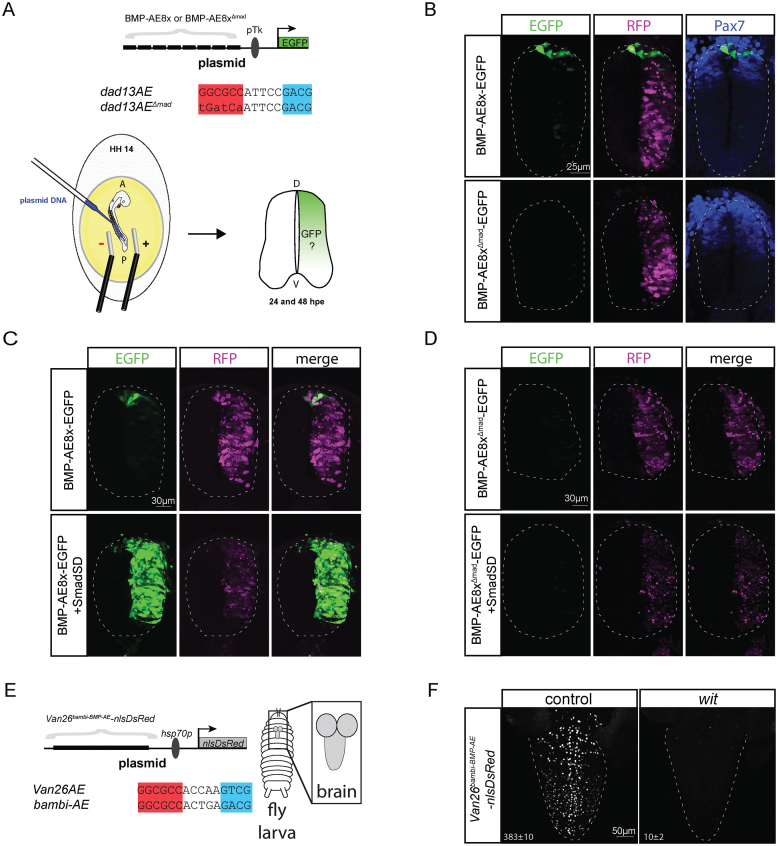
Our analysis identifies a number of cis-regulatory motifs and genes that represent part of the BMP network in neurons of the fly VNC. This prompted us to test the hypothesis that together with pMad and Medea, the BMP-AE forms a fundamental regulatory circuit in the nervous system of distantly related animals. To test this, we used two approaches. First, we generated a reporter containing 8× concatenated BMP-AE sequences taken from the Drosophila dad gene locus (dad13) (Figure 6A), placed upstream of a minimal thymidine kinase (Tk) gene promotor and enhanced GFP (EGFP) reporter gene (termed BMP-AE8x-EGFP). This reporter was electroporated into the developing chick spinal cord at stage HH14 and its activity was analyzed after 24 and 48 h. At 24 h, one day post-electroporation (dpe), the EGFP reporter was robustly expressed in the dorsal-most region of the spinal cord, where BMP signaling is highest during a period of active patterning of the dorsal neuronal subtypes (Figure 6B) (85,86). At 2 dpe, reporter expression was also detected in medial to ventral regions of the developing neural spinal cord, which is consistent with previous observations (85) (Supplementary Figure S14). To test whether the observed dorsal BMP-AE8x-EGFP reporter activity was BMP-dependent, we first tested the activity of a BMP-AE8x-EGFP reporter mutated at each pMad-binding site (BMP-AE8xΔmad-EGFP) (Figure 6A). This eliminated reporter activity at both 1 dpe and 2 dpe (Figure 6B and Supplementary Figure S14). Second, we co-electroporated the BMP-AE8x-EGFP and BMP-AE8xΔmad-EGFP reporters with a phospho-mimetic (constitutively activated) form of Smad1 (Smad1 S/D) (85), which is a vertebrate ortholog of Drosophila Mad. We found that the BMP-AE8x-EGFP reporter expression greatly increased and expanded throughout the neural tube, but the BMP-8xAEΔmad-EGFP reporter expression was unresponsive and exhibited no activity (Figure 6C, D). We conclude that the Drosophila BMP-AE sequence is directly activated by BMP/pSmad1 signaling in the developing chick neural tube.
Figure 6.
The BMP-AE mediates a conserved Smad-responsive function in the Drosophila and vertebrate CNS. (A) Schematic representation of the chick neural tube electroporation experiment with the sequence of the wildtype and mutated dad13-BMP-AE. (B) Expression of an octamerized version of a wildtype and mutated BMP-AE with an EGFP reporter at 24 hpe. The wildtype reporter displays strong EGFP expression in dorsal-most part of the neural tube, whereas the mutant reporter does not show any activity. An RFP control plasmid is co-electroporated with the EGFP reporters to indicate the efficiency of electroporation. Pax7 immunostaining indicates the dorsal region of the spinal cord. (C, D) Co-electroporation of a phosphomimetic form of Smad1 (Smad S/D) strongly increases and expands expression of the wildtype BMP-AE throughout the whole neural tube (C), but does not activate expression of the mutated version (D). (E) Schematic representation of the Drosophila transgenesis experiment where the Van26 genomic fragment has its BMP-AE motif replaced with a BMP-AE taken from the X. laevis bambi enhancer. (F) Nuclear DsRed expression driven from the Van26AEbambi-BMP-AE-nlsDsRed reporter in control and wit mutants third instar larval VNCs. The mean±SEM number of nuclei per VNC that express the reporter is indicated at the bottom of each panel. Reporter activity was significantly reduced in wit mutants (P = 0.0079 two-sided Wilcoxon rank-sum test). At least five VNCs were analyzed for both genotypes.
To further test the hypothesis that functional DNA motifs similar to the BMP-AE are conserved in vertebrates, we replaced the sequence of the BMP-AE1 within the Van26 reporter with a 15 bp motif taken from a BMP-responsive enhancer (BRE) of the Xenopus laevis bambi gene (termed Van26bambi-BMP-AE-nlsDsRed) (Figure 6E) (87). Even though no such exact BMP-AE sequence was tested in our reporter analysis, this motif perfectly matches the Drosophila BMP-AE consensus. Notably, this bambi BMP-AE motif functionally replaced the wildtype BMP-AE1, as strong reporter activity was observed in 383 ± 10 nuclei per VNC (n = 5) The pattern of expression was strikingly similar to that of Van26, which was expressed in 341 ± 15 nuclei per VNC (n = 6) (Figure 6F). Moreover, reporter activity of the Van26bambi-BMP-AE-nlsDsRed reporter was reduced to 10 ± 2 nuclei per VNC (n = 5; P = 0.0079) in wit mutants, which is comparable to the reduction of reporter expression observed for the wildtype Van26 reporter, which was in 4±2 nuclei per VNC (n = 6; P = 0.0022) (Figures 2, 6F).
Taken together, these data show that the fundamental transcriptional mechanisms acting downstream of neuronal BMP signaling are conserved from Drosophila to vertebrates. It further suggests that the BMP-AE motif may be useful in future studies to predict and identify direct BMP-responsive genes and enhancers in vertebrate neurons.
DISCUSSION
BMP signaling acts via the transcriptional activity of pMad to control neuronal differentiation and synaptic function in Drosophila (9–11,13–15,22,26). BMP signaling plays similarly critical roles throughout vertebrate nervous system development, function and repair (88–94). Yet, how BMP-responsive transcription coordinates diverse gene regulatory programs in the nervous system remains poorly defined in any system. To start defining these transcriptional programs in the nervous system, we have demonstrated the utility of a combined computational and transgenic approach in predicting and validating 34 novel direct wit-responsive genomic fragments in motor and neuropeptidergic neurons of the Drosophila VNC. Together with differential transcript profiling, we showed that the BMP-AE motifs within these wit-responsive genomic fragments are enriched around BMP-activated genes, indicative of putative regulatory pairings that we validated for the twit gene. Overall, our data provide insight into the organization of directly BMP-driven gene networks in neurons that control neuronal identity and synaptic function. In addition, we provide the largest verified collection of genomic fragments whose underlying enhancers are targeted by a developmental signaling pathway in neurons; offering a resource for detailed analysis of gene regulatory mechanisms underlying synaptic growth and function.
Prediction and validation of a large collection of direct BMP-activated neuronal enhancers
Since the discovery of a role for retrograde BMP signaling in motor and neuropeptidergic neurons, only two wit/pMad-responsive enhancers have been functionally verified in vivo, namely the Tv4 enhancer of FMRFa and a proximal promotor region of trio (24,26). The Tv4 enhancer is only expressed in the six Tv4 neurons of the VNC (95,96). It contains a 39 bp wit-responsive cis-element with a GGCGCC pMad binding site that is required for enhancer function (26); however, there is no consensus BMP-AE within this wit-responsive cis-element. In addition, the promotor region of trio was immunoprecipitated by Myc::Mad in Drosophila embryonic motor neurons, and also shown to be responsive to BMP signaling in human embryonic kidney 293 cells; however, this region does not contain a BMP-AE motif nor was a specific pMad/Medea-responsive cis-element pinpointed (24). Therefore, it has been uncertain whether canonical pMad/Medea-binding motifs have widespread activity in neurons or whether pMad/Medea-binding motifs are highly diversified in a way that might be expected to reflect the high diversity of neuronal subtypes and their subtype-specific gene expression profiles.
In this study, we chose the BMP-AE for our computational approach because its 15 bp sequence offered a high stringency parameter for genomic searches that had previously been used to successfully identify a number of BMP-responsive genomic fragments throughout development (34). We showed that the BMP-AE is a widely deployed activator of gene expression in neurons, often acting as the single necessary pMad/Medea-responsive cis-element within a genomic fragment. Thus, we have demonstrated that such cis-elements can be exploited for BMP-responsive enhancer discovery in Drosophila neurons. Also, the motif confidence method used here has previously proven a successful approach to the prediction of functional transcription factor and miRNA binding motifs (43,44). In this study, we used high stringency parameters testing BMP-AEs with a motif confidence high score of 0.8 or above. Because the function of the BMP-AE in the developing embryo is tolerant to modest changes in the linker length between the pMad and Medea binding motifs (34,39) and that many poorly conserved or degenerate cis-elements are fully functional (97–99), it will be interesting to test BMP-AEs with lower motif confidence score. This will allow us to assess conservation levels that still efficiently discover functional BMP-responsive motifs, albeit at the potential expense of a lower rate of success in identifying functional BMP-AEs.
In addition to the 34 wit-responsive fragments, we found that 24 of the genomic fragments tested exhibited no apparent wit-responsive expression in late third instar VNC neurons. This lack of wit-responsiveness may be the result of the low affinity of individual BMP-AE motifs for pMad/Medea complexes. Our in vitro analysis demonstrated that BMP-AEs within wit-responsive fragments typically have a higher affinity for an activated Mad/Medea complex than those within wit-non responsive fragments. These differences likely reside in sequence preferences for pMad/Medea recruitment, within the overall BMP-AE consensus. However, while we could not find any sequence signatures that explain the difference in affinity for all the BMP-AE motifs tested, we did observe a significant overrepresentation of specific nucleotides at positions 11 and 15 of functional and non-functional BMP-AEs, respectively. While these data suggest that there are underlying sequence preferences for pMad/Medea recruitment that confer apparent differences in in vivo enhancer activity, additional studies will be required to fully define these. We expect that better definition of the BMP-AE motif would lead to an increase in the efficiency of computational discovery of novel functional BMP-AEs.
Numerous BMP-AEs within the 24 wit-non responsive fragments display high affinity to the pMad/Medea complex. There are numerous lines of evidence or reasoning to suggest that a fraction of these BMP-AEs are indeed functional: (i) The BMP-activated cv-2 gene is flanked by the Van15/CV2-AE5 wit-responsive fragment and the CV2-AE6 wit-non responsive fragment (Figure 4A; 2R:21360767). Interestingly, the CV2-AE6 fragment has tkv-responsive enhancer activity in cells of the dorsal vessel and in an ectodermal stripe in embryos (30). Together with our data in the VNC, this suggests that cv-2 is regulated by two distinct BMP-activated enhancers in the late embryo and third instar VNC. This may be a clear example where a single gene is regulated by an array of autonomous enhancers acting in a modular fashion to confer a variety of cell type- and temporal-specific expression patterns (46,100–102). Correspondingly, we found that several non-functional BMP-AEs are located in close proximity to functional BMP-AEs and a BMP-activated gene(s). Thus, several of these wit-non responsive BMP-AEs may provide pMad/Medea-regulated gene expression in different tissues. (ii) A lack of wit-responsiveness for a number of genomic fragments may also arise as a result of missing critical cis-regulatory elements required for BMP-AE to function. Indeed, cis-regulatory elements that act combinatorially to control gene expression can be spread over large distances that cannot be readily captured within the short 2kb fragments tested here (102). Accordingly, we found that two genomic fragments, Van37 and Van38, only exhibited wit-responsive enhancer activity in the context of a larger fragment, fortuitously available through the Janelia GAL4 collection (60). (iii) It is also possible that additional cis-regulatory elements recognized by other transcription factors, or alternatively multiple linked enhancers within the same genomic fragment, compensate for the loss of BMP signaling in wit mutants to support the retention of genomic fragment activity.
The diversity of BMP-AE enhancer activities in the Drosophila nervous system
The 34 wit-responsive fragments discovered here display highly diverse expression patterns; some expressing broadly and others with restricted subtype-specific expression. Given our demonstration that the BMP-AE itself is necessary for the wit-responsive activity of these reporters, these results strongly suggest that pMad/Medea acting at this common BMP-AE motif must functionally collaborate with other subtype-specific transcription factors bound to flanking cis-elements, in order to generate these diverse expression patterns. We previously showed that the FMRFa gene is activated by a combinatorial code of pMad and a set of subtype-specific transcription factors (10,26,103,104), and reported that this integration occurs through the combined activities of distinct pMad and Apterous-binding cis-elements in the Tv4 enhancer of the FMRFa gene (26). This would be consistent with many other studies showing that the cell-specific activities of BMP-activated Smad complexes are often shaped by their functional interactions with local subtype-specific transcription factors in multiple organisms (105,106). It will be of great interest to define these subtype-specific transcription factors for neurons and to determine how their intersection with pMad/Medea at wit-responsive enhancers shapes BMP-dependent neuronal differentiation and synaptic function.
Interestingly, for all previously known BMP-activated genes that include FMRFa, Pburs, CCAP, Trio, Mip and Ilp7 (10,11,13), we only found Pburs to have conserved BMP-AEs within 50kb (Supplementary Table S2). This suggests that other pMad/Medea-responsive motifs play a role in mediating BMP-dependent gene regulation in the VNC. Indeed, a variety of pMad/Medea-responsive motif sequences have been identified in Drosophila (27–36), including those outside of the BMP-AE or BMP-SE consensus, and this diversity is further increased for motifs that bind complexes of activated Smads with other cell type-specific transcription factors and co-factors (105,106). Such diversity would limit the utility of computational discovery approaches to identify those pMad/Medea-responsive cis-elements.
Although we did not explicitly demonstrate that the functional BMP-AE motifs we identified here are directly bound by pMad/Medea complexes in the CNS, we postulate that this is likely the case. In addition of our in vitro and genetic data, two previous studies in the early Drosophila embryo and Drosophila Kc cells have identified three of the BMP-AEs identified here (for dad13, Van34 and Van36 reporters) to be directly bound by pMad and Medea, using ChIP-seq (36,107). Ongoing studies that aim to confirm that these BMP-AEs are indeed directly bound by pMad/Medea complexes and to identify other additional BMP-responsive motifs will expand our understanding of how BMP signaling and pMad/Medea transcriptional activity controls subtype-specific neuronal differentiation and synaptic function.
Towards identification of a directly-regulated BMP effector gene network in neurons
Identification of the direct target genes of developmental signaling pathways is a key step to understanding and modeling the properties of biological systems. Before this study, only FMRFa and trio have been suggested to be direct targets of BMP-activated Mad transcriptional activity. In addition, a previous microarray study identified 101 genes whose expression is regulated by wit in the whole larval CNS (25). Interestingly, comparing the previous microarray analysis of wit-responsive genes with the RNA-seq analysis performed here, we found notable differences in the two datasets. Of the 101 genes found in the microarray, only 26 genes were also found to be differentially expressed by RNA-seq analysis here, with 7 of those genes being differentially expressed in the opposite direction (Supplementary Table S3). A potential explanation for this discrepancy could reside in the different sensitivities of these two methodologies, as well as the differences in tissues examined; the microarray analysis used the whole CNS and the ring gland, while our RNA-seq analysis study used VNCs with the brain lobes and ring gland removed. Regardless, these genes represent candidate direct target genes for pMad/Medea transcriptional control, yet leave open the possibility that BMP signaling acts indirectly on many genes, perhaps through the activation or repression of unknown transcription factor intermediaries. Examples of this can be drawn from vertebrate models where BMP4-mediated topographic mapping of trigeminal sensory neurons appear to occur via transcription factor intermediaries (108), and also where retrograde GDNF or NT-3 signaling from target cells induces expression of ETS transcription factors, Pea3 or Er81 respectively, to control appropriate motor and sensory neuronal connectivity (109,110). In such cases, pMad/Medea-dependent direct regulation should be limited to a small number of transcription factors. However, our pairing of in vivo reporter analysis and differential transcript profiling identified a large set of BMP-AEs and nearby BMP-activated genes; most likely representing genuine regulatory relationships, as demonstrated for twit.
We found that BMP-AEs embedded in wit-responsive fragments were enriched within 20–50 kb of BMP-activated genes. These distances correspond to those found for pMad-bound ChIP peaks enriched within 40 kb of tkv-responsive genes in the dorsal ectoderm of 2–3.5 h embryos (36). Also, evidence from large scale enhancer analysis indicates that Drosophila enhancers have a bias to regulate the adjacent gene (59), and accordingly we found an enrichment of functional BMP-AE motifs adjacent to BMP-activated genes. However, enhancers also operate over longer distances and we do not exclude the likelihood that this is the case for BMP-AEs. This may account for the wit-responsive activity of 8 genomic fragments whose BMP-AEs are not within 50 kb of a BMP-activated gene, but instead within 200 kb of a BMP-activated gene (Supplementary Table S2), which may represent longer distance regulatory relationships.
Amongst the BMP-activated genes near BMP-AEs, we found two known regulators of BMP signaling, dad and cv-2. Loss of function mutants for dad display synaptic overgrowth at the NMJ, a role that is consistent with a direct neuronal BMP feedback inhibitor (111). Cv-2 encodes a secreted protein that binds the type I BMP receptor Tkv and heparan sulfate proteoglycans (HSPG) on the cell surface to promote BMP signaling during posterior cross-vein formation in the pupal wing (112,113). The wit-responsive expression of this gene in neurons raises the possibility that it plays a BMP signal strength modulatory role at the NMJ. It is intriguing to note that two HSPGs, Syndecan (Sdc) and Dally-like, are both present at the NMJ and influence synaptic growth and active zone form and function, respectively (114). Future studies will shed light on the conservation of molecular mechanisms regulating the binding of the BMP ligands to their receptors between the NMJ and the pupal wing.
A second interesting set of BMP-activated genes are those that control the organization and function of the cytoskeleton. For instance, the stathmin gene encodes a tubulin binding phosphoprotein that destabilizes microtubules (115). Stathmin is required for stability and growth of the NMJ, suggesting that Stathmin may also promote the transport of BMP ligands, or other signaling molecules, along the microtubule (116). If this model proves to be correct, it would indicate that BMP signaling directly activates stathmin expression in a positive feedback loop.
We identified two additional BMP-activated genes (cg14274 and cg7781) that encode glycosylphosphatidylinositol (GPI) anchored proteins of the Ly-6 family. These genes are of particular interest because of the previous identification of the related twit gene as a BMP effector required for the regulation of spontaneous neurotransmitter release at the NMJ (23). All these genes have multiple BMP-AEs within broadly expressed wit-responsive enhancers near their transcription start sites. This suggests that the transcriptional activation of multiple members of this gene family by BMP signaling is mediated by nearby BMP-AEs and that these genes have synergistic, additive or redundant functions to control neurotransmission at the NMJ. It will be interesting to pursue the phenotypic consequences of mutation of this novel family of genes in order to reveal the combined roles of this poorly understood family of proteins in neurotransmission.
DATA AVAILABILITY
The raw sequence data from this study have been submitted to the NCBI GEO under accession number GSE106650.
Supplementary Material
ACKNOWLEDGEMENTS
We are very grateful to Drs Anaïs Bardet and Alexander Stark for performing the motif confidence analysis for the BMP-AE motifs in Drosophila genomes. We gratefully acknowledge members of the Allan laboratory, Drs Stefan Thor, Pejmun Haghighi and Timothy O’Connor for insightful discussion and critical reading of the manuscript. Also, we thank the anonymous referees whose constructive suggestions greatly improved the manuscript. We are very grateful to Drs T. Michael Underhill and Ryan Vander Werff for assistance with RNA-seq, at the BRC-seq Centre. We are very grateful to Dr Artur Kania for crucial advice and reagents required to perform the chick spinal cord electroporation studies, and to Dr Joy Richman and lab members for infrastructure, lab space and training in chick spinal cord electroporation. We gratefully acknowledge Dr Lyubov Veverytsa (Allan lab) for generating the pStingerattBDsRednls plasmid. We thank Dr Michelle Markstein for providing us the merMer software. We also thank Drs Markus Affolter, Vanessa Auld, Christian Boekel, Justin Kumar, Allen Laughon, Gwenvael Le Dréau, Elisa Marti, Laurel Raftery, Guy Tanentzapf, Esther Verheyen, the Bloomington Drosophila Stock Center, the Drosophila Genomics Resource Center and P[acman] Resources for fly stocks and molecular reagents.
Author contributions: R.V. and D.W.A. conceived and designed the experiments. R.V. performed most of the experiments. Z.N.K. assisted R.V. with the chick electroporation and processed samples for confocal imaging. T.L. performed the EMSA band shift experiments and generated the wildtype and mutated 8x concatenated BMP-AE and twit rescue constructs with the help of R.V. G.P. and A.F. initiated this project by establishing collaboration with Alexander Stark to calculate BMP-AE motif confidence, and generated the first reporter constructs to analyze BMP-dependent reporter expression in the developing embryo and imaginal discs tissues. S.F. performed the bioinformatics on the RNA-seq data. R.V., T.L., S.F., Z.N.K. and D.W.A. analyzed data. R.V. and D.W.A wrote the paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institutes of Health Research (CIHR) [MOP-98011, PJT-148905 to D.W.A.]; Deutsche Forschungsgemeinschaft Collaborative Research Centre [SFB592 to G.P.]; Excellence Initiative of the German Federal and State Governments [EXC294 to G.P.]. Funding for open access charge: CIHR [PJT-148905].
Conflict of interest statement. None declared.
REFERENCES
- 1. Allan D.W., Thor S.. Transcriptional selectors, masters, and combinatorial codes: regulatory principles of neural subtype specification. Wiley Interdiscip. Rev. Dev. Biol. 2015; 4:505–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hobert O., Carrera I., Stefanakis N.. The molecular and gene regulatory signature of a neuron. Trends Neurosci. 2010; 33:435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hobert O. Regulation of terminal differentiation programs in the nervous system. Annu. Rev. Cell Dev. Biol. 2011; 27:681–696. [DOI] [PubMed] [Google Scholar]
- 4. Dasen J.S. Transcriptional networks in the early development of sensory-motor circuits. Curr. Top. Dev. Biol. 2009; 87:119–148. [DOI] [PubMed] [Google Scholar]
- 5. da Silva S., Wang F.. Retrograde neural circuit specification by target-derived neurotrophins and growth factors. Curr. Opin. Neurobiol. 2011; 21:61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hippenmeyer S., Kramer I., Arber S.. Control of neuronal phenotype: what targets tell the cell bodies. Trends Neurosci. 2004; 27:482–488. [DOI] [PubMed] [Google Scholar]
- 7. Marqués G. Morphogens and synaptogenesis in Drosophila. J. Neurobiol. 2005; 64:417–434. [DOI] [PubMed] [Google Scholar]
- 8. Layden M.J., Odden J.P., Schmid A., Garces A., Thor S., Doe C.Q.. Zfh1, a somatic motor neuron transcription factor, regulates axon exit from the CNS. Dev. Biol. 2006; 291:253–263. [DOI] [PubMed] [Google Scholar]
- 9. Marques G., Bao H., Haerry T.E., Shimell M.J., Duchek P., Zhang B., O’Connor M.B.. The Drosophila BMP type II receptor wishful thinking regulates neuromuscular synapse morphology and function. Neuron. 2002; 33:529–543. [DOI] [PubMed] [Google Scholar]
- 10. Allan D.W., St Pierre S.E., Miguel-Aliaga I., Thor S.. Specification of neuropeptide cell identity by the integration of retrograde BMP signaling and a combinatorial transcription factor code. Cell. 2003; 113:73–86. [DOI] [PubMed] [Google Scholar]
- 11. Veverytsa L., Allan D.W.. Retrograde BMP signaling controls Drosophila behavior through regulation of a peptide hormone battery. Development. 2011; 138:3147–3157. [DOI] [PubMed] [Google Scholar]
- 12. McCabe B.D., Marques G., Haghighi A.P., Fetter R.D., Crotty M.L., Haerry T.E., Goodman C.S., O’Connor M.B.. The BMP homolog Gbb provides a retrograde signal that regulates synaptic growth at the Drosophila neuromuscular junction. Neuron. 2003; 39:241–254. [DOI] [PubMed] [Google Scholar]
- 13. Miguel-Aliaga I., Thor S., Gould A.P.. Postmitotic specification of Drosophila insulinergic neurons from pioneer neurons. PLoS Biol. 2008; 6:e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Aberle H., Haghighi A.P., Fetter R.D., McCabe B.D., Magalhaes T.R., Goodman C.S.. wishful thinking encodes a BMP type II receptor that regulates synaptic growth in Drosophila. Neuron. 2002; 33:545–558. [DOI] [PubMed] [Google Scholar]
- 15. Goold C.P., Davis G.W.. The BMP ligand Gbb gates the expression of synaptic homeostasis independent of synaptic growth control. Neuron. 2007; 56:109–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. James R.E., Hoover K.M., Bulgari D., McLaughlin C.N., Wilson C.G., Wharton K.A., Levitan E.S., Broihier H.T.. Crimpy enables discrimination of presynaptic and postsynaptic pools of a BMP at the drosophila neuromuscular junction. Dev. Cell. 2014; 31:586–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rawson J.M., Lee M., Kennedy E.L., Selleck S.B.. Drosophila neuromuscular synapse assembly and function require the TGF-beta type I receptor saxophone and the transcription factor Mad. J. Neurobiol. 2003; 55:134–150. [DOI] [PubMed] [Google Scholar]
- 18. Rodal A.A., Blunk A.D., Akbergenova Y., Jorquera R.A., Buhl L.K., Littleton J.T.. A presynaptic endosomal trafficking pathway controls synaptic growth signaling. J. Cell Biol. 2011; 193:201–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vanlandingham P.A., Fore T.R., Chastain L.R., Royer S.M., Bao H., Reist N.E., Zhang B.. Epsin 1 promotes synaptic growth by enhancing BMP signal levels in motoneuron nuclei. PLoS One. 2013; 8:e65997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Smith R.B., Machamer J.B., Kim N.C., Hays T.S., Marques G.. Relay of retrograde synaptogenic signals through axonal transport of BMP receptors. J. Cell Sci. 2012; 125:3752–3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McCabe B.D., Hom S., Aberle H., Fetter R.D., Marques G., Haerry T.E., Wan H., O’Connor M.B., Goodman C.S., Haghighi A.P.. Highwire regulates presynaptic BMP signaling essential for synaptic growth. Neuron. 2004; 41:891–905. [DOI] [PubMed] [Google Scholar]
- 22. Berke B., Wittnam J., McNeill E., Van Vactor D.L., Keshishian H.. Retrograde BMP signaling at the synapse: a permissive signal for synapse maturation and activity-dependent plasticity. J. Neurosci. 2013; 33:17937–17950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim N.C., Marques G.. The Ly6 neurotoxin-like molecule target of wit regulates spontaneous neurotransmitter release at the developing neuromuscular junction in Drosophila. Dev. Neurobiol. 2012; 72:1541–1558. [DOI] [PubMed] [Google Scholar]
- 24. Ball R.W., Warren-Paquin M., Tsurudome K., Liao E.H., Elazzouzi F., Cavanagh C., An B.S., Wang T.T., White J.H., Haghighi A.P.. Retrograde BMP signaling controls synaptic growth at the NMJ by regulating trio expression in motor neurons. Neuron. 2010; 66:536–549. [DOI] [PubMed] [Google Scholar]
- 25. Kim N.C., Marques G.. Identification of downstream targets of the bone morphogenetic protein pathway in the Drosophila nervous system. Dev. Dyn. 2010; 239:2413–2425. [DOI] [PubMed] [Google Scholar]
- 26. Berndt A.J., Tang J.C., Ridyard M.S., Lian T., Keatings K., Allan D.W.. Gene regulatory mechanisms underlying the spatial and temporal regulation of Target-Dependent gene expression in Drosophila neurons. PLos Genet. 2015; 11:e1005754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wharton S.J., Basu S.P., Ashe H.L.. Smad affinity can direct distinct readouts of the embryonic extracellular Dpp gradient in Drosophila. Curr. Biol. 2004; 14:1550–1558. [DOI] [PubMed] [Google Scholar]
- 28. Walsh C.M., Carroll S.B.. Collaboration between Smads and a Hox protein in target gene repression. Development. 2007; 134:3585–3592. [DOI] [PubMed] [Google Scholar]
- 29. Kim J., Johnson K., Chen H.J., Carroll S., Laughon A.. Drosophila Mad binds to DNA and directly mediates activation of vestigial by Decapentaplegic. Nature. 1997; 388:304–308. [DOI] [PubMed] [Google Scholar]
- 30. Rushlow C., Colosimo P.F., Lin M.C., Xu M., Kirov N.. Transcriptional regulation of the Drosophila gene zen by competing Smad and Brinker inputs. Genes Dev. 2001; 15:340–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xu X., Yin Z., Hudson J.B., Ferguson E.L., Frasch M.. Smad proteins act in combination with synergistic and antagonistic regulators to target Dpp responses to the Drosophila mesoderm. Genes Dev. 1998; 12:2354–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin M.C., Park J., Kirov N., Rushlow C.. Threshold response of C15 to the Dpp gradient in Drosophila is established by the cumulative effect of Smad and Zen activators and negative cues. Development. 2006; 133:4805–4813. [DOI] [PubMed] [Google Scholar]
- 33. Gao S., Steffen J., Laughon A.. Dpp-responsive silencers are bound by a trimeric Mad-Medea complex. J. Biol. Chem. 2005; 280:36158–36164. [DOI] [PubMed] [Google Scholar]
- 34. Weiss A., Charbonnier E., Ellertsdottir E., Tsirigos A., Wolf C., Schuh R., Pyrowolakis G., Affolter M.. A conserved activation element in BMP signaling during Drosophila development. Nat. Struct. Mol. Biol. 2010; 17:69–76. [DOI] [PubMed] [Google Scholar]
- 35. Pyrowolakis G., Hartmann B., Muller B., Basler K., Affolter M.. A simple molecular complex mediates widespread BMP-induced repression during Drosophila development. Dev. Cell. 2004; 7:229–240. [DOI] [PubMed] [Google Scholar]
- 36. Deignan L., Pinheiro M.T., Sutcliffe C., Saunders A., Wilcockson S.G., Zeef L.A., Donaldson I.J., Ashe H.L.. Regulation of the BMP Signaling-Responsive Transcriptional Network in the Drosophila Embryo. PLoS Genet. 2016; 12:e1006164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Muller B., Hartmann B., Pyrowolakis G., Affolter M., Basler K.. Conversion of an extracellular Dpp/BMP morphogen gradient into an inverse transcriptional gradient. Cell. 2003; 113:221–233. [DOI] [PubMed] [Google Scholar]
- 38. Vuilleumier R., Springhorn A., Patterson L., Koidl S., Hammerschmidt M., Affolter M., Pyrowolakis G.. Control of Dpp morphogen signalling by a secreted feedback regulator. Nat. Cell Biol. 2010; 12:611–617. [DOI] [PubMed] [Google Scholar]
- 39. Esteves F.F., Springhorn A., Kague E., Taylor E., Pyrowolakis G., Fisher S., Bier E.. BMPs regulate msx gene expression in the dorsal neuroectoderm of Drosophila and vertebrates by distinct mechanisms. PLoS Genet. 2014; 10:e1004625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen D., McKearin D.. Dpp signaling silences bam transcription directly to establish asymmetric divisions of germline stem cells. Curr. Biol. 2003; 13:1786–1791. [DOI] [PubMed] [Google Scholar]
- 41. Stathopoulos A., Levine M.. Localized repressors delineate the neurogenic ectoderm in the early Drosophila embryo. Dev. Biol. 2005; 280:482–493. [DOI] [PubMed] [Google Scholar]
- 42. Crocker J., Erives A.. A Schnurri/Mad/Medea complex attenuates the dorsal-twist gradient readout at vnd. Dev. Biol. 2013; 378:64–72. [DOI] [PubMed] [Google Scholar]
- 43. Stark A., Lin M.F., Kheradpour P., Pedersen J.S., Parts L., Carlson J.W., Crosby M.A., Rasmussen M.D., Roy S., Deoras A.N. et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007; 450:219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kheradpour P., Stark A., Roy S., Kellis M.. Reliable prediction of regulator targets using 12 Drosophila genomes. Genome Res. 2007; 17:1919–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gurdziel K., Lorberbaum D.S., Udager A.M., Song J.Y., Richards N., Parker D.S., Johnson L.A., Allen B.L., Barolo S., Gumucio D.L.. Identification and validation of novel Hedgehog-Responsive enhancers predicted by computational analysis of Ci/Gli binding site density. PLoS One. 2015; 10:e0145225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shlyueva D., Stampfel G., Stark A.. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 2014; 15:272–286. [DOI] [PubMed] [Google Scholar]
- 47. Aerts S., Quan X.J., Claeys A., Sanchez M.N., Tate P., Yan J., Hassan B.A.. Robust target gene discovery through transcriptome perturbations and genome-wide enhancer predictions in drosophila uncovers a regulatory basis for sensory specification. PLoS Biol. 2010; 8:e1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Markstein M., Markstein P., Markstein V., Levine M.S.. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:763–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Celniker S.E., Rubin G.M.. The Drosophila melanogaster genome. Annu. Rev. Genomics Hum. Genet. 2003; 4:89–117. [DOI] [PubMed] [Google Scholar]
- 50. Chintapalli V.R., Wang J., Dow J.A.. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 2007; 39:715–720. [DOI] [PubMed] [Google Scholar]
- 51. mod E.C., Roy S., Ernst J., Kharchenko P.V, Kheradpour P., Negre N., Eaton M.L., Landolin J.M., Bristow C.A., Ma L. et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010; 330:1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Berger C., Harzer H., Burkard T.R., Steinmann J., van der Horst S., Laurenson A.S., Novatchkova M., Reichert H., Knoblich J.A.. FACS purification and transcriptome analysis of drosophila neural stem cells reveals a role for Klumpfuss in self-renewal. Cell Rep. 2012; 2:407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wartlick O., Mumcu P., Kicheva A., Bittig T., Seum C., Julicher F., Gonzalez-Gaitan M.. Dynamics of Dpp signaling and proliferation control. Science. 2011; 331:1154–1159. [DOI] [PubMed] [Google Scholar]
- 54. Tsuneizumi K., Nakayama T., Kamoshida Y., Kornberg T.B., Christian J.L., Tabata T.. Daughters against dpp modulates dpp organizing activity in Drosophila wing development. Nature. 1997; 389:627–631. [DOI] [PubMed] [Google Scholar]
- 55. Haerry T.E., Khalsa O., O’Connor M.B., Wharton K.A.. Synergistic signaling by two BMP ligands through the SAX and TKV receptors controls wing growth and patterning in Drosophila. Development. 1998; 125:3977–3987. [DOI] [PubMed] [Google Scholar]
- 56. Osterwalder T., Yoon K.S., White B.H., Keshishian H.. A conditional tissue-specific transgene expression system using inducible GAL4. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:12596–12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nagarkar-Jaiswal S., Lee P.T., Campbell M.E., Chen K., Anguiano-Zarate S., Gutierrez M.C., Busby T., Lin W.W., He Y., Schulze K.L. et al. A library of MiMICs allows tagging of genes and reversible, spatial and temporal knockdown of proteins in Drosophila. Elife. 2015; 4:e05338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hamburger V., Hamilton H.L.. A series of normal stages in the development of the chick embryo. J. Morphol. 1951; 88:49–92. [PubMed] [Google Scholar]
- 59. Kvon E.Z., Kazmar T., Stampfel G., Yanez-Cuna J.O., Pagani M., Schernhuber K., Dickson B.J., Stark A.. Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature. 2014; 512:91–95. [DOI] [PubMed] [Google Scholar]
- 60. Pfeiffer B.D., Jenett A., Hammonds A.S., Ngo T.T., Misra S., Murphy C., Scully A., Carlson J.W., Wan K.H., Laverty T.R. et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:9715–9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Marmion R.A., Jevtic M., Springhorn A., Pyrowolakis G., Yakoby N.. The Drosophila BMPRII, wishful thinking, is required for eggshell patterning. Dev. Biol. 2013; 375:45–53. [DOI] [PubMed] [Google Scholar]
- 62. Bischof J., Maeda R.K., Hediger M., Karch F., Basler K.. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Marx A., Backes C., Meese E., Lenhof H.P., Keller A.. EDISON-WMW: exact dynamic programing solution of the Wilcoxon-Mann-Whitney test. Genomics Proteomics Bioinforma. 2016; 14:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Uchikawa M., Ishida Y., Takemoto T., Kamachi Y., Kondoh H.. Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev. Cell. 2003; 4:509–519. [DOI] [PubMed] [Google Scholar]
- 65. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L.. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016; 11:1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bray N.L., Pimentel H., Melsted P., Pachter L.. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016; 34:525–527. [DOI] [PubMed] [Google Scholar]
- 68. Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C.. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017; 14:417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Li B., Dewey C.N.. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011; 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. dos Santos G., Schroeder A.J., Goodman J.L., Strelets V.B., Crosby M.A., Thurmond J., Emmert D.B., Gelbart W.M., FlyBase C.. FlyBase: introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res. 2015; 43:D690–D697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pimentel H., Bray N.L., Puente S., Melsted P., Pachter L.. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods. 2017; 14:687–690. [DOI] [PubMed] [Google Scholar]
- 74. Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011; 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wickham H. Ggplot2: Elegant Graphics for Data Analysis. 2009; NY: Springer. [Google Scholar]
- 76. Chen H., Xu Z., Mei C., Yu D., Small S.. A system of repressor gradients spatially organizes the boundaries of Bicoid-dependent target genes. Cell. 2012; 149:618–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. R core team R: A Language and Environment for Statistical Computing. 2017; Vienna: R Found. Stat. Comput; http://www.R-project.org/. [Google Scholar]
- 78. Vacic V., Iakoucheva L.M., Radivojac P.. Two Sample Logo: a graphical representation of the differences between two sets of sequence alignments. Bioinformatics. 2006; 22:1536–1537. [DOI] [PubMed] [Google Scholar]
- 79. Saka Y., Hagemann A.I., Piepenburg O., Smith J.C.. Nuclear accumulation of Smad complexes occurs only after the midblastula transition in Xenopus. Development. 2007; 134:4209–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhang X.E., Cui Z., Wang D.. Sensing of biomolecular interactions using fluorescence complementing systems in living cells. Biosens. Bioelectron. 2016; 76:243–250. [DOI] [PubMed] [Google Scholar]
- 81. Miller K.E., Kim Y., Huh W.K., Park H.O.. Bimolecular fluorescence complementation (BiFC) analysis: advances and recent applications for genome-wide interaction studies. J. Mol. Biol. 2015; 427:2039–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hu C.D., Chinenov Y., Kerppola T.K.. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell. 2002; 9:789–798. [DOI] [PubMed] [Google Scholar]
- 83. Park D., Veenstra J.A., Park J.H., Taghert P.H.. Mapping peptidergic cells in Drosophila: where DIMM fits in. PLoS One. 2008; 3:e1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Landgraf M., Thor S.. Development of Drosophila motoneurons: Specification and morphology. Semin. Cell Dev. Biol. 2006; 17:3–11. [DOI] [PubMed] [Google Scholar]
- 85. Le Dreau G., Garcia-Campmany L., Rabadan M.A., Ferronha T., Tozer S., Briscoe J., Marti E.. Canonical BMP7 activity is required for the generation of discrete neuronal populations in the dorsal spinal cord. Development. 2012; 139:259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Le Dréau G., Martí E.. Dorsal-ventral patterning of the neural tube: a tale of three signals. Dev. Neurobiol. 2012; 72:1471–1481. [DOI] [PubMed] [Google Scholar]
- 87. Karaulanov E., Knöchel W., Niehrs C.. Transcriptional regulation of BMP4 synexpression in transgenic Xenopus. EMBO J. 2004; 23:844–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Osses N., Henriquez J.P.. Bone morphogenetic protein signaling in vertebrate motor neurons and neuromuscular communication. Front. Cell Neurosci. 2014; 8:453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Chou H.J., Lai D.M., Huang C.W., McLennan I.S., Wang H.D., Wang P.Y.. BMP4 is a peripherally-derived factor for motor neurons and attenuates glutamate-induced excitotoxicity in vitro. PLoS One. 2013; 8:e58441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zhong J., Zou H.. BMP signaling in axon regeneration. Curr. Opin. Neurobiol. 2014; 27:127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Xiao L., Michalski N., Kronander E., Gjoni E., Genoud C., Knott G., Schneggenburger R.. BMP signaling specifies the development of a large and fast CNS synapse. Nat. Neurosci. 2013; 16:856–864. [DOI] [PubMed] [Google Scholar]
- 92. Sun M., Thomas M.J., Herder R., Bofenkamp M.L., Selleck S.B., O’Connor M.B.. Presynaptic contributions of chordin to hippocampal plasticity and spatial learning. J. Neurosci. 2007; 27:7740–7750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Rohrer H. The role of bone morphogenetic proteins in sympathetic neuron development. Drug News Perspect. 2003; 16:589–596. [DOI] [PubMed] [Google Scholar]
- 94. Hegarty S.V, O’Keeffe G.W., Sullivan A.M.. BMP-Smad 1/5/8 signalling in the development of the nervous system. Prog. Neurobiol. 2013; 109:28–41. [DOI] [PubMed] [Google Scholar]
- 95. Benveniste R.J., Thor S., Thomas J.B., Taghert P.H.. Cell type-specific regulation of the Drosophila FMRF-NH2 neuropeptide gene by Apterous, a LIM homeodomain transcription factor. Development. 1998; 125:4757–4765. [DOI] [PubMed] [Google Scholar]
- 96. Benveniste R.J., Taghert P.H.. Cell type-specific regulatory sequences control expression of the Drosophila FMRF-NH2 neuropeptide gene. J. Neurobiol. 1999; 38:507–520. [DOI] [PubMed] [Google Scholar]
- 97. Weirauch M.T., Hughes T.R.. Conserved expression without conserved regulatory sequence: the more things change, the more they stay the same. Trends Genet. 2010; 26:66–74. [DOI] [PubMed] [Google Scholar]
- 98. Ramos A.I., Barolo S.. Low-affinity transcription factor binding sites shape morphogen responses and enhancer evolution. Philos. Trans. R Soc. L. B Biol. Sci. 2013; 368:20130018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Crocker J., Noon E.P., Stern D.L.. The soft Touch: Low-Affinity transcription factor binding sites in development and evolution. Curr. Top. Dev. Biol. 2016; 117:455–469. [DOI] [PubMed] [Google Scholar]
- 100. Howard M.L., Davidson E.H.. cis-Regulatory control circuits in development. Dev. Biol. 2004; 271:109–118. [DOI] [PubMed] [Google Scholar]
- 101. Yáñez-Cuna J.O., Kvon E.Z., Stark A.. Deciphering the transcriptional cis-regulatory code. Trends Genet. 2013; 29:11–22. [DOI] [PubMed] [Google Scholar]
- 102. Spitz F., Furlong E.E.M.. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 2012; 13:613–626. [DOI] [PubMed] [Google Scholar]
- 103. Miguel-Aliaga I., Allan D.W., Thor S.. Independent roles of the dachshund and eyes absent genes in BMP signaling, axon pathfinding and neuronal specification. Development. 2004; 131:5837–5848. [DOI] [PubMed] [Google Scholar]
- 104. Allan D.W., Park D., St Pierre S.E., Taghert P.H., Thor S.. Regulators acting in combinatorial codes also act independently in single differentiating neurons. Neuron. 2005; 45:689–700. [DOI] [PubMed] [Google Scholar]
- 105. Blitz I.L., Cho K.W.. Finding partners: how BMPs select their targets. Dev. Dyn. 2009; 238:1321–1331. [DOI] [PubMed] [Google Scholar]
- 106. Ross S., Hill C.S.. How the Smads regulate transcription. Int. J. Biochem. Cell Biol. 2008; 40:383–408. [DOI] [PubMed] [Google Scholar]
- 107. Van Bortle K., Peterson A.J., Takenaka N., O’Connor M.B., Corces V.G.. CTCF-dependent co-localization of canonical Smad signaling factors at architectural protein binding sites in D. melanogaster. Cell Cycle. 2015; 14:2677–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Hodge L.K., Klassen M.P., Han B.X., Yiu G., Hurrell J., Howell A., Rousseau G., Lemaigre F., Tessier-Lavigne M., Wang F.. Retrograde BMP signaling regulates trigeminal sensory neuron identities and the formation of precise face maps. Neuron. 2007; 55:572–586. [DOI] [PubMed] [Google Scholar]
- 109. Patel T.D., Kramer I., Kucera J., Niederkofler V., Jessell T.M., Arber S., Snider W.D.. Peripheral NT3 signaling is required for ETS protein expression and central patterning of proprioceptive sensory afferents. Neuron. 2003; 38:403–416. [DOI] [PubMed] [Google Scholar]
- 110. Haase G., Dessaud E., Garcès A., De Bovis B., Birling M.C., Filippi P., Schmalbruch H., Arber S., DeLapeyrière O.. GDNF acts through PEA3 to regulate cell body positioning and muscle innervation of specific motor neuron pools. Neuron. 2002; 35:893–905. [DOI] [PubMed] [Google Scholar]
- 111. Sweeney S.T., Davis G.W.. Unrestricted synaptic growth in spinster-a late endosomal protein implicated in TGF-beta-mediated synaptic growth regulation. Neuron. 2002; 36:403–416. [DOI] [PubMed] [Google Scholar]
- 112. Conley C.A., Silburn R., Singer M.A., Ralston A., Rohwer-Nutter D., Olson D.J., Gelbart W., Blair S.S.. Crossveinless 2 contains cysteine-rich domains and is required for high levels of BMP-like activity during the formation of the cross veins in Drosophila. Development. 2000; 127:3947–3959. [DOI] [PubMed] [Google Scholar]
- 113. Serpe M., Umulis D., Ralston A., Chen J., Olson D.J., Avanesov A., Othmer H., O’Connor M.B., Blair S.S.. The BMP-binding protein Crossveinless 2 is a short-range, concentration-dependent, biphasic modulator of BMP signaling in Drosophila. Dev. Cell. 2008; 14:940–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Johnson K.G., Tenney A.P., Ghose A., Duckworth A.M., Higashi M.E., Parfitt K., Marcu O., Heslip T.R., Marsh J.L., Schwarz T.L. et al. The HSPGs Syndecan and Dallylike bind the receptor phosphatase LAR and exert distinct effects on synaptic development. Neuron. 2006; 49:517–531. [DOI] [PubMed] [Google Scholar]
- 115. Ozon S., Guichet A., Gavet O., Roth S., Sobel A.. Drosophila stathmin: a microtubule-destabilizing factor involved in nervous system formation. Mol. Biol. Cell. 2002; 13:698–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Graf E.R., Heerssen H.M., Wright C.M., Davis G.W., DiAntonio A.. Stathmin is required for stability of the Drosophila neuromuscular junction. J. Neurosci. 2011; 31:15026–15034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence data from this study have been submitted to the NCBI GEO under accession number GSE106650.