Abstract
Proneural transcription factors (TFs) drive highly efficient differentiation of pluripotent stem cells to lineage‐specific neurons. However, current strategies mainly rely on genome‐integrating viruses. Here, we used synthetic mRNAs coding two proneural TFs (Atoh1 and Ngn2) to differentiate induced pluripotent stem cells (iPSCs) into midbrain dopaminergic (mDA) neurons. mRNAs coding Atoh1 and Ngn2 with defined phosphosite modifications led to higher and more stable protein expression, and induced more efficient neuron conversion, as compared to mRNAs coding wild‐type proteins. Using these two modified mRNAs with morphogens, we established a 5‐day protocol that can rapidly generate mDA neurons with >90% purity from normal and Parkinson's disease iPSCs. After in vitro maturation, these mRNA‐induced mDA (miDA) neurons recapitulate key biochemical and electrophysiological features of primary mDA neurons and can provide high‐content neuron cultures for drug discovery. Proteomic analysis of Atoh1‐binding proteins identified the nonmuscle myosin II (NM‐II) complex as a new binding partner of nuclear Atoh1. The NM‐II complex, commonly known as an ATP‐dependent molecular motor, binds more strongly to phosphosite‐modified Atoh1 than the wild type. Blebbistatin, an NM‐II complex antagonist, and bradykinin, an NM‐II complex agonist, inhibited and promoted, respectively, the transcriptional activity of Atoh1 and the efficiency of miDA neuron generation. These findings established the first mRNA‐driven strategy for efficient iPSC differentiation to mDA neurons. We further identified the NM‐II complex as a positive modulator of Atoh1‐driven neuron differentiation. The methodology described here will facilitate the development of mRNA‐driven differentiation strategies for generating iPSC‐derived progenies widely applicable to disease modeling and cell replacement therapy. stem cells translational medicine 2019;8:112&12
Keywords: Induced pluripotent stem cell, Dopaminergic neuron, Proneural transcription factor, Parkinson's disease, Protein phosphorylation, Myosin, Motor protein
Significance Statement.
Highly efficient induced pluripotent stem cells (iPSCs) differentiation strategies are critical for the application of iPSC technology in regenerative medicine. Traditional methods based on chemical compounds and viruses are relatively slow, variable, or unsafe. In this study, a highly efficient strategy was established based on using synthetic mRNAs coding proneural transcription factors to drive rapid iPSC differentiation to functional and integration‐free mDA neurons. Furthermore, the NM‐II complex was identified as a positive modulator of this neuron induction process. These discoveries will facilitate the applications of iPSC‐derived mDA neurons in Parkinson's disease modeling and therapy, and guide the development of robust methods for generating various lineage‐specific progenies from iPSCs.
Introduction
Pluripotent stem cells (PSCs), including embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs), exhibit unique characteristics, such as indefinite self‐renewal capacity and multilineage differentiation potential. Human PSCs, in particular patient‐derived iPSCs, hold enormous promise for various applications in regenerative medicine, including disease modeling, drug discovery, and cell replacement therapy 1, 2, 3. Broader application of iPSC technology in regenerative medicine relies on highly efficient differentiation strategies for generating lineage‐specific and functional progenies, such as various neuron subtypes.
Traditional protocols for neuron differentiation are based on embryoid body formation, stromal feeder coculture, selective survival conditions, or chemical inhibitors of the SMAD signaling 4, 5, 6, 7, 8. The principle of these methods is to sequentially activate transcription factor (TF) drivers of neural conversion, neuronal lineage specification, and functional maturation. For example, a set of chemical compounds are used to induce TF drivers (e.g., Ngn2, FOXA2, LMX1A, and NURR1) for midbrain dopaminergic (mDA) lineage conversion 9. Current compound‐based iPSC differentiation is relatively slow and variable, as compounds act indirectly by modulating signaling pathways (e.g., the BMP and TGFβ signaling) that induce TF drivers of cell differentiation. Alternatively, direct transgenic expression of TF drivers has been shown to induce more rapid and efficient neuronal conversion in iPSCs. As an example, we previously showed that lentivirus‐mediated expression of the proneural TF Atoh1 potently induces mDA neuron conversion in iPSCs 10. Lentiviral expression of the proneural TF Ngn2 also induces rapid neuronal differentiation in iPSCs 11, 12, 13. However, TF‐based cell differentiation strategies must move beyond genome‐integrating viruses that are cumbersome and associated with safety limitations for clinical translation.
Synthetic mRNAs coding Yamanaka factors have been successfully used to drive efficient cell reprogramming 14, 15. mRNA‐based reprogramming kits are widely used for generating integration‐free iPSCs. In comparison, mRNA‐driven iPSC differentiation is also feasible but more challenging 14, 16, because an optimal iPSC differentiation protocol requires significantly higher efficiency (>80%) than that of cell reprogramming (0.01%–2%). Here, we focus on developing highly efficient strategies for generating mRNA‐induced mDA neurons, hereby referred to as miDA neurons. We previously showed that lentiviral expression of the Atoh1 TF in iPSCs activates downstream neurogenic TFs, including Ngn2 and NeuroD1, and rapidly generates highly pure mDA neurons from normal and Parkinson's disease (PD)‐patient‐derived iPSCs 10. This work intrigued us to develop integration‐free iPSC differentiation strategy based on sequentially delivered synthetic mRNAs coding Atoh1 and likely its downstream targets for rapid mDA differentiation. We further tested mRNAs coding Atoh1 and Ngn2 with defined phosphosite modifications for higher protein expression and more efficient neuronal conversion. Here, we established the first practical strategy for generating miDA neurons with >90% purity and recapitulating the functional properties of primary mDA neurons. Following proteomic analysis further revealed that the nonmuscle myosin II (NM‐II) motor protein complex interacts with Atoh1 in the cell nucleus and promotes miDA neuron generation.
This article provides a novel mRNA‐driven strategy for rapid and efficient iPSC differentiation into lineage‐specific neurons. Our results support that phosphosite modification in TFs and NM‐II complex activation can effectively promote the efficiency of this mRNA‐driven iPSC differentiation process. Therefore, this knowledge will guide the development of various mRNA‐driven strategies for cell identity engineering, such as cell differentiation, reprogramming, and transdifferentiation.
Materials and Methods
Cell Culture
Human iPSC lines were obtained from the Coriell Cell Repositories (Camden, NY, http://ccr.coriell.org). ND1014 and N1 (referred to as iPSC1 and iPSC3, respectively) were derived from normal human skin fibroblasts. ND27760 (referred to as iPSC2) was derived from skin fibroblasts from a PD patient with SNCA triplication. Cell reprogramming was performed using a nonintegrating 4‐factor (SOX2/OCT4/KLF4/MYC) Sendai virus system (CytoTune‐iPS Reprogramming Kit; Thermo Fisher Scientific, Rockville, MD, http://www.thermofisher.com). The pluripotency of these iPSCs has been characterized by immunocytochemistry for pluripotent cell markers (NANOG, OCT4, TRA‐1‐60, and SSEA‐3) and embryoid body formation assay. iPSCs were maintained as feeder‐free cultures in mTESR1 medium (StemCell Technologies, Vancouver, BC, Canada, http://www.stemcell.com) in 5% CO2/95% air condition at 37°C, and were passaged using ReLeSR (StemCell Technologies). Karyotype analysis of G‐banded metaphase chromosomes has been performed to confirm the chromosomal integrity of these iPSCs. All experiments involving human stem cells were performed with the approval of the Johns Hopkins Medicine Institutional Review Boards.
mRNA Synthesis and Transfection
Coding sequences of human Atoh1 and Ngn2 were cloned into a vector containing the T7 promoter and poly(A) tail for in vitro transcription. These vectors were linearized and subjected to mRNA synthesis using the HiScribe T7 ARCA mRNA Kit (New England Biolabs, Ipswich, MA, https://www.neb.com). mRNAs were purified by the MEGAclear Transcription Clean‐Up Kit (Thermo Fisher Scientific). mRNA transfection was performed using the Stem‐In cationic lipid (MTI‐GlobalStem, Gaithersburg, MD, http://www.mti‐globalstem.com). For each well of the 12‐well plate, we incubated 0.25 μg mRNA with 1.5 μl lipid in 50 μl PBS for 15 minutes before adding to cell cultures. All procedures involving recombinant DNA follow the National Institutes of Health guidelines.
iPSC Differentiation
For the 5‐day miDA neuron differentiation protocol, iPSCs were plated at a density of 1.5 × 105 cells per cm2 in the 12‐well plate coated with growth‐factor‐reduced Matrigel (Corning, Corning, NY, https://www.corning.com). iPSCs were transfected with A‐SA mRNA for 3 days followed by 1 day of N‐SA mRNA transfection. Culture medium was changed daily, and gradually shifted from mTeSR1 to N2 (Thermo Fisher Scientific) in 3 days. The medium also contains SHH (100 ng/ml), FGF8b (100 ng/ml), and DAPT (10 μM). Cells at differentiation day 5 were dissociated by Accutase and replated with neuron medium (neurobasal medium with B27 supplement, BDNF[10 ng/ml], GDNF [10 ng/ml], TGFβ‐3 [1 ng/ml], cAMP [0.1 mM], ascorbic acid [0.2 mM], and DAPT [10 μM]) in poly‐d‐lysine/laminin‐coated plates at the density of 1 × 105 cells per cm2. Cells can also be cryopreserved in medium containing 40% neurobasal medium with B27 supplement, 50% fetal bovine serum and 10% DMSO. Neuron culture medium was changed after 48 hours to remove unattached cells followed by half change every 3–4 days during in vitro maturation. Cell proliferation and death analysis use the MTT (Thermo Fisher Scientific) and LDH (Promega, Madison, WI, http://www.promega.com) kit, respectively, following the manufacturer's instruction.
Quantitative Real‐Time Polymerase Chain Reaction (qRT‐PCR)
Total RNA was extracted using the RNeasy Mini kit (Qiagen, Hilden, Germany, http://www1.qiagen.com). Reverse transcription was performed using murine leukemia virus reverse transcriptase (Applied Biosystems, Foster City, CA, http://www.appliedbiosystems.com) and oligo(dT) primer. qRT‐PCR was performed using SYBR Green PCR Master Mix (Applied Biosystems) and the IQ5 RT‐PCR detection system (Bio‐Rad, Hercules, CA, http://www.bio-rad.com). Relative expression of each gene was normalized to the 18S rRNA. Primer sequences are listed in Supporting Information Table S4.
Western Blot
Total cellular proteins were extracted using RIPA buffer (Sigma‐Aldrich, St. Louis, http://www.sigmaaldrich.com) containing protease and phosphatase inhibitors (Calbiochem, San Diego, CA, http://www.emdbiosciences.com). SDS‐polyacrylamide gel electrophoresis was performed with 50 mg of total cellular proteins per lane using 4%–12% gradient Tris‐glycine gels (Lonza, Walkersville, MD, http://www.lonza.com). Western blot was performed using a Quantitative Western Blot System (LI‐COR Biosciences, Lincoln, NE, http://www.licor.com) following the manufacturer's instructions. The primary antibodies are listed in Supporting Information Table S4. Secondary antibodies were labeled with IR dyes, and protein levels were quantified with the Odyssey IR Imaging System (LI‐COR Biosciences).
Immunofluorescence and Cell Counting
Cells were fixed in 4% paraformaldehyde in PBS (pH 7.4) and blocked with 5% normal goat serum and 0.2% Triton X‐100. Primary antibodies (listed in Supporting Information Table S4) were diluted in 5% normal goat serum and incubated with samples overnight at 4°C. Cy3‐ and Alexa Fluor 488‐labeled secondary antibodies were used. Samples were counterstained with 4′,6‐diamidino‐2‐phenylindole (DAPI) and mounted on glass slides using the ProLong antifade kit (Thermo Fisher Scientific). The percentage of marker‐positive cells was determined in samples derived from at least three independent experiments. Images from 10 randomly selected fields were used for counting the number of DAPI‐positive cells expressing a specific marker in the ImageJ software.
Electrophysiological Recordings
Voltage‐clamp recording was performed at 35°C in a chamber perfused with regular artificial cerebrospinal fluid (124 mM NaCl, 2.5 mM KCl, 1.3 mM MgCl2, 2.5 mM CaCl2, 1 mM NaH2PO4, 26.2 mM NaHCO3, 20 mM glucose, pH 7.4, equilibrated with 95% O2 and 5% CO2, ∼310 mosm), which flowed at 3 ml/minute. Patch electrodes were pulled from borosilicate glass and had resistances of 2.0–4.0 MΩ when filled with an intracellular solution (135 mM KMeSO4, 5 mM KCl, 5 mM HEPES, 0.25 mM EGTA‐free acid, 2 mM Mg‐ATP, 0.5 mM GTP, 10 mM phosphocreatine‐tris, pH 7.3, ∼290 mosm).
Neurons were identified using a ×10 objective mounted on an upright microscope with transmitted light, and their neuronal somata were then visualized through a ×40 water immersion objective using IR differential interference contrast optics. The cell somatic recordings were made using an Axopatch 200B amplifier in combination with pClamp 10.7 software (Molecular Devices, Union City, CA, http://www.moleculardevices.com). Neurons were voltage‐clamped at −70 mV. Rseries and Rinput were monitored using a 2.5‐mV 100‐ms depolarizing voltage step in each recording sweep. Current traces were filtered at 5 kHz, digitized at 10 kHz using a Digidata 1440 interface, and stored for off‐line analysis. The resting membrane potential and the action potential were monitored for more than 5 minutes before drug applications. Tetraethylammonium (TEA) and ZD 7288 (Sigma‐Aldrich) were added to the artificial cerebrospinal fluid, to block K+ or I h currents, respectively.
High‐Performance Liquid Chromatography (HPLC)
Neuron culture medium was replaced by Hanks' balanced saline solution buffer with the addition of 56 mM KCl and incubated for 15 minutes at 37°C. The medium was collected and centrifuged to clear cell debris. Neuron pellet was also collected. Samples were immediately frozen in liquid nitrogen and stored at −80°C. For HPLC analysis, samples were thawed and concentrated using a vacuum (Savant SDP 121P, Thermo Fisher Scientific) connected with refrigerated vapor trap (Savant RVT 5105, Thermo Fisher Scientific), and the freeze‐dried samples were resuspended in 10 mM perchloric acid. Monoamines were analyzed by HPLC electrochemical detection by dual channel Coulochem III electrochemical detector (model 5300; ESA Inc., Chelmsford, MA, http://www.esainc.com), and monoamines were separated by using a reverse phase C18 column (3‐mm 3 150‐mm C‐18 RP‐column; Acclaim Polar Advantage II; Thermo Fisher Scientific) with a flow rate of 0.600 ml/minute. Monoamine concentrations were quantified by comparison of the area under the curve to known standard dilutions.
6‐Hydroxydopamine (6‐OHDA) Treatment and Neurite Tracing in miDA Neurons
miDA neurons (3 × 103 per well) were plated in 384‐well plate coated with poly‐d‐lysine and laminin. The calcein AM assay is based on the conversion of the cell permeant nonfluorescent calcein AM dye to the fluorescent calcein dye by intracellular esterase activity in live cells. 6‐OHDA treatment was performed using 6‐OHDA freshly prepared in vehicle solution (0.15% ascorbic acid in H2O). Control cells were treated with vehicle solution alone. After 24 hours, live cell imaging was performed by staining live cells with calcein‐AM (1 μM, Thermo Fisher Scientific) and taking images using a confocal microscope (Olympus). Neurite length was quantified using the high content analysis software (HCA‐Vision V2.2.0. CSIRO). Ngn2‐induced neurons were generated by differentiating iPSCs with three daily transfection of N‐SA mRNAs.
Immunoprecipitation (IP) and Mass Spectrometry
Twenty‐four hours after mRNA transfection, nuclear proteins of iPSCs were extracted using the NE‐PER Nuclear and Cytoplasmic Extraction Kit (Thermo Fisher Scientific). FLAG IP was performed using anti‐FLAG M2 antibody (Sigma‐Aldrich) and Protein G Dynabeads (Thermo Fisher Scientific) following the manufacturer's protocol. Binding proteins were eluted by elution buffer (8 M urea, 1 M NaHCO3 in water). For mass spectrometry analysis, protein samples were denatured in 8 M urea/1 M NH4HCO3 buffer, reduced by 10 mM TCEP at 37°C for 1 hour and alkylated by 15 mM iodoacetamide at room temperature in the dark for 30 minutes. The solutions were diluted fivefold with deionized water. Then, sequencing grade trypsin (Promega; protein: enzyme, 50:1, w/w) was added to the samples and incubated at 37°C overnight with shaking. Samples were centrifuged at 13,000g for 10 minutes to remove any particulate matter and purified by a C18 solid‐phase extraction. Peptides were eluted from the C18 column in 60% ACN/0.1% TFA and the peptide concentrations were measured by the NanoDrop microvolume spectrophotometer (Thermo Fisher Scientific). The peptides were resuspended in 0.2% FA solution, and 1 μg in 6 μl solution were injected for each LC–MS/MS analysis using Velos Orbitrap mass spectrometer with a 120‐minute online LC separation.
The raw data were searched against UniProt human protein database (reviewed, downloaded on 31 August 2016) by Proteome Discoverer (Ver. 1.4) with the following parameters: two missed cleavages were allowed for trypsin digestion, carbamidomethyl (C) was set as a fixed modification, and oxidation (M) and acetyl (protein N‐terminal) were set as variable modifications. We normalized the intensity of each identified proteins to the intensity of their A‐WT or A‐SA proteins, respectively, and further calculated the A‐SA/A‐WT ratio of normalized intensity.
Data Analysis and Statistics
All results reported here represent at least three independent replications. Statistical analysis was performed using Prism software (GraphPad, San Diego, CA, http://www.graphpad.com). Post hoc tests included the Student's t test and the Tukey multiple comparison tests as appropriate. For neurophysiological recordings, the recorded data were first visualized with Clampfit 9.2 and exported to MATLAB (MathWorks, Natick, MA, http://www.mathworks.com) for further analysis and plotting. The recording traces are visualized with Igor Pro 6.0 (WaveMetrics, Portland, OR, http://www.wavemetrics.com). All data are represented as mean ± SEM.
Results
Synthetic mRNAs Coding Atoh1 and Ngn2 with Phosphosite Modification Drive Highly Efficient Neuronal Conversion of iPSCs
We delivered synthetic mRNAs to express Atoh1 followed by its downstream target Ngn2 to recapitulate the sequential induction of proneural TFs during neuron development. Two vectors were constructed for T7‐promoter‐driven in vitro transcription to synthesize mRNAs coding wide‐type Atoh1 and Ngn2 (see details in Materials and Methods). Meanwhile, we also synthesized Atoh1 and Ngn2 gene with defined serine‐to‐alanine modifications (Fig. 1A). Blocking the phosphorylation of these serine sites has been shown to stabilize these TF proteins and/or enhance their transcriptional activity 17, 18.
Figure 1.
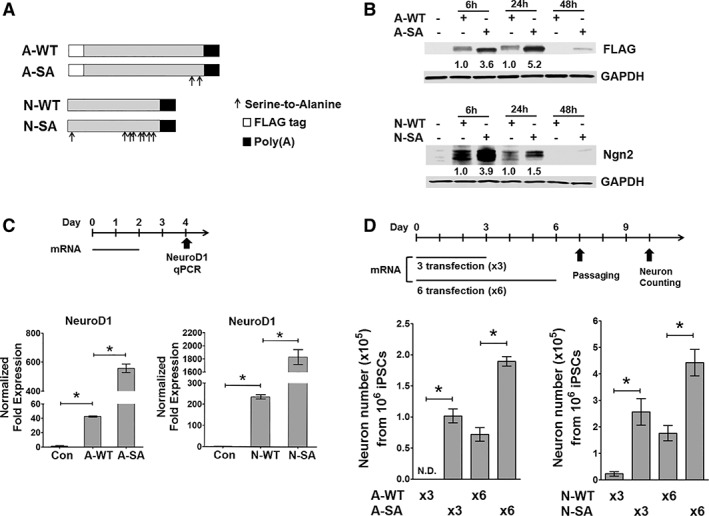
Synthetic mRNAs coding Atoh1 and Ngn2 with phosphosite modification show enhanced protein expression and neuronal induction capability. (A): Diagram of mRNAs coding wild‐type and phosphosite‐modified Atoh1 (A‐WT and A‐SA) and Ngn2 (N‐WT and N‐SA). Arrows: serine‐to‐alanine mutations; Atoh1: at 331 and 342 amino acids; Ngn2: at 24,193, 207, 209, 219, 232, 239, and 242 amino acids. (B): Induced pluripotent stem cells (iPSC1) received a single transfection of mRNAs as indicated, and total proteins were harvested at indicated time points for FLAG (Atoh1) and Ngn2 immunoblotting. Protein fold expression normalized to glyceral‐dehyde‐3‐phosphate dehydrogenase are shown below each lane (A‐WT and N‐WT samples = 1.0). (C): iPSC1 cells received two daily mRNA transfection. Total RNA was isolated for Ngn2 and NeuroD1 quantitative real‐time polymerase chain reaction (qRT‐PCR) at day 2 post transfection (top panel). A‐SA and N‐SA more potently induce their downstream targets than their wild‐type forms (bottom panel, n = 3). (D): iPSC1 cells received three (×3) or six (×6) daily mRNA transfection. Neurons after cell replating were counted at day 10 of differentiation (top panel, n = 6). A‐SA and N‐SA induce more neurons than their wild‐type forms (bottom panel, n = 6). Data represents mean ± SEM; *p < .01.
Before testing these synthetic mRNAs, we first optimized two elements essential for efficient mRNA delivery in iPSCs. First, we identified the mRNA‐In Stem cationic lipids as a highly efficient mRNA transfection reagent for iPSCs. A single transfection of both GFP and Atoh1 mRNAs in iPSCs achieved more than 80% transfection efficiency (Supporting Information Fig. S1A, S1B). Second, we tested the possibility of cell toxicity induced by single‐strand mRNAs (ss‐mRNAs) that would decrease the efficiency of cell reprogramming 14. Cell toxicity induced by ss‐mRNAs is partially mediated by the type I interferon (IFN) response and can be minimized by using mRNAs synthesized with pseudouridine‐ and 5‐methylcytidine‐modified nucleotides 14. We asked if these modified nucleotides are required in mRNA‐induced iPSC differentiation. iPSCs were transfected with GFP and Atoh1 mRNAs synthesized with or without these modified nucleotides, respectively. IFN‐signaling markers at 24 hours after transfection were higher in iPSCs receiving Atoh1 mRNAs synthesized without modified nucleotides as compared to cells receiving GFP mRNAs containing modified nucleotides (Supporting Information Fig. S1D). However, this IFN response was transient as evidenced by its significant downregulation 72 hours after mRNA transfection. More importantly, Atoh1 mRNAs did not increase death of transfected iPSCs compared to both GFP‐mRNA‐transfected and untransfected cells (Supporting Information Fig. S1C). These results supported out strategy using Atoh1 and Ngn2 mRNAs without modified nucleotides to drive the neuronal conversion of iPSCs.
Based on our optimized strategy for mRNA synthesis and transfection, we tested mRNAs coding Atoh1 and Ngn2 in wild‐type and phosphosite‐modified forms (Fig. 1A) to determine which more potently induces protein expression and neuronal conversion. mRNA coding Atoh1 with S331A and S342A phosphosite modifications (referred to as A‐SA) resulted in 2.6‐fold higher protein expression than wild‐type Atoh1 (referred to as A‐WT) as quantified 6 hours post transfection (Fig. 1B). Similarly, mRNA coding Ngn2 with eight serine‐to‐alanine phosphosite mutations (referred to as N‐SA) generated 2.9‐fold higher protein expression than wild‐type Ngn2 (referred to as N‐WT) (Fig. 1B). We asked if A‐SA and N‐SA mRNAs more potently induce their validated downstream targets, such as Ngn2 for Atoh1 and NeuroD1 for Ngn2 10, 17, 19. We transfected iPSCs with either Atoh1 or Ngn2 mRNAs once per day for 2 days, and measured Ngn2 and NeuroD1 level, respectively, at 2 days after the last dose (Fig. 1C, top panel). A‐SA and N‐SA mRNAs induce 3.6‐ and 8‐fold higher Ngn2 or NeuroD1 expression, respectively, as compared to wild‐type mRNAs (Fig. 1C, bottom panel). Next, the strategy shown in Figure 1D (top panel) was followed to compare neuron induction efficiency after three or six daily transfections of Atoh1 or Ngn2 mRNAs. Differentiated cells were replated onto surfaces coated for neuron culture (Supporting Information Fig. S2) for neuron number quantification (Fig. 1D, bottom panel). Neuron numbers from three transfections of A‐WT or N‐WT mRNA were very low. In comparison, three transfections of either A‐SA or N‐SA mRNAs induced efficient neuronal conversion. Six transfections of either A‐SA or N‐SA mRNAs also induced >100% more neurons, as compared to six transfections of A‐WT or N‐WT, respectively. Thus, we conclude that A‐SA and N‐SA mRNAs are better drivers of iPSC neuronal conversion than their respective wild‐type forms.
iPSC Differentiation into mRNA‐Induced mDA (miDA) Neurons
We tested various mRNA transfection schemes using A‐SA and N‐SA mRNAs, and established a 5‐day protocol (outlined in Fig. 2A and also see details in Materials and Methods) including three daily transfections of A‐SA followed by one N‐SA transfection. Two morphogens (SHH and FGF8b) and the Notch signaling inhibitor DAPT were used to enhance mDA lineage conversion 9, 20. iPSCs were induced by A‐SA and N‐SA to form neuronal precursor cells (NPCs) over 5 days (Fig. 2B and Supporting Information Fig. S4, TUJ1+/DAPI+: >90%), and then passaged onto surfaces coated for neuron culture. Three days after cell passaging, mRNA‐induced neurons adhered and formed neuronal processes (Fig. 2B and Supporting Information Fig. S4, TUJ1+/DAPI+: >90%). In contrast, iPSCs receiving GFP mRNAs failed to attach and survive in neuron culture condition (data not shown). We used this protocol to generate miDA neurons from normal iPSC line (iPSC1 and iPSC3) and PD iPSC line (iPSC2) (Fig. 2C).
Figure 2.
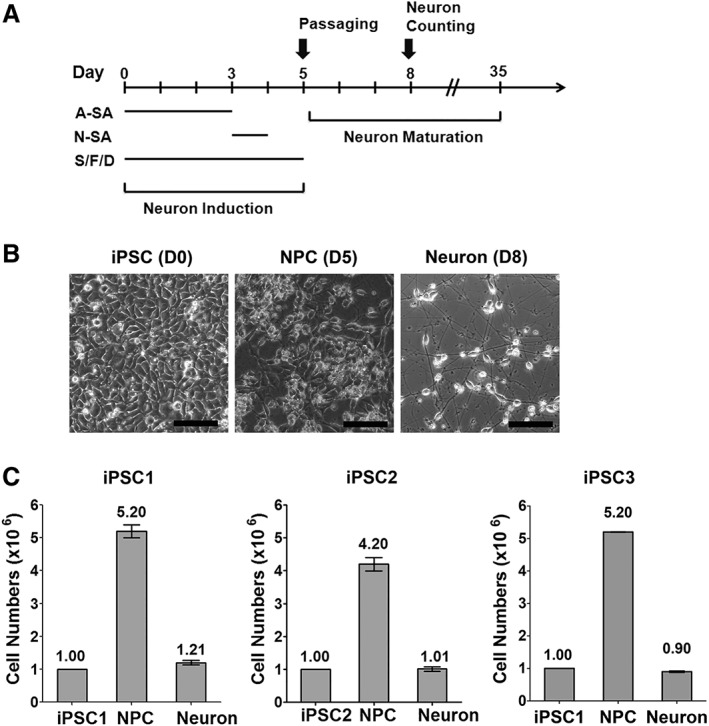
A highly efficient strategy for generating mRNA‐induced midbrain dopaminergic (miDA) neurons from Induced pluripotent stem cells (iPSCs). (A): Diagram of the differentiation protocol (S/F/D: SHH, FGF8b, and DAPT). (B): Brightfield microscopic images show iPSC1 cells at day 0, 5, and 8 of differentiation (scale bar: 100 μm). (C): Three iPSC lines were differentiated following the protocol shown in (A). Cells were counted to calculate the number of neuronal precursor cells (NPCs) and neurons at day 5 and 8 of differentiation, respectively. Data represents mean ± SEM (n = 6).
To examine the need for N‐SA transfection, we compared the efficiency of miDA neuron generation in response to four A‐SA transfections or three A‐SA transfections followed by one N‐SA transfection (Supporting Information Fig. S3A). Including the N‐SA transfection significantly increased NeuroD1 expression and doubled miDA neuron yield without significantly altering the expression of mDA lineage markers LMX1A, NURR1 and TH (Supporting Information Fig. S3B, S3C). This result suggests that the sequential delivery of mRNAs coding different proneural TFs may more closely recapitulate TF activation during neuron development, allowing more efficient neuronal conversion from iPSCs. In addition, we found that iPSCs receiving four doses of A‐SA mRNAs showed significantly higher induction of mDA lineage markers (FOXA2 and LMX1A) than cells receiving four doses of N‐SA mRNAs (Supporting Information Fig. S3D, S3E), indicating that N‐SA alone is not capable to drive efficient DA lineage conversion. This result is consistent with the previous publication showing that Ngn2 induces iPSC differentiation into excitatory cortical neurons 13.
Expression of mDA Lineage Markers in miDA Neurons
We characterized the expression of pan‐neuronal and mDA lineage markers during various stages of miDA neuron induction. TUJ1+ NPCs at day 5 of differentiation coexpress mDA lineage marker FOXA2 and LMX1A (Fig. 3A, 3B, FOXA2+/TUJ1+: 81.3%, LMX1A+/TUJ1+: 78.6%), and these NPCs also show FOXA2 and LMX1A co‐expression (Supporting Information Fig. S5, FOXA2+/LMX1A+: 62.8% and 77.2% in iPSC1 and iPSC2 cells, respectively). TUJ1+ miDA neurons derived from normal and PD iPSCs express mDA marker TH and GIRK2 (Fig. 3C; TH+/TUJ1+: >97%; GIRK2+/TUJ1+: >93%). miDA neurons also co‐express mDA marker NURR1 and TH (Fig. 3D). After in vitro maturation for 45 days, miDA neurons express the dopamine transporter and show robust expression of synaptic vesicle protein synapsin along TUJ1+ nerve fibers (Fig. 3E). These results support that miDA neurons express a panel of markers shared with mDA neurons located in substantia nigra pars compacta (SNpc).
Figure 3.
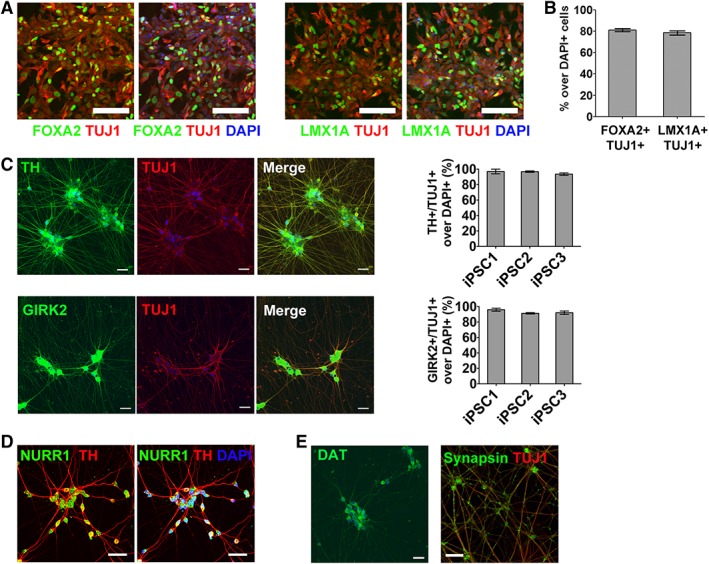
mRNA‐induced midbrain dopaminergic (miDA) neurons express midbrain DA neuron markers. (A, B): FOXA2+/TUJ1+ and LMX1A+/TUJ1+ cells were immunostained and quantified in neuronal precursor cells (NPCs) from iPSC1 cells (day 5 of differentiation, scale bar: 100 μm). (C): TH+/TUJ1+ and GIRK2+/TUJ1+ cells were immunostained and quantified in miDA neurons from iPSC1 and iPSC2 cells (day 20 of differentiation, scale bar: 20 μm). (D, E): miDA neurons after in‐vitro maturation for 45 days were immunostained for midbrain DA neuron markers (NURR1, TH, and dopamine transporter [DAT]) and mature neuron marker synapsin (scale bar: 20 μm). Cell nuclei were counterstained with DAPI. Data represents mean ± SEM (n = 6).
Functional Characterization of miDA Neurons
Primary mDA neurons are well‐known as pacemaker neurons that discharge spontaneously at a rate of 1–10 Hz with an average rate of 4.5 Hz, which functional property has also been reported in iPSC‐derived mDA neurons 10, 21, 22, 23, 24, 25. Here, we performed patch‐clamp recording in miDA neurons (n = 46) during differentiation day 36–49 (Fig. 4A). Fifty‐nine percent of these neurons being tested showed spontaneous spiking activity with a mean frequency of 5.7 Hz (Fig. 4B, 4C, n = 27). Next, we tested the effect of TEA, a K+ current blocker, on the spontaneous activity of miDA neurons. TEA depolarized the resting membrane potentials from −50 to −40 mV, and reversibly blocked the spontaneous spiking of miDA neurons (Fig. 4D), indicating that the delayed rectifier potassium channels as reported previously in mDA neurons 26, 27 were also functionally expressed in miDA neurons. The hyperpolarization‐activated cation channels are critical for the spontaneous activity of DA neurons 27, 28, 29. ZD 7288, a hyperpolarization‐activated current (I h) blocker, significantly attenuated I h in miDA neurons (Fig. 4E), which is consistent with its effects as previously shown in primary and iPSC‐derived mDA neurons 27, 28, 29.
Figure 4.
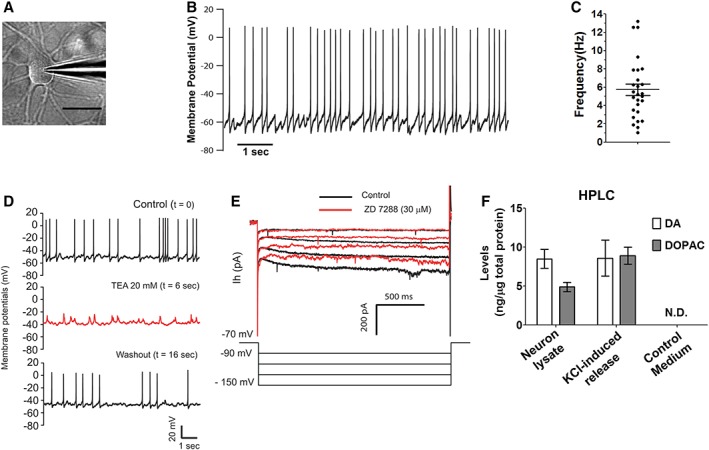
Functional characterization of mRNA‐induced midbrain dopaminergic (miDA) neurons. (A): Differential interference contrast optics image of a patched miDA neuron (scale bar: 20 μm). (B, C): miDA neurons derived from iPSC1 cells showed spontaneous spiking activity. This cell has a resting membrane potential of −60 mV (B). The spontaneous spiking frequencies from 27 miDA neurons were plotted with mean ± SEM marked inside (C). (D): Spontaneous spiking of miDA neurons was blocked by tetraethylammonium (TEA) and returned when TEA was washed away (n = 6). (E): Hyperpolaring the membrane potential of miDA neurons from −90 to −150 mV produced an outward cation current, which was blocked by ZD 7288 (30 μM, n = 4). (F): High‐performance liquid chromatography (HPLC) quantification of DA and its metabolite 3,4‐dihydroxyphenylacetic acid (DOPAC) released from miDA neurons (differentiation day 50) in response to KCl‐evoked depolarization for 15 minutes. Data represents mean ± SEM (n = 2, N.D.: not detected).
HPLC analysis detected the release of DA and the DA metabolite 3,4‐dihydroxyphenylacetic acid (DOPAC) evoked by KCl depolarization in miDA neurons at differentiation day 45 (Fig. 4D). DA and DOPAC were also detected in miDA neuron lysates without depolarization treatment, but not in plain culture medium as control. Overall, these results support that miDA neurons exhibit the key functional features of primary mDA neurons.
6‐OHDA Induces Neurotoxicity in High‐Content Cultures of miDA Neurons
We evaluated the potential to use miDA neurons in a high‐throughput neurotoxicity assay. miDA NPCs at day 5 of differentiation were plated in 384‐well plates precoated for neuron culture (Fig. 5A). These miDA neurons remained viable for more than 15 days and developed robust neurite outgrowth as detected by live cell imaging using the Calcein AM dye (Fig. 5A, 5B). Computer‐aided neurite tracing and quantification of Calcein‐AM‐stained neurons showed greater than eightfold increase of neurite length during days 3 and 5 after cell replating, and neurite length stabilized after day 5 post cell replating (Fig. 5C).
Figure 5.
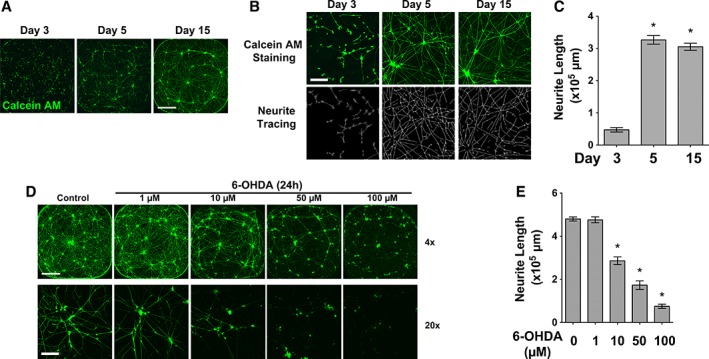
6‐Hydroxydopamine (6‐OHDA) induces neurotoxicity in mRNA‐induced midbrain dopaminergic (miDA) neurons. (A): miDA neurons derived from iPSC1 cells were grown in 384‐well plates and stained with Calcein AM after 3, 5, and 15 days of in vitro maturation (scale bar: 1000 μm). (B, C): Neurite tracing (B) and quantification (C, n = 6) shows increase of neurite length in miDA neurons during in vitro maturation (scale bar: 100 μm). (D, E): miDA neurons after 5 days of maturation received 6‐OHDA or mock treatment for 24 hours. Calcein‐AM‐stained neurons were subjected to neurite length quantification (scale bar: 1,000 μm [×4] and 100 μm [×20]). Data represents mean ± SEM. *p < .01.
We tested the response of miDA neurons to 6‐OHDA, a neurotoxin widely used to induce neurotoxicity both in vivo and in vitro to model mDA neuron loss in PD pathogenesis 30. miDA neurons cultured in 384‐well plates for 5 days received 6‐OHDA treatment (1, 10, 50, and 100 μM for 24 hours). 6‐OHDA at ≥10 μM induced morphological signs of neuron death, including cell body condensation and neurite fragmentation (Fig. 5D). Using image‐based high‐content analysis, we quantified neurite length in 6‐OHDA‐treated and control miDA neurons (Fig. 5E). 6‐OHDA at ≥10 μM significantly decreased neurite length as compared to mock‐treated neurons. Ngn2 has been shown to differentiate human iPSCs into excitatory cortical neurons 13. Using Ngn2‐induced neurons from iPSCs as the non‐mDA control, we showed that 6‐OHDA at 10 μM did not significantly inhibit neurite length, and 6‐OHDA at 50 μM also show less inhibitory effect on neurite length of Ngn2‐induced neurons when compared to that in miDA neurons (Supporting Information Fig. S6, percentage of inhibition at 50 μM: 36.1% vs. 65.5%, p < .01).
The NM‐II Complex Binds to and Promotes the Function of Atoh1 in Driving miDA Neuron Differentiation
To better understand the mechanisms underlying miDA neuron differentiation and identify potential modulators that promote this differentiation, we used proteomic analysis to profile Atoh1‐binding proteins in cytoplasm and nucleus. Following the strategy shown in Figure 6A, FLAG IP was used to pull down FLAG‐tagged Atoh1 and its associated proteins from nuclear and cytoplasmic fractions isolated from iPSCs following one transfection of either A‐WT or A‐SA mRNAs (Fig. 6B). Mass spectrometry analysis identified 253 Atoh1‐binding proteins, of which 114 specifically associated with only cytoplasmic Atoh1 and 54 associated with only nuclear Atoh1. Differential binding to cytoplasmic and nuclear A‐SA and A‐WT was further evaluated. The binding intensity of each Atoh1‐binding protein (Supporting Information Table S1) was first normalized to the intensity of A‐WT or A‐SA proteins, to minimize the variation from the levels of A‐WT and A‐SA expression. We further calculated the A‐SA/A‐WT ratio of normalized intensity to generate four lists of Atoh1‐binding proteins that binds to either A‐WT or A‐SA with higher affinity (referred to as A‐WThigh (ratio ≤ 0.6) or A‐SAhigh (ratio ≥ 1.4), respectively), in either nuclear or cytoplasmic protein fractions (Supporting Information Table S2). Previously reported Atoh1‐binding proteins were identified (Supporting Information Tables S1 and S2). For example, HUWE1, an E3 ubiquitin ligase, was ~100‐fold less associated with A‐SA compared to A‐WT. HUWE1 was found to mediate Atoh1 degradation in mouse cerebellum cells, a process dependent on Atoh1 phosphorylation at Serine 328 and 339 18. These phosphosites are conserved to Serine 331 and 342 sites in human Atoh1 18, which are mutated in A‐SA. Our results showing reduced HUWE1 binding to A‐SA is consistent with the enhanced protein stability and elevated proneural activity of A‐SA as shown in Figure 1.
Figure 6.
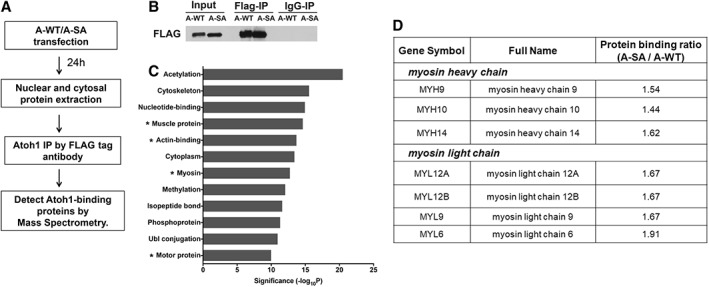
Proteomic analysis identified proteins differentially binding to A‐WT and A‐SA. (A): Scheme of the proteomic analysis using iPSC1 cells transfected by A‐WT or A‐SA mRNAs. (B): Western blotting shows the immunoprecipitation (IP) of A‐WT and A‐SA proteins by the FLAG tag antibody but not mouse IgG. (C): Nuclear A‐SAhigh proteins was analyzed in the DAVID Bioinformatics Database to identify enriched pathways (ranked by −log10[p‐value] calculated by the Fisher's exact test; *pathways containing the nonmuscle myosin II [NM‐II] complex components). (D): The A‐SA/A‐WT binding ratio of the NM‐II complex components identified in the list of nuclear A‐SAhigh proteins.
Four Atoh1‐binding protein lists (cytoplasmic A‐WThigh or A‐SAhigh, nuclear A‐WThigh or A‐SAhigh; Supporting Information Table S2) were subjected to pathway enrichment analysis in the DAVID Bioinformatics Database (Supporting Information Table S3). We focused on signaling pathways enriched in the nuclear A‐SAhigh list to identify positive modulators of Atoh1 transcriptional activity that may drive more efficient miDA neuron conversion. The nuclear A‐SAhigh list was found to contain multiple actin‐binding motor proteins, in particular seven members of the non‐muscle myosin II (NM‐II) complex (Fig. 6C, 6D), indicating a potential role of the NM‐II complex in regulating Atoh1‐driven miDA neuron differentiation.
Based on our working model (Fig. 7A), we used IP and immunoblot analysis to validate two candidate Atoh1‐binding proteins, the NM‐II complex members MYH9 and MYH10. MYH9 and MYH10 were consistently more highly associated with A‐SA than A‐WT, and this protein interaction was only detected in the nuclear fraction (Fig. 7B). The NM‐II complex agonist bradykinin (BK) and antagonist blebbistatin (Ble) were used to determine if the efficiency of miDA neuron generation was modulated by NM‐II activity. BK has been shown to activate myosin light chain by diphosphorylation, and increase the ATPase activity of myosin 31. BK treatment increased the yield of miDA neurons by 45% (Fig. 7C), indicating that the NM‐II complex is a positive regulator of A‐SA activity. In contrast, Ble is a classic NM‐II inhibitor that functions by preventing the formation of actin‐myosin complexes 32. Ble treatment reduced the yield of miDA neurons by 80%, and also attenuated the effect of BK in promoting miDA neuron conversion (Fig. 7C). We further monitored the transcriptional activity of Atoh1 by measuring the induction of Atoh1 target gene Ngn2, in response to A‐SA mRNA transfection along with BK and/or Ble treatment (Fig. 7D). BK significantly promoted Ngn2 induction in response to A‐SA mRNAs. Ble treatment attenuated this Ngn2 induction and the promoting effect of BK on Ngn2 induction. These results suggest that these two NM‐II modulating compounds may alter mDA neuron generation through affecting the transcriptional activity of A‐SA. Cell viability in A‐SA‐transfected iPSCs was not altered by BK or Ble treatment, as determined by the MTT and LDH assay (Supporting Information Fig. S7), indicating that BK and Ble may not modulate Atoh1‐driven neuronal conversion through mechanisms involved in cell death or proliferation. We also found that the purity of TH+/TUJ1+ miDA neurons was not changed by BK or Ble (data not shown), suggesting that these NM‐II modulators regulate the proneural activity of A‐SA but not mDA lineage specification. The nuclear levels of A‐SA were also not affected by BK or Ble (data not shown), indicating that A‐SA expression or nuclear translocation is not changed by NM‐II modulators.
Figure 7.
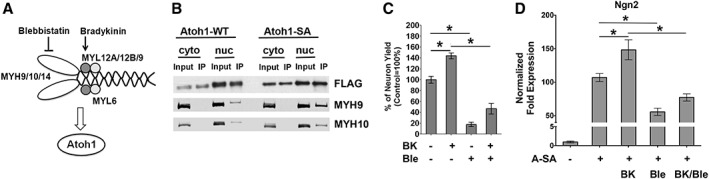
The NM‐II complex binds to Atoh1 and promotes Atoh1‐driven mRNA‐induced midbrain dopaminergic (miDA) neuron generation. (A): Diagram of the NM‐II complex and our working model. The heavy and light chain components of the NM‐II complex as identified in the nuclear A‐SAhigh protein list were marked. The NM‐II agonist bradykinin (BK) and antagonist blebbistatin (Ble) target the light and heavy chain of the NM‐II complex, respectively. (B): iPSC1 cells with A‐WT or A‐SA mRNA transfection for 24 hours were subjected to FLAG immunoprecipitation (IP). IP samples were immunoblotted for components of the NM‐II complex (cyto, cytoplasm; nuc, nuclear). (C): BK and Ble were used along with A‐SA mRNAs during miDA neuron differentiation. Neurons were counted at day 8 of differentiation to calculate the neuron yield from 106 iPSC1 cells and normalized to control condition without BK or Ble treatment. (D): BK and Ble were tested with two daily transfection of A‐SA mRNAs in iPSC1 cells. At 24 hours after transfection, Ngn2 expression was quantified by quantitative real‐time polymerase chain reaction (qRT‐PCR). Data represents mean ± SEM. *p < .01.
Discussion
Synthetic mRNAs are practical vehicles for expressing TF drivers to change cell identity, and this strategy has been widely used in cell reprogramming 14. In contrast, mRNA‐driven iPSC differentiation still requires optimization due to the more stringent efficiency requirement of iPSC differentiation (>80%) as compared with cell reprogramming (<2%). Here, we report a highly efficient strategy for differentiating iPSCs into lineage‐specific neurons. We also provide protocol optimization strategies, including TF phosphosite modification, the sequential use of different TFs, and using the NM‐II agonist BK, to significantly enhance the efficiency of mRNA‐driven neuron generation from iPSCs.
A major discovery of this article is that phosphosite modification of the proneural TF Atoh1 and Ngn2 effectively enhances their protein expression and proneural activity. Traditional differentiation methods based on chemical compounds can only induce endogenous TFs without sequence modification. In comparison, mRNA‐based TF delivery is more flexible, allowing the expression of TFs with modified sequences to promote their expression and transcriptional activity. Phosphosites in various TFs, including Atoh1 18 and Ngn2 17, are essential for their protein stability and proneural activity. Our results support that phosphosite modification is critical for establishing a practical strategy for mRNA‐induced iPSC differentiation. As an example, three transfections of A‐SA mRNA successfully induce neuronal conversion in iPSCs, compared to the failure of neuron induction after the same dose of A‐WT mRNA. The result from Ngn2 mRNA also supports this conclusion. Recently, Goparaju et al. reported a novel mRNA‐based strategy using the cotransfection of five TFs every 8 hours to generate iPSC‐derived motor neurons 16. It is possible that using the same TFs with phosphosite modification may make this protocol more efficient by reducing mRNA amount and/or transfection intervals. This phosphosite modification strategy will likely be applicable to various cell reprogramming and differentiation strategies driven by TF mRNAs. For example, OLIG2 is a potent TF driver of motor neuron differentiation 33. Defined serine sites in OLIG2 are essential for its function in driving motor neuron conversion and maturation 34. Thus, mRNA coding phosphosite‐modified OLIG2 may be used in efficient mRNA‐driven motor neuron production from iPSCs. Phosphosites in the Yamanaka factors (e.g., SOX2) have also been reported to regulate their protein stability, complex formation, and ability in maintaining pluripotency 35, 36. This knowledge will likely allow us to optimize current mRNA‐driven cell reprogramming system.
We also showed that sequentially using Atoh1 and Ngn2 mRNAs is more efficient than using Atoh1 alone, likely due to a more precise control of the transition from pluripotency to neuronal fate. The advantage of mRNA delivery is the ability to specifically and transiently activate defined TF drivers, thus allowing optimal iPSC differentiation strategies that better recapitulate the sequential activation of multiple TF drivers during normal development. In comparison, it is still difficult to precisely control TF expression mediated by viral vectors. Although it is possible to use Tet‐on or Tet‐off vectors for this purpose, this system is more cumbersome and less flexible if multiple TFs are used simultaneously and/or sequentially.
We established a rapid 5‐day protocol for generating miDA neurons with >90% TH+/TUJ1+ purity. Functional assays, including DA release and electrophysiological recording, support the functional similarity between miDA neurons and primary mDA neurons located in the SNpc (A9 DA neurons). We also successfully generated miDA neurons from PD iPSCs with α‐synuclein amplification, thus providing a disease modeling system for mechanistic studies and drug discovery. Large‐scale production of synthetic mRNAs is feasible and cost‐effective through in vitro transcription, thus supporting the scalability of miDA neuron production. We also showed that nucleotides with pseudouridine and 5‐methylcytidine modification are not critical for miDA neuron production, further reducing the cost of this method. We have tested the cryopreservation of miDA neurons and achieved >50% recovery rate (data not shown), supporting the feasibility of banking miDA neurons. No genome‐integration viruses are used in miDA neuron generation, supporting that miDA neurons are applicable to cell replacement therapy. Kikuchi et al. recently reported a group of 11 signature genes that correlate with better therapeutic efficacy of iPSC‐derived mDA neurons transplanted in primate PD models 37. It is possible that TF drivers inducing these signature genes will be potential candidates for mRNA delivery in combination with A‐SA and N‐SA mRNAs. Such combinatorial approaches may produce more potent miDA neurons for cell replacement therapy for PD.
Optimal miDA neuron differentiation relies on mechanic insights of modulators that regulate protein stability and/or proneural activity of Atoh1. Our proteomic analysis established the first profiles of Atoh1‐binding proteins in the nucleus and cytoplasm of iPSCs during differentiation. A‐SA and A‐WT show differential binding affinity to a set of binding partners. The E3 ligase HUWE1 has been identified as a positive modulator of Atoh1 degradation 18. The lower binding affinity between A‐SA and HUWE1, as compared to A‐WT, reveals a potential mechanism leading to the elevated protein level of A‐SA. Here, we focus on nuclear Atoh1‐binding proteins that may regulate the transcriptional activity of Atoh1. We showed for the first time that the NM‐II complex is a functional Atoh1‐binding partner and the NM‐II agonist BK promotes miDA neuron generation. The NM‐II complex is commonly known as cytoplasmic actin‐binding motor proteins. Emerging evidence indicates the involvement of the NM‐II and other myosin complex in transcriptional regulation through various mechanisms, such as generating force to support the assembly of preinitiation complex or RNA polymerase movement, or controlling nuclear translocation of TFs 38, 39, 40, 41. Our results elucidated a novel function of the NM‐II complex in promoting the transcriptional activity of Atoh1 and its proneural function. These results warrant future studies to elucidate the structural basis of the Atoh1/NM‐II interaction, and, based on this knowledge, to optimize miDA neuron production. Our unpublished results also support the interaction between Ngn2 and the NM‐II complex, suggesting that the NM‐II complex may interact with various proneural TFs, and NM‐II agonists may promote iPSC differentiation driven by various proneural TFs, such as Ngn2‐driven motor neuron differentiation 42, 43.
Conclusion
We used synthetic mRNAs coding Atoh1 and Ngn2 to establish a novel strategy for efficient generation of functional and integration‐free mDA neurons from iPSCs. We further identified the NM‐II complex as an Atoh1‐binding partner and a positive modulator of this differentiation process. The methodology established here will facilitate the application of iPSC‐derived mDA neurons in PD modeling and therapy, and will likely be applicable to establish robust methods for generating various lineage‐specific progenies from iPSCs.
Author Contributions
Y.X., X.Z., S.S., and S.S.K.: collection of data, data analysis and interpretation, manuscript writing, final approval of manuscript. V.L.D., T.M.D., and S.X.: provision of study material, manuscript writing, final approval of manuscript. J.L. and J.Z.: conception and design, financial support, manuscript writing, final approval of manuscript. M.Y.: conception and design, financial support, collection of data, data analysis and interpretation, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
M.Y. and J.L. were named on an uncompensated patent application for Atoh1‐driven neuronal conversion methods submitted by the Johns Hopkins University. The other authors indicated no potential conflicts of interest.
Supporting information
Supporting Information Figure S1. Highly efficient mRNA transfection in iPSCs. (A and B) iPSC1 were transfected with GFP or wild‐type Atoh1 (FLAG‐tagged) mRNA as indicated. 24 hours after transfection, brightfield (BF) and fluorescence microscopic images show high percentage of GFP+ or FLAG+ cells (DAPI: nuclei; Bar: 20 μm).(C and D) iPSC1 cells received one dose of GFP or Atoh1 mRNA transfection or lipid alone as the control. The GFP mRNA but not the Atoh1 mRNA was synthesized with 5‐methylcytidine and pseudouridine. Cells were subjected to the lactate dehydrogenase (LDH) assay at 24 h and 48 hours (C). Total RNA was isolated after 24 h and 72 hours to measure markers of the IFN signaling (D).
Data represents Mean ± SEM (n = 3).
Supporting Information Figure S2. Representative microscopic images of neurons induced by Atoh1 and Ngn2 mRNAs from iPSC1. iPSC1 cells were transfected daily with mRNAs as indicated for 3 days (x3) or 6 days (x6), following the protocol shown in Figure 1D (top panel). Differentiated cells were replated in 24‐well plate at the density of 4x105 or 1x105 per well for Atoh1 or Ngn2 mRNA transfection, respectively. Images show neurons at 3 days after cell replating (Bar: 100 μm).
Supporting Information Figure S3. N‐SA mRNA transfection enhances miDA neuron conversion. (A) Diagram of two differentiation conditions with or without N‐SA mRNA (S/F/D: SHH, FGF8b and DAPT). (B) Neuron numbers were quantified at day 8 of differentiation to compare two conditions shown in A. (C) Neuronal and mDA lineage markers were measured at day 5 of differentiation by qRT‐PCR. Data represents Mean ± SEM (n = 3). *: p < .01. (D) Diagram of two differentiation conditions using A‐SA or N‐SA mRNA alone (S/F/D: SHH, FGF8b and DAPT). (E) mDA lineage markers were measured by qRT‐PCR in cells at day 5 of differentiation from the conditions as shown in D. Data are represented as Mean ± SEM (n = 3). *: p < .01.
Supporting Information Figure S4. The expression of neuronal marker TUJ1 in NPCs and neurons. TUJ1 were stained in NPCs and neurons at differentiation day 5 and 8, respectively (also shown in Fig. 2B). Cell nuclei were counterstained with DAPI. The percentage of TUJ1+/DAPI+ cells was quantified. Data represents Mean ± SEM (n = 6). Bar: 100 μm.
Supporting Information Figure S5. FOXA2 and LMX1A co‐expression in NPCs. NPCs at day 5 of differentiation as shown in Figure 3A were subjected to FOXA2 and LMX1A co‐staining. Cell nuclei were counterstained with DAPI (Bar: 100 μm). FOXA2+/LMX1A+ cells were quantified. Data represents Mean ± SEM (n = 6).
Supporting Information Figure S6. 6‐OHDA‐induced neurotoxicity in Ngn2‐induced neurons from iPSCs. iPSC1‐derived neurons were established by 3 daily doses of N‐SA mRNA transfection, following the strategy shown in Figure 1D and Supplemental Figure 2. Neurons after being matured for 5 days received 6‐OHDA or mock treatment for 24 hours. Neurite length was quantified in Calcein‐AM‐stained neurons. Data represents Mean ± SEM. *: p < .01 as compared to mock‐treated cells.
Supporting Information Figure S7. Bradykinin (BK) and Blebbistatin (Ble) showed no effects on cell death and viability of iPSCs transfected with A‐SA mRNA. BK and Ble were used together with A‐SA mRNA in iPSC1 cells. Cells at 24 h after mRNA transfection and compound treatment were subjected to the MTT and LDH assay. Data represents Mean ± SEM.
Supporting Information Table S1. Proteomics analysis results show the A‐SA/A‐WT binding ratio of each proteins identified in mass spectrometry analysis.
Supporting Information Table S2. Four lists of proteins belonging to cytoplasmic A‐WThigh, cytoplasmic A‐SAhigh, nuclear A‐WThigh, or nuclear A‐SAhigh.
Supporting Information Table S3. Proteins from Supplemental Table 2 were subjected to pathway enrichment analysis in the DAVID Bioinformatics Database to identify signaling pathways enriched in proteins belonging to cytoplasmic A‐WThigh, cytoplasmic A‐SAhigh, nuclear A‐WThigh, or nuclear A‐SAhigh.
Supporting Information Table S4. qRT‐PCR primers and antibodies.
Acknowledgments
We thank Dr. Joel Jessee from MTI‐GlobalStem, Inc. for providing mRNA transfection reagents. This work was supported by the Maryland Stem Cell Research Fund (M.Y. and T.M.D.). This work was supported by the CAMS Initiative for Innovative Medicine (2016‐I2M‐1‐008), the National Key Research and Development Program of China (2016YFA0101001, 2016YFC0903900), and CAMS Central Public Welfare Scientific Research Institute Basal Research Expenses (2017PT31004). The authors acknowledge the Adrienne Helis Malvin Medical Research Foundation and its direct engagement in the continuous active conduct of medical research in conjunction with the Johns Hopkins Hospital and the Johns Hopkins University School of Medicine and the Foundation's Parkinson's Disease Program M2015. T.M.D. is the Leonard and Madlyn Abramson Professor in Neurodegenerative Diseases.
Contributor Information
Jianmin Zhang, Email: jzhang42@163.com.
Mingyao Ying, Email: ying@kennedykrieger.org.
References
- 1. Qiang L, Fujita R, Abeliovich A. Remodeling neurodegeneration: Somatic cell reprogramming‐based models of adult neurological disorders. Neuron 2013;78:957–969. [DOI] [PubMed] [Google Scholar]
- 2. Ito D, Okano H, Suzuki N. Accelerating progress in induced pluripotent stem cell research for neurological diseases. Ann Neurol 2012;72:167–174. [DOI] [PubMed] [Google Scholar]
- 3. Jung YW, Hysolli E, Kim KY et al. Human induced pluripotent stem cells and neurodegenerative disease: Prospects for novel therapies. Curr Opin Neurol 2012;25:125–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kawasaki H, Mizuseki K, Nishikawa S et al. Induction of midbrain dopaminergic neurons from ES cells by stromal cell‐derived inducing activity. Neuron 2000;28:31–40. [DOI] [PubMed] [Google Scholar]
- 5. Swistowski A, Peng J, Liu Q et al. Efficient generation of functional dopaminergic neurons from human induced pluripotent stem cells under defined conditions. Stem Cells 2010;28:1893–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Perrier AL, Tabar V, Barberi T et al. Derivation of midbrain dopamine neurons from human embryonic stem cells. Proc Natl Acad Sci U S A 2004;101:12543–12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chambers SM, Fasano CA, Papapetrou EP et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol 2009;27:275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sundberg M, Bogetofte H, Lawson T et al. Improved cell therapy protocols for Parkinson's disease based on differentiation efficiency and safety of hESC‐, hiPSC‐, and non‐human primate iPSC‐derived dopaminergic neurons. Stem Cells 2013;31:1548–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kriks S, Shim JW, Piao J et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson's disease. Nature 2011;480:547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sagal J, Zhan X, Xu J et al. Proneural transcription factor Atoh1 drives highly efficient differentiation of human pluripotent stem cells into dopaminergic neurons. Stem Cells Translational Medicine 2014;3:888–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thoma EC, Wischmeyer E, Offen N et al. Ectopic expression of neurogenin 2 alone is sufficient to induce differentiation of embryonic stem cells into mature neurons. PLoS One 2012;7:e38651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ho SM, Hartley BJ, Tcw J et al. Rapid Ngn2‐induction of excitatory neurons from hiPSC‐derived neural progenitor cells. Methods 2016;101:113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang Y, Pak C, Han Y et al. Rapid single‐step induction of functional neurons from human pluripotent stem cells. Neuron 2013;78:785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Warren L, Manos PD, Ahfeldt T et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 2010;7:618–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yoshioka N, Gros E, Li HR et al. Efficient generation of human iPSCs by a synthetic self‐replicative RNA. Cell Stem Cell 2013;13:246–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goparaju SK, Kohda K, Ibata K et al. Rapid differentiation of human pluripotent stem cells into functional neurons by mRNAs encoding transcription factors. Sci Rep 2017;7:42367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ali F, Hindley C, McDowell G et al. Cell cycle‐regulated multi‐site phosphorylation of Neurogenin 2 coordinates cell cycling with differentiation during neurogenesis. Development 2011;138:4267–4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Forget A, Bihannic L, Cigna SM et al. Shh signaling protects Atoh1 from degradation mediated by the E3 ubiquitin ligase Huwe1 in neural precursors. Dev Cell 2014;29:649–661. [DOI] [PubMed] [Google Scholar]
- 19. Srivastava R, Kumar M, Peineau S et al. Conditional induction of Math1 specifies embryonic stem cells to cerebellar granule neuron lineage and promotes differentiation into mature granule neurons. Stem Cells 2013;31:652–665. [DOI] [PubMed] [Google Scholar]
- 20. Tieng V, Stoppini L, Villy S et al. Engineering of midbrain organoids containing long‐lived dopaminergic neurons. Stem Cells Dev 2014;23:1535–1547. [DOI] [PubMed] [Google Scholar]
- 21. Grace AA, Bunney BS. Nigral dopamine neurons: Intracellular recording and identification with L‐dopa injection and histofluorescence. Science 1980;210:654–656. [DOI] [PubMed] [Google Scholar]
- 22. Grace AA, Bunney BS. The control of firing pattern in nigral dopamine neurons: Burst firing. J Neurosci 1984;4:2877–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grace AA, Bunney BS. The control of firing pattern in nigral dopamine neurons: Single spike firing. J Neurosci 1984;4:2866–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guzman JN, Sanchez‐Padilla J, Chan CS et al. Robust pacemaking in substantia nigra dopaminergic neurons. J Neurosci 2009;29:11011–11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Roeper J. Dissecting the diversity of midbrain dopamine neurons. Trends Neurosci 2013;36:336–342. [DOI] [PubMed] [Google Scholar]
- 26. Sagal J, Zhan X, Xu J et al. Proneural transcription factor Atoh1 drives highly efficient differentiation of human pluripotent stem cells into dopaminergic neurons. Stem Cells Translational Medicine 2014;3:888–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Borgs L, Peyre E, Alix P et al. Dopaminergic neurons differentiating from LRRK2 G2019S induced pluripotent stem cells show early neuritic branching defects. Sci Rep 2016;6:33377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zou L, Xue Y, Jones M et al. The effects of quinine on neurophysiological properties of dopaminergic neurons. Neurotox Res 2018;34(1):62–73. [DOI] [PubMed] [Google Scholar]
- 29. Gambardella C, Pignatelli A, Belluzzi O. The h‐current in the substantia nigra pars compacta neurons: A re‐examination. PLoS One. 2012;7:e52329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bove J, Perier C. Neurotoxin‐based models of Parkinson's disease. Neuroscience 2012;211:51–76. [DOI] [PubMed] [Google Scholar]
- 31. Watanabe H, Takahashi R, Zhang XX et al. An essential role of myosin light‐chain kinase in the regulation of agonist‐ and fluid flow‐stimulated Ca2+ influx in endothelial cells. FASEB J 1998;12:341–348. [DOI] [PubMed] [Google Scholar]
- 32. Bond LM, Tumbarello DA, Kendrick‐Jones J et al. Small‐molecule inhibitors of myosin proteins. Future Med Chem 2013;5:41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ligon KL, Fancy SP, Franklin RJ et al. Olig gene function in CNS development and disease. Glia 2006;54:1–10. [DOI] [PubMed] [Google Scholar]
- 34. Sun Y, Meijer DH, Alberta JA et al. Phosphorylation state of Olig2 regulates proliferation of neural progenitors. Neuron 2011;69:906–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Abulaiti X, Zhang H, Wang A et al. Phosphorylation of threonine343 is crucial for OCT4 interaction with SOX2 in the maintenance of mouse embryonic stem cell pluripotency. Stem Cell Rep 2017;9:1630–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Malak PN, Dannenmann B, Hirth A et al. Novel AKT phosphorylation sites identified in the pluripotency factors OCT4, SOX2 and KLF4. Cell Cycle 2015;14:3748–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kikuchi T, Morizane A, Doi D et al. Human iPS cell‐derived dopaminergic neurons function in a primate Parkinson's disease model. Nature 2017;548:592–596. [DOI] [PubMed] [Google Scholar]
- 38. Schramek D, Sendoel A, Segal JP et al. Direct in vivo RNAi screen unveils myosin IIa as a tumor suppressor of squamous cell carcinomas. Science 2014;343:309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. de Lanerolle P. Nuclear actin and myosins at a glance. J Cell Sci 2012;125:4945–4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li Q, Sarna SK. Nuclear myosin II regulates the assembly of preinitiation complex for ICAM‐1 gene transcription. Gastroenterology 2009;137:1051–1060, 1060 e1051‐1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Percipalle P, Fomproix N, Cavellan E et al. The chromatin remodelling complex WSTF‐SNF2h interacts with nuclear myosin 1 and has a role in RNA polymerase I transcription. EMBO Rep 2006;7:525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Goto K, Imamura K, Komatsu K et al. Simple derivation of spinal motor neurons from ESCs/iPSCs using sendai virus vectors. Mol Ther Methods Clin Dev 2017;4:115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hester ME, Murtha MJ, Song S et al. Rapid and efficient generation of functional motor neurons from human pluripotent stem cells using gene delivered transcription factor codes. Mol Ther 2011;19:1905–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure S1. Highly efficient mRNA transfection in iPSCs. (A and B) iPSC1 were transfected with GFP or wild‐type Atoh1 (FLAG‐tagged) mRNA as indicated. 24 hours after transfection, brightfield (BF) and fluorescence microscopic images show high percentage of GFP+ or FLAG+ cells (DAPI: nuclei; Bar: 20 μm).(C and D) iPSC1 cells received one dose of GFP or Atoh1 mRNA transfection or lipid alone as the control. The GFP mRNA but not the Atoh1 mRNA was synthesized with 5‐methylcytidine and pseudouridine. Cells were subjected to the lactate dehydrogenase (LDH) assay at 24 h and 48 hours (C). Total RNA was isolated after 24 h and 72 hours to measure markers of the IFN signaling (D).
Data represents Mean ± SEM (n = 3).
Supporting Information Figure S2. Representative microscopic images of neurons induced by Atoh1 and Ngn2 mRNAs from iPSC1. iPSC1 cells were transfected daily with mRNAs as indicated for 3 days (x3) or 6 days (x6), following the protocol shown in Figure 1D (top panel). Differentiated cells were replated in 24‐well plate at the density of 4x105 or 1x105 per well for Atoh1 or Ngn2 mRNA transfection, respectively. Images show neurons at 3 days after cell replating (Bar: 100 μm).
Supporting Information Figure S3. N‐SA mRNA transfection enhances miDA neuron conversion. (A) Diagram of two differentiation conditions with or without N‐SA mRNA (S/F/D: SHH, FGF8b and DAPT). (B) Neuron numbers were quantified at day 8 of differentiation to compare two conditions shown in A. (C) Neuronal and mDA lineage markers were measured at day 5 of differentiation by qRT‐PCR. Data represents Mean ± SEM (n = 3). *: p < .01. (D) Diagram of two differentiation conditions using A‐SA or N‐SA mRNA alone (S/F/D: SHH, FGF8b and DAPT). (E) mDA lineage markers were measured by qRT‐PCR in cells at day 5 of differentiation from the conditions as shown in D. Data are represented as Mean ± SEM (n = 3). *: p < .01.
Supporting Information Figure S4. The expression of neuronal marker TUJ1 in NPCs and neurons. TUJ1 were stained in NPCs and neurons at differentiation day 5 and 8, respectively (also shown in Fig. 2B). Cell nuclei were counterstained with DAPI. The percentage of TUJ1+/DAPI+ cells was quantified. Data represents Mean ± SEM (n = 6). Bar: 100 μm.
Supporting Information Figure S5. FOXA2 and LMX1A co‐expression in NPCs. NPCs at day 5 of differentiation as shown in Figure 3A were subjected to FOXA2 and LMX1A co‐staining. Cell nuclei were counterstained with DAPI (Bar: 100 μm). FOXA2+/LMX1A+ cells were quantified. Data represents Mean ± SEM (n = 6).
Supporting Information Figure S6. 6‐OHDA‐induced neurotoxicity in Ngn2‐induced neurons from iPSCs. iPSC1‐derived neurons were established by 3 daily doses of N‐SA mRNA transfection, following the strategy shown in Figure 1D and Supplemental Figure 2. Neurons after being matured for 5 days received 6‐OHDA or mock treatment for 24 hours. Neurite length was quantified in Calcein‐AM‐stained neurons. Data represents Mean ± SEM. *: p < .01 as compared to mock‐treated cells.
Supporting Information Figure S7. Bradykinin (BK) and Blebbistatin (Ble) showed no effects on cell death and viability of iPSCs transfected with A‐SA mRNA. BK and Ble were used together with A‐SA mRNA in iPSC1 cells. Cells at 24 h after mRNA transfection and compound treatment were subjected to the MTT and LDH assay. Data represents Mean ± SEM.
Supporting Information Table S1. Proteomics analysis results show the A‐SA/A‐WT binding ratio of each proteins identified in mass spectrometry analysis.
Supporting Information Table S2. Four lists of proteins belonging to cytoplasmic A‐WThigh, cytoplasmic A‐SAhigh, nuclear A‐WThigh, or nuclear A‐SAhigh.
Supporting Information Table S3. Proteins from Supplemental Table 2 were subjected to pathway enrichment analysis in the DAVID Bioinformatics Database to identify signaling pathways enriched in proteins belonging to cytoplasmic A‐WThigh, cytoplasmic A‐SAhigh, nuclear A‐WThigh, or nuclear A‐SAhigh.
Supporting Information Table S4. qRT‐PCR primers and antibodies.


