Abstract
Scope:
Low fiber intake is associated with increased risk for cardiovascular disease (CVD) and cancer. However, the underlying mechanisms are not well understood. We tested two hypotheses: 1) dietary fiber would be associated with DNA methylation levels; 2) those DNA methylation changes would be associated with visceral adiposity and inflammation. We also explored the possibility that the associations between fiber and DNA methylation levels might be confounded with folic acid intake as sensitivity analysis.
Methods and results:
An epigenome-wide association study was conducted using Illumina 450K Bead-Chip on leukocyte DNA in 284 African American adolescents. Linear regression was performed to identify differentially methylated CpG sites associated with fiber. The methylation levels of 3 CpG sites (cg15200711, cg19462022 and cg07035602) in LPCAT1 and RASA3 genes were associated with fiber (FDR < 0.05) after adjustment for covariates including folic acid. The methylation levels of cg07035602 and cg19462022 were also associated with visceral adiposity and inflammation.
Conclusions:
Our data show that DNA methylation levels at LPCAT1 and RASA3 genes are associated with dietary fiber intake as well as with adiposity and inflammation. Future studies are warranted to determine whether epigenetic regulation may underlie the beneficial effects of fiber intake on adiposity and inflammation.
Keywords: Fiber intake, DNA methylation, African American, adolescents, adiposity, inflammation
1. Introduction
Low dietary fiber intake is associated with increased risk for cardiovascular disease (CVD), coronary artery disease and cancer [1]. We have previously shown that lower fiber intake is associated with higher levels of inflammation and visceral adiposity in adolescents [2]. However, the underlying mechanisms of health benefits of fiber are not well understood.
Emerging studies have suggested that nutrition plays a role in epigenetic regulation. For example, a 2-year randomized, placebo-controlled trial showed that long-term supplementation with folic acid in elderly subjects resulted in effects on DNA methylation of several genes which are implicated in developmental processes [3]. A recent cross-sectional study found significant associations among low folate intakes, lower CAMKK2 gene methylation, and insulin resistance in obese individuals [4]. In a cross-sectional study of Greek preadolescents, dietary fat intake was significantly associated with methylation levels of one CpG island shore and four sites [5]. We have shown that vitamin D deficiency is associated with global DNA hypomethylation, a hallmark of cancer events, and that vitamin D supplementation increases levels of global DNA methylation [6]. In addition, vitamin D deficiency is also associated with locus-specific leukocyte DNA methylation levels [7]. Besides, a study found that the consumption of polyunsaturated fatty acids was also associated with DNA methylation [8].
Very few studies have investigated the role of dietary fiber intake in epigenetic regulation. A small study showed that high fiber intake was associated with TNF-α hypomethylation [9]. However, the relationships between fiber intake and DNA methylation have not been examined in a genome-wide fashion. Given African Americans consume significantly less dietary fiber compared with other race/ethnic groups [10] and tend to have higher cardiometabolic risk [11], we conducted an epigenome-wide association study in African American adolescents to test the hypotheses that dietary fiber intake would be associated with leukocyte DNA methylation levels, and that those methylation levels would be associated with visceral adiposity and inflammation.
2. Materials and methods
2.1. Participants
Participants were previously recruited from local public high schools in Augusta, Georgia area into the Lifestyle, Adiposity and Cardiovascular Health in Youth (LACHY) study [12]. Demographic information obtained from the school systems was used to select schools that enrolled both black and white students. After receiving approval from the county superintendents and school principals, flyers were distributed to all students in the selected schools. Subjects were asked to self-identify their ethnicity [2]. Subjects who identified themselves as being white/Caucasian or black/African American were eligible for the study. Participants currently taking medication, or diagnosed with chronic medical conditions were excluded from the study. An epigenome-wide association study was conducted in a total of 284 African American LACHY participants (age 16.24±1.27). All participants provided written informed consent, and the study protocol was approved by the Institutional Review Board at the Medical College of Georgia, Augusta University (Augusta, GA, USA, protocol #622505). All data were de-identified.
2.2. Dietary intake
Dietary intakes were assessed with individual, non-consecutive, 24-hour recalls covering the period from midnight to midnight using the Nutrition Data System for Research (NDS-R 2006, Nutrition Coordinating Center, University of Minnesota, Minneapolis, MN) by trained dietitians. Seven independent dietary recalls were obtained within a period of 12 weeks for each participant. The first two recalls were performed in person, and the following ones were conducted by telephone. An average value was taken out of the seven recalls for further analysis.
2.3. Physical activity
As described in our previous paper [2], the number of minutes per day spent in moderate and vigorous physical activities were assessed using MTI Actigraph monitors (model 7164; MTIHealth Services, Fort Walton Beach, FL), uniaxial accelerometers that measure vertical acceleration and deceleration. The subjects were instructed to wear the monitor for a period of 7 days. Data from day 1 and 7 were discarded because a full day of information was not available for those days. Movement counts were converted to average minutes per day spent in moderate (3–6 metabolic equivalents) and vigorous (6 metabolic equivalents) physical activity by the software accompanying the device.
2.4. Anthropometry measurements
Height was measured using a wall-mounted stadiometer (Tanita Corporation of American, Arlington Heights, IL); weight was obtained using a calibrated electronic scale (model CN2OL; Cardinal Detecto, Webb City, MO). Body mass index (BMI) was calculated as weight (kg) per square of height (m2). Sexual development of the participants was measured by a five-stage scale, ranging from 1 (prepubertal) to 5 (fully mature). Using a gender-specific questionnaire, the subjects reported their sexual maturation stage (or Tanner stage) by comparing their own physical development to the five stages in standard sets of diagrams. A parent or research coordinator then reviewed the results with the children to make sure they understood the questionnaire. When an individual reported discordant stages of pubic hair and breast or genital development, the higher of the two stages was used [2].
2.5. Laboratory tests
Fasting blood samples were obtained for measurement of serum leptin, plasma adiponectin, plasma C-reactive protein (CRP), and plasma fibrinogen. Leptin was measured using serum that was assayed in duplicate using ELISA (R&D Systems, Minneapolis, MN). The intra-assay coefficient of variation (CV) for leptin was 2%, and the inter-assay CV was 5%. Adiponectin was measured using plasma that was assayed in duplicate using ELISA (Linco Research Inc., St. Charles, MO). Adiponectin had an intra-assay CV at 7.4% and an inter-assay CV at 8.4%. CRP was measured in plasma using high-sensitivity ELISA (ALPCO Diagnostics, Windham, NH). The mean intra- and inter-assay CV for CRP were 10 and 10.2%, respectively [2].
2.6. Body composition and fat distribution measures
Fat-free soft tissue (FFST) mass (kg) and fat mass (kg) were assessed by dual-energy x-ray absorptiometry (QDR-4500W; Hologic Inc., Waltham, MA). For determination of measurement reproducibility, one-way random-effects model, single-measure intra-class correlation coefficients were calculated for participants 15–18 yr of age (n = 219). Each participant was scanned twice within a 7-d period for FFST mass and fat mass (both r ≥ 0.97) [2].
Visceral adipose tissue (VAT) was measured by using magnetic resonance imaging (1.5-T; General Electric Medical Systems, Milwaukee, WI). Briefly, a series of five transverse images was acquired from the lumbar region beginning at the inferior border of the fifth lumbar vertebra and proceeding toward the head; a 2-mm gap between images was used to prevent cross talk. To calculate volume for VAT, the cross-sectional area (square cm) from each slice was multiplied by the slice width (1 cm), and then the individual volumes (cubic cm) were summed. The intra-class correlation coefficients for repeat analyses of the same scans on separate days within a 7-d period was r = 0.98 for VAT [2].
2.7. Genome-wide DNA methylation
DNA was extracted from stored buffy coat using the QIAamp DNA Mini Kit (QIAGEN). Genome-wide DNA methylation levels were analyzed by the Illumina Infinium Human Methylation 450K Beadchip (Illumina Inc.). In the quality control stage, DNA methylation data was processed using the Minfi package [13] and incorporating Control Probe Adjustment and reduction of global CORrelation (CPACOR) package [14]. We excluded CpG sites located on sex chromosomes. Illumina background correction and quantile normalization were applied to all intensity values and β value was further calculated and used as the index of CpG methylation levels. Detectable probes were defined as the probes with detection p-value < 10−16 in more than 95% samples; detectable samples were those with detection p-value < 10−16 in more than 95% CpG sites. Data was further corrected by 30 PCs from control probe intensities and the estimated cell compositions to remove test statistic inflation [14].
2.8. Statistical analysis
General characteristics are presented as mean ± SD for continuous variables. Normality of continuous variables was tested based on a combination test statistics of skewness and kurtosis. Dietary fiber intake was square root transformed to achieve a normal distribution. To test the difference of continuous measurements between different genders; two-tailed t-tests were conducted for variables with normal distribution, while Wilcoxon rank-sum tests were used instead to test for non-normally distributed variables.
For DNA methylation analysis, Limma package was used to identify differentially methylated CpG sites associated with dietary fiber intake [15]. Linear regression was carried out, using the adjusted CpG site methylation level (β values) as dependent variable, fiber intake as independent variable, controlling for age, sex, BMI and energy intake. Age, sex and BMI have been proven to be significantly associated with DNA methylation [16–18]. And as a study of nutrients, controlling for energy intake is suggested [19]. β values of CpGs, which were significantly associated with dietary fiber intake, were extracted and their associations with CRP, leptin, adiponectin, fibrinogen, visceral adipose and fat mass were estimated in linear regressions. CRP, leptin, adiponectin, fibrinogen, visceral adipose tissue and fat mass were log transformed in the regression models. Those models were adjusted for age, sex, sexual development, FFST mass, energy intake and physical activity, which have been proven or assumed to be associated with visceral adiposity [20–23] or inflammation [24, 25]. All statistical analysis was performed using R version 3.4.0 (R Foundation for Statistical Computing Vienna, Austria).
Folic acid is a critical component in the one-carbon metabolism pathway. As dietary folic acid intake may be a potential confounding factor of the association between fiber and DNA methylation, a sensitivity analysis was performed to examine whether the associations between fiber and DNA methylation were independent of folic acid intake. Coefficients and significance levels from models with and without adjustment for folic acid intake were compared between each other.
3. Results
3.1. Participants Characteristics
Characteristics of participants are presented in Table 1. The males were taller, heavier, and consumed more fiber, folic acid and energy (p < 0.001). The females had higher levels of leptin, fibrinogen, VAT and fat mass (p < 0.05).
Table 1.
General characteristics of participants*
| Characteristic | Total (N=284) |
Males (N=142) |
Females (N=142) |
p-value |
|---|---|---|---|---|
| Age (years) | 16.2±1.27 | 16.1±1.28 | 16.3±1.3 | 0.203 |
| Male (%) | 142 (50) | N/A | N/A | N/A |
| BMI (kg/m2) | 24.1±5.6 | 23.7±5.2 | 24.5±5.9 | 0.215 |
| Height (cm) | 168.6±9.2 | 174.5±7.9 | 162.8±6.1 | <0.001 |
| Weight (kg) | 68.6±17.0 | 72.6±17.3 | 64.7±16.0 | <0.001 |
| Energy Intake (kcal/day) | 1868.8±585.2 | 2136.4±628.3 | 1685.8±674.7 | <0.001 |
| Fiber Intake (gram/day) | 9.7±4.2 | 10.8±4.3 | 8.6±3.8 | <0.001 |
| Folic Acid Intake (μg/day) | 284.8±129.3 | 333.8±136.6 | 235.8±100.5 | <0.001 |
| CRP (ng/mL) | 1191.3±2212.3 | 1036.6±1838.5 | 1333.0±2505.3 | 0.288 |
| Leptin (pg/mL) | 13984.1±14121.9 | 7274.7±9989.6 | 20047.4±14589.8 | <0.001 |
| Adiponectin (ug/mL) | 7.8±4.8 | 7.4±5.1 | 8.2±4.5 | 0.179 |
| Fibrinogen (mg/dL) | 288.7±54.9 | 276.4±54.8 | 301.0±52.4 | <0.001 |
| VAT (cm3) | 94.0±69.9 | 81.4±62.4 | 112.5±76.4 | 0.003 |
| Fat Mass (kg) | 16.8±11.0 | 13.8±10.8 | 19.8±10.4 | <0.001 |
Statistics display as mean±SD for continuous variables, and N(%) for categorical variables.
3.2. Linear regression analysis
Figure 1 is a volcano plot to visualize CpGs associated with fiber intake in both adjusted changes and statistical significance (Figure 1). β values represent DNA methylation changes with 1 unit increase in square root of fiber intake. Table 2 lists the top 20 differential methylated CpG sites associated with fiber intakes. The top 4 CpG sites achieved genome-wide significance, all of which were negatively associated with fiber intake (FDR < 0.05, Table 2). Among those, cg15200711 and cg07035602 are both located in the gene body region of LPCAT1 gene on chromosome 5p15.33. cg19462022 is located in the gene body region of RASA3 gene on chromosome 13q34. cg15302376 is located in the 5’UTR region of DNMT3 gene on chromosome 2p23.3 (Figure 2).
Figure 1.
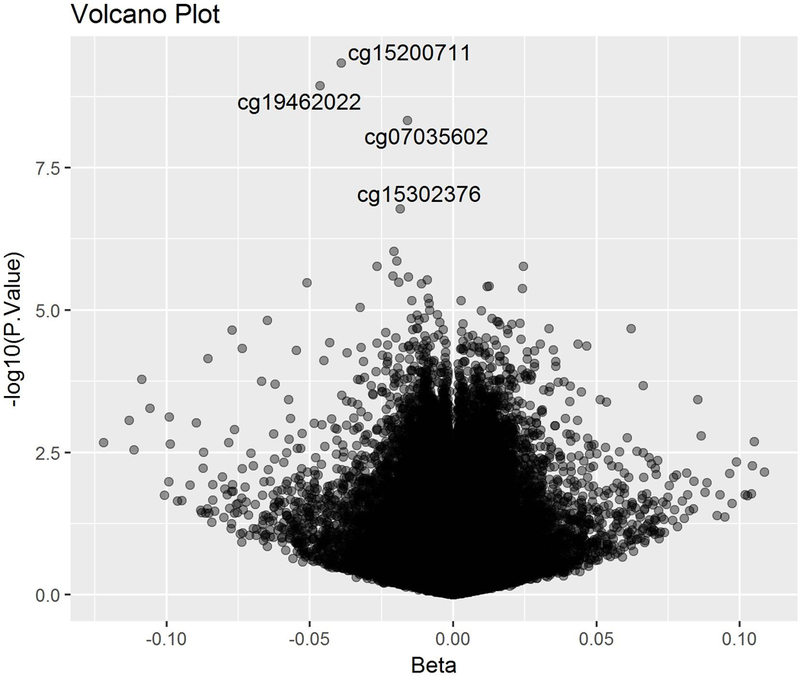
Volcano plot. Beta values represent DNA methylation changes with 1 unit increase in square root of fiber intake, and the vertical axis indicates –log10 transformed observed p-values. Models were adjusted for age, sex, BMI and energy intake.
Table 2.
Top 20 differential methylated CpG sites associated with dietary fiber intakes and sensitivity analysis
| Rank | CpGs | CHR | Gene | Group | Model adjusted for covariates* | Model adjusted for covariates and folic acid* | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| β** | p-value | FDR | β** | p-value | FDR | |||||
| 1 | cg15200711 | 5p15.33 | LPCAT1 | Body | −0.039 | 4.53e–10 | <0.001 | −0.045 | 1.80E–09 | 0.001 |
| 2 | cg19462022 | 13q34 | RASA3 | Body | −0.047 | 1.14e–09 | <0.001 | −0.049 | 8.20E–08 | 0.019 |
| 3 | cg07035602 | 5p15.33 | LPCAT1 | Body | −0.016 | 4.63e–09 | 0.001 | −0.017 | 1.90E–07 | 0.023 |
| 4 | cg15302376 | 2p23.3 | DNMT3A | 5’UTR | −0.019 | 1.65e–07 | 0.019 | −0.016 | 1.00E–04 | 0.505 |
| 5 | cg26391080 | 10q23.1 | SH2D4B | TSS1500 | −0.021 | 9.31e–07 | 0.088 | −0.020 | 5.40E–05 | 0.428 |
| 6 | cg03589898 | −0.020 | 1.36e–06 | 0.101 | −0.019 | 6.60E–05 | 0.428 | |||
| 7 | cg08740107 | 0.024 | 1.68e–06 | 0.101 | 0.032 | 1.90E–07 | 0.023 | |||
| 8 | cg03394700 | 11p11.2 | TP53I11 | 5’UTR | −0.027 | 1.71e–06 | 0.101 | −0.031 | 3.80E–06 | 0.296 |
| 9 | cg03462714 | 10p15.3 | ADARB2 | Body | −0.021 | 2.53e–06 | 0.113 | −0.021 | 7.70E–05 | 0.452 |
| 10 | cg01000812 | 20p13 | LOC100288797 | TSS200 | −0.016 | 2.61e–06 | 0.113 | −0.016 | 9.50E–05 | 0.494 |
| 11 | cg06271327 | 17p13.1 | VAMP2 | 3’UTR | −0.009 | 2.94e–06 | 0.113 | −0.007 | 1.06E–03 | 0.707 |
| 12 | cg09422701 | −0.019 | 3.22e–06 | 0.113 | −0.020 | 3.00E–05 | 0.428 | |||
| 13 | cg25826698 | −0.051 | 3.27e–06 | 0.113 | −0.053 | 4.60E–05 | 0.428 | |||
| 14 | cg25258002 | −0.011 | 3.45e–06 | 0.113 | −0.012 | 5.10E–05 | 0.428 | |||
| 15 | cg21739719 | 15q24.2 | C15orf27 | Body | 0.012 | 3.76e–06 | 0.113 | 0.014 | 2.50E–05 | 0.428 |
| 16 | cg12405742 | 3p24.2 | THRB | 5’UTR | 0.012 | 3.83e–06 | 0.113 | 0.011 | 3.12E–04 | 0.613 |
| 17 | cg11813512 | 0.024 | 4.14e–06 | 0.115 | 0.022 | 5.31E–04 | 0.677 | |||
| 18 | cg26159735 | −0.009 | 6.21e–06 | 0.162 | −0.009 | 2.16E–04 | 0.613 | |||
| 19 | cg19641404 | 2q37.1 | DIS3L2 | TSS200 | −0.014 | 6.74e–06 | 0.162 | −0.015 | 8.10E–05 | 0.452 |
| 20 | cg20257411 | 22q11.1 | CECR5 | Body;TSS200 | 0.003 | 6.86e–06 | 0.162 | 0.003 | 4.50E–05 | 0.428 |
Covariates include age, sex, BMI, energy intake
DNA methylation changes with 1 unit increase in square root transformed fiber intake.
Figure 2.
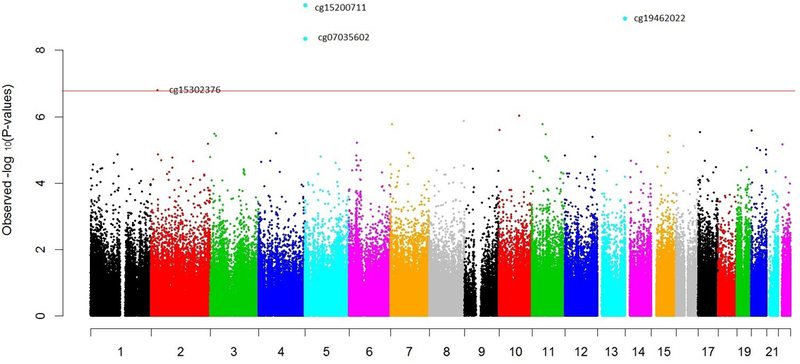
Manhattan plot. The vertical axis indicates –log10 transformed observed p-values. Models were adjusted for age, sex, BMI and energy intake.
3.3. Sensitivity Analysis
Linear regression models were further adjusted for folic acid intake as a sensitivity analysis to eliminate the confounding effect of folic acid (Table 2). The β value for fiber in relation to methylation changed minimally among top 20 CpGs (median change 6.7%, 25–75th percentile 4.0–15.2%). Three out of the four CpGs sites (cg15200711, cg19462022 and cg07035602) remained statistically significant (FDR < 0.05). In addition, we conducted a genome-wide analysis of folic acid intake; none of the CpGs was significantly associated with folic acid intake in linear regressions adjusted for age, sex, BMI and energy intake (Supporting Information Table S1).
3.4. Associations with visceral adiposity and inflammation
We further tested whether the three significant CpG sites were associated with visceral adiposity and inflammation. cg07035602 (LPCAT1) was significantly associated with CRP, leptin, fibrinogen and visceral adiposity (all p-values <0.05). cg19462022 (RASA3) was significantly associated with CRP (p-value = 0.002). No significant association was identified between cg15200711 and selected markers for inflammation and visceral adiposity (Table 3).
Table 3.
Adjusted associations between fiber-related CpGs and markers of inflammation and visceral adiposity*
| CpGs | CRP (N=214) |
Leptin (N=218) |
Adiponectin (N=196) |
Fibrinogen (N=128) |
Visceral adipose (N=158) |
Fat mass (N=238) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β | p-value | β | p-value | β | p-value | β | p-value | β | p-value | β | p-value | |
| cg15200711 | 1.91 | 0.454 | 0.39 | 0.802 | 0.10 | 0.933 | 0.28 | 0.281 | 0.41 | 0.694 | 0.07 | 0.804 |
| cg19462022 | 5.90 | 0.002 | 1.83 | 0.127 | -0.78 | 0.395 | 0.18 | 0.385 | 0.52 | 0.583 | 0.19 | 0.420 |
| cg07035602 | 12.75 | 0.034 | 8.85 | 0.015 | 2.31 | 0.400 | 1.26 | 0.049 | 6.13 | 0.008 | 1.31 | 0.053 |
Leptin, adiponectin, CRP, fibrinogen, visceral adipose and fat mass were log-transformed. Models were adjusted for age, sex, sexual development, FFST mass, energy intake, and physical activity. There are missing values for CRP (31), leptin (27), adiponectin (52), fibrinogen (139), visceral adipose (99), fat mass (5), sexual development (7), FFST mass (6), energy intake (5), and physical activity (41).
4. Discussion
In this epigenome-wide association study, the methylation levels of 3 CpG sites (cg15200711, cg19462022 and cg07035602) in LPCAT1 and RASA3 genes were associated with dietary fiber intakes. In addition, the methylation levels of cg07035602 and cg19462022 were also associated with visceral adiposity and inflammation.
Two out of three significant CpG sites (cg15200711 and cg07035602) are located in the gene body of Lysophosphatidylcholine Acyltransferase 1 gene (LPCAT1). The encoded enzyme plays a role in phospholipid metabolism, specifically in the conversion of lysophosphatidylcholine to phosphatidylcholine in the presence of acyl-CoA [26]. This process is important in the synthesis of lung surfactant and platelet-activating factor. LPCAT1 can localize to the lipid droplet surface, modulate lipid droplet number and size as well as influence the release of lipoprotein from liver cells [27]. Moreover, LPCAT1, an anti-inflammatory agent, plays a key role in platelet-activating factor remodeling [28]. We observed that the methylation level of cg07035602 of LPCAT1 was associated with visceral adiposity and inflammation, which provides a piece of clinical evidence suggesting that LPCAT1 may be involved in inflammatory processes.
LPCAT1 also plays a role in many disease processes. Activity of LPCAT1 was decreased in the liver of cirrhotic animals [28]. LPCAT1 was also down regulated in brain tissue of diabetic mice and was upregulated by anti-diabetic treatment [29]. Dysfunction of LPCAT1 is also involved in breast carcinoma [30], lung cancer [31], liver cancer [32], prostate cancer [33] and renal cell carcinoma [34].
cg19462022 is located in the gene body of Ras GTPase-activating protein 3 gene (RASA3). It is localized to the cell membrane via a pleckstrin homology domain in the C-terminal region [35]. GO annotations related to this gene include GTPase activator activity and calcium-release channel activity. RASA3 encodes a protein that binds inositol 1,3,4,5-tetrakisphosphate and stimulates the GTPase activity of Ras p21. RASA3 is a member of the GAP1 family of GTPase-activing proteins, part of Ras signaling, acting as a suppressor of RAS function and controls cellular proliferation and differentiation [36]. RASA3 is also a critical regulator of Rap1 in endothelial cells which controls adhesions properties and vascular lumen integrity [37]. RASA3 is involved in the pathogenesis of many types of cancers including colorectal, gastric and other types of cancer [38–40]. In our data, the methylation level of cg19462022 was also associated with CRP.
The methylation level of cg15302376 in the 5’UTR region of DNA methyltransferase 3 alpha (DNMT3A) was associated with fiber intake, which may be critical because the DNA methyltransferase family plays a central role in epigenetic gene regulation and whose activity and expression changes may have a dramatic effect on the methylation level of a large number of genes. However, cg15302376 lost its significance in the sensitivity analysis after further adjustment for folic acid intake. The fact that folic acid was associated with the methylation site in the DNA methyltransferase and rather than methylation sites in other genes, suggests that folic acid may affect the expression of certain methyltransferases and thus affect the methylation level of substrate genes rather than directly affect the gene methylation level. As in the sensitivity analysis, by adjusting for folic acid intake, potential bias introduced by both direct and indirect effect of folic acid has been considered.
A small study previously showed that fiber intake was associated with differential TNF-α methylation [9]. Out of 5 CpG sites in TNF- α gene on our 450 K platform, 2 CpG sites (cg21467614 and cg08553327) were significant with p-values < 0.05. The p-values of the remaining 3 CpG sites were between 0.061–0.088 (Supporting Information Table S2). Our data also suggest that fiber intake may influence TNF- α methylation.
Food containing more fiber tends to be rich in folic acid, which is a critical component in the one-carbon metabolism pathway providing the methyl group for DNA methylation [41]. In our dataset, pairwise correlation coefficient between dietary fiber intake and folic acid intake is 0.81 (p-value < 0.001). To address the concern that folic acid intake might confound the associations between fiber intake and DNA methylation, we conducted a sensitivity analysis and showed that further adjustment of folic acid intake in the model did not change the β value of the top 20 CpGs much (median change 6.7%, 25–75th percentile 4.0–15.2%). In addition, three out of the four significant CpGs remained statistically significant. In addition, folic acid intake failed to show any independent associations with DNA methylation in our study (Supporting Information Table S1). Thus, folic acid intake may not likely be a confounding factor of the fiber-methylation associations for these 3 CpG sites identified in our study.
To the best of our knowledge, the present study is the first to examine the influence of dietary fiber intake on DNA methylation in a genome-wide fashion in African American adolescents. We did not replicate the results by splitting the entire sample into discovery and validation sets, but analyzed the full dataset to optimize power by following the best practice [42, 43]. Studies suggest that joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies [42, 43]. This genome-wide association analysis was only done in African American adolescents; thus, whether the identified methylation sites are unique in African American population is not known. Future studies are warranted to replicate our findings in independent cohorts and other ethnic groups. Another shortcoming of this study is the cross-sectional design. We have shown that methylation levels at cg07035602 and cg19462022 were associated with both fiber intake as well as adiposity and inflammation; however, the causal relationships cannot be established. We have explored the causal mediation analysis to estimate whether the methylation levels of the CpG sites mediated the effect of dietary fiber intake on adiposity and inflammation, which was not significant. It may be attributed to the limited power due to some missing values in the inflammation and adiposity measurements. In addition, Maxwell and Cole found that cross-sectional approaches to longitudinal mediation can substantially over- or underestimate longitudinal effects even under the ideal conditions, and researchers who interested in mediational processes were urged to include multiple waves of data in the analyses [44]. Therefore, we were only able to provide the linear regression association results between those identified CpG sites and inflammation/adiposity. Future studies are needed to further investigate the role of methylation in the relationship between dietary fiber intake and inflammation/adiposity.
Our epigenome-wide association study identifies 3 CpG sites that are associated with dietary fiber intake. Those sites are also associated with visceral adiposity and inflammation. Our data suggest that dietary fiber may influence epigenetic regulation. Future studies are warranted to determine whether epigenetic regulation (i.e. DNA methylation) underlies the beneficial effects of fiber intake on adiposity and inflammation.
Supplementary Material
Acknowledgements
LC and HZ conceptualized and designed the study, drafted the initial manuscript, critically reviewed and revised the manuscript. YH, BG and YD collected the data, critically reviewed and revised the manuscript. XW and GH critically reviewed the manuscript. All authors approved the final manuscript as submitted and agree to be accountable for all aspects of the work. Study was funded by National Heart, Lung and Blood Institute (HL105689), and American Heart Association (14GRNT20480211).
Abbreviations
- CVD
cardiovascular disease
- LACHY
Lifestyle, Adiposity, and Cardiovascular Health in Youth study
- BMI
body mass index
- CRP
C-reactive protein
- CV
coefficient of variation
- FFST
fat-free soft tissue
- VAT
visceral adipose tissue
- QIAGEN
QIAamp DNA Mini Kit
- CPACOR
Control Probe Adjustment and reduction of global CORrelation
- BH
Benjamini-Hochberg
- FDR
false discovery rate
- DNMT3A
DNA methyltransferase 3 alpha
- LPCAT1
Lysophosphatidylcholine Acyltransferase 1
- RASA3
Ras GTPase-activating protein 3
Footnotes
Conflict of Interest
The authors have declared no conflict of interest.
References
- [1].Veronese N, Solmi M, Caruso MG, Giannelli G, Osella AR, Evangelou E, Maggi S, Fontana L, Stubbs B, Tzoulaki I, Dietary fiber and health outcomes: an umbrella review of systematic reviews and meta-analyses. Am J Clin Nutr 2018, 107, 436–444. [DOI] [PubMed] [Google Scholar]
- [2].Parikh S, Pollock NK, Bhagatwala J, Guo D-H, Gutin B, Zhu H, Dong Y, Adolescent Fiber Consumption Is Associated with Visceral Fat and Inflammatory Markers. The Journal of Clinical Endocrinology and Metabolism 2012, 97, E1451–E1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kok DE, Dhonukshe-Rutten RA, Lute C, Heil SG, Uitterlinden AG, van der Velde N, van Meurs JB, van Schoor NM, Hooiveld GJ, de Groot LC, Kampman E, Steegenga WT, The effects of long-term daily folic acid and vitamin B12 supplementation on genome-wide DNA methylation in elderly subjects. Clin Epigenetics 2015, 7, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ramos-Lopez O, Samblas M, Milagro FI, Zulet MA, Mansego ML, Riezu-Boj JI, Martinez JA, Association of low dietary folate intake with lower CAMKK2 gene methylation, adiposity, and insulin resistance in obese subjects. Nutr Res 2018, 50, 53–62. [DOI] [PubMed] [Google Scholar]
- [5].Voisin S, Almen MS, Moschonis G, Chrousos GP, Manios Y, Schioth HB, Dietary fat quality impacts genome-wide DNA methylation patterns in a cross-sectional study of Greek preadolescents. Eur J Hum Genet 2015, 23, 654–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhu HD, Bhagatwala J, Huang Y, Pollock NK, Parikh S, Raed A, Gutin B, Harshfield GA, Dong YB, Race/Ethnicity-Specific Association of Vitamin D and Global DNA Methylation: Cross-Sectional and Interventional Findings. PloS one 2016, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhu HD, Wang XL, Shi HD, Su SY, Harshfield GA, Gutin B, Snieder H, Dong YB , A Genome-Wide Methylation Study of Severe Vitamin D Deficiency in African American Adolescents. J Pediatr-Us 2013, 162, 1004–U1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Arpon A, Milagro FI, Razquin C, Corella D, Estruch R, Fito M, Marti A, Martinez-Gonzalez MA, Ros E, Salas-Salvado J, Riezu-Boj JI, Martinez JA, Impact of Consuming Extra-Virgin Olive Oil or Nuts within a Mediterranean Diet on DNA Methylation in Peripheral White Blood Cells within the PREDIMED-Navarra Randomized Controlled Trial: A Role for Dietary Lipids. Nutrients 2017, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Carraro JCC, Hermsdorff HHM, Mansego ML, Zulet MA, Milagro FI, Bressan J, Martinez JA, Higher Fruit Intake Is Related to TNF-alpha Hypomethylation and Better Glucose Tolerance in Healthy Subjects. J Nutrigenet Nutrige 2016, 9, 95–105. [DOI] [PubMed] [Google Scholar]
- [10].Storey M, Anderson P, Income and race/ethnicity influence dietary fiber intake and vegetable consumption. Nutr Res 2014, 34, 844–850. [DOI] [PubMed] [Google Scholar]
- [11].Ski CF, King-Shier KM, Thompson DR, Gender, socioeconomic and ethnic/racial disparities in cardiovascular disease: a time for change. Int J Cardiol 2014, 170, 255–257. [DOI] [PubMed] [Google Scholar]
- [12].Gutin B, Johnson MH, Humphries MC, Hatfield-Laube JL, Kapuku GK, Allison JD, Gower BA, Daniels SR, Barbeau P, Relationship of visceral adiposity to cardiovascular disease risk factors in black and white teens. Obesity (Silver Spring) 2007, 15, 1029–1035. [DOI] [PubMed] [Google Scholar]
- [13].Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA, Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014, 30, 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lehne B, Drong AW, Loh M, Zhang W, Scott WR, Tan ST, Afzal U, Scott J, Jarvelin MR, Elliott P, McCarthy MI, Kooner JS, Chambers JC, A coherent approach for analysis of the Illumina HumanMethylation450 BeadChip improves data quality and performance in epigenome-wide association studies. Genome Biol 2015, 16, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Smyth GK, Michaud J, Scott HS, Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 2005, 21, 2067–2075. [DOI] [PubMed] [Google Scholar]
- [16].Jones MJ, Goodman SJ, Kobor MS, DNA methylation and healthy human aging. Aging Cell 2015, 14, 924–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hall E, Volkov P, Dayeh T, Esguerra JL, Salo S, Eliasson L, Ronn T, Bacos K, Ling C, Sex differences in the genome-wide DNA methylation pattern and impact on gene expression, microRNA levels and insulin secretion in human pancreatic islets. Genome Biol 2014, 15, 522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dick KJ, Nelson CP, Tsaprouni L, Sandling JK, Aissi D, Wahl S, Meduri E, Morange PE, Gagnon F, Grallert H, Waldenberger M, Peters A, Erdmann J, Hengstenberg C, Cambien F, Goodall AH, Ouwehand WH, Schunkert H, Thompson JR, Spector TD, Gieger C, Tregouet DA, Deloukas P, Samani NJ, DNA methylation and body-mass index: a genome-wide analysis. Lancet 2014, 383, 1990–1998. [DOI] [PubMed] [Google Scholar]
- [19].Willett W, Stampfer MJ, Total energy intake: implications for epidemiologic analyses. Am J Epidemiol 1986, 124, 17–27. [DOI] [PubMed] [Google Scholar]
- [20].Siervo M, Lara J, Celis-Morales C, Vacca M, Oggioni C, Battezzati A, Leone A, Tagliabue A, Spadafranca A, Bertoli S, Age-related changes in basal substrate oxidation and visceral adiposity and their association with metabolic syndrome. Eur J Nutr 2016, 55, 1755–1767. [DOI] [PubMed] [Google Scholar]
- [21].Staiano AE, Katzmarzyk PT, Ethnic and sex differences in body fat and visceral and subcutaneous adiposity in children and adolescents. Int J Obes (Lond) 2012, 36, 1261–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kondoh T, Takase H, Yamaguchi TF, Ochiai R, Katashima M, Katsuragi Y, Sakane N, Association of dietary factors with abdominal subcutaneous and visceral adiposity in Japanese men. Obesity research & clinical practice 2014, 8, e16–25. [DOI] [PubMed] [Google Scholar]
- [23].Keating SE, Parker HM, Pavey TG, Baker MK, Caterson ID, George J, Johnson NA, Objectively Quantified Physical Activity and Sedentary Behavior in Predicting Visceral Adiposity and Liver Fat. J Obes 2016, 2016, 2719014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Adams SA, Wirth MD, Khan S, Murphy EA, Heiney SP, Davis LC, Davis B, Drayton RF, Hurley TG, Blair SN, Hebert JR, The association of C-reactive protein and physical activity among a church-based population of African Americans. Preventive medicine 2015, 77, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Shanahan L, Copeland WE, Worthman CM, Erkanli A, Angold A, Costello EJ, Sex-differentiated changes in C-reactive protein from ages 9 to 21: the contributions of BMI and physical/sexual maturation. Psychoneuroendocrinology 2013, 38, 2209–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Moessinger C, Kuerschner L, Spandl J, Shevchenko A, Thiele C, Human lysophosphatidylcholine acyltransferases 1 and 2 are located in lipid droplets where they catalyze the formation of phosphatidylcholine. J Biol Chem 2011, 286, 21330–21339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Moessinger C, Klizaite K, Steinhagen A, Philippou-Massier J, Shevchenko A, Hoch M, Ejsing CS, Thiele C, Two different pathways of phosphatidylcholine synthesis, the Kennedy Pathway and the Lands Cycle, differentially regulate cellular triacylglycerol storage. BMC cell biology 2014, 15, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Stanca E, Serviddio G, Bellanti F, Vendemiale G, Siculella L, Giudetti AM, Down-regulation of LPCAT expression increases platelet-activating factor level in cirrhotic rat liver: potential antiinflammatory effect of silybin. Biochim Biophys Acta 2013, 1832, 2019–2026. [DOI] [PubMed] [Google Scholar]
- [29].Cheng L, Han X, Shi Y, A regulatory role of LPCAT1 in the synthesis of inflammatory lipids, PAF and LPC, in the retina of diabetic mice. Am J Physiol Endocrinol Metab 2009, 297, E1276–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Abdelzaher E, Mostafa MF, Lysophosphatidylcholine acyltransferase 1 (LPCAT1) upregulation in breast carcinoma contributes to tumor progression and predicts early tumor recurrence. Tumor Biol 2015, 36, 5473–5483. [DOI] [PubMed] [Google Scholar]
- [31].Ellis B, Kaercher L, Snavely C, Zhao Y, Zou C, Lipopolysaccharide triggers nuclear import of Lpcat1 to regulate inducible gene expression in lung epithelia. World J Biol Chem 2012, 3, 159–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Morita Y, Sakaguchi T, Ikegami K, Goto-Inoue N, Hayasaka T, Hang VT, Tanaka H, Harada T, Shibasaki Y, Suzuki A, Fukumoto K, Inaba K, Murakami M, Setou M, Konno H, Lysophosphatidylcholine acyltransferase 1 altered phospholipid composition and regulated hepatoma progression. J Hepatol 2013, 59, 292–299. [DOI] [PubMed] [Google Scholar]
- [33].Zhou XC, Lawrence TJ, He Z, Pound CR, Mao JH, Bigler SA, The expression level of lysophosphatidylcholine acyltransferase 1 (LPCAT1) correlates to the progression of prostate cancer. Exp Mol Pathol 2012, 92, 105–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Du Y, Wang Q, Zhang X, Wang X, Qin C, Sheng Z, Yin H, Jiang C, Li J, Xu T, Lysophosphatidylcholine acyltransferase 1 upregulation and concomitant phospholipid alterations in clear cell renal cell carcinoma. J Exp Clin Cancer Res 2017, 36, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Schurmans S, Polizzi S, Scoumanne A, Sayyed S, Molina-Ortiz P, The Ras/Rap GTPase activating protein RASA3: from gene structure to in vivo functions. Advances in biological regulation 2015, 57, 153–161. [DOI] [PubMed] [Google Scholar]
- [36].Hancock JF, Ras proteins: different signals from different locations. Nature reviews. Molecular cell biology 2003, 4, 373–384. [DOI] [PubMed] [Google Scholar]
- [37].Molina-Ortiz P, Orban T, Martin M, Habets A, Dequiedt F, Schurmans S, Rasa3 controls turnover of endothelial cell adhesion and vascular lumen integrity by a Rap1-dependent mechanism. Plos Genet 2018, 14, e1007195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Tang J, Li Y, Lyon K, Camps J, Dalton S, Ried T, Zhao S, Cancer driver-passenger distinction via sporadic human and dog cancer comparison: a proof-of-principle study with colorectal cancer. Oncogene 2014, 33, 814–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Qamra A, Xing M, Padmanabhan N, Kwok JJT, Zhang S, Xu C, Leong YS, Lee Lim AP, Tang Q, Ooi WF, Suling Lin J, Nandi T, Yao X, Ong X, Lee M, Tay ST, Keng ATL, Gondo Santoso E, Ng CCY, Ng A, Jusakul A, Smoot D, Ashktorab H, Rha SY, Yeoh KG, Peng Yong W, Chow PKH, Chan WH, Ong HS, Soo KC, Kim KM, Wong WK, Rozen SG, Teh BT, Kappei D, Lee J, Connolly J, Tan P, Epigenomic Promoter Alterations Amplify Gene Isoform and Immunogenic Diversity in Gastric Adenocarcinoma. Cancer Discov 2017, 7, 630–651. [DOI] [PubMed] [Google Scholar]
- [40].Choi YJ, Lee SH, Kim MS, Jung SH, Hur SY, Chung YJ, Lee SH, Whole-exome sequencing identified the genetic origin of a mucinous neoplasm in a mature cystic teratoma. Pathology 2016, 48, 372–376. [DOI] [PubMed] [Google Scholar]
- [41].Joubert BR, den Dekker HT, Felix JF, Bohlin J, Ligthart S, Beckett E, Tiemeier H, van Meurs JB, Uitterlinden AG, Hofman A, Haberg SE, Reese SE, Peters MJ, Andreassen BK, Steegers EA, Nilsen RM, Vollset SE, Midttun O, Ueland PM, Franco OH, Dehghan A, de Jongste JC, Wu MC, Wang T, Peddada SD, Jaddoe VW, Nystad W, Duijts L, London SJ, Maternal plasma folate impacts differential DNA methylation in an epigenome-wide meta-analysis of newborns. Nat Commun 2016, 7, 10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Skol AD, Scott LJ, Abecasis GR, Boehnke M, Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet 2006, 38, 209–213. [DOI] [PubMed] [Google Scholar]
- [43].Hutter CM, Mechanic LE, Chatterjee N, Kraft P, Gillanders EM, Gene-environment interactions in cancer epidemiology: a National Cancer Institute Think Tank report. Genetic epidemiology 2013, 37, 643–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Maxwell SE, Cole DA, Bias in cross-sectional analyses of longitudinal mediation. Psychological methods 2007, 12, 23–44. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


