Abstract
Context:
Endometriosis is a chronic inflammatory disease that causes pain and infertility in women of reproductive age.
Objective:
To investigate the pathologic pathways in endometrial stromal and epithelial cells that contribute to the manifestation of endometriosis.
Design:
In vitro cellular and molecular analyses of isolated eutopic endometrial stromal and epithelial cells.
Methods:
Eutopic stromal and epithelial cells from endometriotic and normal patients were isolated by fluorescence-activated cell sorting for paired sibling RNA sequencing and microRNA microarray. Aberrant pathways were identified using ingenuity pathway analysis networks and confirmed with in vitro modulation of the affected pathways in stromal and epithelial cell cultures.
Results:
Both stromal versus epithelial cell types and paired endometriotic versus normal samples exhibited distinct hierarchical clustering. Compared to normal samples, there were 151 and 215 differentially expressed genes in the endometriotic stromal and epithelial populations, respectively, and concomitantly 9 and 16 differentially expressed microRNAs. Overall, endometriotic stromal and epithelial cells revealed distinct defects. In endometriotic stromal cells, key decidualization genes Zinc finger E-box Binding protein 1 (ZEB1), Heart And Neural crest Derivatives expressed 2 (HAND2), WNT4, and Interleukin 15 (IL-15) were found to be downregulated and Periostin (POSTN) and Matrix Metallopeptidase 7 (MMP7) were upregulated. Specifically, ZEB1 was downregulated in stromal cells by aberrant elevation in miR-200b. In contrast, ZEB1 was found to be upregulated in endometriotic epithelial cells through associated upregulation of transforming growth factor β1 (TGFβ1), inducer of the TGFβ1–Bone Morphogenetic Protein 2 (BMP2)–MMP2–Prostaglandin-endoperoxide Synthase 2 (COX2)–ZEB1 pathway, which activates epithelial–mesenchymal transition.
Conclusion:
Manifestation of endometriosis involves dysregulation of unique molecular pathways within the diseased endometrial stromal and epithelial cells in the endometrium. Targeting the cell type–specific defects may offer a novel approach to treating endometriosis.
Keywords: decidualization, epithelial–mesenchymal transition, microRNA, TGFβ1, ZEB1
Introduction
The extrauterine invasion and growth of estrogen-dependent endometrial cells is the clinical hallmark of patients with endometriosis.1–3 It is an inflammatory disease that affects approximately 10% of women of reproductive-age,4,5 with symptoms such as chronic pelvic pain, dysmenorrhea, and infertility, which often precede the diagnosis of endometriosis by an average of 11 years.3,6,7 Even when diagnosed, there is no current effective and definitive treatment beyond temporizing surgical or medical intervention.
Retrograde menstruation of eutopic endometrial tissues, stem cell origin, coelomic metaplasia, or developmental origins during fetal development have all been hypothesized to be potential causes of endometriosis; however, the precise pathophysiology of endometriosis remains to be understood.8–16 Recently, molecular profiling studies examining the disease from a variety of approaches have offered critical insights into the basic understanding of endometriosis.17–22 Microarray studies have revealed differential gene expression in the eutopic endometrium between endometriotic and normal, between ectopic and eutopic tissues, and comparisons made between the phases of the menstrual cycle.17,23–27 Similarly, epigenetic mechanisms, which often mediate environmental factors,28–31 such as DNA methylation,32–37 histone modification,38–41 long noncoding RNA,22,42 and microRNA,43–49 have also been investigated.
However, there are inconsistencies and a general lack of consensus among studies for specific genes and microRNAs involved in the reported aberrant molecular pathways.17,45 Perhaps this lack of clarity is due to the complex nature of this disease and the diversity of the endometrial tissues used in these studies.17 Specifically, molecular studies often use tissues collected from different phases of the menstrual cycle and predominantly utilize whole endometrial biopsies consisting of mixed populations of endothelial, stromal, and epithelial cells for analyses.25,46,50–52 While informative, it is challenging to resolve the specific impact of the menstrual cycle and how individual cell types within the endometrium contribute to the overall pathophysiology of the disease.53–55 Since endometrium from patients was shown to share similar defects to ectopic endometrium (although there are some differences), the studies of isolated stromal and epithelial cells from the eutopic endometrium could minimize the heterogeneity seen among current studies.23,51
Endometrial stromal and epithelial cells have separate yet important interacting functions. Stromal cells are fibroblast-like cells that differentiate into larger, rounder decidual cells during the secretory phase of the menstrual cycle in order to be receptive and nourish an embryo ready to implant inside the endometrium.56,57 Failure of stromal cells to decidualize properly has consequences such as impaired fertility, placenta accreta, poorly developed placenta, or miscarriage.58–60 Meanwhile, epithelial cells form a protective lining of the lumen interface of the endometrium and glandular ducts and are the point of attachment for an embryo to implant.61 A 2-way paracrine interaction exists between stromal and epithelial cells, in particular, retinoic acid signaling from stromal to epithelial cells, and conversely, the Hedgehog pathway paracrine signaling from epithelial cells to stromal cells that regulates WNT family member 4 (WNT4), a critical protein for decidualization.62–66
In this study, paired eutopic endometrial stromal and epithelial cells from diseased (endometriotic) and normal endometrial biopsies were used to investigate the cell type–specific aberrant molecular pathways involved in the pathogenesis of endometriosis.
Materials and Methods
Tissue Collection
Endometrial tissue was collected, after informed written consent, from normal patients and patients with endometriosis (Supplementary Table 1) under a protocol approved in writing by the Committee on Human Research of the University of California, San Francisco (UCSF). All patients had regular menstrual cycles and were in the proliferative phase. Endometriotic tissue was confirmed by laparoscopy, with histologic evaluation of normal and stage I to IV endometriosis. Staging of endometriosis was defined according to the revised American Fertility Society classification system. Exclusion criteria were tissues from patients with fibroids, endometrial polyps, pregnancy, or any hormonal medications known to interfere with the menstrual cycle either directly on the uterus or centrally for at least 3 months before tissue collection.
Tissue Digestion
Endometrial tissue from more than 29 patients was digested according to established protocol. The fresh tissue was minced and digested with 3.15 mg/mL collagenase I (Worthington Biochemical, Lakewood, New Jersey) and 40 µg/mL deoxyribonuclease I (DNase I; Sigma Aldrich, St Louis, Missouri) in digestion media (Dulbecco’s minimum essential medium [DMEM]/F12 phenol red-free supplemented with 10% fetal bovine serum [FBS]; Gibco, Life Technologies, Carlsbad, California) and 1% antimycotic (penicillin, streptomycin, fungizone [amphotericin B]; Cell Culture Facility, UCSF) on a rotator at 37°C for 1 hour. The digestion was filtered through a 40-μm cell strainer. Both the flow-through fraction and the backwash fraction were collected in 10 mL digestion media and centrifuged. The first fraction pellet was reconstituted and the second fraction incubated with 0.8 mg/mL collagenase II (Worthington) and 40 µg/mL DNase I at 37°C for 20 minutes and centrifuged, after which the pellet was incubated with 2 mL Accutase (Life Technologies) at 37°C for 10 minutes. This fraction was filtered through a 70-μm cell strainer, centrifuged, and the pellet reconstituted.
Fluorescence-Activated Cell Sorting of Human Endometrial Stromal and Epithelial Cells
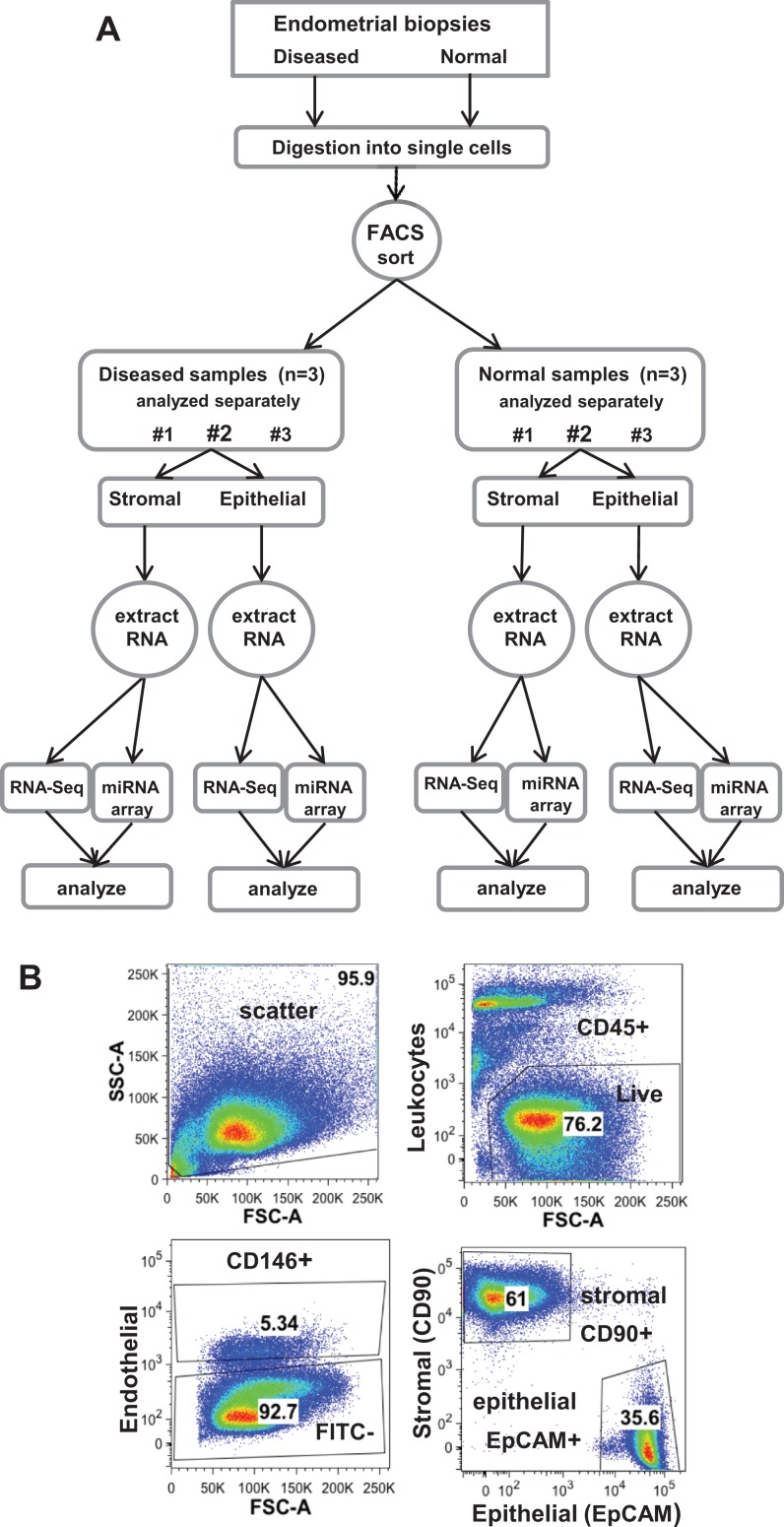
To sort into stromal fibroblast and epithelial cells, digested cells were labeled with antibody staining, incubated for 30 minutes at 37°C, and sorted by fluorescence-activated cell sorting (FACS) AriaII UV, with FACSDiva software (BD Biosciences, San Jose, California; DRC Center Grant [NIH P30 DK063720] Parnassus Flow Cytometry Core, UCSF). Otherwise, all cells were kept on ice during processing and after the FACS sorting. Antibody staining to isolate stromal cells was CD90 (BioLegend, San Diego, California) and epithelial cells was EpCAM PerCP-Cy5.5 (CD326, cat #347199; Becton Dickinson, Franklin Lakes, New Jersey). All cells were labeled with FITC CD146 (cat # 560846; BD Pharmingen, San Jose, California) to remove endothelial cells, Pacific Blue CD45 (cat # 304029; BioLegend) to remove leukocytes, and Sytox Blue (Life Technologies) to remove dead cells. The FACS cell sort was analyzed using FlowJo v9.6 (FlowJo, Ashland, Oregon). (Fluorescence-activated cell sorting selects large quantities of cells purely compared to laser capture of cells).
Isolation of Stromal and Epithelial Cells
To compare diseased versus normal endometrial stromal and epithelial cells, these 2 cell types were isolated from endometrial biopsies from both diseased and normal patients (Figure 1A). Only live nonleukocytic endometrial cells were gated for initial analysis. Subsequently, only enriched stromal fibroblast cells (CD90+) and epithelial cells (EPCAM+) were collected after the exclusion of endothelial cells (Figure 1B). Immunocytochemistry confirmed the purity of stromal and epithelial cells with vimentin staining of stromal cells (C9080; Sigma Aldrich) and pan-cytokeratin of epithelial cells (M0821; DakoCytomation, Santa Clara, California). RNA was collected from the sibling stromal and epithelial population from each patient sample, and each RNA sample was further divided equally for paired RNA and microRNA analyses with RNA sequencing (RNA-Seq) and microRNA microarray, respectively (Figure 1A).
Figure 1.
A, Isolation of stromal and epithelial cells from endometriotic and normal tissue for sibling-paired RNA-Seq and micro-RNA microarray analyses; each sample was independently sorted and analyzed. B, Isolation of stromal fibroblasts (CD90+) and epithelial (EpCAM+) cells by fluorescence-activated cell sorting (FACS) and analyzed by FlowJo v9.6. Only single, CD45−/CD146− live cells were gated for stromal fibroblast and epithelial cell isolation (leukocytes and endothelial cells were excluded). FSC-A indicates forward scatter area; SSC-A, side scatter area.
Cell Culture
Stromal cells were cultured in DMEM/F12 phenol red-free 10% FBS media (Gibco, Life Technologies) with 1% antimycotic (penicillin/streptomycin/fungizone). Epithelial cells were cultured in defined keratinocyte serum-free media supplemented with l-glutamine and initially 28% FBS (Life Technologies). All cells were incubated at humidified 37°C and 5% CO2. Cell cultures of both stromal (n = 4) and epithelial cells (n = 6) were used for transient transfection studies of microRNA and their target genes.
RNA Isolation
Total RNA was isolated for RNA-Seq and microRNA microarray by RNeasy Plus Mini Kit (Qiagen, Valencia, California). High-quality RNA for both assays was confirmed on a Bioanalyzer 2100 (Bioanalyzer; Agilent Technologies, Santa Clara, California), and the samples had a minimum RNA integrity number of 8.8. Otherwise, total RNA for quantitative polymerase chain reaction (qPCR) was isolated by TRI-reagent (Sigma-Aldrich) following the manufacturer’s instructions.
RNA Sequencing Expression Profiling
Endometrial cells from 3 patients with endometriosis and 3 normal participants were sorted by FACS into stromal and epithelial cells (Figure 1A). Cell type–specific diseased and normal samples were processed individually during RNA isolation. Each sample was divided equally for RNA-Seq and microRNA microarray. TopHat and Cufflinks were used to process genome-wide sequencing data and Cuffdiff to obtain values for FPKM (fragments per kilobase of exon per million fragments mapped). Significant messenger RNA (mRNA) differential expression was defined as >2-fold change and Cuffdiff P < .05.
Pathway Analysis of Different Gene Networks in Endometriotic Stromal and Epithelial Cells
Pathway analysis of the differentially expressed genes profiled by RNA-Seq from stromal and epithelial cells was performed using ingenuity pathway analysis (IPA; Qiagen, Redwood City, California, www.qiagen.com/ingenuity). The DAVID Bioinformatics Resource (v6.7) was also used to analyze the stromal and epithelial data sets (Database for Annotation, Visualization and Integrated Discovery. [NIAID] NIH. Huang et al 2009).
microRNA Microarray Expression Profiling
The microarray was conducted on an Affymetrix GeneChip miRNA 3.0 Array (Affymetrix, Santa Clara, California). Unique reads were aligned to human microRNA sequences from miRBaseGv17 (www.mirbase.org). The microarray detected more than 1300 microRNAs (Supplementary Table 2). Significant microRNA differential expression was defined as ≥1.5-fold change and Students t test P ≤ .05.
Real-Time qPCR for mRNA and microRNA
Validation of RNA-Seq and microarray was performed by qPCR of stromal cell differential mRNA (n = 4), microRNA (n = 3), and epithelial mRNA (n = 5), and microRNA (n = 6). Total RNA was converted to complementary DNA (cDNA) by qScript SuperMix (Quanta Biosciences, Gaithersburg, Maryland) for mRNA expression and qScript microRNA cDNA Synthesis (Quanta Biosciences) for microRNA by following the manufacturer’s instructions. FastStart SYBR-Green ROX (Roche Diagnostics, Indianapolis, Indiana) was used for mRNA expression and PerfeCTa SYBR Green Supermix Low-ROX (Quanta Biosciences) used for microRNA expression. Quantitative PCR was performed in an Applied Biosystems ViiA7 real-time PCR system (Life Technologies). Primers used for qPCR are listed in Supplementary Table 3. The ΔΔCT method was used to calculate the relative quantity of transcripts. The reference genes for mRNA qPCR were selected as suitable for 2 different cell types from our previous studies: RNA18S5 for stromal, Glyceraldehyde 3-Phosphate Dehydrogenase (GAPDH) for epithelial cells, and Small Nucleolar RNA C/D box 44 (SNORD44) for microRNA qPCR.
microRNA Target Genes
The selection of predicted mRNA target genes of miR-200b and miR-204 for stromal cells and miR-504 and miR-1827 for epithelial cells from our microarray data was based on DIANA-lab MicroT-CDS (www.diana.imis.athena-innovation.gr/DianaTools/index.php) and TargetScanHuman v6.2 (www.targetscan.org). The predicted targets of microRNAs were matched with our RNA-Seq data set of differentially expressed mRNA (Supplementary Table 4). Transfection conditions for mimics (overexpression) and antagomirs (inhibition) of selected microRNAs (Supplementary Table 5) were 15 nM for mimics, incubated for 48 hours, and 50 nM for antagomirs, incubated for 72 hours. The transfection reagent DharmaFECT 1 (T-2001-03; GE Dharmacon, Layfayette, Colorado) was applied at 1 µL per transfection in 6-well plates. Nontargeting mimic and hairpin inhibitor were negative controls.
Statistical Analysis
Data were analyzed and graphed using GraphPad Prism v6.0.5 for Windows (GraphPad Software, La Jolla, California, www.graphpad.com). Results are expressed as mean (standard deviation). Statistical significance was determined by Student unpaired, 2-tailed t test and for groups of 3 or more by 1-way analysis of variance with Benjamini-Hochberg multiple testing correction for false discovery rate: for RNA-Seq and microarray assays; n ≥ 3 samples for each experiment.
Results
Differential Expression of Genes in Endometriotic Stromal and Epithelial Cells
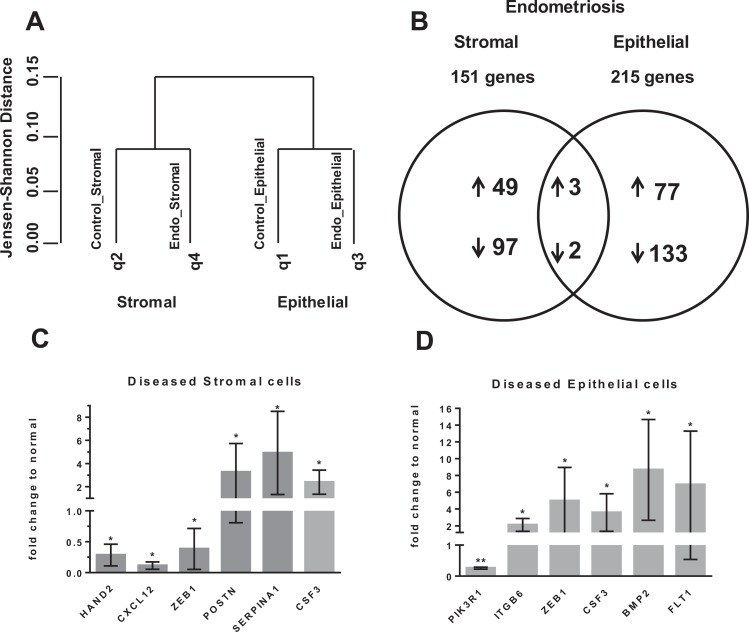
To determine the differential gene expression between endometriotic and normal stromal cells, and similarly for epithelial cells, RNA-Seq was performed on paired sibling samples of stromal and epithelial cells. On average, there were 51 501 530 and 45 364 780 total reads of >23 000 genes from stromal and epithelial samples, respectively. Hierarchical clustering analyses revealed that there were distinct, differentially expressed genes between stromal and epithelial cells, regardless of whether they were endometriotic or normal (Figure 2A). Additionally, distinct clustering was also observed between endometriotic and normal stromal and epithelial endometrial cells (Figure 2A). Endometriotic stromal and epithelial cells had 151 (52 upregulated) and 215 (80 upregulated) differentially expressed genes, respectively (Figure 2B; Table 1). There were only 5 common differentially regulated genes between diseased stromal and epithelial cells (upregulated: Colony Stimulating Factor 3 (CSF3), Serpin family A member 1 (SERPINA1), ASH1 Like histone lysine methyltransferase (ASH1L)67; downregulated: Phosphoinositide-3-Kinase Regulatory subunit 1 (PIK3R1), latent TGFβ1-binding protein 4 (LTBP4); Figure 2B). Zinc finger E-Box Binding homeobox 1 (ZEB1), Heart And Neural crest Derivatives expressed 2 (HAND2), and C-X-C motif Chemokine Ligand 12 (CXCL12) were confirmed to be downregulated, while Periostin (POSTN), SERPINA1, and CSF3 were upregulated in diseased stromal cells by qPCR, consistent with RNA-Seq data (Figure 2C). The qPCR also confirmed upregulation of ZEB1, Integrin subunit beta 6 (ITGB6), CSF3, Bone Morphogenetic Protein 2 (BMP2), and Fms related tyrosine kinase 1 (FLT1) and downregulation of PIK3R1 in diseased epithelial cells (Figure 2D). Interestingly, ZEB1 was downregulated in diseased stromal cells, while it was upregulated in diseased epithelial cells.
Figure 2.
A, Dendrogram of hierarchical clustering analysis (HCA) of messenger RNA (mRNA) data from RNA-Seq; q1-q4 are sample queries. B, Venn diagram of mRNA expression from RNA-Seq of endometriotic compared to normal stromal and epithelial cells; selection by >2-fold change, CuffDiff, P < .05. C, Endometriotic stromal genes from RNA-Seq data set validated by quantitative polymerase chain reaction (qPCR), normalized with 18SRNA, n = 4. D, Endometriotic epithelial genes from RNA-Seq data set validated by PCR, normalized with glyceraldehyde 3-phosphate dehydrogenase (GAPDH), n = 5. Mean (Standard deviation). *P ≤ .05, **P ≤ .01.
Table 1.
Differentially Expressed Genes in Endometriosis From RNA-Seq: 151 Endometriotic Stromal Genes and 215 Epithelial Genes.a
| Stromal Genes | Log2 Change |
|---|---|
| ABCA8 | −2.593 |
| ABCC8 | −3.511 |
| ADAMTS15 | −2.024 |
| ADAMTS5 | −1.818 |
| ADCY1 | −1.983 |
| AHNAK2 | −1.364 |
| ALDH1A3 | −1.548 |
| ALDH1L2 | −1.405 |
| ALPK3 | 2.736 |
| ANKRD13B | −2.648 |
| AP2A1 | −1.517 |
| APBB1IP | −3.899 |
| ASH1L | 2.699 |
| ATP8A2 | −2.044 |
| BCAT1 | −1.321 |
| BMPER | −2.231 |
| BRINP1 | −3.483 |
| BTBD2 | −1.770 |
| BTBD3 | −1.359 |
| C10orf128 | 4.444 |
| C1orf95 | −3.335 |
| C2 | −2.545 |
| C20orf27 | −2.313 |
| C3 | −1.644 |
| CBS | −2.425 |
| CD93 | 1.683 |
| CDH6 | 1.873 |
| CFD | −4.148 |
| CILP | −3.583 |
| CLDN11 | 2.847 |
| CLEC3B | −3.926 |
| COL17A1 | −6.048 |
| COL24A1 | 1.537 |
| COL4A5 | −1.956 |
| COL4A6 | −4.199 |
| COLEC11 | −2.198 |
| CPE | 2.253 |
| CRABP2 | 1.616 |
| CREB5 | 1.143 |
| CSF3 | 2.672 |
| CXCL12 | −1.642 |
| CYP1A1 | 4.355 |
| DEPTOR | −3.093 |
| DIO3 | −3.555 |
| DLL4 | 1.922 |
| DUSP10 | 1.735 |
| EMILIN2 | −1.241 |
| EMR2 | 1.452 |
| EPHA7 | 1.962 |
| ETV5 | 1.466 |
| FAM134B | −1.702 |
| FBLN2 | −1.631 |
| FERMT1 | −2.387 |
| FGF10 | −3.295 |
| FMNL3 | 1.528 |
| FNDC1 | 2.163 |
| FOXF1 | 1.999 |
| G0S2 | 1.782 |
| GABRA2 | −2.857 |
| GALNT13 | −2.554 |
| GFRA2 | 3.334 |
| GLB1L2 | −4.410 |
| GNG4 | −1.477 |
| GPBAR1 | −2.955 |
| GPR116 | 2.633 |
| GPR133 | −2.057 |
| GPR64 | −2.186 |
| GRIA4 | −2.102 |
| H6PD | −1.248 |
| HAND2 | −1.388 |
| HSDL2 | −1.681 |
| HSPB6 | −2.728 |
| IGBP1P1 | 4.444 |
| IGFBP2 | −1.445 |
| IL15 | −2.158 |
| ITGB8 | −1.196 |
| KAL1 | −2.639 |
| KCND3 | −3.827 |
| KCNH1 | −2.149 |
| KCNK3 | −2.210 |
| LAPTM5 | 2.705 |
| LGI2 | −2.298 |
| LIMCH1 | −1.817 |
| LRRC15 | −2.763 |
| LTBP4 | −1.659 |
| LYNX1 | −3.025 |
| MAOB | −1.831 |
| MICAL1 | −1.303 |
| MMP7 | 2.081 |
| MPZL2 | 3.159 |
| MROH6 | −2.577 |
| MTR | −1.293 |
| MYD88 | −1.295 |
| MYLK3 | −1.826 |
| MYOCD | −1.867 |
| NMB | 1.925 |
| NPW | −3.533 |
| NRARP | 2.329 |
| NRG1 | 1.751 |
| NRXN1 | −2.261 |
| OAS2 | −1.956 |
| PAPLN | −1.291 |
| PCDH20 | −3.091 |
| PDE3A | 1.825 |
| PDGFA | 1.847 |
| PDLIM3 | 2.191 |
| PFKFB4 | 2.181 |
| PI15 | 3.574 |
| PIK3R1 | −1.145 |
| PILRA | −3.592 |
| PIM2 | 2.249 |
| PLA2G7 | −2.310 |
| PLCB1 | 3.325 |
| PLCD1 | −2.120 |
| PLCL1 | −1.446 |
| PLXND1 | −1.507 |
| POSTN | 1.973 |
| PPM1H | −1.617 |
| PPP1R14A | −2.849 |
| PPP4R4 | 3.947 |
| PROK1 | −5.321 |
| PTCH1 | −1.619 |
| PTGDS | −1.830 |
| PTPRB | 1.853 |
| PYGL | −1.741 |
| RAPGEF4 | 2.370 |
| RASD1 | −1.462 |
| RASGRP3 | 2.266 |
| REXO1 | 3.576 |
| RPRM | −3.173 |
| S100A7 | 4.444 |
| SCARA5 | −6.773 |
| SDK1 | −1.382 |
| SELE | 1.910 |
| SERPINA1 | 3.799 |
| SHB | 1.492 |
| SLC2A8 | −2.747 |
| SLC40A1 | −1.515 |
| SMIM3 | 1.509 |
| SPATA18 | −2.621 |
| STEAP1 | 2.559 |
| SULF2 | −1.292 |
| TMEM132B | −1.692 |
| TMEM196 | −3.447 |
| TMEM37 | −2.290 |
| UBA7 | −5.969 |
| UCN2 | 1.774 |
| WNT4 | −2.081 |
| XIRP1 | 2.145 |
| ZC3H12C | 1.229 |
| ZNF703 | −1.630 |
| Epithelial genes | Log2 Change |
| ABCA6 | −2.541 |
| ABCD1 | −2.951 |
| ABL2 | 1.543 |
| ACPP | −2.450 |
| ACTG2 | −3.924 |
| ADAMTS1 | −1.221 |
| ADAMTS8 | −6.935 |
| AKR1C2 | 3.146 |
| ALDH3B2 | −4.806 |
| ALOX5 | 1.839 |
| ALPL | −1.593 |
| ANO6 | 1.160 |
| AOX1 | −2.827 |
| AQP9 | 2.792 |
| ARHGAP15 | −4.444 |
| ARNTL2 | 1.206 |
| ASH1L | 2.792 |
| ASS1 | 1.359 |
| ATP10B | −4.313 |
| ATP6V0E2 | −2.908 |
| AXL | 2.646 |
| BMP2 | 3.687 |
| C14orf183 | 4.444 |
| C1orf64 | −4.444 |
| C2orf54 | 2.703 |
| CABYR | −2.156 |
| CAMP | −4.444 |
| CAPN14 | 2.627 |
| CAPN6 | −2.525 |
| CBX4 | −1.776 |
| CCBL1 | −3.111 |
| CCDC144B | 1.692 |
| CD36 | −4.850 |
| CD40 | 3.180 |
| CDK6 | 2.611 |
| CENPF | −2.349 |
| CES4A | −2.668 |
| CHST10 | −2.045 |
| CIC | −1.832 |
| CLGN | −1.896 |
| CMYA5 | −2.098 |
| COL1A2 | −3.050 |
| COL27A1 | 2.500 |
| COL6A2 | 3.924 |
| COL6A3 | 2.396 |
| CREB3L1 | −2.739 |
| CSF3 | 2.628 |
| CST6 | 2.768 |
| CXCL5 | 2.128 |
| CXorf27 | 4.444 |
| CYP26A1 | −4.647 |
| DFNB31 | −1.719 |
| DLX5 | −1.397 |
| DNAJA4 | −1.319 |
| DPP4 | −1.609 |
| DPYSL3 | 2.766 |
| DUOX1 | −2.608 |
| DUOXA1 | −2.589 |
| DYNC1I1 | −2.271 |
| ECI2 | −2.016 |
| EDNRB | −2.450 |
| EEF2K | −1.403 |
| ELK3 | 1.259 |
| ELMSAN1 | −1.384 |
| ELP3 | −2.000 |
| EML5 | −2.105 |
| EMP1 | 1.247 |
| EMP3 | 2.144 |
| EPN3 | −3.353 |
| ETV1 | −2.130 |
| FAM129A | 1.802 |
| FAM171B | −2.418 |
| FAM172A | 3.709 |
| FGB | 2.721 |
| FGF2 | 2.273 |
| FILIP1 | −3.850 |
| FJX1 | 2.023 |
| FLT1 | 2.257 |
| FMOD | 2.986 |
| FN1 | 2.144 |
| GCNT1 | 1.529 |
| GJB4 | 3.413 |
| GLYATL2 | −5.294 |
| GLYATL3 | −4.444 |
| GMPR | −2.515 |
| GPR141 | 4.444 |
| GRAMD1C | −1.629 |
| GREM2 | −2.130 |
| HEY1 | −2.369 |
| HGD | −2.045 |
| HPGD | −1.925 |
| HS6ST1 | −1.659 |
| HSD11B2 | −2.567 |
| HSD17B2 | −2.184 |
| HSPA6 | −1.878 |
| HYAL1 | −2.535 |
| IDH1 | −2.126 |
| IGF2 | 2.123 |
| IGFBP5 | 2.024 |
| IL1B | 2.942 |
| IL1R2 | 2.076 |
| IL1RN | 1.675 |
| IL20RA | −2.481 |
| ITGB6 | 1.352 |
| KAT2B | −1.876 |
| KCTD12 | 2.115 |
| KLF2 | −2.404 |
| KRT13 | 2.134 |
| KRT17 | 1.444 |
| KRT7 | 1.294 |
| LSAMP | 2.916 |
| LTBP4 | −2.107 |
| MAOA | −1.668 |
| MAP2K6 | −2.859 |
| MARCKS | 1.322 |
| MESP1 | −4.444 |
| MMP2 | 2.872 |
| MMP26 | −1.956 |
| MSX1 | −1.273 |
| MT1G | −2.416 |
| MT1H | −3.334 |
| MT1M | −2.949 |
| MTSS1 | −2.023 |
| MUC5B | 3.391 |
| MXRA5 | 1.560 |
| NAV3 | 2.895 |
| NDC80 | −3.144 |
| NEK6 | −1.323 |
| NLRP5 | −4.444 |
| NME4 | −1.641 |
| NOS2 | 4.861 |
| NPR3 | −4.013 |
| NQO1 | −1.331 |
| NRP2 | 2.272 |
| OGDHL | −2.263 |
| OPRK1 | −2.528 |
| PAPSS1 | −1.229 |
| PAX2 | −1.976 |
| PCSK5 | 1.925 |
| PEBP4 | −4.444 |
| PER1 | −1.715 |
| PIK3R1 | −1.931 |
| PIK3R2 | −1.558 |
| PKHD1L1 | −2.633 |
| PLLP | −1.501 |
| PLXNA4 | −4.199 |
| PRICKLE1 | −2.119 |
| PROL1 | −4.444 |
| PSG9 | −5.337 |
| PTGS2 | 1.792 |
| RAB20 | −2.004 |
| RAB31 | 1.925 |
| RAP1GAP | −2.171 |
| RBP1 | 1.920 |
| RNASE1 | −3.146 |
| RYR3 | −2.010 |
| SAPCD2 | −2.872 |
| SCGB1D2 | −2.090 |
| SCGB1D4 | −4.296 |
| SCNN1B | 2.356 |
| SERPINA1 | 1.201 |
| SERPINA5 | −3.252 |
| SERPINB2 | 5.797 |
| SERPINB5 | 3.364 |
| SERPINE2 | 2.326 |
| SFTPA2 | −4.444 |
| SHISA2 | 2.114 |
| SHISA8 | −5.245 |
| SLC15A2 | −2.851 |
| SLC25A1 | −1.564 |
| SLC25A15 | −3.775 |
| SLC25A38 | −1.395 |
| SLC25A48 | −4.444 |
| SLC26A9 | 2.495 |
| SLC39A8 | −2.066 |
| SLC43A1 | −3.262 |
| SLC46A2 | −4.623 |
| SLC47A1 | 2.041 |
| SLC6A14 | 2.618 |
| SLC7A4 | −3.340 |
| SLC7A8 | −1.826 |
| SLCO2A1 | 2.901 |
| SMAD9 | −1.745 |
| SPARC | 2.106 |
| SPDEF | −2.606 |
| SPOCD1 | 2.612 |
| SPOCK1 | 1.774 |
| SPON1 | −1.533 |
| SQLE | −1.358 |
| ST3GAL3 | −2.045 |
| ST6GALNAC1 | −2.143 |
| STEAP4 | −1.486 |
| STRA6 | 3.899 |
| SUFU | −2.606 |
| TCEA3 | −3.842 |
| TEX36 | 4.444 |
| TGFBI | 4.850 |
| TGM2 | 1.806 |
| THEM4 | −1.444 |
| TMEM101 | −1.617 |
| TMEM132C | −2.257 |
| TMEM252 | −3.294 |
| TMEM42 | 2.413 |
| TOP2A | −2.279 |
| TPD52L1 | −1.420 |
| TRPC6 | −2.375 |
| TRPM8 | −2.202 |
| TSPAN12 | −1.461 |
| TTLL1 | −3.297 |
| UBAP2 | 3.475 |
| USP54 | −1.223 |
| WFS1 | −1.482 |
| ZEB1 | 2.103 |
| ZNF319 | −2.254 |
| ZNF589 | −1.718 |
aSelection by >2-fold change, P < .05.
Differential Expression of microRNAs in Endometriotic Stromal and Epithelial Cells
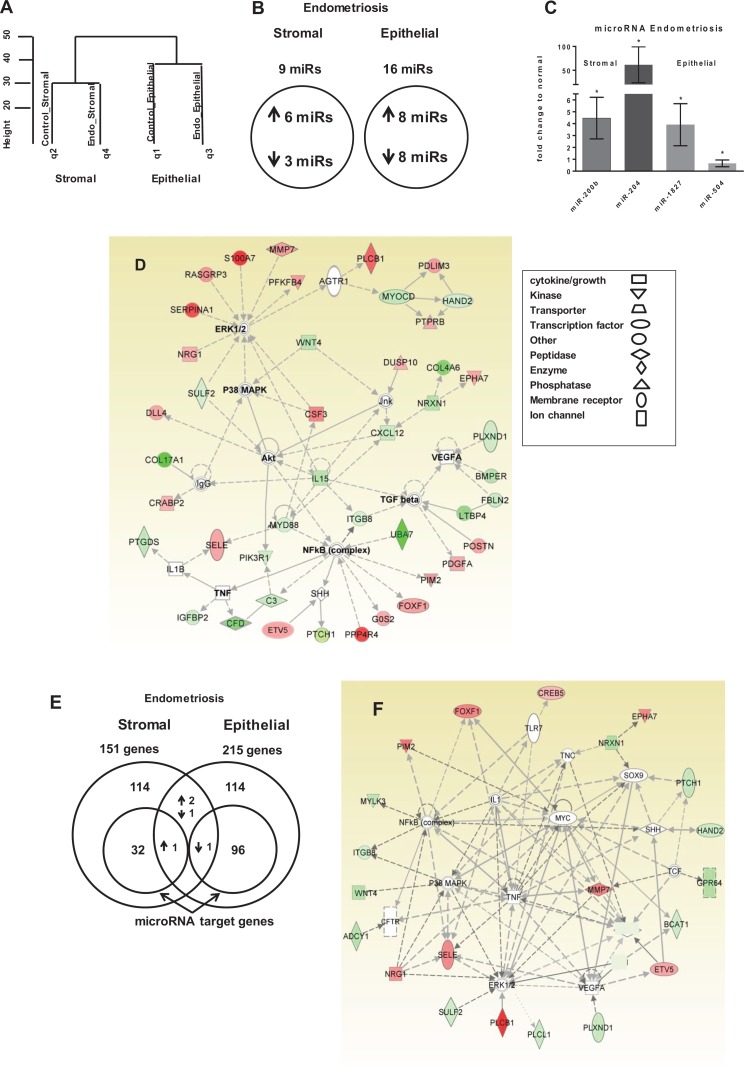
To determine the specific microRNA expression between diseased and normal stromal and epithelial cells, the same RNA samples that were used for RNA-Seq were also used for microRNA microarray for paired microRNA–mRNA analyses. Hierarchical clustering of these data revealed that differentially expressed microRNAs in stromal cells were distinct from microRNAs in epithelial cells and that differentially expressed microRNAs in endometriotic samples were distinct from microRNAs in normal samples (Figure 3A). Compared to normal samples, there were 9 (6 upregulated) and 16 (8 upregulated) differentially expressed microRNAs in diseased stromal cells and epithelial cells, respectively (Figure 3B; Table 2). Specifically, miR-200b and miR-204 were confirmed to be upregulated in diseased stromal cells, while miR-1827 was confirmed to be upregulated and miR-504 downregulated in diseased epithelial cells by qPCR (Figure 3C).
Figure 3.
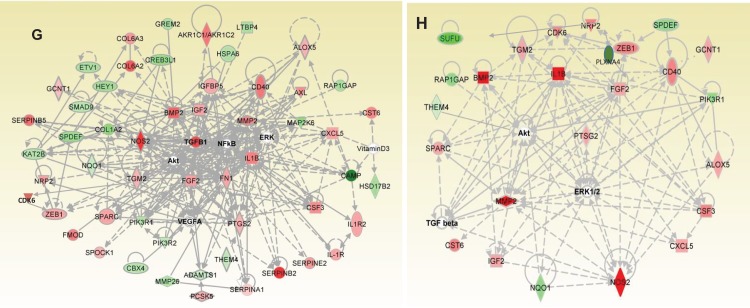
A, Dendrogram of hierarchical clustering analysis of microRNA data from microRNA microarray, q1 to q4 are sample queries. B, Venn diagram of microRNA expression from microRNA microarray of endometriotic stromal and epithelial cells; selection by >1.5-fold change, P ≤ .05. C, MicroRNAs from microRNA microarray of endometriotic stromal (n = 3) and epithelial cells (n = 6) validated by quantitative polymerase chain reaction (qPCR), normalized with SNORD44. D, Ingenuity pathway analysis (IPA) of Akt/ERK/P38MAPK/TNF/NF-κB network of endometriotic stromal messenger RNA (mRNA). E, Venn diagram of mRNA expression from RNA-Seq of endometriotic stromal and epithelial genes with subsets of predicted microRNA target genes. F, The IPA analysis of microRNA predicted target gene network in endometriotic stromal cells. G, The IPA analysis of mRNA transforming growth factor β1 (TGFβ1)/interleukin 1B (IL-1B) network in endometriotic epithelial cells. H, The IPA analysis of microRNA predicted target gene network in endometriotic epithelial cells. Shaded genes are statistically significant. Shading intensity of a gene corresponds with the degree of up- or downregulation in endometriosis. Red-colored genes are upregulated in endometriosis and green-colored genes are downregulated. Mean (standard deviation). *P ≤ .05.
Table 2.
microRNA Differential Expression in Endometriotic Stromal and Epithelial Cells.a
| Stromal | Epithelial | ||
|---|---|---|---|
| Up | Fold Change | Up | Fold Change |
| miR-200b | 2.93 | miR-936 | 1.57 |
| miR-204 | 2.19 | miR-1827 | 1.98 |
| miR-629 | 1.77 | miR-3141 | 3.76 |
| miR-3926 | 1.49 | miR-3938 | 1.51 |
| miR-3928 | 1.69 | miR-4266 | 1.59 |
| miR-4670 | 1.66 | miR-4450 | 1.83 |
| miR-4522 | 1.64 | ||
| miR-4792 | 2.27 | ||
| Down | Down | ||
| miR-548al | 0.28 | miR-375 | 0.21 |
| miR-1297 | 0.52 | miR-504 | 0.53 |
| miR-4795 | 0.57 | miR-548n | 0.54 |
| miR-601 | 0.61 | ||
| miR-603 | 0.56 | ||
| miR-3136 | 0.51 | ||
| miR-3176 | 0.59 | ||
| miR-4476 | 0.49 | ||
aSelection by >1.5-fold change, P ≤ .05.
Aberrant Pathways Involved in Endometriosis
Potential molecular pathways involved in both endometriotic stromal and epithelial cells were evaluated by IPA software and by DAVID Bioinformatics (Table 3). Based on the differentially expressed genes between the endometriotic and the normal stromal populations, IPA identified 44 genes involved in the Protein Kinase B (Akt)/Extracellular signal-Regulated Kinases (ERK)/Mitogen Activated Protein Kinase 14 (P38MAPK)/Tumor Necrosis Factor (TNF)/Nuclear Factor kappa B (NF-κB) decidualization pathways, such as WNT4, HAND2, POSTN, Interleukin 15 (IL-15), Complement Factor D (CFD, SERPINA1, and Matrix Metallopeptidase 7 (MMP7) (Figure 3D). The IPA identified the differentially expressed genes within the diseased stromal population that function to interfere with normal immune cell trafficking (z = −2.190), with myeloid differentiation primary response 88 (MYD88) as the master upstream regulator together with IL-15, Complement C3 (C3), Plexin D1 (PLXND1), and CXCL12. Additionally, the differentially expressed MMP7, POSTN, SERPINA1, and Insulin like Growth Factor Binding Protein 2 (IGFBP2) genes also function to modulate cell migration (2.186; Table 3). DAVID classified the main altered functional groups of stromal cells into signal (64 genes), extracellular (29 genes), cell adhesion (17 genes), vasculature development (12 genes), and neuropeptide signal (7 genes) (Table 3; Supplementary Table 6). Diseased stromal cells have 5 differentially expressed microRNAs that have 33 predicted target mRNAs within the same differentially regulated genes found in RNA-Seq (Supplementary Table 4), represented as a subset in the Venn diagram (Figure 3E). The predicted TNF/ERK/p38MAPK/NF-κB pathways based on the differentially expressed microRNAs and their target mRNAs in endometriotic stromal cells (Figure 3F) suggests that upstream microRNA regulation plays an important role in impaired decidualization, with key genes Myosin Light chain Kinase 3 (MYLK3), ITGB8, Patched 1 (PTCH1), and WNT4 being downregulated. WNT4 regulates the critical Forkhead box O1 (FOXO1) transcription factor via the canonical β-catenin signaling pathway.66
Table 3.
Network and Gene Analysis of Differentially Expressed mRNA, from RNA-Seq of Endometriotic Stromal and Epithelial Samples, by Ingenuity Pathway Analysis (IPA) and DAVID.
| Ingenuity Pathway Analysis | ||||
|---|---|---|---|---|
| z Score | Function | Genes | ||
| Stromal | ||||
| −2.190 | Immune cell trafficking decrease | MYD88 upstream | ||
| −2.366 | Antigen-presenting cell movement decrease | C3, CXCL12, IL15, MYD88, PLXND1, PTGDS, SELE | ||
| −2.000 | Mast cell migration decrease | C3, CXCL12, IL15, PIK3R1 | ||
| −2.216 | Macrophage cell movement decrease | C3, CXCL12, IL15, PLXND1, PTGDS | ||
| −2.177 | Phagocyte maturation decrease | CSF3, IL-15, MYD88, PIK3R1, SERPINA1 | ||
| −2.219 | Inflammatory disease decrease | C3, CXCL12, DLL4, MYD88, PIK3R1 | ||
| 2.321 | Cell morphology increases cell size | CBS, DEPTOR, PIM2, PLCB1, UCN2, IGFBP2 | ||
| 2.186 | Muscle cell migration increase | IGFBP2, MMP7, MYOCD, POSTN, SERPINA1 | ||
| Epithelial | ||||
| 2.268 | Cancer increase | TGFβ1 upstream | ||
| 2.537 | Tumor cell proliferation increase | AXL, CST6, IL1RN, MMP2, NOS2, PTGS2 (COX2), ZEB1 | ||
| 2.420 | Vascularization increase | ADAMTS1, ALOX5, FGF2, IGF2, IL1B, NOS2, NRP2, SPARC | ||
| 2.803 | Cell migration increase | ALOX5, AXL, CDK6, CHST10, CXCL5, FGF2, FMOD, GCNT1, HEY1, IGF2, IL1B, NOS2, NQO1, NRP2, RAP1GAP, SPARC, SPDEF, TGM2 | ||
| 2.608 | Leukocyte migration increase | ALOX5, AQP9, CXCL5, FGF2, GCNT1, IGF2, IL1B, NOS2, NQO1, NRP2, RAP1GAP, SPARC, SPDEF, TGM2 | ||
| DAVID | ||||
| Function | Gene Number | Enrichment | P Value | Benjamini |
| Stromal | 150 genes | |||
| Signal | 64 | 8.34 | 2.3 × 10−13 | 5.9 × 10−11 |
| Extracellular region | 29 | 5.03 | 4.9 × 10−8 | 4.6 × 10−6 |
| Cell adhesion | 17 | 3.54 | 4.5 × 10−4 | 7.4 × 10−2 |
| Vasculature development | 12 | 2.66 | 1.4 × 10−5 | 4.2 × 10−3 |
| EGF-like region | 10 | 2.17 | 7.5 × 10−4 | 1.5 × 10−1 |
| Neuropeptide signal | 7 | 1.88 | 1.7 × 10−4 | 4.1 × 10−2 |
| Carbohydrate binding | 11 | 1.83 | 7.2 × 10−4 | 1.2 × 10−1 |
| Hormone regulation | 8 | 1.78 | 3.9 × 10−4 | 7.4 × 10−2 |
| Epithelial | 213 genes | |||
| Extracellular | 35 | 5.41 | 8.1 × 10−8 | 1.9 × 10−5 |
| Signal | 48 | 5.37 | 1.6 × 10−9 | 5.6 × 10−7 |
| Vasculature development | 11 | 2.44 | 1.1 × 10−3 | 1.3 × 10−1 |
| Oxidoreductase | 19 | 2.27 | 4.7 × 10−5 | 3.3 × 10−3 |
Of the 215 differentially regulated genes from the diseased epithelial cells, there were 87 genes identified from the transforming growth factor β1 (TGFβ1) family, involved in the TGFβ1/interleukin 1B (IL-1B) pro-inflammatory pathways (Figure 3G). Additionally, IPA identified an overall increase associated with cancer development (z = 2.268), with TGFβ1 as an upstream regulator, which encompassed tumor cell proliferation (2.537), leukocyte migration (2.608), cell migration (2.803), and vascularization (2.420; Table 3). The critical genes TGFβ1, ZEB1, BMP2, MMP2, and Prostaglandin-endoperoxide Synthase 2 (PTGS2, COX2), which were all upregulated in RNA-Seq, activate the epithelial–mesenchymal transition (EMT) pathway (Table 1). DAVID classified the 215 differentially regulated genes into the main functional gene groups of signal, (48 genes) extracellular, (35 genes) and vasculature development, (11 genes) which is similar to stromal cells but with a different set of genes (Table 3; Supplementary Table 6). The differentially expressed microRNAs in the diseased epithelial cells shared 97 predicted target genes found to be differentially expressed based on RNA-Seq analyses of the diseased epithelial cells (Supplementary Table 4). Of these 97 target genes, 24 target genes represent the main functional groups from the IPA analysis of the TGFβ1/IL-1B/Akt/ERK pathway (Figure 3H).
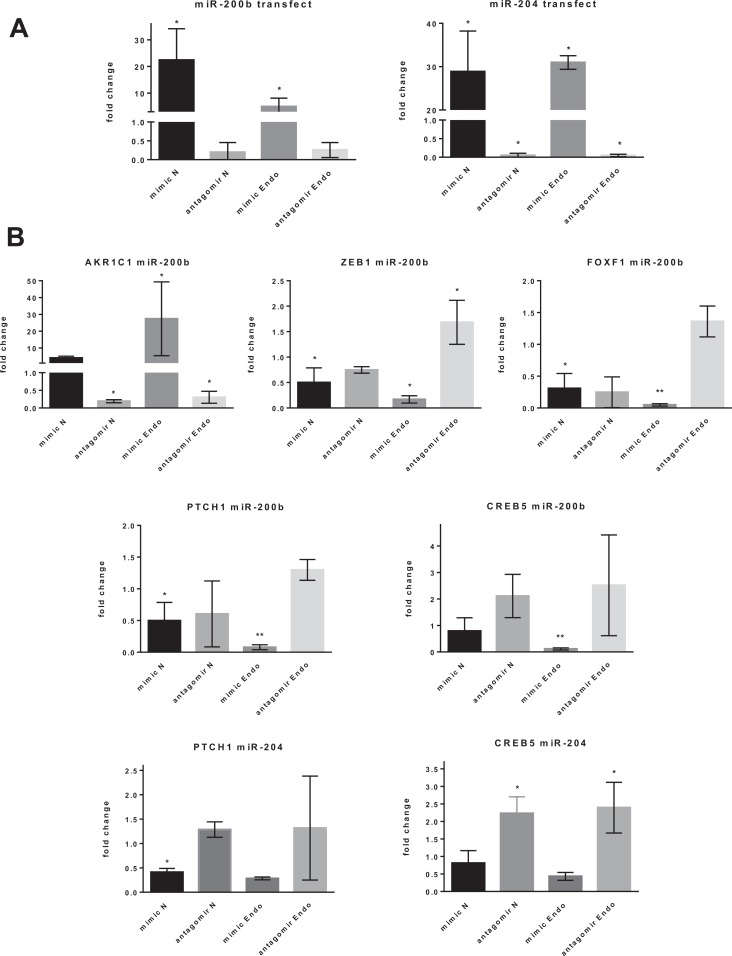
Target Genes of miR-200b and miR-204 in Stromal Cells
To investigate and confirm the target genes of differentially expressed microRNAs involved in the aberrant pathways described, the levels of miR-200b and miR-204 within the diseased and normal stromal cells were modulated with specific mimics and antagomirs by transient transfection. The qPCR confirmed that mimics and antagomirs could modulate the levels of miR-200b and miR-204 within the stromal cells in vitro as expected (Figure 4A). Upregulation of miR-200b by mimic downregulated the expression of target genes ZEB1, PTCH1, Forkhead box F1 (FOXF1), and CAMP Responsive Element Binding protein 5 (CREB5); however, the sequestration of miR-200b through antagomir upregulated only ZEB1 (Figure 4B). Conversely, Aldo-Ketose Reductase family 1 member C1 (AKR1C1) was upregulated by miR-200b mimic and downregulated by miR-200b antagomir. The miR-204 mimic and antagomir downregulated PTCH1 and upregulated CREB5 expression, respectively (Figure 4B).
Figure 4.
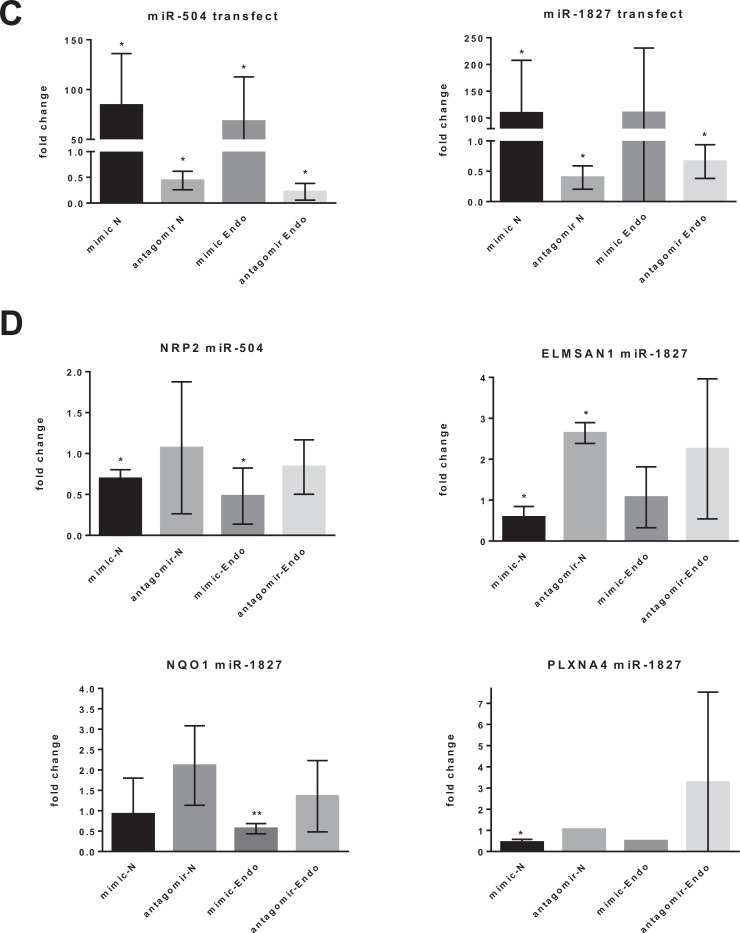
A, miR-200b and miR-204 expression after transfection of mimics and antagomirs into endometriotic and normal stromal cells validated by quantitative polymerase chain reaction (qPCR), normalized with SNORD44. B, miR-200b and miR-204 target gene expression analyzed by qPCR for endometriotic compared to normal stromal cells, normalized with 18SRNA, n = 4. C, miR-504 and miR-1827 expression after transfection of mimics and antagomirs into endometriotic and normal epithelial cells validated by qPCR, normalized with SNORD44. D, miR-504 and miR-1827 target gene expression analyzed by qPCR for endometriotic compared to normal epithelial cells, normalized with glyceraldehyde 3-phosphate dehydrogenase (GAPDH), n = 6; Mean (standard deviation). *P ≤ .05, **P ≤ .01. antagomir Endo indicates antagomir endometriosis; antagomir N, antagomir normal; mimic N, mimic normal; mimic Endo, mimic endometriosis; AKR1C1, Aldo-Keto Reductase family 1 member C1; CREB5, CAMP Responsive Element Binding Protein 5; ELMSAN1, ELM2 And Myb/SANT Domain containing 1; FOXF1, Forkhead box F1; NQO1, NAD(P)H Quinone dehydrogenase 1; NRP2, Neuropilin 2; PLXNA4, Plexin A4; PTCH1, Patched 1; ZEB1, Zinc Finger E-Box Binding homeobox 1.
Target Genes of miR-504 and miR-1827 in Epithelial Cells
Similarly, miR-504 and miR-1827 mimics and antagomirs were transiently transfected into diseased and normal epithelial cells, and qPCR confirmed their altered expression following modulation (Figure 4C). Selected predicted target genes of miR-504 and miR-1827 in the epithelial cell population were analyzed (Figure 4D). The miR-504 mimic downregulated Neuropilin 2 (NRP2) in endometriotic and normal epithelial cells. The miR-1827 mimic downregulated ELM2 And Myb/SANT domain containing 1 (ELMSAN1) and PLXNA4 in normal epithelial cells and NAD(P)H Quinone dehydrogenase 1 (NQO1) in endometriotic epithelial cells. Conversely, miR-1827 antagomir upregulated ELMSAN1 in normal epithelial cells.
Discussion
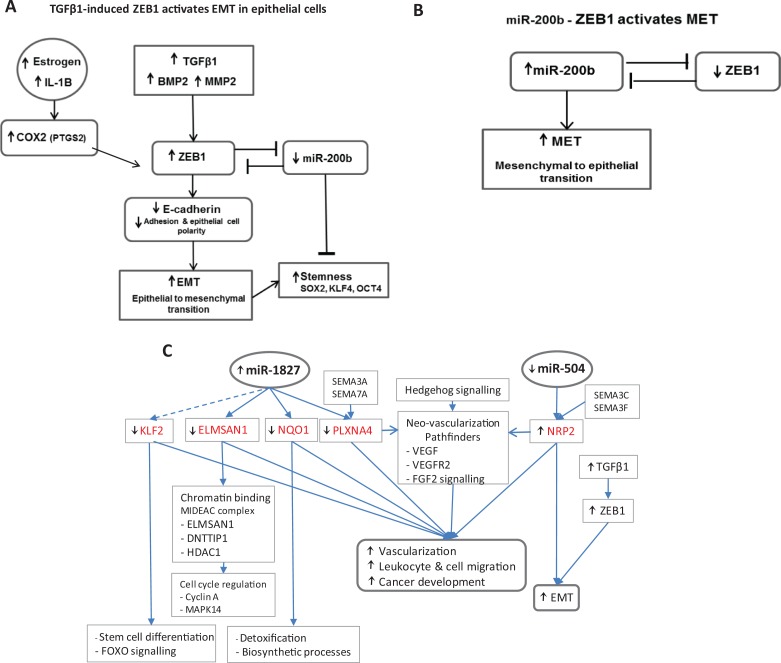
This study demonstrates that endometriotic stromal and epithelial cells are associated with uniquely aberrant molecular pathways that were regulated by microRNAs. In endometriotic stromal cells, these associated aberrant pathways interfere with immune cell trafficking and impair decidualization through the Akt/ERK/P38MAPK/TNF/NF-κB pathway, and specific target gene members were modulated by miR-200b and miR-204. In contrast, dysregulation of the IL-1B/Akt/ERK/NF-κB/Vascular Endothelial Growth Factor (VEGF) and TGFβ1/BMP2/MMP2/COX2/ZEB1 pathways associated with endometriotic epithelial cells may induce inflammation and potential activation of EMT, respectively (Figure 5A).
Figure 5.
A, Transforming growth factor β1 (TGFβ1)/ZEB1/miR-200b feedback loop for endometriotic epithelial cells in EMT. B, miR-200b/ZEB1 double-negative feedback loop for endometriotic stromal cells in MET. C, miR-1827/miR-504 model of target genes and pathways in endometriotic epithelial cells. BMP2 indicates Bone Morphogenetic Protein 2; COX2 (PTGS2), Prostaglandin-endoperoxide Synthase 2; DNTTIP1, deoxynucleotidyltransferase terminal interacting protein 1; ELMSAN1, (MIDEAS) ELM2 and Myb/SANT domain containing 1; EMT, epithelial–mesenchymal transition; FGF2, Fibroblast Growth Factor 2; FOXO, Forkhead Box; HDAC1, histone deacetylase 1; IL-1B, Interleukin 1B; KLF2/4, Kruppel Like Factor 2/4; MET, mesenchymal–epithelial transition; MMP2, Matrix Metallopeptidase 2; NQO1, NAD(P)H Quinone dehydrogenase 1; NRP2, neuropilin 2; OCT4 (POU5F1), POU class 5 homeobox 1; PLXNA4, plexin A4; SEMA, semaphorin; SOX2, SRY-box 2; MMP2, Matrix Metallopeptidase 2; TGFβ1, transforming growth factor beta 1; VEGF, Vascular Endothelial Growth Factor; ZEB1, Zinc finger E-box Binding protein 2.
Impaired decidualization and aberrant immune cell trafficking of stromal cells in endometriosis often result in infertility or poor embryo implantation.68–70 Diseased stromal cells potentially modulate immune cell trafficking mainly by downregulation of the cytokines IL-15, CXCL12 and the mediator of IL-1, MYD88, that decrease movement of mast and antigen-presenting dendritic and macrophage cells (Table 3).71–73 Decreased immune cell trafficking has 2 consequences, first, reduced inflammatory response to infectious pathogens, such as bacteria,74 and second, the reduced influx of dendritic, uterine natural killer (uNK), mast, and macrophage cells leads to several reproductive disorders, such as embryo implantation failure, recurrent miscarriages, and preeclampsia.69,75 Interleukin 15 both responds to infectious pathogens and stimulates proliferation and differentiation of uNK cells in the endometrium during decidualization.76,77 Upon decidualization, dendritic and uNK cells crucially remodel endometrial spiral arteries ready for trophoblast invasion that will convert the arteries into wide-diameter, high-volume blood vessels for placental development.78,79 Decidual cells act as a physical barrier to prevent the excessive invasion of the columns of extravillous trophoblast cells.59,80,81 However, if decidualization is impaired, then trophoblasts will invade too deeply, and the life-threatening disorders of choriocarcinoma or placenta accreta may occur.58,60,82
Key decidualization genes were found to be aberrantly expressed and may contribute to impaired decidualization in endometriotic stromal cells, such as upregulation of POSTN, SERPINA1, MMP7, and miR-200b and downregulation of HAND2, IL-15, ZEB1, and WNT4 (Figure 2C).66,83,84 Although the stromal cells analyzed came from the proliferative phase, it has been demonstrated by other studies that aberrant expression of key decidualization genes in the proliferative phase, such as POSTN, FK506 Binding Protein 4 (FKBP4), progesterone receptor chaperone, Corticotropin Releasing Hormone (CRH), and Urocortin (UCN), have persisted into the secretory phase and adversely affect decidualization by delaying the phase transition due to progesterone resistance of endometriosis.24,84–87 During decidualization, the miR-200 family is downregulated; however, miR-200b is upregulated in endometriotic stromal cells during the proliferative phase (Figure 3C), potentially affecting decidualization by downregulating CREB5, Sulfatase 2 (SULF2), and ZEB1, which are normally upregulated during decidualization.84,88 CREB5 is a member of the important cyclic adenosine monophoshate/protein kinase A induction pathway of decidualization.89 Impaired decidualization presages that the embryo will have difficulty implanting into the endometrium and, if successful, may result in poor placental development or miscarriage.90,91 A poorly developed placenta can lead to preeclampsia, preterm birth, or intrauterine growth restriction.82,92,93
Interestingly, in endometriotic stromal cells, the downregulation of ZEB1 through aberrant upregulation of miR200b (Figure 4B)94–97 potentially activates a mesenchymal–epithelial transition (MET) pathway by a double-negative feedback loop.98–100 Double-negative feedback loops consist of 2 molecular interactions that reciprocally repress each other, depending on their relative levels, and act as a reversible molecular switch that can maintain bistable cellular states, for example, in this case, either stromal or epithelial cells, by switching between either EMT or MET depending on the prevailing molecular factors. Activation of MET has been demonstrated by Wang et al to mediate early-stage induction of fibroblast dedifferentiation into induced pluripotent stem cells (Figure 5B).101 It is not clear what properties of MET-transformed stromal cells contribute to endometriosis other than, perhaps, being prone to form lesions ectopically.102
Progesterone-resistant dysregulation of decidualization has been shown to play a significant role in the pathogenesis of endometriosis.50,103–105 Increased POSTN upregulates Akt (protein kinase B), which downregulates the progesterone receptors (PRA/B), so that increased Akt activation in endometriosis inhibits decidualization.106,107 Decreased HAND2 in endometriotic stromal cells, demonstrated here, may impact proper metabolism of estradiol and instead downregulate critical genes for decidualization of IL-15, TIMP Metallopeptidase Inhibitor 3 (TIMP3), and FOXO1. 108 Progesterone resistance is also due to aberrant expression of steroid biosynthesis enzymes in endometriotic stromal and epithelial cells.63,105 The aldo-keto reductases AKR1C1/2/3 are progesterone and steroid metabolizers that are upregulated in endometriosis.109 Increased AKR1C1 (20α-hydroxysteroid dehydrogenase) inactivates progesterone, and in stromal cells, miR-200b overexpression upregulated AKR1C1 (Figure 4B). AKR1C2 inactivates potent 5-alpha-Dihydrotestosterone (5α-DHT) and is upregulated in endometriotic epithelial cells (Table 1).109 Other metabolizing enzymes, such as 17β-hydroxysteroid dehydrogenase 2 (HSD17B2), metabolize estradiol to weakly estrogenic estrone and HSD11B2, which inactivates cortisol. HSD17B2 and HSD11B2 were both downregulated in endometriotic epithelial cells (Table 1).63 HSD17B2 in epithelial cells is regulated by stromal paracrine factors, especially retinoic acid, which in normal stromal cells is upregulated by progesterone.65,110 However, in endometriotic stromal cells, progesterone resistance disrupts the paracrine retinoic acid signaling and downregulates HSD17B2 in epithelial cells so that estradiol is not properly metabolized and abnormally accumulates.63,65,106
Progesterone resistance from stromal cells also disrupts the 2-way paracrine interaction between stromal and epithelial cells.64,111 Decreased progesterone signaling in the stromal cells downregulates Indian Hedgehog (IHH) in epithelial cells, which is downregulated in endometriosis.64,112,113 Not only is IHH downregulated in diseased epithelial cells with decreased signal to stromal cells, but also the Hedgehog receptor, PTCH1, in stromal cells, is downregulated by miR-200b and miR-204 (Figure 4B). Hedgehog signaling in stromal cells also involves CREB5 and FOXF1, the target genes of miR-200b and miR-204, and NRP2, the target gene of miR-504 in epithelial cells.114,115 The Hedgehog pathway and miR-204 in endometriotic stromal cells separately downregulate WNT4, essential for decidualization.66 So the dysregulated Hedgehog signaling pathway from endometriotic epithelial cells impairs decidualization in stromal cells.62
Furthermore, in diseased epithelial cells, inflammation-related genes are upregulated, such as TGFβ1, IL-1B, BMP2, PTGS2 (COX2), SERPINA1, and CSF3. 116 (SERPINA1 and CSF3 are in common with diseased stromal cells as well as downregulated LTBP4, which releases increased TGFβ1.) Increased TGFβ1 expression activates the ZEB1 molecular pathway to EMT and may increase pluripotency and stemness (Figure 5A) as well as increase the homing of mononuclear leukocytes (Table 3).94,117–121 An increased expression of TGFβ1, MMP2, COX2, and BMP2 upregulates ZEB1.122–124 ZEB1 overexpression promotes metastasis in cancer and marks aggressive endometrial carcinoma.117,125–127 ZEB1 activates EMT by repressing E-cadherin (CDH1), which dissociates cell adhesion and disrupts epithelial cell polarity.96 ZEB1 represses stemness-inhibiting microRNAs, especially miR-200b, to induce stem cell-like properties in epithelial cells, notably in cancer, including endometrial carcinoma (Figure 5A).95,117,128,129 Although the downregulation of miR-200b was not discernible in this study of endometriotic epithelial cells, other studies have found that miR-200b is downregulated in endometriotic endometrium.18,21,130 Perhaps the role of miR-200b in the ZEB1/miR-200b feedback loop in endometriotic epithelial cells is for attachment at an ectopic site, so that upregulated miR-200b inhibits ZEB1, reverses EMT, and induces MET.129 (Similarly, downregulated ZEB1 in diseased stromal cells could activate an MET pathway in eutopic endometrium.) The EMT in endometriotic epithelial cells might also be activated by Hedgehog signaling indirectly via Fibroblast Growth Factor 2 (FGF2), NOTCH, and TGFβ1 signaling pathways or by TGFβ1, ZEB1, and miR-504 upregulating NRP2 (Figures 3H and 5C).114,131 In turn, EMT strongly induces AXL receptor tyrosine kinase (AXL), which was upregulated in endometriotic epithelial cells in this study (Table 1). AXL increases cell migration and metastasis and is a stem cell regulator.132–136
When potentially invasive, endometriotic, stem cell–like EMT cells are refluxed during menstruation and attach in ectopic sites, these EMT cells are ready primed by IL-1B for neovascularization with altered expression of Nitric Oxide Synthase 3 (NOS), FGF2, MMP2, TGFβ1, BMP2, and FLT1 to enable invasion.118,128 The target genes of miR-504 and miR-1827, NRP2 and NQO1, increase vascularization, leukocyte, and cell migration in endometriotic epithelial cells (Table 3; Figure 5C). NRP2 and PLXNA4 are neovascularization pathfinders for ectopic sites.137 However, ectopic lesions, compared to eutopic endometrium, can have reversed molecular expression of critical genes. Other studies have shown that when the refluxed EMT cells are implanted at an ectopic site, ZEB1 is downregulated, E-cadherin and miR-200b are upregulated, the cells transform back into epithelial cells by MET, and these cells establish, vascularize, and grow into lesions.102,138
In conclusion, aberrant molecular pathways associated with diseased stromal cells decrease immune cell trafficking and impair decidualization, while diseased epithelial cells are associated with TGFβ1 and IL-1B inflammation-driven pathways that increase vascularization, cell proliferation, leukocyte and cell migration, and pathological EMT. The target genes of the differentially expressed microRNAs, such as miR-200b, miR-204, miR-504, and miR-1827, play a prominent role in the main aberrant functions associated with endometriotic stromal and epithelial cells. Understanding these cell type–specific microRNA—mRNA molecular pathways and interactions will contribute to the targeted treatment of this complex and devastating disease.
Supplementary Material
Acknowledgments
The authors thank Kim Chi Vo and Jacquelyn Hoffman, from the National Institutes of Health (NIH)/University of California, San Francisco (UCSF) Human Endometrial Tissue and DNA Bank, for collecting the endometrial tissue samples.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Institutes of Health (Grant number NIH-U54HD055764).
ORCID iD: Philip C. Logan, PhD http://orcid.org/0000-0001-5656-6538.
Supplemental Material: Supplementary material for this article is available online.
References
- 1. Giudice LC. Endometriosis. N Engl J Med. 2010;362(25):2389–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Burney RO, Giudice LC. Pathogenesis and pathophysiology of endometriosis. Fertil Steril. 2012;98(3):511–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Agarwal N, Subramanian A. Endometriosis—morphology, clinical presentations and molecular pathology. J Lab Physicians. 2010;2(2):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brosens I, Puttemans P, Benagiano G. Endometriosis: a life cycle approach? Am J Obstet Gynecol. 2013;209(4):307–316. [DOI] [PubMed] [Google Scholar]
- 5. Brosens I, Brosens J, Benagiano G. Neonatal uterine bleeding as antecedent of pelvic endometriosis. Hum Reprod. 2013;28(11):2893–2897. [DOI] [PubMed] [Google Scholar]
- 6. Lessey B, Lebovic D, Taylor R. Eutopic endometrium in women with endometriosis: ground zero for the study of implantation defects. Semin Reprod Med. 2013;31(2):109–124. [DOI] [PubMed] [Google Scholar]
- 7. Hadfield R, Mardon H, Barlow D, Kennedy S. Delay in the diagnosis of endometriosis: a survey of women from the USA and the UK. Hum Reprod. 1996;11(4):878–880. [DOI] [PubMed] [Google Scholar]
- 8. Maruyama T, Yoshimura Y. Stem cell theory for the pathogenesis of endometriosis. Front Biosci. 2012;4E(8):2754–2763. [DOI] [PubMed] [Google Scholar]
- 9. Aznaurova Y, Zhumataev M, Roberts T, Aliper A, Zhavoronkov A. Molecular aspects of development and regulation of endometriosis. Reprod Biol Endocrinol. 2014;12(1):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gargett CE, Schwab KE, Brosens JJ, Puttemans P, Benagiano G, Brosens I. Potential role of endometrial stem/progenitor cells in the pathogenesis of early-onset endometriosis. Mol Hum Reprod. 2014;20(7):591–598. [DOI] [PubMed] [Google Scholar]
- 11. Du H, Taylor HS. Contribution of bone marrow-derived stem cells to endometrium and endometriosis. Stem Cells. 2007;25(8):2082–2086. [DOI] [PubMed] [Google Scholar]
- 12. Zanatta A, Rocha A, Carvalho F, et al. The role of the Hoxa10/HOXA10 gene in the etiology of endometriosis and its related infertility: a review. J Assist Reprod Genet. 2010;27(12):701–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sampson J. Peritoneal endometriosis due to the menstrual dissemination of endometrial tissue into the peritoneal cavity. Am J Obstet Gynecol. 1927;14(4):422–469. [Google Scholar]
- 14. Marsh EE, Laufer MR. Endometriosis in premenarcheal girls who do not have an associated obstructive anomaly. Fertil Steril. 2005;83(3):758–760. [DOI] [PubMed] [Google Scholar]
- 15. Matsuura K, Ohtake H, Katabuchi H, Okamura H. Coelomic metaplasia theory of endometriosis: evidence from in vivo studies and an in vitro experimental model. Gynecol Obstet Invest. 1999;47(suppl 1):18–22. [DOI] [PubMed] [Google Scholar]
- 16. Figueira PGM, Abrão MS, Krikun G, Taylor H. Stem cells in endometrium and their role in the pathogenesis of endometriosis. Ann NY Acad Sci. 2011;1221(1):10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wren JD, Wu Y, Guo SW. A system-wide analysis of differentially expressed genes in ectopic and eutopic endometrium. Hum Reprod. 2007;22(8):2093–2102. [DOI] [PubMed] [Google Scholar]
- 18. Ohlsson Teague EMC, Van der Hoek KH, Van der Hoek MB, et al. MicroRNA-regulated pathways associated with endometriosis. Mol Endocrinol. 2009;23(2):265–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Matsuzaki S. DNA microarray analysis in endometriosis for development of more effective targeted therapies. Front Biosci. 2011;3E(3):1139–1153. [DOI] [PubMed] [Google Scholar]
- 20. Painter JN, Anderson CA, Nyholt DR, et al. Genome-wide association study identifies a locus at 7p15.2 associated with endometriosis. Nat Genet. 2011;43(1):51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Filigheddu N, Gregnanin I, Porporato PE, et al. Differential expression of microRNAs between eutopic and ectopic endometrium in ovarian endometriosis. J Biomed Biotechnol. 2010;2010:369549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sun PR, Jia SZ, Lin H, Leng JH, Lang JH. Genome-wide profiling of long noncoding ribonucleic acid expression patterns in ovarian endometriosis by microarray. Fertil Steril. 2014;101(4):1038–1046.e7. [DOI] [PubMed] [Google Scholar]
- 23. Matsuzaki S, Canis M, Vaurs-Barrière C, Boespflug-Tanguy O, Dastugue B, Mage G. DNA microarray analysis of gene expression in eutopic endometrium from patients with deep endometriosis using laser capture microdissection. Fertil Steril. 2005;84(suppl 2):1180–1190. [DOI] [PubMed] [Google Scholar]
- 24. Burney RO, Talbi S, Hamilton AE, et al. Gene expression analysis of endometrium reveals progesterone resistance and candidate susceptibility genes in women with endometriosis. Endocrinology. 2007;148(8):3814–3826. [DOI] [PubMed] [Google Scholar]
- 25. Graham A, Falcone T, Nothnick WB. The expression of microRNA-451 in human endometriotic lesions is inversely related to that of macrophage migration inhibitory factor (MIF) and regulates MIF expression and modulation of epithelial cell survival. Hum Reprod. 2015;30(3):642–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sherwin JRA, Sharkey AM, Mihalyi A, Simsa P, Catalano RD, D’Hooghe TM. Global gene analysis of late secretory phase, eutopic endometrium does not provide the basis for a minimally invasive test of endometriosis. Hum Reprod. 2008;23(5):1063–1068. [DOI] [PubMed] [Google Scholar]
- 27. Meola J, Rosa e Silva JC, Dentillo DB, et al. Differentially expressed genes in eutopic and ectopic endometrium of women with endometriosis. Fertil Steril. 2010;93(6):1750–1773. [DOI] [PubMed] [Google Scholar]
- 28. Rosser MD, Haris PI, Ankrett DN, Konje JC. The emerging role of epigenetics and miRNAs in endometriosis. Expert Rev Obstet Gynecol. 2011;6(4):431–450. [Google Scholar]
- 29. Kennedy SH. The genetics of endometriosis. Eur J Obstet Gynecol Reprod Biol. 1999;82(2):129–133. [DOI] [PubMed] [Google Scholar]
- 30. Guo SW. Epigenetics of endometriosis. Mol Hum Reprod. 2009;15(10):587–607. [DOI] [PubMed] [Google Scholar]
- 31. Baccarelli A, Bollati V. Epigenetics and environmental chemicals. Curr Opin Pediatr. 2009;21(2):243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee B, Du H, Taylor HS. Experimental murine endometriosis induces DNA methylation and altered gene expression in eutopic endometrium. Biol Reprod. 2009;80(1):79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xue Q, Lin Z, Cheng YH, et al. Promoter methylation regulates estrogen receptor 2 in endometrium and endometriosis. Biol Reprod. 2007;77(4):681–687. [DOI] [PubMed] [Google Scholar]
- 34. Dyson MT, Roqueiro D, Monsivais D, et al. Genome-wide DNA methylation analysis predicts an epigenetic switch for GATA factor expression in endometriosis. PLoS Genet. 2014;10(3):e1004158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu Y, Strawn E, Basir Z, Halverson G, Guo SW. Aberrant expression of deoxyribonucleic acid methyltransferases DNMT1, DNMT3A, and DNMT3B in women with endometriosis. Fertil Steril. 2007;87(1):24–32. [DOI] [PubMed] [Google Scholar]
- 36. Nasu K, Kawano Y, Tsukamoto Y, et al. Aberrant DNA methylation status of endometriosis: epigenetics as the pathogenesis, biomarker and therapeutic target. J Obstet Gynaecol Res. 2011;37(7):683–695. [DOI] [PubMed] [Google Scholar]
- 37. Naqvi H, Ilagan Y, Krikun G, Taylor HS. Altered genome-wide methylation in endometriosis. Reprod Sci. 2014;21(10):1237–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Colon-Diaz M, Baez-Vega P, Garcia M, et al. HDAC1 and HDAC2 are differentially expressed in endometriosis. Reprod Sci. 2012;19(5):483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kawano Y, Nasu K, Li H, et al. Application of the histone deacetylase inhibitors for the treatment of endometriosis: histone modifications as pathogenesis and novel therapeutic target. Hum Reprod. 2011;26(9):2486–2498. [DOI] [PubMed] [Google Scholar]
- 40. Nasu K, Kawano Y, Kai K, et al. Aberrant histone modification in endometriosis. Front Biosci. 2014;19:1202–1214. [DOI] [PubMed] [Google Scholar]
- 41. Monteiro JB, Colón-Díaz M, García M, et al. Endometriosis is characterized by a distinct pattern of histone 3 and histone 4 lysine modifications. Reprod Sci. 2014;21(3):305–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2010;220(2):126–139. [DOI] [PubMed] [Google Scholar]
- 43. Santamaria X, Taylor H. MicroRNA and gynecological reproductive diseases. Fertil Steril. 2014;101(6):1545–1551. [DOI] [PubMed] [Google Scholar]
- 44. Petracco R, Grechukhina O, Popkhadze S, Massasa E, Zhou Y, Taylor HS. MicroRNA 135 regulates HOXA10 expression in endometriosis. J Clin Endocrinol Metab. 2011;96(12):E1925–E1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ohlsson TE, Print CG, Hull ML. The role of microRNAs in endometriosis and associated reproductive conditions. Hum Reprod Update. 2010;16(2):142–165. [DOI] [PubMed] [Google Scholar]
- 46. Hawkins SM, Creighton CJ, Han DY, et al. Functional microRNA involved in endometriosis. Mol Endocrinol. 2011;25(5):821–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pan Q, Chegini N. MicroRNA signature and regulatory functions in the endometrium during normal and disease states. Semin Reprod Med. 2008;26(6):479–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Braza-Boïls A, Marí-Alexandre J, Gilabert J, et al. MicroRNA expression profile in endometriosis: its relation to angiogenesis and fibrinolytic factors. Hum Reprod. 2014;29(5):978–988. [DOI] [PubMed] [Google Scholar]
- 49. Okamoto M, Nasu K, Abe W, et al. Enhanced miR-210 expression promotes the pathogenesis of endometriosis through activation of signal transducer and activator of transcription 3. Hum Reprod. 2015;30(3):632–641. [DOI] [PubMed] [Google Scholar]
- 50. Kao LC, Germeyer A, Tulac S, et al. Expression profiling of endometrium from women with endometriosis reveals candidate genes for disease-based implantation failure and infertility. Endocrinology. 2003;144(7):2870–2881. [DOI] [PubMed] [Google Scholar]
- 51. Burney RO, Hamilton AE, Aghajanova L, et al. MicroRNA expression profiling of eutopic secretory endometrium in women with versus without endometriosis. Mol Hum Reprod. 2009;15(10):625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zafrakas M, Tarlatzis BC, Streichert T, et al. Genome-wide microarray gene expression, array-CGH analysis, and telomerase activity in advanced ovarian endometriosis: a high degree of differentiation rather than malignant potential. Int J Mol Med. 2008;21(3):335–344. [PubMed] [Google Scholar]
- 53. Oger F, Gheeraert C, Mogilenko D, et al. Cell-specific dysregulation of MicroRNA expression in obese white adipose tissue. J Clin Endocrinol Metab. 2014;99(8):2821–2833. [DOI] [PubMed] [Google Scholar]
- 54. Jovičić A, Roshan R, Moisoi N, et al. Comprehensive expression analyses of neural cell-type-specific miRNAs identify new determinants of the specification and maintenance of neuronal phenotypes. J Neurosci. 2013;33(12):5127–5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. McCall MN, Kent OA, Yu J, Fox-Talbot K, Zaiman AL, Halushka MK. MicroRNA profiling of diverse endothelial cell types. BMC Med Genomics. 2011;4(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gellersen B, Brosens IA, Brosens JJ. Decidualization of the human endometrium: mechanisms, functions, and clinical perspectives. Semin Reprod Med. 2007;25(6):445–453. [DOI] [PubMed] [Google Scholar]
- 57. Guzeloglu-Kayisli O, Kayisli UA, Taylor HS. The role of growth factors and cytokines during implantation: endocrine and paracrine interactions. Semin Reprod Med. 2009;27(1):62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Graham C, Lala P. Mechanisms of placental invasion of the uterus and their control. Biochem Cell Biol. 1992;70(10-11):867–874. [DOI] [PubMed] [Google Scholar]
- 59. Lockwood C, Krikun G, Schatz F. The decidua regulates hemostasis in human endometrium. Semin Reprod Endocrinol. 1999;17(1):45–51. [DOI] [PubMed] [Google Scholar]
- 60. Norwitz ER. Defective implantation and placentation: laying the blueprint for pregnancy complications. Reprod Biomed Online. 2006;13(4):591–599. [DOI] [PubMed] [Google Scholar]
- 61. Aplin JD. Embryo implantation: the molecular mechanism remains elusive. Reprod Biomed Online. 2006;13(6):833–839. [DOI] [PubMed] [Google Scholar]
- 62. Kurihara I, Lee DK, Petit FG, et al. COUP-TFII mediates progesterone regulation of uterine implantation by controlling ER activity. PLoS Genetics. 2007;3(6):e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bulun SE, Cheng YH, Pavone ME, et al. 17Beta-hydroxysteroid dehydrogenase-2 deficiency and progesterone resistance in endometriosis. Semin Reprod Med. 2010;28(1):44–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lee KY, Jeong JW, Tsai SY, Lydon JP, DeMayo FJ. Mouse models of implantation. Trend Endocrinol Metab. 2007;18(6):234–239. [DOI] [PubMed] [Google Scholar]
- 65. Bulun SE. Endometriosis. N Engl J Med. 2009;360(3):268–279. [DOI] [PubMed] [Google Scholar]
- 66. Li Q, Kannan A, Das A, et al. WNT4 acts downstream of BMP2 and functions via β-catenin signaling pathway to regulate human endometrial stromal cell differentiation. Endocrinology. 2013;154(1):446–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gregory GD, Vakoc CR, Rozovskaia T, et al. Mammalian ASH1L is a histone methyltransferase that occupies the transcribed region of active genes. Mol Cell Biol. 2007;27(24):8466–8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Klemmt PA, Carver JG, Kennedy SH, Koninckx PR, Mardon HJ. Stromal cells from endometriotic lesions and endometrium from women with endometriosis have reduced decidualization capacity. Fertil Steril. 2006;85(3):564–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lash GE, Bulmer JN. Do uterine natural killer (uNK) cells contribute to female reproductive disorders? J Reprod Immunol. 2011;88(2):156–164. [DOI] [PubMed] [Google Scholar]
- 70. Salamonsen L, Hannan N, Dimitriadis E. Cytokines and chemokines during human embryo implantation: roles in implantation and early placentation. Semin Reprod Med. 2007;25(6):437–444. [DOI] [PubMed] [Google Scholar]
- 71. Waldmann TA, Tagaya Y. The multifaceted regulation of interleukin-15 expression and the role of this cytokine in NK cell differentiation and host response to intracellular pathogens. Ann Rev Immunol. 1999;17(1):19–49. [DOI] [PubMed] [Google Scholar]
- 72. Sozzani S, Allavena P, Vecchi A, Mantovani A. The role of chemokines in the regulation of dendritic cell trafficking. J Leuk Biol. 1999;66(1):1–9. [DOI] [PubMed] [Google Scholar]
- 73. Schiraldi M, Raucci A, Muñoz LM, et al. HMGB1 promotes recruitment of inflammatory cells to damaged tissues by forming a complex with CXCL12 and signaling via CXCR4. J Exp Med. 2012;209(3):551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kalesnikoff J, Galli SJ. New developments in mast cell biology. Nat Immunol. 2008;9(11):1215–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Thiruchelvam U, Dransfield I, Saunders PTK, Critchley HOD. The importance of the macrophage within the human endometrium. J Leukoc Biol. 2013;93(2):217–225. [DOI] [PubMed] [Google Scholar]
- 76. Okada H, Nakajima T, Sanezumi M, Ikuta A, Yasuda K, Kanzaki H. Progesterone enhances interleukin-15 production in human endometrial stromal cells in vitro. J Clin Endocrinol Metab. 2000;85(12):4765–4770. [DOI] [PubMed] [Google Scholar]
- 77. Godbole G, Modi D. Regulation of decidualization, interleukin-11 and interleukin-15 by homeobox A 10 in endometrial stromal cells. J Reprod Immunol. 2010;85(2):130–139. [DOI] [PubMed] [Google Scholar]
- 78. Lee JY, Lee M, Lee SK. Role of endometrial immune cells in implantation. Clin Exp Reprod Med. 2011;38(3):119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Cartwright JE, Balarajah G. Trophoblast interactions with endothelial cells are increased by interleukin-1[beta] and tumour necrosis factor [alpha] and involve vascular cell adhesion molecule-1 and [alpha]4[beta]1. Exp Cell Res. 2005;304(1):328–336. [DOI] [PubMed] [Google Scholar]
- 80. Gellersen B, Reimann K, Samalecos A, Aupers S, Bamberger AM. Invasiveness of human endometrial stromal cells is promoted by decidualization and by trophoblast-derived signals. Hum Reprod. 2010;25(4):862–873. [DOI] [PubMed] [Google Scholar]
- 81. Fisher SJ, Damsky CH. Human cytotrophoblast invasion. Semin Cell Biol. 1993;4(3):183–188. [DOI] [PubMed] [Google Scholar]
- 82. Brosens I, Pijnenborg R, Vercruysse L, Romero R. The Great Obstetrical Syndromes are associated with disorders of deep placentation. Am J Obstet Gynecol. 2011;204(3):193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Duncan WC, Shaw JLV, Burgess S, McDonald SE, Critchley HOD, Horne AW. Ectopic pregnancy as a model to identify endometrial genes and signaling pathways important in decidualization and regulated by local trophoblast. PLoS One. 2011;6(8):e23595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Aghajanova L, Tatsumi K, Horcajadas JA, et al. Unique transcriptome, pathways, and networks in the human endometrial fibroblast response to progesterone in endometriosis. Biol Reprod. 2011;84(4):801–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Novembri R, Borges LE, Carrarelli P, et al. Impaired CRH and urocortin expression and function in eutopic endometrium of women with endometriosis. J Clin Endocrinol Metab. 2011;96(4):1145–1150. [DOI] [PubMed] [Google Scholar]
- 86. Shen L, Liu P, Zhang P, Zhang X, Cui J. Characterization of periostin expression in human endometrium and endometriotic lesions. Gynecol Endocrinol. 2012;28(10):815–818. [DOI] [PubMed] [Google Scholar]
- 87. Yang H, Zhou Y, Edelshain B, Schatz F, Lockwood CJ, Taylor HS. FKBP4 is regulated by HOXA10 during decidualization and in endometriosis. Reproduction. 2012;143(4):531–538. [DOI] [PubMed] [Google Scholar]
- 88. Estella C, Herrer I, Moreno-Moya JM, et al. miRNA signature and Dicer requirement during human endometrial stromal decidualization in vitro . PLoS One. 2012;7(7):e41080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Gellersen B, Kempf R, Telgmann R. Human endometrial stromal cells express novel isoforms of the transcriptional modulator CREM and up-regulate ICER in the course of decidualization. Mol Endocrinol. 1997;11(1):97–113. [DOI] [PubMed] [Google Scholar]
- 90. Teklenburg G, Salker M, Heijnen CJ, Macklon NS, Brosens JJ. The molecular basis of recurrent pregnancy loss: impaired natural embryo selection. Mol Hum Reprod. 2010;16(12):886–895. [DOI] [PubMed] [Google Scholar]
- 91. Lockwood C, Krikun G, Rahman M, Caze R, Buchwalder L, Schatz F. The role of decidualization in regulating endometrial hemostasis during the menstrual cycle, gestation, and in pathological states. Semin Thromb Hemost. 2007(01):111–117. [DOI] [PubMed] [Google Scholar]
- 92. Kaufmann P, Black S, Huppertz B. Endovascular trophoblast invasion: implications for the pathogenesis of intrauterine growth retardation and preeclampsia. Biol Reprod. 2003;69(1):1–7. [DOI] [PubMed] [Google Scholar]
- 93. Lucas ES, Dyer NP, Murakami K, et al. Loss of endometrial plasticity in recurrent pregnancy loss. Stem cells. 2016;34(2):346–356. [DOI] [PubMed] [Google Scholar]
- 94. Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop—a motor of cellular plasticity in development and cancer? EMBO Rep. 2010;11(9):670–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Renthal NE, Chen CC, Williams KC, Gerard RD, Prange-Kiel J, Mendelson CR. miR-200 family and targets, ZEB1 and ZEB2, modulate uterine quiescence and contractility during pregnancy and labor. Proc Natl Acad Sci. 2010;107(48):20828–20833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Romero-Perez L, Lopez-Garcia MA, Diaz-Martin J, et al. ZEB1 overexpression associated with E-cadherin and microRNA-200 downregulation is characteristic of undifferentiated endometrial carcinoma. Mod Pathol. 2013;26(11):1514–1524. [DOI] [PubMed] [Google Scholar]
- 97. Haraguchi H, Saito-Fujita T, Hirota Y, et al. MicroRNA-200a locally attenuates progesterone signaling in the cervix, preventing embryo implantation. Mol Endocrinol. 2014;28(7):1108–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10(5):593–601. [DOI] [PubMed] [Google Scholar]
- 99. Hill L, Browne G, Tulchinsky E. ZEB/miR-200 feedback loop: at the crossroads of signal transduction in cancer. Int J Cancer. 2013;132(4):745–754. [DOI] [PubMed] [Google Scholar]
- 100. Dykxhoorn DM. MicroRNAs and metastasis: little RNAs go a long way. Cancer Res. 2010;70(16):6401–6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wang G, Guo X, Hong W, et al. Critical regulation of miR-200/ZEB2 pathway in Oct4/Sox2-induced mesenchymal-to-epithelial transition and induced pluripotent stem cell generation. Proc Natl Acad Sci. 2013;110(8):2858–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Yao D, Dai C, Peng S. Mechanism of the mesenchymal–epithelial transition and its relationship with metastatic tumor formation. Mol Cancer Res. 2011;9(12):1608–1620. [DOI] [PubMed] [Google Scholar]
- 103. Barragan F, Irwin JC, Balayan S, et al. Human endometrial fibroblasts derived from mesenchymal progenitors inherit progesterone resistance and acquire an inflammatory phenotype in the endometrial niche in endometriosis. Biol Reprod. 2016;94(5):118, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Bulun SE, Cheng YH, Pavone ME, et al. Estrogen receptor-beta, estrogen receptor-alpha, and progesterone resistance in endometriosis. Semin Reprod Med. 2010;28(1):36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Bulun SE, Cheng YH, Yin P, et al. Progesterone resistance in endometriosis: link to failure to metabolize estradiol. Mol Cell Endocrinol. 2006;248(1-2):94–103. [DOI] [PubMed] [Google Scholar]
- 106. Eaton JL, Unno K, Caraveo M, Lu Z, Kim JJ. Increased AKT or MEK1/2 activity influences progesterone receptor levels and localization in endometriosis. J Clin Endocrinol Metab. 2013;98(12):e1871–e1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Morra L, Moch H. Periostin expression and epithelial-mesenchymal transition in cancer: a review and an update. Virchows Arch. 2011;459(5):465–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Shindoh H, Okada H, Tsuzuki T, Nishigaki A, Kanzaki H. Requirement of heart and neural crest derivatives–expressed transcript 2 during decidualization of human endometrial stromal cells in vitro. Fertil Steril. 2014;101(6):1781–1790.e5. [DOI] [PubMed] [Google Scholar]
- 109. Hevir N, Vouk K, Šinkovec J, Ribič-Pucelj M, Rižner TL. Aldo-keto reductases AKR1C1, AKR1C2 and AKR1C3 may enhance progesterone metabolism in ovarian endometriosis. Chem Biol Interact. 2011;191(1-3):217–226. [DOI] [PubMed] [Google Scholar]
- 110. Cheng YH, Utsunomiya H, Pavone ME, Yin P, Bulun SE. Retinoic acid inhibits endometrial cancer cell growth via multiple genomic mechanisms. J Mol Endocrinol. 2011;46(2):139–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Wetendorf M, DeMayo FJ. The progesterone receptor regulates implantation, decidualization, and glandular development via a complex paracrine signaling network. Mol Cell Endocrinol. 2011;357(1-2):108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Smith K, Alnifaidy R, Wei Q, Nieman LK. Endometrial Indian Hedgehog expression is decreased in women with endometriosis. Fertil Steril. 2011;95(8):2738–2741.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Large MJ, DeMayo FJ. The regulation of embryo implantation and endometrial decidualization by progesterone receptor signaling. Mol Cell Endocrinol. 2012;358(2):155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Katoh Y, Katoh M. Hedgehog signaling, epithelial-to-mesenchymal transition and miRNA (review). Int J Mol Med. 2008;22(3):271–275. [PubMed] [Google Scholar]
- 115. Hillman RT, Feng BY, Ni J, et al. Neuropilins are positive regulators of Hedgehog signal transduction. Genes Dev. 2011;25(22):2333–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Fu L, Lockhart MG, Amir MM, Winnall WR, Rogers PAW, Girling JE. Differential TGFB1-signalling in endometrium from women with endometriosis: importance of appropriate housekeeping genes. J Endometr Pelvic Pain Disord. 2014;6(1):41–54. [Google Scholar]
- 117. Gregory PA, Bracken CP, Smith E, et al. An autocrine TGF-β/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Mol Biol Cell. 2011;22(10):1686–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Proestling K, Birner P, Gamperl S, et al. Enhanced epithelial to mesenchymal transition (EMT) and upregulated MYC in ectopic lesions contribute independently to endometriosis. Reprod Biol Endocrinol. 2015;13(1):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Bartley J, Jülicher A, Hotz B, Mechsner S, Hotz H. Epithelial to mesenchymal transition (EMT) seems to be regulated differently in endometriosis and the endometrium. Arch Gynecol Obstet. 2013;289(4):871–881. [DOI] [PubMed] [Google Scholar]
- 120. Fuxe J, Karlsson MCI. TGF-β-induced epithelial-mesenchymal transition: a link between cancer and inflammation. Semin Cancer Biol. 2012;22(5-6):455–461. [DOI] [PubMed] [Google Scholar]
- 121. Moustakas A, Heldin CH. Induction of epithelial–mesenchymal transition by transforming growth factor β. Semin Cancer Biol. 2012;22(5-6):446–454. [DOI] [PubMed] [Google Scholar]
- 122. Hurt E, Saykally J, Anose B, Kalli K, Sanders M. Expression of the ZEB1 (δEF1) transcription factor in human: additional insights. Mol Cell Biochem. 2008;318(1-2):89–99. [DOI] [PubMed] [Google Scholar]
- 123. Abe W, Nasu K, Nakada C, Kawano Y, Moriyama M, Narahara H. miR-196b targets c-myc and Bcl-2 expression, inhibits proliferation and induces apoptosis in endometriotic stromal cells. Hum Reprod. 2013;28(3):750–761. [DOI] [PubMed] [Google Scholar]
- 124. Anose BM, Sanders MM. Androgen receptor regulates transcription of the ZEB1 transcription factor. Int J Endocrinol. 2011;2011:903918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Singh M, Spoelstra NS, Jean A, et al. ZEB1 expression in type I vs type II endometrial cancers: a marker of aggressive disease. Mod Pathol. 2008;21(7):912–923. [DOI] [PubMed] [Google Scholar]
- 126. Gregory PA, Bracken CP, Bert AG, Goodall GJ. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle. 2008;7(20):3112–3117. [DOI] [PubMed] [Google Scholar]
- 127. Zhang GJ, Zhou T, Tian HP, Liu ZL, Xia SS. High expression of ZEB1 correlates with liver metastasis and poor prognosis in colorectal cancer. Oncol Lett. 2013;5(2):564–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Lebovic DI, Baldocchi RA, Mueller MD, Taylor RN. Altered expression of a cell-cycle suppressor gene, Tob-1, in endometriotic cells by cDNA array analyses. Fertil Steril. 2002;78(4):849–854. [DOI] [PubMed] [Google Scholar]
- 129. Wellner U, Schubert J, Burk UC, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11(12):1487–1495. [DOI] [PubMed] [Google Scholar]
- 130. Eggers JC, Martino V, Reinbold R, et al. microRNA miR-200b affects proliferation, invasiveness and stemness of endometriotic cells by targeting ZEB1, ZEB2 and KLF4. Reprod BioMed Online. 2016;32(4):434–445. [DOI] [PubMed] [Google Scholar]
- 131. Nasarre P, Gemmill RM, Potiron VA, et al. Neuropilin-2 is upregulated in lung cancer cells during TGFβ1–induced epithelial–mesenchymal transition. Cancer Res. 2013;73(23):7111–7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Axelrod H, Pienta KJ. Axl as a mediator of cellular growth and survival. Oncotarget. 2014;5(19):8818–8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Asiedu MK, Beauchamp-Perez FD, Ingle JN, Behrens MD, Radisky DC, Knutson KL. AXL induces epithelial-to-mesenchymal transition and regulates the function of breast cancer stem cells. Oncogene. 2014;33(10):1316–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Gjerdrum C, Tiron C, Høiby T, et al. Axl is an essential epithelial-to-mesenchymal transition-induced regulator of breast cancer metastasis and patient survival. Proc Natl Acad Sci. 2010;107(3):1124–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Sun WS, Misao R, Iwagaki S, Fujimoto J, Tamaya T. Coexpression of growth arrest-specific gene 6 and receptor tyrosine kinases, Axl and Sky, in human uterine endometrium and ovarian endometriosis. Mol Hum Reprod. 2002;8(6):552–558. [DOI] [PubMed] [Google Scholar]
- 136. Honda H, Barrueto FF, Gogusev J, Im DD, Morin PJ. Serial analysis of gene expression reveals differential expression between endometriosis and normal endometrium. Possible roles for AXL and SHC1 in the pathogenesis of endometriosis. Reprod Biol Endocrinol. 2008;6(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Torres-Vázquez J, Gitler AD, Fraser SD, et al. Semaphorin-plexin signaling guides patterning of the developing vasculature. Dev Cell. 2004;7(1):117–123. [DOI] [PubMed] [Google Scholar]
- 138. Saare M, Rekker K, Laisk-Podar T, et al. High-throughput sequencing approach uncovers the miRNome of peritoneal endometriotic lesions and adjacent healthy tissues. PLoS One. 2014;9(11):e112630. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.