Protecting the innocents—What will become of the twin girls whose DNA was recklessly mutated to destroy the CCR5 gene?
Like almost everyone, I was stunned by the birth announcement of genetically modified twins, Lulu and Nana. A Tweet pointed me to the MIT Technology Review scoop1 that, along with a subsequent Associated Press article confirming the births,2 revealed something shocking: Chinese researcher He Jiankui (Southern University of Science and Technology) had used CRISPR*-Cas9 gene editing technology to mutate (sic) the DNA of human embryos—embryos that were then implanted and brought to term. Though CRISPR pioneer Jennifer Doudna (HHMI/UC Berkeley) warned us this might happen,3 I had trusted that my fellow scientists would ensure that human trials involving embryonic genome editing would be done transparently, ethically, and with strong moral purpose.
Alas, it quickly became clear this was not the case. The first direct comments from Dr. He were released in an unusual way: through a series of YouTube videos. He explained his choice to mutate CCR5—the gene encoding a highly conserved chemokine receptor implicated in human immunodeficiency virus (HIV) uptake into T cells—by claiming that “protecting against HIV requires the simplest gene surgery, which is to remove just a few DNA letters,” and because “100 million people naturally have a genetic variation that disables CCR5” (see https://youtu.be/aezxaOn0efE). As such, his goal was not to correct a disease-causing variant to save a life. Instead, it was to destroy a normal gene using the simplest editing strategy, hoping to improve upon an otherwise healthy embryo. This set off the “designer baby” warning alarms and raised one particularly critical question: what exactly was done to these children?
When I found out (again through Twitter) that Dr. He planned to present his work at the International Summit on Human Genome Editing in Hong Kong (see the accompanying First Cut by Kevin Davies), and that the National Academies of Science, Engineering, and Medicine would be streaming the talk live, I joined as many as 1.8 million people (the figure reported by summit co-host Lap-Chee Tsui) watching the webcast around the world. We learned that He had indeed injected CRISPR-Cas9 with a guide RNA targeting CCR5 into embryos. He assessed the extent of editing using pre-implantation genetic diagnosis, implanted those embryos into the mother (named Grace), and monitored the pregnancy through birth. He then sequenced DNA from their placenta, umbilical cord, and cord blood to assess on- and off-target mutations.
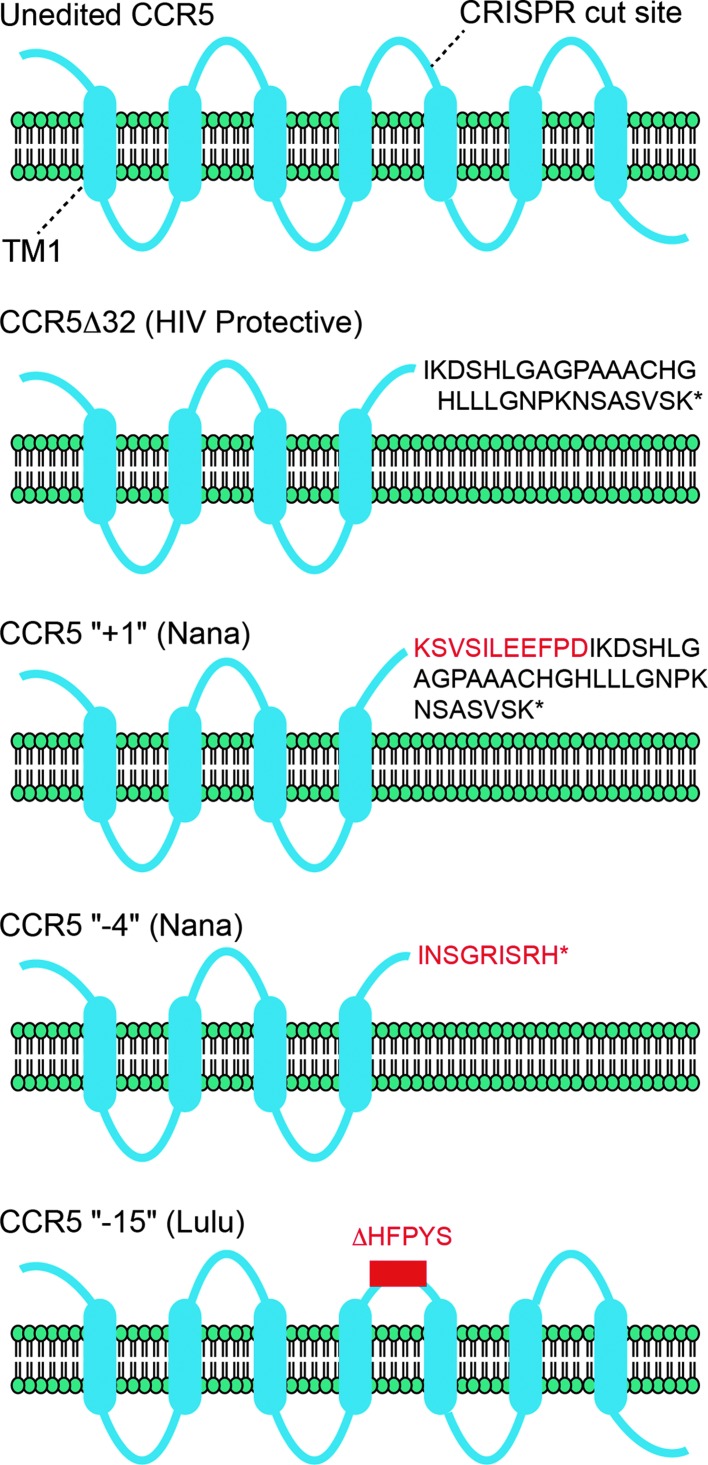
The unpublished data, as yet unverified, indicated that Lulu carries a non-edited allele of CCR5, as well as a variant that has a 15 bp in-frame deletion (“−15”). Nana carries a 4 bp deletion (“−4”) and a single base insertion (“+1”). Moreover, Lulu has evidence of an off-target mutation in an intergenic region, and both babies appear to exhibit mosaicism in sequencing chromatograms (as pointed out on Twitter by Gaetan Burgio [Australian National University]). Importantly, neither baby carries the well-studied CCR5Δ32 variant protective against HIV (see Fig. 1).4–6
FIG. 1.
The membrane topology of CCR5 and variants Δ32, Nana “+1,” Nana “−4”, and Lulu “−15” are shown relative to a membrane bilayer. Loops are not drawn to scale. Sequences present in the variants that are not present in the unedited version of CCR5 are shown. Sequences in red are unique to the given variant. TM1 represents the first transmembrane helix. The position of CRISPR targeting is marked.
As noted in his YouTube video, He relied upon non-homologous end joining to introduce imprecise insertions and deletions near the position where the Δ32 mutation appears. Both of Nana's frameshift mutations, “−4” and “+1”, are predicted to produce non-functional truncated proteins. The “+1” mutation is most similar to Δ32: it produces a similar C-terminus but adds 11 additional amino acids not found in the unedited or the Δ32 variants. The “−4” mutation shifts to a different frame, resulting in a shorter variant that has a different amino-acid sequence at the C-terminus. And the “−15” mutation in Lulu is in-frame, leading to the deletion of five amino acids near the HIV binding site. He predicts this will inhibit HIV uptake, but that hasn't been demonstrated in any model system, let alone humans. His clear assumption is that any mutation that inactivates CCR5 will confer a protective benefit.
Is that assumption reasonable? Fyodor Urnov (Altius Institute) points out that Carl June et al. demonstrated that editing of CCR5 in CD4+ T cells protects against HIV infection in vitro in a NOG mouse model of HIV infection and in HIV patients.7,8 Here, zinc finger nucleases were targeted to the first transmembrane span of CCR5 (see Fig. 1), leading to a wide variety of indels that would almost certainly inactivate the protein in a way that would prevent membrane insertion. By contrast, Nana's mutations retain four transmembrane spans, and Lulu's all seven.
But are the new mutations safe? He justified his work by saying that ∼100 million people have the non-functioning CCR5Δ32 allele. Yet, while Δ32 is relatively abundant in the population (minor allele frequency ∼0.073 in the ExAC database),9,10 other naturally occurring frameshifting indels are not (1 × 10−5 to 9 × 10−4 in ExAC). Moreover, Δ32 may cause harm, as the allele seems to increase risk for West Nile virus infection (reviewed in Lim et al.11).
To be clear, the mutations that He made are not Δ32. They are never-before-seen variants of unknown significance. They might inactivate CCR5 activity and block HIV uptake, but they might also incur new risks. It is not reasonable to rely upon the work of June et al. to justify the safety of these new variants because (1) he targeted a different region of CCR5, and (2) edited T cells, not a zygote.7 What is safe in a CD4+ T cell may not be safe in a developing embryo, or a baby, or in other tissues. He could have assessed the safety of these mutants in a mouse model, as he did for a different mutation, “−23,” but he did not. He could have followed the strategy of June et al. to assess efficacy of the mutations,7,8 but he did not. Instead, he appears to have relied on hunches and hopes. As a result, Lulu and Nana—living, human babies—are now the experiment.
It is my fervent hope that Lulu and Nana will lead long healthy lives. It is certainly possible, even plausible, that they will remain healthy based upon what is known about CCR5. However, there are enough uncertainties about the function of the mutations to raise clear scientific objections to the work, in addition to the moral and medical objections. I pray that Lulu and Nana are never exposed to HIV. I hope we never have a chance to find out if the experiment “worked.” To me, the best outcome is that the babies are unaffected by the procedures of the He lab. Worse outcomes are possible.
Based upon the mutations present, Dr. He reversed the names of the babies in his pre- and post-birth data slides. As such, it is impossible to assign a name to a specific genotype without ambiguity. I've used the identities listed on his post-birth data slide throughout this manuscript.
Acknowledgments
I thank Jen Phillips, Gaetan Burgio, Fyodor Urnov, Paul Knoepfler, and several other Twitter users for helpful discussions and explanations; Bob Matthews for encouragement; and Erik Sontheimer, Nick Rhind, and Brian Kelch for comments on a draft of this article. Work in the Ryder Lab is funded by NIH Grant GM117237 to S.P.R. and GM117008 to Francesca Massi and S.P.R.
Author Disclosure Statement
The author declares that apart from using CRISPR-Cas9 genome editing technology in his own research on Caenorhabditis elegans embryogenesis. He has no conflicts of interest, financial or otherwise.
Clustered Regularly Interspaced Short Palindromic Repeats.
References
- 1. Regalado A. Chinese scientists are creating CRISPR babies. MIT Technology Review. November 25, 2018. Availiable online at https://www.technologyreview.com/s/612458/exclusive-chinese-scientists-are-creating-crispr-babies/ (accessed December6, 2018)
- 2. Marchione M. Chinese researcher claims first gene-edited babies. Washington Post, November 26, 2018. Available online at: https://www.washingtonpost.com/world/asia_pacific/ap-exclusive-first-gene-edited-babies-claimed-in-china/2018/11/25/bb9b74de-f124-11e8-99c2-cfca6fcf610c_story.html?utm_term=.8e546caa068a (accessed December6, 2018)
- 3. Doudna JA, Sternberg SH. A Crack In Creation. New York: Houghton Mifflin Harcourt, 2017 [Google Scholar]
- 4. Samson M, Libert F, Doranz BJ, et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 1996;382:722–725. DOI: 10.1038/382722a0 [DOI] [PubMed] [Google Scholar]
- 5. Liu R, Paxton WA, Choe S, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 1996;86:367–377 [DOI] [PubMed] [Google Scholar]
- 6. Dean M, Carrington M, Winkler C, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science 1996;273:1856–1862 [DOI] [PubMed] [Google Scholar]
- 7. Tebas P, Stein D, Tang WW, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. New Engl J Med 2014;370:901–910. DOI: 10.1056/NEJMoa1300662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Perez EE, Wang J, Miller JC, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol 2008;26:808–816. DOI: 10.1038/nbt1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–291. DOI: 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Genomes Project C, Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature 2015;526:68–74. DOI: 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lim JK, Glass WG, McDermott DH, et al. CCR5: no longer a “good for nothing” gene—chemokine control of West Nile virus infection. Trends Immunol 2006;27:308–312. DOI: 10.1016/j.it.2006.05.007 [DOI] [PubMed] [Google Scholar]