One Sentence Summary:
New machine-learning techniques entering medical practice are often both opaque and changeable, raising challenges in validation, regulation, and integration into practice.
Recently, a deep neural network was able to identify skin cancer based solely on images of skin lesions, performing as well as board-certified dermatologists.1 A different algorithm identifies trauma patients in need of intervention to reduce the chance of hemorrhage, increasing the chance of prompt intervention without the need for consistent expert monitoring.2 Machine-learning algorithms have been predicted to come into widespread use in the areas of prognosis, radiology, and pathology within the next several years, and diagnosis within the next decade, substantially increasing the power and ubiquity of existing clinical decision support software.3 Consumers already have access to some machine-learning algorithms, such as smartphone apps that aim to identify developmental disorders in young children.4 Further afield from basic medical practice, algorithms can guide the allocation of scarce resources across health systems.5 How should algorithms like these be validated, regulated, and integrated into medical practice to ensure that they perform well in different populations at different times?
In a challenge for medical researchers and professionals, machine-learning techniques are typically opaque. They provide black-box algorithms for prediction or recommendation but do not explain or justify those results.5 Some techniques do not identify any explicit relationships; a neural network trained to identify tumors can identify them, but uses opaque hidden layers to do so.6 Other techniques may be able to list predictive relationships—but those relationships are so complex as to defy human understanding or explicit verification in any feasible timeframe. While not all medical algorithms are black-box, black-box algorithms can allow the health system to leverage complex biological relationships well before those relationships are understood.5 This raises a challenging question for the medical profession: should machine-learning models be pushed towards those implementations that are more interpretable, more mechanistically modeled, and ultimately aimed at increased understanding, or should acceptable models include fundamentally black-box algorithms that are practically useful but provide little scientific insight? The former privileges scientific understanding; the latter privileges immediate patient benefits. The question whether interpretability necessarily sacrifices performance is itself hotly contested, and the question of algorithmic interpretability in general is the subject of an expansive literature (https://arxiv.org/abs/1702.08608, https://arxiv.org/abs/1606.03490). Nevertheless, it is already time to consider how to deal with rapidly developing algorithms, many of which are, at least for now, opaque—though questions raised may need to be revisited as the field evolves.
Black-box medical algorithms are also often plastic, changing in response to new data. This form of frequent updating is relatively common in software but relatively rare in the context of other medical interventions, such as drugs, that are identified, verified, and then used for year, decades, or centuries. These two aspects of black-box medical algorithms—opacity and plasticity—require hard thinking about how such algorithms should be validated and regulated.
Validation
How can providers, developers, regulators, and insurers be sure that black-box algorithms are accurate and useful? Diagnostic tests, a useful if imperfect parallel, are evaluated for analytical validity, clinical validity, and clinical utility; roughly speaking, does the test accurately measure what it purports to, does that measurement accurately track clinical quantities of interest, and can the results usefully guide clinical care? An ideal genetic test, for instance, accurately identifies which alleles a patient has (analytical validity) of a gene that is linked to a medical condition (clinical validity) for which knowledge of the allele can be used to direct and improve treatment (clinical utility). Both the instrumentation and the interpretation are key; without accurate instruments, the allele cannot be identified accurately, but without interpretation, the test lacks any medical use for that knowledge. For black-box algorithms, these traditional validation tools break down, because neither developers nor users knows precisely what an algorithm measures—or, more precisely, what constellation of already-measured characteristics it takes into account—or what biomedical quantities it tracks, but only what it predicts or recommends. While scientific understanding provides only limited assurance of external validity, it provides some, and that assurance is missing for black-box algorithms. To make validation more complicated, those predictions and recommendations may also change over time as the algorithm is updated to account for new data. This lack of explicit knowledge means that black-box algorithms often cannot rely on scientific understanding to provide baseline confidence in their efficacy, and do not themselves enhance understanding.
Clinical trials may also be substantially harder to undertake, depending on the nature of the algorithm in question. For some black-box algorithms, clinical trials may be feasible. For instance, algorithms to identify indolent versus aggressive prostate tumors could be evaluated in clinical trial settings, as could algorithms suggesting differential drug response by disease class. Other algorithms—like a currently speculative one that used many factors from a large dataset predict a truly individualized increased stroke risk and to recommend individualized off-label drug use to reduce that risk—would be much more difficult to evaluate through clinical trials in part because of small sample sizes.7 Demonstrating robust external validity through clinical trials is challenging even in more straightforward contexts.8 Perhaps more importantly, to the extent that an ideal black-box algorithm is plastic and frequently updated, the clinical-trial validation model breaks down further, since the model depends on a static product subject to stable validation. Finally, traditional clinical trials are often slow, expensive, and limited in size, which limits the types of algorithms that can be feasibly developed. New clinical trial models that randomize algorithmic support embedded in electronic health records could help, but even these models face challenges in the ongoing validation of changing algorithms.
Instead, validating black-box algorithms will turn on computation and data in three related steps (Fig. 1). The first is procedural: ensuring that algorithms are developed according to well-vetted techniques and trained on high-quality data. A second is harder: demonstrating that an algorithm reliably finds patterns in data. This type of validation depends on what the algorithm is trying to do. Some algorithms are trained to measure what we already know about the world, just more quickly, cheaply, or accurately than current methods; analyzing skin lesions to identify cancer falls into this category. Showing that the this type of algorithm performs at the desired level is relatively straightforward; the initial developer and, ideally, independent third parties should test predictions against held-back, independently-created, or later-generated test datasets. Other algorithms optimize based purely on patient data and self-feedback without developers providing a “correct” answer, such as an insulin pump program that measures patient response to insulin and self-adjusts over time. This type of algorithm cannot be validated with test datasets.
Fig. 1.
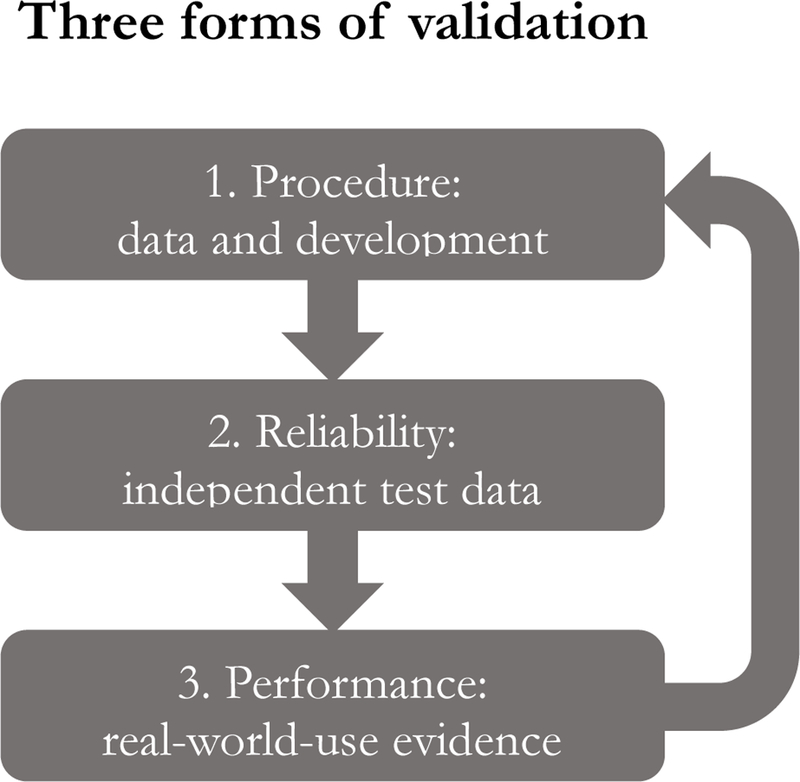
Validation of black-box algorithms. Computational validation of black-box algorithms involves three related steps: 1) ensuring basic quality of training data and development procedures; 2) testing algorithm performance against independent test data; and 3) evaluating performance in ongoing use. Data from real-world use can be used to improve further iterations of the algorithm.
The third and most important step of validation applies to all sorts of black-box algorithms: they should be continuously validated by tracking successes and failures as they are actually implemented in care settings. The learning health-care system (https://www.nap.edu/catalog/11903/the-learning-healthcare-system-workshop-summary) is thus a crucial enabler of black-box algorithms, using clinical experience to enable retrospective or contemporary analysis of algorithm-driven outcomes to confirm algorithm quality. Newly collected data not only can validate existing algorithms, they also can—and should—improve algorithms by enabling dynamic updates and improvements.
Algorithmic validation of these types would require increased transparency of details about algorithmic development: What techniques and datasets were used to develop the algorithm? Enabling the rapid deployment of high-quality algorithms thus suggests a preference for making algorithmic development open and public rather than proprietary, given the inherent opacity, uncertainty, and plasticity of their actual predictive mechanisms. Such openness necessarily complicates the mix of incentives available for algorithm developers, because it eliminates the possibility of trade secrecy; patents are already difficult to acquire for black-box algorithms based on the recent Supreme Court cases of Mayo v. Prometheus and Alice v. CLS Bank, raising other questions for policymakers seeking to facilitate algorithmic development.5 Nevertheless, demonstrating algorithmic validity is key to patient safety and the provision of high-quality medical care, and transparency is key to that effort.
Regulation
Regulatory oversight is closely linked to validation. In the United States, medical algorithms fall within the purview of the Food & Drug Administration, which regulates the broad category of medical devices.8 FDA’s default approach relies on controlling market entry by requiring pre-entry demonstrations of safety and efficacy, typically through clinical trials unless the product is low-risk or an equivalent product is already approved. The 21st Century Cures Act allows FDA to regulate medical software if it analyzes images or does not allow a provider to understand and review the basis for its conclusions. FDA recently stated that it intends to exercise this authority for clinical decision support software with non-reviewable decision mechanisms, a category which includes most black-box medical algorithms (https://www.fda.gov/downloads/medicaldevices/deviceregulationandguidance/guidancedocuments/ucm587819.pdf). Although FDA’s statement included little detail on the agency’s thinking or how much evidence it plans to require, a different, relatively recent draft guidance suggests fairly strict premarket scrutiny is likely (https://www.fda.gov/downloads/medicaldevices/deviceregulationandguidance/guidancedocuments/ucm416685.pdf). Unfortunately, this approach could keep a broad swath of black-box medical algorithms from use, since traditional clinical trials will often be infeasible and developers frequently will be unable to point to an antecedent already-approved product, not least because the algorithms involved are opaque. When algorithms evolve in response to new data, strict pre-market gatekeeping, which assumes a static product, becomes even less appropriate.
Instead, regulators could recognize that black-box medical algorithms—and potentially algorithms more broadly—should be regulated with methods that require validation while allowing room for flexibility and innovation. At the time of initial deployment, procedural checks could verify competent development and, in some circumstances, independently reproducible results. FDA’s Digital Health Innovation Action Plan, announced in 2017, adopts at a pilot level a flexible approach focused on trusted developers and more postmarket review rather than product-level premarket review, and could be adapted for black-box algorithms as appropriate (https://www.fda.gov/downloads/MedicalDevices/DigitalHealth/UCM568735.pdf). More flexible pre-market evaluation, however, should be only part of the picture; more robust continuing oversight would be needed as algorithms are deployed and evolve. Oversight could be graduated based on the risks implicated by the algorithm, an approach FDA has recognized as appropriate for medical software (https://www.fda.gov/downloads/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/UCM524904.pdf). Ideally, such oversight would involve collaboration with other health-care stakeholders, and would incorporate continued feedback from data collected in learning health systems.7
This combination may seem risky, and parallels calls for relaxed standards for drug approval that themselves raise substantial concerns about patient safety and drug efficacy.9 Ensuring that postmarket surveillance is robust thus represents a real and ongoing challenge. Such oversight can be improved by continued involvement of other stakeholders in the health care system, facilitated by FDA-mediated data sharing and transparency and by the ongoing development of learning health systems and real-world evidence strategies.10
Moving forward
Actually putting black-box algorithms into practice presents substantial challenges, but those challenges need to be addressed soon. A key task will be informing providers who may implement black-box algorithms in practice and setting the incentives and oversight mechanisms appropriately for those providers. Especially during the early stages of black-box algorithms, provider knowledge and expertise can provide an invaluable line of experience-based review. Providers can serve to evaluate the balance between a recommended intervention’s risk and the level of confidence and evidence in that recommendation. If, for instance, an algorithm suggests a hidden risk of lung cancer that calls only for further testing or watchful waiting, even relatively low levels of validation might justify that recommendation. On the other hand, if an algorithm recommends forgoing a standard treatment, or treating an unknown indication with a powerful drug, provider experience could judge such a recommendation too risky in the absence of very strong validation. Providers can also help balance an algorithm’s goal of average, overall accuracy with the realities of individual patient experiences and the possibilities of rare diseases outside algorithmic training sets. Of concern is the possibility that such provider second-guessing could swamp the added value of algorithms; black-box medical algorithms should not be artificially limited to only those applications that confirm what providers already know. All of these considerations will take place within the confines of a medical malpractice system that often unfortunately promotes conservative practice over carefully adopting new technology.
Black-box medical algorithms provide tremendous possibilities for using big health data in ways that are not merely incremental but transformative. While potential benefits are significant, so are the hurdles to the development and deployment of high-quality, validated, usable algorithms. The technology is coming quickly, and the groundwork for its arrival should be laid now. The current path is worrisome; an easy default is for algorithm developers to keep everything as secret as they can, for FDA— once it weighs in on this question specifically—to crack down by demanding clinical trials and to provoke a corresponding backlash from industry, and for doctors and patients to lose out on potential benefits. Needless to say, this is an outcome to avoid. A more engaged approach would combine regulatory flexibility, transparency, and broader involvement by different actors across the health system. Researchers, providers, and regulators alike should take notice of this rapidly approaching technology and begin to engage with challenging questions about how it should best be validated, regulated, and deployed.
Acknowledgements:
I thank Sam Handelman and Greg Curfman for helpful comments on earlier drafts.
Funding: This work was supported by the National Cancer Institute (1-R01-CA-214829-01-A1; The Lifecycle of Health Data: Policies and Practices) and by the Novo Nordisk Foundation (NNF17SA0027784; CeBIL: Collaborative Research Program in Biomedical Innovation Law).
Footnotes
Competing interests: The author declares no competing interests.
References
- 1.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S, Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu NT, Holcomb JB, Wade CE, Batchinsky AI, Cancio LC, Darrah MI, Salinas J, Development and validation of a machine learning algorithm and hybrid system to predict the need for life-saving interventions in trauma patients. Med. Biol. Eng. Comput 52, 193–203 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Obermeyer Z, Emanuel EJ, Predicting the Future — Big Data, Machine Learning, and Clinical Medicine. N. Engl. J. Med 375, 1216–1219 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liehr T, Acquarola N, Pyle K, St-Pierre S, Rinholm M, Bar O, Wilhelm K, Schreyer I, Next generation phenotyping in Emanuel and Pallister Killian Syndrome using computer-aided facial dysmorphology analysis of 2D photos. Clin. Genet 93, 378–381 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Price WN 2nd, Black-box Medicine. Harv J. L. & Tech 28, 419–467 (2015). [Google Scholar]
- 6.Burrell J, How the machine ‘thinks’: Understanding opacity in machine learning algorithms. Big Data Soc 3, 2053951715622512 (2016). [Google Scholar]
- 7.Price WN 2nd, Regulating Black-box Medicine. Mich. L. Rev 116, 421–474 (2017). [PubMed] [Google Scholar]
- 8.Rothwell PM, External validity of randomised controlled trials: “to whom do the results of this trial apply?”. Lancet 365, 82–93 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Darrow JJ, Avorn J, Kesselheim AS, New FDA Breakthrough-Drug Category — Implications for Patients. N. Engl. J. Med 370, 1252–1258 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Sherman RE, Anderson SA, Dal Pan GJ, Gray GW, Gross T, Hunter NL, LaVange L, Marinac-Dabic D, Marks PW, Robb MA, Shuren J, Temple R, Woodcosk J, Yue LQ, Califf RM, Real-world evidence—what is it and what can it tell us. N. Engl. J. Med 375, 2293–2297 (2016). [DOI] [PubMed] [Google Scholar]


