Abstract
Variegation is a frequently observed genetic phenomenon in landscaping. In this study, an ethyl methanesulfonate induced variegated leaf (Csvl) mutant in cucumber (Cucumis sativus L.) was identified. The Csvl mutant displayed green-yellow-white variegation phenotype throughout the whole growth cycle, while the leaf of wild type plants was normal green. The photosynthetic pigment contents and photosynthetic parameters of Csvl was significantly lower than wild type. The cytology observation results showed that the mesophyll cells of Csvl mutant contained defective chloroplasts. Genetic analysis indicated that variegated leaf phenotype was monogenic recessive inheritance. MutMap and genotyping results revealed that Csa6G405290 (Cscs), encoding chorismate synthase, was the candidate gene for variegated leaf mutant in cucumber. The expression level of Cscs was similar between wild type and variegated leaf mutant leaves. Transcriptome profile analysis of leaves of Csvl mutant identified 183 candidate genes involved in variegated leaf development in cucumber, including genes that encode heat shock protein, zinc finger protein. Cscs may regulate variegated leaf in cucumber by interacting with these genes. In a word, these results revealed that Cscs might regulate the variegated leaf phenotype in cucumber.
Keywords: chorismate synthase, cucumber, leaf variegation, MutMap, RNA-seq
Introduction
Leaf is a significant organ for photosynthesis, respiration, and nutrient transformation in plants, the change in leaf color is determined by genotype and growth environment (Esteban et al. 2008, Sakamoto et al. 2009). Leaf variegation, forming different colored sectors in the same leaf, is a frequently observed genetic phenomenon in landscaping (Yu et al. 2007). In nature, the variegated plants usually occur in tropical and subtropical forests (Cui and Guan 2013). Some adaptive advantages of variegated leaves caused by the genotype can play a role in protecting themselves so that they can persist in the local populations. For example, the brightly colored patches can be alert to predators and attract insects to pollinate (Chen et al. 2017b, Sheue et al. 2012). Some previous studies have been conducted on chemical reagent induced leaf variegation mutants. At present, extensive studies have been conducted on the molecular mechanism of leaf variegation. The Arabidopsis var2 mutant phenotype was attributed to a mutation in the subunit of the filamentation temperature-sensitive (FtsH) complex located in the thylakoid, which may participate in chloroplast development and biogenesis (Chen et al. 2000). Complete loss of the FtsH complex caused the chloroplast to develop stagnant, while slight loss led to plant leaf variegation (Putarjunan et al. 2013). Recent studies on rice stripe mutants showed that stripe1–2 (st1–2) encoded a small subunit of ribonucleotide reductase 1 (RNRS1), and the mutation of Val-171 residue in st1–2 impacted the attachment of RNRS1 to iron, leading to defective chloroplast development (Chen et al. 2015). The tomato gh gene encoded a plastid terminal oxidase, which was involved in carotenoid biosynthesis as a phytoene desaturase cofactor, and its deficiency led to photobleaching in leaves under high-light, mainly due to the ability to synthesize colored carotenoids in gh mutant (Shahbazi et al. 2007).
Cucumber (Cucumis sativus L.) is one of the important horticultural crop worldwide, whose genome was sequenced successfully (Huang et al. 2009). However, the narrow hereditary basis of cucumber has seriously restricted its molecular breeding progress. In order to increase the genetic resources of cucumbers, several research groups have constructed cucumber mutant libraries, and some important mutation traits have been obtained, such as dwarf plant, fruit peel color and fruit thorn and tumor (Boualem et al. 2014, Fraenkel et al. 2012, Wang et al. 2014, Xue et al. 2016). Researchers have performed cucumber gene function analysis by genomic information and techniques combined with large-scale construction of cucumber mutant library, such as super compact (SCP) mutant C257 (Wang et al. 2017), virescent yellow leaf mutant vyl (Song et al. 2018), male sterile cucumber mutant (Han et al. 2018), male flower mutant 406a (Chen et al. 2016), light green peel mutant lgp (Zhou et al. 2015) and tendril-less mutant td-1 (Chen et al. 2017a). Spontaneous mutants of cucumber were also used to identify the glabrous genes gl-2, csgl1/Mict (Li et al. 2015), csgl3 (Cui et al. 2016, Pan et al. 2015), Tril (Wang et al. 2016a, Zhao et al. 2015), white peel gene w (Liu et al. 2016), short hypocotyl1 gene SH1 (Bo et al. 2016) and CsDET2 involving in BR biosynthesis (Hou et al. 2017) respectively. The screening of these mutagenic and spontaneous mutants and the identification of target genes promote the molecular breeding in cucumber.
Currently, MutMap method has been successfully used to precisely locate genetic loci that cause mutations from different species. And it greatly reduced the number of genetic crosses and the number of mutant progeny populations (Takagi et al. 2015). The MutMap method was first applied to the re-sequencing of seven mutants of the rice cultivar ‘Hitomebore’ crossing with their parents. The SNP molecular markers were then used to quickly locate the gene that caused the target phenotype (Abe et al. 2012). Abe et al. (2012) mapped the trait related mutation regions by analyzing the differences in the SNP frequency of the offspring DNA pool and wild-type parent, and quickly located four rice semi-dwarf, two light green leaves, and one male sterility related candidate mutation sites (Abe et al. 2012). Besides, MutMap technology was applied to the identification of salt tolerance gene in rice (Takagi et al. 2015) and root development gene CTR1 in Arabidopsis (Tabata et al. 2013). MutMap method has also successfully identified some important genes in cucumber, such as gl2 encoding a C-type lectin receptor-like tyrosine protein kinase (Xu et al. 2015), Ycf54 gene controlling the cucumber light green leaf and fruit (Lun et al. 2016), plant dwarf gene CsaVFB1 (Lin et al. 2016), light green peel gene CsaARC5 (Zhou et al. 2015) and yellow green peel gene CsMYB36 (Hao et al. 2018).
In our previous study, we isolated an ethyl methanesulfonate (EMS) induced variegated leaf (Csvl) mutant in cucumber (Cucumis sativus L.) (Xue et al. 2016). The wild type leaves showed normal green, while the leaves of the Csvl mutant displayed variegation. Csvl mutant had lower photosynthetic pigment content and net photosynthetic rate than wild type plants. The morphology of chloroplasts was defective in Csvl mutant. Csa6G405290 (Cscs), encoding chorismate synthase, was identified to be the candidate gene for variegated leaf mutant in cucumber by conducting MutMap and kompetitive allele specific PCR (KASP) genotyping analysis. The expression level of Cscs was similar between wild type and Csvl mutant. Transcriptome sequencing also identified some potential target genes which might interact with Cscs to regulated the variegated leaf phenotype in cucumber. Over all, the results obtained in the present study are beneficial to further dissect the molecular mechanism leaf variegation formation in cucumber.
Materials and Methods
Plant materials and growth condition
In our previous study, we isolated some ethyl methanesulfonate (EMS) induced important mutants in cucumber, such as few spines mutant, fruit color mutant, short fruits mutant, round leaf mutant, dwarf mutant, no flower mutant, variegated leaf mutant, long fruits mutant and leaf crimple mutant (Xue et al. 2016). In the present study, the Csvl mutant was investigated. The F2 population was crossed between wild type and Csvl mutant. The Csvl mutant, wild type plants, and F2 population were used for phenotypic characterization, casual gene identification and genotyping. All plants were grown in the greenhouse of Xiangyang farm of Northeast Agricultural University, Harbin, China.
Phenotype identification of Csvl mutant in cucumber
The color of the cotyledons and leaves were observed since the seeds emergency. Other traits observation and comparison between Csvl and wild type plants were performed at 60 d after sowing. Plant height was measured by using a tape. Leaf area of the top 7th node leaf was measured using a leaf area meter. Five plants of each type were measured, each measurement was repeated three times, and t test was conducted for statistical analysis.
Transmission electron microscopy analysis
To investigate morphology of chloroplast, the leaves of the top 4th node of wild type and Csvl plants grown for 50 d were sampled. The yellow, white, light green, normal green leaf tissues were treated for transmission electron microscopy analysis. In short, five different buffers were used in sequence, namely 2.5% glutaraldehyde, 0.1 M phosphate, 1% osmate acid, a series of ethyl alcohol, and a variety of acetone. The prepared samples were cut and stained with uranyl acetate and lead citrate. Finally, transmission electron microscopy (H-765, Hitachi Ltd., Japan) was used for observation. Three replicates were performed for each sample.
Photosynthetic pigment content quantification
Photosynthetic pigment was extracted from leaves of 5-week-old wild type and Csvl mutant plants. 0.2 g fresh leaf sample was collected from a fully expanded leaf (variegated or green) of the top 3rd or 4th node, and then steeped in 80% acetone for 48 h. The quantification of pigments was conducted using a microplate spectrophotometer (Epoch, Bio Tek Instruments Inc., USA) according to the method reported previously (Garcia and Nicolas 1998, Wellburn 1994). The photosynthetic pigment content of 10 plants of each type was measured for a total of three measurements. It’s worth noting that all measurement operations should be carried out in the dark to avoid photosynthetic pigment degradation.
Photosynthetic parameter determination
Photosynthetic rate (Pn), transpiration rate (Tr), stomatal conductance (Gs), and intercellular CO2 molar fraction (Ci) of fully expanded true leaf from wild type and Csv1 mutant plants were determined using portable photosynthesis system (LI-6400XT, LI-COR Inc., USA). A total of 10 measurements were taken for each type of leaf on sunny day 9:00–11:00.
Genetic analysis
The formation of variegation in F2 population were observed after the obvious period of variegation phenotype. The number of plants in the wild type and the variegation leaf phenotype was counted and subjected to χ2 test.
RNA extraction
Total RNA of root, stem, leaf, tendril, female flower and male flower in wild type plants and leaf in Csvl were extracted using TRIzol reagent (Invitrogen, USA), Dried RNA samples were dissolved in H2O treated with diethylpyrocarbonate. UV-spectrophotometer was used to measure RNA concentration and purity. Its quality was detected by 1% agarose gel electrophoresis. Reverse transcription was carried out using the ReverTra Ace qPCR RT Kit (TOYOBO) in accordance with the manufacturer’s instruction.
Whole genome re-sequencing, SNP calling and filtering
DNA of leaves of wild type and F2 population plants were extracted according to CTAB method (Murray and Thompson 1980) respectively. The mutant DNA bulk from 17 F2 plants with variegation phenotype and DNA of one wild type were re-sequenced respectively as a commercial service at Annoroad Genomics (Beijing).
MutMap technique (Takagi et al. 2015, Xu et al. 2015) was used to identify candidate genes of Csvl mutant. The mixed pool data of mutant bulk and the data of wild type were compared with the reference genome (ftp://www.icugi.org/pub/genome/cucumber/Chinese_long/v2/) (Huang et al. 2009) to obtain the single nucleotide polymorphism (SNP) data set after the sequencing was completed. Then, the SNPs identified in both wild type and mutant DNA pool were removed, SNPs unique to mutant DNA pool were obtained. The SNP-index is calculated by dividing the frequency of the mutation sequence at this locus by the total depth of the locus (Li et al. 2009).
KASP genotyping
KASP (Saxena et al. 2014, Semagn et al. 2014), was applied for SNP genotyping. The DNA samples from the variegation phenotype and normal leaf color phenotype plants in F2 population were selected. KASP Primer mix containing two different, allele specific, competing forward primers with unique tail sequences and one reverse primer and universal KASP Master mix containing the commonly used FRET cassette fluorescent primers, ROX internal reference dyes, Klear Taq DNA polymerase, dNTP and MgCl2 were added into the sample DNA for PCR amplification. Following completion of the KASP PCR, the test results were read and the SNP site genotype data of the sample was analyzed.
Quantitative reverse transcription PCR (qRT-PCR) analysis
To identify the expression of candidate gene in different tissues, gene-specific primers were designed on-line (https://www.genscript.com/tools/real-time-pcr-tagman-primerdesign-tool) based on candidate gene CDS sequence information (Supplemental Table 1). The CsEF1α gene was amplified as the reference gene. The Taq SYBR® Green qPCR Premix (Yugong Biolabs, Inc., CN) and iCycler iQTM5 real-time PCR detection system (Bio-Rad, Hercules, CA, USA) were used to perform the qRT-PCR. The program used in this assay was as follows: 96°C for 1 min, 95°C for 15 s, 56°C for 15 s, 72°C for 45 s, 45 cycles. Four technical replicates were set for each sample and Tukey’s test and t test were conducted for statistical analysis.
Bioinformatics analysis
The protein sequences of the candidate gene and its homologues were obtained from GenBank (http://www.ncbi.nlm.nih.gov). The structure of the gene and the full length of DNA were searched in the Cucurbit Genomics Database (http://cucurbitgenomics.org/organism/2). Multiple sequence alignment analysis was performed using DNAMAN version 8.0. Prediction of subcellular localization of eukaryotic proteins was conducted by TargetP on line (http://www.cbs.dtu.dk/services/TargetP).
RNA-seq analysis
To further probe the mechanism of variegated leaf development, the differentially expressed genes from Csvl and wild type were screened by RNA-seq. Leaves of wild type and Csvl plants, with length equal to 3 cm, were harvested at 53 d after sowing for RNA extraction. At least 20 μg of total RNA (≥600 ng/μL) was prepared for Solexa sequencing. The Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) was used to evaluate total RNA quantitation. A total quantity of 1 μg RNA per sample with RIN values above 7 was used for library preparation. Then constructed libraries were tested to qualitative analysis and sequenced using Illumina HiSeq instrument in line with manufacturer’s instructions (Illumina, San Diego, CA, USA).
Genome website (http://www.icugi.org/cgi-bin/ICuGI/index.cgi) was consulted for the reference genome, and gene model annotation files. Hisat2 (v2.0.14) was used for indexing of the reference genome and clean data was employed for comparison (Kim et al. 2015). The read numbers mapped to each gene were calculated using Integrative Genomics Viewer. Gene expression levels were calculated using software Htseq (v0.6.1). Analysis of the overall expression of the transcriptome using the standard RPKM method (Reads per kilobase per million mapped reads) to calculate the expression levels of genes (Mortazavi et al. 2008).
Analysis of differential expression in wild type and Csvl was conducted using the DEGSeq Bioconductor package (Robinson and Oshlack 2010). The p values were adjusted using the Benjamini and Hochberg method. Corrected p value of 0.05 was set as the threshold for significantly differential expression.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of differential expression genes
The gene length bias was corrected by GO enrichment analysis implementing the GO-Term Finder. The GO terms with corrected p values of less than 0.05 were considered significantly enriched after correction by Bonferroni. KEGG is the main public database on pathway (Kanehisa et al. 2008). Pathway significance enrichment can identify the major biochemical metabolic and signal transduction pathways that differentially expressed genes are involved. After the data correction, statistical enrichment of differential expression genes in the KEGG pathways were tested (Mao et al. 2005).
Results
Phenotypic characterization of a cucumber variegated leaf (Csv1) mutant
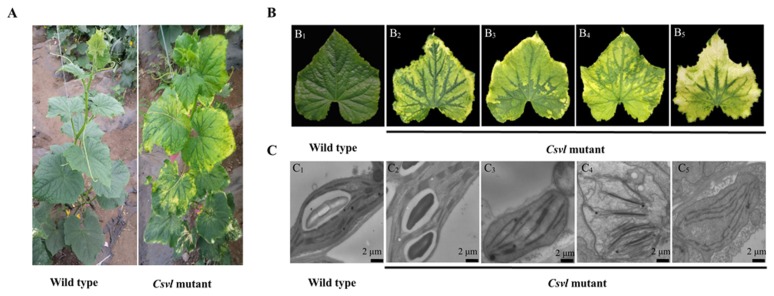
The variegated leaf Csvl mutant in cucumber was identified in our previous study (Xue et al. 2016). Phenotype identification results showed that the color of wild type leaves of each node appeared normal green throughout the growth cycle, while the color of each leaf of Csvl plants appeared variegation when the leaves grew to 3 cm during 2–3 true leaves stage (Fig. 1A). The typical character of leaf variegation was the formation of yellow or white and green sectors in the same leaf. Yellow sectors were mainly observed in the central region of the leaves, randomly, or near from leaf margins, and the white sectors were mainly at the leaf margins (Fig. 1B2–B5). The green sector of the leaves of Csvl plants showed normal green similar to those of the wild type (Fig. 1B1–B5). Furthermore, the plant height and leaf area of Csvl mutant plants were significantly reduced compared to those of wild type plants (Supplemental Fig. 1). The plant height of wild type was 191 ± 19.67 cm, which was significantly higher than Csvl, whose height was 146 ± 6.52 cm (Supplemental Fig. 1A). The leaf area of Csvl was 174.92 ± 17.37 cm2, which was significantly less than that of wild type (442.67 ± 69.92 cm2) (Supplemental Fig. 1B). In addition, the fertility, leaf angle, and leaf shape of the Csvl mutant were not significantly different from wild type plants (data not shown) (p < 0.05).
Fig. 1.
Comparison of the phenotype of wild type and variegated leaf mutant Csvl in cucumber. A: Wild type and variegated leaf mutant Csvl plants grown for 60 d. B: Wild type leaf (B1) and the main variegated leaf phenotype in Csvl mutant (B2, B3, B4 and B5). C: Observation of chloroplast ultrastructure of wild type leaf and mutant with different color sectors. (C1) Wild type. (C2) Green sector in Csvl mutant. (C3) Light green sector in Csvl mutant. (C4) Yellow sector in Csvl mutant. (C5) White sector in Csvl mutant. Magnification times: 25000 ×.
It is speculated that the formation of variegated leaf may be related to the development of chloroplasts. Therefore the chloroplast morphology of wild type and mutant was observed and compared. The result showed that the chloroplast of wild type leaf included well-organized thylakoids and starch grain. The mesophyll cell of the Csvl mutant contained a visible chloroplast but no structural feature of normal chloroplast, including the loose grana lamellae, unclear membrane structure and irregular shape (Fig. 1C1–C5). The thylakoid packed into grana and starch grain were present in mesophyll cell in the green part of the Csvl mutant (Fig. 1C2). In contrast, plastid hypoplasia in light green mesophyll cell had no starch grain and less grana lamellae (Fig. 1C3). The plastids of yellow and white leaves were hypoplastic, lacked starch grains and contained unstacked and loose thylakoids (Fig. 1C4, 1C5).
Comparison of photosynthetic pigment content and photosynthetic parameter in wild type and Csvl mutant
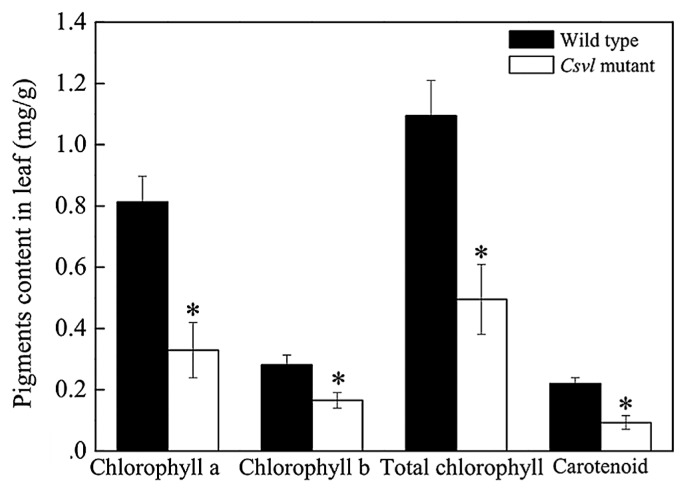
Abnormal chloroplast morphology may affect chlorophyll synthesis, so the contents of photosynthetic pigments in Csvl mutant and wild type were determined. The result showed that the content of chlorophyll a, chlorophyll b, total chlorophyll and carotenoid from mutant was significantly decreased by 59.54%, 41.14%, 54.80% and 57.71% than in wild type (Fig. 2). The analysis of photosynthetic pigment content showed that chloroplast did not completely lose the ability to synthesize chlorophyll, but the content of photosynthetic pigment decreased.
Fig. 2.
Contents of chlorophyll a, chlorophyll b, total chlorophyll and carotenoid in Csvl mutant and wild type leaves. ‘*’ indicate the significant difference of target between wild type and Csvl mutant with p value < 0.05. Data were presented as means ± SD of five replicates. t test was conducted for statistical analysis.
The leaf color is closely related to photosynthesis, and the photosynthetic rate in the leaf variegation mutant may be decreased. Therefore, the photosynthetic parameters in Csvl mutant and wild type were measured at 50 d after planting. The result showed that photosynthetic rate of reduction in Csvl mutant was 59.05% compared to in wild type (Supplemental Table 2). The stomatal conductance, intercellular CO2 concentration and transpiration rate of mutant significantly increased by 47.73%, 11.59% and 28.01% than that of wild type respectively (Supplemental Table 2).
Identification of candidate genes for Csvl mutant
In order to identify the genetic characteristics of the variegated leaf phenotype in cucumber, Csv1 was crossed with wild type. The F1 plants had the same phenotype as the wild type with no variegated leaves. However, the significant phenotypic segregation was occurred in the F2 population. The result of χ2 test (χ2 = 0.59, p > 0.05) showed that the ratio of wild phenotype and variegation phenotype plants in F2 population was 3:1 (WT:Csvl = 65:17), which revealed that the phenotype of Csvl mutant was regulated by a single recessive gene.
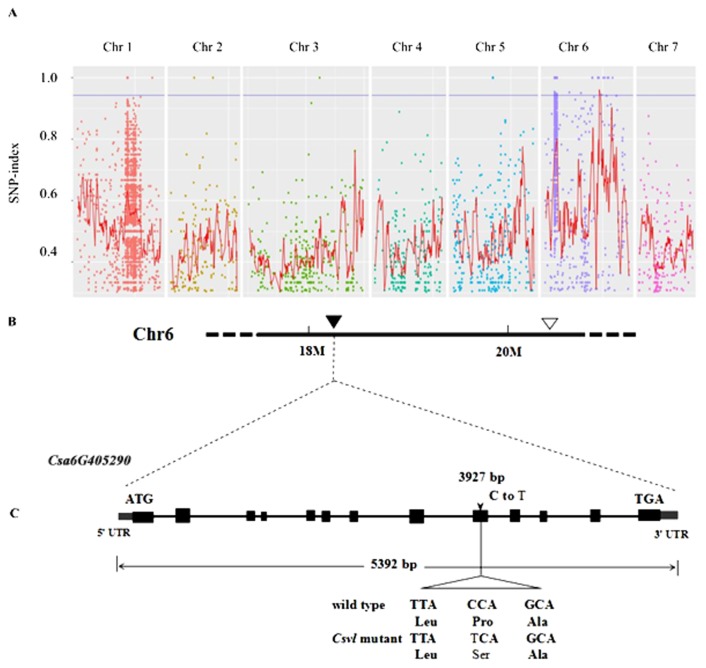
MutMap technique was then used to identify the candidate gene of the Csvl mutant. The DNA bulk from 17 F2 plants showing leaf variegation phenotype (mutant pool) and DNA of one wild type was re-sequenced respectively. A total of 6408 pool-specific SNPs were detected in Csv1 mutant (Supplemental Table 3), and the distribution map of SNP site for all seven chromosomes of cucumber was plotted by calculating the SNP-index (Fig. 3A). Candidate SNPs were selected in accordance with; following norms: first, SNP-index equal to 1; second, the mutation mode is homozygous, meaning that mutation should occur in the mutant instead of the wild type; third, mutation should be the typical EMS mutagenized type, including mutation from C to T or from G to A; fourth, mutation should be located in the exon region with nonsynonymous mutation or in the alternative splicing site of the intron of the gene. Then six SNPs with SNP-index of 1 in the bulk of mutant were screened (Table 1). Among the six candidate SNPs, SNP 18506237 was located in the intronic region without an alternative splice site and can be eliminated. Among the remaining five SNPs, three SNPs (18670038, 22022883, and 21578651) were located in downstream and UTR3. SNP 18277305 was located in the exon with an non-synonymous amino acid change, and the SNP-23376757 were located in splicing site of intron (Table 1, Fig. 3B), which indicated that genes harboring these two SNPs might be the candidate gene of Csvl mutant in cucumber.
Fig. 3.
Scatter plot of SNP-index for Csvl mutant and the structure diagram of Cscs gene. A: Distribution of SNP-index on seven chromosomes of cucumber in mutant pool. The red line indicates the distribution of average SNP-index; the blue line represents the threshold. B: Two SNPs located on cucumber chromosome 6. Black triangles indicate non-synonymous SNP-18277305; white triangles indicate SNP-23376757 located in the splicing site. C: The structure of predicted Csa6G405290. Rectangles represent exons; solid lines represent introns; arrow indicates the position of SNP 18277305.
Table 1.
Analysis of the candidate SNPs for the Csvl mutant
| chromosome | Position | Gene | Reference | Alteration | Wild type | Reference depth | Alteration depth | SNP_index | Type | Amino acid change | Annotation |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr. 6 | 18506237 | Csa6G406550 | C | T | 0/0 | 0 | 15 | 1 | intron | ||
| Chr. 6 | 18670038 | Csa6G408170 | C | T | 0/0 | 0 | 15 | 1 | downstream | ||
| Chr. 6 | 21578651 | Csa6G452640 | C | T | 0/0 | 0 | 12 | 1 | UTR3 | ||
| Chr. 6 | 22022883 | Csa6G476670 | G | A | 0/0 | 0 | 20 | 1 | downstream | ||
| Chr. 6 | 23376757 | Csa6G490810 | G | A | 0/0 | 0 | 11 | 1 | splicing | Nodulation protein H | |
| Chr. 6 | 18277305 | Csa6G405290 | C | T | 0\0 | 0 | 12 | 1 | exon | P to S | Chorismate synthase |
To further validate the candidate SNP, the genotyping analysis in F2 population plants was conducted by using the mutation information of two SNPs (18277305 and 23376757) with KASP technique. The results indicated that SNP 18277305 of Csa6G405290 was co-segregated with phenotyping data in the F2 population. The genotype of plants with mutant phenotype was T: T, and plants with wild type phenotype was C:T or C:C (Supplemental Table 4). However, SNP 23376757 was not co-segregated with the phenotype in F2 population plants (Supplemental Table 4). These results demonstrated that Csa6G405290, encoding chorismate synthase, which contains SNP 18277305, was identified to be the candidate gene of the cucumber Csvl mutant. In this study, the candidate gene of cucumber variegated leaf mutant is named Cscs.
Sequence and expression analysis of Cscs gene
The DNA sequence of Cscs (Csa6G405290) was 5392 bp in length containing 13 exons and 12 introns by consulting Cucurbit Genomics Database (http://cucurbitgenomics.org/organism/2) (Fig. 3C). A transition from proline (Pro) to serine (Ser) was occurred in the ninth exon of Cscs gene due to a C-to-T substitution (Fig. 3C). Cscs encoded the chorismate synthase catalyzed the dephosphorylation of 5-enolpyruvylshikimate 3-phosphate (EPSP) to generate the chorismate (Braus 1991).
Cscs protein contains three characteristic domains, ranging from 70th to 85th amino acids, 177th to 193th amino acids and 383th to 399th amino acids, respectively (Supplemental Fig. 2). Most species have these three characteristic regions, which are mainly used for binding to the substrates EPSP and NADPH in higher plants, bacteria and other microorganisms. Mutations occured at certain site in the conserved domains can result in changes in enzyme activity (Ahn et al. 2004). Chloroplast transit peptide was found in Cscs protein by the prediction of TargetP (http://www.cbs.dtu.dk/services/TargetP).
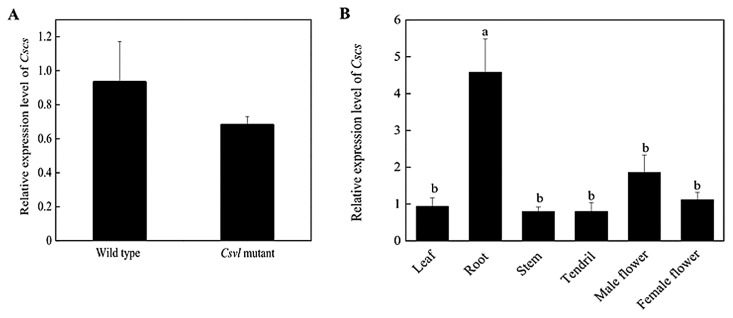
Gene mutation may lead to changes in its expression. In order to detect whether the expression of Cscs was changed in mutant, its relative expression level was detected by qRT-PCR. The results show that the expression of Cscs in mutant was similar to wild type (Fig. 4A). In addition, Cscs expression in root of wild type plants was significantly higher than that of leaf, stem, female flower, male flower and tendril (Fig. 4B).
Fig. 4.
The relative expression of Cscs in cucumber. A: The relative expression level of Cscs in leaves of wild type and Csvl mutant. t test was conducted for statistical analysis (p value < 0.05). B: The relative expression of Cscs in different organs of wild type. Letters indicate the significant difference of traits between wild type and Csvl mutant with p value < 0.05. Data were presented as means ± SD of four replicates. Tukey’s test was conducted for statistical analysis.
Transcriptome profile analysis of Csvl mutant in cucumber
We tried to explore the molecular mechanism of variegation related genes from the transcriptional level and lay a foundation for the further study of variegation mutant in cucumber. A total of 183 differentially expressed genes related to variegated leaf in cucumber were identified by RNA-seq, within which 128 genes were significantly up-regulated and 55 genes were significantly down-regulated in the leaves of Csvl mutant as compared to wild type (Supplemental Table 5).
qRT-PCR were performed to verity the differentially expressed genes identified by RNA-Seq in the present study. The expression changes of the 18 differentially expressed genes showed that the results of qRT-PCR analysis were consistent with RNA-Seq data (Supplemental Table 6), indicating that the RNA-Seq data were reliable.
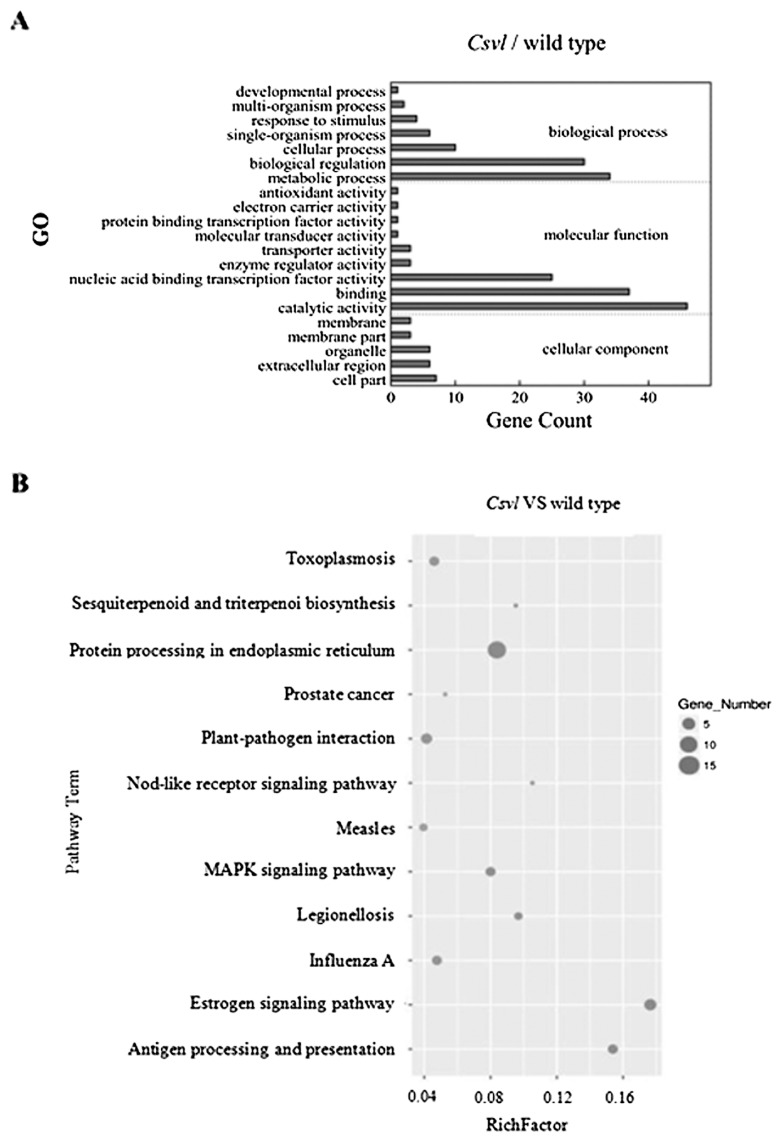
GO terms are used for functional classifications of differentially expressed genes obtained in this study, which include biological processes, cellular components, and molecular functions. Pathways analysis are derived from the KEGG database. Results showed that these genes were significantly enriched in 21 GO terms and 12 KEGG pathways (Csvl versus wild type). The major biological groups of differentially expressed genes were functionally enriched in nucleic acid binding transcription factor activity, cellular process, catalytic activity, metabolic process, biological regulation, and binding (Fig. 5A). The main pathways of differentially expressed genes were enriched in estrogen signaling pathway, protein processing in endoplasmic reticulum, plant-pathogen interaction, antigen processing and presentation pathways, and MAPK signaling pathway (Fig. 5B).
Fig. 5.
The analysis of GO classification and KEGG enrichment of variegated leaf related genes in cucumber. A: The main GO classification of variegated leaf responsive genes in cucumber. The abscissa indicates the number of different genes in terms, and the ordinate indicates the enriched GO term (p values < 0.05). B: KEGG enrichment scatter plot of differentially expressed genes related to leaf variegation in cucumber. Ordinate represents the pathway name; the abscissa represents the Rich factor, and the count of differentially expressed genes is represented by the size of the dots in this pathway.
Based on known function and functional predictions, some candidate important genes related to leaf variegation in cucumber were selected, such as genes encoding heat shock protein and zinc finger protein (Supplemental Tables 7, 8). Under normal growth and stress conditions, plants can produce heat shock proteins with molecular weight of 10–200 kD. Heat shock proteins were divided into five classes according to molecular weight, including HSP100, HSP90, HSP70, HSP60 and sHSPs (Waters et al. 1996). Some heat shock protein-related genes were identified as differentially expressed genes related to variegated leaf development in cucumber (Supplemental Table 7). These genes were predominantly up-regulated in Csvl mutant, such as HSP70 (Csa2G070310, Csa5G514500, Csa5G512930, Csa7G312930), DNAJ HSP (Csa3G710780, Csa2G264570), HSP90 (Csa3G183950), Hsp100 (Csa1G039100) and smHSPs (Csa5G198120, Csa5G190530, Csa3G020090, Csa3G020080, Csa7G072770, Csa1G527870, Csa5G591720, Csa5G190560, Csa1G527860). Zinc finger protein (ZFP) transcription factor, one of the important family of transcription factors in plants, acts a pivotal part in many biological processes, such as plant morphogenesis, transcriptional activation and stress (Laity et al. 2001, Takatsuji 1999). Some up-regulated genes related to ZFP were found as differentially expressed genes related to variegated leaf development in cucumber (Supplemental Table 8), such as Csa6G303740 (C2H2 type), Csa6G405920 (GATA type), Csa6G499220 (C3HC4-type), Csa6G152950 (AN1-like) and salt tolerance zinc finger (Csa2G354820, Csa4G642460, Csa5G207940).
Discussion
Leaf color variation is mainly due to gene mutation affects the synthesis and degradation of chlorophyll, which in turn leads to changes in pigment content, composition and proportion (Beale 2005). In this study, reduced chlorophyll, carotenoid content (Fig. 2) and defective chloroplast (Fig. 1C2–C5) were contained in Csvl mutant. The Cscs gene involved in the formation of cucumber leaf variegation was successfully identified, which encoded chorismate synthase. Chorismate synthase catalyzed the dephosphorylation of EPSP to generate the chorismate, which was an important intermediary metabolism substance (Chen et al. 2012, Herrmann 1995). Salicylic acid (SA), produced with chorismate as precursor is a phenolic plant hormone that involves in the regulation of photosynthetic pigment and photosynthetic machinery (Kusumi et al. 2006, Wildermuth et al. 2001). Under the environmental stress, moderate salicylic acid can avoid the decrease of the photosynthetic pigment content and maintain a high photosynthetic rate to ensure the normal growth in plant. Chloroplast is one of the major sites for ROS production in cells and ROS accumulation can cause oxidative damage to photosynthetic membranes (Apel and Hirt 2004, Wang et al. 2010). SA also mediately affects photosynthetic electron transport, indicating that SA plays a significant part in regulating the balance of redox reaction in the chloroplast (Janda et al. 2012). A benzoquinone derivative, plastoquinone, was also produced by a series of reactions to chorismate (Ksas et al. 2015). Plastoquinone acted as the electron mediator in the photosynthetic chain, and its absence in thylakoid caused the electron transfer of the photosynthetic chain to be blocked, and eventually the plastid was photooxidized (Norris et al. 1995).
Based on previous research, Cscs gene encoding chorismate synthase, which is the key enzyme in the chloroplast SA and plastoquinone biosynthetic pathway. Blocked SA and plastoquinone synthesis pathways due to Cscs gene mutation can affect the biogenesis of chloroplasts and photosynthetic pigments, and destroy the balance of redox reactions in the chloroplasts. Therefore, we hypothesized that mutation in Cscs caused insufficiency of chorismate, and then SA and plastoquinone level reduced in the chloroplasts of the Csvl mutant. SA protects the PSII reaction center and regulates the turnover of D1 protein in chloroplasts and reduce the damage of photosynthetic apparatus in leaves under high temperature and light stress (Sahu et al. 2002). PSII can not be repaired under excessive light, causing damage to the chloroplasts and rapid degradation of photosynthetic pigments. Meanwhile, the lack of plastoquinone may block the transfer of electrons from PS II to the cytochrome b6/f complex in photosynthetic chain. The photoreaction proceed abnormally caused photooxidation damage of the PS II in the chloroplast, and abnormal photosynthetic pigment metabolism. In summary, a new mechanistic insight has been provided that Cscs gene mutation can lead to the formation of the variegated leaves in cucumber.
Transcriptional sequencing experiment also provided a theoretical basis for the molecular mechanism variegation formation. We analyzed differentially expressed genes from leaves of wild type and Csvl, which were heat shock protein and zinc finger protein. Heat shock proteins can be expressed in peroxidation (Wang et al. 2004). Previous studies indicated that chloroplast Hsp70 was involved in photoprotection and repair of photodamaged photosystem II (PS II) (Schroda et al. 1999). DNAJ heat shock protein was a helper protein of HSP70 (Kampinga and Craig 2010). Some of the DnaJ-like zinc finger domain proteins were involved in the development or maintenance of plastid and regulate the redox reactions in PS I (Wang et al. 2016b). For example, the AtJ8 protein was found to be nearly absent from the Arabidopsis variegation (var1) mutant (Chen et al. 2011). Zinc finger proteins were also involved in photosynthetic physiology and pigment regulation. Salt-tolerant zinc finger protein gene (STZ) was found in Arabidopsis, which encode a C2H2 zinc finger protein with double zinc finger structure (Sakamoto et al. 2000). C2H2-type zinc finger protein also take part in the response to photooxidative stress and internal changes in leaves, such as palisade tissue, anthocyanin and chlorophyll content increased significantly (Lida et al. 2000). Cscs may also regulate the variegated leaf development in cucumber by interacting with these genes.
In conclusion, we can assume that the formation of variegated leaf is due to mutation of Cscs gene. In Csvl, mutation in the Cscs encoding the chorismate synthase hindered the synthesis and metabolism of the chorismate. Decreased levels of chorismate derivatives, such as SA and plastoquinone, result in abnormal redox reaction in chloroplast. Photo-oxidation damaged the construction of chloroplast and insufficient photosynthetic pigment content in Csvl mutant, and also induced down-regulation of photosynthesis-related genes in the nucleus, thus resulting in variegated leaf phenotype in Csvl mutant plants. In addition, Cscs may also regulate the variegated leaf development in cucumber by interacting with differentially expressed genes obtained from RNA-seq. The results obtained in the present study are important to explore the formation of the leaf variegation in cucumber from the perspective of Cscs encoding chorismate synthase.
Supplementary Information
Acknowledgements
We are thankful to University Nursing Program for Young Scholars with Creative Talents in Heilongjiang Province [grant number UNPYSCT-2015001]; National Natural Science Foundation of China [grant number 31701934]; ‘Academic backbone’ Project of Northeast Agricultural University [grant number 16XG05].
Literature Cited
- Abe, A., Kosugi, S., Yoshida, K., Natsume, S., Takagi, H., Kanzaki, H., Matsumura, H., Yoshida, K., Mitsuoka, C., Tamiru, M.et al. (2012) Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 30: 174–178. [DOI] [PubMed] [Google Scholar]
- Ahn, H.J., Yoon, H.J., Lee, B. and Suh, S.W. (2004) Crystal structure of chorismate synthase: A novel FMN-binding protein fold and functional insights. J. Biol. Chem. 336: 903–915. [DOI] [PubMed] [Google Scholar]
- Apel, K. and Hirt, H. (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55: 373–399. [DOI] [PubMed] [Google Scholar]
- Beale, S.I. (2005) Green genes gleaned. Trends. Plant Sci. 10: 309–312. [DOI] [PubMed] [Google Scholar]
- Bo, K., Wang, H., Pan, Y., Behra, T., Pandey, S., Wen, C., Wang, Y., Simon, P.W., Li, Y., Chen, J.et al. (2016) SHORT HYPOCOTYL 1 encodes a SMARCA3-like chromatin remodeling factor regulating elongation. Plant Physiol. 172: 1273–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boualem, A., Fleurier, S., Troadec, C., Audigier, P., Kumar, A.P., Chatterjee, M., Alsadon, A.A., Sadder, M.T., Wahb-Allah, M.A., Al-Doss, A.A.et al. (2014) Development of a Cucumis sativus TILLinG platform for forward and reverse genetics. PLoS ONE 9: e97963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braus, G.H. (1991) Aromatic amino acid biosynthesis in the yeast Saccharomyces cerevisiae, a model system for the regulation of a eukaryotic biosynthetic pathway. Microbiol. Rev. 55: 349–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, F., Fu, B., Pan, Y., Zhang, C., Wen, H., Weng, Y., Chen, P. and Li, Y. (2017a) Fine mapping identifes CsGCN5 encoding a histone acetyltransferase as putative candidate gene for tendril-less1 mutation (td-1) in cucumber. Theor. Appl. Genet. 130: 1549–1558. [DOI] [PubMed] [Google Scholar]
- Chen, H., Sun, J., Li, S., Cui, Q., Zhang, H., Xin, F., Wang, H., Lin, T., Gao, D., Wang, S.et al. (2016) An ACC oxidase gene essential for cucumber carpel development. Mol. Plant 9: 1315–1327. [DOI] [PubMed] [Google Scholar]
- Chen, K., Dou, J., Tang, S.R., Yang, Y.S., Wang, H., Fang, H.Q. and Zhou, C.L. (2012) Deletion of the aroK gene is essential for high shikimic acid accumulation through the shikimate pathway in E. coli. Bioresour. Technol. 119: 141–147. [DOI] [PubMed] [Google Scholar]
- Chen, K.M., Piippo, M., Holmström, M., Nurmi, M., Pakula, E., Suorsa, M. and Aro, E.M. (2011) A chloroplast-targeted DnaJ protein AtJ8 is negatively regulated by light and has rapid turnover in darkness. J. Plant Physiol. 168: 1780–1783. [DOI] [PubMed] [Google Scholar]
- Chen, M., Choi, Y., Voytas, D.F. and Rodermel, S. (2000) Mutations in the Arabidopsis VAR2 locus cause leaf variegation due to the loss of chloroplast FtsH protease. Plant J. 22: 303–313. [DOI] [PubMed] [Google Scholar]
- Chen, X.Q., Zhu, L., Xin, L., Du, K.X., Ran, X.H., Cui, X.Y., Xiang, Q.J., Zhang, H.Y., Xu, P.Z. and Wu, X.Z. (2015) Rice stripe1–2 and stripe1–3 mutants encoding the small subunit of ribonucleotide reductase are temperature sensitive and are required for chlorophyll biosynthesis. PLoS ONE 10: e0130172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.S., Chesson, P., Wu, H.W., Pao, S.H., Liu, J.W., Chien, L.F., Yong, W.H.J. and Sheue, C.H. (2017b) Leaf structure affects a plant’s appearance: combined multiple-mechanisms intensify remarkable foliar variegation. J. Plant Res. 130: 311–325. [DOI] [PubMed] [Google Scholar]
- Cui, J.Y., Miao, H., Ding, L.H., Wehner, T.C., Liu, P.N., Wang, Y., Zhang, S.P. and Gu, X.F. (2016) A new glabrous gene (csgl3) identified in trichome development in cucumber (Cucumis sativus L.). PLoS ONE 11: e0148422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, W.H. and Guan, K.Y. (2013) Diversity of leaf variegation in Chinese begonia. Plant Divers. Resour. 35: 119–127. [Google Scholar]
- Esteban, R., Fernandez-Marin, B., Becerril, J.M. and Garcia-Plazaola, J.I. (2008) Photoprotective implications of leaf variegation in E. denscanis L. and P. officinalis L. J. Plant Physiol. 165: 1255–1263. [DOI] [PubMed] [Google Scholar]
- Fraenkel, R., Kovalski, I., Troadec, C., Bendahmane, A. and Perl-Treves, R. (2012) A TILLING population for cucumber forward and reverse genetics. Cucurbitaceae Xth Eucarpia Meeting on Genetics & Breeding of Cucurbitaceae 2012: 598–603. [Google Scholar]
- Garcia, A.L. and Nicolas, N. (1998) Influence of the degree of solvent impurity on the spectrophotometric determination of chlorophylls in 80% aqueous acetone and dimethyl form amide application to nonabrasive extraction of leaves of Citrus aurantium. Photosynthetica 35: 545–550. [Google Scholar]
- Han, Y., Zhao, F., Gao, S., Wang, X., Wei, A., Chen, Z., Liu, N., Tong, X., Fu, X., Wen, C.et al. (2018) Fine mapping of a male sterility gene ms-3 in a novel cucumber (Cucumis sativus L.) mutant. Theor. Appl. Genet. 131: 449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, N., Du, Y., Li, H., Wang, C., Wang, C., Gong, S., Zhou, S. and Wu, T. (2018) CsMYB36 is involved in the formation of yellow green peel in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 131: 1659–1669. [DOI] [PubMed] [Google Scholar]
- Herrmann, K.M. (1995) The shikimate pathway: Early steps in the biosynthesis of aromatic compounds. Plant Cell 7: 907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, S., Niu, H., Tao, Q., Wang, S., Gong, Z., Li, S., Weng, Y. and Li, Z. (2017) A mutant in the CsDET2 gene leads to a systemic brassinosteriod deficiency and super compact phenotype in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 130: 1–11. [DOI] [PubMed] [Google Scholar]
- Huang, S., Li, R., Zhang, Z., Li, L., Gu, X., Fan, W., Lucas, W.J., Wang, X., Xie, B., Ni, P.et al. (2009) The genome of the cucumber, Cucumis sativus L. Nat. Genet. 41: 1275–1281. [DOI] [PubMed] [Google Scholar]
- Janda, K., Hideg, E., Szalai, G., Kovács, L. and Janda, T. (2012) Salicylic acid may indirectly influence the photosynthetic electron transport. J. Plant Physiol. 169: 971–978. [DOI] [PubMed] [Google Scholar]
- Kampinga, H.H. and Craig, E.A. (2010) The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11: 579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., Katayama, T., Kawashima, S., Okuda, S., Tokimatsu, T.et al. (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36: 480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D., Langmead, B. and Salzberg, S.L. (2015) HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksas, B., Becuwe, N., Chevalier, A. and Havaux, M. (2015) Plant tolerance to excess light energy and photooxidative damage relies on plastoquinone biosynthesis. Sci. Rep-UK. 5: 10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi, K., Yaeno, T., Kojo, K., Hirayama, M., Hirokawa, D., Yara, A. and Iba, K. (2006) The role of salicylic acid in the glutathione-mediated protection against photooxidative stress in rice. Physiol. Plant. 128: 651–661. [Google Scholar]
- Laity, J.H., Lee, B.M. and Wright, P.E. (2001) Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol. 11: 39–46. [DOI] [PubMed] [Google Scholar]
- Li, Q., Cao, C., Zhang, C., Zheng, S., Wang, Z., Wang, L. and Ren, Z. (2015) The identification of Cucumis sativus Glabrous 1 (CsGL1) required for the formation of trichomes uncovers a novel function for the homeodomain-leucine zipper I gene. J. Exp. Bot. 66: 2515–2526. [DOI] [PubMed] [Google Scholar]
- Li, R.Q., Li, Y.R., Fang, X.D., Yang, H.M., Wang, J., Karsten, K. and Wang, J.X. (2009) SNP detection for massively parallel whole-genome resequencing. Genome Res. 19: 1124–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lida, A., Kazuoka, T., Torikai, S., Kikuchi, H. and Oeda, K. (2000) A zinc finger protein RHL41 mediates the light acclimatization response in Arabidops. Plant J. 24: 191–203. [DOI] [PubMed] [Google Scholar]
- Lin, T., Wang, S.H., Zhong, Y., Gao, D.L., Cui, Q.Z., Chen, H.M., Zhang, Z.H., Shen, H.L., Weng, Y.Q. and Huang, S.W. (2016) A truncated F-box protein confers the dwarfism in cucumber. J. Genet. Genomics 43: 223–226. [DOI] [PubMed] [Google Scholar]
- Liu, H., Jiao, J., Liang, X., Liu, J., Meng, H., Chen, S., Li, Y. and Cheng, Z. (2016) Map-based cloning, identification and characterization of the w gene controlling white immature fruit color in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 129: 1247–1256. [DOI] [PubMed] [Google Scholar]
- Lun, Y.Y., Wang, X. and Zhang, C.Z. (2016) A Cs Ycf54 variant conferring light green coloration in cucumber. Euphytica 208: 509–517. [Google Scholar]
- Mao, X.Z., Cai, T., Olyarchuk, J.G. and Wei, L.P. (2005) Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21: 3787–3793. [DOI] [PubMed] [Google Scholar]
- Mortazavi, A., Williams, B.A., Mccue, K., Schaeffer, L. and Wold, B. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Murray, M.G. and Thompson, W.F. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8: 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris, S.R., Barrette, T.R. and Della, P.D. (1995) Genetic dissection of carotenoid synthesis in a rabidopsis defining plastoquinone as an essential component of phytoene desaturation. Plant Cell 7: 2139–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, Y., Bo, K., Cheng, Z. and Weng, Y. (2015) The loss-of-function GLABROUS 3 mutation in cucumber is due to LTR-retrotransposon insertion in a class IV HD-ZIP transcription factor gene CsGL3 that is epistatic over Cs GL1. BMC Plant Biol. 15: 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putarjunan, A., Liu, X.Y., Nolan, T., Yu, F. and Rodermel, S. (2013) Understanding chloroplast biogenesis using second-site suppressors of immutans and var2. Photosyn. Res. 116: 437–453. [DOI] [PubMed] [Google Scholar]
- Robinson, M.D. and Oshlack, A. (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu, G.K., Kar, M. and Sabat, S.C. (2002) Electron transport activities of isolated thylakoids from wheat plants grown in salicylic acid. Plant Biol. 4: 321–328. [Google Scholar]
- Sakamoto, H., Araki, T., Meshi, T. and Iwabuchi, M. (2000) Expression of a subset of the Arabidopsis Cys2/His2-type zinc-finger protein gene family under water stress. Gene 248: 23–32. [DOI] [PubMed] [Google Scholar]
- Sakamoto, W., Uno, Y., Zhang, Q., Miura, E., Kato, Y. and Sodmergen (2009) Arrested differentiation of proplastids into chloroplasts in variegated leaves characterized by plastid ultrastructure and nucleoid morphology. Plant Cell Physiol. 50: 2069–2083. [DOI] [PubMed] [Google Scholar]
- Saxena, R.K., von Wettberg, E., Upadhyaya, H.D., Sanchez, V., Sonqok, S., Saxena, K., Kimurto, P. and Varshney, R.K. (2014) Genetic diversity and demographic history of Cajanus spp. illustrated from genome-wide SNPs. PLoS ONE 9: e88568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda, M., Vallon, O., Wollman, F.A. and Beck, C.F. (1999) A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell 11: 1165–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semagn, K., Babu, R., Hearne, S. and Olsen, M. (2014) Single nucleotide polymorphism genotyping using kompetitive allele specific PCR (KASP): overview of the technology and its application in crop improvement. Mol. Breed. 33: 1–14. [Google Scholar]
- Shahbazi, M., Gilbert, M., Labouré, A.M. and Kuntz, M. (2007) Dual role of the plastid terminal oxidase in tomato. Plant Physiol. 145: 691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheue, C.R., Pao, S.H., Chien, L.F., Chesson, P. and Peng, C.I. (2012) Natural foliar variegation without costs? The case of Begonia. Ann. Bot. 109: 1065–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, M., Wei, Q., Wang, J., Fu, W., Qin, X., Lu, X., Cheng, F., Yang, K., Zhang, L., Yu, X.et al. (2018) Fine mapping of CsVYL, conferring virescent leaf through the regulation of chloroplast development in cucumber. Front. Plant Sci. 9: 432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata, R., Kamiya, T., Shigenobu, S., Yamaguchi, K., Yamada, M., Hasebe, M., Fujiwara, T. and Sawa, S. (2013) Identification of an EMS-induced causal mutation in a gene required for boron-mediated root development by low-coverage genome re-sequencing in Arabidopsis. Plant Signal. Behav. 8: e22534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi, H., Tamiru, M., Abe, A., Yoshida, K., Uemura, A., Yaegashi, H., Obara, T., Oikawa, K., Utsushi, H., Kanzaki, E.et al. (2015) MutMap acelerates breeding of a salt-tolerant rice cultivar. Nat. Biotechnol. 33: 445–449. [DOI] [PubMed] [Google Scholar]
- Takatsuji, H. (1999) Zinc-finger proteins: the classical zinc finger emerges in contemporary plant science. Plant Mol. Biol. 39: 1073–1078. [DOI] [PubMed] [Google Scholar]
- Wang, H., Li, W., Qin, Y., Pan, Y., Wang, X., Weng, Y., Chen, P. and Li, Y. (2017) The cytochrome P450 gene CsCYP85A1 is a putative candidate for super compact-1 (Scp-1) plant architecture mutation in cucumber (Cucumis sativus L.). Front. Plant Sci. 8: 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L.J., Fan, L., Loescher, W., Duan, W., Liu, G.J., Cheng, J.S., Luo, H.B. and Li, S.H. (2010) Salicylic acid alleviates decreases in photosynthesis under heat stress and accelerates recovery in grapevine leaves. BMC Plant Biol. 10: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L.N., Zhang, B., Li, J.R., Yang, X.Y. and Ren, Z.H. (2014) Ethyl methanesulfonate (EMS)-mediated mutagenesis of cucumber (Cucumis sativus L.). Agric. Sci. 5: 716–721. [Google Scholar]
- Wang, W.X., Vinocur, B., Shoseyov, O. and Altman, A. (2004) Role of plant heat shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9: 244–252. [DOI] [PubMed] [Google Scholar]
- Wang, Y.L., Nie, J., Chen, H.M., Guo, C., Pan, J., He, H.L., Pan, J.S. and Cai, R. (2016a) Identification and mapping of Tril, a homeodomain-leucine zipper gene involved in multicellular trichome initiation in Cucumis sativus. Theor. Appl. Genet. 129: 305–316. [DOI] [PubMed] [Google Scholar]
- Wang, Y.W., Chen, S.M., Wang, W.J., Huang, X.Q., Zhou, C.F., Zhuang, Z. and Lu, S. (2016b) The DnaJ-like zinc finger domain protein PSA2 affects light acclimation and chloroplast development in Arabidopsis thaliana. Front. Plant Sci. 7: 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters, E.R., Lee, G.J. and Vierling, E. (1996) Evolution, structure and function of the small heat shock proteins in plants. J. Exp. Bot. 47: 325–338. [Google Scholar]
- Wellburn, A.R. (1994) The spectral determination of chlorophyll a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J. Plant Physiol. 144: 307–313. [Google Scholar]
- Wildermuth, M.C., Dewdney, J., Wu, G. and Ausubel, F.M. (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565. [DOI] [PubMed] [Google Scholar]
- Xu, M.N., Wang, S.H., Zhang, S., Cui, Q.Z., Gao, D.L., Chen, H.M. and Huang, S.W. (2015) A new gene conferring the glabrous trait in cucumber identified using MutMap. Hortic. Plant J. 1: 29–34. [Google Scholar]
- Xue, C.B., Wang, C., Fan, L.X., Hao, N., Zou, D.D., Zhang, Q., Chen, H.H. and Wu, T. (2016) Ethylmethanesulfonate mutagenesis of cucumber for large-scale mutant screens. Pak. J. Bot. 48: 2261–2266. [Google Scholar]
- Yu, F., Fu, A., Aluru, M., Park, S., Xu, Y., Liu, H., Liu, X., Foudree, A., Nambogga, M. and Rodermel, S. (2007) Variegation mutants and mechanisms of chloroplast biogenesis. Plant Cell Environ. 30: 350–365. [DOI] [PubMed] [Google Scholar]
- Zhao, J.L., Pan, J.S., Guan, Y., Zhang, W.W., Bie, B.B., Wang, Y.L., He, H.L., Lian, H.L. and Cai, R. (2015) Micro-trichome as a class I homeodomain-leucine zipper gene regulates multicellular trichome development in Cucumis sativus. J. Integr. Plant Biol. 57: 925–935. [DOI] [PubMed] [Google Scholar]
- Zhou, Q. and Wang, S. and Hu, B. and Chen, H. and Zhang, Z. and Huang, S. (2015) An ACCUMULATION AND REPLICATION OF CHLOROPLASTS 5 gene mutation confers light green peel in cucumber. J. Integr. Plant Biol. 57: 936–942. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.