Abstract
Grain number per panicle is a major component of rice yield that is typically controlled by many quantitative trait loci (QTLs). The identification of genes controlling grain number per panicle in rice would be valuable for the breeding of high-yielding rice. The Oryza glaberrima chromosome segment substitution line 9IL188 had significantly smaller panicles compared with the recurrent parent 9311. QTL analysis in an F2 population derived from a cross between 9IL188 and 9311 revealed that qgnp7(t), a major QTL located on the short arm of chromosome 7, was responsible for this phenotypic variation. Fine mapping was conducted using a large F3 population containing 2250 individuals that were derived from the F2 heterozygous plants. Additionally, plant height, panicle length, and grain number per panicle of the key F4 recombinant families were examined. Through two-step substitution mapping, qgnp7(t) was finally localized to a 41 kb interval in which eight annotated genes were identified according to available sequence annotation databases. Phenotypic evaluation of near isogenic lines (NIL-qgnp7 and NIL-qGNP7) indicated that qgnp7(t) has pleiotropic effects on rice plant architecture and panicle structure. In addition, yield estimation of NILs indicated that qGNP7(t) derived from 9311 is the favorable allele. Our results provide a foundation for isolating qgnp7(t). Markers flanking this QTL will be a useful tool for the marker-assisted selection of favorable alleles in O. glaberrima improvement programs.
Keywords: African rice (Oryza glaberrima S.), grain number per panicle, chromosome segment substituted lines, fine mapping
Introduction
Rice (Oryza sativa L.) is one of the most important grain crops worldwide with increasing rice per unit yield and total yield playing an important role in ensuring global food security. Three components largely determine rice yield: effective tillers per plant, grain number per panicle, and grain weight (Sakamoto and Matsuoka 2008). As with the other components, grain number per panicle is controlled by many quantitative trait loci (QTLs) and is affected by environmental factors. As grain number per panicle is a typical quantitative trait, characterizing it using conventional methods is difficult. Molecular markers are a useful tool for investigating the genetic basis of complex quantitative traits (Kurata et al. 1994, McCouch et al. 1988, 2002). With QTL analysis based on molecular markers, grain yield has been extensively investigated and numerous QTLs have been reported over the last decade. According to the Gramene database information, there are 353 QTLs associated with grain number per panicle (www.gramene.org). Most of the materials used in mapping were, however, primary segregating populations such as F2, recombinant inbred lines (RILs), and doubled haploid lines (DHs). The excessive genetic background noise in these lines means that results obtained from each are variable and there is limited comparability among lines, because background genes can suppress or exacerbate phenotypes which are ubiquitous and highly polymorphic.
For fine mapping of a QTL, the effect of genetic background on the expression of the QTL should be eliminated. Thus, advanced populations such as chromosome segment substituted lines (CSSLs) and near-isogenic lines (NILs) containing one or a small number of introgressed fragments from a donor parent into a recipient parent genetic background are ideal materials for the fine mapping and isolation of target QTLs (Howell et al. 1996). Secondary segregation populations, which include F2 and F3 populations developed from a cross between one NIL with the target QTL and the recurrent parent, can be used to identify recombinants with the introgressed segment using flanking markers. Based on this strategy, several QTLs for rice grain number per panicle have been finely mapped (Chen et al. 2014, Deshmukh et al. 2010, Liu et al. 2009, Xie et al. 2008, Xing et al. 2008) and even isolated in recent years (Ashikari et al. 2005, Bai et al. 2010, Huang et al. 2009, Huo et al. 2017, Song et al. 2007, Wu et al. 2016, Xue et al. 2008, Zhao et al. 2015).
In this study, we report our research on the quantitative trait locus, qgnp7(t) for grain number per panicle on chromosome 7. A CSSL, 9IL188, with O. glaberrima chromosome segments in the indica rice 9311 background, showed significantly smaller panicles containing fewer grains compared with the recurrent parent 9311. We found that the qgnp7(t) allele from O. glaberrima was responsible for this variation. We then carried out fine mapping and localized qgnp7(t) to a 41 kb region on the short arm of chromosome 7. These results provide a foundation for isolating qgnp7(t) and investigating its molecular mechanism.
Materials and Methods
Plant materials and growth conditions
9IL188 is a CSSL developed by introgressing chromosome segments from a cultivar of African cultivated rice (IRGC102305, O. glaberrima) into Asian Oryza sativa 9311 background based on four generations of backcrossing and four generations of selfing. A small F2 population consisting of 152 plants derived from 9IL188 × 9311 was grown in a paddy field in Sanya (109°E, 18°N), Hainan Province, China, in the winter of 2013. Heterozygous F2 plants in the target region were selected to produce a larger F3 population for fine mapping of qgnp7(t). The F3 population containing 2,258 plants was grown in a paddy field in Taicang (121°E, 31°N), Jiangsu province, China, in the summer of 2014. The key F4 recombinant lines derived from the F3 population were grown in Sanya as above and used for progeny testing.
Phenotypic evaluation
Quantitative analysis of agronomic traits including plant height, tillers per plant, grains per panicle, seed setting rate, and 1,000-grain weight for the two parents, 9IL188 and 9311, were performed using 10 plants. The number of grains per panicle was evaluated for the 152 plants of the F2 population by dividing total number of grains per plant by tillers per plant. Quantitative analysis of plant height and grains per panicle for the two parents and the key recombinant lines derived from the F3 population were performed using 30 plants each. Quantitative analysis of agronomic traits including plant height, first-six internode from top (IN1–IN6), panicle length, number of primary and secodary branches per panicle (PB and SB), grains per panicle, seed setting rate, 1,000-grain weight, tillers per plant, and grain yield per plant for the two NILs, NIL-qgnp7 and NIL-qGNP7, were performed using 10 plants.
DNA extraction and molecular marker analysis
Micro-quantities of DNA were extracted from fresh rice leaves following a published method (Murray and Thompson 1980) with minor modification. Polymerase chain reaction (PCR) amplification was performed as previously described (Panaud et al. 1996). PCR products were separated by 6% polyacrylamide gel electrophoresis and detected by silver staining (Ji et al. 2007). Simple sequence repeat (SSR) markers in the qgnp7(t) region were identified from the Gramene database for grasses (http://www.gramene.org/). Besides the public molecular markers, four insertion/ deletion (indel) markers containing polymorphisms between 9IL188 and 9311 were newly developed according to the publicly available rice genome sequence (http://www.gramene.org/) for fine mapping. The primer information for the molecular markers used in substitution mapping is listed in Table 1.
Table 1.
PCR-based molecular markers used in qgnp7(t) mapping
| Markers | Type | Position (kb)a | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|---|---|
| RM436 | SSR | 2,523 | ATTCCTGCAGTAAAGCACGG | CTTCGTGTACCTCCCCAAAC |
| RM427 | SSR | 2,655 | TCACTAGCTCTGCCCTGACC | TGATGAGAGTTGGTTGCGAG |
| RM298 | SSR | 2,745 | CTGATCACTGGATCGATCATG | CATGCCAAGATGCAACAG |
| 7Sui21 | Indel | 2,763 | GAAGCAGCCAAGAAACAAAC | ATCGAGGGGATCTAGCTAG |
| SM2 | Indel | 2,773 | AAAGAATAAGCGGTGGCT | AGCTCTGCACGTTCTCCA |
| 7Sui24 | Indel | 2,804 | ATTATAGGGGAGGGGTGTAG | TCTCTTAGCCATTATATTTTGCA |
| 7Sui25 | Indel | 2,855 | TTGAGAACAGAAAGAGTAG | ATCCCGTTGTTGAATTGGTT |
| RM82 | SSR | 3,010 | TGCTTCTTGTCAATTCGCC | CGACTCGTGGAGGTACGG |
| RM4986 | SSR | 3,172 | GTCTTTAATGGATTGGATTC | CTGGTACATGCTAAATTTCA |
Physical distance of markers in 9311 genome sequences.
Data analysis
Likelihood ratio chi-square test was used to test the degree of significance, and the correlation degree was detected by P value in SPSS v17.0. Mapmaker/Exp v3.0 (Lincoln et al. 1992) was used for linkage analysis based on the 157-plant F2 population. The Kosambi function was used to calculate the genetic distance. Map Manager QTXb17 (Manly et al. 2001) was used to determine the QTL positions, the expected additive and dominance effects, and the phenotypic variance explained by individual QTLs. The genotypes of all markers in the target region were determined for the recombinant plants in the F3 family and were further confirmed by progeny testing.
Sequence analysis of candidate genes
Full genomes of Oryza sativa indica and Oryza glaberrima were downloaded from the website: http://www.gramene.org/, and full genome of Oryza glaberrima was also sequenced using Illumina/Solexa technology by BGI (Beijing Genome Institute, China). Candidate genes in the target region were amplified from 9311 and O. glaberrima using PCR with LA-Taq polymerase (Takara, Otsu, Japan). PCR products were purified with a PCR purification kit (Axygen, USA), introduced into a pGEM-T Easy Vector (Promega, Madison, WI, USA), and transformed into E.coli strain DH5α. The resulting plasmid was sequenced with an ABI Analyzer. Sequence alignment was performed using GENtle.
RNA extraction and qRT-PCR
Total mRNA was isolated from fresh young panicle using a RNAprep Pure Plant Kit (Tiangen, Beijing, China). First-strand cDNA was generated using a Primescript RT Master Mix kit (Perfect Real Time) (TaKaRa, Japan). qRT-PCR analysis was carried out using a CFX96 TouchTM Real-Time PCR Detection System (Bio-Rad, CA, USA) and a SYBR Premix Ex Tap II kit (TaKaRa, Japan). The primer information used in qRT-PCR is listed in Table 2.
Table 2.
Primers used in qRT-PCR
| Name | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| ACTIN1 | AGCAACTGGGATGATATGGA | CAGGGCGATGTAGGAAAGC |
| BG164 | TCGTACGGTGGATTTGGCAATA | ACCATAAGAGCCTCCCAACCCA |
| BG165 | GATTGCTCCCTCTGCCATGC | CCACCACCCTCCTGTCTTTG |
| BG807 | CGGATGTGCGCC GTCAA | TCTCGCCCTGGTGGTAGTGG |
| BG806 | ATAATGCCTCCCAAGCCACCAATA | TTTCAGCGCGATCTTTCGACCC |
| BG166 | GCCAACATTACAGTTCCATTTTGC | TACTCAATCCTTGCCTCATCTCCA |
| BG805 | TCTCCTGACGATTCCCAAGAGC | CGACGAGATCAGAGCCTGGTTG |
| BG804 | TCATCCCACCAATCTCCAGTCC | AGGCCAGGTATGAGGGGTGTC |
| BG167 | CAAAGAAGGTGGCGATGTGG | GCGATGACGACGCTGACAAT |
Sample paraffin sectioning and microscopy
Plant materials were fixed in FAA (50% ethanol, 5% glacial acetic acid and 2% formaldehyde) for 16 h, dehydrated in an ethanol series, and embedded in paraffin. Tissue sections (10 mm thick) were cut with a rotary microtome (Leica RM2235) and then mounted on glass slides. Sections were scanned using a Panoramic P250 FLASH II.
Results
Phenotypic characterization and validation of qgnp7(t)
The 9IL188 plants were dwarfed and had obviously small panicles (Fig. 1A, 1B) when compared with the 9311 parent line. Phenotypic evaluation of agronomic traits showed that the differences between 9IL188 and 9311 in plant height, panicle length, grains per panicle and tillers per plant were significant (P < 0.01) (Fig. 1C–1F), while there were no significant difference in seed setting rate and 1,000-grain weight (Fig. 1G, 1H).
Fig. 1.
Phenotypic performance of 9311 and 9IL188 at Taicang experimental station. A. 9311 and 9IL188 plants at the mature stage. Scale bar = 10 cm, B. Panicle phenotypes of 9311 and 9IL188 at the mature stage, Scale bar = 3 cm. Comparison of plant height (C), grains per panicle (D), panicle length (E), tillers per plant (F), seed setting rate (G), and grain weight (H). Data presented are means with SD (n = 10 plants). ***, P = 0.001; NS, not significant at P = 0.05.
A total of 326 SSR markers, which are polymorphic between the two parents and evenly distributed on 12 chromosomes, were used to detect the substitution sites in 9IL188. Six O. glaberrima chromosome segments of approximately 55 cM in total were detected in the 9311 background and were located on chromosomes 1, 6, 7, and 12 (Fig. 2).
Fig. 2.
The graphic genotype of the 9IL188 plant. The black and white chromosome segments were the O. glaberrima homozygote and 9311 homozygote, respectively. Only four chromosomes contained segments of donor parent, O. glaberrima. The other eight chromosomes were fixed with 9311.
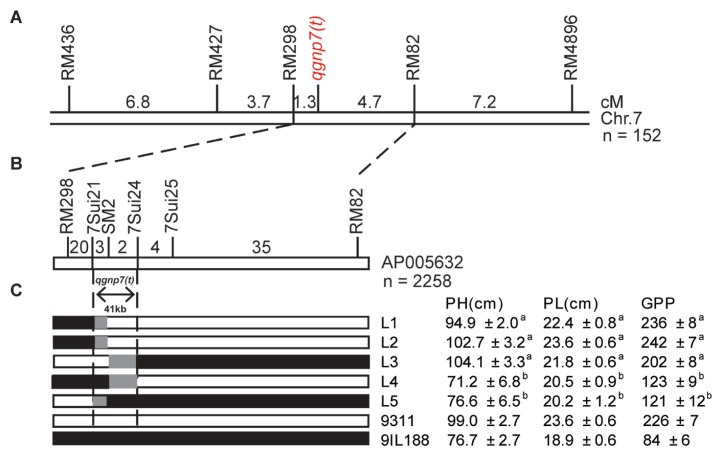
The 152 plants of the F2 population were phenotyped for GPP at the grain filling stage and genotyped using 10 SSR markers distributed on the six substitution sites. Of these, there were 33 plants showing small panicles (<100 spikeles) and dwarfing (<80 cm). Progeny testing further confirmed that 46 and 44 families showed identical small and large panicles, respectively, whereas 62 families showed varied panicles (Fig. 3). The segregation ratio agreed with the Mendelian ratio (1:2:1) for single locus segregation (χ2 = 4.67 < χ2 0.05,2 = 5.99). These data indicated that the phenotypic difference between 9IL188 and 9311 was mainly controlled by a single gene. QTL analysis revealed that one QTL with a LOD value of 64.21 for GPP was located in the O. glaberrima interval between RM298 and RM82 on chromosome 7. We tentatively named this QTL qgnp7(t) and preliminarily mapped it to a 6.0 cM region between SSR markers RM298 and RM82 (Fig. 4A). The genetic distances from the target gene to two SSR markers were 1.3 cM and 4.7 cM, respectively. Phenotypic variance explained by this QTL was 63%, and the O. glaberrimaderived allele contributed a decreasing effect on GPP.
Fig. 3.
Frequency distribution of grains per panicle in the F2 population. The three genotypes of homozygous 9311 (9), heterozygote (H), and homozygous O. glaberrima (g) at qgnp7(t) were identified by progeny testing.
Fig. 4.
Substitution mapping of qgnp7(t). A. Genetic linkage map of the qgnp7(t) region on chromosome 7 constructed using 152 F2 plants. qgnp7(t) was mapped to the interval between markers RM298 and RM82. Numbers above the line indicate the genetic distance between adjacent markers, B. High-resolution linkage map of the qgnp7(t) region (AP005632) constructed using 2,258 F3 plants. The number of recombinants between adjacent markers is indicated above the linkage map, C. Progeny testing of homozygous recombinants localized the qgnp7(t) locus to the region between markers 7Sui21 and 7Sui24. On the right, the phenotypic value (mean ± SD; n = 30 plants) of each F4 families and two parents were indicated. PH plant height, PL panicle length, GPP grains per panicle.
Substitution mapping of qgnp7(t)
To further refine the position of qgnp7(t), a larger F3 population containing 2,258 plants derived from the F2 heterozygous plants was constructed and genotyped with SSR markers RM298 and RM82. Sixty-two recombinants were identified and further genotyped with four additional Indel markers 7Sui21, SM2, 7Sui24, and 7Sui25 (Table 1). There were 35 and 4 recombination events on the RM82 side and 20, 3 and 2, recombination events on the RM298 side (Fig. 4B). The mean phenotypic values of recombinant lines (L1–3) for PH, PL, and GPP were compared with that of recombinant lines (L4–5) at P < 0.001 level. Finally, according to their marker genotypes and phenotypic values, the position of qgnp7(t) was mapped to a 41 kb interval defined by 7Sui21 and 7Sui24 (Fig. 4C).
Candidate genes in the qgnp7(t) location interval
Based on the published sequence annotation for Oryza sativa indica and Oryza glaberrima (http://www.gramene.org/), eight predicted genes were identified in the qgnp7(t) location interval. Of these, one gene is a Glycolate oxidase gene (GLO5) and the others are still of unknown function. We comparatively aligned the coding region of the common genes between 9311 and O. glaberrima. Compared with the alleles in 9311, all the genes in O. glaberrima showed differences except for BGIOSGA025165 (ORGLA07G0029000). Potential genes and alignment results are listed in Table 3.
Table 3.
Information and comparative analysis of candidate genes in the qgnp7(t) location interval
| 9311 Gene ID | Gene annotation | O. glaberrima gene ID | Comparison between 9311 gene and O. glaberrima gene | ID%a |
|---|---|---|---|---|
| BGIOSGA025164 | protein coding | ORGLA07G0028900 | 6 SNP | 99 |
| BGIOSGA025165 | Peroxisomal (S)-2-hydroxy-acid oxidase, GLO5 | ORGLA07G0029000 | same | 100 |
| BGIOSGA024807 | protein coding | ORGLA07G0029100 | 1 SNP | 99 |
| BGIOSGA024806 | protein coding | No annotation | 19 SNP and 24 deletion | 96 |
| BGIOSGA025166 | protein coding | No annotation | 9 SNP and 2 insertion | 53 |
| BGIOSGA024805 | protein coding | ORGLA07G0029200 | 3 SNP | 99 |
| BGIOSGA024804 | protein coding | ORGLA07G0029300 | 5 SNP | 99 |
| BGIOSGA025167 | protein coding | ORGLA07G0029400 | 12 SNP | 93 |
9311 was used as the reference.
Amino acid sequence identity derived from the O. glaberrima and the 9311 qgnp7(t) candidate genes.
We also checked the expression levels of these genes in the young panicles of two parents at booting stage. Compared to the 9311 plants, five genes were down-regulated and two gene was up-regulated in the 9IL188 plants (Fig. 5). Besides, the expression of BGIOSGA025166 was not detected in both parents (Fig. 5).
Fig. 5.
Relative expression of the qgnp7(t) candidate genes in two parents (9311 and 9IL188). Panicle materials were collected at Booting Stage. Data presented are means with SE (n = 3 plants). *, P = 0.05; ***, P = 0.001; NS, not significant at P = 0.05.
Characterization of qgnp7(t)
To validate the role of qgnp7(t) in rice development, we developed the following NILs from the F4 generation: NIL-qgnp7, which carries the O. glaberrima-derived allele, and its matching line NIL-qGNP7, which carries the homologous segment from 9311. Correspondingly, the phenotypic evaluation of agronomic traits was investigated between the NILs at maturity. The largest differences detected between the two lines were for plant height and grains per panicle related traits (Table 4).
Table 4.
Phenotypic performance of NIL-qGNP7 and NIL-qgnp7
| Agronomic traits | NIL-qGNP7 | NIL-qgnp7 |
|---|---|---|
| Plant height (cm) | 99.3 ± 1.75a | 77.4 ± 1.51b |
| First internode from top (cm) | 29.9 ± 0.90a | 22.9 ± 0.64b |
| Second internode from top (cm) | 18.5 ± 0.64a | 12.8 ± 0.52b |
| Third internode from top (cm) | 14.8 ± 0.42a | 12.5 ± 0.37b |
| Fourth internode from top (cm) | 8.0 ± 0.52a | 5.9 ± 0.35b |
| Fifth internode from top (cm) | 3.6 ± 0.35a | 2.5 ± 0.34b |
| Sixth internode from top (cm) | 1.8 ± 0.20a | 1.5 ± 0.12a |
| Panicle length (cm) | 25.0 ± 0.70a | 20.8 ± 1.01b |
| Primary branches per panicle | 15.3 ± 0.67a | 11.0 ± 0.94b |
| Secondary branches per panicle | 64.4 ± 1.90a | 24.6 ± 1.35b |
| Grains per panicle | 277.4 ± 12.63a | 130.9 ± 9.01b |
| Seed setting rate (%) | 73.0 ± 0.56a | 74.5 ± 0.60a |
| 1,000-grain weight | 27.9 ± 0.47a | 27.5 ± 0.62a |
| Tillers per plant | 5.4 ± 0.97a | 6.8 ± 0.79b |
| Grain yield per plant (g) | 27.8 ± 0.90a | 15.6 ± 1.01b |
A different letter indicates a significant difference at P < 0.001 (the same as below).
NIL-qgnp7 had a dwarf phenotype. The six internodes (from top to bottom) in NIL-qgnp7 were reduced by about 23.4, 30.6, 16.0, 26.2, 29.8, and 17.4% respectively, compared with NIL-qGNP7 (Table 4). The elongation mode of each internode in NIL-qgnp7 was similar to that in NIL-qGNP7 and the internodes were just proportionally shortened (Fig. 6). According to the six modes of internode elongation for rice dwarf mutants (Takeda 1977), we classified the mode of internode elongation in qgnp7(t) genotypes into the dn-type dwarf mutant.
Fig. 6.
The proportion of each internode to stem in NIL-qGNP7 and NIL-qgnp7. Data presented are means with SD (n = 10 plants). IN1–6 internode 1–6.
Furthermore, NIL-qgnp7 had a small panicle phenotype. The primary and secondary branches per panicle in NIL-qgnp7 were reduced by about 26.6 and 62.5% respectively, compared with NIL-qGNP7 (Table 4). In NIL-qGNP7, the primary and secondary branches per panicle contributed 19.2 and 80.8%, respectively, to the total branches per panicle, while in NIL-qgnp7, the proportion was 30.9 and 69.1%, respectively (Fig. 7). The analysis showed that the small panicle phenotype of NIL-qgnp7 was mainly caused by the decrease in secondary branches per panicle.
Fig. 7.

The proportion of primary and secondary branches per panicle to total branches per panicle in NIL-qGNP7 and NIL-qgnp7. Data presented are means with SD (n = 10 plants). PB primary branches per panicle, SB secondary branches per panicle.
To understand whether the phenotype difference can be attributed to the difference in cell division or cell enlargement, we examined the stem cell morphology in the heading stage using paraffin sections. The transverse section showed that the cell size in the same stem tissue site of the two NILs was similar (Fig. 8A, 8B), however, the stem cells of NIL-qGNP7 were significantly larger than NIL-qgnp7 in the longitudinal direction (Fig. 8C, 8D). These results showed that qgnp7(t) affected PH potentially by regulating cell enlargement of internode in the longitudinal direction.
Fig. 8.
Stem cell morphology of NIL-qGNP7 and NIL-qgnp7. A–B. Transverse sections of stems in the heading stage. Sampling site was in the middle of the uppermost internode of the main culm of the plant. Scale bar = 1000 μm and 100 μm; C–D. Longitudinal sections of stems in the heading stage. Sampling site was in the middle of the uppermost internode of the main culm of the plant. Scale bar = 1000 μm and 100 μm.
Taken together, the above results indicate that qgnp7(t) has pleiotropic effects on rice plant architecture and panicle structure, suggesting that qgnp7(t) plays an important role in the growth and development of rice.
Discussion
Oryza sp. includes two cultivated species (O. sativa and O. glaberrima) that originated from O. rufipogon and O. barthii, respectively (Wu et al. 2017). O. sativa has a high yield and is cultivated worldwide while O. glaberrima has a low yield and is only cultivated in the south of the Sahara desert in West Africa. To gain insight into the genetic basis for complex traits in O. glaberrima such as yield, plant architecture, stress response, and domestication, we developed a set of 159 chromosomal segment substitution lines (CSSLs) by repetitive backcross and marker-assisted selection derived from a cross between African Oryza glaberrima and Asian Oryza 9311 with 9311 as the recurrent parent. In this study, a major QTL for GPP, qgnp7(t), was detected in an F2 population derived from a cross between 9IL188 (a member of the 159 CSSLs) and 9311. Through two-step substitution mapping, qgnp7(t) was finally localized to a 41 kb interval which defined by Indel markers 7Sui21 and 7Sui24.
According to available sequence annotation for Oryza sativa indica and Oryza glaberrima (http://www.gramene.org/), there are eight annotated genes in the qgnp7(t) location interval (Table 3). Of these, only one gene (BGIOSGA025165/ORGLA07G0029000) has a definite function with the others being of unknown function. Glycolate oxidase (GLO) is an important enzyme in photorespiration. BGIOSGA025165 (LOC_Os07g05820 in Oryza sativa japonica), which is one of the four GLO genes in rice, is predominantly expressed in the leaf and has enzymatic activity (Zhang et al. 2012). Sequence comparison and analysis revealed that the coding sequence of this gene is the same between 9311 and 9IL188. Genetic analysis indicated that qgnp7(t) could be treated as a single recessive allele mutation and that the small panicle phenotype of 9IL188 plants may be caused by the lack of function of a single gene. We comparatively aligned the amino acid sequence encoded by the common genes between 9311 and O. glaberrima. This revealed that the mutations in O. glaberrima genes only caused amino acid changes or small partial deletions in the amino acid sequences except for BGIOSGA025166 (Table 3). BGIOSGA025166 has 2bp insertional mutation in O. glaberrima, which leads to premature termination of protein translation. The results of qRT-PCR showed that the expression level of seven genes varied in the qgnp7(t) candidate genes between two parents, while the expression of BGIOSGA025166 was not detected (Fig. 5). Among the seven genes, the expression level of BGIOSGA025165 (ORGLA07G0029000) was reduced by about five times in the 9IL188 plants, and that of BGIOSGA025167 (ORGLA07G0029400) was increased by about four times (Fig. 5). The changes of expression levels of these two genes were extremely significant. Further high-resolution mapping and genetic transformation experiments are required to confirm the candidate gene for qgnp7(t).
In previous reports, locations on chromosome 7 containing or overlapping the qgnp7(t) interval were reported to be an important target during Asian rice domestication selection for plant architecture and panicle structure (Li et al. 2006, Tian et al. 2006). Unlike Li et al. (2006) and Tian et al. (2006), our study was conducted based on a CSSL (9IL188) with introgression fragments derived from Africa’s Oryza glaberrima. Considering this, it is possible that the qgnp7(t) interval was not selected during the related domestication of African rice. In our study, the yield estimation of NILs (NIL-qgnp7 and NIL-qGNP7) showed that the yield of NIL-qGNP7 plants was significantly improved by selecting the 9311 allele (Table 4). Thus, the identification of qGNP7(t) would provide novel gene resource for O. glaberrima improvement programs. Over the past 30 years, New Rice for Africa (NERICA) varieties were developed from crosses between improved tropical japonica and Oryza glaberrima, which possess the fine characters of Asian cultivated rice and African cultivated rice, and have carved a special niche for itself among upland rice farmers in sub-Saharan Africa (Nassirou and He 2011, Saito et al. 2018). During this process, refined method of conventional breeding, specifically-developed anther culture and double-haploidization techniques were used to overcome sterility and to hasten the breeding process. According to our previous strategy for constructing chromosomal segment substitution lines, the qGNP7(t) interval could be directly introduced from 9311 into O. glaberrima through repetitive backcross and marker-assisted selection. Comparing with NERICA breeding programs, this can greatly improve breeding efficiency and shorten breeding cycle for related traits.
In summary, qgnp7(t) is a major QTL for grains per panicle. Understanding the molecular mechanisms underlying this locus would be valuable for identification and transference of favorable alleles in African rice improvement programs. Studies of the genetic basis and function of qgnp7(t) are underway.
Acknowledgments
This research was supported by grants from the National Natural Science Foundation of China (No. 31471461, 31671655), the Basic Application Research Program from the Shanghai Municipal Agriculture Commission (2014-7-1-2) and the Program of Shanghai Technology Research Leader (18XD1424300).
We thank Emma Tacken, PhD, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Abbreviations
- CSSL
chromosome segment substitution line
- DHs
doubled haploid lines
- IN1-IN6
first-six internode from top
- Indel
insertion/deletion
- GPP
grains per panicle
- LOD
log of odds
- NILs
near-isogenic lines
- PCR
polymerase chain reaction
- PB
primary branch
- PH
plant height
- PL
panicle length
- QTL
quantitative trait locus
- RILs
recombinant inbred lines
- SB
secondary branch
- SSR
simple sequence repeat
Literature Cited
- Ashikari, M., Sakakibara, H., Lin, S., Yamamoto, T., Takashi, T., Nishimura, A., Angeles, E.R., Qian, Q., Kitano, H. and Matsuoka, M. (2005) Cytokinin oxidase regulates rice grain production. Science 309: 741–745. [DOI] [PubMed] [Google Scholar]
- Bai, X.F., Luo, L.J., Yan, W.H., Kovi, M.R., Zhan, W. and Xing, Y.Z. (2010) Genetic dissection of rice grain shape using a recombinant inbred line population derived from two contrasting parents and fine mapping a pleiotropic quantitative trait locus qGL7. BMC Genet. 11: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J.Y., Guo, L., Ma, H., Chen, Y.Y., Zhang, H.W., Ying, J.Z. and Zhuang, J.Y. (2014) Fine mapping of qHd1, a minor heading date QTL with pleiotropism for yield traits in rice (Oryza sativa L.). Theor. Appl. Genet. 127: 2515–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh, R., Singh, A., Jain, N., Anand, S., Gacche, R., Singh, A., Gaikwad, K., Sharma, T., Mohapatra, T. and Singh, N. (2010) Identification of candidate genes for grain number in rice (Oryza sativa L.). Funct. Integr. Genomics 10: 339–347. [DOI] [PubMed] [Google Scholar]
- Howell, P.M., Lydiate, D.J. and Marshall, D.F. (1996) Towards developing intervarietal substitution lines in Brassica napus using marker-assisted selection. Genome 39: 348–358. [DOI] [PubMed] [Google Scholar]
- Huang, X.Z., Qian, Q., Liu, Z.B., Sun, H.Y., He, S.Y., Luo, D., Xia, G.M., Chu, C.C., Li, J.Y. and Fu, X.D. (2009) Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 41: 494–497. [DOI] [PubMed] [Google Scholar]
- Huo, X., Wu, S., Zhu, Z.F., Liu, F.X., Fu, Y.C., Cai, H.W., Sun, X.Y., Gu, P., Xie, D.X., Tan, L.B.et al. (2017) NOG1 increases grain production in rice. Nat. Commun. 8: 1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, Y.T., Qu, C.Q. and Cao, B.Y. (2007) An optimal method of DNA silver staining in polyacrylamide gels. Electrophoresis 28: 1173–1175. [DOI] [PubMed] [Google Scholar]
- Kurata, N., Nagamura, Y., Yamamoto, K., Harushima, Y., Sue, N., Wu, J., Antonio, B.A., Shomura, A., Shimizu, T., Lin, S.Y.et al. (1994) A 300 kilobase interval genetic map of rice including 883 expressed sequences. Nat. Genet. 8: 365–372. [DOI] [PubMed] [Google Scholar]
- Li, C.B., Zhou, A.L. and Sang, T. (2006) Genetic analysis of rice domestication syndrome with the wild annual species, Oryza nivara. New Phytol. 170: 185–194. [DOI] [PubMed] [Google Scholar]
- Lincoln, S., Daly, M. and Lander, E. (1992) Constructing genetics maps with MAPMAKER/EXP 3.0. Whitehead Institute Technical Report. Whitehead Institute, Cambridge. [Google Scholar]
- Liu, T.M., Mao, D.H., Zhang, S.P., Xu, C.G. and Xing, Y.Z. (2009) Fine mapping SPP1, a QTL controlling the number of spikelets per panicle, to a BAC clone in rice (Oryza sativa). Theor. Appl. Genet. 118: 1509–1517. [DOI] [PubMed] [Google Scholar]
- Manly, K.F., Cudmore, R.H. and Meer, J.M. (2001) Map Manager QTX: cross-platform software for genetic mapping. Mamm. Genome 12: 930–932. [DOI] [PubMed] [Google Scholar]
- McCouch, S.R., Kochert, G., Yu, Z.H., Wang, Z.Y., Khush, G.S., Coffman, W.R. and Tanksley, S.D. (1988) Molecular mapping of rice chromosomes. Theor. Appl. Genet. 76: 815–829. [DOI] [PubMed] [Google Scholar]
- McCouch, S.R., Teytelman, L., Xu, Y.B., Lobos, K.B., Clare, K., Walton, M., Fu, B.Y., Maghirang, R., Li, Z.K., Xing, Y.Z.et al. (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res. 9: 199–207. [DOI] [PubMed] [Google Scholar]
- Murray, M.G. and Thompson, W.F. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8: 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassirou, T.Y. and He, Y. (2011) NERICA: A hope for fighting hunger and poverty in Africa. Mol. Plant Breed. 2: 75–82. [Google Scholar]
- Panaud, O., Chen, X. and McCouch, S.R. (1996) Development of micro-satellite markers and characterization of simple sequence length polymorphism (SSLP) in rice (Oryza sativa L.). Mol. Gen. Genet. 252: 597–607. [DOI] [PubMed] [Google Scholar]
- Saito, K., Asai, H., Zhao, D., Laborte, A.G. and Grenier, C. (2018) Progress in varietal improvement for increasing upland rice productivity in the tropics. Plant Prod. Sci. 21: 145–158. [Google Scholar]
- Sakamoto, T. and Matsuoka, M. (2008) Identifying and exploiting grain yield genes in rice. Curr. Opin. Plant Biol. 11: 209–214. [DOI] [PubMed] [Google Scholar]
- Song, X.J., Huang, W., Shi, M., Zhu, M.Z. and Lin, H.X. (2007) A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39: 623–630. [DOI] [PubMed] [Google Scholar]
- Takeda, K. (1977) Internode elongation and dwarfism in some gramineous plants. Gamma Field Symposia 16: 1–18. [Google Scholar]
- Tian, F., Zhu, Z.F., Zhang, B.S., Tan, L.B., Fu, Y.C., Wang, X.K. and Sun, C.Q. (2006) Fine mapping of a quantitative trait locus for grain number per panicle from wild rice (Oryza rufipogon Griff.). Theor. Appl. Genet. 113: 619–629. [DOI] [PubMed] [Google Scholar]
- Wu, W.G., Liu, X.Y., Wang, M.H., Meyer, R.S., Luo, X.J., Ndjiondjop, M.N., Tan, L.B., Zhang, J.W., Wu, J.Z., Cai, H.W.et al. (2017) A single-nucleotide polymorphism causes smaller grain size and loss of seed shattering during African rice domestication. Nat. Plants 3: 17064–17071. [DOI] [PubMed] [Google Scholar]
- Wu, Y., Wang, Y., Mi, X.F., Shan, J.X., Li, X.M., Xu, J.L. and Lin, H.X. (2016) The QTL GNP1 encodes GA20ox1, which increases grain number and yield by increasing cytokinin activity in rice panicle meristems. PLoS Genet. 12: e1006386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, X.B., Jin, F.X., Song, M.H., Suh, J.P., Hwang, H.G., Kim, Y.G., McCouch, S.R. and Ahn, S.N. (2008) Fine mapping of a yield-enhancing QTL cluster associated with transgressive variation in an Oryza sativa × O. rufipogon cross. Theor. Appl. Genet. 116: 613–622. [DOI] [PubMed] [Google Scholar]
- Xing, Y.Z., Tang, W.J., Xue, W.Y., Xu, C.G. and Zhang, Q.F. (2008) Fine mapping of a major quantitative trait loci, qSSP7, controlling the number of spikelets per panicle as a single Mendelian factor in rice. Theor. Appl. Genet. 116: 789–796. [DOI] [PubMed] [Google Scholar]
- Xue, W.Y., Xing, Y.Z., Weng, X.Y., Zhao, Y., Tang, W.J., Wang, L., Zhou, H.J., Yu, S.B., Xu, C.G., Li, X.H.et al. (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40: 761–767. [DOI] [PubMed] [Google Scholar]
- Zhang, Z.D., Lu, Y.S., Zhai, L.G., Deng, R.S., Jiang, J., Li, Y., He, Z.H. and Peng, X.X. (2012) Glycolate oxidase isozymes are coordinately controlled by GLO1 and GLO4 in rice. PLoS ONE 7: e39658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, L. and Tan, L.B. and Zhu, Z.F. and Xiao, L.T. and Xie, D.X. and Sun, C.Q. (2015) PAY1 improves plant architecture and enhances grain yield in rice. Plant J. 83: 528–536. [DOI] [PMC free article] [PubMed] [Google Scholar]