Abstract
Super-resolution methods allow the visualization of phenomena thus far elusive to light microscopy. While current implementations are technically approaching true molecular-scale resolution, this does not translate to imaging biological specimen, due to the unavailability of small affinity reagents. Here, we introduce SOMAmers as labeling reagents for DNA-PAINT. We demonstrate the achievable resolution and specificity by labeling and imaging of transmembrane as well as intracellular targets in fixed and live cell-specimen.
Optical super-resolution techniques1–4 make it possible to image biological processes well below the classical diffraction limit of light and are starting to provide novel insights in thus far hidden biological phenomena5 with recent technical developments approaching true biomolecular resolution6–8.
DNA Points Accumulation in Nanoscale Topography (DNA-PAINT)9 is a simple implementation of single-molecule localization microscopy (SMLM) using transient binding of dye-labeled DNA strands to complementary target-bound strands, enabling spatial resolution better than 5 nm as recently demonstrated using artificial DNA nanostructures6, 8.
However, this high spatial resolution does not translate to molecular-scale imaging of cellular targets, since commonly used labeling probes are currently one of the major limitations in high-resolution optical microscopy research, due to their relatively large size (~150 kDa in the case of antibodies). Furthermore, fully stoichiometric, quantitative labeling via site-specific conjugation is not yet readily available for a large number of targets, thus preventing the analysis of complex biosystems in a quantitative manner, one of the promises of quantitative biology.
The “ideal” labeling probe should therefore satisfy several requirements: (1) Smallest possible size for maximal labeling efficiency and minimal linkage error, (2) quantitative labeling (i.e. 1:1 stoichiometry for protein targeting), (3) rapid selection procedure for novel targets (or ideally an already available library of well-characterized binders). While nanobodies10 satisfy some of these criteria, they are not readily available for many cellular targets. Aptamers11, 12 have the potential to fulfill most of these requirements: They allow for rapid in vitro selection, are comparably small (few tens of kDa or less) and can be quantitatively labeled. However, their wide-spread application to fluorescence and super-resolution imaging has thus far been limited by mainly three reasons: (1) restricted availability of specific aptamers to a wide range of targets, (2) concerns about the compatibility with fixation procedures, and (3) the limited ability to label intracellular targets.
We here introduce Slow Off-rate Modified Aptamer (SOMAmer) reagents13, 14 as small (7–30 kDa), quantitative, and versatile labeling probes for high-resolution in situ DNA-PAINT imaging. SOMAmer reagents represent a unique class of aptamers that contain modified bases employing hydrophobic residues, similar to the amino acid residues abundantly found in antibody epitopes for high-specificity and high-affinity binding of proteins. These base modifications increase the range of protein targets for which high-affinity ligands can be selected15.
We successfully assayed seven (Supplementary Table 1) different SOMAmers (21–28 kDa) as probes for DNA-PAINT to quantify proteins in different cellular compartments: against the transmembrane receptor EGFR (Fig. 1), GFP (Fig. 2), catalase proteins localizing to peroxisomes (Fig. 3), ErbB2 and HSP90 (Supplementary Figure 1), and finally a lysosomal membrane protein LIMP-2 and mitochondrial HSP60 (Supplementary Figure 2).
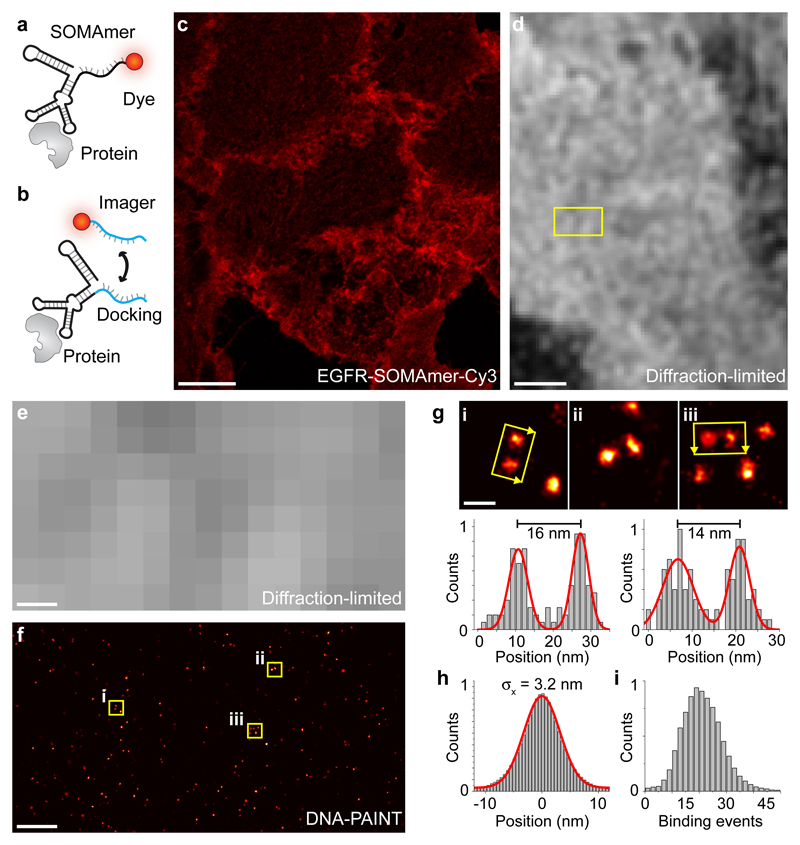
Figure 1. SOMAmers as labeling probes for quantitative, high resolution DNA-PAINT imaging of membrane receptors.
(a) Labeling scheme using SOMAmers for diffraction-limited imaging with a fixed dye. (b) Labeling scheme using SOMAmers for DNA-PAINT super-resolution imaging. Transient binding of a dye-labeled strand to a complementary docking strand is enabled by single-stranded extension of the SOMAmer sequence. (c) SOMAmer against EGFR labeled with a fixed Cy3 dye (SL1069, see scheme in a) allows specific detection of EGFR in A549 cells using confocal microscopy. (d) Diffraction-limited DNA-PAINT image using a complementary dye-labeled imager strand against a docking-site-modified SOMAmer (see scheme in b) against EGFR (standard deviation image) in A431 cells. (e) Zoom-in of highlighted area in the diffraction-limited image in d. (f) Corresponding DNA-PAINT super-resolution image of the highlighted area in d. (g, top) Zoom-ins of highlighted areas (i, ii, and iii) in f. (g, bottom) Cross-sectional histogram analysis in i, and iii, respectively, demonstrates high-resolution DNA-PAINT imaging of single EGFR proteins labeled using SOMAmers. (h) Fitting a Gaussian distribution to the center-of-mass-aligned single-molecule localizations of ~34000 SOMAmer-labeled EGFR proteins yields a localization precision of 3.2 nm. (i) qPAINT analysis of single EGFR proteins yields a unimodal distribution of binding events, confirming quantitative 1:1 labeling of EGFR proteins using SOMAmers. Scale bars: 10 µm (c), 2 µm (d), 200 nm (e, f), 20 nm (g). Experiments were repeated at least three times with similar results; representative data are shown.
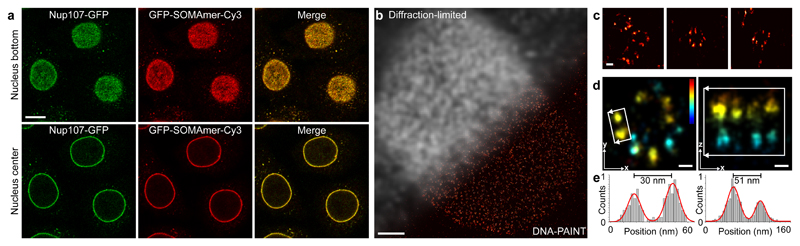
Figure 2. Intracellular labeling of GFP-tagged Nup107 for DNA-PAINT imaging using GFP-SOMAmers.
(a) Confocal images of GFP-tagged Nup107 nucleoporins labeled with Cy3-modified GFP-SOMAmers. GFP (SL1070, left, green), SOMAmer-Cy3 (center, red), and merged signal (right) at the bottom of the nucleus (top) and center (bottom) showing specific binding of the SOMAmer to the GFP-tagged Nup107. (b) Diffraction-limited and corresponding DNA-PAINT super-resolution image of GFP-tagged Nup107 using a docking-strand-extended SOMAmer against GFP. (c) Representative zoom-ins of single nuclear pore complexes (NPCs). (d) xy-projection (left) and xz-projection (right) of a single NPC shows well-resolvable clusters of Nup107 in the xy-projection (left) and the nuclear and cytoplasmic rings in the xz-projection (right). Color indicates height. (e) Cross-sectional histogram analysis of highlighted areas in d reveals a distance of ~30 nm between Nup107 in xy (left) as well as a ~51 nm distance in z of the nuclear and cytoplasmic rings (right). Scale bars, 10 µm (a), 2 µm (b), 50 nm (c), 25 nm (d). Height scale, -100 nm to 400 nm (blue to red). Experiments were repeated at least three times with similar results; representative data are shown.
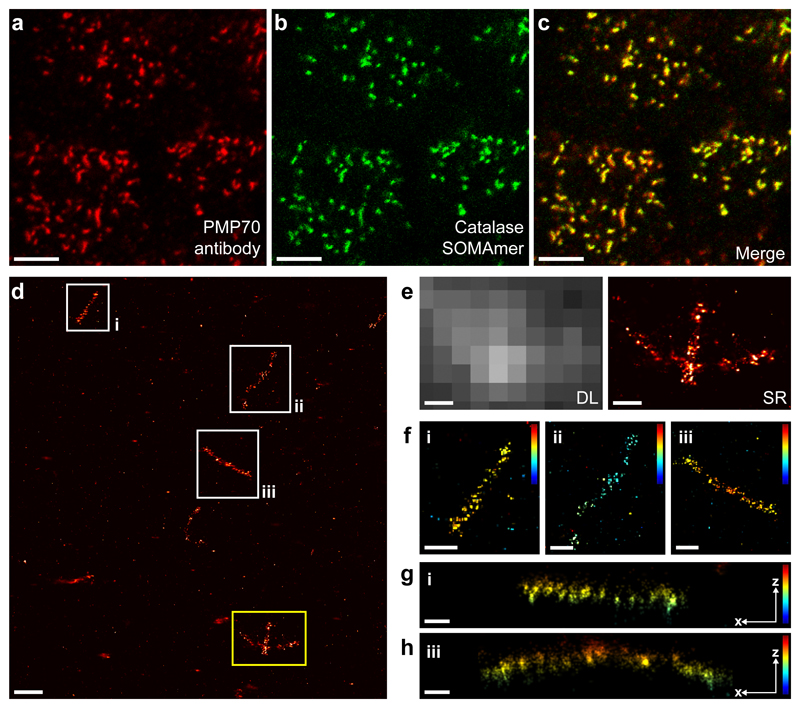
Figure 3. Intracellular labeling and DNA-PAINT imaging of catalase proteins in peroxisomes using SOMAmers.
(a) Confocal micrograph of PMP70 proteins in the peroxisomal membrane of A431 cells labeled using primary and Alexa647-conjugated secondary antibodies. (b) Confocal micrograph of catalase proteins labeled using Fluorescein-conjugated SOMAmers. (c) Overlay of PMP70 antibody and catalase SOMAmer (SL1071) signal demonstrates co-localization of both proteins to peroxisomes. (d) DNA-PAINT super-resolution image of catalase molecules in peroxisomes using docking-strand-extended SOMAmers. (e) Comparison of diffraction-limited (left) and super-resolved (right) zoom-in of the yellow-highlighted area in d reveals single catalase molecules in peroxisomes in the SR image. (f) Zoom-ins of areas highlighted in white in d. 3D localization information is color-coded, revealing distinct z localizations of peroxisomes. (g) xz-projection of catalase molecules in a single peroxisome from area i in d and f. (h) xz-projection of catalase molecules in a single peroxisome from area iii in d and f. Scale bars, 5 µm (a, b, c), 500 nm (d), 200 nm (e, f), 100 nm (g, h). Height scale, -100 nm to 400 nm (blue to red). Experiments were repeated at least three times with similar results; representative data are shown.
To initially evaluate labeling specificity, we conjugated a dye to the SOMAmer (Fig. 1a) for diffraction-limited confocal microscopy. For subsequent DNA-PAINT imaging, the SOMAmer sequence was extended with a single-stranded docking site (Fig. 1b, either on the 3’- or 5’-end). First, we labeled EGFR in fixed A549 cells using a Cy3-conjugated SOMAmer and evaluated the labeling specificity with confocal microscopy (Fig. 1a, c, and Supplementary Figures 3–5). We then performed DNA-PAINT imaging in A431 cells using the same EGFR-SOMAmer with a docking-site extension instead of the fixed dye (Fig. 1b). Comparing the diffraction-limited (Fig. 1d and e) and DNA-PAINT image (Fig. 1f) reveals sub-diffraction spatial resolution and specific targeting of EGFR proteins in the plasma membrane. Zoom-ins (Fig. 1g) of three highlighted areas in Figure 1f underline the achievable high resolution obtained due to the small size of the SOMAmer in combination with the high localization precision of DNA-PAINT. This is furthermore exemplified by our ability to resolve EGFR molecules spaced only ~14 nm apart (see Fig. 1g). To obtain an average measure of achievable resolution, we quantified the localization precision of SOMAmer-targeted EGF receptors by overlaying the localizations of ~34,000 EGFR proteins by their center-of-mass. We achieved an average localization precision of ~3.2 nm (Fig. 1h), translating to a full width at half maximum (FWHM)-limited resolution of less than 8 nm (also highlighted by the clearly separable distributions in the cross-sectional histograms in Fig. 1g). Using classical primary and DNA-conjugated secondary antibody labeling and downstream DNA-PAINT imaging of EGFR, we see an approx. two-fold larger apparent size of single EGFR proteins when compared to SOMAmer labeling (Supplementary Figure 6).
As SMLM methods provide quantitative information from localization datasets16, 17, it becomes important to label target molecules in a quantitative fashion to harness the precision of such methods in biological systems. Ideally, a single label should be attached to a target. SOMAmers could deliver on this promise as they are single-stranded nucleic acid molecules that can be easily modified during or after chemical synthesis.
Here, we examine the ability of SOMAmers to serve as quantitative labeling probes for qPAINT18 by labeling EGFR proteins and subsequently performing a qPAINT analysis on presumably single EGF receptors. We quantified the same ~34,000 EGFR molecules (used above to determine localization precision) and calculated the number of binding events during the time of our image acquisition. The measure for binding events is directly linked to the number of available DNA strands per target molecule (i.e. two strands will exhibit twice the number of binding events compared to a single site). The resulting analysis revealed a unimodal distribution (Fig. 1i), thus confirming quantitative 1:1 labeling of EGFR by the SOMAmers (i.e. either one or no SOMAmer was bound). Using classical primary and DNA-conjugated secondary antibody labeling and subsequent qPAINT analysis we see a clear multimodal distribution of binding events, highlighting the broader distribution of binding sites in the antibody case compared to the unimodal distribution in the SOMAmer case (Supplementary Figure 7).
Next, we turned our attention to the labeling and imaging of an intracellular target. We chose GFP-labeled Nup107 to demonstrate the widespread applicability of SOMAmers for intracellular DNA-PAINT. First, we confirmed the specific labeling of GFP-Nup107 with confocal imaging, yielding good co-localization of the GFP and Cy3-SOMAmer signal (Fig. 2a). Subsequent DNA-PAINT imaging revealed super-resolved single Nup107 clusters in nuclear pore complexes (NPCs, Fig. 2b and c). Using astigmatism-based point-spread-function-shaping to obtain 3D super-localization19, we were able to spatially separate the cytoplasmic and nuclear rings of the NPCs (Fig. 2d and e). Cross-sectional histogram analysis of both xy- and xz-projections yield the expected distances (Fig. 2d and e).
Next, we performed labeling of intracellular targets without relying on GFP, to further illustrate the flexibility and intracellular specificity of SOMAmers. Here, we chose to label catalase proteins localizing to peroxisomes and assayed the binding specificity of a dye-labeled SOMAmer against catalase with confocal microscopy. We co-stained peroxisomal membrane proteins PMP70 using dye-labeled antibodies (Fig. 3a) in combination with fluorescein-labeled SOMAmers against catalase (Fig. 3b). The merged overlay reveals co-localization of PMP70 and catalase signals, again suggesting high labeling specificity of SOMAmers (Fig. 3c). Subsequent DNA-PAINT imaging reveals resolvable single catalase molecules in peroxisomes (Fig. 3d and e). 3D DNA-PAINT micrographs furthermore reveal distinct z localizations of peroxisomes and catalase molecules within them (Fig. 3f–h). We furthermore performed Exchange-PAINT20 (Supplementary Figure 8) visualizing antibody-stained PMP70 and SOMAmer-labeled catalase molecules to demonstrate the ability of co-labeling with antibodies and SOMAmers. To demonstrate the compatibility of simultaneously labeling multiple cellular targets using SOMAmer reagents, we performed two additional Exchange-PAINT experiments: (1) Simultaneous labeling and sequential imaging of HSP60 and LIMP-2 (Supplementary Figure 2), as well as (2) EGFR, ErbB2, and HSP90 in single cells (Supplementary Figure 1).
We next evaluated the role of fixation (Supplementary Figures 9 and 10) and anionic competitors (Supplementary Figures 11 and 12) on the labeling specificity as well as the possibility of direct membrane protein staining on living cells. Aptamers in general and SOMAmers specifically are selected against native protein targets. The fixation conditions typically used for immunostaining of tissues and single cells most likely disrupt the native structure of proteins to some extent, thus potentially decreasing or (in severe cases) preventing labeling. However, successful SOMAmer staining demonstrated here against EGFR, Nup107-GFP, and catalase after a typical paraformaldehyde fixation suggests that SOMAmers can still specifically bind their epitopes in formaldehyde-fixed samples. Nevertheless, we expect that the fixation conditions have to be slightly adjusted for SOMAmers against other targets in order to achieve optimal labeling specificity (see also Supplementary Table 2). Standard fixation conditions used for conventional immunostaining such as 4% paraformaldehyde fixation have proven to be a good starting point for this optimization process. We also note that in fixed cells, the polyanionic competitors dextran sulfate and Z-Block successfully quenched both non-specific nuclear- and cellular organelle-binding (Supplementary Note 1).
Finally, we performed EGFR staining without prior fixation on living cells with the EGFR SOMAmer (Supplementary Note 2 and Supplementary Figure 13), enabling (for the first time) live-cell DNA-PAINT imaging and tracking of membrane targets in their native state (Supplementary Figure 14 and Supplementary Video 1).
In conclusion, we introduce SOMAmers as small, efficient, quantitative, and universal labeling probes for DNA-PAINT, directly translating its high achievable resolution and quantitative imaging capability to a diverse variety of cellular targets. SOMAmers with their enhanced affinity and readily available library of thousands of cellular targets are poised to replace antibodies and nanobodies as labels, thus potentially becoming the preferred affinity reagent for super-resolution microscopy. Together with the spectrally-unlimited multiplexing capabilities in Exchange-PAINT20, SOMAmers should make it possible to eventually image tens to hundreds of cellular targets in single cells with single-molecule spatial resolution in a quantitative fashion and furthermore allow for live labeling and imaging of membrane-bound proteins. However, we note that there is – apart from synthesizing and assaying the large number of additional SOMAmers – a considerable challenge to achieve this amount of multiplexing: Finding compatible fixation conditions for a large variety of targets. This might be potentially alleviated by the possibility of performing live labeling followed by subsequent fixation, however, future assays are necessary in this direction. Taken together, quantitative SOMAmer labeling for DNA-PAINT (due to the easy modification with exactly one single docking site per SOMAmer) might have far-reaching implications with the potential to deliver on one of the ultimate promises of SMLM: Enabling system-wide biological studies with quantitative single-protein resolution. Possible applications could include the study of the interplay of homo- and heterodimerization of membrane receptors after different stimulation treatments on the single-protein-level, which could lead to new insights into their nanoscale organization and physiological function.
Online Methods
Materials
Cy3b-modified and thiolated DNA oligonucleotides were purchased from MWG Eurofins. Tris 1M pH 8.0 (cat: AM9856), EDTA 0.5M pH 8.0 (cat: AM9261), magnesium 1M (cat: AM9530G), sodium chloride 5M (cat: AM9759), and ultrapure water (cat: 10977-035) were ordered from Thermo Fisher Scientific. Potassium chloride (cat: 6781.1), Triton-X-100 (cat: 6683.1), and EGTA (cat: 3054) were ordered from Roth. Sodium hydroxide (cat: 31627.290) was purchased from VWR. Tween-20 (cat: P9416-50ML), glycerol (cat: G5516-500ML) and methanol (cat: 32213-2.5L) were ordered from Sigma Aldrich. Protocatechuate 3,4-Dioxygenase pseudomonas (PCD) (cat: P8279), 3,4-Dihydroxybenzoic acid (PCA) (cat: 37580-25G-F) and (+-)-6-Hydroxy-2,5,7,8-tetra-methylchromane-2-carboxylic acid (Trolox) (cat: 238813-5G) were purchased from Sigma-Aldrich. PIPES (cat: 5625-37-6) was ordered from Sigma-Aldrich. Sucrose (cat: 57-50-1) was obtained from Merck. Dextran sulfate sodium salt from Leuconostoc spp. (6.5–10 kDa, cat: D4911-10G) was purchased from Sigma-Aldrich. Salmon sperm DNA, sheared (10 mg/ml) (cat: AM9680) was ordered from Thermo Fisher Scientific. A431, A549 and SK-BR-3 cells were purchased from ATCC. mEGFP-Nup107 HeLa Kyoto was obtained from the Ellenberg lab21. Tissue Culture Treated Flasks (cat: 353136) were ordered from Falcon. Dulbecco’s Modified Eagle medium (DMEM) with high glucose, GlutaMAX™ and sodium pyruvate (cat: 31966-021), McCoy’s5A Medium (cat: 26600-023), Fetal Bovine Serum (FBS) (cat: 10500-064), 1× Phosphate Buffered Saline (PBS) pH 7.2 (cat: 20012-019),10× PBS pH 7.4 (cat: 70011036), 0.05% Trypsin–EDTA (cat: 25300-054), and Leibovitz’s L-15 medium (cat: 11415064) was purchased from Thermo Fisher Scientific. 0.25% Trypsin-EDTA (T4049-100ML) was ordered from Sigma-Aldrich. 16% (w/v) Formaldehyde solution (cat: 28906) was purchased from Thermo Fisher Scientific. Glutaraldehyde (cat: 16220) was obtained from Electron Microscopy Sciences. NH4Cl (cat: 254134-25G) was ordered from Sigma-Aldrich. Bovine Serum Albumin (cat: A4503-10G) was ordered from Sigma-Aldrich. Eppendorf 8-well chambered coverglass (cat: 0030742036) was purchased from Eppendorf and glass-bottomed 8-well µ-slides (cat: 80827) were ordered from ibidi. Human EGF (cat: E9644-.2MG) was purchased from Sigma Aldrich. 90 nm standard gold nanoparticles (G-90-10) were ordered from Cytodiagnostics. Primary monoclonal Anti-EGFR antibody (cat: 4267S) was purchased from Cell Signaling. Secondary polyclonal anti-rabbit antibody conjugated to Alexa Fluor 647 (cat: ab150075) was ordered from Abcam. Primary anti-PMP70 antibody (ab211533) was purchased from Abcam. Secondary AffiniPure Anti-Mouse IgG antibody (cat: 115-005-003) was obtained from Jackson ImmunoResearch.
Buffers
The following buffers were used for sample preparation and imaging22: SOMAmer staining buffer. 1×PBS pH 7.4, 5 mM MgCl2, 0.05% Tween-20, 1% BSA, 1 mM dextran sulfate, 10–100 µM Z-Block, 0.1–0.2 mg/ml sheared salmon sperm DNA Imaging buffer. 1×PBS pH 7.4, 500 mM NaCl, pH 8, 1×Trolox, 1×PCA, 1×PCD.
100×Trolox: 100 mg Trolox, 430 µL 100% Methanol, 345 µl of 1 M NaOH in 3.2 mL H2O. 40×PCA: 154 mg PCA, 10 mL water and NaOH were mixed and adjusted to pH 9.0. 100×PCD: 9.3 mg PCD, 13.3 mL of buffer (100 mM Tris-HCl pH 8.0, 50 mM KCl, 1 mM EDTA, 50% glycerol).
SOMAmer reagents
SOMAmer design with docking strand. Each SOMAmer construct is described in Supplementary Table 1.
Protein Targets Used in SELEX
Green fluorescent protein (GFP) with a His-tag was purchased from Millipore Sigma (cat: 14-392). Catalase purified from human erythrocyte was purchased from Athens Research and Technology (cat: 16-05-030000). HSP90 with a His-tag and HSP60 were purchased from VWR (cat: 102036-254 (HSP90), 80059-208 (HSP60)). LIMP2/SR-B2 Fc chimera, ErbB2/Her2 Fc chimera, CF and EGFR Fc chimera, CF were ordered from R & D Systems (cat: 1966-LM (LIMP2), 1129-ER (ErbB2), 344-ER (EGFR)).
Modified Aptamer Discovery and Synthesis
Aptamers were discovered using the Systematic Evolution of Ligands by EXponential Enrichment (SELEX) method as described in Gold et al., 201023. Selections were performed using modified DNA libraries with a 40N random region containing either 5-(N-benzylcarboxamide)-2’-deoxyuridine (BndU) or 5-[N-(1-naphthylmethyl)carboxamide]- 2’-deoxyuridine (NapdU) in place of dT. Oligonucleotides were synthesized by solid phase synthesis with modified deoxyuridine-5-carboxamide amidite reagents as described earlier24, using phosphoramidite chemistry25. Both modified aptamer sequences were 50 nucleotides in length (40 nucleotides from the originally random sequence, plus 5 flanking nucleotides on each end from the fixed sequences) and were synthesized with cyanine-3 from Glen Research (cat: 10-5913-02) and P1 handle (10 nucleotides) at the 5’end of the modified aptamer. Each modified aptamer was cleaved and deprotected from solid support with 20% diethylamine (Sigma-Aldrich, cat: 471216)/acetonitrile (Honeywell, cat: CS017-56) followed by gaseous methylamine (Sigma-Aldrich, cat: 295531) at 40°C for 90 minutes, washed with 90% acetonitrile/water, and eluted with deionized water. Product was purified by HPLC with 100 mM triethylammonium bicarbonate with 5% acetonitrile (A) and 100 mM triethylammonium bicarbonate with 70% acetonitrile (B)26 and characterized by standard methods for purity (UPLC), identity (LC/MS), quantity (UV spectrophotometry), and activity (solution binding affinity).
Z-block, Polyanionic Competitor
Z-block was designed as a non-specific, polyanionic competitor for SOMAmer reagent target interactions. This synthetic molecule has the same modified BndU nucleotides2 incorporated during solid phase synthesis using phosphoramidite chemistry3. After solid phase synthesis, the product is cleaved and deprotected with t-butylamine:methanol:water (1:1:2) at 37°C for 24 hours and evaporated to dryness27. The re-constituted product is purified by HPLC and characterized by standard methods for purity, identity, and quantity.
Equilibrium Binding Constants (Kd)
Equilibrium binding constants of aptamers were measured in solution at 37°C as described in Gold et al., 201023. Recombinant EGFR (cat: 344-ER-050), GFP_AEQVI (cat:14392), and Catalase (cat: 16-05-030000) proteins were purchased from R&D Systems, EMD Millipore, and Athens Research & Technology, respectively. Briefly, heat-cooled 5’-32P-radiolabeled DNA SOMAmers (heating to 90°C for 5 min and cooling down to room temperature over the course of 20–30 min) were mixed with different concentrations of target protein in binding buffer (40 mM HEPES [pH 7.5], 102 mM NaCl, 5 mM KCl, 1 mM EDTA, 0.05% Tween-20). SOMAmer-protein complexes were captured with Zorbax beads (Agilent, cat: 899999-777) and quantified with a phosphoimager.
Cell culture
A431 and A549 cells were used for EGFR and Catalase imaging. SK-BR-3 cells were used for exchange-PAINT experiments. For GFP SOMAmer staining, a Hela-Kyoto-2×ZFN-mEGFP cell line was used. A431, A549 and Hela cells were grown in high glucose (4.5 g/L) DMEM supplemented with GlutaMAX™, 1 mM sodium pyruvate and 10% FBS. SK-BR-3 cells were grown in McCoy’s5A medium supplemented with 15% FBS. Cells were seeded into 8-well-chambered cover glasses and grown to 50–70% confluency.
SOMAmer preparation and folding
Lyophilized SOMAmer reagents were reconstituted in TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0) and stored in 20 µM aliquots at -20°C. Working aliquots were stored at 4°C until used. SOMAmer reagents were heat-cooled in PBS at a concentration of 0.2–2 µM and used for labeling on the same day.
GFP-Nup-107 SOMAmer staining
Cells were rinsed with PBS and fixed in pre-warmed (to 37°C) 2.4% paraformaldehyde in PBS for 15 min at room temperature. Then, the cells were washed twice with PBS followed by an incubation in 0.1 M NH4Cl for 10 min. Cells were permeabilized with 0.25% Triton X-100 in PBS for 5 min and subsequently blocked using 3% BSA in PBS for 15–30 min. SOMAmer against GFP (100 nM in SOMAmer staining buffer containing 100 µM Z-Block and 1 mM dextran sulfate) was incubated with the cells for 1 h at room temperature. After incubation with the SOMAmers, the cells were washed three times with PBS supplemented with 5 mM MgCl2.
EGFR (ErbB1) SOMAmer staining
Prior to fixation A431 cells were serum depleted overnight. Cells were rinsed with PBS and fixed in pre-warmed (to 37°C) 4% paraformaldehyde in cytoskeleton buffer28 (500 mM NaCl, 50 mM PIPES, 15 mM MgCl2, 5 mM EGTA, and 5 mM sucrose) in PBS for 30 min at room temperature. Free aldehyde groups were quenched using 0.1 M glycine and 3% BSA in PBS for 15 min. The folded SOMAmer against EGFR was diluted to 100 nM in SOMAmer staining buffer (1×PBS, 5 mM MgCl2, 1% BSA, 0.05% Tween-20, 50 µM Z-Block, 1 mM dextran sulfate) and incubated with the cells for 1 h at room temperature. After SOMAmer incubation, the cells were washed three times with PBS supplemented with 5 mM MgCl2. Post-fixation was performed using 4% paraformaldehyde and 0.1% glutaraldehyde in PBS with 5 mM MgCl2 for 10 min at room temperature. Afterwards, cells were washed 3 times with PBS and incubated in 3% BSA, 0.1 M glycine and 5 mM MgCl2 in PBS for 10 min.
EGFR immunostaining
Cells were fixed in the same way as described for EGFR SOMAmer staining. A monoclonal anti-EGFR antibody against intracellular EGFR in blocking buffer (3% BSA, 0.05% Tween-20, 1×PBS) was used to stain the cells for 90 min at room temperature or overnight at 4°C. Cells were briefly rinsed with PBS and washed three times with PBS with an incubation time of 5 min for each washing step. Secondary anti-rabbit antibody conjugated to Alexa 647 was added in blocking buffer (1:200 dilution) for 60 min at room temperature. Cells were briefly rinsed with PBS and washed three times with PBS with an incubation time of 5 min for each washing step.
PMP70 immunostaining and Catalase SOMAmer staining
Cells were rinsed with PBS and fixed in pre-warmed (to 37°C) 4% paraformaldehyde in PBS for 15 min at room temperature. Then the cells were washed twice with PBS and permeabilized with 0.25% Triton X-100 in PBS for 10 min. After an additional washing step with PBS cells were blocked with 3% BSA in PBS for 60 min. Primary anti-PMP70 antibody was incubated in 3% BSA in PBS overnight at 4°C. Cells were briefly rinsed with PBS and washed three times with PBS with an incubation time of 5 min for each washing step. Secondary anti-mouse antibody conjugated to Alexa 647 or P1 DNA-PAINT docking site was added in blocking buffer for 60 min at room temperature. Cells were briefly rinsed and washed three times with PBS (5 min incubation time each). Catalase SOMAmer reagent was added in staining buffer supplemented with 1 mM dextran sulfate, 10 uM Z-Block and 0.2 mg/ml sheared salmon sperm DNA and incubated overnight at 4°C. After SOMAmer incubation, the cells were washed three times with PBS supplemented with 5 mM MgCl2.
Live-cell staining with EGFR SOMAmers
Cell medium was aspirated and the cells were briefly rinsed in phenol-red free Leibovitz’s L-15 medium. The SOMAmer live-cell labeling solution (100 nM EGFR SOMAmer reagent and 10 µM Z-Block in Leibovitz’s L-15 medium) was added and incubated for 10 min at room temperature or 20 min at 4°C. The cell staining solution was removed and cells were washed three times with L-15 medium. Finally, cells were fixed for 30 min using 4% paraformaldehyde and 0.1% glutaraldehyde in PBS with 5 mM MgCl2. After fixation, cells were washed 3 times with PBS and incubated in 3% BSA and 0.1 M glycine in PBS for 10 min.
Confocal imaging
The confocal imaging was carried out on a ZEISS (Jena, Germany) LSM780 confocal laser scanning microscope equipped with a ZEISS Plan-APO 63×/NA1.46 oil immersion objective. GFP, Cy3 and Alexa Fluor 647 excitation was performed with 488 nm, 561 nm and 633 nm diode lasers, respectively. The pinhole size was adjusted to 1 AU. Laser power was used at 4–10% and detector gain was set to 700–900. Imaging conditions were kept constant for each experiment.
DNA-PAINT imaging
Before imaging, 90 nm gold nanoparticle fiducial markers were added and incubated for 5 min (diluted 1:5 in PBS + 5 mM MgCl2). After rinsing with PBS with 5 mM MgCl2, imaging buffer containing DNA-PAINT imaging strands was added. DNA-PAINT imaging was carried out on an inverted Nikon Eclipse Ti microscope (Nikon Instruments) with the Perfect Focus System, applying an objective-type TIRF configuration with an oil-immersion objective (CFI Apo TIRF 100×, NA 1.49, Oil). Samples were excited using a 561 nm laser (200 mW nominal, Coherent Sapphire). The laser beam was passed through a cleanup filter (ZET561/10, Chroma Technology) and coupled into the microscope objective using a beam splitter (ZT561rdc, Chroma Technology). Fluorescence light was spectrally filtered with two emission filters (ET600/50m and ET575lp, Chroma Technology) and imaged on an sCMOS camera (Zyla 4.2, Andor Technologies). Imaging was performed without additional magnification in the detection path and 2×2 camera binning resulting in a pixel size of 130 nm.
DNA-PAINT single-particle-tracking
Cells were stained with 100 nM EGFR SOMAmer in L-15 medium and 10 µM Z-Block. After labeling, cells were washed two times with L-15. Imaging was performed using 1 nM P1-5’ Cy3b imager. EM Gain was set to 300, laser power density to ~0.08 kW/cm2, and 50 ms integration was used. Particle traces were analyzed using ImageTracker from the Mosaic group in ImageJ29. Detection settings were as follows: Radius: 4, Percentile: 0.8, Cutoff radius: 0.0001, Displacement: 1.5, Link range: 2.
DNA-PAINT imaging conditions
For imaging, the following DNA-PAINT imager were used: P1-5’-Cy3b: 5’-Cy3b-TAGATGTAT-3’, P1-3’-Cy3b: 5’-CTAGATGTAT-Cy3b-3’, P3-Cy3b: 5’-TAATGAAGA-Cy3b-3’, P5-Cy3b: 5’-CATACATTGA-Cy3b-3’. P6-Cy3b: 5’-CTTTACCTAA-Cy3b-3’. All DNA-PAINT measurements were performed in imaging buffer (1×PBS pH 8, 500 mM NaCl, 1×Trolox, 1×PCA, 1×PCD).
Figure 1d–g
Imaging was performed using 150 ms integration time for 40,000 frames with a P1-5’-Cy3b imager strand concentration of 1 nM. Laser power was set to ~1.8 kW/cm2 before the Back Focal Plane (BFP) of the objective.
Figure 2c–e
Imaging was performed using 300 ms integration time for 60,000 frames with a P1-5’-Cy3b imager strand concentration of 0.8 nM. Laser power was set to ~0.3 kW/cm2 before the BFP of the objective.
Figure 3d-h
Imaging was performed using 250 ms integration time for 20,000 frames with a P5-Cy3b imager strand concentration of 0.6 nM. Laser power was set to ~0.8 kW/cm2 before the BFP of objective
Image analysis
Super-resolution images were reconstructed using the ‘Picasso’ software package as described before22, 30.
Data availability statement
All raw data are available upon request from the authors.
Supplementary Material
Acknowledgements
We thank M. Spitaler and the imaging facility of the MPI of Biochemistry for confocal imaging support and A. Auer and F. Schueder for super-resolution microscopy support. This research was funded by the German Research Foundation through the Emmy Noether Program (DFG JU 2957/1-1), the European Research Council through an ERC Starting Grant (MolMap, Grant agreement number 680241), the Allen Distinguished Investigator Program through The Paul G. Allen Frontiers Group, the Max Planck Society, and the Max Planck Foundation. S.S. acknowledges support by the QBM graduate school. M.T.S. acknowledges support by the IMPRS-LS graduate school. We thank our SomaLogic colleagues in discovery, process chemistry and analytical chemistry for their support of this project. SOMAmer_reagent is a registered trademark of SomaLogic, Inc.
Footnotes
Author Contributions
S.S. designed and performed the experiments, analyzed the data, and wrote the manuscript. P.N. designed experiments and wrote the manuscript. M.T.S. analyzed data and wrote software. V.J.S. and J.E. created the Nup107-GFP cell line. J.D.C., S.G. and N.J. contributed to generation of SOMAmer reagents and provided guidance on their use. R.J. conceived of and supervised the study, analyzed the data, and wrote the manuscript. All authors reviewed and approved the manuscript.
Competing Interests
The authors declare the following competing interests: J.D.C., S.G. and N.J. are SomaLogic, Inc. employees and stakeholders.
References
- 1.Hell SW, Wichmann J. Opt Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 2.Betzig E, et al. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 3.Hess ST, Girirajan TP, Mason MD. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rust MJ, Bates M, Zhuang X. Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sahl SJ, Hell SW, Jakobs S. Nat Rev Mol Cell Biol. 2017;18:685–701. doi: 10.1038/nrm.2017.71. [DOI] [PubMed] [Google Scholar]
- 6.Dai M, Jungmann R, Yin P. Nat Nanotechnol. 2016;11:798–807. doi: 10.1038/nnano.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balzarotti F, et al. Science. 2017;355:606–612. doi: 10.1126/science.aak9913. [DOI] [PubMed] [Google Scholar]
- 8.Schnitzbauer J, Strauss MT, Schlichthaerle T, Schueder F, Jungmann R. Nat Protoc. 2017;12:1198–1228. doi: 10.1038/nprot.2017.024. [DOI] [PubMed] [Google Scholar]
- 9.Jungmann R, et al. Nano Lett. 2010;10:4756–4761. doi: 10.1021/nl103427w. [DOI] [PubMed] [Google Scholar]
- 10.Ries J, Kaplan C, Platonova E, Eghlidi H, Ewers H. Nat Methods. 2012;9:582–584. doi: 10.1038/nmeth.1991. [DOI] [PubMed] [Google Scholar]
- 11.Opazo F, et al. Nat Methods. 2012;9:938–939. doi: 10.1038/nmeth.2179. [DOI] [PubMed] [Google Scholar]
- 12.Gomes de Castro MA, Hobartner C, Opazo F. PLoS One. 2017;12:e0173050. doi: 10.1371/journal.pone.0173050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gold L, et al. PLoS One. 2010;5:e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta S, et al. Appl Immunohisto M M. 2011;19:273–278. doi: 10.1097/PAI.0b013e3182008c29. [DOI] [PubMed] [Google Scholar]
- 15.Rohloff JC, et al. Mol Ther-Nucl Acids. 2014;3 doi: 10.1038/mtna.2014.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Endesfelder U, Heilemann M. Nat Methods. 2014;11:235–238. doi: 10.1038/nmeth.2852. [DOI] [PubMed] [Google Scholar]
- 17.Culley S, et al. Nat Methods. 2018 [Google Scholar]
- 18.Jungmann R, et al. Nat Methods. 2016;13:439–442. doi: 10.1038/nmeth.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang B, Wang W, Bates M, Zhuang X. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jungmann R, et al. Nat Methods. 2014;11:313–318. doi: 10.1038/nmeth.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otsuka S, et al. Elife. 2016;5 doi: 10.7554/eLife.19071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schnitzbauer J, Strauss MT, Schlichthaerle T, Schueder F, Jungmann R. Nat Protoc. 2017;12:1198–1228. doi: 10.1038/nprot.2017.024. [DOI] [PubMed] [Google Scholar]
- 23.Gold L, et al. PLoS One. 2010;5:e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta S, et al. J Biol Chem. 2014;289:8706–8719. doi: 10.1074/jbc.M113.532580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beaucage SL, Caruthers MH. Tetrahedron Letters. 1981;22:1859–1862. [Google Scholar]
- 26.Carlson M, Carter JD, Rohloff J. Green Chem Lett Rev. 2015;8:37–39. [Google Scholar]
- 27.Mullah B, Livak K, Andrus A, Kenney P. Nucleic Acids Res. 1998;26:1026–1031. doi: 10.1093/nar/26.4.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whelan DR, Bell TD. Sci Rep. 2015;5:7924. doi: 10.1038/srep07924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sbalzarini IF, Koumoutsakos P. J Struct Biol. 2005;151:182–195. doi: 10.1016/j.jsb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 30.Schueder F, et al. Nat Commun. 2017;8:2090. doi: 10.1038/s41467-017-02028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data are available upon request from the authors.