Abstract
Usually, cells balance their growth with their division. Coordinating growth inputs with cell division ensures the proper timing of division when sufficient cell material is available and affects the overall rate of cell proliferation. At a very fundamental level, cellular replicative lifespan—defined as the number of times a cell can divide, is a manifestation of cell cycle control. Hence, control of mitotic cell divisions, especially when the commitment is made to a new round of cell division, is intimately linked to replicative aging of cells. In this chapter, we review our current understanding, and its shortcomings, of how unbalanced growth and division, can dramatically influence the proliferative potential of cells, often leading to cellular and organismal aging phenotypes. The interplay between growth and division also underpins cellular senescence (i.e., inability to divide) and quiescence, when cells exit the cell cycle but still retain their ability to divide.
Keywords: Cell senescence, Quiescence, Aging, Cell growth, Protein translation, mTOR signaling, Asymmetric division, Hypertrophy and Cdk
8.1. Introduction
The two cells generated at the end of the cell cycle usually inherit sufficient amounts of essential constituents, ensuring their survival. Moreover, the composition of proliferating cells varies very little from generation to generation, implying that proliferating cells somehow balance their growth (increase in biomass) with their division. Since different levels of nutrients and growth factors sustain different rates of cell proliferation, cells have elaborate mechanisms to sense nutrient and growth signals, adjusting their metabolic and proliferative activity accordingly. A detailed mechanistic understanding of this coupling between growth and division has remained elusive. Nonetheless, properly coupling growth with division is thought to determine the rate at which cells proliferate [1–6].
Nutrient and growth factor limitations do not delay all cell cycle transitions uniformly. Instead, transit through some cell cycle phases is delayed disproportionately. Overwhelmingly, poor growth conditions prolong the G1 phase of the cell cycle, preceding initiation of DNA synthesis, while transit through the remaining cell cycle phases is not delayed significantly [1, 2, 7–12]. In yeast, the point of commitment to a new round of cell division is called START [2] and in animal cells the Restriction Point [6]. Once cells pass through these points in late G1 phase, they will initiate and complete their division even if they encounter growth limitations [1, 2, 6, 12]. Hence, although there may be some nutrient and growth factor inputs in later stages of the cell cycle [13, 14], it is in G1 that cells delay committing to a new round of cell division in the face of weak growth prospects.
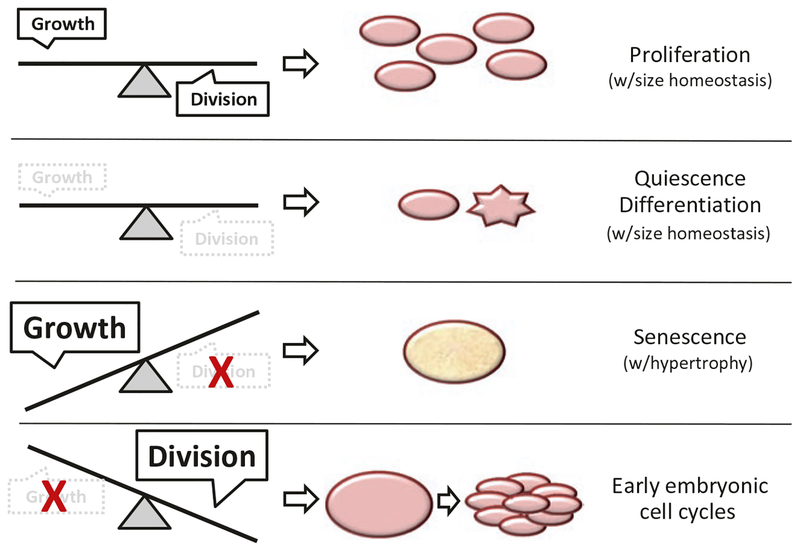
The consequences of uncoupling growth from division are profound and accompanied by changes in the size of cells (see Fig. 8.1 for a schematic). In this Chapter, we describe possible outcomes when growth and division are not balanced. We also discuss models that envision imbalances between growth and division as a critical component of senescence and aging mechanisms [15, 16]. Our discussion will include examples from animal models systems and unicellular organisms, especially the budding yeast S. cerevisiae.
Fig. 8.1.
Schematic representation of all possible outcomes when growth and division are balanced or unbalanced
8.2. Growth and Division: A Tight Balancing Act
Intuitively, it makes sense that for a cell to successfully divide and give rise to two viable cells, it must produce enough macromolecules, membranes, and organelles for the two cells that will arise at the end of cytokinesis. Since these cellular components are determinants of the cell’s volume, it is not surprising that cell size has often been used as an “umbrella” metric for cell growth [3, 5, 14, 17, 18]. Lately, there is renewed interest in the development of methodologies that report accurately and precisely on the size of animal cells [5, 19–21]. In addition, asymmetric segregation of cellular constituents between two products of a mitotic cell division is also tightly regulated and will be discussed below.
Although cell size is a very useful “growth” metric, the events most closely associated with cell proliferation are anabolic processes that yield the macromolecules necessary to build new cells. Among those macromolecules, proteins are often considered the most important component of growth, and for good reason. The protein fraction of dry mass is ≈55% in E. coli [22] and ≈40–45% in the budding yeast S. cerevisiae [22, 23]. The protein content of mammalian cells varies in different tissues. For commonly used cell lines, such as mouse fibroblasts (NIH3T3 cells) or human HeLa cells, the protein molecules per unit volume is roughly the same as in budding yeast cells (1–2E + 06 proteins/fL; [22]). Also, a significant fraction of the proteome (>20%) is dedicated to making ribosomal proteins and translation factors that will, in turn, promote the synthesis of more proteins [24].
Making ribosomal components and assembling them into functional ribosomes involves a broad repertoire of cellular constituents and processes [25–27]. In budding yeast, the cytoplasmic ribosome contains 78 ribosomal proteins encoded by the RP regulon of 138 genes. Note that 59 of the 78 yeast ribosomal proteins are encoded by pairs of very similar paralogs [28, 29]. The ribosomal proteins together with the four rRNAs (5S, 5.8S, 18S, and 25S) make up the ribosome. The rRNA genes are encoded by rDNA tandem repeats, whose number is dynamic (usually ≈100–200) and varies with growth conditions. Greater than 200 protein assembly and accessory factors are needed at many stages to put a functional ribosome together. Their expression is thought to be regulated coordinately, through the ribosome biogenesis (Ribi) regulon. In the Ribi regulon, one also finds the various tRNA synthetases, rRNA processing and modifying enzymes, and translation factors, which collectively control translational capacity [30, 31]. Most of the cell’s transcriptional activity is devoted to building and maintaining the translational machinery. Of all the RNA in the cell, 80% is rRNA, 15% is tRNA, and 5% is mRNA, and a large fraction of mRNA is devoted to ribosome synthesis [25, 32].
Transcription of RP genes alone is responsible for approximately 50% of all RNA PolII-mediated transcription initiation events. The energetic cost of making the translation machinery is astounding, consuming as much as ≈90% of the total energy of fast-proliferating yeast cells [25]. Estimates of the ribosome content of cells give an even more impressive view of the centrality of ribosome biogenesis in governing the growth of cells. From super-resolution, single-molecule imaging techniques, it seems that E. coli cells contain 30,000–50,000 ribosomes per fL [33]. Analogous quantitative measurements are lacking in eukaryotes, but prior estimates in yeast put the number at about 200,000 ribosomes per cell [25]. On average then, during one cell cycle lasting ≈100 min, a yeast cell must produce ≈2000 ribosomes per minute. Based on these metrics of the cellular economy, one can easily see why for decades protein synthesis has been viewed as the fundamental measure of cell growth in considerations of balancing growth with cell division [34].
Building and maintaining the ability to synthesize proteins is such a costly process that would be expected to influence if, and when, cells commit to a new round of cell division. The earliest evidence for specific effects on the cell cycle due to translational control was the isolation of budding conditional yeast cdc (cell division cycle) mutants in what turned out to be translation factors [2]. Hypomorphic mutations in translation initiation factors impair the capacity of cells to initiate a new round of cell division [12, 35–40]. Moreover, signaling pathways that control initiation of division, such as the Target of Rapamycin (TOR) pathway, may do so, at least in part, by regulating translation initiation. Loss of TOR function causes G1 arrest in mammalian cells [41, 42] and yeast [43, 44]. Conversely, overexpression of translation initiation factor eIF4E in mammals is oncogenic [45], and the translational output of TOR signaling is critical for cancer initiation [46]. Moreover, inhibiting translation elongation with cycloheximide also prolongs the G1 phase of the cell cycle [12, 47]. In budding yeast, cycloheximide reduces the newborn cell size [12, 47] and the rate at which cells increase in size [48]. It also increases the critical size threshold for START [47, 48]. Together, these results support the notion that a critical rate of protein synthesis is required for G1 transit and completion of START in budding yeast [49] and animal cells [50, 51].
If ribosome biogenesis and protein synthesis are such integral parts of cell growth, propelling cells to divide, how do cells control ribosome biogenesis? In all eukaryotes, the principal regulator of catabolic processes leading to energy production is protein kinase A (PKA), while the analogous “master” regulator of anabolic, biosynthetic processes is the TOR kinase (the TORC1 complex). As we will describe in subsequent sections, these two pathways have overlapping roles as determinants of cellular and organismal replicative potential and aging. In rapidly proliferating cells, however, for maximal growth and rates of cell division, both PKA and TORC1 are active, and they are both needed to activate ribosome biogenesis fully by derepressing the Ribi regulon [44, 52, 53].
In yeast, replicative aging is defined by the number of buds that can be produced by one mother cell, indicative of the number of times it can progress through the cell cycle and undergo mitosis [54]. An increase in cell size (i.e., cellular hypertrophy) has been linked to replicative aging. The cellular hypertrophy model of aging was formulated to account for the yeast replicative aging, invoking the existence of a maximal cell size beyond which cells could not maintain division [55, 56]. According to the cellular hypertrophy model, the large cell size of old yeast mother cells is incompatible with some cellular function that is necessary for cell division. Every time mother cells divide, they increase in size until they reach the terminal, large size, at which point they will enter a state of proliferative arrest. The hypertrophy model predicts that small cells would be able to divide more times before reaching the terminal size, resulting in longer replicative lifespan. On the other hand, large cells will reach the terminal size after fewer divisions, having a shorter replicative lifespan. Overall, the model predicted that changes in replicative lifespan and cell size ought to be proportional. However, based on genome-wide measurements of cell size, we recently reported that the mean cell size of long-lived yeast mutants was not significantly different from the size of mutants that were not long-lived [57]. This finding is incompatible with the key prediction of the hypertrophy model that long-lived mutants would have a small overall cell size, allowing these cells to divide more times until they reach the terminal size and enter senescence. Therefore, based on these experiments in yeast, it seems that the cellular enlargement is not a primary determinant of aging.
The factors linked to cell growth and protein translation definitely affect aging, however. Reduced mTOR signaling leads to lifespan extension is yeast [58, 59], worms [60], flies [61], mice [62, 63], and initial studies suggest possibly humans [64, 65]. The mechanisms that underlie lifespan extension remain to be fully determined, partly due to the complexity of these signaling pathways, but evidence exists both for altered protein synthesis and enhanced protein turnover through autophagy [66]. The former comes from findings that reduced expression of several translation initiation factors and ribosomal components also lead to lifespan extension in a range of organisms [67]. It is clear at least for replicative lifespan in yeast (the number of times one cell can divide to produce daughter cells), however, that globally and uniformly inhibiting protein synthesis in insufficient to slow aging since cycloheximide is unable to mediate this effect [68]. This finding indicates that translational changes to specific mRNAs are likely conferring, at least in part, longevity phenotypes in mTORC1 and translation factor mutants.
Interestingly, one downstream factor linked to lifespan extension is GCN4, a transcription factor regulated itself by translation and dependent on the presence of small upstream open reading frames in its mRNA [68, 69]. Inhibition of mTORC1, reduced 60S ribosomal subunit levels and calorie restriction all lead to enhanced GCN4 translation [70–74], which in yeast induces expression of stress and nutrient response pathways [69]. Loss of GCN4 at least partially abrogates lifespan extension by these interventions. The mammalian ortholog of GCN4, ATF4, is also induced in cells and mice from a range of interventions conferring long lifespan [75, 76], arguing for conservation of this pathway.
Reduced mTORC1 signaling also leads to enhanced autophagy, which has been linked to lifespan extension as well in a range of conditions [77]. In non-vertebrates, for instance, inhibition of autophagy is sufficient to block lifespan extension by mutants that affect translation [78–81]. Moreover, at least in flies and mice, induction of autophagy through overexpression of autophagy components is reported to lead to lifespan, and sometimes healthspan, extension [82–85].
The mTORC1 complex regulates transcription as well and this phenomenon has been studied extensively in yeast. For chronological aging, defined as survival in a post-replicative state, reduced mTORC1 signaling enhances longevity [59, 86], likely through mechanisms leading to enhanced transcription of a stress response transcription factor network driven by Msn2/4 and Gis1 [54]. Interestingly, reduced PKA signaling promotes chronological lifespan extension through overlapping mechanisms [87, 88].
Protein kinase A signaling has been connected to aging in multiple organisms [89]. In yeast, under maximal growth conditions in rich media, where both PKA and mTOR collaborate to drive protein synthesis, mutations leading to reduced PKA activity promote replicative and chronological lifespan extension [90, 91]. Although not studied as extensively in multi-cellular organisms, this phenomenon may also be conserved as mice lacking the protein kinase subunit RIIβ are long-lived [92, 93], as well as those lacking ADCY5, encoding type 5-adenylyl cyclase (AC5) that converts ATP into cAMP in turn activating PKA. These mice are stress resistant and experience a 30% increase in median lifespan [94, 95]. In addition, genetic variants leading to reduced production of the adenylyl cyclase-activating β2-adrenergic receptor, are prevalent in men from a Chinese centenarian population [95].
In conclusion, in balanced growth and division, the increased ribosome biogenesis and protein synthesis is coupled to cell division, maintaining the overall cellular homeostasis and macromolecular composition. These processes also have robust effects on aging, although the links are far from straightforward.
8.3. Asymmetric Segregation During Cell Division
When cells divide, their cellular constituents have to be divided between the two offspring and studies have started to address mechanisms underlying this partitioning. In mammalian cells, for instance primary fibroblasts in culture, division is symmetric and while partitioning may occur, it is hard to distinguish morphologically. Interestingly the culture of primary fibroblasts senescences at a similar number of population doublings, suggesting that with respect to cellular aging damaged molecules may not be partitioned specifically to one cell after division.
Yeast, being a single-celled organism, has to maintain continuous division in the colony in the face of the challenges of aging. By virtue of their division by budding, which produces a larger mother cell and a smaller bud that are easily distinguishable, yeast offers a great opportunity to detect differential segregation of cellular materials. While the mother cell ages, the daughter remains youthful, suggesting that damaged cellular constituents driving aging may remain in the mother cell [96]. Some components of the daughter cell, such as the cell wall are largely the result of new synthesis during division, representing one method of segregating old material to mothers. However, many cytoplasmic factors partition and several aging factors are reported to remain in mothers. For instance, extrachromosomal rDNA circles (ERCs), small episomes containing rDNA repeats that drive aging possibly by competing for replication factors with chromosomal origins [97], are heavily partitioned toward mothers [98]. The mechanism likely relates to the closed mitosis of yeast, which maintain a nuclear structure. Originally nuclear pore association was proposed as a mechanism by which ERCs were retained in the mother, with the assertion that nuclear pores have very restricted access to daughters [99]. Later reports called that into question [100], and suggested that ERCs may simply not diffuse efficiency through the bud neck [101–103]. Thus geometry drives asymmetry.
Damaged aggregated proteins are also retained in the mother cells, likely through restriction of access to daughters and also by active transport of damaged molecules from daughters to back to mothers [96]. The former process again has been reported to involved both active retention and limits to passive diffusion to daughters, while the latter likely involves the actin cytoskeleton and requires Sir2, a protein deacetylase linked to aging [104]. The mechanisms underlying these processes remain to be fully elaborated.
Mitochondria are reported to undergo asymmetric inheritance in yeast, with fitter mitochondria finding their way to daughters [105]. More work is required to access whether and how partition occurs in this and other organelles. In fact, cellular processes to maintain asymmetry may be broader that we currently appreciate as recent single cell based screens have identified hundreds of asymmetrically partitioned proteins during budding. One screen identified 74 proteins partitioned to mothers and 60 to daughters [106]. Interestingly, strains lacking genes for the mother-specific proteins are more likely to have an enhanced lifespan. Whether it comes to individual proteins, damaged protein aggregates, extrachromosomal rDNA circles or organelles, evidence suggests that asymmetry breaks down with the age of the mother and this is consistent with observations that daughters from old mothers do not enjoy a full replicative lifespan.
Of course, asymmetry in cell division has massive impacts during development and cell differentiation throughout the mammalian organism. A classic example is an adult stem cell that divides to produce another stem cell and a cell committed to a differentiation pathway. Asymmetry of cellular constituents plays a role in defining cell fate in this context and it is highly likely that damaged molecules are partitioned to the more committed cell [107]. Clearly cell- autonomous and–non autonomous mechanisms are in play and it will be intriguing to determine to what extent the more elaborated mechanisms in yeast are conserved with the cell autonomous mechanisms.
8.4. Quiescence: Not Dividing, but Keep on Ticking
Cells can enter a quiescent state, in response to a range of signals, in which they do not divide, but maintain a metabolically active state and can resume the capacity to divide, when conditions permit. When cells adopt a differentiated state they exit the cell cycle, sometimes permanently [108]. In all eukaryotes, cyclin-dependent kinase (Cdk) protein complexes are at the core of the cell division machinery [109]. Initiation of cell division requires an increase in Cdk activity. Later cell cycle transitions also need high Cdk activity, while a drop in Cdk activity triggers exit from mitosis. Cdks are Ser/Thr protein kinases, similar in structure to most kinases [110]. However, all Cdks are active only when they bind other activating proteins, such as cyclins. Cdk activity is further regulated by phosphorylation or binding of additional protein subunits. These layers of control can raise or lower overall Cdk activity, depending on the phosphorylated Cdk residue, or the interacting protein, in each case.
Changes in Cdk activity underlie transitions from resting to proliferative cellular states in health and disease. Indeed, high Cdk activity contributes to most proliferative disorders, including cancer cell development [111, 112]. On the other hand, low Cdk activity is associated with terminal differentiation [113], and accompanies poor organ regeneration, for example, in hepatic [114], cardiac [115], neuronal [116], or appendage tissues [117].
It is clear that in quiescent cells there is a strong albeit potentially reversible block in cell division. Maintaining the potential to divide, however, is a key feature that distinguishes quiescent from senescent cells. This concept was put to the test almost two decades ago, in a particularly lucid experiment. Microinjection of pre-formed active Cdk protein complexes was sufficient to initiate cell division in quiescent human fibroblasts, in the absence of growth factors [118].
In quiescent cells, the block in cell division is also accompanied by a profound reprogramming of cellular metabolism. The cells remain metabolically active, enabling them to stay alive (e.g., quiescent yeast cells) or perform the functions prescribed by their terminally differentiated state. Interestingly, balanced downregulation of the master “growth” signaling pathways we described above, the PKA and the TOR pathways, is observed in quiescent yeast cells [119], and this is important for chronological lifespan extension [119], which is the period of time a cell can remain viable in a non-proliferative state.
More recently, it was reported that quiescent cells have a massively re-organized chromatin structure [120]. In yeast cells entering quiescence, the conserved lysine deacetylase Rpd3p establishes a repressive transcriptional state, reducing by ≈30-fold steady-state mRNA levels [120]. Cells lacking Rpd3p also have a 2–3 fold reduction in their mean chronological lifespan [120]. The replicative lifespan of these cells, however, is not affected [121]. This is not surprising since there is no significant overlap of gene deletions that extend lifespan in both the chronological and replicative lifespan assays [122], at least under the assay conditions tested.
Interestingly, however, there are connections between the two types of yeast aging, as chronologically aged cells have reduced replicative lifespan when returned to the cell cycle [123–125]. This is clearly linked to metabolic state, as dietary restriction during the replicative phase of this experiment results in suppression of the short lifespan [126]. Quiescent cells certainly accumulate damage, but once a quiescent cell reenters the cell cycle, this damage may stay with the mother cell [127]. In that scenario, the proliferative capacity and fitness of the population as a whole would be maintained. While this nice model needs further testing, what is clear is that growth and division are still balanced in the quiescent state, and homeostasis is maintained (see Fig. 8.1, the second case from top).
In quiescence, the down-regulation of TOR and PKA leads to significantly reduced ribosome biogenesis and overall protein synthesis [128]. Cell growth and metabolic activity is generally low in quiescent cells [129]. But because this is happening in the context of cell cycle arrest [129, 130], the general properties and macromolecular composition of quiescent cells remain stable and they are easily recognized [16, 128]. Overall, quiescence likely represents a physiological extreme in the normal range of balancing growth with division, a case where both growth and division are coordinately downregulated.
8.5. Senescence: Growing Desperately, with No Possibility of Ever Dividing Again
It has become clearer in recent years that cell cycle arrest can come in different flavors, especially in the context of unabated cell growth. If a cell continues to make proteins and other macromolecules at a high rate in the face of a cell cycle block, then there are only a few possible outcomes. (1) The cell must find ways to get rid of the large excess (e.g., lysosomal degradation, secretion). (2) The cell must somehow accommodate the extra macromolecular amounts within its boundaries, inevitably leading to increased cell size. In fact, the above are typical properties of senescent cells [128, 131] (described in more detail below) and exemplify a clear case of unbalanced growth and division (see Fig. 8.1, the third case from top). The strong growth of senescent cells (often the result of oncogenic stimulation), is not balanced with cell division. Instead, it persists in the face of stable cell cycle blocks.
An important component in the cell cycle arrest of quiescent and senescent cells is the accumulation of Cdk inhibitor molecules. The kinds of Cdk inhibitors employed in each case, however, are different. The cell cycle arrest of quiescent or fully differentiated cells is usually imposed by members of the p27KIP1 family of Cdk inhibitors, which inhibit multiple cyclin/Cdk complexes by interacting with both the cyclin and the Cdk. On the other hand, in senescent cells there is a buildup of p16INK4 Cdk inhibitors, which bind to monomeric Cdk4/6 and reduce cyclin binding affinity [128, 131].
Likewise, while both in quiescent and senescent cells there is an accumulation of tumor suppressors that broadly inhibit transcription associated with entry into the cell cycle, the molecular players are different in each case. Quiescence is associated with the pRB-like proteins p107 and p130, which interact with the transcription factor E2F during G1 phase to inhibit G1/S transcription and commitment to division. Instead, senescent cells have high levels of pRB, and there is also a buildup of p53, a regulator of multiple processes (e.g., DNA damage response) that impinge on the cell cycle [128, 131]. Hence, the molecular effectors of the cell cycle arrest are different. Furthermore, the exit point of the cell cycle may be different in quiescent vs. senescent cells. Quiescent cells uniformly exit the cell cycle before initiation of DNA replication in G1 phase [128, 131]. G1 arrest is also common in senescence. Surprisingly, however, in several cases senescent cells appear to have a permanent G2 phase block in later stages of the cell cycle [128, 132–135].
The unbalanced growth and division observed in senescence is associated with a variety of phenotypes typical of extremely stressed cells. The exact signatures are still a matter of debate [128]. In addition to the cell cycle markers we mentioned above, other traits associated with senescence often include: short or dysfunctional telomeres, lysosomal stress and expression of β-galactosidase, DNA damage response, stress granule formation, hyper-secretory functions, formation of heterochromatic foci, and the senescence-associated secretory phenotype (SASP) [128, 131, 136]. Overall, senescent cells have been aptly compared to automobiles that simultaneously attempt to accelerate (i.e., hyperactive growth pathways) and stop (i.e., strong, permanent cell cycle block), putting the cell on its way to a highly stressed, irreversibly aged state [15, 16, 137].
The phenomenon of cell senescence was discovered more than 50 years ago and it was almost immediately hypothesized to be associated with organismal aging [138]. While it has been clearly established that cell senescence serves as a tumor suppressive mechanism [131], support for the aging theory has waxed and waned over the years. Currently, it is buoyed by a series of recent studies linking cell senescence to aging in mice.
A principle argument against a role for cell senescence in aging has been that even in old individuals, only a small fraction of cells within a tissue appear to be senescent. How could a phenomenon affecting only a small percentage of cells seriously impair an entire tissue? This question has been potentially resolved with the discovery and characterization of the SASP, wherein senescent cells secrete a novel panel of factors in part comprised of inflammatory cytokines that can have potent paracrine and endocrine effects on non-senescent cells [139, 140]. Moreover, a better understanding has emerged regarding the events that can drive cellular senescence. These now include a wide range of cellular stresses [131], which are associated with chronic diseases of aging, suggesting that aging events may drive cell senescence that in turn promote increased aging.
Senescent cells do accumulate with aging and the Cdk inhibitor, p16INK4, has been proposed as a biomarker of aging [128]. Indeed, in selected T cell populations, p16INK4 levels do show a statistically significant predictive value for human age. In addition, panels of inflammatory cytokines have been proposed as aging biomarkers and these may be at least in part related to the SASP. Several recent studies have reported mechanistic insights into SASP induction in senescent cells. Several pathways appear to be involved, including those related to cell growth, such as mTOR, and cell proliferation, such as p53. Rapamycin suppresses aspects of the SASP, but must be delivered continuously to have this effect [141]. This is in contrast to organismal aging, where a transient three-month exposure to rapamycin in middle age is sufficient to extend the lifespan of mice [142, 143].
To test the role of senescence in aging, two different strategies were employed to conditionally ablate senescent cells, both related to the specificity of p16INK4 expression in this cellular condition. Findings in these mice appear promising as ablation of senescent cells is linked to partial suppression of pathology in a mouse progeria model, the BubR1 mice [144], and can extend the lifespan and some healthspan parameters in normal mice [145].
The connection between BubR1 and progeria is interesting in its own right as the gene encodes a component of the mitotic spindle assembly checkpoint, which prevents cells from initiating anaphase if one or more kinetochores are not attached to the mitotic spindle [146]. Mice hypomorphic for BubR1 rapidly develop aging features, including kyphosis, cachexia, and cataracts [147, 148]. They also have a severely reduced lifespan. With age, BubR1 expression declines in a number of tissues, suggesting that reduced expression of the protein late in life may contribute to normal aging [147]. Moreover, overexpression of BubR1 delays aspects of aging [149]. A potential unifying model is that reduced BubR1 expression leads to mitotic defects, driving cell senescence and that the senescent cells drive aging phenotypes through the SASP or other mechanisms [144]. Ablation of senescent cells improves a range of healthspan parameters.
The promise of research in senescence has led to drug discovery approaches designed to specifically kill senescent cells. Several candidates have already emerged, and these compounds have shown efficacy in preclinical models of chronic disease states [150–153]. While the clinical work remains to be done, the last 10 years have seen cell senescence emerge as one pathway likely to modulate organismal aging and many new pathways of therapeutics for age-associated diseases.
8.6. Division Without Growth
In the classic experiments by Hartwell and colleagues, it was established that in most cases growth controls cell division and not the other way around [1, 2, 12]. Stopping cell growth will also stop cell division, but stopping cell division does not usually stop cell growth (as displayed in senescent cells, see discussion in the previous section). From these principles, it follows then that cell division in the absence of growth is untenable, at least when the mass of the daughter cells is reduced below a threshold necessary to sustain their viability. This is precisely what happens during the early embryonic cell cycles after fertilization until the mid-blastula transition ([154]; see Fig. 8.1, last case). At the mid-blastula transition, before the re-establishment of the normal somatic cell cycles, the block in cell division is imposed by Cdk inhibitors. In mutants lacking these inhibitors, cells usually undergo just one extra division [154–156]. Overall, these early embryonic cell cycles do not necessarily violate the fundamental need to balance growth with division. They just reflect the fact that growth needs have been satisfied during oogenesis.
8.7. Outlook
In yeast, it is implicit that aging, both replicative and chronological, must be linked to critical cell cycle decisions. Balancing cell growth with division to maintain cellular homeostasis is a critical component of this process, whether cells are dividing or in a non-proliferative state. The key pathways that coordinate cell growth signals are intimately linked to aging in yeast, and considerable evidence suggests that they have conserved effects on aging in multicellular organisms. Therefore, continued efforts to understand yeast aging in the context of cell growth and division are likely to continue to inform about human aging. A major challenge now in yeast is to understand aging at the systems level, taking a holistic approach to integrate the contributions of different aging mechanisms and pathways in order to model the aging condition. This approach involves combining large-scale studies, including transcriptomics and epistasis network analysis, with directed studies with the goal of establishing as complete as possible a picture of single cell aging that can set the stage for similar studies in multicellular organisms.
In the multi-cellular context, a major challenge has been to understand the links between aging at the level of the organism and (causal?) changes to cells in the aging body. In that context, cell senescence has emerged as a major candidate driver of aging processes. Major insights in this arena have led to the identification of candidate therapeutics to kill senescent cells as means of offsetting or treating age-related chronic diseases. The next few years will help define the merits of this new therapeutic route based on aging studies.
More broadly, aging is linked to several pathways involved in cell growth and specifically in protein synthesis and turnover. It is clear for instance that reduced mTORC1 signaling leads to lifespan extension, but further work needs to be done to identify whether aging benefits come from altered protein translation, increased turnover of damaged macromolecules, suppression of the SASP, or some other mechanism. Moreover, it is important to identify in what tissues reduced mTOR signaling, and other pathways such as PKA, promotes longevity. With dramatic increases in the aging population and new insights from research on aging and longevity, the promise is there for major new advances that could refocus medical care toward interventions that slow aging and keep people healthy longer. Understanding links between cell growth, division and aging are integral to achieving this goal.
Contributor Information
Michael Polymenis, Department of Biochemistry and Biophysics, Texas A&M University, 2128 TAMU, College Station, TX 77845, USA.
Brian K. Kennedy, The Buck Institute for Research on Aging, 8001 Redwood Blvd, Novato, CA 94945, USA
References
- 1.Johnston GC, Pringle JR, Hartwell LH (1977) Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp Cell Res 105(1):79–98 [DOI] [PubMed] [Google Scholar]
- 2.Pringle JR, Hartwell LH (1981) The Saccharomyces cerevisiae cell cycle In: The molecular and cellular biology of the yeast saccharomyces, vol 1 Cold Spring Harbor Laboratory Press, New York, pp 97–142 [Google Scholar]
- 3.Turner JJ, Ewald JC, Skotheim JM (2012) Cell size control in yeast. Curr Biol 22(9):R350–R359. doi: 10.1016/j.cub.2012.02.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jorgensen P, Tyers M (2004) How cells coordinate growth and division. Curr Biol 14(23):R1014–R1027. doi: 10.1016/j.cub.2004.11.027 [DOI] [PubMed] [Google Scholar]
- 5.Ginzberg MB, Kafri R, Kirschner M (2015) Cell biology. On being the right (cell) size. Science 348(6236):1245075. doi: 10.1126/science.1245075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blagosklonny MV, Pardee AB (2002) The restriction point of the cell cycle. Cell Cycle 1(2):103–110 [PubMed] [Google Scholar]
- 7.Carter BL, Jagadish MN (1978) Control of cell division in the yeast Saccharomyces cerevisiae cultured at different growth rates. Exp Cell Res 112(2):373–383 [DOI] [PubMed] [Google Scholar]
- 8.Jagadish MN, Carter BL (1977) Genetic control of cell division in yeast cultured at different growth rates. Nature 269(5624):145–147 [DOI] [PubMed] [Google Scholar]
- 9.Jagadish M, Carter B (1978) Effects of temperature and nutritional conditions on the mitotic cell cycle of Saccharomyces cerevisiae. J Cell Sci 31(1):71–78 [DOI] [PubMed] [Google Scholar]
- 10.Johnston GC, Singer RA, McFarlane S (1977) Growth and cell division during nitrogen starvation of the yeast Saccharomyces cerevisiae. J Bacteriol 132(2):723–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lillie SH, Pringle JR (1980) Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J Bacteriol 143(3):1384–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartwell LH, Unger MW (1977) Unequal division in Saccharomyces cerevisiae and its implications for the control of cell division. J Cell Biol 75(2 Pt 1):422–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Messier V, Zenklusen D, Michnick SW (2013) A nutrient-responsive pathway that determines M phase timing through control of B-cyclin mRNA stability. Cell 153(5):1080–1093 [DOI] [PubMed] [Google Scholar]
- 14.Wood E, Nurse P (2015) Sizing up to divide: mitotic cell-size control in fission yeast. Annu Rev Cell Dev Biol 31:11–29. doi: 10.1146/annurev-cellbio-100814-125601 [DOI] [PubMed] [Google Scholar]
- 15.Blagosklonny MV (2012) Answering the ultimate question “what is the proximal cause of aging?”. Aging 4(12):861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blagosklonny MV (2014) Geroconversion: irreversible step to cellular senescence. Cell Cycle 13(23):3628–3635. doi: 10.4161/15384101.2014.985507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmoller KM, Skotheim JM (2015) The biosynthetic basis of cell size control. Trends Cell Biol 25(12):793–802. doi: 10.1016/j.tcb.2015.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson A, Skotheim JM (2013) Start and the restriction point. Curr Opin Cell Biol. doi: 10.1016/j.ceb.2013.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Son S, Tzur A, Weng Y, Jorgensen P, Kim J, Kirschner MW, Manalis SR (2012) Direct observation of mammalian cell growth and size regulation. Nat Methods 9(9):910–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Son S, Kang JH, Oh S, Kirschner MW, Mitchison TJ, Manalis S (2015) Resonant microchannel volume and mass measurements show that suspended cells swell during mitosis. J Cell Biol 211(4):757–763. doi: 10.1083/jcb.201505058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Godin M, Delgado FF, Son S, Grover WH, Bryan AK, Tzur A, Jorgensen P, Payer K, Grossman AD, Kirschner MW (2010) Using buoyant mass to measure the growth of single cells. Nat Methods 7(5):387–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milo R (2013) What is the total number of protein molecules per cell volume? A call to rethink some published values. Bioessays 35(12):1050–1055. doi: 10.1002/bies.201300066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lange HC, Heijnen JJ (2001) Statistical reconciliation of the elemental and molecular biomass composition of Saccharomyces cerevisiae. Biotechnol Bioeng 75(3):334–344 [DOI] [PubMed] [Google Scholar]
- 24.Liebermeister W, Noor E, Flamholz A, Davidi D, Bernhardt J, Milo R (2014) Visual account of protein investment in cellular functions. Proc Natl Acad Sci U S A 111(23):8488–8493. doi: 10.1073/pnas.1314810111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warner JR (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24(11):437–440 [DOI] [PubMed] [Google Scholar]
- 26.Fatica A, Tollervey D (2002) Making ribosomes. Curr Opin Cell Biol 14(3):313–318 [DOI] [PubMed] [Google Scholar]
- 27.Venema J, Tollervey D (1999) Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet 33(1):261–311 [DOI] [PubMed] [Google Scholar]
- 28.Wapinski I, Pfiffner J, French C, Socha A, Thompson DA, Regev A (2010) Gene duplication and the evolution of ribosomal protein gene regulation in yeast. Proc Natl Acad Sci 107(12):5505–5510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Planta RJ, Mager WH (1998) The list of cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Yeast 14(5):471–477. doi: [DOI] [PubMed] [Google Scholar]
- 30.Wade CH, Umbarger MA, McAlear MA (2006) The budding yeast rRNA and ribosome biosynthesis (RRB) regulon contains over 200 genes. Yeast 23(4):293–306. doi: 10.1002/yea.1353 [DOI] [PubMed] [Google Scholar]
- 31.Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M (2002) Systematic identification of pathways that couple cell growth and division in yeast. Science 297(5580):395–400. doi: 10.1126/science.1070850 [DOI] [PubMed] [Google Scholar]
- 32.Velculescu VE, Zhang L, Zhou W, Vogelstein J, Basrai MA, Bassett DE, Hieter P, Vogelstein B, Kinzler KW (1997) Characterization of the yeast transcriptome. Cell 88(2):243–251 [DOI] [PubMed] [Google Scholar]
- 33.Bakshi S, Siryaporn A, Goulian M, Weisshaar JC (2012) Superresolution imaging of ribosomes and RNA polymerase in live Escherichia coli cells. Mol Microbiol 85(1):21–38. doi: 10.1111/j.1365-2958.2012.08081.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitchison JM (1971) Cell growth and protein synthesis In: The biology of the cell cycle. Cambridge University Press, New York, p 129 [Google Scholar]
- 35.Brenner C, Nakayama N, Goebl M, Tanaka K, Toh-e A, Matsumoto K (1988) CDC33 encodes mRNA cap-binding protein eIF-4E of Saccharomyces cerevisiae. Mol Cell Biol 8(8):3556–3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanic-Joyce PJ, Johnston GC, Singer RA (1987) Regulated arrest of cell proliferation mediated by yeast prt1 mutations. Exp Cell Res 172(1):134–145 [DOI] [PubMed] [Google Scholar]
- 37.Bedard DP, Johnston GC, Singer RA (1981) New mutations in the yeast Saccharomyces cerevisiae affecting completion of “start”. Curr Genet 4(3):205–214. doi: 10.1007/BF00420500 [DOI] [PubMed] [Google Scholar]
- 38.Unger MW, Hartwell LH (1976) Control of cell division in Saccharomyces cerevisiae by methionyl-tRNA. Proc Natl Acad Sci U S A 73(5):1664–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu L, Pena Castillo L, Mnaimneh S, Hughes TR, Brown GW (2006) A survey of essential gene function in the yeast cell division cycle. Mol Biol Cell 17(11):4736–4747. doi: 10.1091/mbc.E06-04-0368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Polymenis M, Aramayo R (2015) Translate to divide: сontrol of the cell cycle by protein synthesis. Microbial Cell 2(4):94–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wicker LS, Boltz RC, Matt V, Nichols EA, Peterson LB, Sigal NH (1990) Suppression of B cell activation by cyclosporin a, FK506 and rapamycin. Eur J Immunol 20(10):2277–2283 [DOI] [PubMed] [Google Scholar]
- 42.Zoncu R, Efeyan A, Sabatini DM (2010) mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 12(1):21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heitman J, Movva NR, Hall MN (1991) Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253(5022):905–909 [DOI] [PubMed] [Google Scholar]
- 44.Loewith R, Hall MN (2011) Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189(4):1177–1201. doi: 10.1534/genetics.111.133363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lazaris-Karatzas A, Montine KS, Sonenberg N (1990) Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature 345(6275):544–547. doi: 10.1038/345544a0 [DOI] [PubMed] [Google Scholar]
- 46.Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, Wang S, Ren P, Martin M, Jessen K, Feldman ME, Weissman JS, Shokat KM, Rommel C, Ruggero D (2012) The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485(7396):55–61. doi: 10.1038/nature10912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Popolo L, Vanoni M, Alberghina L (1982) Control of the yeast cell cycle by protein synthesis. Exp Cell Res 142(1):69–78 [DOI] [PubMed] [Google Scholar]
- 48.Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M (2004) A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev 18(20):2491–2505. doi: 10.1101/gad.1228804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moore SA (1988) Kinetic evidence for a critical rate of protein synthesis in the Saccharomyces cerevisiae yeast cell cycle. J Biol Chem 263(20):9674–9681 [PubMed] [Google Scholar]
- 50.Rossow PW, Riddle VG, Pardee AB (1979) Synthesis of labile, serum-dependent protein in early G1 controls animal cell growth. Proc Natl Acad Sci U S A 76(9):4446–4450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brooks R (1977) Continuous protein synthesis is required to maintain the probability of entry into S phase. Cell 12(1):311–317 [DOI] [PubMed] [Google Scholar]
- 52.Lippman SI, Broach JR (2009) Protein kinase a and TORC1 activate genes for ribosomal biogenesis by inactivating repressors encoded by Dot6 and its homolog Tod6. Proc Natl Acad Sci 106(47):19928–19933. doi: 10.1073/pnas.0907027106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Broach JR (2012) Nutritional control of growth and development in yeast. Genetics 192(1):73–105. doi: 10.1534/genetics.111.135731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Longo VD, Shadel GS, Kaeberlein M, Kennedy B (2012) Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab 16(1):18–31. doi: 10.1016/j.cmet.2012.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bilinski T, Zadrag-Tecza R, Bartosz G (2012) Hypertrophy hypothesis as an alternative explanation of the phenomenon of replicative aging of yeast. FEMS Yeast Res 12(1):97–101. doi: 10.1111/j.1567-1364.2011.00759.x [DOI] [PubMed] [Google Scholar]
- 56.Zadrag R, Kwolek-Mirek M, Bartosz G, Bilinski T (2006) Relationship between the replicative age and cell volume in Saccharomyces cerevisiae. Acta Biochim Pol 53(4):747–751 [PubMed] [Google Scholar]
- 57.He C, Tsuchiyama SK, Nguyen QT, Plyusnina EN, Terrill SR, Sahibzada S, Patel B, Faulkner AR, Shaposhnikov MV, Tian R, Tsuchiya M, Kaeberlein M, Moskalev AA, Kennedy BK, Polymenis M (2014) Enhanced longevity by ibuprofen, conserved in multiple species, occurs in yeast through inhibition of tryptophan import. PLoS Genet 10(12):e1004860. doi: 10.1371/journal.pgen.1004860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaeberlein M, Powers RW 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK (2005) Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310(5751):1193–1196. doi: 10.1126/science.1115535 [DOI] [PubMed] [Google Scholar]
- 59.Powers RW 3rd, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S (2006) Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev 20(2):174–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F (2003) Genetics: influence of TOR kinase on lifespan in C. elegans. Nature 426:620. [DOI] [PubMed] [Google Scholar]
- 61.Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S (2004) Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol 14:885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandez E, Miller RA (2009) Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460(7253):392–395. doi: 10.1038/nature08221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen C, Liu Y, Zheng P (2009) mTOR regulation and therapeutic rejuvenation of aging hematopoietic stem cells. Sci Signal 2(98):ra75. doi: 10.1126/scisignal.2000559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mannick JB, Del Giudice G, Lattanzi M, Valiante NM, Praestgaard J, Huang B, Lonetto MA, Maecker HT, Kovarik J, Carson S, Glass DJ, Klickstein LB (2014) mTOR inhibition improves immune function in the elderly. Sci Transl Med 6(268):268ra179. doi: 10.1126/scitranslmed.3009892 [DOI] [PubMed] [Google Scholar]
- 65.Passtoors WM, Beekman M, Deelen J, van der Breggen R, Maier AB, Guigas B, Derhovanessian E, van Heemst D, de Craen AJ, Gunn DA, Pawelec G, Slagboom PE (2013) Gene expression analysis of mTOR pathway: association with human longevity. Aging Cell 12(1):24–31. doi: 10.1111/acel.12015 [DOI] [PubMed] [Google Scholar]
- 66.Kennedy BK, Lamming DW (2016) The mechanistic target of rapamycin: the grand conductor of metabolism and aging. Cell Metab 23(6):990–1003. doi: 10.1016/j.cmet.2016.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Steffen KK, Dillin A (2016) A ribosomal perspective on proteostasis and aging. Cell Metab 23(6):1004–1012. doi: 10.1016/j.cmet.2016.05.013 [DOI] [PubMed] [Google Scholar]
- 68.Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, Pak DN, Fields S, Kennedy BK, Kaeberlein M (2008) Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell 133(2):292–302. doi: 10.1016/j.cell.2008.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hinnebusch AG (2005) Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59:407–450 [DOI] [PubMed] [Google Scholar]
- 70.Foiani M, Cigan AM, Paddon CJ, Harashima S, Hinnebusch AG (1991) GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol Cell Biol 11(6):3203–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cherkasova VA, Hinnebusch AG (2003) Translational control by TOR and TAP42 through dephosphorylation of eIF2alpha kinase GCN2. Genes Dev 17(7):859–872. doi: 10.1101/gad.1069003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kubota H, Obata T, Ota K, Sasaki T, Ito T (2003) Rapamycin-induced translational derepression of GCN4 mRNA involves a novel mechanism for activation of the eIF2 alpha kinase GCN2. J Biol Chem 278(23):20457–20460 [DOI] [PubMed] [Google Scholar]
- 73.Valenzuela L, Aranda C, Gonzalez A (2001) TOR modulates GCN4-dependent expression of genes turned on by nitrogen limitation. J Bacteriol 183(7):2331–2334. doi: 10.1128/JB.183.7.2331-2334.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martin-Marcos P, Hinnebusch AG, Tamame M (2007) Ribosomal protein L33 is required for ribosome biogenesis, subunit joining and repression of GCN4 translation. Mol Cell Biol. 27(17):5968–5985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li W, Li X, Miller RA (2014) ATF4 activity: a common feature shared by many kinds of slow-aging mice. Aging Cell 13(6):1012–1018. doi: 10.1111/acel.12264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li W, Miller RA (2014) Elevated ATF4 function in fibroblasts and liver of slow-aging mutant mice. J Gerontol A Biol Sci Med Sci. doi: 10.1093/gerona/glu040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lapierre LR, Kumsta C, Sandri M, Ballabio A, Hansen M (2015) Transcriptional and epigenetic regulation of autophagy in aging. Autophagy 11(6):867–880. doi: 10.1080/15548627.2015.1034410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alvers AL, Wood MS, Hu D, Kaywell AC, Dunn WA Jr, Aris JP (2009) Autophagy is required for extension of yeast chronological life span by rapamycin. Autophagy 5(6):847–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Toth ML, Sigmond T, Borsos E, Barna J, Erdelyi P, Takacs-Vellai K, Orosz L, Kovacs AL, Csikos G, Sass M, Vellai T (2008) Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy 4(3):330–338 [DOI] [PubMed] [Google Scholar]
- 80.Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C (2008) A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet 4(2):e24. doi: 10.1371/journal.pgen.0040024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, Partridge L (2010) Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab 11(1):35–46. doi: 10.1016/j.cmet.2009.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pyo JO, Yoo SM, Ahn HH, Nah J, Hong SH, Kam TI, Jung S, Jung YK (2013) Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat Commun 4:2300. doi: 10.1038/ncomms3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD (2008) Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy 4(2):176–184 [DOI] [PubMed] [Google Scholar]
- 84.Bai H, Kang P, Hernandez AM, Tatar M (2013) Activin signaling targeted by insulin/dFOXO regulates aging and muscle proteostasis in Drosophila. PLoS Genet 9(11):e1003941. doi: 10.1371/journal.pgen.1003941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ulgherait M, Rana A, Rera M, Graniel J, Walker DW (2014) AMPK modulates tissue and organismal aging in a non-cell-autonomous manner. Cell Rep 8(6):1767–1780. doi: 10.1016/j.celrep.2014.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD (2001) Regulation of longevity and stress resistance by Sch9 in yeast. Science 292:288–290 [DOI] [PubMed] [Google Scholar]
- 87.Longo VD (1999) Mutations in signal transduction proteins increase stress resistance and longevity in yeast, nematodes, fruit flies, and mammalian neuronal cells. Neurobiol Aging 20(5):479–486 [DOI] [PubMed] [Google Scholar]
- 88.Fabrizio P, Liou LL, Moy VN, Diaspro A, SelverstoneValentine J, Gralla EB, Longo VD (2003) SOD2 functions downstream of Sch9 to extend longevity in yeast. Genetics 163(1):35–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Enns LC, Ladiges W (2010) Protein kinase A signaling as an anti-aging target. Ageing Res Rev 9(3):269–272. doi: 10.1016/j.arr.2010.02.004 [DOI] [PubMed] [Google Scholar]
- 90.Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L (2002) Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature 418(6895):344–348 [DOI] [PubMed] [Google Scholar]
- 91.Gendron CM, Minois N, Longo VD, Pletcher SD, Vaupel JW (2003) Biodemographic trajectories of age-specific reproliferation from stationary phase in the yeast Saccharomyces cerevisiae seem multiphasic. Mech Ageing Dev 124:1059–1063 [DOI] [PubMed] [Google Scholar]
- 92.Enns LC, Morton JF, Mangalindan RS, McKnight GS, Schwartz MW, Kaeberlein MR, Kennedy BK, Rabinovitch PS, Ladiges WC (2009) Attenuation of age-related metabolic dysfunction in mice with a targeted disruption of the Cbeta subunit of protein kinase a. J Gerontol A Biol Sci Med Sci 64(12):1221–1231. doi: 10.1093/gerona/glp133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Enns LC, Morton JF, Treuting PR, Emond MJ, Wolf NS, McKnight GS, Rabinovitch PS, Ladiges WC (2009) Disruption of protein kinase a in mice enhances healthy aging. PLoS One 4(6):e5963. doi: 10.1371/journal.pone.0005963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yan L, Vatner DE, O’Connor JP, Ivessa A, Ge H, Chen W, Hirotani S, Ishikawa Y, Sadoshima J, Vatner SF (2007) Type 5 adenylyl cyclase disruption increases longevity and protects against stress. Cell 130(2):247–258. doi: 10.1016/j.cell.2007.05.038 [DOI] [PubMed] [Google Scholar]
- 95.Vatner SF, Park M, Yan L, Lee GJ, Lai L, Iwatsubo K, Ishikawa Y, Pessin J, Vatner DE (2013) Adenylyl cyclase type 5 in cardiac disease, metabolism, and aging. Am J Phys Heart Circ Phys 305(1):H1–H8. doi: 10.1152/ajpheart.00080.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Higuchi-Sanabria R, Pernice WM, Vevea JD, Alessi Wolken DM, Boldogh IR, Pon LA (2014) Role of asymmetric cell division in lifespan control in Saccharomyces cerevisiae. FEMS Yeast Res 14(8):1133–1146. doi: 10.1111/1567-1364.12216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kwan EX, Foss EJ, Tsuchiyama S, Alvino GM, Kruglyak L, Kaeberlein M, Raghuraman MK, Brewer BJ, Kennedy BK, Bedalov A (2013) A natural polymorphism in rDNA replication origins links origin activation with calorie restriction and lifespan. PLoS Genet 9(3):e1003329. doi: 10.1371/journal.pgen.1003329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sinclair DA, Guarente L (1997) Extrachromosomal rDNA circles--a cause of aging in yeast. Cell 91(7):1033–1042 [DOI] [PubMed] [Google Scholar]
- 99.Shcheprova Z, Baldi S, Frei SB, Gonnet G, Barral Y (2008) A mechanism for asymmetric segregation of age during yeast budding. Nature 454(7205):728–734. doi: 10.1038/nature07212 [DOI] [PubMed] [Google Scholar]
- 100.Khmelinskii A, Keller PJ, Lorenz H, Schiebel E, Knop M (2010) Segregation of yeast nuclear pores. Nature 466(7305):E1. doi: 10.1038/nature09255 [DOI] [PubMed] [Google Scholar]
- 101.Kennedy BK, McCormick MA (2011) Asymmetric segregation: the shape of things to come? Curr Biol 21(4):R149–R151. doi: 10.1016/j.cub.2011.01.018 [DOI] [PubMed] [Google Scholar]
- 102.Khmelinskii A, Meurer M, Knop M, Schiebel E (2011) Artificial tethering to nuclear pores promotes partitioning of extrachromosomal DNA during yeast asymmetric cell division. Curr Biol 21(1):R17–R18. doi: 10.1016/j.cub.2010.11.034 [DOI] [PubMed] [Google Scholar]
- 103.Gehlen LR, Nagai S, Shimada K, Meister P, Taddei A, Gasser SM (2011) Nuclear geometry and rapid mitosis ensure asymmetric episome segregation in yeast. Curr Biol 21(1):25–33. doi: 10.1016/j.cub.2010.12.016 [DOI] [PubMed] [Google Scholar]
- 104.Erjavec N, Nystrom T (2007) Sir2p-dependent protein segregation gives rise to a superior reactive oxygen species management in the progeny of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 104(26):10877–10881. doi: 10.1073/pnas.0701634104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McFaline-Figueroa JR, Vevea J, Swayne TC, Zhou C, Liu C, Leung G, Boldogh IR, Pon LA (2011) Mitochondrial quality control during inheritance is associated with lifespan and mother-daughter age asymmetry in budding yeast. Aging Cell 10(5):885–895. doi: 10.1111/j.1474-9726.2011.00731.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang J, McCormick MA, Zheng J, Xie Z, Tsuchiya M, Tsuchiyama S, El-Samad H, Ouyang Q, Kaeberlein M, Kennedy BK, Li H (2015) Systematic analysis of asymmetric partitioning of yeast proteome between mother and daughter cells reveals “aging factors” and mechanism of lifespan asymmetry. Proc Natl Acad Sci U S A 112(38):11977–11982. doi: 10.1073/pnas.1506054112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen C, Fingerhut JM, Yamashita YM (2016) The ins(ide) and outs(ide) of asymmetric stem cell division. Curr Opin Cell Biol 43:1–6. doi: 10.1016/j.ceb.2016.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Raff M (2006) The mystery of intracellular developmental programmes and timers. Biochem Soc Trans 34(Pt 5):663–670. doi: 10.1042/BST0340663 [DOI] [PubMed] [Google Scholar]
- 109.Malumbres M (2014) Cyclin-dependent kinases. Genome Biol 15(6):122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pavletich NP (1999) Mechanisms of cyclin-dependent kinase regulation: structures of Cdks, their cyclin activators, and Cip and INK4 inhibitors. J Mol Biol 287(5):821–828. doi: 10.1006/jmbi.1999.2640 [DOI] [PubMed] [Google Scholar]
- 111.Fisher RP (2010) Coming full circle: cyclin-dependent kinases as anti-cancer drug targets. Subcell Biochem 50:1–15. doi: 10.1007/978-90-481-3471-7_1 [DOI] [PubMed] [Google Scholar]
- 112.Asghar U, Witkiewicz AK, Turner NC, Knudsen ES (2015) The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat Rev Drug Discov 14(2):130–146. doi: 10.1038/nrd4504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chandler H, Peters G (2013) Stressing the cell cycle in senescence and aging. Curr Opin Cell Biol 25(6):765–771. doi: 10.1016/j.ceb.2013.07.005 [DOI] [PubMed] [Google Scholar]
- 114.Menjo M, Ikeda K, Nakanishi M (1998) Regulation of G1 cyclin-dependent kinases in liver regeneration. J Gastroenterol Hepatol 13(Suppl):S100–S105 [DOI] [PubMed] [Google Scholar]
- 115.Tamamori-Adachi M, Takagi H, Hashimoto K, Goto K, Hidaka T, Koshimizu U, Yamada K, Goto I, Maejima Y, Isobe M, Nakayama KI, Inomata N, Kitajima S (2008) Cardiomyocyte proliferation and protection against post-myocardial infarction heart failure by cyclin D1 and Skp2 ubiquitin ligase. Cardiovasc Res 80(2):181–190. doi: 10.1093/cvr/cvn183 [DOI] [PubMed] [Google Scholar]
- 116.Han IS, Seo TB, Kim KH, Yoon JH, Yoon SJ, Namgung U (2007) Cdc2-mediated Schwann cell migration during peripheral nerve regeneration. J Cell Sci 120(Pt 2):246–255. doi: 10.1242/jcs.03322 [DOI] [PubMed] [Google Scholar]
- 117.Bedelbaeva K, Snyder A, Gourevitch D, Clark L, Zhang XM, Leferovich J, Cheverud JM, Lieberman P, Heber-Katz E (2010) Lack of p21 expression links cell cycle control and appendage regeneration in mice. Proc Natl Acad Sci U S A 107(13):5845–5850. doi: 10.1073/pnas.1000830107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Connell-Crowley L, Elledge SJ, Harper JW (1998) G1 cyclin-dependent kinases are sufficient to initiate DNA synthesis in quiescent human fibroblasts. Curr Biol 8:65–68 [DOI] [PubMed] [Google Scholar]
- 119.Wei M, Fabrizio P, Hu J, Ge H, Cheng C, Li L, Longo VD (2008) Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, tor, and Sch9. PLoS Genet 4(1):e13. doi: 10.1371/journal.pgen.0040013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.McKnight JN, Boerma JW, Breeden LL, Tsukiyama T (2015) Global promoter targeting of a conserved lysine deacetylase for transcriptional shutoff during quiescence entry. Mol Cell 59(5):732–743. doi: 10.1016/j.molcel.2015.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.McCormick MA, Delaney JR, Tsuchiya M, Tsuchiyama S, Shemorry A, Sim S, Chou AC, Ahmed U, Carr D, Murakami CJ, Schleit J, Sutphin GL, Wasko BM, Bennett CF, Wang AM, Olsen B, Beyer RP, Bammler TK, Prunkard D, Johnson SC, Pennypacker JK, An E, Anies A, Castanza AS, Choi E, Dang N, Enerio S, Fletcher M, Fox L, Goswami S, Higgins SA, Holmberg MA, Hu D, Hui J, Jelic M, Jeong KS, Johnston E, Kerr EO, Kim J, Kim D, Kirkland K, Klum S, Kotireddy S, Liao E, Lim M, Lin MS, Lo WC, Lockshon D, Miller HA, Moller RM, Muller B, Oakes J, Pak DN, Peng ZJ, Pham KM, Pollard TG, Pradeep P, Pruett D, Rai D, Robison B, Rodriguez AA, Ros B, Sage M, Singh MK, Smith ED, Snead K, Solanky A, Spector BL, Steffen KK, Tchao BN, Ting MK, Vander Wende H, Wang D, Welton KL, Westman EA, Brem RB, Liu XG, Suh Y, Zhou Z, Kaeberlein M, Kennedy BK (2015) A comprehensive analysis of replicative lifespan in 4,698 single-gene deletion strains uncovers conserved mechanisms of aging. Cell Metab. doi: 10.1016/j.cmet.2015.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Burtner CR, Murakami CJ, Olsen B, Kennedy BK, Kaeberlein M (2011) A genomic analysis of chronological longevity factors in budding yeast. Cell Cycle 10(9):1385–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ashrafi K, Sinclair D, Gordon JI, Guarente L (1999) Passage through stationary phase advances replicative aging in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 96(16):9100–9105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Piper PW, Harris NL, MacLean M (2006) Preadaptation to efficient respiratory maintenance is essential both for maximal longevity and the retention of replicative potential in chronologically ageing yeast. Mech Ageing Dev 127(9):733–740. doi: 10.1016/j.mad.2006.05.004 [DOI] [PubMed] [Google Scholar]
- 125.Polymenis M, Kennedy BK (2012) Chronological and replicative lifespan in yeast: do they meet in the middle? Cell Cycle 11(19):3531–3532. doi: 10.4161/cc.22041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Delaney JR, Murakami C, Chou A, Carr D, Schleit J, Sutphin GL, An EH, Castanza AS, Fletcher M, Goswami S, Higgins S, Holmberg M, Hui J, Jelic M, Jeong KS, Kim JR, Klum S, Liao E, Lin MS, Lo W, Miller H, Moller R, Peng ZJ, Pollard T, Pradeep P, Pruett D, Rai D, Ros V, Schuster A, Singh M, Spector BL, Wende HV, Wang AM, Wasko BM, Olsen B, Kaeberlein M (2013) Dietary restriction and mitochondrial function link replicative and chronological aging in Saccharomyces cerevisiae. Exp Gerontol 48(10):1006–1013. doi: 10.1016/j.exger.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Murakami C, Delaney JR, Chou A, Carr D, Schleit J, Sutphin GL, An EH, Castanza AS, Fletcher M, Goswami S, Higgins S, Holmberg M, Hui J, Jelic M, Jeong KS, Kim JR, Klum S, Liao E, Lin MS, Lo W, Miller H, Moller R, Peng ZJ, Pollard T, Pradeep P, Pruett D, Rai D, Ros V, Schuster A, Singh M, Spector BL, Vander Wende H, Wang AM, Wasko BM, Olsen B, Kaeberlein M (2012) pH neutralization protects against reduction in replicative lifespan following chronological aging in yeast. Cell Cycle 11(16):3087–3096. doi: 10.4161/cc.21465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sharpless NE, Sherr CJ (2015) Forging a signature of in vivo senescence. Nat Rev Cancer 15(7):397–408. doi: 10.1038/nrc3960 [DOI] [PubMed] [Google Scholar]
- 129.Pardee AB (1989) G1 events and regulation of cell proliferation. Science 246(4930): 603–608 [DOI] [PubMed] [Google Scholar]
- 130.Sherr CJ, Roberts JM (1995) Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev 9(10):1149–1163 [DOI] [PubMed] [Google Scholar]
- 131.Campisi J (2013) Aging, cellular senescence, and cancer. Annu Rev Physiol 75:685–705. doi: 10.1146/annurev-physiol-030212-183653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mao Z, Ke Z, Gorbunova V, Seluanov A (2012) Replicatively senescent cells are arrested in G1 and G2 phases. Aging 4(6):431–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Baus F, Gire V, Fisher D, Piette J, Dulic V (2003) Permanent cell cycle exit in G2 phase after DNA damage in normal human fibroblasts. EMBO J 22(15):3992–4002. doi: 10.1093/emboj/cdg387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gire V, Dulic V (2015) Senescence from G2 arrest, revisited. Cell Cycle 14(3):297–304. doi: 10.1080/15384101.2014.1000134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sherwood SW, Rush D, Ellsworth JL, Schimke RT (1988) Defining cellular senescence in IMR-90 cells: a flow cytometric analysis. Proc Natl Acad Sci U S A 85(23):9086–9090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rodier F, Campisi J (2011) Four faces of cellular senescence. J Cell Biol 192(4):547–556. doi: 10.1083/jcb.201009094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Blagosklonny MV (2012) Cell cycle arrest is not yet senescence, which is not just cell cycle arrest: terminology for TOR-driven aging. Aging 4(3):159–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hayflick L (1965) The limited in vitro lifetime of human diploid cell strains. Exp Cell Res 37:614–636 [DOI] [PubMed] [Google Scholar]
- 139.Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, Nelson PS, Desprez PY, Campisi J (2008) Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol 6(12):2853–2868. doi: 10.1371/journal.pbio.0060301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, Aarden LA, Mooi WJ, Peeper DS (2008) Oncogene-induced senescence relayed by an interleukin- dependent inflammatory network. Cell 133(6):1019–1031. doi: 10.1016/j.cell.2008.03.039 [DOI] [PubMed] [Google Scholar]
- 141.Laberge RM, Sun Y, Orjalo AV, Patil CK, Freund A, Zhou L, Curran SC, Davalos AR, Wilson-Edell KA, Liu S, Limbad C, Demaria M, Li P, Hubbard GB, Ikeno Y, Javors M, Desprez PY, Benz CC, Kapahi P, Nelson PS, Campisi J (2015) MTOR regulates the pro-tumorigenic senescence-associated secretory phenotype by promoting IL1A translation. Nat Cell Biol 17(8):1049–1061. doi: 10.1038/ncb3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bitto A, Ito TK, Pineda VV, LeTexier NJ, Huang HZ, Sutlief E, Tung H, Vizzini N, Chen B, Smith K, Meza D, Yajima M, Beyer RP, Kerr KF, Davis DJ, Gillespie CH, Snyder JM, Treuting PM, Kaeberlein M (2016) Transient rapamycin treatment can increase lifespan and healthspan in middle-aged mice Elife 5. doi: 10.7554/eLife.16351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chen C, Liu Y, Liu Y, Zheng P (2009) mTOR regulation and therapeutic rejuvenation of aging hematopoietic stem cells. Science Signal 2(98):ra75. doi: 10.1126/scisignal.2000559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, Kirkland JL, van Deursen JM (2011) Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 479(7372):232–236. doi: 10.1038/nature10600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Baker DJ, Childs BG, Durik M, Wijers ME, Sieben CJ, Zhong J, Saltness RA, Jeganathan KB, Verzosa GC, Pezeshki A, Khazaie K, Miller JD, van Deursen JM (2016) Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature 530(7589):184–189. doi: 10.1038/nature16932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kapanidou M, Lee S, Bolanos-Garcia VM (2015) BubR1 kinase: protection against aneuploidy and premature aging. Trends Mol Med 21(6):364–372. doi: 10.1016/j.molmed.2015.04.003 [DOI] [PubMed] [Google Scholar]
- 147.Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, Kumar R, Jenkins RB, de Groen PC, Roche P, van Deursen JM (2004) BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet 36(7):744–749. doi: 10.1038/ng1382 [DOI] [PubMed] [Google Scholar]
- 148.Matsumoto T, Baker DJ, d’Uscio LV, Mozammel G, Katusic ZS, van Deursen JM (2007) Aging-associated vascular phenotype in mutant mice with low levels of BubR1. Stroke 38(3):1050–1056. doi: 10.1161/01.str.0000257967.86132.01 [DOI] [PubMed] [Google Scholar]
- 149.Baker DJ, Dawlaty MM, Wijshake T, Jeganathan KB, Malureanu L, van Ree JH, Crespo-Diaz R, Reyes S, Seaburg L, Shapiro V, Behfar A, Terzic A, van de Sluis B, van Deursen JM (2013) Increased expression of BubR1 protects against aneuploidy and cancer and extends healthy lifespan. Nat Cell Biol 15(1):96–102. doi: 10.1038/ncb2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Roos CM, Zhang B, Palmer AK, Ogrodnik MB, Pirtskhalava T, Thalji NM, Hagler M, Jurk D, Smith LA, Casaclang-Verzosa G, Zhu Y, Schafer MJ, Tchkonia T, Kirkland JL, Miller JD (2016) Chronic senolytic treatment alleviates established vasomotor dysfunction in aged or atherosclerotic mice. Aging Cell doi: 10.1111/acel.12458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhu Y, Tchkonia T, Fuhrmann-Stroissnigg H, Dai HM, Ling YY, Stout MB, Pirtskhalava T, Giorgadze N, Johnson KO, Giles CB, Wren JD, Niedernhofer LJ, Robbins PD, Kirkland JL (2016) Identification of a novel senolytic agent, navitoclax, targeting the Bcl-2 family of anti-apoptotic factors. Aging Cell 15(3):428–435. doi: 10.1111/acel.12445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhu Y, Tchkonia T, Pirtskhalava T, Gower AC, Ding H, Giorgadze N, Palmer AK, Ikeno Y, Hubbard GB, Lenburg M, O’Hara SP, LaRusso NF, Miller JD, Roos CM, Verzosa GC, LeBrasseur NK, Wren JD, Farr JN, Khosla S, Stout MB, McGowan SJ, Fuhrmann-Stroissnigg H, Gurkar AU, Zhao J, Colangelo D, Dorronsoro A, Ling YY, Barghouthy AS, Navarro DC, Sano T, Robbins PD, Niedernhofer LJ, Kirkland JL (2015) The Achilles’ heel of senescent cells: from transcriptome to senolytic drugs. Aging Cell 14(4):644–658. doi: 10.1111/acel.12344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chang J, Wang Y, Shao L, Laberge RM, Demaria M, Campisi J, Janakiraman K, Sharpless NE, Ding S, Feng W, Luo Y, Wang X, Aykin-Burns N, Krager K, Ponnappan U, Hauer-Jensen M, Meng A, Zhou D (2016) Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat Med 22(1):78–83. doi: 10.1038/nm.4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Farrell JA, O’Farrell PH (2014) From egg to gastrula: how the cell cycle is remodeled during the Drosophila mid-blastula transition. Annu Rev Genet 48:269–294. doi: 10.1146/annurev-genet-111212-133531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.de Nooij JC, Letendre MA, Hariharan IK (1996) A cyclin-dependent kinase inhibitor, Dacapo, is necessary for timely exit from the cell cycle during Drosophila embryogenesis. Cell 87(7):1237–1247 [DOI] [PubMed] [Google Scholar]
- 156.Lane ME, Sauer K, Wallace K, Jan YN, Lehner CF, Vaessin H (1996) Dacapo, a cyclin-dependent kinase inhibitor, stops cell proliferation during Drosophila development. Cell 87(7):1225–1235 [DOI] [PubMed] [Google Scholar]