Abstract
Dengue virus Type 2 (DENV-2) is predominant serotype causing major dengue epidemics. There are a number of studies carried out to find its effective antiviral, however to date, there is still no molecule either from peptide or small molecules released as a drug. The present study aims to identify small molecules inhibitor from National Cancer Institute database through virtual screening. One of the hits, D0713 (IC50 = 62 μM) bearing thioguanine scaffold was derivatised into 21 compounds and evaluated for DENV-2 NS2B/NS3 protease inhibitory activity. Compounds 18 and 21 demonstrated the most potent activity with IC50 of 0.38 μM and 16 μM, respectively. Molecular dynamics and MM/PBSA free energy of binding calculation were conducted to study the interaction mechanism of these compounds with the protease. The free energy of binding of 18 calculated by MM/PBSA is -16.10 kcal/mol compared to the known inhibitor, panduratin A (-11.27 kcal/mol), which corroborates well with the experimental observation. Results from molecular dynamics simulations also showed that both 18 and 21 bind in the active site and stabilised by the formation of hydrogen bonds with Asn174.
Introduction
Dengue, caused by Dengue Virus (DENV), is the most important mosquito-borne viral disease affecting the tropics and subtropics [1]. Endemic in more than 100 countries [2,3], the virus is estimated to cause 390 million infections each year [4]. DENV infections can result in serious diseases including dengue fever, dengue hemorrhagic fever (DHF), dengue shock syndrome (DSS) and even death [5]. There are no approved antiviral drugs for these diseases and currently, patients are treated with supportive care to relieve fever, pain, and dehydration [6]. A tetravalent dengue vaccine (CYD-TDENV or Dengvaxia), the first dengue vaccine has recently been registered in several countries [7]. Despite being a leading cause of hospitalisation and death among children in some Asian and Latin American countries [8], this vaccine is not recommended for use in children under 9 years of age due to safety concerns [7]. Therefore, there exists an urgent need for antiviral therapies to treat dengue.
DENV carries a positive single strand RNA in its genome and five serotypes (DENV-1 to 5) have been identified. DENV-2 is the most prevalent type in dengue epidemic, especially in the South East Asian region and has been associated with severe dengue cases [9]. The new serotype (DENV-5) [10] was discovered in 2013 in Sarawak, Malaysia further complicates the prevention and treatment of the disease. The virus genome is encoded by three structural proteins (C, prM, E) as well as seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5) [11]. Of these proteins, NS2B/NS3protease (NS2B/NS3pro) has been well studied and regarded as a promising target in anti-dengue discovery [12–15].
NS3 is a trypsin like serine protease which plays a role in post-translation in the virus maturation. This domain has a catalytic triad made up of His51, Asp75 and Ser135 and its activity is enhanced by NS2B as the cofactor [16,17]. This cofactor contributes to the NS3 activity through its hydrophilic region which is responsible for holding and promoting the activation of NS3 while the hydrophobic region takes part in membrane association upon the cleavage process [18,19].
Previous discoveries of dengue inhibitors by targeting NS2B/NS3pro activity were mainly based on the non-prime substrates which were identified by profiling dengue virus using tetrapeptides [20–22]. Significant challenges arise as the protein possesses a solvent-exposed, topologically shallow active site and dependent on the selectivity for substrates containing basic amino acids (arginine and lysine) at P1 and P2 positions [23]. Nevertheless, many peptidic or modified peptidic molecules have been discovered to have good NS2B/NS3pro inhibition activities [15, 24–30]. In addition, there are also reports of potent small molecule NS2B/NS3pro inhibitors from natural products (panduratin[31], agathisflavone and quercetin [32]), from synthetic medicinal chemistry (dehydronaphthalene [33], benzimidazole [34], and thiadiazoloacrylamide [35]) or from the utilisation of computational methods [36–39].
Here, we report the discovery of potential NS2B/NS3pro inhibitors designed based on thioguanine scaffold identified through the virtual screening of compounds library from National Cancer Institute (NCI) diversity set II. Twenty-four compounds were found as in silico hits based on the free energy of binding (ΔGbind) ranking from a total of 1990 compounds. Twenty of them were obtained from NCI for in vitro assay, out of which four demonstrated moderate inhibition towards DENV-2 NS2B/NS3pro (IC50 = 29–77 μM). Although Diversity0713 (D0713) is not the most potent compound identified in the virtual screening, the structure contains thioguanine (TG or 6-thioguanine) scaffold can be used as a template to develop a series of analogues as its simple chemical structure benefits feasible synthetic steps. TG showed 56% inhibition at 200 μg/mL (1.2 mM) indicating that even without any modification to the structure, the scaffold itself is able to inhibit the protease activity. This compound is a drug classed as anti-neoplastic agent and used with other compounds in treating leukemia [40] and has been investigated in many pharmacological activities such as immunomodulators in autoimmune diseases [41] and transplant graft rejection [42]. From this initial study, we are interested to utilise thioguanine scaffold in the design of DENV-2 NS2B/NS3pro inhibitor(s). Thus, in this study, we designed, synthesised thioguanine analogues and investigated their possible DENV-2 NS2B/NS3pro inhibition activity. We hope that this study could contribute to the efforts in discovering novel and potent anti-dengue agents.
Materials and methods
Virtual screening
Virtual screening was carried out using AutoDockVina [43] (www.autodock.scripps.edu). The DENV-2 NS2B/NS3pro model was taken from published article [44], where the model was built based on the DENV-2 complex cofactor-protease using the crystal structure of NS2B/NS3pro West Nile Virus (WNV) as the template. The docking procedure was initiated by the preparation of NS2B/NS3pro as a macromolecule using AutoDock Tools (version 1.5.6) with default parameters for docking with AutoDock Vina. The exhaustiveness was set to 8 and other parameters were unchanged. The centre of the grid box was set at 30.71, 50.48 and 4.10 Å in x, y, z coordinates, respectively, with a box size of 25 x 25 x 25 points. The internal validation was done by re-docking the tetrapeptide inhibitor (Bz-Nle-Lys-Arg-Arg-H) with the RMSD value not greater than 2Å. The external validation was done using panduratin A (a competitive inhibitor), that dock at the same binding site of tetrapeptide inhibitor. The in silico screening of 1990 ligands from NCI diversity set (II) was carried out using the docking parameters above. The hit compounds were ranked according to the free energy of binding (ΔGbind) and analysis of their binding modes were performed using Discovery Studio 3.5 (www.accelrys.com).
DENV-2 NS2B/NS3pro expression and purification, optimum activity and inhibition assay
The DENV-2 NS2B/NS3pro expression was carried out according to the established method by Yusof et al., (2000) [45] with minor modifications according to published articles [46–50]. The plasmid encoding the NS2B/NS3 protease sequence from DENV-2 was transformed into Escherichia coli strain XL1-Blue transformed with pQE30.CF40.gly(T).NS3pro expression plasmid were grown in LB medium containing 10 μg/ml ampicillin at 37°C until the OD600 reached 0.6. First, the cells were incubated at 37°C, 200 rpm until OD600 reached ~0.6. One ml of isopropyl-β-D-thiogalactopyranoside (0.5 mM in LB medium) was added to the bacterial cells for 2 hours to induce protein expression. Expression of the recombinant protein was induced by the addition of 0.5 M IPTG and the culture was incubated for 2 hours.
The cells were harvested by centrifugation at 8000 rpm (Sorvall RC-5B Refrigerated Superspeed centrifuge) for 15 minutes at 80°C. The cell pellets were thawed (1 g) and resuspended in lysis buffer (5 mL) followed by mixing them using vortex until milky. For purification, cells were lysed by sonication conducting (6 times 15-second pulse, duty cycle 10%, output control no 3) using Ultrasonic Cell Disruptor, Branson Sonifier 450, Germany. The lysate was incubated on ice for 1 hour and then centrifuged at 8000 rpm for 1 hour at 4°C. The soluble 6x-His-NS2B/NS3protease in its native form was filtered (45 μm), batch-bound to 2 x 2 ml Ni2+-NTA (nickel-nitrilotriacetic acid) resin (pre-equilibrated with column buffer) and incubated overnight at 4°C. The resin was cleaned up from the unbound fraction by centrifugation and the resin with bound protein was collected and loaded into columns (Bio-Rad; 1 x 3 cm). The gradient technique columns were washed extensively with 3 x 15 ml of wash buffer and further eluted with 10 ml of elution buffer for each column while being monitored using Bio-Rad Bradford protein assay. The purified protein was then analysed with 12% SDS-PAGE, pooled and stored at -80°C for further use in the dengue protease activity and inhibition studies. The dengue protease activity assay was developed as previously described [51] with a slight modification [46,47,50]. Briefly, the assay system comprised of 200 mM Tris-HCl (pH 8.5) buffer, DENV-2 NS2B/NS3pro and Boc-GRR-MCA as the substrate. Protease optimum assay was executed to ascertain maximum protease activity at constant concentration of the substrate (25 μM). The protease concentrations were varied within 0–10 μM. The 7-Amino-4-methylcoumarin (AMC) [51] produced was measured as fluorescence intensity at λexcitation of 340 nm and λemission of 440 nm by using ELISA modulus microplate reader.
The compounds were also checked for its requirement to pass the pan assay interference compounds (PAINS) (http://cbligand.org/PAINS/) before tested for their inhibition activities against DENV-2 NS2B/NS3pro at a range of concentrations (0 to 300 μg/mL). The result shows that all the tested compounds, except for D1855 passed the PAINS filter. The concentration of the protease being used was 3 μM initially incubated with the compound for 10 minutes at 37°C with 200 rpm of rotation. Subsequently, 25 μM of the substrate was added and the incubation was further prolonged for 60 minutes. The fluorescence intensity at 340/ 440 nm was measured as the AMC byproduct was released upon the peptide substrate cleavage by the protease. The experiments were triplicated and Panduratin A was used as the positive control.
Synthesis of thioguanine derivatives
All reactions were carried out using standard techniques for the exclusion of moisture [52,53] except those in aqueous media. The progress of reaction was monitored using TLC on 0.25 mm silica F254 and detected under UV light, or iodine vapor. 1H-NMR and 13C-NMR spectra were determined using Bruker Avance 500 spectrometer with TMS as an internal standard and the mass spectra were determined using XEVO-G2TOF #YCA153. Melting points were obtained using a STUART SMP electro-thermal apparatus and were uncorrected. Anhydrous reactions were carried out in over-dried glassware under a nitrogen atmosphere. The detail method of synthesis, the compounds’ structures as well as the numbering due to NMR assignation can be seen in S1 Text.
Molecular docking simulation
To obtain the bound pose of the thioguanine analogues for MD simulation, molecular docking was carried out. 3D structures of the two most active compounds (18 and 21) were constructed and minimised using Hyperchem 8.0 [54] with 1000 steps of steepest descent followed by 1000 steps of conjugate gradient. The minimised structures were than subjected to molecular docking using AutoDock4.2 [55]. The same DENV-2 NS2B/NS3pro model [43] used in the virtual screening was used here. The protein and its inhibitors (compounds 18 and 21) were subsequently prepared using AutoDockTools 1.5.6. Polar hydrogen and Kollman charges were added into the DENV NS2B/NS3pro. In preparing both the ligands, the non-polar hydrogen atoms were merged and Gasteiger charges were assigned. The grid box size and the grid spacing were set around the catalytic triad to 60×60×60 dimension and 0.375 Å respectively, with the centre set at x = 21.517, y = 43.428 and z = -1.743. AutoDock4.2 was used to run docking with the Lamarckian Genetics Algorithm (GA) search program applied to generate 100 runs. The conformations with the ones of lowest free energy of binding and of the most populated cluster were selected. The docked conformation for each compound (18 and 21) was used as the starting structure for the subsequent dynamics studies. The interaction analysis was conducted using ligplus+ [56] and VMD 1.9. Panduratin A was used as the positive control and prepared for the simulation, in a similar way as with compounds 18 and 21.
Molecular dynamics simulation
To gain insight into the binding interaction of compounds 18, 21 and panduratin A with NS2B/NS3pro, molecular dynamics simulations were carried out using Amber 14 [57]. The following is the description of the setups of NS2B/NS3pro-18 (compound 18), NS2B/NS3pro-21 (compound 21) and NS2B/NS3pro-panduratin. All systems were prepared using LEaP program. Amber generalised force filed (GAFF) were assigned to the three compounds whilst amber ff14SB to NS2B/NS3pro. Each system was neutralised with sodium ions and solvated using TIP3P water in a 53.57, 46.51, 44.05 Å truncated octahedral water box. To eliminate the steric clashes, each system was subjected to a total of 500 stepwise minimisation using steepest descent followed by conjugated gradient. The solvent of the system was first heated to 100 K with an NVT ensemble, followed by heating of the whole system to 300 K with an NPT ensemble. Throughout the MD simulation, a 0.2 fs time step, SHAKE algorithm, periodic boundary, and 10 Å cutoffs were applied. The analysis for RMSD and gyration was taken from the beginning. The equilibration phase was 1 ns, and the production phase of the simulation was 69 ns, with a total simulation time of 70 ns for each system. Analysis was conducted using CPPTRAJ modules in AmberTools 14 and visualised through VMD.
MM/PBSA calculation
A total of 100 frames were extracted from the last nanosecond of the 70 ns simulation trajectory files. MM/PBSA calculations were performed on each frame using MMPBSA.py module in AMBER 14. Energy compositions of compounds 18, 21 as well as panduratin A with NS2B/NS3pro was dissected accordingly.
Results and discussion
Virtual screening and the confirmation of activity of the in silico hits
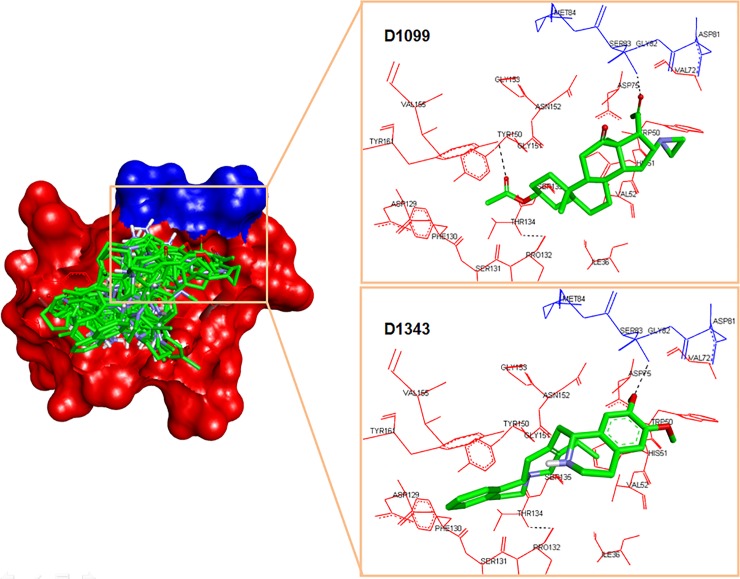
In this study, 1990 compounds from NCI Diversity Set II were docked into the active site pocket of DENV-2 NS2B/NS3pro. The active site is made up of important amino acid residues in S1-S4 subpockets such as Asp129_NS3, Ser135_NS3, Tyr150_NS3 and Tyr161_NS3 (S1 pocket); Asp81_NS2B, Gly82_NS2B, Ser83_NS2B, Asp75_NS3 and Asn152_NS3 (S2); Ser85_NS2B, Ile86_NS2B and Lys87_NS2B (S3); Val154_NS3 and Ile155_NS3 (S4). The 24 top hit compounds were investigated for their interaction with the active site’s residues and ranked in term of the free energy of binding (ΔGbind = -10 kcal/mol to -5 kcal/mol, see S1 Table. We identified that there are more than 50 ligands docked into the active site with that ΔGbind range, but only 24 of them shows interesting interactions with the essential amino acid residues. The selection of ΔGbind range is adopted from the study reported by Shityakov (2014) that Gibbs free energy of binding < 6.0 kcal/mol is clustered as active when this prediction is highly correlated with the experimental results with R2 = 0.880; F = 692.4 standard error of estimate = 0.775 and p-value = 0.0001 [58]. This virtual screening study has a limitation in which no decoy database is being used to validate the docking protocol. The non-binding molecule could be selected over the true ligand called as false positive. Decoy database is the false positive hits which may improve the validity of the test by generating the true positive rate as well as enrichment factor. Unfortunately, the decoy for this DENV-2 NS2B/NS3pro has not been available in a database of useful decoy (DUD dude.docking.org), therefore, the validation of this virtual screening only relies on internal control docking and by in vitro assay [59]. Fig 1 shows the overlay of all 24 compounds docked into the active site of the protease. Most of the ligands bound to NS3pro, but there are two ligands (D1099 and D1343) also interacting with NS2B, particularly at Ser83.
Fig 1. Docked conformation of top 24 NCI compounds in the active pocket of DENV-2 NS2B/NS3pro.
The NS2B and NS3 domains are presented as surface form (blue area = NS2B, red area = NS3pro). Insets are the two ligands that bound to the NS2B, instead of NS3pro.
Dengue protease inhibition assay
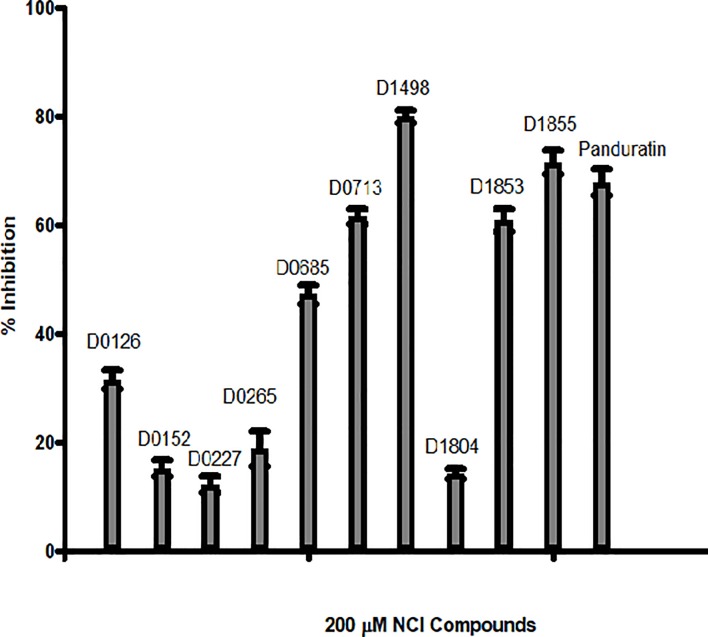
From the 24 hits identified in the virtual screening, only 20 were available for the inhibition assay. Of these 20 compounds, only 10 compounds exhibited significant inhibition activity towards DENV-2 NS2B/NS3pro activity (Fig 2). The other 10 NCI compounds exhibited negligible inhibition (<5% inhibition at 200 μM) towards the protease activity.
Fig 2. In vitro DENV-2 NS2B/NS3pro inhibition assay of Panduratin A and selected NCI compounds; NCI code D0265, D0685, D0227, D0152, D0126, D1804, D1855, D1498, D0713 and D1853 with D in the code stands for Diversity.
The assays contained 0 μM NCI compounds were taken as 0% protease inhibition.
Four NCI compounds with the percentage of inhibition greater than 50% were selected for further assay to determine their IC50 (Fig 3). D1855 showed the strongest inhibition towards the protease activity with IC50 = 29 μM followed by D1498 (48 μM), D0713 (62 μM) and D1853 (77 μM) (Table 1). Panduratin A was used as a control in this experiment. Previously, panduratin A was isolated from finger root (Boesenbergia rotunda (L.)) demonstrating competitive inhibition toward DENV-2 NS2B/NS3pro with Ki = 25 μM [31].
Fig 3. Plot of % DENV-2 NS2B/NS3pro inhibition vs log concentration of the four NCI compounds.
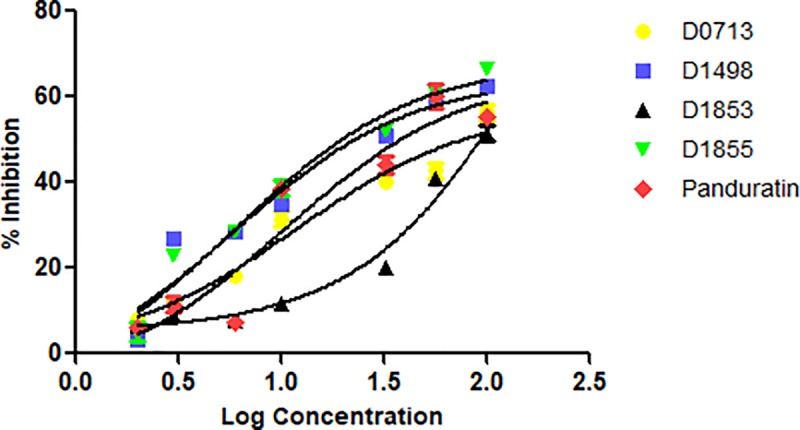
Panduratin A was used as a control in this experiment.
Table 1. Top in vitro hits from NCI diversity set compounds towards both S1 and S2 pockets of DENV-2 NS2B/NS3pro.
| Ligands | ΔGbind (Kcal/ mol) | Experimental IC50 (μM) |
|---|---|---|
| D1498 | -7.40 | 48 |
| D1853 | -9.90 | 77 |
| D1855 | -8.90 | 29 |
| D0713 | -7.10 | 62 |
| Panduratin A | -6.30 | 56 |
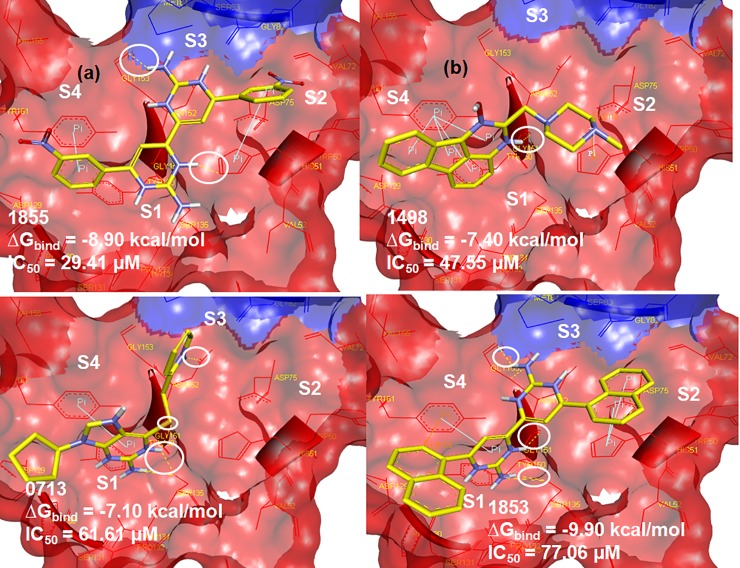
D1855 demonstrated the best inhibition towards the protease activity at concentration less than 50 μM as the relative protease activity dramatically decreased from 100 to 30% (IC50 = 29 μM). This inhibition is probably provided by the hydrogen bonding with Gly153 and His51 and the phenyl ring of the compound formed π-π- interactions with Tyr161 and His51 (Fig 4(A)). D1498 (IC50 = 48 μM) is the second most potent of these four NCI compounds where the activity might be contributed by H-bond interaction with Gly151 of NS3, π—π interaction between its anthracene ring with Tyr161 and π-cation interaction with His51 (Fig 4(B)). D0713 also demonstrated significant inhibition towards the protease with IC50 = 62 μM. The docked pose shows the formation of H-bonds between its amino groups with Asn152, Gly151, and Ser135 and π—π interaction between its guanine ring and Tyr161 (Fig 4(C)). Although the IC50 of D1853 is approximately 77 μM, this compound showed nearly 70% inhibition of the protease activity at 200 μM. As with the above NCI compounds, this ligand also formed π- π as well as π-cation interactions with Tyr161 and His51, respectively, in addition to the H-bond interaction between its amino groups with Gly151, Ser135 and Gly153 (Fig 4(D)).
Fig 4.
Docked poses of (a) D1855, (b) D1498, (c) D0713 and (d) D1853. The NS2B and NS3pro domains are presented as surface form (blue area = NS2B, red area = NS3pro). The H-bond interactions are assigned as yellow dots inside the white circles.
In our quest to identify suitable scaffold for designing potential novel and potent dengue protease inhibitors, we took into consideration the structure of D0713. Although this is not the most potent compound identified in the virtual screening, the structure contains thioguanine (TG or 6-thioguanine) scaffold. TG, a known drug used with other compounds in treating leukemia [40] has also been investigated in other pharmacological activities such as in autoimmune diseases [41] and transplant graft rejection [42]. In addition, its simple chemical structure also benefits feasible synthetic steps, thus, TG is selected as a template to develop a series of analogues. Therefore, in order to test our hypothesis, TG was subjected to the protease inhibition assay and it was found that the molecule showed 56% inhibition at 200 μg/mL (1 mM) indicating that even without any modification to the structure, the scaffold itself is able to inhibit the protease activity. Thus, TG was chosen as the lead structure in the design and synthesis of potential anti-dengue compounds.
Design, synthesis and protease inhibition activity of thioguanine derivatives
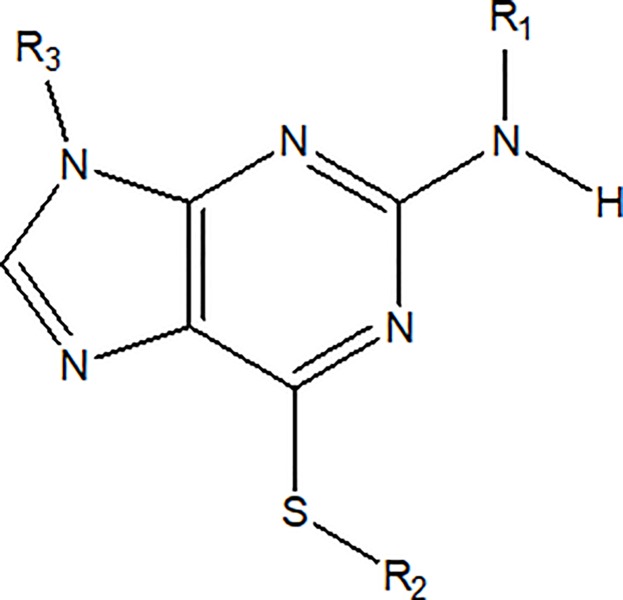
The various thioguanine derivatives designed are shown in Table 2 with the scaffold being illustrated in Fig 5. The design was initiated by connecting the aromatic hydrophilic group as hydrogen bond acceptor (HBA) (1–3) with the amino group; and aromatic hydrophobic group with sulfonyl in 4–6 at R1. In 7–13, acyl groups such as acetyl, butanoyl, isobutanoyl, pentanoyl, isopentanoyl and hexanoyl groups were placed at the same position, while benzoyl group was instead placed in 14. Replacing aliphatic hydrophobic group at R3 with cyclopentyl group, as present in D0713, yielded 15.
Table 2. The list of TG derivatives with their experimental IC50 against DENV -2 NS2B/NS3pro.
| Ligands | R1 | R2 | R3 | IC50 (μM) |
|---|---|---|---|---|
| 1 | (4-nitrophenyl)methanimine | H | H | 1995 |
| 2 | 4-(iminomethyl)benzoic acid | H | H | 1367 |
| 3 | 4-(iminomethyl)-2-methoxy-6-nitrophenol | H | H | 28 |
| 4 | 3-methoxybenzenesulfinic acid | H | H | 68 |
| 5 | 4-methoxybenzenesulfinic acid | H | H | 55 |
| 6 | 3-methylbenzenesulfinic acid | 3-methylbenzenesulfinic acid | H | 63 |
| 7 | acetyl | H | H | 151 |
| 8 | butanoyl | H | H | 57 |
| 9 | isobutanoyl | H | H | 3893 |
| 10 | pentanoyl | H | H | 1037 |
| 11 | 3-methylbutanoyl | H | H | 80 |
| 12 | hexanoyl | H | H | 168 |
| 13 | palmitoyl | H | H | 132 |
| 14 | benzoyl | H | H | 370 |
| 15 | acetyl | H | cyclopentyl | 556 |
| D0713 | H | 2-methylpyridine | cyclopentyl | 62 |
| TG | H | H | H | 753 |
| Panduratin A | - | - | - | 56 |
Fig 5. The structure of thioguanine scaffold.
Compounds 1–3 are imine derivatives of TG. Imine itself has an electron withdrawing nature, albeit weakly. Attaching another EWG such as NO2 (1) and COOH (2) to the benzimine (in the para position) as R1 rendered the compounds to be inactive (IC50> 1000 μM). Interestingly, attaching an electron donating group (EDG) to benzimine at R1 (3) showed high protease inhibition (IC50 = 28 μM). These raised the idea to maintain the EWG group as a linker between the two aromatic rings of TG and the phenyl group which was modified to have EDG character.
Compounds 4 and 5 were designed to have sulfonyl (EWG) with CH3 and/ or OCH3 (EDG) on benzenesulfonyl as R1. These compounds demonstrated lower activities than 3 with IC50 of 68 and 55 μM, respectively. In 6, the same group attached to R1 was also placed at R2 during the synthesis but changes in activity were insignificant. Placing at R1 with acetyl or extended alkyl chain (7–13), in general resulted in marked reduction in protease inhibition, with the exception noted for 11 which showed IC50 = 80 μM.
Placing a simple aromatic hydrophobic group (benzoyl) as seen in 14 did not increase the activity. Exploring placement at R3 with an aliphatic hydrophobic group (15) also did not produce a compound with any improvement in the protease inhibition activity. At this point, it was decided to synthesize six more compounds with expected higher activities.
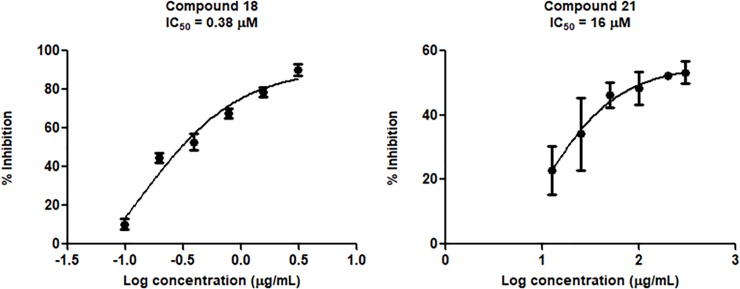
Inspired by 7, 8 and 11 where the compounds have amide group with promising inhibitory activity, six compounds (16–21, see Table 3) having amide group with diverse alkyl chains were designed. As the natural substrate of the protease is peptide, it made sense to incorporate amide groups into the inhibitor structure in order to mimic the peptide character. High activity of 18, 19 and 21 towards DENV-2 NS2B/NS3pro (IC50 of 0.38, 54 and 16 μM, respectively) could be due to this mimic of peptide character. Compound 18 with pentanamide chains at both R1 and R2 possessed the highest activity against the protease. However, attaching benzyl group at R2 (19) dramatically decreased the activity from 0.38 to 54 μM. Interestingly, attaching benzyl group at R3 significantly increased the activity to 16 μM. Compounds 16 and 17 have amide group extended by shorter alkyl chains showed moderate activity with IC50 = 97 and 80 μM, respectively. To confirm the essentiality of amide group, in compound 20, the amide group at R1was omitted and at R2, it was replaced with methylnaphthalene whilst at R3 with isopropyl chain. As predicted, the activity of 20 significantly decreased with IC50 = 258 μM. The activity-dose curve of compounds 18 and 21 are presented in Fig 6.
Table 3. The structures of six newly designed compounds and their activity against DENV-2 NS2B/NS3pro.
| Ligands | R1 | R2 | R3 | IC50 (μM) |
|---|---|---|---|---|
| 16 | propanoyl | propanoyl | H | 97 |
| 17 | isopropanoyl | isopropanoyl | H | 80 |
| 18 | pentanoyl | pentanoyl | H | 0.38 |
| 19 | pentanoyl | benzyl | H | 54 |
| 20 | H | 2-methylnaphtyl | isopropyl | 258 |
| 21 | pentanoyl | benzyl | benzyl | 16 |
Fig 6. The log dose dependent curve of Compound 18 and 21 against DENV-2 NS2B/NS3pro.
Molecular docking simulation
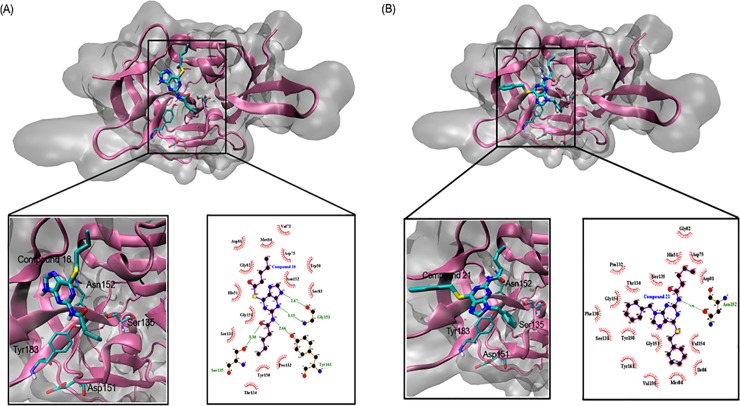
From the docking results, several interactions were identified between NS2B/NS3pro with 18 and 21. For 18, interacting residues such as Ser157(135), Tyr183(161) and Gly175(153) indicated potential hydrogen bonding interactions. The numbering in the bracket indicates the numbering in Wichapong model. Binding stability is contributed by all the hydrophobic interactions surrounding the entire binding site with the free energy of binding– 7.49 kcal/mol (Fig 7). In addition, 21 demonstrated binding affinity (ΔG = -8.10 kcal/mol) with the hydrogen bonding also observed with Asn174.
Fig 7.
Binding orientation and interaction mode of compound with compound 18 (A) and compound 21 (B) in molecular docking simulation.
Stability and conformational changes in MD complexes
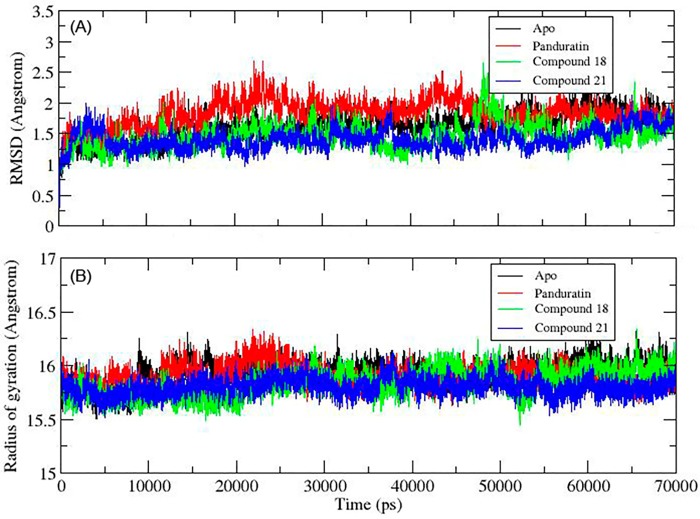
To further substantiate the stability and conformational changes, MD simulation was carried out to understand the dynamic features of the two best compounds with respect to time at nanosecond scale. Overall the backbone RMSD values for Apo, NS2B/NS3pro-18, NS2B/NS3pro-21 are <2 Å throughout the 70 ns simulation time reflecting the stability of the systems. NS2B/NS3pro-Panduratin adopted a higher RMSD with the range of 2.0 to 2.5 Å at 20 to 50 ns simulation time, however, it stabilised at <2 Å from 50 to 70 ns of simulation time (Fig 8).
Fig 8.
(A) Time evolution of RMSD of NS2B/NS3pro backbone CA unbounded (Apo) (black), bounded with panduratin A (red), 18 (green) and 21 (blue). (B) Time evolution of radius of gyration of NS2B/NS3pro backbone CA unbounded (Apo) (black), bounded with panduratin A (red), 18 (green) and 21 (blue).
The gyration value indicates the compactness of the protein which in turn reflects directly on the folding and unfolding of the protein where unfolding of the protein would affect the kinetic of the protein activity. The gyrations of all the four systems (apoenzyme, 18, 21 and Panduratin A) are fluctuating in less than 1 Å at 15.5 to 16.5 Å. No unfolding is observed with a low fluctuation on the gyration.
Hydrogen bonding analysis
Protein-ligand interaction in general is stabilised by various types of weak interactions with hydrogen bonding interaction is potentially one of the most important interactions. Hydrogen bond formation is observed with several important interacting residues such as His73(51), Asp151(129), Ser157(135), Asn174(152) and Tyr183(161) in both systems (Table 4) (The number in parenthesis is according to Wichapong model). Asn174 in NS2B/NS3pro played an important role in forming hydrogen bonds which occupied more than 60% of the simulation time with 18. Asp97(75) was also found to form hydrogen bonds with both nitrogen groups of 18 with the total occupancy of 8.22% simulation time. It is interesting to note that Tyr183(161) and Ser157(135) which are important in the protease activity also demonstrated weak hydrogen bonds formation with the occupancy of 2.85% and 2.73%, respectively. It is thus postulated that these hydrogen network together with the hydrophobic clusters contributed to the binding affinity of 18.
Table 4. Hydrogen bonds between compound 18 and compound 21 with NS2B/NS3pro that found with at least 0.1% occupancy throughout 70ns simulation time.
| Complex | Hydrogen bond formation | Distance (Å) | Occupancy (%) |
|---|---|---|---|
| NS2B/NS3-18 | Gly173@O-Comp18@H5/Comp18@N4 | 2.85 | 27.19 |
| Asn174@OD1-Comp18@H25/Comp18@N2 | 2.85 | 21.16 | |
| Comp18@N1-Asn174@HD21/Asn174@ND2 | 2.91 | 11.21 | |
| Comp18@N1-Tyr183@HH/Tyr183@OH | 2.83 | 8.33 | |
| Gly175@O-Comp18@H25/Comp18@N2 | 2.85 | 6.41 | |
| Asp97@OD1-Comp18@H25/Comp18@N2 | 2.82 | 4.46 | |
| Tyr183@OH-Comp18@H5/Comp18@N4 | 2.90 | 2.74 | |
| Comp18@O2-Ser157@HG/Ser157@OG | 2.77 | 2.19 | |
| Asp97@OD2-Comp18@H5/Comp18@N4 | 2.83 | 1.93 | |
| Asp97@OD2-Comp18@H25/Comp18@N2 | 2.84 | 1.83 | |
| Gly35@O-Comp18@H5/Comp18@N4 | 2.86 | 1.15 | |
| Ser157@OG-Comp18@H5/Comp18@N4 | 2.89 | 0.54 | |
| Ser36@OG-Comp18@H5/Comp18@N4 | 2.88 | 0.51 | |
| Comp18@O2-His73@HE2/His73@NE2 | 2.86 | 0.45 | |
| Asp34@O-Comp18@H5/Comp18@N4 | 2.85 | 0.36 | |
| Gly175@O-Comp18@H5/Comp18@N4 | 2.85 | 0.23 | |
| Comp18@O2-Ser36@HG/Ser36@OG | 2.78 | 0.23 | |
| Met37@O-Comp18@H25/Comp18@N2 | 2.84 | 0.22 | |
| Ser36@OG-Comp18@H25/Comp18@N2 | 2.90 | 0.15 | |
| Comp18@N9-Ser36@HG/Ser36@OG | 2.87 | 0.13 | |
| Comp18@O2-Tyr183@HH/Tyr183@OH | 2.81 | 0.12 | |
| Gly35@O-Comp18@H25/Comp18@N2 | 2.88 | 0.12 | |
| Tyr183@OH-Comp18@H25/Comp18@N2 | 2.92 | 0.11 | |
| Asn174@OD1-Comp18@H5/Comp18@N4 | 2.94 | 0.10 | |
| Comp18@O1-Arg76@HH22/Arg76@NH2 | 2.89 | 0.10 | |
| NS2B/NS3-21 | His73@ND1-Comp21@HN/Comp21@N | 2.89 | 27.40 |
| Ser157@OG- Comp21@H1/Comp21@N1 | 2.86 | 5.12 | |
| Comp21@O-Ser157@HG/Ser157@OG | 2.79 | 4.58 | |
| His73@ND1-Comp21@H/Comp21@N4 | 2.82 | 0.96 | |
| Asp97@OD2-Comp21@HN/Comp21@N | 2.84 | 0.39 | |
| Asp97@OD1-Comp21@HN/Comp21@N | 2.82 | 0.25 | |
| Gly175@O-Comp21@HN/ Comp21@N | 2.85 | 0.15 | |
| Asn174@OD1- Comp21@H/Comp21@N4 | 2.82 | 0.12 | |
| NS2B/NS3-panduratin | Phe152@O-Pandu@H10/Pandu@O3 | 2.68 | 9.59 |
| His73@O-Pandu@H9/Pandu@O1 | 2.71 | 5.74 | |
| Phe152@O-Pandu@H9/Pandu@O1 | 2.71 | 1.50 | |
| Pandu@O-Tyr183@HH/Tyr183@OH | 2.79 | 1.29 | |
| Asp151@OD2-Pandu@H9/Pandu@O1 | 2.65 | 1.13 | |
| Asp151@OD1-Pandu@H9/Pandu@O1 | 2.67 | 0.57 | |
| Pandu@O2-Tyr172@HH/Tyr172@OH | 2.86 | 0.41 | |
| Pandu@O2-Val177@H/Val177@N | 2.89 | 0.40 | |
| Pandu@o2-Ser157@HG/Ser157@OG | 2.83 | 0.28 | |
| Pandu@O3-Tyr183@HH/Tyr183@OH | 2.86 | 0.22 | |
| Pandu@O2-Tyr183@H/Tyr183@N | 2.91 | 0.19 | |
| Asp151@OD1-Pandu@H10/Pandu@O3 | 2.66 | 0.16 | |
| Val74@O-Pandu@H9/Pandu@O1 | 2.80 | 0.13 | |
| His73@ND1-Pandu@H9/Pandu@O1 | 2.85 | 0.13 | |
| Pandu@O3-Phe152@H/Phe152@N | 2.91 | 0.12 | |
| Gly173@O-Pandu@H9/Pandu@O1 | 2.79 | 0.10 | |
| Pandu@O-Tyr183@H/Tyr183@N | 2.90 | 0.10 |
The number of hydrogen bond formation observed with 21 is lesser as compared to 18. His73 formed a hydrogen bond with nitrogen group in 21 with 27.4% occupancy of the total 70ns simulation time. Two hydrogen bonds between Ser157 and the nitrogen and oxygen groups of 21 with 5.12% and 4.58% occupancy while very weak hydrogen bonds were observed with other interacting residues such as Asp97, Asn174 and Tyr183 with less than 1% occupancies.
In the case of panduratin A, only weak hydrogen bonds with less than 10% occupancies throughout 70ns were found with residues such as Phe152, His73, Asp151 and Tyr183. The formation of hydrogen bonding is significantly weaker as compared to 18 and 21.
Comparison of free energy of binding between panduratin A, 18 and 21
The interaction and decomposition energies of the interactions between panduratin A, 18 and 21 were calculated using MM/PBSA. A total of 100 snapshots were extracted every 10 ps interval from the last nanosecond. The absolute binding free energy (Table 5) for panduratin A, 18 and 21 are-11.27 ±2.99, -16.10 ± 2.70 and -18.24 ± 4.66 kcal/mol, respectively. Non-polar interaction between the binding site residues as well as the aliphatic chain (pentanoyl) of 18 and 21 provides major contribution to the binding energy. The binding of panduratin A against NS2B/NS3pro is less favourable as compared to 18 and 21. Similarly, major contribution of panduratin A binding is mainly from the non-polar interaction. This can be inferred from the fact that only weak hydrogen bonds were found between panduratin A and NS2B/NS3pro with low occupancies. This reflects that the binding of panduratin A is stabilised mainly by van der Waals and hydrophobic interactions.
Table 5. Binding free energy predicted using MM/PBSA calculation for 18, 21 and panduratin A.
| Energy Component | Binding Free Energy (kcal/mol) | ||
|---|---|---|---|
| Compound 18 | Compound 21 | Panduratin A | |
| Polar component electrostatics | -30.05 ± 8.77 | -264.23 ± 17.87 | -1.90 ±6.37 |
| Electrostatics solvation | 42.54 ± 8.73 | 292.37 ± 17.63 | 14.21 ± 4.22 |
| Non-polar component van der Waal | -26.22 ± 2.6 | -44.36 ± 2.83 | -21.18 ± 2.54 |
| Non-polar solvation | -2.66 ± 0.15 | -4.73 ± 0.10 | -2.40 ±0.17 |
| TOTAL Binding Free energy | -16.37 ± 3.22 | -20.95 ± 4.12 | -11.27 ± 2.99 |
The binding free energy estimated by MMPBSA calculation indicated that 21 has better binding as compared to 18 but the IC50 for 18 is better. 18 and 21 have identical pentanoyl R1 group but 21 has two aromatic benzyl groups at R2 and R3 instead of a carbonyl chain and hydrogen for 18 at R2 and R3, respectively. Higher van der Waals interaction was observed from MM/PBSA calculation for 21 due to the presence of these two benzyl groups at R2 and R3. However, these interactions might be overestimated using MM/PBSA approach [60].
Further discussion
Thioguanine was identified as a potential scaffold for DENV-2 NS2B/NS3pro inhibitor based on virtual screening, in vitro assay and molecular modelling. The thioguanine scaffold is composed of pyramidine and imidazole rings attached to an amine and a thiol group which may contribute to the compound’s activity. As far as we are aware, no report of pyrimidine inhibition to DENV-2 NS2B/NS3pro, however, 2-(benzylthio)-6-oxo-4-phenyl-1,6-dihydropyrimidine has been reported to demonstrate an activity against SARS-CoV-3C like protease [61]. In addition, pyrimidine has also been shown to actively stop the growth of DENV-2 but it relies on the capability of pyrimidine to inhibit dihydroorotate dehydrogenase (DHODH), an enzyme required for viral pyrimidine biosynthesis [62]. Imidazoles have also been investigated against dengue virus [63] however the exact molecular mechanism or which protein they target is still unknown. The 6-thioguanine scaffold has been reported to non-competitively inhibit ubiquitin specific protease in various cancers [64]. With this background, we postulated that thioguanine derivatives might potentially be good inhibitors against DENV-2 NS2B/NS3pro.
Two most active compounds (18 and 21) against DENV2-NS2B/NS3pro have an amide functional group (instead of an amino group). Amide is a part of peptide which is recognised as the key point for the protease substrate. In the docking study, the amide group is marked as a HBD with N-amide interacts with O-phenol of Tyr161 (in 18) and Asn152 (in 21) via H-bond. Previously, amide, such as α-ketoamide [65], arylcyanoacrylamide [66], and aminobenzamide [67] has been incorporated in the design of DENV NS2B/NS3pro inhibitors. The alkyl chain of 18 and phenyl-benzyl group of 21 demonstrate hydrophobic features and agreeable to docking results which show that these groups bound at the hydrophobic pocket surrounded by Val154, Ile86, Met84 and Val155. These also correlate with the decomposition energies as computed by MM/PBSA which highlight that the major contribution to the interaction with DENV-2 NS2B/NS3pro is from non-polar interaction.
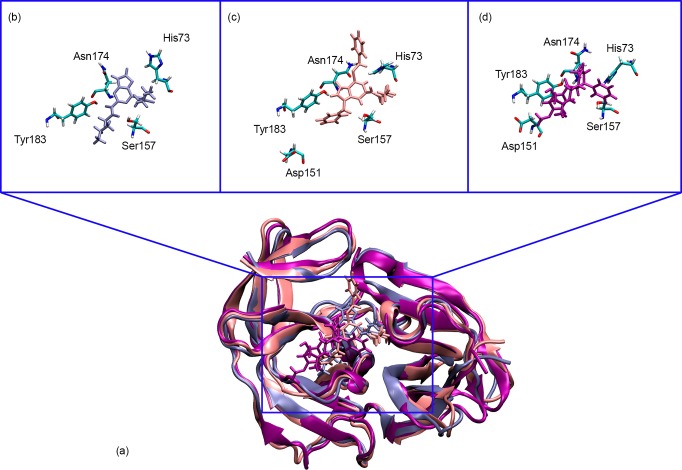
It is interesting to note that the binding orientations from the MD simulations for both compounds are different despite sharing similar thioguanine (Fig 9). The thioguanine moiety is facing toward His73 and Tyr183 for 18 and 21, respectively. Compound 18 has an acyl group at R2 which provides additional electron lone pair enabling the formation of hydrogen bonds that can strengthen its binding towards NS2B/NS3pro. This reflected clearly in the hydrogen bond analysis where 18 formed more hydrogen bonds as compared to 21. Regardless of the slightly lower absolute binding energy from the MM/PBSA prediction, the binding interaction analysis of 18 with NS2B/NS3pro agrees well with experimental results. The absolute free energies of binding for both two active compounds are also more negative than panduratin A supporting the in vitro results in which compound 18 and 21 better IC50 value (0.38 and 16 μM, respectively) than panduratin A (56 μM).
Fig 9.
Binding orientation and interaction mode of compound with 18 (A), 21 (B) and panduratin A (C) in molecular dynamics simulation.
Conclusions
We presented here the computational design of potential DENV-2 NS2B/NS3pro inhibitors. From the virtual screening of National Cancer Institute Database, four compounds including D0713 were observed to have moderate inhibition activity on the protease. D0713 has a thioguanine scaffold in its structure prompting us to consider the scaffold for designing the new thioguanine derivatives as potential DENV-2 NS2B/NS3pro inhibitors. Fifteen compounds were synthesised and the bioactivity against dengue protease showed variation in inhibition activity from inactive to moderately active (1000>IC50>18 μM). Based on this information, a further design of six new compounds was conducted by concentrating on the attachment of amide group. All the compounds showed inhibition activity against the dengue protease with compound 18 being the most potent (IC50 of 0.38 μM). This result agrees well with the MM/PBSA calculation which showed that the interactions are mainly contributed by polar and non-polar interactions. Hydrogen bonding analysis demonstrates the importance of amino acid residues Asn174 (occupancy 60%), Asp75 (8.22%), Tyr183 and Ser157 (2.85 and 2.73%). This is further supported by the experimental results which showed that 18 could be further developed as DENV-2 NS2B/NS3pro inhibitor. It is hoped that the results obtained from this study could be used in designing more active compounds as potential dengue protease inhibitors.
Supporting information
General procedure for synthesis of Schiff base-thioguanine compounds, acetamide-thioguanine compounds, N-alkylation of acetamide-thioguanine, and S/N-benzylation of acetamide-thioguanine.
(PDF)
The numbering system is according to the NMR characterisation.
(PDF)
The NS2B-NS3pro is presented in a surface form. The ligands are presented in a stick form in which carbons are grey while oxygens are red. The ligand is docked at the active site of the protease with the hydrophilic moieties surrounded by catalytic site(His51, Asp75 and Ser135) whereas the nonpolar part of the ligand closes nearly to hydrophobic area of the protease wherein phenyl ring posseses H-bond as well as π-π interactions with Tyr161.
(PDF)
The NCI compounds are named in two codes and the free energy of binding upon DENV-2 NS2B/NS3pro binding site is expressed in kcal/mol.
(PDF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work is funded under Science Fund Grant No. 02-05-23-SF0019 and Research University Grant for Team (RUT) No.1001/PKIMIA/855006. MH thanked Universiti Sains Malaysia for the USM Fellowship Award (2011-2012) and the Malaysian Ministry of Science and Technology Innovation for research assistantship under ScienceFund Grant No. 02-05-23-SF0019 (2014-2015). We thank National Cancer Institute, US for the complementary samples of the 20 NCI hit compounds.
References
- 1.Murray NE, Quam MB, Wilder-Smith A. Epidemiology of dengue: past, present and future prospects. Clin Epidemiol. 2013; 5: 299–309. 10.2147/CLEP.S34440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brady OJ, Gething PW, Bhatt S, Messina JP, Brownstein JS, Hoen AG, et al. Refining the global spatial limits of dengue virus transmission by evidence-based consensus. PLoSNegl Trop Dis. 2012; 6: e1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guzman MG, Harris E. Dengue. Lancet. 2015; 385: 453–465. 10.1016/S0140-6736(14)60572-9 [DOI] [PubMed] [Google Scholar]
- 4.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, et al. The global distribution and burden of dengue. Nature. 2013; 496: 504–507. 10.1038/nature12060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pang T, Cardosa MJ, Guzman MG. Of cascades and perfect storms: the immunopathogenesis of dengue haemorrhagic fever-dengue shock syndrome (DHF/DSS). Immunol Cell Biol. 2007; 85: 43–45. 10.1038/sj.icb.7100008 [DOI] [PubMed] [Google Scholar]
- 6.Simmons CP, McPherson K, Van Vinh Chau N, Hoai Tam DT, Young P, Mackenzie J, et al. Recent advances in dengue pathogenesis and clinical management. Vaccine. 2015; 33: 7061–7068. 10.1016/j.vaccine.2015.09.103 [DOI] [PubMed] [Google Scholar]
- 7.WHO, Dengue vaccine: WHO position paper—July 2016.Wkly Epidemiol Rec. 2016; 91: 349–64. [PubMed] [Google Scholar]
- 8.WHO, Dengue: Guidelines for diagnosis, treatment, prevention and control: New Edition 2013/06/14 ed. World Health Organisation: Geneva: 2009. [PubMed] [Google Scholar]
- 9.Galula JU, Shen WF, Chuang ST, Chang GJ, Chao DY. Virus-like particle secretion and genotype-dependent immunogenicity of dengue virus serotype 2 DNA vaccine. J Virol. 2014; 88: 10813–10830. 10.1128/JVI.00810-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Normile D. First new dengue virus type in 50 years. Health. 2013. [Google Scholar]
- 11.Henchal EA, Putnak JR. The dengue viruses. Clin Microbiol Rev. 1990; 3: 376–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Natarajan S. NS3 protease from flavivirus as a target for designing antiviral inhibitors against dengue virus. Genet Mol Biol. 2010; 33: 214–219. 10.1590/S1415-47572010000200002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wahab HA, Yusof R, Rahman NA. A search for vaccines and therapeutic for dengue: A review. Curr Comput-Aid Drug. 2007; 3:, 101–112. [Google Scholar]
- 14.Behnam MA, Nitsche C, Boldescu V, Klein CD. The medicinal chemistry of dengue virus. J Med Chem. 2016; 59: 5622–5649. 10.1021/acs.jmedchem.5b01653 [DOI] [PubMed] [Google Scholar]
- 15.Behnam MA, Graf D, Bartenschlager R, Zlotos DP, Klein CD. Discovery of nanomolar dengue and west nile virus protease inhibitors containing a 4-benzyloxyphenylglycine residue. J Med Chem. 2015; 58: 9354–9370. 10.1021/acs.jmedchem.5b01441 [DOI] [PubMed] [Google Scholar]
- 16.Frimayanti N, Zain SM, Lee VS, Wahab HA, Yusof R, Rahman NA. Fragment-based molecular design of new competitive dengue Den2 Ns2b/Ns3 inhibitors from the components of fingerroot (Boesenbergia rotunda). In Silico Biol. 2012; 11: 29–37. [DOI] [PubMed] [Google Scholar]
- 17.Yin Z, Patel SJ, Wang WL, Chan WL, Ranga Rao KR, Wang G, et al. Peptide inhibitors of dengue virus NS3 protease. Part 2: SAR study of tetrapeptide aldehyde inhibitors. Bioorg Med Chem Lett. 2006; 16: 40–43. 10.1016/j.bmcl.2005.09.049 [DOI] [PubMed] [Google Scholar]
- 18.Chanprapaph S, Saparpakorn P, Sangma C, Niyomrattanakit P, Hannongbua S, Angsuthanasombat C, et al. Competitive inhibition of the dengue virus NS3 serine protease by synthetic peptides representing polyprotein cleavage sites. Biochem Biophys Res Commun. 2005; 330: 1237–1246. 10.1016/j.bbrc.2005.03.107 [DOI] [PubMed] [Google Scholar]
- 19.Steuer C, Heinonen KH, Kattner L, Klein CD. Optimization of assay conditions for dengue virus protease: Effect of various polyols and nonionic detergents. J Biomol Screen. 2009; 14: 1102–1108. 10.1177/1087057109344115 [DOI] [PubMed] [Google Scholar]
- 20.Frecer V, Miertus S. Design, structure-based focusing and in silico screening of combinatorial library of peptidomimetic inhibitors of Dengue virus NS2B-NS3 protease. J Comput Aided Mol Des. 2010; 24: 195–212. 10.1007/s10822-010-9326-8 [DOI] [PubMed] [Google Scholar]
- 21.Yang CC, Hsieh YC, Lee SJ, Wu SH, Liao CL, Tsao CH, et al. Novel dengue virus-specific NS2B/NS3 protease inhibitor, BP2109, discovered by a high-throughput screening assay. Antimicrob Agents Chemother. 2011; 55: 229–238. 10.1128/AAC.00855-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katzenmeier G. Inhibition of the NS2B–NS3 protease: towards a causative therapy for dengue virus diseases. Dengue Bull. 2004; 28: 58–67. [Google Scholar]
- 23.Yotmanee P, Rungrotmongkol T, Wichapong K, Choi SB, Wahab HA, Kungwan N, et al. Binding spesificity of polypeptide substrates in NS2B/NS3pro serine protease of dengue virus type 2: A molecular dynamics study. J Mol Graph Model 2015, 60, 24–33. 10.1016/j.jmgm.2015.05.008 [DOI] [PubMed] [Google Scholar]
- 24.Nitsche C, Behnam MA, Steuer C, Klein CD. Retro peptide-hybrids as selective inhibitors of the dengue virus NS2B-NS3 protease. Antiviral Res. 2012; 94: 72–79. 10.1016/j.antiviral.2012.02.008 [DOI] [PubMed] [Google Scholar]
- 25.Behnam MA, Nitsche C, Vechi SM, Klein CD. C-terminal residue optimization and fragment merging: discovery of a potent peptide-hybrid inhibitor of dengue protease. ACS Med Chem Lett. 2014;, 5: 1037–1042. 10.1021/ml500245v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nitsche C, Schreier VN, Behnam MA, Kumar A, Bartenschlager R, Klein CD. Thiazolidinone–peptide hybrids as dengue virus protease inhibitors with antiviral activity in cell culture. J Med Chem. 2013; 56: 8389–8403. 10.1021/jm400828u [DOI] [PubMed] [Google Scholar]
- 27.Rothan HA, Han HC, Ramasamy TS, Othman S, Rahman NA, Yusof R. Inhibition of dengue NS2B-NS3 protease and viral replication in Vero cells by recombinant retrocyclin-1. BMC Infect Dis. 2012; 12:314 10.1186/1471-2334-12-314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rothan HA, Abdulrahman AY, Sasikumer PG, Othman S, Rahman NA, Yusof R. Protegrin-1 inhibits dengue NS2B-NS3 serine protease and viral replication in MK2 cells. J Biomed Biotechnol. 2012; 2514–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prusis P, Junaid M, Petrovska R, Yahorava S, Yahorau A, Katzenmeier G, et al. Design and evaluation of substrate-based octapeptide and non substrate-based tetrapeptide inhibitors of dengue virus NS2B-NS3 proteases. Biochem Biophys Res Commun. 2013; 434: 767–772. 10.1016/j.bbrc.2013.03.139 [DOI] [PubMed] [Google Scholar]
- 30.Xu S, Li H, Shao X, Fan C, Ericksen B, Liu J, et al. Critical effect of peptide cyclization on the potency of peptide inhibitors against Dengue virus NS2B-NS3 protease. J Med Chem. 2012; 55: 6881–6887. 10.1021/jm300655h [DOI] [PubMed] [Google Scholar]
- 31.Kiat TS, Pippen R, Yusof R, Ibrahim H, Khalid N, Rahman NA. Inhibitory activity of cyclohexenyl chalcone derivatives and flavonoids of fingerroot, Boesenbergia rotunda (L.), towards dengue-2 virus NS3 protease. Bioorg Med Chem Lett. 2006; 16: 3337–40. 10.1016/j.bmcl.2005.12.075 [DOI] [PubMed] [Google Scholar]
- 32.de Sousa LRF, Wu H, Nebo L, Fernandes JB, Kiefer W, Kanitz M, et al. Flavonoids as noncompetitive inhibitors of dengue virus NS2B-NS3 protease: Inhibition kinetics and docking studies. Bioorg Med Chem Lett. 2015; 23: 466–470. [DOI] [PubMed] [Google Scholar]
- 33.Viswanathan U, Tomlinson SM, Fonner JM, Mock SA, Watowich SJ. Identification of a novel inhibitor of dengue virus protease through use of a virtual screening drug discovery Web portal. J Chem Inf Model. 2014; 54: 2816–2825. 10.1021/ci500531r [DOI] [PubMed] [Google Scholar]
- 34.Raut R, Beesetti H, Tyagi P, Khanna I, Jain SK, Jeankumar VU, et al. A small molecule inhibitor of dengue virus type 2 protease inhibits the replication of all four dengue virus serotypes in cell culture. Virol J. 2015; 12: 1 10.1186/s12985-014-0235-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu H, Wu R, Sun Y, Ye Y, Chen J, Luo X, et al. Identification of novel thiadiazoloacrylamide analogues as inhibitors of dengue-2 virus NS2B/NS3 protease. Bioorg Med Chem. 2014; 22: 6344–6352. 10.1016/j.bmc.2014.09.057 [DOI] [PubMed] [Google Scholar]
- 36.Yusof R, Rahman NA, Wahab HA, Othman R, Padmanabhan R. Design of Chemotherapeutic Agent for Dengue Virus Protease. 2008. [Google Scholar]
- 37.Lee YK, Tan SK, Wahab HA, Yusof R, Rahman NA. Nonsubstrate based inhibitors of dengue virus serine protease: a molecular docking approach to study binding interactions between protease and inhibitors. Asia Pac J Mol Biol Biotechnol. 2007; 15: 53–59. [Google Scholar]
- 38.Cabarcas-Montalvo M, Maldonado-Rojas W, Montes-Grajales D, Bertel-Sevilla A, Wagner-Dobler I, Sztajer H, et al. Discovery of antiviral molecules for dengue: In silico search and biological evaluation. Eur J Med Chem. 2016; 110: 87–97. 10.1016/j.ejmech.2015.12.030 [DOI] [PubMed] [Google Scholar]
- 39.Othman R, Wahab HA, Yusof R, Rahman NA. Analysis of secondary structure predictions of dengue virus type 2 NS2B/NS3 against crystal structure to evaluate the predictive power of the in silico methods. In Silico Biol. 2007; 7: 215–224. [PubMed] [Google Scholar]
- 40.Adamson PC, Poplack DG, Balis FM. The cytotoxicity of thioguanine vs mercaptopurine in acute lymphoblastic leukemia. Leuk Res. 1994; 18: 805–810. [DOI] [PubMed] [Google Scholar]
- 41.Osterman MT, Kundu R, Lichtenstein GR, Lewis JD. Association of 6-thioguanine nucleotide levels and inflammatory bowel disease activity: a meta-analysis. Gastroenterology. 2006; 130: 1047–53. 10.1053/j.gastro.2006.01.046 [DOI] [PubMed] [Google Scholar]
- 42.Elion GB. The purine path to chemotherapy. Biosci Rep. 1989; 9: 509–529. [DOI] [PubMed] [Google Scholar]
- 43.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010; 31: 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wichapong K, Pianwanit S, Sippl W, Kokpol S. Homology modeling and molecular dynamics simulations of Dengue virus NS2B/NS3 protease: insight into molecular. J Mol Recognit. 2010; 23: 283–300. 10.1002/jmr.977 [DOI] [PubMed] [Google Scholar]
- 45.Yusof R, Clum S, Wetzel M, Murthy HM, Padmanabhan R. Purified NS2B/NS3 serine protease of dengue virus type 2 exhibits cofactor NS2B dependence for cleavage of substrates with dibasic amino acids in vitro. J Biol Chem. 2000; 275: 9963–9969. [DOI] [PubMed] [Google Scholar]
- 46.Heh CH, Othman R, Buckle MJ, Sharifuddin Y, Yusof R, Rahman NA. Rational discovery of dengue type 2 non-competitive inhibitors. Chem Biol Drug Des. 2013; 82: 1–11. 10.1111/cbdd.12122 [DOI] [PubMed] [Google Scholar]
- 47.Rahman NA, Hadinur MS, Rashid N, Muhamad M, Yusof R. Studies on quercuslusitanica extracts on DENV-2 replication. Dengue Bull. 2006; 30: 260–269. [Google Scholar]
- 48.Qiagen, The QIA expressionist—A handbook for high-level expression and purification of 6xHis-tagged proteins. 5th ed ed.; 2003.
- 49.Leung D, Schroder K, White H, Fang NX, Stoermer MJ, Abbenante G, et al. Activity of recombinant dengue 2 virus NS3 protease in the presence of a truncated NS2B co-factor, small peptide substrates, and inhibitors. J Biol Chem. 2001; 276: 45762–45771. 10.1074/jbc.M107360200 [DOI] [PubMed] [Google Scholar]
- 50.Zandi K, Lim TH, Rahim NA, Shu MH, Teoh BT, Sam SS, et al. Extract of Scutellariabaicalensis inhibits dengue virus replication. BMC Complement Altern Med. 2013; 13: 91 10.1186/1472-6882-13-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prakash C, Vijay IK. A new fluorescent tag for labeling of saccharides. Anal Biochem. 1983; 128: 41–6. [DOI] [PubMed] [Google Scholar]
- 52.Hariono M, Wahab HA, Tan ML, Rosli MM, Razak IA. 9-Benzyl-6-benzylsulfanyl-9H-purin-2-amine. Acta Cryst E. 2014; 70: o288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hariono M, Ngah N, Wahab HA, Abdul Rahim AS. 2-Bromo-4-(3, 4-dimethyl-5-phenyl-1, 3-oxazolidin-2-yl)-6-methoxyphenol. Acta Cryst E. 2012; 68: o35–o36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.HyperChem (TM) Profesional 8.0, Hypercube, Inc.
- 55.Morris GM, Lindstrom RHW, Sanner MF, Belew RK, Goodsell DS, Olson AJ, AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009; 28: 73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011; 51: 2778–2786. 10.1021/ci200227u [DOI] [PubMed] [Google Scholar]
- 57.Le Grand S, Götz AW, Walker RC. SPFP: Speed without compromise—A mixed precision model for GPU accelerated molecular dynamics simulations. Comput Phys Commun. 2013; 184: 374–380. [Google Scholar]
- 58.Shityakov S, Förster C. In silico predictive model to determine vector-mediated transport properties for the blood–brain barrier choline transporter. Adv Appl Bioinform Chem.2014; 7: 23 10.2147/AABC.S63749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chaput L, Martinez-Sanz J, Saettel N, Mouawad L. Benchmark of four popular virtual screening programs: construction of the active/decoy dataset remains a major determinant of measured performance. J Cheminform. 2016; 8: 56 10.1186/s13321-016-0167-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Genheden S, Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discov. 2015; 10: 449–461. 10.1517/17460441.2015.1032936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramajayam R, Tan K-P, Liu H-G, Liang P-H. Synthesis, docking studies, and evaluation of pyrimidines as inhibitors of SARS-CoV 3CL protease, Bioorg Med Chem Lett. 2010; 20: 3569–3572. 10.1016/j.bmcl.2010.04.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang QY, Bushell S, Qing M, Xu HY, Bonavia A, Nunes S, et al. Inhibition of dengue virus through suppression of host pyrimidine biosynthesis. J Virol. 2011; 85: 6548–6556. 10.1128/JVI.02510-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saudi M, Zmurko J, Kaptein S, Rozenski J, Neyts J, Van Aerschot A. Synthesis and evaluation of imidazole-4,5- and pyrazine-2,3-dicarboxamides targeting dengue and yellow fever virus. Eur J Med Chem. 2014; 87: 529–539. 10.1016/j.ejmech.2014.09.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chuang S-J, Cheng S-C, Tang H-C, Sun C-Y, Chou C-Y. 6-Thioguanine is noncompetitive and slow binding inhibitor of human deubiquitinating protease USP2. Sci Rep. 2018; 8: 1–9. 10.1038/s41598-017-17765-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Steuer C, GeGe C, Fischl W, Heinonen KH, Bartenschlager R, Klein CD. Synthesis and biological evaluation of α-ketoamides as inhibitors of the Dengue virus protease with antiviral activity in cell-culture. Bioorg Med Chem. 2011; 19: 4067–4074. 10.1016/j.bmc.2011.05.015 [DOI] [PubMed] [Google Scholar]
- 66.Nitsche C, Steuer C, Klein CD. Arylcyanoacrylamides as inhibitors of the dengue and west nile virus proteases. Bioorg Med Chem. 2011; 19: 7318–7337. 10.1016/j.bmc.2011.10.061 [DOI] [PubMed] [Google Scholar]
- 67.Aravapalli S, Lai H, Teramoto T, Alliston KR, Lushington GH, Ferguson EL, et al. Inhibitors of Dengue virus and West Nile virus proteases based on the aminobenzamide scaffold. Bioorg Med Chem. 2012; 20: 4140–4148. 10.1016/j.bmc.2012.04.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
General procedure for synthesis of Schiff base-thioguanine compounds, acetamide-thioguanine compounds, N-alkylation of acetamide-thioguanine, and S/N-benzylation of acetamide-thioguanine.
(PDF)
The numbering system is according to the NMR characterisation.
(PDF)
The NS2B-NS3pro is presented in a surface form. The ligands are presented in a stick form in which carbons are grey while oxygens are red. The ligand is docked at the active site of the protease with the hydrophilic moieties surrounded by catalytic site(His51, Asp75 and Ser135) whereas the nonpolar part of the ligand closes nearly to hydrophobic area of the protease wherein phenyl ring posseses H-bond as well as π-π interactions with Tyr161.
(PDF)
The NCI compounds are named in two codes and the free energy of binding upon DENV-2 NS2B/NS3pro binding site is expressed in kcal/mol.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.