Abstract

Itaconic acid is an important platform chemical that can easily be incorporated into polymers and has the potential to replace petrochemical-based acrylic or methacrylic acid. A number of microorganisms have been developed for the biosynthesis of itaconate including Aspergillus terreus, Escherichia coli, and Saccharomyces cerevisiae. However, the number of strains and conditions that can be tested for increased itaconate titers are currently limited because of the lack of high-throughput screening methods. Here we identified itaconate-inducible promoters and their corresponding LysR-type transcriptional regulators from Yersinia pseudotuberculosis and Pseudomonas aeruginosa. We show that the YpItcR/Pccl inducible system is highly inducible by itaconic acid in the model gammaproteobacterium E. coli and the betaproteobacterium Cupriavidus necator (215- and 105-fold, respectively). The kinetics and dynamics of the YpItcR/Pccl inducible system are investigated, and we demonstrate, that in addition to itaconate, the genetically encoded biosensor is capable of detecting mesaconate, cis-, and trans-aconitate in a dose-dependent manner. Moreover, the fluorescence-based biosensor is applied in E. coli to identify the optimum expression level of cadA, the product of which catalyzes the conversion of cis-aconitate into itaconate. The fluorescence output is shown to correlate well with itaconate concentrations quantified using high-performance liquid chromatography coupled with ultraviolet spectroscopy. This work highlights the potential of the YpItcR/Pccl inducible system to be applied as a biosensor for high-throughput microbial strain development to facilitate improved itaconate biosynthesis.
Keywords: itaconic acid, inducible gene expression, fluorescence-based biosensor, Yersinia pseudotuberculosis, Pseudomonas aeruginosa, macrophage infection
The use of biological processes for the production of chemicals and fuels is a promising alternative to the traditional approach of chemical manufacture.1 They offer the opportunity to convert renewable or waste feedstocks into higher value compounds of industrial interest.2 Although many biological processes have the potential to replace synthetic chemistry, product titers and productivity often remain to be optimized in order to achieve economically competitive conversion rates.1,3 To facilitate and expedite the implementation of biocatalysts with improved performance, low-cost and high-throughput microbial engineering strategies need to be developed.
Itaconic acid is an attractive platform chemical with a wide range of industrial applications, such as in rubber, detergents, or surface active agents.4 In 2004, it was reported by the U.S. Department of Energy to be one of the top 12 building block chemicals from biomass.5 The C5-dicarboxylic acid can be converted into poly(acrylamide-co-itaconic acid) which is used as a superabsorbent for aqueous solutions, or poly(methyl methacrylate), also known as Plexiglas.6
Itaconate is a naturally occurring metabolite formed by decarboxylation of aconitate, an intermediate of the citric acid cycle. A number of microorganisms, including Aspergillus terreus,7Ustilago maydis (also known as U. zeae),8 and Candida sp.,9 have been described as natural producers of itaconic acid. It is also produced as an antimicrobial compound by macrophages, mammalian immune cells.10,11 In A. terreus and macrophages, itaconate is synthesized from the tricarboxylic acid cycle intermediate cis-aconitate through the action of a cis-aconitate decarboxylase (CadA). In contrast, in U. maydis it is produced via the unusual intermediate trans-aconitate.12 Heterologous expression of the A. terreus cadA gene has demonstrated that the biosynthesis of itaconic acid can be achieved in different host organisms than the natural producer.13 So far, the highest titer of biotechnologically produced itaconate has been obtained by fermentation of A. terreus.14−16 However, due to feedback inhibition of itaconate biosynthesis at higher concentrations,17 considerable research efforts have been directed toward developing alternative microbial biocatalysts. Other microorganisms that have been investigated for the biosynthesis of itaconic acid include Pseudozyma antarctica, Corynebacterium glutamicum, Escherichia coli, Saccharomyces cerevisiae, Yarrowia lipolytica, and species of Candida and Ustilago.9,18−24 Although some of these microorganisms exhibit beneficial traits, such as a high tolerance to itaconate and a low pH,19,22 production titers need to be considerably improved.
Genetically encoded biosensors have gained increasing interest as molecular tools enabling high-throughput strain development.25 They are composed of transcription factor-based inducible gene expression systems linked to a reporter or an antibiotic resistance gene.26,27 By using a fluorescent reporter gene, changes in intracellular metabolite concentrations can easily be monitored by a fluorescence output enabling the screen of millions of single-cells in a rapid manner.25 Biosensors have been successfully applied to increase products titers of platform chemicals such as acrylate, 3-hydroxypropionate (3-HP), and glucarate.26,28 To date, no itaconate biosensor has been developed which could facilitate the screening process for both metabolically engineered strains and alternative feedstocks, such as biomass hydrolysates, to improve yields and decrease production costs.29
This study was aimed to identify an itaconate-inducible gene expression system and construct a fluorescence-based biosensor. Several natural compounds were screened for biosensor induction and induction kinetics measured. Moreover, the developed biosensor was exploited in the optimization of itaconate production in E. coli, and its output was compared to analytically determined itaconate titers.
Results and Discussion
Identification of an Itaconic Acid-Inducible System
To build an itaconate biosensor, which can be applied across different species, both elements of a transcription-based inducible system, a transcriptional regulator (TR), and the corresponding inducible promoter, are needed. Bacterial degradation pathways, which are often activated exclusively in the presence of the compound to be degraded, represent a rich source of inducible promoters. Even though the pathway for itaconate catabolism had been known for more than 50 years,30 and a few bacteria including Pseudomonas spp., Salmonella spp., and Micrococcus sp. have been shown to possess enzymatic activities for itaconate degradation,31 the genes encoding these enzymes have only recently been identified in Yersinia pestis and Pseudomonas aeruginosa.(32) The pathway comprises three enzymatic reactions (Figure 1A). The first reaction is catalyzed by itaconate CoA transferase (Ict) which converts itaconate to itaconyl-CoA. The CoA ester is subsequently hydrated to (S)-citramalyl-CoA by itaconyl-CoA hydratase (Ich) which is then cleaved into acetyl-CoA and pyruvate by (S)-citramalyl-CoA lyase (Ccl). The production of the Ict and Ich homologues (RipA and RipB, respectively) by Salmonella enterica was shown to be strongly induced after macrophage infection.33 The upregulation of ripA and ripB was suggested by Sasikaran and co-workers to result from macrophagic itaconate secretion as part of the defense mechanism against pathogenic bacteria.32,34 Most likely, the promoters of the gene clusters encoding the enzymes for itaconate catabolism in Y. pestis and P. aeruginosa harbor regulatory elements required for transcription of these genes in the presence of itaconate. Interestingly, a gene encoding a LysR-type transcriptional regulator (LTTR, here termed ItcR) is located in the opposite direction of both the Y. pestis ccl-ich-ict operon (also referred to as ripABC operon) and the P. aeruginosa putative six-gene operon encoding Ich, Ict, Ccl, and three other proteins (Figure 1B). The genes coding for LTTRs are occasionally transcribed in divergent orientation with respect to the cluster of genes they regulate,35 which led to the hypothesis that transcription of the Y. pestis and P. aeruginosa itaconate degradation pathway genes is mediated by their corresponding divergently oriented LTTR genes from an inducible promoter located in their intergenic regions.
Figure 1.

Bacterial itaconate degradation pathway. (A) The enzymes involved in bacterial itaconate degradation include itaconate CoA transferase (Ict), itaconyl-CoA hydratase (Ich), (S)-citramalyl-CoA lyase (Ccl). (B) The gene clusters in Y. pestis, Y. pseudotuberculosis, and P. aeruginosa encoding the enzymes required for itaconate catabolism. Divergently oriented LTTR genes (itcR) and putative itaconate-inducible promoters are depicted. Gene names and locus tags are shown under the schematic illustration of each gene cluster.
Itaconic Acid-Inducible Gene Expression Is Mediated by a LysR-Type Transcriptional Regulator
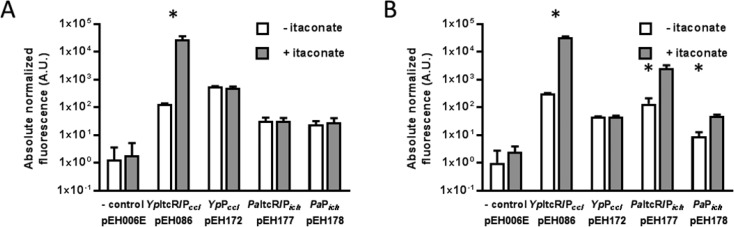
To test our hypothesis that the itaconate degradation pathway is controlled by the transcriptional regulator and corresponding inducible promoter, we cloned both the P. aeruginosa PAO1 and the Yersinia pseudotuberculosis YPIII DNA fragments with a putative itaconate-inducible system, containing an intergenic region with promoters Pich and Pccl, respectively, and gene of the transcriptional regulator (itcR) (Figure 1B), into the reporter plasmid pEH006. The latter plasmid has previously been demonstrated to be suitable for the analysis of inducible systems (Table 1).36 The nucleotide sequence of the Y. pseudotuberculosis itaconate-inducible system is identical to the Y. pestis one, except for three single nucleotide polymorphisms in ItcR coding sequence (YPK_2265) resulting in one amino acid difference. The nucleotide sequences of the intergenic regions containing putative itaconate-inducible promoters are provided in Figure S1. To investigate the potential applicability of the two putative itaconate-inducible systems across different species, red fluorescent protein (RFP) reporter gene expression in response to itaconate was measured by fluorescence output in the model gammaproteobacterium E. coli MG1655 and the betaproteobacterium Cupriavidus necator H16. The latter is a model chemolithoautotroph with the ability to produce energy and chemicals from carbon dioxide and is therefore of interest in biotechnological applications. Single time point fluorescence measurements for E. coli and C. necator harboring the putative itaconate-inducible systems, composed of transcriptional regulator and inducible promoter (ItcR/P), were performed in the absence and presence of itaconate (Figure 2). In both microorganisms, reporter gene expression from the Y. pseudotuberculosis (Yp) inducible system (pEH086) is induced significantly (p < 0.01) 6 h after supplementation with 5 mM itaconate (215-fold in E. coli and 105-fold in C. necator, Figure 2 panels A and B, respectively). In contrast, the P. aeruginosa (Pa) inducible system PaItcR/Pich (pEH177) does not mediate reporter gene expression in response to itaconate in E. coli, whereas in C. necator it demonstrates an 18.5-fold induction. In comparison, in E. coli MG1655, the level of induction mediated by the Y. pseudotuberculosis itaconate-inducible system is considerably higher than the commonly used l-arabinose-inducible system which is subject to catabolite repression. A culture of E. coli MG1655 harboring pEH006 demonstrated a 39-fold increase in RFP expression 6 h after addition of l-arabinose to a final concentration of 0.1% (w/v) in minimal medium.
Table 1. Plasmids Used and Generated in This Study.
| plasmid | characteristic | reference or source |
|---|---|---|
| pBBR1MCS-2-PphaC-eyfp-c1 | Kanr; broad host range vector used to amplify the origin of replication | (37) |
| pEH006 | Cmr; modular vector for the evaluation of inducible systems; ParaC-araC-TrrnB1 and ParaBAD-T7sl-EcRBS-rfp-Tdbl | (36) |
| pEH006E | Cmr; promoterless pEH006 | (36) |
| pEH086 | Cmr; PitcR-itcR-TrrnB1 and Pccl-rfp-Tdbl from Y. pseudotuberculosis YPIII genomic DNA | this study |
| pEH164 | Cmr; ParaC-araC-Tdbl, ParaBAD-T7sl-EcRBS-TrrnB2, YpPitcR-YpitcR-TrrnB1 and YpPccl-rfp-Tdbl | this study |
| pEH165 | Cmr; ParaC-araC-Tdbl, ParaBAD-T7sl-EcRBS-cadA-TrrnB2, YpPitcR-YpitcR-TrrnB1 and YpPccl-rfp-Tdbl | this study |
| pEH172 | Cmr; Pccl-rfp-Tdbl from Y. pseudotuberculosis YPIII genomic DNA | this study |
| pEH177 | Cmr; PitcR-itcR-TrrnB1 and Pich-rfp-Tdbl from P. aeruginosa PAO1 genomic DNA | this study |
| pEH178 | Cmr; Pich-rfp-Tdbl from P. aeruginosa PAO1 genomic DNA | this study |
Figure 2.
Influence of ItcR on inducible gene expression. Absolute normalized fluorescence (in arbitrary units) of (A) E. coli MG1655 and (B) C. necator H16 harboring the Y. pseudotuberculosis (Yp) and P. aeruginosa (Pa) itaconate-inducible systems composed of promoter and transcriptional regulator (ItcR/P), and promoter-only (P) implementation in the absence and presence of 5 mM itaconate. Single time-point fluorescence measurements were taken 6 h after inducer addition. The promoterless reporter plasmid pEH006E was employed as negative control. Error bars represent standard deviations of three biological replicates. Asterisks indicate statistically significant induction values for p < 0.01 (unpaired t test).
To confirm that itaconate-inducible reporter gene expression is indeed controlled by the episomally encoded ItcR, their coding sequences were removed from the vectors containing YpItcR/Pccl and PaItcR/Pich. Single time point fluorescence measurements were repeated for E. coli and C. necator solely harboring the itaconate-inducible promoters in the absence and presence of itaconate (Figure 2). Without YpItcR, induction of reporter gene expression from the Y. pseudotuberculosis itaconate-inducible promoter (YpPccl, pEH172) is abolished in both microorganisms. This confirms that transcription of the Y. pseudotuberculosis itaconate degradation pathway genes is mediated by their divergently oriented itcR gene and that neither of the two tested microorganisms encodes cross-activating TR homologues. In E. coli, the level of normalized fluorescence from PaPich (pEH178) and PaItcR/Pich (pEH177) is higher than the negative control, indicating that the promoter itself is active. However, the normalized fluorescence levels are of equal height, suggesting that the TR might not be produced or able to interact with its cognate operator sequence to activate gene expression in the presence of the effector. Interestingly, in C. necator, even though the coding sequence of PaItcR was removed from the plasmid, reporter gene expression from PaPich (pEH178) is induced significantly (p < 0.01) after the addition of itaconate. A PaItcR homology search in C. necator revealed the presence of several chromosomally encoded LTTRs exhibiting 40–50% protein sequence identity (96–98% coverage). One of the LTTR genes is located within close proximity to the cluster that includes genes potentially involved in itaconate degradation similar to P. aeruginosa (Figure S2). C. necator ItcR homologues can potentially activate gene expression from the heterologous P. aeruginosa itaconate-inducible promoter even in the absence of its corresponding LTTR. However, since both the induction level, and the absolute normalized fluorescence in the presence of itaconate, are higher in the plasmid carrying PaItcR/Pich (pEH177) than the one carrying PaPich (pEH178) alone (by 3.5- and 52-fold, respectively), it can be concluded that PaItcR is involved in activation of gene expression of the itaconate degradation cluster of genes in P. aeruginosa and therefore enables persistence in macrophages. The finding that expression of the genes encoding enzymes involved in itaconate catabolism is mediated by their divergently oriented LTTR genes may aid in developing new antimicrobial agents.
Sensor Characterization
Because of its functionality in both tested microorganisms, regulator-dependent orthogonality and high level of induction, the itaconate-inducible system from Y. pseudotuberculosis was selected to be further characterized. The sensor was evaluated for its kinetics—the time that is required for the system to respond to a change in itaconate levels; dynamics—the range of inducer concentration that mediates a linear fluorescence output; and inducer-dependent orthogonality—the specificity toward itaconate.
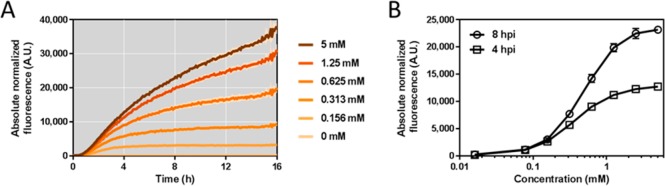
E. coli MG1655 was transformed with the plasmid harboring the YpItcR/Pccl inducible system (pEH086), cultivated in M9 minimal medium, and fluorescence output was monitored over time after supplementation with different concentrations of itaconate. As can be seen from the fluorescence curve of induction kinetics, reporter gene expression is activated immediately after inducer addition, taking into account the time that is required for RFP maturation (Figure 3A).38 This immediate response suggests that the system is solely controlled by ItcR and that it is not affected by host-originating TRs. Furthermore, it suggests that itaconate is a primary inducing molecule, which starts instantly to be uptaken by or diffused into the E. coli cells in minimal medium. It should be noted that the growth was similar for all itaconate concentrations tested.
Figure 3.
Kinetics and dynamics of the YpItcR/Pccl inducible system. (A) Absolute normalized fluorescence of E. coli MG1655 harboring the YpItcR/Pccl inducible system (pEH086) in response to different concentrations of itaconate added at time zero. The standard deviation of three biological replicates is shown as a lighter color ribbon displayed lengthwise of the induction kinetics curve. For the lower concentrations, the standard deviation is too small to be visible. (B) Dose response curve of the YpItcR/Pccl inducible system in E. coli MG1655, illustrating the correlation between inducer concentration and fluorescence output 4 and 8 h post-induction (hpi) with itaconate. Error bars represent standard deviations of three biological replicates.
The correlation between extracellular inducer concentration and fluorescence output, 4 and 8 h after itaconate supplementation, is illustrated in the dose response curve (Figure 3B). It indicates that gene expression can be tuned in the range of approximately 0.07 to 0.7 mM for a linear fluorescence output. The minimum concentration of exogenously added itaconate required for activation of the system is approximately 0.016 mM. The dose response curve indicates a saturation of the YpItcR/Pccl inducible system for itaconate levels above 2.5 mM. However, in order for this system to be applied as biosensor for concentrations of more than 2.5 mM, its elements require modification. This is commonly accomplished by promoter or protein engineering, both strategies aiming to alter the binding affinity of the TR for either the operator sequence or the ligand itself.39−41 Notably, the concentration of exogenously added itaconate required to induce the system in E. coli MG1655 is lower in LB medium than in M9 minimal medium. Four hours after the addition of 0.016 mM itaconate, reporter gene expression is induced 7.7-fold in LB medium compared to a culture without itaconate (Figure S3). This is in contrast to a 1.4-fold induction in M9 minimal medium. The dose response curve indicates that the itaconate concentration, required for a linear fluorescence output in LB medium, ranges between approximately 0.016–0.16 mM (Figure S3). Despite a 5-fold reduced induction threshold for itaconate, the linear output range of the YpItcR/Pccl inducible system spans 1 order of magnitude, similar to what is observed in M9 minimal medium. This suggests that different growth conditions can contribute to the variation of both lower and upper induction thresholds, whereas the magnitude of system response is likely to remain constant.
In addition, the analysis of extracellular and intracellular itaconate by using high-performance liquid chromatography (HPLC) coupled with ultraviolet (UV) spectroscopy shows no significant change in the itaconate concentration during the 12 h period in the actively growing E. coli culture (Table 2). This demonstrates that itaconate is not metabolized and therefore is a primary inducing molecule. Moreover, the analysis confirms that itaconate is taken up by or diffuses into the E. coli cell and reaches a relatively high concentration of at least 1.3 mM after 6 h. It should be noted that the actual intracellular molar concentration could be even higher, since our approximation uses assumption that the intracellular cell volume is equal to the total cell volume including the space occupied by cell membranes, lipids, etc. Interestingly, the intracellular itaconate concentration becomes reduced when E. coli cells reach the stationary phase (12-h time point, Table 2); however, the total itaconate concentration in the culture remains unchanged.
Table 2. Extracellularly Added and Intracellularly Produced Itaconate Distribution between Supernatant and Cells in E. coli Culture Grown in LB Medium.
| itaconate
extracellularly added | |||||
|---|---|---|---|---|---|
| molar
concentration (mM)a |
concentration
in cell culture (mg/L) |
||||
| time (h) | extracellular | intracellular | resulting from supernatant | resulting from cells | Total |
| 0 | 2.5b | ndc | 325.253 | nd | 325.253 |
| 6 | 2.454 ± 0.050 | 1.309 ± 0.132 | 319.242 ± 6.437 | 0.685 ± 0.067 | 319.927 ± 6.437 |
| 12 | 2.462 ± 0.059 | 0.551 ± 0.058 | 320.243 ± 7.715 | 0.411 ± 0.074 | 320.654 ± 7.715 |
| itaconate
intracellularly produced | |||||
|---|---|---|---|---|---|
| molar
concentration (mM) |
normalized
concentration in cell culture (mg/L/OD) |
||||
| time (h) | extracellular | intracellular | resulting from supernatant (% of total) | resulting from cells (% of total) | total |
| 0 | nd | nd | nd | nd | nd |
| 18 | 0.071 ± 0.021 | 0.145 ± 0.008 | 2.149 ± 0.638 (98.67) | 0.029 ± 0.003 (1.33) | 2.178 |
| 36 | 0.242 ± 0.117 | 0.241 ± 0.181 | 7.010 ± 2.506 (99.38) | 0.044 ± 0.028 (0.62) | 7.054 |
Arithmetic mean ± standard deviation is derived using data of three biological replicates.
Itaconate concentration added to cell culture at 0 h time point.
Not detected (nd).
Sensor Specificity
The YpItcR/Pccl inducible system was analyzed for cross-induction by metabolites that may activate reporter gene expression in the absence of the primary inducing molecule itaconate. These can be exogenously added compounds or intermediates naturally involved in cellular metabolism. Compounds that were investigated for cross-induction mainly include citric acid cycle intermediates and structurally similar variants thereof (Figure 4A). Evaluation of these molecules may shed light on structural features required for TR-binding and TR affinity toward itaconate. Furthermore, screening potential candidate compounds might expand the list of metabolites to be detected by TR-based controllable systems and offer the possibility to be utilized as analogue inducers to control gene expression.
Figure 4.
Inducer-dependent orthogonality of the YpItcR/Pccl inducible system. (A) Compounds that were investigated for cross-induction with the YpItcR/Pccl inducible system: itaconic acid (1), succinic acid (2), d-malic acid (3), l-malic acid (4), fumaric acid (5), oxaloacetic acid (6), l-aspartic acid (7), methylsuccinic acid (8), mesaconic acid (9), citraconic acid (10), α-ketoglutaric acid (11), l-glutamic acid (12), acetic acid (13), propionic acid (14), butyric acid (15), 3-butenoic acid (16), valeric acid (17), acrylic acid (18), methacrylic acid (19), tiglic acid (20), citric acid (21), cis-aconitic acid (22), trans-aconitic acid (23), tricarballylic acid (24), isocitric acid (25). (B) Normalized fluorescence (in %) of E. coli MG1655 harboring the YpItcR/Pccl inducible system 12 hours after addition of different compounds at a final concentration of 5 mM, relative to the fluorescence output obtained by adding 5 mM itaconate. (−) uninduced sample. Error bars represent standard deviations of three biological replicates. Asterisks indicate statistically significant induction values for p < 0.01 (unpaired t test).
The fluorescence output from cultures of E. coli MG1655 harboring the YpItcR/Pccl inducible system, and cultivated in M9 minimal medium, was monitored over time after individual addition of each compound at a final concentration of 5 or 10 mM. Normalized fluorescence levels (in %), relative to the output obtained by adding 5 mM itaconate, were determined 12 hours after compound supplementation. In addition to the primary inducer itaconate and under the assumption that all tested metabolites are able to enter the cell, the compounds succinate (2), methylsuccinate (8), mesaconate (9), α-ketoglutarate (11), propionate (14), butyrate (15), 3-butenoate (16), acrylate (18), methacrylate (19), cis-aconitate (22), and trans-aconitate (23) induce reporter gene expression at a final concentration of 5 mM with high statistical significance (p < 0.01) (Figure 4B). Of these 11 compounds, succinate, mesaconate, propionate, butyrate, 3-butenoate, cis-aconitate, and trans-aconitate demonstrated a significant increase in RPF expression at a final concentration of 10 mM (Figure S4). Increased activation of reporter gene expression suggests that these inducers may exhibit a weak binding to TR inducing the system to some extent. The highest level of cross-induction is mediated by trans-aconitate. At a concentration of 10 mM, it reached 9.9% of the absolute normalized fluorescence that was achieved by using 5 mM itaconate. Since E. coli has not been reported to encode a trans-aconitate decarboxylase, converting trans-aconitate into itaconate, induction of reporter gene expression from YpPccl is more likely to be caused by ItcR promiscuity rather than by decarboxylation of trans-aconitate forming itaconate.
cis-Aconitate and trans-aconitate showed more than a 2-fold change in induction level when inducer concentration was 2-fold increased from 5 to 10 mM suggesting that these compounds may activate the system at higher concentrations. To obtain a more accurate resolution of their dose responses, the YpItcR/Pccl inducible system was subjected to a range of concentrations of cis-aconitate, and trans-aconitate. Since mesaconate has been previously shown to act as CoA acceptor by YpIct, with second lowest Km after itaconate,32 this compound was also included in the dose response experiment.
A saturation in fluorescence output when using mesaconate, cis-aconitate, or trans-aconitate as inducer was not possible to obtain. All three inducers demonstrated some degree of toxicity inhibiting cell growth at higher concentrations. However, on the basis of a phenomenological model for metabolite biosensors,41 it can be postulated that the maximal dynamic range of an inducible system, which is the maximal level of expression relative to basal promoter activity, is not affected by metabolite-TR affinity. Therefore, the maximal dynamic range calculated for itaconate as inducer was employed to fit the dynamic range data for mesaconate, cis-aconitate, and trans-aconitate using a Hill function (Figure S5). The resulting Ki, the extracellularly added inducer concentration which mediates half-maximal RFP expression, is different for each of these compounds. They reveal that mesaconate, cis-, and trans-aconitate Ki values are higher (45.2 mM, 31.1 mM, and 13.2 mM, respectively) and therefore activate the YpItcR/Pccl inducible system at much higher extracellular concentrations than itaconate (Ki = 0.43 mM). The structural characteristics may contribute to the ability of metabolites to interact with ItcR and act as inducers. Indeed, mesaconate, cis-aconitate, and trans-aconitate have structural similarities to itaconate, with last two harboring the complete itaconate element. However, the observation that all three compounds have a much higher Ki than itaconate suggests, that for maximal activation of the YpItcR/Pccl inducible system, the unmodified itaconate structure is indispensable. It also suggests that the binding affinity of the TR to a specific ligand may play an important role. Consequently, protein engineering of ItcR may be used to change the binding affinity for itaconate. On the other hand, it cannot be excluded that the change in inducer dynamic range is affected by the differential uptake of these compounds by the E. coli cell.
It should be noted that acetate, propionate, butyrate, methylsuccinate, and mesaconate have previously been demonstrated to act as CoA acceptors by YpIct, albeit at a much higher Km than itaconate,32 suggesting that these compounds might be secondary inducers of the YpItcR/Pccl inducible system. Interestingly, their level of induction correlates with their ability to act as CoA acceptors, with acetate, propionate, and butyrate having a higher, and mesaconate having a lower Km.32 Furthermore, the catalytic efficiency (kcat/Km) of YpIct with itaconate, mesaconate, methylsuccinate, butyrate, propionate, and acetate,32 shows a high level of direct correlation with level of induction by these compounds. This suggests there might be a structural evolutionary link between enzyme (YpIct) and transcriptional regulator (ItcR), where both proteins have coevolved enabling a hierarchical ranking of metabolites as enzyme substrates and TR activators in the following order: itaconate > mesaconate > methylsuccinate > butyrate > propionate > acetate. The direct correlation between catalytic efficiency and level of induction potentially ensures that the hierarchy is supported at the gene expression and enzyme activity levels by securing the highest level of YpIct synthesis and highest catalytic efficiency when itaconate is present in the environment. Overall, the YpItcR/Pccl inducible system demonstrates a high specificity toward itaconate and may therefore be used in combination with other inducible systems to orthogonally control gene expression in biosynthetic pathways composed of multiple genes.
Biosensor-Assisted Optimization of Itaconic Acid Production
Itaconic acid can be synthesized by decarboxylation of the citric acid cycle intermediate cis-aconitic acid. This reaction is catalyzed by cis-aconitate decarboxylase (CadA). The A. terreus cadA gene has previously been expressed in E. coli for the biosynthesis of itaconate by using either a constitutive promoter, or an inducible T7 polymerase-based expression system.13,20,42 Overexpression of cadA was reported to impair cellular growth,42 suggesting that fine-tuning of CadA levels is essential to ensure optimal metabolic flux. Even though the pathway for itaconate biosynthesis in E. coli solely requires the introduction of one additional gene, balancing its expression and quantitatively evaluating its impact on itaconate production can be laborious when using standard analytical techniques. We decided to apply the YpItcR/Pccl inducible system to monitor itaconate production by fluorescence output in response to different levels of CadA.
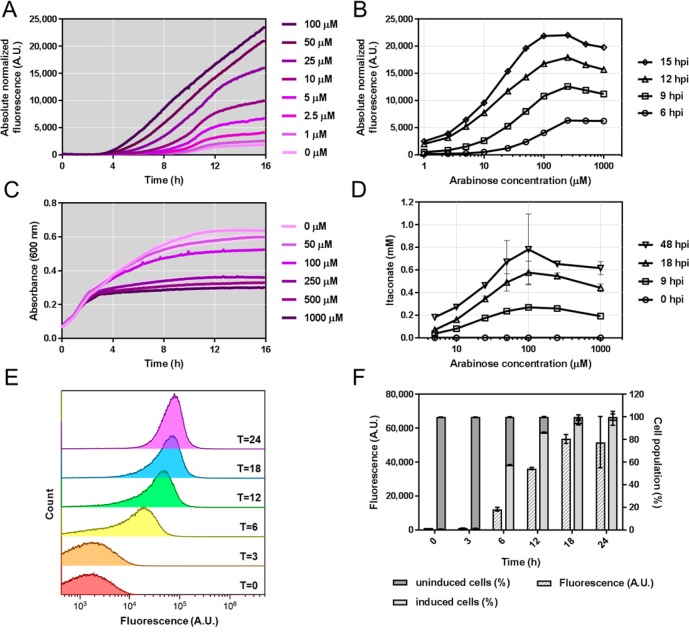
A single plasmid (pEH165) was constructed that contains two modules: one for itaconate production and one for itaconate sensing (Figure 5). The A. terreus cadA (ATEG_09971) coding sequence was cloned downstream of the arabinose-inducible system and a T7 mRNA stem-loop structure sequence, which was incorporated to enhance cadA mRNA stability.43 The itaconate sensing module contains the YpItcR/Pccl inducible system in combination with the rfp reporter gene. The addition of l-arabinose to cells harboring this plasmid was expected to initiate cadA expression, resulting in biosynthesis of itaconate and subsequent activation of reporter gene expression. E. coli TOP10 was transformed with plasmid pEH165, and cells in early exponential growth phase were transferred to a 96-well microtiter plate. Subsequently, growth and fluorescence were monitored over time after supplementation with different concentrations of l-arabinose ranging from 1 to 1000 μM. As it can be seen in the fluorescence curve of induction kinetics, higher concentrations of l-arabinose mediate a faster fluorescence output (Figure 6A). Reporter gene expression above background levels can be observed 150 min after the addition of 100 μM l-arabinose, whereas 10 μM require about 1 h more. The dose response curve indicates that maximum absolute normalized fluorescence is achieved by supplementation with 250 μM l-arabinose (Figure 6B). This suggests that expression of cadA can be fine-tuned when using inducer concentrations in the range between 1 and 100 μM. l-Arabinose concentrations of 0.5 and 1 mM, however, appear to negatively impact reporter gene expression, indicating a drop in itaconate levels. The negative effect of high inducer levels becomes even more evident from the absorbance data, showing that l-arabinose concentrations of 250 μM and more reduce cell density considerably (Figure 6C). Most likely, this behavior results from an increased metabolic burden caused by overproduction of CadA, as mentioned earlier.42
Figure 5.

Schematic illustration of the plasmid containing both an itaconate production and sensing module. Exogenous addition of l-arabinose initiates synthesis of the cis-aconitate decarboxylase CadA which converts cis-aconitate into itaconate. RFP reporter gene expression is subsequently mediated by ItcR in the presence of itaconate.
Figure 6.
Biosensor-assisted optimization of itaconate production. (A) Absolute normalized fluorescence of E. coli TOP10 harboring pEH165, grown in microtiter plates, in response to 1–100 μM of l-arabinose supplemented at time zero. The means of three biological replicates are presented. Error bars are too small to be visible. (B) Dose response curve of E. coli TOP10 harboring pEH165, grown in microtiter plates, 6, 9, 12, and 15 h post induction (hpi) with 1–1000 μM of l-arabinose. The means of three biological replicates are presented. Error bars are too small to be visible. (C) Absorbance at 600 nm of E. coli TOP10 harboring pEH165, grown in microtiter plates, in response to 50–1000 μM of l-arabinose supplemented at time zero. The means of three biological replicates are presented. The standard deviation for 50 μM of inducer is illustrated as lighter color ribbon displayed lengthwise of the growth curve. The error bars for the other inducer concentrations are too small to be visible. (D) Itaconate titers of E. coli TOP10 harboring pEH165, grown in small-volume cultures, 0, 9, 18, and 48 h post induction with 5, 10, 25, 50, 100, 250, and 1000 μM of l-arabinose. Error bars represent standard deviations of three biological replicates. (E) Flow cytometric analysis of E. coli TOP 10 harboring pEH165, grown in small-volume cultures, in response to 100 μM of l-arabinose. Samples were taken 0, 3, 6, 12, 18, and 24 h after inducer addition. For the time points T = 6, T = 12, T = 18 and T = 24, fluorescence from more than 99% of cells are displayed in the histogram, whereas for time points T = 0 and T = 3, less than 25% of cells are below 429 A.U. fluorescence threshold in the histogram. (F) Fluorescence intensity (median) and percentage of uninduced and induced cells corresponding to the data presented in panel E. Error bars represent standard deviations of three biological replicates.
To quantitatively validate the data which was generated from cultures grown in microtiter plates, the experiment was repeated in small culture volumes. E. coli TOP10 pEH165 was grown in 50 mL culture tubes, and expression of cadA was initiated by supplementation with different concentrations of l-arabinose. To determine itaconate titers, samples were subjected to analysis using HPLC-UV. The highest itaconate concentration was achieved in cultures containing 100 μM l-arabinose, resulting in 0.78 ± 0.31 mM itaconate 48 h after inducer addition (Figure 6D). This represents a 4.3-fold improvement over cultures containing only 5 μM l-arabinose. It should be noted that these and data in Table 2 demonstrate that the intracellularly synthesized itaconate was actively excreted or diffused into the media.
The addition of an excessive amount of 1 mM inducer also decreased itaconate levels by 1.3-fold. Therefore, the quantitative data obtained from the small-volume cultures match well with the fluorescence output measured in the microtiter plate (compare Figure 6B and 6D). Particularly when itaconate titers are OD-normalized, 250 μM l-arabinose results in the highest OD-normalized itaconate titer (Figure S6). This experiment illustrates that cadA expression needs to be carefully fine-tuned to guarantee both optimal metabolic flux and viability of cells.
Moreover, using 100 μM of l-arabinose yields itaconate concentrations of 0.24, 0.56, and 0.78 mM after 9, 18, and 48 h post-induction, respectively (Figure 6D). These itaconate concentrations fall within the linear range of dose response (Figure 3B) and result in a fluorescence output with a unimodal distribution suggesting that almost all cells in the population were activated (Figure 6E,F). As demonstrated here, the itaconate biosensor can be employed to facilitate a fluorescence-based high-throughput screen to evaluate various conditions for their impact on itaconate biosynthesis.
Correlation between Biosensor Output and Itaconate Concentration
In addition to HPLC-UV analysis, the samples from the small-volume cultures of E. coli TOP10 pEH165 were analyzed for fluorescence output. The obtained data were used to evaluate whether quantitatively determined itaconate titers correlate with reporter gene expression from the biosensor. The five tested inducer concentrations that did not impair bacterial growth produced a 59-fold range in fluorescence after 6 h (Figure 7A). The addition of 25, 50, and 100 μM l-arabinose resulted in itaconate titers that were sufficiently high to be detected by the biosensor. Notably, in the linear response range of the YpItcR/Pccl inducible system, the fluorescence output shows a high level of correlation with HPLC-UV measured extracellular itaconate titers (Figure 7A) and unimodal fluorescence distribution in the cell population (Figure 7B). l-Arabinose concentrations of 5 and 10 μM result in a bimodal fluorescence response, suggesting an all-or-none induction in which intermediate inducer concentrations give rise to subpopulations. However, when different levels of itaconate are synthesized in the range between 0.1 and 0.78 mM, which corresponds to the linear response range, the fluorescence output becomes unimodal (Figures 6D,E and 7A). This confirms that for itaconate levels in the linear range, the YpItcR/Pccl inducible system mediates a homogeneous induction of cells, exemplifying its potential to fine-tune gene expression across cell populations and to be utilized as a quantitatively reliable biosensor.
Figure 7.
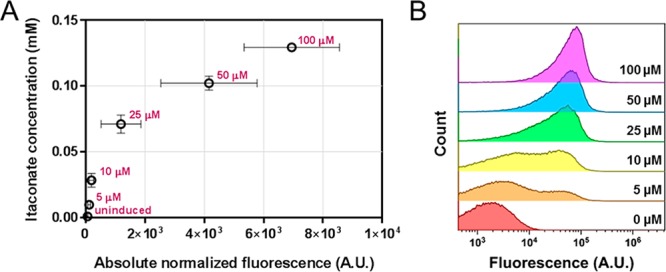
Correlation between biosensor output and itaconate concentration. (A) Absolute normalized fluorescence values of E. coli TOP10 pEH165 are correlated with their corresponding itaconate concentration in the culture supernatant. Samples were taken 6 h after inducer addition. The different concentrations of exogenously added l-arabinose, ranging from 5 to 100 μM, are highlighted. Error bars represent standard deviations of three biological replicates. (B) Flow cytometric analysis of samples from panel A. For l-arabinose (inducer) concentrations of 25, 50, and 100 μM, fluorescence from more than 99% of cells are displayed in the histogram, whereas for concentrations of 10, 5, and 0 μM, less than 2, 10, and 25% of cells, respectively, are below 429 A.U. fluorescence threshold in the histogram.
Methods
Base Strains and Media
E. coli TOP10 (Invitrogen) was used for cloning, plasmid propagation, and biosynthesis of itaconate. RFP fluorescence assays for biosensor characterization were performed in wild type E. coli MG1655 (DSMZ 18039) and C. necator H16 (ATCC 17699). Bacterial strains were propagated in LB medium. For reporter gene assays, E. coli MG1655 was cultivated in M9 minimal medium44 supplemented with 1 μg/L thiamine, 20 μg/mL uracil45 and 0.4% (w/v) glucose, unless otherwise indicated. C. necator reporter gene assays were performed in minimal medium46 containing 0.4% (w/v) sodium gluconate. Antibiotics were added to the growth medium at the following concentrations: 25 μg/mL, or 50 μg/mL chloramphenicol for E. coli, or C. necator, respectively. E. coli TOP10 was grown at 30 or 37 °C. For comparison, both E. coli MG1655 and C. necator were cultivated at 30 °C.
Cloning and Transformation
Plasmid minipreps were carried out using the New England BioLabs (NEB) Monarch Plasmid Miniprep Kit. Microbial genomic DNA was extracted employing the GenElute Bacterial Genomic DNA Kit (Sigma). For cloning, DNA was amplified by PCR using Phusion High-Fidelity DNA polymerase from NEB in 50 μL reactions under recommended conditions. Restriction enzymes and NEBuilder Hifi DNA assembly master mix were purchased from NEB, and reactions were set up according to the manufacturer’s protocol. The NEB Monarch DNA Gel Extraction Kit was used to extract gel-purified linearized DNA which was subsequently used for cloning.
Chemical competent E. coli were prepared and transformed by heat shock as previously described.44 Electrocompetent C. necator were prepared and transformed as reported by Ausubel et al.47
Plasmid Construction
Oligonucleotide primers were synthesized by Sigma-Aldrich (Table S1). Plasmids were constructed by employing either the NEBuilder Hifi DNA assembly method according to the manufacturer’s protocol or by restriction enzyme-based cloning procedures.44 Constructs were verified by DNA sequencing (Source BioScience, Nottingham, UK). The nucleotide sequences of pEH086 and pEH177 have been deposited in the public version of the ACS registry (https://acs-registry.jbei.org) under the accession number ACS_000716 and ACS_000717, respectively.
The itaconate-inducible systems YpItcR/Pccl and PaItcR/Pich were amplified with oligonucleotide primers EH191_f and EH190_r, EH312_f and EH311_r, respectively, from Y. pseudotuberculosis YPIII (Yp) and P. aeruginosa PAO1 (Pa) genomic DNA and cloned into pEH006 by AatII and NdeI restriction sites (resulting in plasmids pEH086 and pEH177). The itaconate-inducible promoters YpPccl and PaPich were amplified with oligonucleotide primers EH191_f and EH302_r, EH312_f and EH313_r, respectively, from Y. pseudotuberculosis YPIII and P. aeruginosa PAO1 genomic DNA and cloned into pEH006 by AatII and NdeI restriction sites (resulting in plasmids pEH172 and pEH178).
Vector pEH164 contains both the itaconate-reporter system composed of YpItcR-Pccl-rfp and the l-arabinose-inducible system including restriction sites for subsequent integration of the cis-aconitate decarboxylase cadA gene (ATEG_09971) downstream of ParaBAD. It was constructed by employing the NEBuilder Hifi DNA assembly method. Oligonucleotide primers EH011_f and EH075_r, EH015_f and EH012_r, EH078_f and EH190_r, EH083_f and EH079_r were used to amplify the replication origin and the chloramphenicol resistance gene, YpItcR-Pccl-rfp, and the l-arabinose-inducible system from pBBR1MCS-2-PphaC-eyfp-c1, pEH086, and pEH006, respectively.36,37
Vector pEH165 contains both the itaconate-production system AraC-ParaBAD-cadA and the itaconate-reporter system YpItcR-Pccl-rfp. Oligonucleotide primers EH294_f and EH293_r, EH296_f and EH295_r were used to amplify exon 1 and exon 2 of ATEG_09971 from A. terreus NIH2642 genomic DNA. The PCR products were combined with BglII/SbfI digested pEH164 and constructed by employing the NEBuilder Hifi DNA assembly method.
RFP Fluorescence Assay
RFP fluorescence was measured with an Infinite M1000 PRO (Tecan) microplate reader using 585 nm as excitation and 620 nm as emission wavelength. The gain factor was set manually to 100%. Absorbance was determined at 600 nm to normalize fluorescence by optical density. Fluorescence and absorbance readings at a single time point, and over time, were performed as described previously.36 The absolute normalized fluorescence was calculated by dividing the absolute fluorescence values by their corresponding absorbance values. Prior to normalization, both values were corrected by the autofluorescence and autoabsorbance of the culture medium.
Production of Itaconate and HPLC-UV Analysis
Real-time biosynthesis of itaconate was monitored quantitatively by high-performance liquid chromatography (HPLC) in combination with ultraviolet (UV) absorbance at 210 nm and by fluorescence output in E. coli T10 harboring pEH165. Single colonies of freshly transformed cells were used to inoculate five mL of LB medium. The preculture was incubated for 18 h at 37 °C and 200 rpm. Subsequently, it was diluted 1:100 in 6 mL of fresh LB medium. The main cultures were grown in 50 mL Falcon tubes at 30 °C and 225 rpm. At an OD600 of 0.5, 50 μL of l-arabinose stock solutions were added to achieve the final concentrations of 5, 10, 25, 50, 100, 250, and 1000 μM. One sample per biological replicate remained uninduced. Samples of 0.5 mL were taken immediately, 6, 9, 12, 18, 24, and 48 h after inducer supplementation. They were directly used for evaluation by flow cytometry, OD600, and fluorescence measurement. The remaining sample was centrifuged for 5 min at 16 000g, and the cell-free supernatant was subjected to HPLC-UV analysis as reported previously.36
Metabolite Extraction
To determine intracellular itaconate concentrations when added extracellularly or synthesized intracellularly, cultures of E. coli TOP10 harboring pEH164 or pEH165 were grown overnight to saturation and diluted 1:100 in 200 mL LB medium. The main cultures were grown in 1-L shake flasks at 30 °C and 225 rpm. At an OD600 of 0.5, inducers were added at final concentrations of 2.5 mM itaconate or 100 μM l-arabinose to cultures of E. coli TOP10 harboring pEH164 or pEH165, respectively. Samples of cells containing pEH164 were taken 0, 6, and 12 h after addition of itaconate. Samples of cells containing pEH165 were taken 0, 18, and 36 h after addition of l-arabinose. Each time, the culture volume corresponding to an OD600 of 50 was centrifuged for 10 min at 16 000g. The supernatant was removed and stored at −80 °C for HPLC analysis. Subsequently, the cell pellet was washed once in 1 mL of phosphate buffered saline (PBS), transferred to a microcentrifuge tube and centrifuged as before. The supernatant was completely removed, and the pellet was weighed using fine balance and frozen overnight at −80 °C.
The extraction of intracellular metabolites including itaconate was performed as described previously48 with modifications as described below. Briefly, 250 μL of −40 °C cold methanol–water solution (60% v/v) was added to the wet cell pellet with the volume of 50–70 μL. Subsequently, the sample was mixed vigorously using vortex until completely resuspended. The cell suspension was frozen at −80 °C for 30 min, thawed on ice, and vortexed vigorously for 1 min. This step was repeated three times before the sample was centrifuged at −10 °C and 26 000g for 20 min. The supernatant was collected and kept at −80 °C. To the pellet, another 250 μL of −40 °C cold methanol–water solution (60% v/v) was added. The cells were resuspended completely using vortex, three freeze–thaw cycles were performed as above, and then the cells were centrifuged as before. The supernatant was pooled with the first collection and stored at at −80 °C until subjected to HPLC analysis.
Calculation of Intracellular Itaconate Concentration in Cell Culture
The total cell volume (Vpellet) in the sample was calculated by dividing the weight of wet cell pellet by the cell density of 1.105 g/mL.49 Together with the volume of extraction solvent added to the sample, Vpellet was used to calculate the dilution factor required to determine the intracellular molar concentration of itaconate. Subsequently, the intracellular itaconate concentration in the cell culture (Cintracellular/CC) was calculated using equation:
The remaining parameters correspond to the formula weight of itaconic acid (FWitaconic acid), the intracellular molar concentration of itaconate determined by HPLC-UV (Cmolar) and the culture volume sampled (Vculture).
Flow Cytometry
Cells were analyzed for induction homogeneity by flow cytometry. The culture sample was centrifuged for 4 min at 5000g. Subsequently, the cell pellet was resuspended in cold and sterile filtered PBS to an OD600 of 0.01 and kept on ice until analyzed using an Astrios EQ flow cytometer (Beckman Coulter) equipped with a 561 nm laser and a 614/20 nm emission band-pass filter. The voltage of photomultiplier tube (PMT) was set to 400 V. The area and height gain was adjusted to 1.0. For each sample, at least 100 000 events were collected. The data was analyzed using the software Kaluza 1.5 (Beckman Coulter). To determine the percentage of induced cells, gating was performed on the uninduced sample to include 99% of cells. The same gate was subsequently applied to each induced sample.
Calculation of Half-Maximal RFP Expression
Because of toxicity at higher levels, the concentrations of mesaconate, cis-aconitate, and trans-aconitate, which mediate half-maximal RFP expression (Ki), were predicted using a phenomenological model as described previously.41 The model describes the change in dynamic range of an inducible system as a function of inducer concentration. It assumes that (1) the maximum dynamic range of a biosensor (μmax) remains constant as long as the genetic context does not change, and (2) Ki is dependent on metabolite-TR affinity.
The dynamic range (μ, also referred to as induction factor) for each concentration of itaconate was calculated using the absolute normalized fluorescence values from the time course experiment 6 h after itaconate addition. After subtraction of the basal output, the resulting dynamic range was fit to the corresponding inducer concentration using the Hill function:
The remaining parameters correspond to concentration of inducer (I), and the Hill coefficient (h). Subsequently, the itaconate μmax was used as fixed parameter to calculate Ki for mesaconate, cis-aconitate, and trans-aconitate employing the same Hill function. The fitted data are illustrated in Figure S5. Calculations were performed using Prism GraphPad software version 7.03.
Acknowledgments
This work was supported by the Biotechnology and Biological Sciences Research Council [grant number BB/L013940/1] (BBSRC); and the Engineering and Physical Sciences Research Council (EPSRC) under the same grant number. We thank University of Nottingham for providing SBRC-DTProg Ph.D. studentship to E.H., Swathi Alagesan for preliminary work on itaconate production in C. necator H16, Matthew Abbott and David Onion for assistance with HPLC analysis and flow cytometry, Thomas Millat for discussion on data analysis, Amy Slater, Carolina Paiva, Matthias Brock, and Elena Geib for gifting Y. pseudotuberculosis, P. aeruginosa and A. terreus genomic DNA and all members of SBRC who helped to carry out this research.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acssynbio.8b00057.
Further experimental details for plasmid construction and additional figures described in the main text (PDF)
Author Contributions
E.H. and N.M designed the study. E.H. performed the experiments. E.H., N.M., and N.P.M. analyzed the data and wrote the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Clomburg J. M.; Crumbley A. M.; Gonzalez R. (2017) Industrial biomanufacturing: The future of chemical production. Science 355 (6320), aag0804. 10.1126/science.aag0804. [DOI] [PubMed] [Google Scholar]
- Latif H.; Zeidan A. A.; Nielsen A. T.; Zengler K. (2014) Trash to treasure: production of biofuels and commodity chemicals via syngas fermenting microorganisms. Curr. Opin. Biotechnol. 27, 79–87. 10.1016/j.copbio.2013.12.001. [DOI] [PubMed] [Google Scholar]
- Keasling J. D. (2010) Manufacturing molecules through metabolic engineering. Science 330 (6009), 1355–1358. 10.1126/science.1193990. [DOI] [PubMed] [Google Scholar]
- Okabe M.; Lies D.; Kanamasa S.; Park E. Y. (2009) Biotechnological production of itaconic acid and its biosynthesis in Aspergillus terreus. Appl. Microbiol. Biotechnol. 84 (4), 597–606. 10.1007/s00253-009-2132-3. [DOI] [PubMed] [Google Scholar]
- Werpy T., Petersen G., Aden A., Bozell J., Holladay J., White J., Manheim A., Eliot D., Lasure L., and Jones S. (2004) Top value added chemicals from biomass. Vol. 1-Results of screening for potential candidates from sugars and synthesis gas, Department of Energy, Washington DC. [Google Scholar]
- Choi S.; Song C. W.; Shin J. H.; Lee S. Y. (2015) Biorefineries for the production of top building block chemicals and their derivatives. Metab. Eng. 28, 223–239. 10.1016/j.ymben.2014.12.007. [DOI] [PubMed] [Google Scholar]
- Bentley R.; Thiessen C. P. (1957) Biosynthesis of itaconic acid in Aspergillus terreus: I. Tracer studies with C14-labeled substrates. J. Biol. Chem. 226 (2), 673–687. [PubMed] [Google Scholar]
- Haskins R.; Thorn J.; Boothroyd B. (1955) Biochemistry of the Ustilaginales: XI. Metabolic products of Ustilago zeae in submerged culture. Can. J. Microbiol. 1 (9), 749–756. 10.1139/m55-089. [DOI] [PubMed] [Google Scholar]
- Tabuchi T.; Sugisawa T.; Ishidori T.; Nakahara T.; Sugiyama J. (1981) Itaconic acid fermentation by a yeast belonging to the genus Candida. Agric. Biol. Chem. 45 (2), 475–479. 10.1080/00021369.1981.10864534. [DOI] [Google Scholar]
- Strelko C. L.; Lu W.; Dufort F. J.; Seyfried T. N.; Chiles T. C.; Rabinowitz J. D.; Roberts M. F. (2011) Itaconic acid is a mammalian metabolite induced during macrophage activation. J. Am. Chem. Soc. 133 (41), 16386–16389. 10.1021/ja2070889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes T.; Michelucci A.; Hiller K. (2015) Itaconic acid: the surprising role of an industrial compound as a mammalian antimicrobial metabolite. Annu. Rev. Nutr. 35, 451–473. 10.1146/annurev-nutr-071714-034243. [DOI] [PubMed] [Google Scholar]
- Geiser E.; Przybilla S. K.; Friedrich A.; Buckel W.; Wierckx N.; Blank L. M.; Bölker M. (2016) Ustilago maydis produces itaconic acid via the unusual intermediate trans-aconitate. Microb. Biotechnol. 9 (1), 116–126. 10.1111/1751-7915.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A.; van Luijk N.; Ter Beek M.; Caspers M.; Punt P.; van der Werf M. (2011) A clone-based transcriptomics approach for the identification of genes relevant for itaconic acid production in Aspergillus. Fungal Genet. Biol. 48 (6), 602–611. 10.1016/j.fgb.2011.01.013. [DOI] [PubMed] [Google Scholar]
- Kuenz A.; Gallenmüller Y.; Willke T.; Vorlop K.-D. (2012) Microbial production of itaconic acid: developing a stable platform for high product concentrations. Appl. Microbiol. Biotechnol. 96 (5), 1209–1216. 10.1007/s00253-012-4221-y. [DOI] [PubMed] [Google Scholar]
- Hevekerl A.; Kuenz A.; Vorlop K.-D. (2014) Filamentous fungi in microtiter plates—an easy way to optimize itaconic acid production with Aspergillus terreus. Appl. Microbiol. Biotechnol. 98 (16), 6983–6989. 10.1007/s00253-014-5743-2. [DOI] [PubMed] [Google Scholar]
- Krull S.; Hevekerl A.; Kuenz A.; Prüße U. (2017) Process development of itaconic acid production by a natural wild type strain of Aspergillus terreus to reach industrially relevant final titers. Appl. Microbiol. Biotechnol. 101 (10), 4063–4072. 10.1007/s00253-017-8192-x. [DOI] [PubMed] [Google Scholar]
- Kanamasa S.; Dwiarti L.; Okabe M.; Park E. Y. (2008) Cloning and functional characterization of the cis-aconitic acid decarboxylase (CAD) gene from Aspergillus terreus. Appl. Microbiol. Biotechnol. 80 (2), 223–229. 10.1007/s00253-008-1523-1. [DOI] [PubMed] [Google Scholar]
- Levinson W. E.; Kurtzman C. P.; Kuo T. M. (2006) Production of itaconic acid by Pseudozyma antarctica NRRL Y-7808 under nitrogen-limited growth conditions. Enzyme Microb. Technol. 39 (4), 824–827. 10.1016/j.enzmictec.2006.01.005. [DOI] [Google Scholar]
- Otten A.; Brocker M.; Bott M. (2015) Metabolic engineering of Corynebacterium glutamicum for the production of itaconate. Metab. Eng. 30, 156–165. 10.1016/j.ymben.2015.06.003. [DOI] [PubMed] [Google Scholar]
- Harder B.-J.; Bettenbrock K.; Klamt S. (2016) Model-based metabolic engineering enables high yield itaconic acid production by Escherichia coli. Metab. Eng. 38, 29–37. 10.1016/j.ymben.2016.05.008. [DOI] [PubMed] [Google Scholar]
- Blazeck J.; Miller J.; Pan A.; Gengler J.; Holden C.; Jamoussi M.; Alper H. S. (2014) Metabolic engineering of Saccharomyces cerevisiae for itaconic acid production. Appl. Microbiol. Biotechnol. 98 (19), 8155–8164. 10.1007/s00253-014-5895-0. [DOI] [PubMed] [Google Scholar]
- Blazeck J.; Hill A.; Jamoussi M.; Pan A.; Miller J.; Alper H. S. (2015) Metabolic engineering of Yarrowia lipolytica for itaconic acid production. Metab. Eng. 32, 66–73. 10.1016/j.ymben.2015.09.005. [DOI] [PubMed] [Google Scholar]
- Guevarra E. D.; Tabuchi T. (1990) Accumulation of itaconic, 2-hydroxyparaconic, itatartaric, and malic acids by strains of the genus Ustilago. Agric. Biol. Chem. 54 (9), 2353–2358. 10.1271/bbb1961.54.2353. [DOI] [Google Scholar]
- Zambanini T.; Tehrani H. H.; Geiser E.; Merker D.; Schleese S.; Krabbe J.; Buescher J. M.; Meurer G.; Wierckx N.; Blank L. M. (2017) Efficient itaconic acid production from glycerol with Ustilago vetiveriae TZ1. Biotechnol. Biofuels 10 (1), 131. 10.1186/s13068-017-0809-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J. K.; Taylor N. D.; Church G. M. (2016) Biosensor-based engineering of biosynthetic pathways. Curr. Opin. Biotechnol. 42, 84–91. 10.1016/j.copbio.2016.03.005. [DOI] [PubMed] [Google Scholar]
- Rogers J. K.; Guzman C. D.; Taylor N. D.; Raman S.; Anderson K.; Church G. M. (2015) Synthetic biosensors for precise gene control and real-time monitoring of metabolites. Nucleic Acids Res. 43 (15), 7648–7660. 10.1093/nar/gkv616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S.; Rogers J. K.; Taylor N. D.; Church G. M. (2014) Evolution-guided optimization of biosynthetic pathways. Proc. Natl. Acad. Sci. U. S. A. 111 (50), 17803–17808. 10.1073/pnas.1409523111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J. K.; Church G. M. (2016) Genetically encoded sensors enable real-time observation of metabolite production. Proc. Natl. Acad. Sci. U. S. A. 113 (9), 2388–2393. 10.1073/pnas.1600375113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klement T.; Büchs J. (2013) Itaconic acid–a biotechnological process in change. Bioresour. Technol. 135, 422–431. 10.1016/j.biortech.2012.11.141. [DOI] [PubMed] [Google Scholar]
- Cooper R.; Kornberg H. (1964) The utilization of itaconate by Pseudomonas sp. Biochem. J. 91 (1), 82. 10.1042/bj0910082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W. R.; Frigan F.; Bergman E. H. (1961) Noninductive metabolism of itaconic acid by Pseudomonas and Salmonella species. J. Bacteriol. 82 (6), 905–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasikaran J.; Ziemski M.; Zadora P. K.; Fleig A.; Berg I. A. (2014) Bacterial itaconate degradation promotes pathogenicity. Nat. Chem. Biol. 10 (5), 371–377. 10.1038/nchembio.1482. [DOI] [PubMed] [Google Scholar]
- Shi L.; Adkins J. N.; Coleman J. R.; Schepmoes A. A.; Dohnkova A.; Mottaz H. M.; Norbeck A. D.; Purvine S. O.; Manes N. P.; Smallwood H. S.; et al. (2006) Proteomic analysis of Salmonella enterica serovar Typhimurium isolated from RAW 264.7 macrophages: identification of a novel protein that contributes to the replication of serovar Typhimurium inside macrophages. J. Biol. Chem. 281 (39), 29131–29140. 10.1074/jbc.M604640200. [DOI] [PubMed] [Google Scholar]
- Michelucci A.; Cordes T.; Ghelfi J.; Pailot A.; Reiling N.; Goldmann O.; Binz T.; Wegner A.; Tallam A.; Rausell A.; et al. (2013) Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc. Natl. Acad. Sci. U. S. A. 110 (19), 7820–7825. 10.1073/pnas.1218599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver P.; Peralta-Gil M.; Tabche M.-L.; Merino E. (2016) Molecular and structural considerations of TF-DNA binding for the generation of biologically meaningful and accurate phylogenetic footprinting analysis: the LysR-type transcriptional regulator family as a study model. BMC Genomics 17 (1), 686. 10.1186/s12864-016-3025-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanko E. K. R.; Minton N. P.; Malys N. (2017) Characterisation of a 3-hydroxypropionic acid-inducible system from Pseudomonas putida for orthogonal gene expression control in Escherichia coli and Cupriavidus necator. Sci. Rep. 7 (1), 1724. 10.1038/s41598-017-01850-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer D.; Jendrossek D. (2011) Interaction between poly (3-hydroxybutyrate) granule-associated proteins as revealed by two-hybrid analysis and identification of a new phasin in Ralstonia eutropha H16. Microbiology 157 (10), 2795–2807. 10.1099/mic.0.051508-0. [DOI] [PubMed] [Google Scholar]
- Campbell R. E.; Tour O.; Palmer A. E.; Steinbach P. A.; Baird G. S.; Zacharias D. A.; Tsien R. Y. (2002) A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. U. S. A. 99 (12), 7877–7882. 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazeck J.; Alper H. S. (2013) Promoter engineering: recent advances in controlling transcription at the most fundamental level. Biotechnol. J. 8 (1), 46–58. 10.1002/biot.201200120. [DOI] [PubMed] [Google Scholar]
- Taylor N. D.; Garruss A. S.; Moretti R.; Chan S.; Arbing M. A.; Cascio D.; Rogers J. K.; Isaacs F. J.; Kosuri S.; Baker D.; et al. (2016) Engineering an allosteric transcription factor to respond to new ligands. Nat. Methods 13 (2), 177–183. 10.1038/nmeth.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannan A. A.; Liu D.; Zhang F.; Oyarzún D. A. (2017) Fundamental design principles for transcription-factor-based metabolite biosensors. ACS Synth. Biol. 6 (10), 1851–1859. 10.1021/acssynbio.7b00172. [DOI] [PubMed] [Google Scholar]
- Vuoristo K. S.; Mars A. E.; Sangra J. V.; Springer J.; Eggink G.; Sanders J. P.; Weusthuis R. A. (2015) Metabolic engineering of itaconate production in Escherichia coli. Appl. Microbiol. Biotechnol. 99 (1), 221–228. 10.1007/s00253-014-6092-x. [DOI] [PubMed] [Google Scholar]
- Bi C.; Su P.; Müller J.; Yeh Y.-C.; Chhabra S. R.; Beller H. R.; Singer S. W.; Hillson N. J. (2013) Development of a broad-host synthetic biology toolbox for ralstonia eutropha and its application to engineering hydrocarbon biofuel production. Microb. Cell Fact. 12 (1), 1–10. 10.1186/1475-2859-12-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., and Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd ed., Cold Spring Harbor Laboratory Press, New York. [Google Scholar]
- Jensen K. F. (1993) The Escherichia coli K-12″ wild types″ W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J. Bacteriol. 175 (11), 3401–3407. 10.1128/jb.175.11.3401-3407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel H.; Kaltwasser H.; Gottschalk G. (1961) Ein Submersverfahren zur Kultur wasserstoffoxydierender Bakterien: Wachstumsphysiologische Untersuchungen. Arch. Microbiol. 38 (3), 209–222. 10.1007/BF00422356. [DOI] [Google Scholar]
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., and Struhl K. e. (2003) Current Protocols in Molecular Biology, John Wiley & Sons. [Google Scholar]
- Duportet X.; Aggio R. B. M.; Carneiro S.; Villas-Bôas S. G. (2012) The biological interpretation of metabolomic data can be misled by the extraction method used. Metabolomics 8 (3), 410–421. 10.1007/s11306-011-0324-1. [DOI] [Google Scholar]
- Martinez-Salas E.; Martin J.; Vicente M. (1981) Relationship of Escherichia coli density to growth rate and cell age. J. Bacteriol. 147 (1), 97–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.