Abstract
a) Purpose of Review:
The intervertebral discs (IVD) are an essential component of the spine. Degeneration of the discs, commonly due to age or injury, is a leading cause of chronic lower back pain. Despite its high prevalence, there is no effective treatment for disc disease due to limited understanding of disc at the cellular and molecular level.
b) Recent Findings:
Recent research has demonstrated the importance of the intracellular developmental pathway sonic hedgehog (Shh) during the formation and postnatal maintenance of the IVD. Recent studies corroborate that the down-regulation of SHH expression is associated with pathological changes in the IVDs and demonstrate the reactivation of the hedgehog pathway as a promising avenue for rescuing health disc structure and function.
c) Summary:
Understanding the role of developmental signaling pathways that regulate disc formation and maintenance may help develop strategies to recapitulate the same mechanism for disc treatment and hence improve the quality and longevity of patient lives.
Keywords: intervertebral disc development, intervertebral disc degeneration, Sonic hedgehog signaling, Brachyury, intervertebral disc regeneration, disc therapy
Introduction:
The intervertebral IVDs (IVDs or discs) form the fibrocartilaginous joints between vertebral bodies. They represent critical components of the vertebral column. By maintaining separation between adjacent vertebrae, each disc plays an essential role in protecting the spinal nerves, while reducing the stress of tension and compressive forces on the spine during movement (1). The IVDs are the most massive avascular and aneural tissue in the body (2). Furthermore, IVDs uniquely exhibit degenerative and aging properties early in life due to the limited diffusion of nutrients into the tissue (3, 4). Degenerative Disc Disease (DDD), a common pathological condition that adversely affects approximately 1 in 7 individuals worldwide, is associated with severe neurological consequences and is a significant cause of radiating chronic lower back pain and paresthesia in the back and extremities (5–7). Up to 45% of cases of lower back pain with or without radiculopathy can be attributed to DDD (8).
Disc Structure, Function, and Pathology:
Structurally, each IVD consists of a soft, hydrated inner structure called the nucleus pulposus (NP), and a firm, collagenous outer annulus fibrosus (AF) made of concentric lamellae surrounding the nucleus pulposi. The NP and AF are sandwiched between cartilaginous endplate (EP) that connects the disc to the adjacent vertebra (Fig. 1A and B). The NP and AF develop embryologically from the notochord and the somitocoele respectively (9–13). Extracellular matrix (ECM) proteins, such as collagens and proteoglycans comprise a significant portion of the IVD and are essential for maintenance of its structure and function. More specifically, proteoglycans consisting of a protein core made of aggrecan (ACAN) or perlecan covalently attach to highly sulfated glycosaminoglycan chains, keeping the IVD hydrated. With aging and degeneration, the IVD becomes fibrous and its water and proteoglycan content diminishes, limiting the ability of IVDs to serve their mechanical functions (14). In addition, disc degeneration is associated with hypocellularity, loss of disc height, neovascularization, and innervation (15–19).
Figure 1.
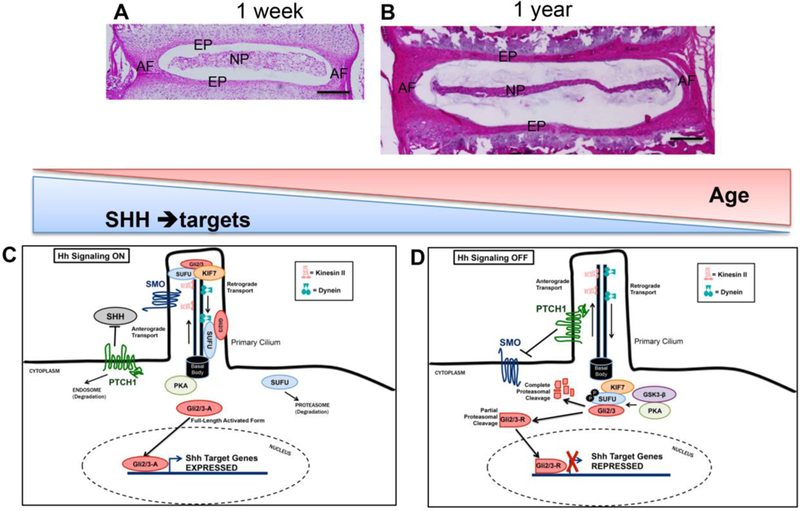
Hematoxylin and Eosin stained mid-coronal sections of lumbar intervertebral disc from one-week-old (A) and one-year-old mice show age-related structural changes. In the young, healthy disc, the Hh signaling pathway is turned on by Shh ligand produced by NP cells, resulting in the activation of the signal transduction pathway as illustrated in (C). Shh expression reduces in NP cells with age, and thus the Hh signaling pathway in (B) would be turned off, as shown in (D). Scale bar in (A) and (B) is 200 µm. NP=nucleus pulposus; AF=annulus fibrosus; EP=end plate
Recent studies have shown that several developmental cell signaling pathways including sonic hedgehog (Shh) (20, 21), Wnt (21–24), transforming growth factor beta (TGFβ) (21, 25–27), bone morphogenetic protein (BMP) (21, 25, 28), and fibroblast growth factor (FGF) (29) are also active in the postnatal IVD. Shh is of particular interest due to its essential role in patterning during embryogenesis. The local signaling mechanism is critical for maintenance of the disc microenvironment given that IVDs are avascular and thus rely on the slow diffusion of systematic signals through circulation for regulation. Hence, understanding the role of intracellular signaling pathways during physiological growth, differentiation and aging of the IVDs will provide critical insight for the development of approaches for its therapy. Biological approaches for the treatment of disc disease and associated back pain would provide an alternate to current palliative therapies, namely surgical repair and pain relievers that solely mitigate symptoms. Hence, the focus of this review is to examine the current literature on the role of Shh and the Hedgehog (Hh) signaling pathway in the formation and maintenance of the IVDs and to discuss its potential in disc therapeutics.
Hedgehog Signaling Pathway:
The Hh signaling pathway plays a crucial role in embryonic patterning and development. Thus, Hh pathway is also widely investigated for its role in postnatal tissue development, regeneration, and repair (30). Shh is one of three Hh family member proteins to activate the Hh signaling pathway, the others being Desert Hedgehog (Dhh) and Indian Hedgehog (Ihh) [reviewed by (31)]. Amongst them, Shh is unique in its involvement in regulating notochord patterning and IVD growth and development. Growth plate chondrocytes of vertebral bodies express Ihh (25, 32, 33).
The Hh signaling cascade is initiated by the binding of a Hh ligand to the twelve-pass transmembrane receptor Patched 1 (Ptch1) that is complexed with the GPCR-like Smoothened (Smo) receptor. Although Ptch1 inhibits Smo while the pathway is inactive, the binding of the Hh ligand to Ptch1 causes phosphorylation of the Smo receptor, which results in localization of activated Smo onto cell surface, through inhibition of ubiquitination-mediated endocytosis and increased presence of its active conformation (Fig. 1C) (34, 35). Additionally, a required step in the transduction of the Hh pathway is the enrichment of the Smo receptor through intraflagellar transport to the primary cilium, a tightly controlled and activation-dependent event (36–38). The primary cilium is a non-motile, flagella-like protrusion present in almost all mammalian cell types in the interphase. The proper accumulation of Smo to the cilia is necessary for signal transduction, but not sufficient for activation of the receptor, given that both inactive and active Smo configurations are found enriched to the ciliary tip. The specific mechanism of Smo activation is not fully understood. Studies involving small molecules like Smoothened Agonist (SAG) that directly bind a heptahelical ligand-binding domain of the Smo receptor have expanded our understanding of the activation process, demonstrating the critical roles of β-arrestin and kinesin motors during transduction (39). Once activated and enriched to the ciliary tip, Smo generates intracellular signals that regulate various protein kinases that in turn activate the transcriptional effectors of the Hh pathway. These target transcription factors are conserved across species and are known as Cubitus interruptus (Ci) in Drosophila and glioma-associated oncogene 1, 2, and 3 (Gli1, Gli2, Gli3) in mammals (40). In the absence of active Hh signaling Gli2 and Gli3 undergo proteolysis on the C-termini to act as repressor (Gli2R, and Gli3R) (41–45), while Gli1 is not modified post-translationally and thus serves primarily as a transcriptional activator (41). Members of the Gli/Ci protein family contain a zinc-finger DNA-binding domain that recognizes a common DNA element (40). Intraflagellar transport to the ciliary tip is necessary for generating truncated Gli2 and Gli3 molecules (38, 43, 46). Upon activation, the Smo receptor cues the eventual stabilization of the full-length forms and localization of Gli proteins to the ciliary tip. However, it is unclear as to how the Hh signaling pathway components traffic “in” and “out” of the cilium, and also how ciliary enrichment regulates their activity.
There are several negative regulators of the Hh signaling pathway. In the absence of Hh ligand, the Ptch1 receptor is localized to the tip of the cilium and inhibits Smo activation and ciliary enrichment (Fig. 1D) (36, 39). Inactive Smo remains restricted to the base of the ciliary structure (47). The binding of Hh ligand induces the export of Ptch1 from the cilium, which alleviates the inhibition of Smo and permits the rapid enrichment of Smo receptors to the tip of the cilium. Suppressor of fused (Sufu) is another negative regulator of the Hh signaling cascade. Sufu inhibits the Hh pathway by directly binding to the Gli transcriptional factors, anchoring them in the cytoplasm and preventing the activation of the Gli target genes (48–50). SUFU is involved in a tetrameric complex involving Gli/Ci, Cos2, Fu, and Sufu in which Cos2 binds to the microtubules in the cytoplasm in the absence of the ligand. This complex plays a critical role in regulating the transcription activity of Gli/Ci proteins (51).
Shh signaling during intervertebral disc formation
The current research on the role of SHH in the IVD is using mouse models, and will be discussed in this review. Shh expression is initially detected in the node (52) and stays “on” in the notochord as shown in the mouse embryo (53, 54). SHH secreted by the notochord regulates patterning of the surrounding structures including floor-plate (55, 56), neural tube (57, 58) and somitocoele (59, 60). Also, SHH from notochord induces Shh expression by the floor-plate (56). Mutations in Shh or other components of the Hh signaling pathway in mice cause defects in patterning of face, skull, limbs and axial skeleton (37, 53, 61, 62). Mutations in the Shh gene are associated with defects in development of limbs, buds, digits, spine, ribs, face and skull and facial abnormalities such as microcephaly, mild hypotelorism, cyclopia, a primitive nasal structure (proboscis) and/or midfacial clefting in human (63–65). Mutations in Smo causes notochord degeneration in mouse embryos that are lethal before E9.5 (61). However, Smo mouse mutants carrying a N223K point mutation, described as Smo cabbie mutant (Smocbb) mutants, survive untill birth but display craniofacial, skeletal and neural tube patterning defects (37). Further, in the Smocbb mutants, the notochord was intact although the floor plate is not correctly specified. These findings confirm that the floor plate, rather than the notochord, requires the highest level of SHH activity for proper formation (37).
Previous studies demonstrated that SHH is crucial for maintenance of notochord, but not for its formation (53). Choi et al. demonstrated the role of SHH in the formation of the IVD (66). Also, it was found that SHH from the notochord and not floor-plate is sufficient for the formation of the IVD (67). Interestingly, loss of Shh was not crucial for the formation of the node. And the Shh null embryos show that the caudal notochord formed initially, but then disintegrated (53). Conditional targeting of Shh using tamoxifen-inducible Shhflox/CreERT2 allele at E8.5 to E10.5 in mouse embryos revealed the critical role of Shh in the formation of the notochord sheath and proper migration of the notochord cells to form nucleus pulposi of the disc (66). Although conditional targeting of Shh using Shhfiox/CreERT2 alleles at E8.5 or E9.5 resulted in defects in the formation of the IVDs and vertebrae, Shh targeting at E11.5 did not affect the formation of the disc or the spine (66). These findings suggest that Shh signaling is crucial for the formation of the intervertebral disc at the early embryonic stage.
Role of Shh in postnatal disc maintenance
The notochord descendant nucleus pulposus continues to express Shh during the postnatal stages (20, 21). Conditional targeting of Shh in vivo, and blockade of Hh signaling using a small molecule inhibitor cyclopamine on cultured IVD in vitro demonstrated the importance of Shh signaling in the development and maintenance of the postnatal IVDs (25). Blockade of Shh signaling resulted in dramatic histological and molecular changes in the NP and AF cells of the neonatal mouse IVD. These studies revealed that in the absence of Shh signaling, NP cells lose their reticular structure and are clumped together in the center of the disc space, while the AF demonstrates reduced polarity and organization. Blockade of Hh signaling was validated by the loss of GLI1 and PTCH1 in NP and AF cells, suggesting that SHH secreted by the NP cells not only has autocrine action on the NP cells but also has paracrine action on the surrounding AF, suggesting the inductive property of NP similar to its precursor notochord. Besides, blockade of Shh signaling resulted in the loss of proliferation and loss of Brachyury (BRA or T) expression by NP cells. BRA is a crucial transcription factor during early embryogenesis and is also a molecular marker of the notochord. In addition, blockade of Shh signaling resulted in reduced expression of differentiation markers, including transcription factors SOX9 and extracellular matrix components including Collagen Ia1 (COL1a1), Collagen IIa1 (COL2), and Chondroitin sulfate (ChSO4) in the NP, AF, and EP indicating that these molecules are downstream targets of Hh signaling in the postnatal IVD (25). Furthermore, the addition of recombinant SHH to the cyclopamine-treated IVDs rescued the effects caused by the blockade of Shh signaling at the histological and molecular level, validating the specificity to loss of response of Shh signaling. Besides, conditionally targeting of Ihh (33) or dissecting out the IHH expressing growth plate before culture (25) did not affect any aspect of the IVD at the cellular or molecular level demonstrating that SHH is the crucial Hh ligand regulating the growth and differentiation of postnatal IVD.
Shh expression by NP cells reduces with age (21, 24). Recently Peck et al., 2017 compared the transcriptome by RNAseq of mouse notochord at E12.5 to that of NP at P0 and found 87.82-fold reduction in Shh mRNA levels at birth. Interestingly, the mRNA expression of Hh targets Ptch1 and Gli1 reduced by 6.72 and 7.99 fold respectively (68). These results suggest that though NP at P0 express less Shh, it is sufficient to activate the Hh targets in P0 mouse IVDs. The extent of this difference may also be due to differences in methodology when preparing the two sets of mRNA for sequencing. Notochord collected at E12.5 was immediately processed for RNA isolation, while NP cells from P0 went through a lengthy procedure of fluorescence assisted cell sorting (FACS) before RNA isolation. Expression of Shh and its targets continues to reduce during the postnatal stage. Components of Shh signaling and its targets are down-regulated in the one-year-old mouse NP cells compared to that from P4 IVDs (21, 24). These markers include transcriptional factors Bra, Sox9, cytokeratin 19 (Krt19), Col1a1, Col2a1, ACAN, and ChSO4. The reduction in Shh signaling is associated with histological and structural changes in the disc such as clumping of NP cells and thinning of AF layers. By two years of age, the reticular or clumped NP cells are absent, and instead, the NP space has cells that resemble chondrocyte-like cells in morphology (24). These findings indicate that the mouse IVDs also degenerate with physiological aging.
Shh acts upstream of other developmental signaling pathways
In addition to its role in growth and differentiation, Shh signaling regulates other major cell signaling pathways in the postnatal mouse IVD. In vivo and in vitro Shh blockade studies have demonstrated that Shh signaling inhibits canonical Wnt Signaling and BMP signaling, while positively regulating TGFβ signaling in neonatal mouse IVDs. Although Shh signaling inhibits Canonical Wnt signaling, in vitro studies using small molecule activator of Wnt signaling BIO, an inhibitor of Wnt XAV939, showed that Wnt signaling acts as an activator of the Shh signaling pathway in the neonatal mouse IVD (24). Treatment of neonatal mouse IVDs with BIO positively regulates downstream targets of Shh, including cell proliferation, and expression of GLI1, BRA, and SOX9, and ECM markers without changing the gene expression of SHH ligand itself. These observations were validated by in vivo targeting of Wntless (Evi/ Wls), a protein involved in secretion of Wnt ligands, at E18.5 in ShhCre; Wlsflx/flx mouse embryos (24). Interestingly, although the response to both Shh and Wnt signaling pathways is reduced by one year of age, in vitro treatment of one-year-old mouse IVDs with Wnt activator BIO rescued the aging phenotype and re-activated the expression of molecular markers of a healthy IVD that were otherwise lost with aging (24). However, the molecular mechanism of interaction between the Shh, Wnt, BMP and TGFβ pathways is not well understood and requires further investigation.
Potential for Hedgehog signaling in disc regeneration:
Future studies are required to examine the potential of critical developmental pathways like Shh that are crucial for disc growth and maintenance in biological treatments for DDD. These therapies would aim to re-activate the Shh signaling pathway in degenerated IVDs to restore differentiation markers like ECM proteins and normal disc histology. Studies using amphibian models like Axolotl larva and Xenopus tadpoles have examined the role of Hh signaling in tail regeneration (69, 70). Although in the Axolotl larva the notochord did not form in the regenerating tail, the Xenopus tadpole tail showed notochord formation that was thought to be due to Shh secreted by the regenerating notochord having an autocrine action (70). The differences in notochord regeneration may be due to the source of Shh in the two model systems. Shh was expressed exclusively by notochord in the Xenopus and spinal cord in Axolotl embryos.
The potential for Hh pathway reactivation in adult mice has been demonstrated in vitro 3D organ culture of one-year-old mouse IVDs where the reactivation of the Wnt signaling pathway, using BIO re-activated Hh signaling pathway and its targets within three days (24). Cultured IVDs showed increased expression of BRA, CHSO4, and ACAN, and Shh targets Gli1 and Ptch1 demonstrating that Wnt signaling can re-activate the Shh pathway downstream of the Shh ligand. Recent studies have shown that Shh expression and its signaling reduces more rapidly and by 12 weeks of age in the sacral IVDs of the mouse. Formation of sacrum by skeletal maturity is a normal developmental process. However, reactivation of Hh signaling using a constitutive active R26LSL-SmoM2-YFP/LSLSmoM2-YFP (71) allele only in a subset of NP cells using NP-specific CK19CreERT2 allele at 12 weeks restored the sacral disc structure and Hh targets like PTCH1 and ECM proteins two and a half weeks later (72). Also, the reticular structure of the NP cells and layers of AF were restored along with decreased vascularization of the sacral IVD. These effects can be attributed to clonal expansion of the recombined NP cells as shown by immunostaining for the reporter YFP, and higher expression of SHH, which may, in turn, have activated the NP cells that did not have the constitutive active SmoM2 allele.
Conclusions:
Degeneration of the IVD is a major cause of lower back pain, which now is considered a global burden (6), still with no option available for its cure. Prevalence of disc degeneration increases with age. Improving our understanding about the cellular and molecular process of how the IVD is formed during the embryonic stages, what are they key regulators for its growth and health during the postnatal stages, and determining whether loss of these key regulators is associated with degeneration of the disc may provide information to develop therapies for IVD regeneration in the way it was originally formed. Shh is one such key regulator that is essential for disc formation and its postnatal maintenance (25, 66, 72). Shh expression and its targets, including ECM proteins that are important for IVD structure and function, as reduced with age. Recent studies suggest that the Hh signaling pathway can be reactivated in adult and aged mouse IVDs, and initial activation of only a subset of cells may be sufficient for reactivation and regeneration of the entire disc (72). Thus, investigating the role of Shh in disc provides a promising avenue for the development of biological therapies for DDD.
Acknowledgements
Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number RO1AR065530 awarded to CLD. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The research award from Gerstner Family Foundation and S & L Marx Foundation made to CLD also supported the research.
Footnotes
Conflict of Interest
Diviya Rajesh and Chitra Lekha Dahia each declare no potential conflicts of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
References:
Papers of particular interest, published recently, have been highlighted as:
* Of importance
** Of major importance
- 1.Urban JPG, Roberts S, Ralphs JR. The Nucleus of the Intervertebral Disc from Development to Degeneration. American Zoologist. 2000;40:53–061. [Google Scholar]
- 2.Melrose J Disc structure function and its potential for repair. APLAR Journal of Rheumatology. 2008;5:A7–A8. [Google Scholar]
- 3.Boos N, Weissbach S, Rohrbach H, Weiler C, Spratt KF, Nerlich AG. Classification of age-related changes in lumbar intervertebral discs: 2002 Volvo Award in basic science. Spine (Phila Pa 1976). 2002. December 1;27(23):2631–44. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 4.Buckwalter JA. Aging and degeneration of the human intervertebral disc. Spine (Phila Pa 1976). 1995. June 1;20(11):1307–14. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 5.Collaborators GBDDH. Global, regional, and national disability-adjusted life years (DALYs) for 306 diseases and injuries and healthy life expectancy (HALE) for 188 countries, 1990–2013: quantifying the epidemiological transition. Lancet. 2015. November 28;386(10009):2145–91. PubMed PMID: . Pubmed Central PMCID: 4673910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartvigsen J, Hancock MJ, Kongsted A, Louw Q, Ferreira ML, Genevay S, et al. What low back pain is and why we need to pay attention. Lancet. 2018. March 20 PubMed PMID: .*This review discusses the cause and consequence of lower back pain as one of the top Global Burden of Disease, and argues the importance of research initiatives to address this major public health problem.
- 7.Taher F, Essig D, Lebl DR, Hughes AP, Sama AA, Cammisa FP, et al. Lumbar degenerative disc disease: current and future concepts of diagnosis and management. Advances in orthopedics. 2012;2012:970752 PubMed PMID: . Pubmed Central PMCID: 3335178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rea W, Kapur S, Mutagi H. Intervertebral disc as a source of pain. Continuing Education in Anaesthesia Critical Care & Pain. 2012;12:279–82. [Google Scholar]
- 9.Brent AE, Schweitzer R, Tabin CJ. A somitic compartment of tendon progenitors. Cell. 2003. April 18;113(2):235–48. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 10.Choi KS, Cohn MJ, Harfe BD. Identification of nucleus pulposus precursor cells and notochordal remnants in the mouse: implications for disk degeneration and chordoma formation. Dev Dyn. 2008. December;237(12):3953–8. PubMed PMID: . Pubmed Central PMCID: PMC2646501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCann MR, Tamplin OJ, Rossant J, Seguin CA. Tracing notochord-derived cells using a Noto-cre mouse: implications for intervertebral disc development. Disease models & mechanisms. 2012. January;5(1):73–82. PubMed PMID: . Pubmed Central PMCID: 3255545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Risbud MV, Schoepflin ZR, Mwale F, Kandel RA, Grad S, Iatridis JC, et al. Defining the phenotype of young healthy nucleus pulposus cells: recommendations of the Spine Research Interest Group at the 2014 annual ORS meeting. J Orthop Res. 2015. March;33(3):283–93. PubMed PMID: . Pubmed Central PMCID: 4399824.*This review discusses the current literature on the molecular and phenotypic markers of nucleus pulposus cells that can be used by researchers for experimental design and better interpretation of their research findings.
- 13.Sugimoto Y, Takimoto A, Akiyama H, Kist R, Scherer G, Nakamura T, et al. Scx+/Sox9+ progenitors contribute to the establishment of the junction between cartilage and tendon/ligament. Development. 2013. June;140(11):2280–8. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 14.Singh K, Masuda K, Thonar EJ, An HS, Cs-Szabo G. Age-related changes in the extracellular matrix of nucleus pulposus and anulus fibrosus of human intervertebral disc. Spine (Phila Pa 1976). 2009. January 1;34(1):10–6. PubMed PMID: . Pubmed Central PMCID: 2837353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson WE, Caterson B, Eisenstein SM, Roberts S. Human intervertebral disc aggrecan inhibits endothelial cell adhesion and cell migration in vitro. Spine (Phila Pa 1976). 2005. May 15;30(10):1139–47. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 16.Kauppila LI. Ingrowth of blood vessels in disc degeneration. Angiographic and histological studies of cadaveric spines. J Bone Joint Surg Am. 1995. January;77(1):26–31. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 17.Nerlich AG, Schaaf R, Walchli B, Boos N. Temporo-spatial distribution of blood vessels in human lumbar intervertebral discs. Eur Spine J. 2007. April;16(4):547–55. PubMed PMID: . Pubmed Central PMCID: PMC2229818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Urban JP, Roberts S. Degeneration of the intervertebral disc. Arthritis Res Ther. 2003;5(3):120–30. PubMed PMID: . Pubmed Central PMCID: PMC165040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vergroesen PP, Kingma I, Emanuel KS, Hoogendoorn RJ, Welting TJ, van Royen BJ, et al. Mechanics and biology in intervertebral disc degeneration: a vicious circle. Osteoarthritis and cartilage. 2015. July;23(7):1057–70. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 20.Dahia CL, Mahoney EJ, Durrani AA, Wylie C. Postnatal growth, differentiation, and aging of the mouse intervertebral disc. Spine (Phila Pa 1976). 2009. March 1;34(5):447–55. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 21.Dahia CL, Mahoney EJ, Durrani AA, Wylie C. Intercellular signaling pathways active during intervertebral disc growth, differentiation, and aging. Spine (Phila Pa 1976). 2009. March 1;34(5):456–62. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 22.Hiyama A, Sakai D, Risbud MV, Tanaka M, Arai F, Abe K, et al. Enhancement of intervertebral disc cell senescence by WNT/beta-catenin signaling-induced matrix metalloproteinase expression. Arthritis and rheumatism. 2010. October;62(10):3036–47. PubMed PMID: . Pubmed Central PMCID: 3622204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kondo N, Yuasa T, Shimono K, Tung W, Okabe T, Yasuhara R, et al. Intervertebral disc development is regulated by Wnt/beta-catenin signaling. Spine (Phila Pa 1976). 2011. April 15;36(8):E513–8. PubMed PMID: . Pubmed Central PMCID: 3072453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winkler T, Mahoney EJ, Sinner D, Wylie CC, Dahia CL. Wnt signaling activates Shh signaling in early postnatal intervertebral discs, and re-activates Shh signaling in old discs in the mouse. PLoS One. 2014;9(6):e98444 PubMed PMID: . Pubmed Central PMCID: PMC4043533.*This study shows that although the respsone to key developmental signals like Shh and Wnt decreases with age, the nucleus pulposus cells of aged discs have the capability to re-activate by small molecule agonists of these pathways to a heathier phenotype.
- 25.Dahia CL, Mahoney E, Wylie C. Shh signaling from the nucleus pulposus is required for the postnatal growth and differentiation of the mouse intervertebral disc. PLoS One. 2012;7(4):e35944 PubMed PMID: . Pubmed Central PMCID: PMC3338762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin H, Shen J, Wang B, Wang M, Shu B, Chen D. TGF-beta signaling plays an essential role in the growth and maintenance of intervertebral disc tissue. FEBS letters. 2011. April 20;585(8):1209–15. PubMed PMID: . Pubmed Central PMCID: 3090135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Risbud MV, Di Martino A, Guttapalli A, Seghatoleslami R, Denaro V, Vaccaro AR, et al. Toward an optimum system for intervertebral disc organ culture: TGF-beta 3 enhances nucleus pulposus and anulus fibrosus survival and function through modulation of TGF-beta-R expression and ERK signaling. Spine (Phila Pa 1976). 2006. April 15;31(8):884–90. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 28.Than KD, Rahman SU, Vanaman MJ, Wang AC, Lin CY, Zhang H, et al. Bone morphogenetic proteins and degenerative disk disease. Neurosurgery. 2012. April;70(4):996–1002; discussion PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 29.Li X, An HS, Ellman M, Phillips F, Thonar EJ, Park DK, et al. Action of fibroblast growth factor-2 on the intervertebral disc. Arthritis Res Ther. 2008;10(2):R48 PubMed PMID: . Pubmed Central PMCID: 2453768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petrova R, Joyner AL. Roles for Hedgehog signaling in adult organ homeostasis and repair. Development. 2014. September;141(18):3445–57. PubMed PMID: . Pubmed Central PMCID: 4197719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beachy PA, Hymowitz SG, Lazarus RA, Leahy DJ, Siebold C. Interactions between Hedgehog proteins and their binding partners come into view. Genes & development. 2010. September 15;24(18):2001–12. PubMed PMID: . Pubmed Central PMCID: 2939362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dahia CL, Mahoney EJ, Durrani AA, Wylie C. Intercellular signaling pathways active during and after growth and differentiation of the lumbar vertebral growth plate. Spine (Phila Pa 1976). 2011. June 15;36(14):1071–80. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 33.Maeda Y, Nakamura E, Nguyen MT, Suva LJ, Swain FL, Razzaque MS, et al. Indian Hedgehog produced by postnatal chondrocytes is essential for maintaining a growth plate and trabecular bone. Proceedings of the National Academy of Sciences of the United States of America. 2007. April 10;104(15):6382–7. PubMed PMID: . Pubmed Central PMCID: 1851055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alcedo J, Zou Y, Noll M. Posttranscriptional regulation of smoothened is part of a self-correcting mechanism in the Hedgehog signaling system. Molecular cell. 2000. August;6(2):457–65. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 35.Jiang K, Jia J. Smoothened regulation in response to Hedgehog stimulation. Frontiers in biology. 2015. December 1;10(6):475–86. PubMed PMID: . Pubmed Central PMCID: 4787298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF. Vertebrate Smoothened functions at the primary cilium. Nature. 2005. October 13;437(7061):1018–21. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 37.Gigante ED, Long AB, Ben-Ami J, Caspary T. Hypomorphic Smo mutant with inefficient ciliary enrichment disrupts the highest level of vertebrate Hedgehog response. Developmental biology. 2018. May 15;437(2):152–62. PubMed PMID: . Pubmed Central PMCID: 5903954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003. November 6;426(6962):83–7. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 39.Kovacs JJ, Whalen EJ, Liu R, Xiao K, Kim J, Chen M, et al. Beta-arrestin-mediated localization of smoothened to the primary cilium. Science. 2008. June 27;320(5884):1777–81. PubMed PMID: . Pubmed Central PMCID: 2587210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruppert JM, Kinzler KW, Wong AJ, Bigner SH, Kao FT, Law ML, et al. The GLI-Kruppel family of human genes. Molecular and cellular biology. 1988. August;8(8):3104–13. PubMed PMID: . Pubmed Central PMCID: 363537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aza-Blanc P, Lin HY, Ruiz i Altaba A, Kornberg TB. Expression of the vertebrate Gli proteins in Drosophila reveals a distribution of activator and repressor activities. Development. 2000. October;127(19):4293–301. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 42.Dai P, Akimaru H, Tanaka Y, Maekawa T, Nakafuku M, Ishii S. Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. The Journal of biological chemistry. 1999. March 19;274(12):8143–52. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 43.Haycraft CJ, Banizs B, Aydin-Son Y, Zhang Q, Michaud EJ, Yoder BK. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS genetics. 2005. October;1(4):e53 PubMed PMID: . Pubmed Central PMCID: 1270009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sasaki H, Nishizaki Y, Hui C, Nakafuku M, Kondoh H. Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development. 1999. September;126(17):3915–24. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 45.Villavicencio EH, Walterhouse DO, Iannaccone PM. The sonic hedgehog-patched-gli pathway in human development and disease. American journal of human genetics. 2000. November;67(5):1047–54. PubMed PMID: . Pubmed Central PMCID: 1288546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu A, Wang B, Niswander LA. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development. 2005. July;132(13):3103–11. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 47.Milenkovic L, Weiss LE, Yoon J, Roth TL, Su YS, Sahl SJ, et al. Single-molecule imaging of Hedgehog pathway protein Smoothened in primary cilia reveals binding events regulated by Patched1. Proceedings of the National Academy of Sciences of the United States of America. 2015. July 7;112(27):8320–5. PubMed PMID: . Pubmed Central PMCID: 4500289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng SY, Bishop JM. Suppressor of Fused represses Gli-mediated transcription by recruiting the SAP18-mSin3 corepressor complex. Proceedings of the National Academy of Sciences of the United States of America. 2002. April 16;99(8):5442–7. PubMed PMID: . Pubmed Central PMCID: 122788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kogerman P, Grimm T, Kogerman L, Krause D, Unden AB, Sandstedt B, et al. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nature cell biology. 1999. September;1(5):312–9. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 50.Paces-Fessy M, Boucher D, Petit E, Paute-Briand S, Blanchet-Tournier MF . The negative regulator of Gli, Suppressor of fused (Sufu), interacts with SAP18, Galectin3 and other nuclear proteins. The Biochemical journal. 2004. March 1;378(Pt 2):353–62. PubMed PMID: . Pubmed Central PMCID: 1223961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruel L, Therond PP. Variations in Hedgehog signaling: divergence and perpetuation in Sufu regulation of Gli. Genes & development. 2009. August 15;23(16):1843–8. PubMed PMID: . Pubmed Central PMCID: 2725945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jeong Y, Epstein DJ. Distinct regulators of Shh transcription in the floor plate and notochord indicate separate origins for these tissues in the mouse node. Development. 2003. August;130(16):3891–902. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 53.Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, et al. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996. October 3;383(6599):407–13. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 54.Echelard Y, Epstein DJ, St-Jacques B, Shen L, Mohler J, McMahon JA, et al. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993. December 31;75(7):1417–30. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 55.Ericson J, Morton S, Kawakami A, Roelink H, Jessell TM. Two critical periods of Sonic Hedgehog signaling required for the specification of motor neuron identity. Cell. 1996. November 15;87(4):661–73. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 56.Teillet MA, Lapointe F, Le Douarin NM. The relationships between notochord and floor plate in vertebrate development revisited. Proceedings of the National Academy of Sciences of the United States of America. 1998. September 29;95(20):11733–8. PubMed PMID: . Pubmed Central PMCID: 21709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dessaud E, McMahon AP, Briscoe J. Pattern formation in the vertebrate neural tube: a sonic hedgehog morphogen-regulated transcriptional network. Development. 2008. August;135(15):2489–503. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 58.Wilson L, Maden M. The mechanisms of dorsoventral patterning in the vertebrate neural tube. Developmental biology. 2005. June 1;282(1): 1–13. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 59.Resende TP, Ferreira M, Teillet MA, Tavares AT, Andrade RP, Palmeirim I. Sonic hedgehog in temporal control of somite formation. Proceedings of the National Academy of Sciences of the United States of America. 2010. July 20;107(29):12907–12. PubMed PMID: . Pubmed Central PMCID: 2919945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Teillet M, Watanabe Y, Jeffs P, Duprez D, Lapointe F, Le Douarin NM. Sonic hedgehog is required for survival of both myogenic and chondrogenic somitic lineages. Development. 1998. June;125(11):2019–30. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 61.Caspary T, Garcia-Garcia MJ, Huangfu D, Eggenschwiler JT, Wyler MR, Rakeman AS, et al. Mouse Dispatched homolog1 is required for long-range, but not juxtacrine, Hh signaling. Current biology : CB. 2002. September 17;12(18):1628–32. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 62.Zhang XM, Ramalho-Santos M, McMahon AP. Smoothened mutants reveal redundant roles for Shh and Ihh signaling including regulation of L/R symmetry by the mouse node. Cell. 2001. July 27;106(2):781–92. PubMed PMID: . [PubMed] [Google Scholar]
- 63.Belloni E, Muenke M, Roessler E, Traverso G, Siegel-Bartelt J, Frumkin A, et al. Identification of Sonic hedgehog as a candidate gene responsible for holoprosencephaly. Nature genetics. 1996. November;14(3):353–6. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 64.Kelley RL, Roessler E, Hennekam RC, Feldman GL, Kosaki K, Jones MC, et al. Holoprosencephaly in RSH/Smith-Lemli-Opitz syndrome: does abnormal cholesterol metabolism affect the function of Sonic Hedgehog? American journal of medical genetics. 1996. December 30;66(4):478–84. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 65.Roessler E, Belloni E, Gaudenz K, Jay P, Berta P, Scherer SW, et al. Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nature genetics. 1996. November;14(3):357–60. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 66.Choi KS, Harfe BD. Hedgehog signaling is required for formation of the notochord sheath and patterning of nuclei pulposi within the intervertebral discs. Proceedings of the National Academy of Sciences of the United States of America. 2011. June 7;108(23):9484–9. PubMed PMID: . Pubmed Central PMCID: 3111270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Choi KS, Lee C, Harfe BD. Sonic hedgehog in the notochord is sufficient for patterning of the intervertebral discs. Mechanisms of development. 2012. Sep-Dec; 129(9–12):255–62. PubMed PMID: . Pubmed Central PMCID: 3478436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peck SH, McKee KK, Tobias JW, Malhotra NR, Harfe BD, Smith LJ. Whole Transcriptome Analysis of Notochord-Derived Cells during Embryonic Formation of the Nucleus Pulposus. Scientific reports. 2017. September 5;7(1):10504 PubMed PMID: . Pubmed Central PMCID: 5585380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schnapp E, Kragl M, Rubin L, Tanaka EM. Hedgehog signaling controls dorsoventral patterning, blastema cell proliferation and cartilage induction during axolotl tail regeneration. Development. 2005. July;132(14):3243–53. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 70.Taniguchi Y, Watanabe K, Mochii M. Notochord-derived hedgehog is essential for tail regeneration in Xenopus tadpole. BMC developmental biology. 2014. June 18;14:27 PubMed PMID: . Pubmed Central PMCID: 4074850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jeong J, Mao J, Tenzen T, Kottmann AH, McMahon AP. Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes & development. 2004. April 15;18(8):937–51. PubMed PMID: . Pubmed Central PMCID: PMC395852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bonavita R, Vincent K, Pinelli R, Dahia CL. Formation of the sacrum requires down-regulation of sonic hedgehog signaling in the sacral intervertebral discs. Biology open. 2018. May 21 PubMed PMID: .**This study for the first time not only provide a biological basis for disc degeneration and collapse but it also provides a ground for therapeutic intervention for the regeneration of the intervertebral disc by activation of a sub-set of nucleus pulposus cells using Hedgehog signaling.


