Abstract
This review focuses on recent progress in noncomputational methods to introduce catalytic function into proteins, peptides, and peptide assemblies. We discuss various approaches to creating catalytic activity and classification of noncomputational methods into rational and combinatorial classes. The section on rational design covers recent progress in the development of short peptides and oligomeric peptide assemblies for various natural and unnatural reactions. The section on combinatorial design describes recent advances in the discovery of catalytic peptides. We present the future prospects of these and other new approaches in a broader context, including implications for functional material design.
Keywords: protein design, phage display, catalysis, supramolecular assemblies, metalloenzymes
1. INTRODUCTION
Enzymes catalyze a number of chemical reactions with high efficiency and selectivity under ambient conditions. Much effort has been dedicated to developing new enzymes and changing the scope and selectivity of existing enzymes. Over the past 30 years, many different approaches have emerged to design new catalysts for various chemical reactions. Most recently, the design of a novel enzymatic function was complemented with efficient computational techniques (1). However, despite advances in design methodologies, we are still unable to create an efficient catalyst from scratch for a given chemical transformation (2). In addition to computational approaches to enzyme design that have been the focus of many recent reviews (3, 4), the “old school” rational designs, as well as advanced high-throughput combinatorial techniques, have had many recent successes. Here we review the latest progress in noncomputational methods to introduce catalytic function into proteins, peptides, and peptide assemblies.
Rational design utilizes basic fundamental knowledge of enzymatic function, often in combination with structural information, to create new activity or alter the stereochemistry and regioselectivity of the existing enzymes by introducing a fairly small number of mutations (5, 6). Although it is hard to draw a clear line between the rational and computational approaches (as computational approaches are rational by definition), for the purpose of this review we define the computational approach as the one that engages complex algorithms to quantitatively predict the effects of mutations on the overall stability and function of the protein scaffold.
The combinatorial approach mimics natural evolution, in which mutation and selection cycles are repeated and appropriate candidates propagate and improve through trial-and-error loops. Therefore, structural information about the target enzyme and its reaction mechanism are not required for this approach. A large library of random mutations is created; subsequently, mutant enzymes are screened for altered activity and/or specificity. The selected enzymes are subjected to several cycles of mutagenesis and screening, yielding enzymes with better properties. This design method requires that the link between the phenotype (enzymatic properties) and its genotype (DNA sequence encoding the enzyme) be maintained throughout the many cycles of the selection process.
Combinatorial phage display uses bacterial phage templates (e.g., M13, M9, and lambda) to display randomly mutated peptides or proteins on the phage surface by means of genetic engineering. The DNA sequence encoding the displayed proteins is encapsulated in the viral protein shells; therefore, the phenotype and genotype are not separated. Additionally, it is easy to amplify the selected phage to generate a new library in the subsequent round. Because the selection process is based on the binding properties and is quite easy to perform, this system has been widely used to identify artificial antibody-like proteins (7), ligands (8), enzyme substrates(9), and peptides bound to inorganic material (10). In contrast, direct selection based on catalytic properties is hard to perform using the phage-display method. Conjugation of an enzyme and its substrate together on phages is one of the proposed strategies to overcome this difficulty (11). Substrates that are covalently (12) or noncovalently (13) bound to the enzyme-displaying phages are converted to specific products, and subsequent selection steps are performed with affinity supports that specifically bind the product but do not have affinity toward the original substrate. Although these phage-display systems are suitable for handling huge libraries of mutants, a critical drawback is that the screening process often results in nonspecific binding and, thus, false-positive selection. The size of the enzymes that can be displayed on phages represents another limitation of this approach. If the fused enzymes are too bulky, then self-assembly of the phage shell can be disrupted.
Selection based on fluorescence-activated cell sorting (FACS) has recently been used as an alternative method for high-throughput screening of catalytically improved enzymes (14). In this method, a target enzyme and its substrate are conjugated on/in micrometer-sized objects (microcompartments), such as yeast cells (15, 16), emulsion droplets (17), or polyelectrolyte shells(18), which also contain a DNA sequence encoding the mutated enzyme in order to preserve a phenotype–genotype link. Catalytic conversion of substrates provides fluorescent readout that is used to select for microcompartments exhibiting high catalytic activity. Modern cell sorters enable the screening of enzyme libraries as large as 108 variants (14, 17). Because formation of the fluorescent product is directly related to the catalytic activity of the target enzyme, FACS-based screening is a straightforward way to select enzymes with desired properties.
The combinatorial screening systems described above are powerful tools used to evolve enzymatic activities. However, it remains impossible to completely explore all possible enzyme sequences in the search for the best catalytic properties (e.g., when a target enzyme is composed of 230 amino acid residues, a typical size of a TIM barrel protein, its peptide sequence space is 20230 ≈ 10300 combinations). In order to circumvent the difficulty of covering a huge space, only the limited domains that are essential to enzymatic functions are randomized (6). The amino acid positions to be randomized are predicted on the basis of deep knowledge of the target enzyme, such as features of peptide sequence and three-dimensional (3D) structure (19). This strategy combines the advantages of both rational and combinatorial approaches and is therefore referred to as a semirational approach. Knowledge-based restriction of randomized sequences can minimize the size of the mutation library to 103 clones (19), allowing investigators to efficiently redesign the target enzymes.
In contrast to enzymes, catalytic peptides have only recently emerged as biomolecular catalysts. They have potential for broad applications ranging from materials to drug development. In this review, we define catalytic peptides as short oligopeptides with catalytic functions. Therefore, this review does not cover catalytic antibodies that bind to and stabilize the transition state in order to catalyze chemical reactions (20, 21), catalytic peptide dendrimers that are artificially synthesized peptidic dendrimers composed of densely packed catalytic amino acid residues (22,23), or ultrashort catalytic peptides capable of asymmetric catalysis (24). We describe methods currently used to design catalytic peptides and discuss the advantages and future prospective of catalytic peptides as compared with enzymes.
2. RATIONAL DESIGN
In the broadest sense, the rational design of enzymatic activity uses a priori knowledge of protein/peptide structure and/or a basic mechanistic picture of the chemical reaction being catalyzed. Such a broad definition includes several very general bioengineering and directed evolution techniques (25–27); thus, our review focuses on rational approaches that either utilize de novo designed scaffolds or completely alter the function of existing proteins.
2.1. Rational Designs That Utilize Metal Ions as Cofactors
Historically, rational designs featuring metal cofactors have been the most successful. One can easily introduce metal ions into proteins by making very few mutations; moreover, proteins can tolerate the incorporation of complex cofactors and even entire small-molecule metal complexes already preconfigured to catalyze chemical reactions. The past several years have witnessed significant improvements in established approaches to rational design, as well as the emergence of peptide self-assembly as a versatile tool for creating catalytic activity. Overall, rationally designed metalloenzymes can be placed into two broad (and partially overlapping) categories discussed in the following two subsections.
2.1.1. Introduction of metal cofactors into de novo designed scaffolds.
De novo designed helical bundles present a robust and well-controlled environment for functionalization (28). Recent progress in this field has led to more diverse functional designs and an improved ability to control the design.
Ghirlanda and colleagues (29) introduced eight cysteine residues into the hydrophobic core of a de novo designed homodimeric helical protein, DSD, to create two binding sites for iron–sulfur clusters. The resulting protein, DSD-Fdm, binds two [4Fe–4S] clusters with high yield and can transfer electrons to cytochrome c. These authors subsequently changed the selectivity of the protein to bind [3Fe–4S], a different type of iron–sulfur cluster (30).
Pecoraro and colleagues (31) recently reported an efficient, de novo designed catalyst of carbon dioxide hydration with greater activity than that of any small-molecule catalyst for this reaction. Although the activity of this metalloenzyme is about an order of magnitude lower than that of a different enzyme created earlier by the same group (32), the scaffold used in the newer design requires no extra structural sites and offers greater potential for subsequent evolution. The simplicity of the metal coordination spheres formed by peptides that self-assemble into helical bundles allowed these authors to utilize the trimeric coiled-coil scaffold to design Cu(TRIL23H)3, a functional copper nitrite reductase model (33). The authors further optimized the redox potential of the copper site in Cu(TRIL23H)3 by rationally engineering charge patterns around the active site (34).
Even the simplest scaffolds offer opportunities for multiple functionalities. Farid et al. (35) designed a self-assembling four-α-helix protein capable of light-activated intraprotein energy transfer and charge separation, approximating the core reactions of photosynthesis, cryptochrome, and photolyase.
2.1.2. Introduction of metal cofactors into existing proteins.
The reprogramming of an existing metalloprotein to achieve new functions allows access to highly specialized metal folds and often provides a wealth of information about the structure and function of the existing protein. For example, Lu and coworkers (36) redesigned the oxygen carrier myoglobin (Mb) into a heme–copper oxidase by introducing a copper-binding site into a distal pocket of Mb. Later, the same group engineered favorable electrostatic interactions between this functional oxidase model and the native redox partner of the oxidase enzyme (37). The oxygen reduction rate of the resulting Mb-based oxidase (52 s−1) is comparable to that of a native cytochrome c oxidase (50 s−1) under identical conditions.
Bren and colleagues (38) created a molecular electrocatalyst that reduces protons to dihydrogen in neutral water under aerobic conditions with near quantitative Faradaic efficiency. The biomolecular catalyst was created by substitution of iron for cobalt in a water-soluble heme-undecapeptide derived from the horse cytochrome c protein.
Kuhlman and colleagues (39) showed that rational design can create catalytic sites on protein–protein interfaces. Song & Tezcan (40) perfected this approach by constructing an artificial metallo-β-lactamase via self-assembly of a monomeric redox protein into a tetrameric assembly with catalytic zinc sites in its interfaces. This protein is functional in the Escherichia coli periplasm and enables the bacteria to survive treatment with ampicillin.
Direct engineering of new sites in proteins is not the only way to create new functional activity. Highly advanced small-molecule metal catalysts can be tethered to proteins using an array of techniques. This approach combines metal complexes’ high reactivity with proteins’ regio- and enantioselectivity and compatibility with water. Several groups recently used rational protein design to develop water-compatible catalysts for olefin ring-closing metathesis, illustrating advances in these techniques. For instance, Mayer et al. (41) introduced a cysteine residue onto the rim of the heat-shock protein MjHSP, and Okuda and colleagues (42, 43) engineered a cysteine residue at the entry of the pore of the β-barrel protein FhuA. In both cases, small-molecule catalysts bearing a thiol reactive group (e.g., maleimide) were then covalently linked to the proteins. In a different approach, Ward and colleagues (44) attached a Grubbs–Hoveyda catalyst to avidin by linking the catalyst to D-biotin, and Hirota and colleagues (45) used a so-called Trojan horse coupling strategy to covalently attach a ruthenium catalyst to α-chymotrypsin. In all of these cases, the resulting protein–catalyst conjugates catalyzed the desired reaction with only a modest effect on regioselectivity. Although these results are encouraging, they demonstrate a need for improvement in a priori predictions of efficient tethering sites in different protein scaffolds.
Various tethering techniques have been applied to other types of scaffolds and chemical reactions. Ward and colleagues (46, 47) used sulfonamide inhibitors to attach iridium complexes to various proteins to create enantioselective hybrid hydrogenation catalysts, and investigators have recently reported protein–small molecule hybrid catalysts for other reactions, such as rhodiumcatalyzed polymerization (48, 49) and olefin cyclopropanation (50), manganese-promoted epoxidation (51, 52) and sulfoxidation (53), and epoxide ring opening catalyzed by scandium (54).
2.2. Supramolecular Approach to the Rational Design of Catalysts
Even very small peptides have enantioselective catalytic properties (55) and can self-assemble into “protein-like” large aggregates. For these reasons, self-assembling peptides present an exciting opportunity for rational design of catalytic activity.
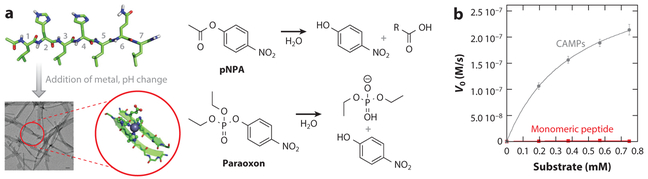
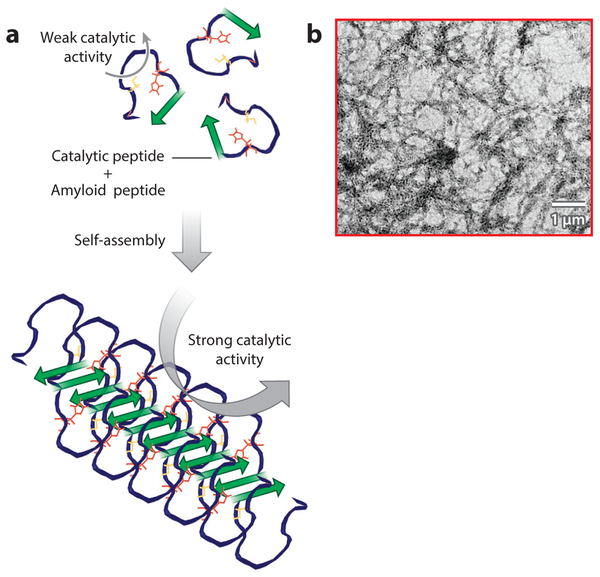
We recently showed that small seven-residue amyloid-forming peptides can be designed from first principles to form efficient catalysts of ester hydrolysis whose activity is comparable to that of the best small-molecule and peptide catalysts reported to date (Figure 1) (56). These results represent the first demonstration of substantial catalytic activity in simple peptide amyloids and, from a practical perspective, will enable the design of highly stable, robust, and easily varied enzyme-like catalysts. Moreover, by mixing different peptides we observed synergistic interactions that increased activity even further. The ability to screen multiple stable arrangements of functional groups in a single fibril provides essentially limitless opportunities for high-throughput screening for functional activity by mixing peptides with different sequences. We showed that this catalytic system can hydrolyze even highly challenging substrates, such as paraoxon (Figure 1). Friedmann et al. (57) combined rational design with high-throughput screening to identify several peptide sequences capable of catalyzing hydrolysis in a mechanistically different fashion; these results suggest that even very simple amyloid-like peptide assemblies can promote different chemical reactivities. Zhang et al. (58) and Huang et al. (59) used an analogous approach to develop self-assembling hydrolases for peptides of various lengths without the aid of metal.
Figure 1.
Catalysis assisted by catalytic amyloids. (a) Overall approach to the design of peptide-based fibrils and the chemical reactions they catalyze. (b) p-Nitrophenyl acetate (pNPA) hydrolysis by catalytic amyloid peptides (CAMPs) (gray circles) compared with a monomeric control peptide (red squares) (56).
The ability of self-assembled peptidic structures to promote catalysis is not limited to hydrolytic reactions. Escuder and colleagues (60) recently developed proline-containing peptide assemblies for an enantioselective aldol reaction. Liu and colleagues (61) created a copper-binding bolaamphiphile that self-assembles in water to catalyze the Diels–Alder reaction. Moreover, supramolecular assemblies can be designed to maintain multiple functionalities. Weingarten et al. (62) created a hydrogel that combines a light-absorbing chromophore with a small-molecule nickel catalyst for light-driven hydrogen production. Fry et al. (63, 64) showed that carefully designed self-assembling peptides can support charge transfer between light-harvesting metalloporphyrins and titanium dioxide nanoparticles.
3. COMBINATORIAL APPROACHES TO IDENTIFY CATALYTIC PEPTIDES
3.1. Screening of Catalytic Peptides That Promote Growth of Inorganic Nanocrystals
The phage-display system has become a powerful tool to perform screening for functional peptides. Pioneering research in this field employed high-throughput screening to identify the peptides promoting inorganic nanocrystal growth, using the ability to bind to target inorganic materials for the selection of positive clones. Investigators discovered peptides that exhibit selective affinity to inorganic surfaces (10), some of which catalytically promote growth of noble-metal nanocrystals [e.g., silver (65, 66), nickel (67, 68), and copper (67, 69)], metal alloys [e.g., cobalt–platinum (66)], metal oxides [e.g., zinc oxide (ZnO) (70)], and metal sulfides [e.g., zinc sulfide (71)]. However, the peptides selected on the basis of binding are not necessarily the best promoters of the target inorganic nanocrystal nucleation. Therefore, it is desirable to establish a direct method to isolate catalytic peptides on the basis of their nanocrystal-growing property rather than their binding property.
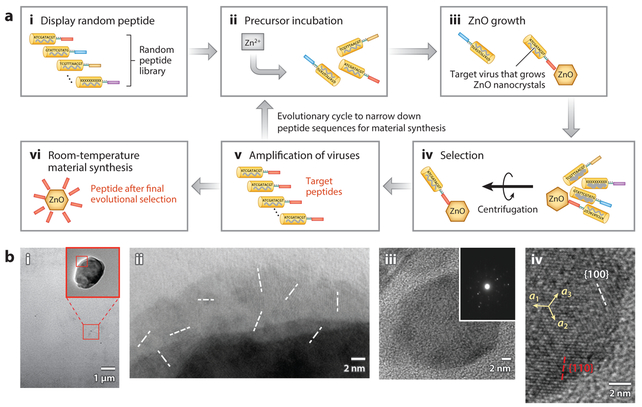
The Matsui group (72) developed a novel strategy to isolate catalytic peptides by utilizing the so-called anchor effect of nanocrystals. Figure 2a highlights the basic strategy of this method, in which ZnO crystallization was used as a proof of concept. The phage-displayed library and the zinc precursor were coincubated, and some of the peptides on the phage template accelerated the growth of ZnO nanocrystals. These crystals served as anchors and increased the total weight of the phage–ZnO complexes. Thereafter, the phage–ZnO complexes were retrieved from the library through centrifugation. After washing to remove nonspecific phages, the residual phage viruses were released from the ZnO anchor with an acid treatment and amplified. After the selection step was repeated three times, the sequences of the catalytic peptides displayed on the phage template were identified by DNA sequencing. One of the two peptides identified (ZP-1 peptide; GAMHLPWHMGTL) was determined to be the predominant sequence for room-temperature growth. Note that completely soluble precursors must be used. The use of zinc precursors that tend to form amorphous zinc hydroxide will yield mainly the Zn(OH)x intermediate and its affinity phage, rather than the target peptide capable of promoting nanocrystal formation.
Figure 2.
Combinatorial phage-display approach to the identification of peptides that crystallize zinc oxide (ZnO) at room temperature.(a) Screening of a phage-display peptide library. (i,ii) The phage-displayed library and the zinc precursor (10 mg/mL zinc nitrate solution) were coincubated for 3 days at room temperature. (iii) Some of the peptides on the phage template accelerated the growth of ZnO nanocrystals. (iv) The phage–ZnO complexes were retrieved from the library by centrifugation. (v) After washing to remove nonspecific phages, the residual phage viruses were released from the ZnO anchor with an acid treatment and amplified. (vi) The selected peptide ZP-1 can catalyze ZnO nanocrystal growth without a phage template by incubating the same zinc nitrate precursor and pure peptide. (b) Microscopy studies of ZnO mineralization process promoted by ZP-1 peptide. (i) Transmission electron microscopy (TEM) image of ZnO nanoparticles after 4 days’ incubation in the precursor solution. (Inset) Magnified image of the area within the red square. (ii) High-resolution TEM (HRTEM) image of the square region in panel i, showing nanoparticle domains with (100) faces (white dashed lines) oriented in random directions. (iii) TEM image of ZnO nanocrystals after 3 weeks’ incubation in the precursor solution with the peptide. (Inset) Nanobeam electron diffraction pattern of this nanocrystal showing single crystallinity, with a [0001] transmission direction. (iv) HRTEM image of panel iii, resolving (100) faces (white dashed lines) and (110) faces (red dashed lines). Faces are indicated by arrows.
Peptide ZP-1 was confirmed to catalyze ZnO nanocrystal growth without a phage template by incubating the same zinc nitrate precursor and pure peptide for 4 days. High-resolution transmission electron microscopy (HRTEM) and selected area electron diffraction (SAED) analyses demonstrated that these nanocrystals were wurtzite ZnO with a diameter of 20–100 nm (Figure 2b). In the absence of the ZP-1 peptide, no particles were observed. These results indicate that the biopanning method can indeed isolate catalytic peptides that promote inorganic nanocrystal growth. Interestingly, the ZP-1 peptide facilitated ZnO crystallization in an unusual direction; the nonpolar (100) and (110) faces were preferentially developed, in contrast to the conventional development of polar (001) and (00–1) faces. The tendency of ZnO to grow along unusual directions had previously been observed when the precursor’s access to the crystallization sites was insufficient (73). Although the mechanistic details of the peptide-promoted development of nonpolar (100) and (110) faces have yet to be determined, the charged groups on the ZP-1 can preferentially attach to the polar (001) and (00–1) faces of wurtzite, which would quench the anisotropic growth along these directions.
3.2. Hydrogel-Based Screening of Catalytic Peptides
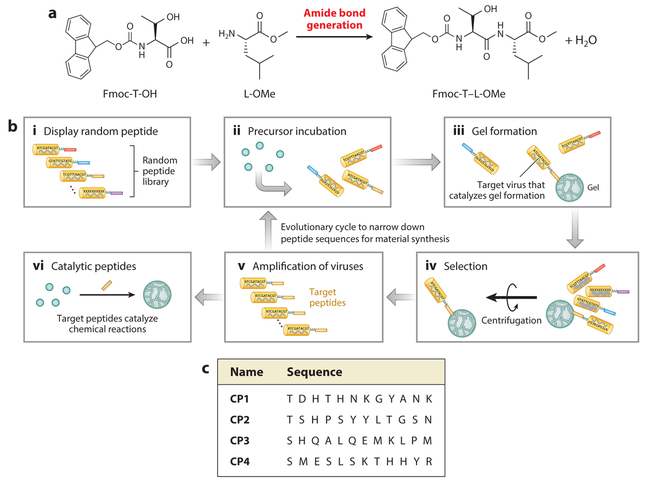
The key step in the screening method described above is the deposition of anchor materials, which enables separation of the specific phage-displaying catalytic peptides from the peptide library. This strategy could be used to discover catalytic peptides promoting a wide range of chemical reactions if those reactions could promote anchor aggregation. In collaboration with Ulijn’s group (74–76), we used peptide hydrogel as an anchor material. This hydrogel is formed by condensation of 9-fluorenylmethoxycarbonyl-threonine (Fmoc-T-OH) and leucine–methyl ester (L-OMe), which are amphiphilic peptide hydrogelators that are completely soluble prior to enzymatic reaction (Figure 3a). The addition of protease catalyzes reverse hydrolysis of a peptide bond driven by thermodynamically favorable self-assembly of the resulting dipeptide product, Fmoc-TL-OMe (Figure 3a). The self-assembly of the peptide hydrogelators is stabilized by hydrogen bonds and a π–π stacking network, which favor peptide synthesis over hydrolysis. Finally, the Fmoc-TLOMe peptide product assembles into nanofibers, thereby producing a hydrogel (74). Instead of a protease, a phage-display peptide library was used in the same system (Figure 3b). Some of the catalytic peptides mimicking protease function generated a peptide bond between Fmoc-T-OH and L-OMe, and the resulting hydrogel was deposited at the catalytic site (Figure 3b). The phages with the hydrogel were centrifuged, and the phage from the deposited hydrogel was eluted through the addition of an esterase that hydrolyzed the ester bond in Fmoc-TL-OMe. The hydrolysis of ester bond generated a hydrophilic carboxyl group, causing disassembly of the hydrogel. After this biopanning process was repeated, catalytic peptides exhibiting protease function were identified (77, 78).
Figure 3.
Hydrogel-based phage-display approach to the identification of enzyme-mimicking catalytic peptides. (a) A hydrogelation process triggered by an amide bond formation between 9-fluorenylmethoxycarbonyl-threonine (Fmoc-T) and leucine–methyl ester (L-OMe). (b) Identification of catalytic peptides using a phage-display peptide library. (c) Catalytic peptides displayed on the selected phage templates.
Although there is no obvious sequence similarity among the identified catalytic peptides, most of the selected peptides contain amino acids that are typically associated with nucleophilic catalysis (e.g., histidine, serine, aspartate, and glutamate) (Figure 3c), as observed in natural proteases(79), and they could play a role in catalytic function. Additionally, natural proteases demonstrate esterase activity because these two types of enzymes share the same catalytic center, which is referred to as a catalytic triad (79). This fact encouraged us to analyze whether the protease-mimicking peptides (CP1–CP4) could catalyze ester hydrolysis. A colorimetric assay using p-nitrophenyl acetate (pNPA) revealed that all of the protease-mimicking peptides tested (CP1–CP4) have esterase properties, with CP4 peptide (SMESLSKTHHYR) being the most active.
A critical advantage of this biopanning strategy for screening catalytic peptides is that it allows one to examine numerous catalytic activities by changing the types of hydrogel precursors for which different chemical reactions can trigger the gelation. In addition to condensation/hydrolysis of the peptide bond, ester hydrolysis (76) and dephosphorylation (80) reactions also trigger hydro-gelation and can be used to screen esterase- and phosphatase-mimicking peptides, respectively. Improvements to the molecular design of hydrogel anchors will likely allow the identification of peptides with desirable catalytic functions.
As described above, our biopanning method represents a general and promising strategy to isolate catalytic peptides with desired functions. However, the catalytic performance of the peptides selected using this genetic evolution method is still lower than that of enzymes developed through natural evolution. For example, the protease and esterase activities of the peptides identified in the hydrogel-based phage approach exhibited catalytic activities that were seven orders of magnitude lower than those of natural enzymes (77). This might be due to the lack of well-defined structures in the peptides. Thus, in order to make practical catalysts it is necessary to develop strategies that address the inherent shortcomings of peptides relative to larger proteins.
3.3. Catalytic Peptides for Material Synthesis in Nonaqueous Environments
A potential way to turn this weakness into strength involves the stability of peptides in harsh environments, where enzymes cannot maintain their functional properties. Enzymes are excellent catalysts in biological aqueous environments; however, their functional conformations tend to become denatured in abiotic nonaqueous environments, such as organic solvents. By contrast, flexible catalytic oligopeptides with no rigid 3D structures could maintain catalytic activities in such harsh environments. To test this hypothesis, investigators examined the catalytic ability of CP4 peptide (Figure 3c) to crystallize an oxide semiconductor material (82). Ester elimination(83) can synthesize various nanocrystalline materials, such as ZnO (83–85), CoO (86), CuO (87), Fe3O4/Fe2O3, and CoFe2O4 (88). Equation 1 shows the formation of ZnO via the solvothermal esterification reaction (83).
| (1) |
Metal acetate is dissolved in alcohol, followed by generation of a hydroxide intermediate and subsequent crystallization of the metal oxide. Although several combinations of metal acetate precursors and alcohol systems have been explored in order to create various functional metal oxides, the heating process [to ~100–300◦C (83)] is still necessary to facilitate the reaction and growth of the metal oxide. Biomolecular catalysts that promote this ester elimination at room temperature can aid the synthesis of the oxide semiconductor. However, the growth of the nanocrystals should be concomitant with the reverse hydrolysis of the ester bond, as shown in Equation 1, and such reverse hydrolysis reactions are favored and more efficient in organic solvents. Therefore, it is necessary to develop biomolecular catalysts that can tolerate organic solvents for the growth of highly crystalline metal oxide nanocrystals via reverse hydrolysis.
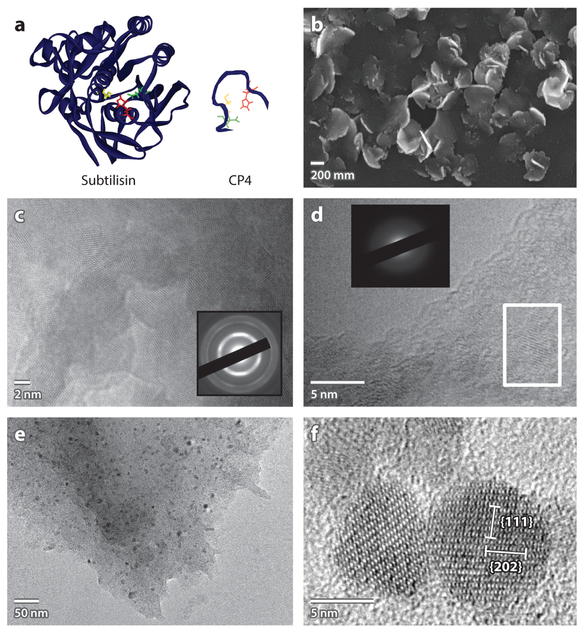
To achieve this goal, we tested the ability of CP4 peptide to promote ZnO formation in methanol. Zinc acetate precursor dissolved in methanol was mixed with CP4 peptide (Figures 3c and 4a) (82). After incubation for 3 days at room temperature, flake-shaped ZnO formed (Figure 4b). HRTEM and SAED analyses revealed that the resulting ZnO nanocrystals were polycrystalline (Figure 4c). Without the peptide, no crystal formation was observed, indicating that CP4 peptide has a catalytic function. A by-product, methyl acetate (Equation 1), was detected with gas chromatography–mass spectrometry, supporting the idea that ZnO was formed through an ester elimination reaction. To examine the advantages of oligopeptide biocatalysts over natural enzymes in organic solvents, we reacted the same precursor solution with a pro-tease/esterase enzyme, subtilisin (Figure 4a). Subtilisin-crystallized ZnO has inferior crystalline properties compared with ZnO crystallized in the presence of CP4 (Figure 4d). The result clearly indicates that CP4 peptide, which was evolved through hydrogel-based combinatory screening, catalyzed better growth of ZnO nanocrystals with higher crystallinity in an organic solvent in comparison to the protease enzyme. The ester elimination reaction catalyzed by CP4 peptide can be a general way to synthesize industrially important metal oxides. CP4 peptide also catalyzes the crystallization of the spinel ferrite MnFe2O4 (Figure 4e,f) (88a), which is used in electrode materials in lithium ion batteries (89, 90), magnetic storage (91), ferrofluids (92, 93), catalysts (94), and biomedical applications (95; Y. Maeda, J. Fang, Y. Kezoe, D. Pike, V. Nanda & H. Matsui, manuscript submitted). In a methanol–benzyl alcohol solvent system (50% volume ratio), CP4 peptide can crystallize MnFe2O4 nanoparticles under ambient temperature. According to super-conducting quantum interference device (SQUID) analysis, these MnFe2O4 nanoparticles exhibit superparamagnetism. The blocking temperature at which the transition between superparamagnetic and paramagnetic states occurs in the MnFe2O4 nanoparticles produced using CP4 peptide is similar to that of the conventional spinel ferrite nanoparticles synthesized at high temperature (96).
Figure 4.
Metal oxide crystallization mediated by catalytic peptide in methanol. (a) (Left) The natural protein subtilisin and (right) the catalytic oligopeptide CP4. Microscopic studies of ZnO nanocrystals synthesized through ester elimination with (b,c) CP4 peptide and (d) subtilisin at room temperature. (e,f) Transmission electron microscope images of crystalline MnFe2O4 prepared using an ester elimination reaction catalyzed by CP4.
3.4. Catalytic Peptides That Assist Self-Assembly
A combination of rational design and combinatorial selection of oligopeptide catalysts is another possible strategy to improve catalytic properties. The lack of a rigid 3D structure and a defined substrate-binding pocket lined by hydrophobic groups (97, 98) could be responsible for low catalytic function in aqueous environments. To address this issue, we fused a fragment (FFKLVFF) of the amyloid-β (Aβ) peptide to CP4 to simulate a hydrophobic pocket adjacent to the CP4 active site. In addition to providing hydrophobicity, the Aβ peptide led to self-assembly into amyloid fibrils (Figure 5a) (99–101). Such crowding effects could stabilize catalytically active conformations of the oligopeptide. We confirmed that CP4–Aβ (SMESLSKTHHYRFFKLVFF) assembled into a nanofibril structure (Figure 5b), and its esterase activity was enhanced fourfold. These results suggest that both hydrophobicity and crowding effects benefit the catalytic activity (Y. Maeda,J. Fang, Y. Ikezoe, D. Pike, V. Nanda & H. Matsui, manuscript submitted). Note that fusion to collagen (a different type of macromolecular crowding assembly system) did not improve the esterase activity. Therefore, the mode of Aβ self-assembly might specifically induce the formation of a catalytically active conformation. Indeed, detailed all-atom simulations suggest that CP4–Aβ has a greater propensity to form incipient catalytic triads in comparison to CP4 alone, a finding that is consistent with experimental results.
Figure 5.
Self-assembling strategy to enhance the esterase activity of CP4 peptide. (a) A schematic illustrationand (b) a transmission electron microscope image of a de novo designed fusion peptide (SMESLSKTHHYRFFKLVFF), where the catalytic peptide CP4 is fused with the amyloid-β (Aβ) peptide.
The combinatorial approach using a phage-display peptide library is a powerful tool for the design of catalytic peptides for both organic and inorganic reactions. An advantage of this approach is its versatility, allowing one to explore various types of catalytic peptides simply by changing hydrogel precursors. Once promising catalytic peptide candidates are discovered in peptide libraries, their activity could be further enhanced by varying the solvent or folding them into catalytically active conformations. These new strategies could lead to the creation of de novo catalytic peptides for use in various fields. A disadvantage of the combinatorial approach is that the selected peptides still have a lower catalytic activity than that of natural enzymes. However, self-assembly of catalytic peptides into larger assemblies with the appropriate conformation can give rise to practical peptide catalysts.
Designed catalytic peptides are not the first constructs to mimic protein functions using high-throughput methods. Some of the most successful examples are artificial antibodies (7, 102), which are engineered to stabilize a reaction’s transition state. For instance, investigators have engineered affibodies, composed of α-helical Z domain scaffolds decorated with affinity residues, by passing them through the screening cycles that randomize affinity residues and selecting the ones showing higher affinity (7). This approach, which utilizes rigid scaffolds, resembles our amyloid fusion strategy, and numerous scaffolds are available (103). Combination of screening via multiple selection rounds with approaches that rely on amino acid patterning (104) might produce catalysts with even better properties.
4. FUTURE PROSPECTS FOR NONCOMPUTATIONAL DESIGN OF CATALYTIC FUNCTION
The de novo design of efficient catalysts is a long and difficult process. Despite decades of effort and the development of sophisticated tools, we are still unable to design highly efficient catalysts that rival those created by nature. Nonetheless, progress in our understanding of enzymatic function has been significant. Even the introduction of a strategically placed single mutation into a nonenzymatic protein scaffold can lead to nascent catalytic activities (105–108). Thus, the application of even crude design concepts can lead to significant levels of catalytic activity, enough to use it as a starting point for subsequent directed evolution. From a practical standpoint several major challenges for protein design remain. Ability to simultaneously sample a large number of potential candidates is one of them. Indeed, screening (computationally or otherwise) the space of all 10300 possible sequence combinations for a TIM barrel protein is impossible. But is investigating this enormous and mostly empty sequence space in detail even necessary? Why can’t we limit our search to rationally preselected catalyst candidates to identify new catalysts? Unfortunately, neither our ability to identify potential enzyme candidates with an adequate degree of accuracy nor the technology to experimentally evaluate large enough protein libraries is currently sufficient.
The use of short oligopeptides as alternatives to large folded proteins has recently emerged as an efficient catalyst design tool. Not only are oligopeptides the building blocks of enzymes; they can serve as catalysts that promote a number of chemical reactions. A critical advantage of catalytic peptides is their small size in comparison to conventional enzymes, enabling easy molecular design. Moreover, existing high-throughput screening methods such as phage display can cover enough sequence space to completely probe all sequence of peptides containing as many as 8–10 residues. Although the catalytic enzymes’ properties are less favorable than those of their natural enzyme counterparts, several strategies can turn this weakness into strength, as described above. The ability of short peptides to assemble into large supramolecular structures provides additional advantages from a standpoint of practical applicability. Additionally, self-assembly allows one to simultaneously screen a large number of possible arrangements of functional groups by simply mixing peptides with different amino acid sequences, thereby accelerating the discovery process.
The near future will undoubtedly bring novel high-throughput combinatorial and computational techniques together with rational design to evolve peptide-based catalysts for practical use. In particular, recent advances in applying traditional high-throughput screening tools (e.g., ribosome display) to searches for catalytic activities could greatly expand our ability to design new catalytic functions (109).
ACKNOWLEDGMENTS
The writing of this review was supported in part by the US Department of Energy, Office of Basic Energy Sciences, Division of Materials Sciences and Engineering (award DE-FG-02–01ER45935) and by the Emerging Frontiers in Research and Innovation program of the National Science Foundation (grant 1332349), an Oak Ridge Associated Universities Ralph E. Powe Junior Faculty Enhancement award, and a Humboldt Fellowship to I.V.K. Hunter College infrastructure is supported by the National Institute on Minority Health and Health Disparities of the National Institutes of Health (MD007599).
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Swiderek K, Tunon I, Moliner V, Bertran J. 2015. Computational strategies for the design of new enzymatic functions. Arch. Biochem. Biophys 582:68–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Korendovych IV, DeGrado WF. 2014. Catalytic effciency of designed catalytic proteins. Curr. Opin. Struct. Biol 27:113–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wijma HJ, Janssen DB. 2013. Computational design gains momentum in enzyme catalysis engineering. FEBS J. 280:2948–60 [DOI] [PubMed] [Google Scholar]
- 4.Kries H, Blomberg R, Hilvert D. 2013. De novo enzymes by computational design. Curr. Opin. Chem. Biol 17:221–28 [DOI] [PubMed] [Google Scholar]
- 5.Cedrone F, Ménez A, Quéméneur E. 2000. Tailoring new enzyme functions by rational redesign. Curr. Opin. Struct. Biol 10:405–10 [DOI] [PubMed] [Google Scholar]
- 6.Chica RA, Doucet N, Pelletier JN. 2005. Semi-rational approaches to engineering enzyme activity: combining the benefits of directed evolution and rational design. Curr. Opin. Biotechnol 16:378–84 [DOI] [PubMed] [Google Scholar]
- 7.Nord K, Gunneriusson E, Ringdahl J, Ståhl S, Uhlén M, Nygren PA. 1997. Binding proteins selected from combinatorial libraries of an α-helical bacterial receptor domain. Nat. Biotechnol 15:772–77 [DOI] [PubMed] [Google Scholar]
- 8.Koivunen E, Arap W, Rajotte D, Lahdenranta J, Pasqualini R. 1999. Identification of receptor ligands with phage display peptide libraries. J. Nucl. Med 40:883–88 [PubMed] [Google Scholar]
- 9.Slavoff SA, Chen I, Choi YA, Ting AY. 2008. Expanding the substrate tolerance of biotin ligase through exploration of enzymes from diverse species. J. Am. Chem. Soc 130:1160–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarikaya M, Tamerler C, Jen AK, Schulten K, Baneyx F. 2003. Molecular biomimetics: nanotechnology through biology. Nat. Mater 2:577–85 [DOI] [PubMed] [Google Scholar]
- 11.Forrer P, Jung S, Plückthun A. 1999. Beyond binding: using phage display to select for structure, folding and enzymatic activity in proteins. Curr. Opin. Struct. Biol 9:514–20 [DOI] [PubMed] [Google Scholar]
- 12.Pedersen H, Holder S, Sutherlin DP, Schwitter U, King DS, Schultz PG. 1998. A method for directed evolution and functional cloning of enzymes. PNAS 95:10523–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demartis S, Huber A, Viti F, Lozzi L, Giovannoni L, et al. 1999. A strategy for the isolation of catalytic activities from repertoires of enzymes displayed on phage. J. Mol. Biol 286:617–33 [DOI] [PubMed] [Google Scholar]
- 14.Yang G, Withers SG. 2009. Ultrahigh-throughput FACS-based screening for directed enzyme evolution. ChemBioChem 10:2704–15 [DOI] [PubMed] [Google Scholar]
- 15.Chen I, Dorr BM, Liu DR. 2011. A general strategy for the evolution of bond-forming enzymes using yeast display. PNAS 108:11399–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lam SS, Martell JD, Kamer KJ, Deerinck TJ, Ellisman MH, et al. 2015. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 12:51–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zinchenko A, Devenish SR, Kintses B, Colin PY, Fischlechner M, Hollfelder F. 2014. One in a million: flow cytometric sorting of single cell-lysate assays in monodisperse picolitre double emulsion droplets for directed evolution. Anal. Chem 86:2526–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischlechner M, Schaerli Y, Mohamed MF, Patil S, Abell C, Hollfelder F. 2014. Evolution of enzyme catalysts caged in biomimetic gel-shell beads. Nat. Chem 6:791–96 [DOI] [PubMed] [Google Scholar]
- 19.Lutz S 2010. Beyond directed evolution—semi-rational protein engineering and design. Curr. Opin. Biotechnol 21:734–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pollack SJ, Jacobs JW, Schultz PG. 1986. Selective chemical catalysis by an antibody. Science 234:1570–73 [DOI] [PubMed] [Google Scholar]
- 21.Tramontano A, Janda KD, Lerner RA. 1986. Catalytic antibodies. Science 234:1566–70 [DOI] [PubMed] [Google Scholar]
- 22.Esposito A, Delort E, Lagnoux D, Djojo F, Reymond JL. 2003. Catalytic peptide dendrimers. Angew. Chem. Int. Ed. Engl 42:1381–83 [DOI] [PubMed] [Google Scholar]
- 23.Douat-Casassus C, Darbre T, Reymond JL. 2004. Selective catalysis with peptide dendrimers. J. Am. Chem. Soc 126:7817–26 [DOI] [PubMed] [Google Scholar]
- 24.Colby Davie EA, Mennen SM, Xu Y, Miller SJ. 2007. Asymmetric catalysis mediated by synthetic peptides. Chem. Rev 107:5759–812 [DOI] [PubMed] [Google Scholar]
- 25.Kazlauskas RJ, Bornscheuer UT. 2009. Finding better protein engineering strategies. Nat. Chem. Biol 5:526–29 [DOI] [PubMed] [Google Scholar]
- 26.Reetz MT. 2013. Biocatalysis in organic chemistry and biotechnology: past, present, and future. J. Am. Chem. Soc 135:12480–96 [DOI] [PubMed] [Google Scholar]
- 27.Makhlynets OV, Raymond EA, Korendovych IV. 2015. Design of allosterically regulated protein catalysts. Biochemistry 54:1444–56 [DOI] [PubMed] [Google Scholar]
- 28.Yu F, Cangelosi VM, Zastrow ML, Tegoni M, Plegaria JS, et al. 2014. Protein design: toward functional metalloenzymes. Chem. Rev 114:3495–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roy A, Sommer DJ, Schmitz RA, Brown CL, Gust D, et al. 2014. A de novo designed 2[4Fe–4S] ferredoxin mimic mediates electron transfer. J. Am. Chem. Soc 136:17343–49 [DOI] [PubMed] [Google Scholar]
- 30.Sommer DJ, Roy A, Astashkin A, Ghirlanda G. 2015. Modulation of cluster incorporation specificity in a de novo iron–sulfur cluster binding peptide. Biopolymers 104:412–18 [DOI] [PubMed] [Google Scholar]
- 31.Cangelosi VM, Deb A, Penner-Hahn JE, Pecoraro VL. 2014. A de novo designed metalloenzyme for the hydration of CO2. Angew. Chem. Int. Ed. Engl 53:7900–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zastrow ML, Peacock AF, Stuckey JA, Pecoraro VL. 2012. Hydrolytic catalysis and structural stabilization in a designed metalloprotein. Nat. Chem 4:118–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tegoni M, Yu F, Bersellini M, Penner-Hahn JE, Pecoraro VL. 2012. Designing a functional type 2 copper center that has nitrite reductase activity within α-helical coiled coils. PNAS 109:21234–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu F, Penner-Hahn JE, Pecoraro VL. 2013. De novo–designed metallopeptides with type 2 copper centers: modulation of reduction potentials and nitrite reductase activities. J. Am. Chem. Soc 135:18096–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farid TA, Kodali G, Solomon LA, Lichtenstein BR, Sheehan MM, et al. 2013. Elementary tetrahelical protein design for diverse oxidoreductase functions. Nat. Chem. Biol 9:826–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miner KD, Mukherjee A, Gao Y-G, Null EL, Petrik ID, et al. 2012. A designed functional metalloenzyme that reduces O2 to H2O with over one thousand turnovers. Angew. Chem. Int. Ed 51:5589–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu Y, Cui C, Liu X, Petrik ID, Wang J, Lu Y. 2015. A designed metalloenzyme achieving the catalytic rate of a native enzyme. J. Am. Chem. Soc 137:11570–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleingardner JG, Kandemir B, Bren KL. 2014. Hydrogen evolution from neutral water under aerobic conditions catalyzed by cobalt microperoxidase 11. J. Am. Chem. Soc 136:4–7 [DOI] [PubMed] [Google Scholar]
- 39.Der BS, Edwards DR, Kuhlman B. 2012. Catalysis by a de novo zinc-mediated protein interface: implications for natural enzyme evolution and rational enzyme engineering. Biochemistry 51:3933–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song WJ, Tezcan FA. 2014. A designed supramolecular protein assembly with in vivo enzymatic activity. Science 346:1525–28 [DOI] [PubMed] [Google Scholar]
- 41.Mayer C, Gillingham DG, Ward TR, Hilvert D. 2011. An artificial metalloenzyme for olefin metathesis. Chem. Commun 47:12068–70 [DOI] [PubMed] [Google Scholar]
- 42.Philippart F, Arlt M, Gotzen S, Tenne SJ, Bocola M, et al. 2013. A hybrid ring-opening metathesis polymerization catalyst based on an engineered variant of the β-barrel protein FhuA. Chemistry 19:13865–71 [DOI] [PubMed] [Google Scholar]
- 43.Sauer DF, Bocola M, Broglia C, Arlt M, Zhu LL, et al. 2015. Hybrid ruthenium ROMP catalysts based on an engineered variant of β-barrel protein FhuA ΔCVFtev: effect of spacer length. Chem. Asian J. 10:177–82 [DOI] [PubMed] [Google Scholar]
- 44.Lo C, Ringenberg MR, Gnandt D, Wilson Y, Ward TR. 2011. Artificial metalloenzymes for olefin metathesis based on the biotin–(strept)avidin technology. Chem. Commun 47:12065–67 [DOI] [PubMed] [Google Scholar]
- 45.Matsuo T, Imai C, Yoshida T, Saito T, Hayashi T, Hirota S. 2012. Creation of an artificial metalloprotein with a Hoveyda–Grubbs catalyst moiety through the intrinsic inhibition mechanism of α-chymotrypsin. Chem. Commun 48:1662–64 [DOI] [PubMed] [Google Scholar]
- 46.Genz M, Koehler V, Krauss M, Singer D, Hoffmann R, et al. 2014. An artificial imine reductase based on the ribonuclease S scaffold. ChemCatChem 6:736–40 [Google Scholar]
- 47.Monnard FW, Nogueira ES, Heinisch T, Schirmer T, Ward TR. 2013. Human carbonic anhydrase II as host protein for the creation of artificial metalloenzymes: the asymmetric transfer hydrogenation of imines. Chem. Sci 4:3269–74 [Google Scholar]
- 48.Fukumoto K, Onoda A, Mizohata E, Bocola M, Inoue T, et al. 2014. Rhodium-complex-linked hybrid biocatalyst: stereo-controlled phenylacetylene polymerization within an engineered protein cavity. ChemCatChem 6:1229–35 [Google Scholar]
- 49.Onoda A, Fukumoto K, Arlt M, Bocola M, Schwaneberg U, Hayashi T. 2012. A rhodium complex–linked β-barrel protein as a hybrid biocatalyst for phenylacetylene polymerization. Chem. Commun 48:9756–58 [DOI] [PubMed] [Google Scholar]
- 50.Srivastava P, Yang H, Ellis-Guardiola K, Lewis JC. 2015. Engineering a dirhodium artificial metal-loenzyme for selective olefin cyclopropanation. Nat. Commun 6:7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang C, Srivastava P, Ellis-Guardiola K, Lewis JC. 2014. Manganese terpyridine artificial metalloenzymes for benzylic oxygenation and olefin epoxidation. Tetrahedron 70:4245–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Allard M, Dupont C, Muñoz Robles V, Doucet N, Lledós A, et al. 2012. Incorporation of manganese complexes into xylanase: new artificial metalloenzymes for enantioselective epoxidation. ChemBioChem 13:240–51 [DOI] [PubMed] [Google Scholar]
- 53.Sansiaume-Dagousset E, Urvoas A, Chelly K, Ghattas W, Maréchal J-D, et al. 2014. Neocarzinostatin-based hybrid biocatalysts for oxidation reactions. Dalton Trans. 43:8344–54 [DOI] [PubMed] [Google Scholar]
- 54.Inaba H, Kanamaru S, Arisaka F, Kitagawa S, Ueno T. 2012. Semi-synthesis of an artificial scandium(III) enzyme with a β-helical bio-nanotube. Dalton Trans. 41:11424–27 [DOI] [PubMed] [Google Scholar]
- 55.Lewandowski B, Wennemers H. 2014. Asymmetric catalysis with short-chain peptides. Curr. Opin. Chem. Biol 22:40–46 [DOI] [PubMed] [Google Scholar]
- 56.Rufo CM, Moroz YS, Moroz OV, Stohr J, Smith TA, et al. 2014. Short peptides self-assemble to produce catalytic amyloids. Nat. Chem 6:303–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Friedmann MP, Torbeev V, Zelenay V, Sobol A, Greenwald J, et al. 2015. Towards prebiotic catalytic amyloids using high throughput screening. PLOS ONE 10:e0143948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang CQ, Xue XD, Luo Q, Li YW, Yang KN, et al. 2014. Self-assembled peptide nanofibers designed as biological enzymes for catalyzing ester hydrolysis. ACS Nano 8:11715–23 [DOI] [PubMed] [Google Scholar]
- 59.Huang Z, Guan S, Wang Y, Shi G, Cao L, et al. 2013. Self-assembly of amphiphilic peptides into bio-functionalized nanotubes: a novel hydrolase model. J. Mater. Chem. B 1:2297–304 [DOI] [PubMed] [Google Scholar]
- 60.Berdugo C, Miravet JF, Escuder B. 2013. Substrate selective catalytic molecular hydrogels: the role of the hydrophobic effect. Chem. Commun 49:10608–10 [DOI] [PubMed] [Google Scholar]
- 61.Jin Q, Zhang L, Cao H, Wang T, Zhu X, et al. 2011. Self-assembly of copper(II) ion–mediated nanotube and its supramolecular chiral catalytic behavior. Langmuir 27:13847–53 [DOI] [PubMed] [Google Scholar]
- 62.Weingarten AS, Kazantsev RV, Palmer LC, McClendon M, Koltonow AR, et al. 2014. Self-assembling hydrogel scaffolds for photocatalytic hydrogen production. Nat. Chem 6:964–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fry HC, Garcia JM, Medina MJ, Ricoy UM, Gosztola DJ, et al. 2012. Self-assembly of highly ordered peptide amphiphile metalloporphyrin arrays. J. Am. Chem. Soc 134:14646–49 [DOI] [PubMed] [Google Scholar]
- 64.Fry HC, Liu Y, Dimitrijevic NM, Rajh T. 2014. Photoinitated charge separation in a hybrid titanium dioxide metalloporphyrin peptide material. Nat. Commun 5:8. [DOI] [PubMed] [Google Scholar]
- 65.Naik RR, Stringer SJ, Agarwal G, Jones SE, Stone MO. 2002. Biomimetic synthesis and patterning of silver nanoparticles. Nat. Mater 1:169–72 [DOI] [PubMed] [Google Scholar]
- 66.Naik RR, Jones SE, Murray CJ, McAuliffe JC, Vaia RA, Stone MO. 2004. Peptide templates for nanoparticle synthesis derived from polymerase chain reaction–driven phage display. Adv. Funct. Mater 14:25–30 [Google Scholar]
- 67.Pappalardo G, Impellizzeri G, Bonomo RP, Campagna T, Grasso G, Saita MG. 2002. Copper(II) and nickel(II) binding modes in a histidine-containing model dodecapeptide. New J. Chem 26:593–600 [Google Scholar]
- 68.Yu L, Banerjee IA, Shima M, Rajan K, Matsui H. 2004. Size-controlled Ni nanocrystal growth on peptide nanotubes and their magnetic properties. Adv. Mater 16:709–12 [Google Scholar]
- 69.Banerjee IA, Yu L, Matsui H. 2003. Cu nanocrystal growth on peptide nanotubes by biomineralization: size control of Cu nanocrystals by tuning peptide conformation. PNAS 100:14678–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Umetsu M, Mizuta M, Tsumoto K, Ohara S, Takami S, et al. 2005. Bioassisted room-temperature immobilization and mineralization of zinc oxide—the structural ordering of ZnO nanoparticles into a flower-type morphology. Adv. Mater 17:2571–75 [Google Scholar]
- 71.Mao C, Flynn CE, Hayhurst A, Sweeney R, Qi J, et al. 2003. Viral assembly of oriented quantum dot nanowires. PNAS 100:6946–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wei Z, Maeda Y, Matsui H. 2011. Discovery of catalytic peptides for inorganic nanocrystal synthesis by a combinatorial phage display approach. Angew. Chem. Int. Ed. Engl 50:10585–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kisailus D, Schwenzer B, Gomm J, Weaver JC, Morse DE. 2006. Kinetically controlled catalytic formation of zinc oxide thin films at low temperature. J. Am. Chem. Soc 128:10276–80 [DOI] [PubMed] [Google Scholar]
- 74.Toledano S, Williams RJ, Jayawarna V, Ulijn RV. 2006. Enzyme-triggered self-assembly of peptide hydrogels via reversed hydrolysis. J. Am. Chem. Soc 128:1070–71 [DOI] [PubMed] [Google Scholar]
- 75.Williams RJ, Smith AM, Collins R, Hodson N, Das AK, Ulijn RV. 2009. Enzyme-assisted self-assembly under thermodynamic control. Nat. Nanotechnol 4:19–24 [DOI] [PubMed] [Google Scholar]
- 76.Das AK, Collins R, Ulijn RV. 2008. Exploiting enzymatic (reversed) hydrolysis in directed self-assembly of peptide nanostructures. Small 4:279–87 [DOI] [PubMed] [Google Scholar]
- 77.Maeda Y, Javid N, Duncan K, Birchall L, Gibson KF, et al. 2014. Discovery of catalytic phages by biocatalytic self-assembly. J. Am. Chem. Soc 136:15893–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Matsui H, Maeda Y, Ulijn RV. 2013. Method for screening catalytic peptides using phage display technology. US Patent WO2015002649A1
- 79.Ekici OD, Paetzel M, Dalbey RE. 2008. Unconventional serine proteases: variations on the catalytic Ser/His/Asp triad configuration. Protein Sci. 17:2023–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sadownik JW, Leckie J, Ulijn RV. 2011. Micelle to fibre biocatalytic supramolecular transformation of an aromatic peptide amphiphile. Chem. Commun 47:728–30 [DOI] [PubMed] [Google Scholar]
- 81.Deleted in proof.
- 82.Maeda Y, Wei Z, Ikezoe Y, Matsui H. 2015. Enzyme-mimicking peptides to catalytically grow ZnO nanocrystals in non-aqueous environments. ChemMatNano 1:319–23 [Google Scholar]
- 83.Du H, Yuan F, Huang S, Li J, Zhu Y. 2004. A new reaction to ZnO nanoparticles. Chem. Lett 33:770–71 [Google Scholar]
- 84.Demir MM, Muñoz-Espí R, Lieberwirth I, Wegner G. 2006. Precipitation of monodisperse ZnO nanocrystals via acid-catalyzed esterification of zinc acetate. J. Mater. Chem 16:2940–47 [Google Scholar]
- 85.Joo J, Kwon SG, Yu JH, Hyeon T. 2005. Synthesis of ZnO nanocrystals with cone, hexagonal cone, and rod shapes via non-hydrolytic ester elimination sol-gel reactions. Adv. Mater 17:1873–77 [Google Scholar]
- 86.Ye Y, Yuan F, Li S. 2006. Synthesis of CoO nanoparticles by esterification reaction under solvothermal conditions. Mater. Lett 60:3175–78 [Google Scholar]
- 87.Hong Z, Cao Y, Deng J. 2002. A convenient alcohothermal approach for low temperature synthesis of CuO nanoparticles. Mater. Lett 52:34–38 [Google Scholar]
- 88.Baldi G, Bonacchi D, Franchini MC, Gentili D, Lorenzi G, et al. 2007. Synthesis and coating of cobalt ferrite nanoparticles: a first step toward the obtainment of new magnetic nanocarriers. Langmuir 23:4026–28 [DOI] [PubMed] [Google Scholar]
- 88a.Maeda Y, Wei Z, Ikezoe Y, Tam E, Matsui H. 2015. Biomimetic crystallization of MnFe2O4 mediated by peptide-catalyzed esterification at low temperature. ChemNanoMat 2:419–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chu YQ, Fu ZW, Qin QZ. 2004. Cobalt ferrite thin films as anode material for lithium ion batteries. Electrochim. Acta 49:4915–21 [Google Scholar]
- 90.Zhang D, Zhang X, Ni X, Song JM, Zheng H. 2006. Low-temperature fabrication of MnFe2O4 octahedrons: magnetic and electrochemical properties. Chem. Phys. Lett 426:120–23 [Google Scholar]
- 91.Lee DK, Kim YH, Kang YS, Stroeve P. 2005. Preparation of a vast CoFe2O4 magnetic monolayer by Langmuir–Blodgett technique. J. Phys. Chem. B 109:14939–44 [DOI] [PubMed] [Google Scholar]
- 92.Auzans E, Zins D, Blums E, Massart R. 1999. Synthesis and properties of Mn-Zn ferrite ferrofluids.J. Mater. Sci 34:1253–60 [Google Scholar]
- 93.Tourinho FA, Franck R, Massart R. 1990. Aqueous ferrofluids based on manganese and cobalt ferrites.J. Mater. Sci 25:3249–54 [Google Scholar]
- 94.Kanazawa A, Kanaoka S, Yagita N, Oaki Y, Imai H, et al. 2012. Biologically synthesized or bioinspired process-derived iron oxides as catalysts for living cationic polymerization of a vinyl ether. Chem. Commun 48:10904–6 [DOI] [PubMed] [Google Scholar]
- 95.Lee JH, Huh YM, Jun YW, Seo JW, Jang JT, et al. 2007. Artificially engineered magnetic nanoparticles for ultra-sensitive molecular imaging. Nat. Med 13:95–99 [DOI] [PubMed] [Google Scholar]
- 96.Vestal CR, Song Q, Zhang ZJ. 2004. Effects of interparticle interactions upon the magnetic properties of CoFe2O4 and MnFe2O4 nanocrystals. J. Phys. Chem. B 108:18222–27 [Google Scholar]
- 97.Sorensen SB, Bech LM, Meldal M, Breddam K. 1993. Mutational replacements of the amino acid residues forming the hydrophobic S4 binding pocket of subtilisin 309 from Bacillus lentus. Biochemistry 32:8994–99 [DOI] [PubMed] [Google Scholar]
- 98.Carey C, Cheng Y-K, Rossky PJ. 2000. Hydration structure of the α-chymotrypsin substrate binding pocket: the impact of constrained geometry. Chem. Phys 258:415–25 [Google Scholar]
- 99.Hamley I, Krysmann M. 2008. Effect of PEG crystallization on the self-assembly of PEG–peptide copolymers containing amyloid peptide fragments. Langmuir 24:8210–14 [DOI] [PubMed] [Google Scholar]
- 100.Krysmann M, Castelletto V, Hamley I. 2007. Fibrillisation of hydrophobically modified amyloid peptide fragments in an organic solvent. Soft Matter 3:1401–6 [DOI] [PubMed] [Google Scholar]
- 101.Castelletto V, Zhu N, Hamley I, Noirez L. 2010. Self-assembly of PEGylated peptide conjugates containing a modified amyloid β-peptide fragment. Langmuir 26:9986–96 [DOI] [PubMed] [Google Scholar]
- 102.Suzuki N, Fujii I. 1999. Optimization of the loop length for folding of a helix-loop-helix peptide. Tetrahedron Lett. 40:6013–17 [Google Scholar]
- 103.Duncan KL, Ulijn RV. 2015. Short peptides in minimalistic biocatalyst design. Biocatalysis 1:67–81 [Google Scholar]
- 104.Patel SC, Hecht MH. 2012. Directed evolution of the peroxidase activity of a de novo–designed protein. Protein Eng. Des. Sel 25:445–52 [DOI] [PubMed] [Google Scholar]
- 105.Korendovych IV, Kulp DW, Wu Y, Cheng H, Roder H, DeGrado WF. 2011. Design of a switchable eliminase. PNAS 108:6823–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Moroz OV, Moroz YS, Wu Y, Olsen AB, Cheng H, et al. 2013. A single mutation in a regulatory protein produces evolvable allosterically regulated catalyst of unnatural reaction. Angew. Chem. Int. Ed 52:6246–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Moroz YS, Dunston TT, Makhlynets OV, Moroz OV, Wu Y, et al. 2015. New tricks for old proteins: Single mutations in a non-enzymatic protein give rise to various catalytic activities. J. Am. Chem. Soc 137:14905–11 [DOI] [PubMed] [Google Scholar]
- 108.Raymond EA, Mack KL, Yoon JH, Moroz OV, Moroz YS, Korendovych IV. 2015. Design of an allosterically regulated retroaldolase. Protein Sci. 24:561–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Seelig B, Szostak JW. 2007. Selection and evolution of enzymes from a partially randomized non-catalytic scaffold. Nature 448:828–31 [DOI] [PMC free article] [PubMed] [Google Scholar]