Abstract
Carbonic anhydrase VI (CA6) catalyses the reversible hydration of carbon dioxide in saliva with possible pH regulation, taste perception, and tooth formation effects. This study assessed effects of variation in the CA6 gene on oral microbiota and specifically the acidophilic and caries-associated Streptococcus mutans in 17-year old Swedish adolescents (n = 154). Associations with caries status and secreted CA6 protein were also evaluated. Single Nucleotide Polymorphisms (27 SNPs in 5 haploblocks) and saliva and tooth biofilm microbiota from Illumina MiSeq 16S rDNA (V3-V4) sequencing and culturing were analysed. Haploblock 4 (rs10864376, rs3737665, rs12138897) CCC associated with low prevalence of S. mutans (OR (95% CI): 0.5 (0.3, 0.8)), and caries (OR 0.6 (0.3, 0.9)), whereas haploblock 4 TTG associated with high prevalence of S. mutans (OR: 2.7 (1.2, 5.9)) and caries (OR: 2.3 (1.2, 4.4)). The TTG-haploblock 4 (represented by rs12138897(G)) was characterized by S. mutans, Scardovia wiggsiae, Treponema sp. HOT268, Tannerella sp. HOT286, Veillonella gp.1 compared with the CCC-haploblock 4 (represented by rs12138897(C)). Secreted CA6 in saliva was weakly linked to CA6 gene variation. In conclusion, the results indicate that CA6 gene polymorphisms influence S. mutans colonization, tooth biofilm microbiota composition and risk of dental caries in Swedish adolescents.
Introduction
Carbonic anhydrases (CAs) are zinc enzymes that catalyse the reversible hydration of carbon dioxide in the reaction CO2 + H2O ⇔ HCO3− + H+ and are important for pH homeostasis in body tissues and fluids and for removal of intracellular carbon dioxide1. In mammalian cells, CAs are involved in several biological processes, including HCO3− dependent metabolic processes, secretion of electrolytes, respiration, pH regulation, bone resorption, biomineralization, and odontogenesis2. Today, 16 different CA isozymes have been identified in humans1, of which 13 have been shown to be enzymatically active. Some isozymes are expressed in most tissues, while others are tissue- or organ-specific. Five isozymes are cytosolic (I, II, III, VII and XIII), five are membrane-bound (IV, IX, XII, XIV and XV), two are present in the mitochondria (VA and VB), and one (VI, CA6) is a secreted isozyme3,4.
CA6 is secreted into saliva with considerable individual variation in concentration and activity5, and incorporated in the protein pellicle on tooth enamel and in dental biofilms6–8. CA6 supports neutralization of lactic and other bacteria-produced acids through conversion of saliva HCO3− to water and carbon dioxide and pH maintenance through the subsequent phase buffering9. CA6 is therefore suggested to be an important enzyme in oral physiology and tooth tissues integrity, i.e., resistance to caries and dental erosion6,10. However, data are conflicting as both high and low concentrations and activities of CA6 have been associated with caries4,11,12.
Single nucleotide polymorphisms (SNPs) and multisite haploblocks in the CA6 gene have been linked to CA6 concentration and activity in saliva and to caries status13–17, but findings are not consistent18. However, no study has confirmed the association between CA6 in saliva and S. mutans colonization in humans and no study has investigated CA6 polymorphism and levels of caries associated bacteria or overall oral microbiota.
The primary aim of this study was to evaluate the effects of genetic variation in the CA6 gene region on oral microbiota and specifically the acidophilic and caries-associated S. mutans in Swedish adolescents. Secondary aims were to evaluate associations between variation in the CA6 gene region and secreted CA6 protein and caries status.
Results
Participant characteristics
Using next-generation sequencing (NGS), 75.3% of participants had detectable S. mutans, with significantly higher proportions among the caries-affected than caries-free subjects (p < 0.001; Table 1). Overall, the median caries score (DeFS) was 2.0 tooth surfaces and 67% of participants were affected by caries (DeFS ≥ 1) and 33% were caries-free (DeFS = 0) (Table 1). There were no significant differences between those with detectable S. mutans versus not or those who were caries-affected versus not with respect to sex, smoking, sweet snacking, BMI, and number of tooth surfaces. The proportion who reported brushing the teeth twice a day did not differ between those with S. mutans versus not, but it was significantly lower among caries-affected than caries-free participants (p < 0.001; Table 1).
Table 1.
Characteristics of the 17-year-old participants all together and by S. mutans and caries status.
| All participants | S. mutans status by NGS | Caries status | |||||
|---|---|---|---|---|---|---|---|
| S. mutans free | S. mutans present | p-value | Caries-free | Caries-affected | p-value | ||
| (n = 154) | (n = 38) | (n = 116) | (n = 51) | (n = 102) | |||
| Sex, % male | 44.2 | 50.0 | 42.2 | 0.403 | 47.1 | 42.2 | 0.565 |
| Smoker, % | 2.6 | 0 | 3.5 | 0.244 | 2.0 | 2.9 | 0.600 |
| Tooth brushing twice daily, % | 83.8 | 89.5 | 81.9 | 0.272 | 98.0 | 76.5 | <0.001 |
| Sweet snack frequency/day | 1.1 (0.5, 2.7) | 1.2 (0.6, 3.8) | 0.9 (0.4, 3.6) | 0.182 | 1.1 (0.5, 2. 7) | 1.0 (0.4, 2.6) | 0.763 |
| BMIc, kg/m2 | 21.6 (18.5, 26.0) | 22.4 (18.2, 26.7) | 21.4 (18.6, 25.9) | 0.533 | 21.3 (18.4, 25.3) | 22.0 (18.7, 26.3) | 0.169 |
| Dental status | |||||||
| Number of tooth surfaces | 128 (108, 128) | 128 (128, 129) | 128 (108, 128) | 0.010 | 128 (118, 132) | 128 (108, 128) | 0.049 |
| DeFS | 2.0 (0, 17.0) | 0.0 (0.0, 7.0) | 3.0 (0.0, 18.4) | <0.001 | 0 (0, 0) | 4.0 (1.0, 19.7) | — |
| S. mutans | |||||||
| Proportion with S. mutans by NGS, % | 75.3 | — | — | — | 58.8 | 84.3 | <0.001 |
| Mutans streptococci | |||||||
| CFU/mL, log10-value | 2.8 (0, 4.8) | 0.0 (0.0, 3.2) | 3.1 (0.0, 5.1) | <0.001 | 2.27 (0, 4.17) | 2.92 (0, 5.16) | 0.035 |
| Proportion with detection by culture, % | 61.4a | 64.9 | 69.8 | 52.0 | 65.7 | 0.104 | |
| Saliva | |||||||
| Protein concentration, mg/mL | 0.6 (0.3, 0.9) | 0.7 (0.3, 1.4) | 0.5 (0.3, 0.9) | 0.971 | 0.5 (0.3, 0.8) | 0.6 (0.3, 1.0) | 0.702 |
| Flow rate, ml/min | 1.5 (0.7, 2.5) | 1.5 (0.2, 3.2) | 1.4 (0.6, 2.5) | 0.925 | 1.6 (0.8, 2.7) | 1.4 (0.5, 2.5) | 0.066 |
| CA6 protein in saliva | |||||||
| Proportion (%) of total protein in saliva | 1.6 (0.1, 3.7) | 1.8 (0.1, 3.8) | 1.6 (0.1, 3.6) | 0.430 | 2.0 (0.03, 4.2) | 1.5 (0.08, 3.8) | 0.146 |
| Concentration, µg/mL | 8.4 (0.3, 17.0) | 11.5 (0.03, 35.6) | 7.7 (0.3, 16.7) | 0.351 | 9.8 (0.2, 19.4) | 7.8 (0.3, 17.0) | 0.203 |
| Concentration, µg/mL high tertile, % | 33.3 | 41.2 | 31.5 | 0.446 | 50.0 | 25.8 | 0.024 |
| Secreted amounts, µg/min | 11.0 (0.4, 25.4) | 12.8 (1.1, 30.5) | 10.6 (4.1, 25.7) | 0.518 | 17.1 (0.2, 54.2) | 8.3 (0.6, 20.2) | 0.012 |
| Secreted amounts, µg/min, high tertile, % | 33.7 | 35.3 | 33.3 | 0.878 | 53.6 | 24.6 | 0.007 |
Caries information was missing for 1 participant who was excluded. Continuous measures are presented as medians (10, 90 percentiles) and group differences were tested with Mann Whitney U test. Differences between group numbers (presented as %) were tested with Chi2 or Fisher’s exact test. P-values were considered significant at FDR < 0.25 but significant differences at FDR ≤ 0.06 are also indicated in bold. CFU for colony forming units and NGS for Next Generation Sequencing of DNA extracted from saliva (n = 152) or tooth biofilm (n = 139).
CA6 single nucleotide polymorphic sites and gene structure
After quality control measures, 27 out of 30 Single Nucleotide Polymorphisms (SNPs) were retained for analyses. Two SNP (rs2274333 and rs17032942) were excluded due to sample call rate of 0%. An additional SNP (rs6697763) was excluded from analysis for deviation from Hardy-Weinberg equilibrium (p = 0.033), leaving 27 SNPs for further analyses.
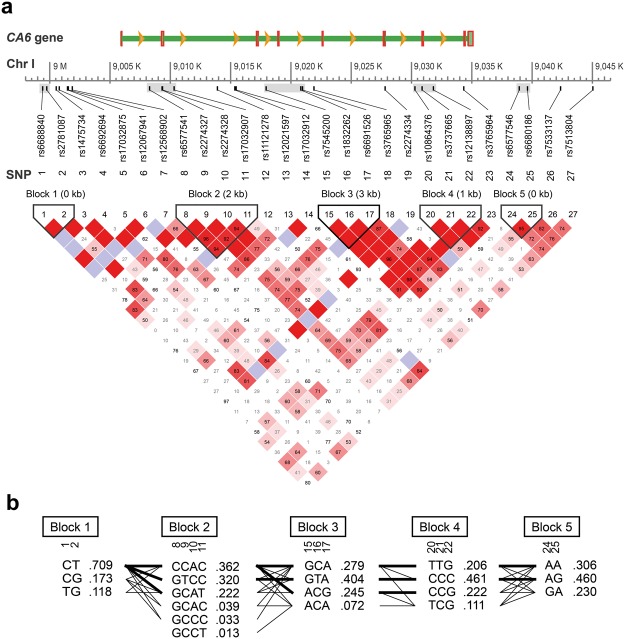
Evaluating potential multisite haploblocks within or around the CA6 gene resulted in five haploblocks (Fig. 1). Haploblock 1, comprised two SNPs (rs6688840 and rs2781087) and was located upstream of the first coding exon (+6387 to +6050 bp), whereas Haploblock 5 comprised rs6577546 and rs6680186 and was located downstream of the stop codon (−3871 to −4553 bp) of the CA6 gene. Haploblocks 2, 3 and 4 where within the CA6 gene; haploblock 2 with four SNPs (rs6577541, rs2274327, rs2274328, and rs17032907) covered the 2477 to 4512 bp region; haploblock 3 with three SNPs (rs7545200, rs1832262, and rs6691526) covered the 12,141 to 15,160 bp region; and haploblock 4 with three SNPs (rs10864376, rs3737665, and rs12138897) covered the 24,479 to 26,010 bp region of the CA6 gene.
Figure 1.
Haplotype map of CA6 locus. (a) The CA6 gene structure and distribution of selected and quality-controlled tag single nucleotide polymorphic sites (SNP). Graphics illustrates pairwise linkage disequilibrium (LD) between polymorphisms and each square display coefficient of linkage disequilibrium D′ value (%) between marker pairs. In the event of D′ = 100% where no recombination events are observed, the boxes are empty. (b) Haploblocks are estimated using Haploview as described in the method section. Uncommon haplotypes (observed in <1% of participants) are not shown. Genetically linked haploblocks are connected with (inherited together with >1% (thin line) or >10% (thick line) together) lines.
CA6 gene variation and S. mutans colonization by NGS
Five SNPs, i.e., rs10864376 (T), rs3737665 (T), rs12138897 (G), rs3765964 (A), and rs6680186 (A) were associated with increased odds of detectable S. mutans in saliva or tooth biofilm by NGS (Table 2). Different genetic models (allelic-, dominant- and recessive model) yielded qualitatively similar conclusion, but with some variation in test statistics (Fig. 2).
Table 2.
CA6 SNP and haploblock associations with S. mutans detection in both saliva and tooth biofilm by NGS.
| SNP | Haploblock | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP number | Name | Allele | Detected vs. Not detected | Haplo-block number | Allele | Frequency | Detected vs. Not detected | ||
| OR (95%CI) | pchi2 | OR (95%CI) | pchi2 | ||||||
| 1 | rs6688840 | C/T | 1.2 (0.5, 2.6) | 0.664 | Block 1 | CT | 0.709 | 1.1 (0.6, 1.9) | 0.794 |
| 2 | rs2781087 | T/G | 1.2 (0.6, 1.9) | 0.794 | CG | 0.173 | 1.0 (0.5, 2.0) | 0.954 | |
| 3 | rs1475734 | T/G | 1.1 (0.5, 2.4) | 0.737 | TG | 0.118 | 0.8 (0.4, 1.8) | 0.663 | |
| 4 | rs6692694 | C/T | 1.0 (0.5, 2.1) | 0.906 | |||||
| 5 | rs17032875 | C/T | 1.1 (0.6, 1.8) | 0.873 | |||||
| 6 | rs12067941 | C/A | 1.1 (0.5, 2.6) | 0.807 | |||||
| 7 | rs12568902 | A/T | 1.3 (0.5, 3.0) | 0.584 | |||||
| 8 | rs6577541 | G/C | 1.4 (0.8, 2.3) | 0.251 | Block 2 | CCAC | 0.362 | 0.7 (0.4, 1.2) | 0.222 |
| 9 | rs2274327 | T/C | 1.4 (0.8, 2.4) | 0.279 | GTCC | 0.320 | 1.4 (0.8, 2.6) | 0.216 | |
| 10 | rs2274328 | C/A | 1.5 (0.9, 2.6) | 0.158 | GCAT | 0.222 | 0.7 (0.4, 1.3) | 0.319 | |
| 11 | rs17032907 | C/T | 1.4 (0.8, 2.4) | 0.230 | GCAC | 0.039 | 8.5 (0.5, 145.7) | 0.047 | |
| 12 | rs11121278 | G/C | 1.4 (0.5, 3.8) | 0.528 | GCCC | 0.033 | 1.3 (0.3, 6.4) | 0.691 | |
| 13 | rs12021597* | A/G | 1.1 (0.6, 1.9) | 0.708 | GCCT | 0.013 | 1.0 (0.1, 9.7) | 0.980 | |
| 14 | rs17032912 | G/C | 1.1 (0.5, 2.2) | 0.835 | |||||
| 15 | rs7545200 | A/G | 1.1 (0.6, 1.9) | 0.756 | Block 3 | GTA | 0.404 | 0.6 (0.4, 1.1) | 0.089 |
| 16 | rs1832262 | C/T | 1.6 (0.9, 2.6) | 0.092 | GCA | 0.279 | 1.6 (0.9, 3.0) | 0.124 | |
| 17 | rs6691526 | A/G | 1.1 (0.6, 2.1) | 0.673 | ACG | 0.245 | 0.9 (0.5, 1.6) | 0.672 | |
| 18 | rs3765965 | T/C | 1.0 (0.5, 2.0) | 0.908 | ACA | 0.072 | 2.2 (0.6, 7.6) | 0.206 | |
| 19 | rs2274334 | G/T | 1.2 (0.4, 4.0) | 0.741 | |||||
| 20 | rs10864376 * | T/C | 3.5 (1.8, 7.1) | 2.0E-4 | Block 4 | CCC* | 0.461 | 0.5 (0.3, 0.8) | 0.003 |
| 21 | rs3737665* | T/C | 2.7 (1.2, 5.9) | 0.012 | CCG | 0.222 | 0.8 (0.4, 1.5) | 0.501 | |
| 22 | rs12138897* | G/C | 2.2 (1.3, 3.7) | 0.004 | TTG* | 0.206 | 2.7 (1.2, 5.9) | 0.012 | |
| 23 | rs3765964 | A/G | 1.9 (1.1, 3.4) | 0.018 | TCG | 0.111 | 3.8 (1.1, 12.8) | 0.022 | |
| 24 | rs6577546 | G/A | 1.2 (0.6, 2.2) | 0.584 | Block 5 | AG | 0.460 | 0.6 (0.3, 0.9) | 0.035 |
| 25 | rs6680186 | A/G | 1.9 (1.1, 3.1) | 0.021 | AA | 0.306 | 1.7 (0.9, 3.1) | 0.079 | |
| 26 | rs7533137 | G/A | 1.5 (0.8, 2.9) | 0.369 | GA | 0.230 | 1.3 (0.7, 2.5) | 0.411 | |
| 27 | rs7513804 | T/C | 1.3 (0.8, 2.3) | 0.283 | |||||
Alleles in bold indicate significant positive associations and in italic negative associations at FDR of < 0.25 (bold underline for differences significant at FDR ≤0.06 too). * indicates SNPs also associated with caries (see Table 4).
Figure 2.
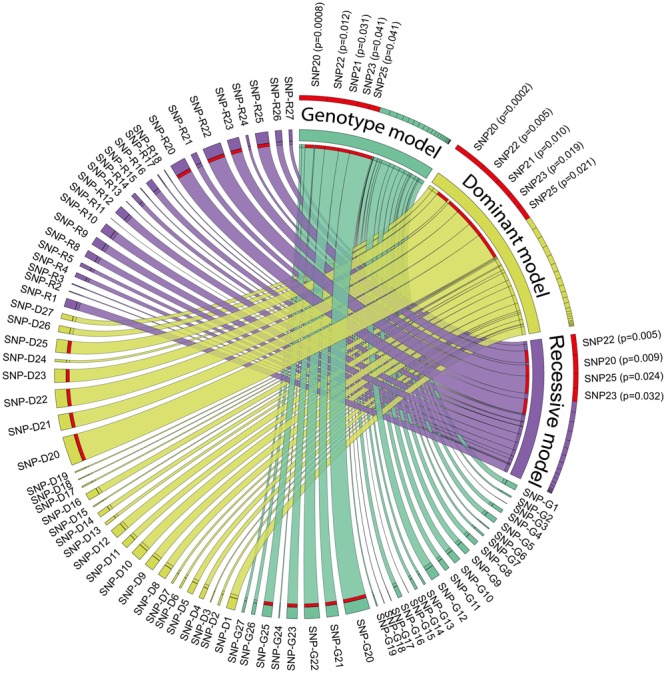
Associations between CA6 gene variation and having detectable S. mutans in saliva or tooth biofilm by NGS. The association between 27 CA6 SNPs and detectable S. mutans in tooth biofilm and saliva by NGS was estimated using a Chi2 test under 3 models; genotype model (green, SNPs marked with G), dominant model (yellow, SNPs marked with D) and recessive model (purple, SNPs marked with R). For each model, the width of the bar connecting the model name to a SNP indicates the relative–log10 p-value testing the null hypothesis of no association with detectable S. mutans. The red bar highlights SNPs where the null hypothesis was rejected after adjustment of p-values for multiple testing (FDR < 0.25).
Further, the CCC-haploblock 4 and the AG-haploblock 5 associated with reduced odds to have S. mutans, whereas the GCAC-haploblock 2, and TCG and TTG haploblock 4 associated with increased odds (Table 2). The odds ratio (95% CI) to have S. mutans in saliva or tooth biofilm if carrying the TCG allele versus the CCC haploblock 4 was 5.0 (1.5–17.2) (p = 0.011), and if carrying the TTG allele versus the CCC haploblock 4 it was 3.3 (1.5–7.6) (p = 0.004).
CA6 gene variation and viable mutans streptococci in saliva
Four SNPs, rs10864376 (T), rs12138897 (G), rs6680186 (A), and rs7513804 (T), were associated with having mutans streptococci by saliva culturing (Table 3), as was the TCG-haploblock 4, whereas, the CCC-haploblock 4 and AG-haploblock 5 were associated with being S. mutans free (Table 3). The odds ratio (95% CI) to have mutans streptococci by culturing if carrying the TCG allele versus the CCC haploblock 4 was 4.3 (1.5–12.2) (p = 0.006). Notably, the association pattern with CA6 gene variation was similar for having S. mutans by NGS and having viable S. mutans by culture.
Table 3.
CA6 SNP and haploblock associations with detection of viable mutans streptococci by saliva culturing.
| SNP | Haploblock | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP number | Name | Allele | Detected vs. Not detected | Haplo-block number | Haplo-type | Frequency | Detected vs. Not detected | ||
| OR (95%CI) | pchi2 | OR (95%CI) | pchi2 | ||||||
| 1 | rs6688840 | C/T | 1.1 (0.5, 2.3) | 0.709 | Block 1 | CT | 0.709 | 1.1 (0.7, 1.8) | 0.707 |
| 2 | rs2781087 | T/G | 1.1 (0.7, 1.8) | 0.707 | CG | 0.173 | 1.0 (0.5, 1.8) | 0.895 | |
| 3 | rs1475734 | T/G | 1.9 (1.0, 3.7) | 0.069 | TG | 0.118 | 0.9 (0.4, 1.8) | 0.709 | |
| 4 | rs6692694 | C/T | 1.2 (0.6, 2.2) | 0.613 | |||||
| 5 | rs17032875 | C/T | 1.5 (0.9, 2.5) | 0.107 | |||||
| 6 | rs12067941 | C/A | 1.5 (0.7, 3.3) | 0.272 | |||||
| 7 | rs12568902 | A/T | 1.6 (0.7, 3.6) | 0.213 | |||||
| 8 | rs6577541 | C/G | 1.3 (0.8, 2.1) | 0.275 | Block 2 | CCAC | 0.362 | 1.3 (0.8, 2.1) | 0.320 |
| 9 | rs2274327 | C/T | 1.0 (0.6, 1.7) | 0.886 | GTCC | 0.320 | 0.9 (0.6, 1.5) | 0.736 | |
| 10 | rs2274328 | C/A | 1.1 (0.7, 1.8) | 0.686 | GCAT | 0.222 | 0.7 (0.4, 1.2) | 0.158 | |
| 11 | rs17032907 | C/T | 1.3 (0.8, 2.2) | 0.369 | GCAC | 0.039 | 1.5 (0.4, 6.0) | 0.761 | |
| 12 | rs11121278 | C/G | 1.7 (0.8, 3.7) | 0.202 | GCCC | 0.033 | 1.5 (0.4, 6.0) | 0.494 | |
| 13 | rs12021597* | A/G | 1.3 (0.8, 2.2) | 0.252 | GCCT | 0.013 | 1.9 (0.2, 18.7) | 0.588 | |
| 14 | rs17032912 | C/G | 1.2 (0.6, 2.2) | 0.655 | |||||
| 15 | rs7545200 | A/G | 1.5 (0.9, 2.6) | 0.093 | Block 3 | GTA | 0.404 | 0.6 (0.4, 1.0) | 0.076 |
| 16 | rs1832262 | C/T | 1.6 (1.0, 2.5) | 0.069 | GCA | 0.279 | 1.0 (0.6, 1.8) | 0.846 | |
| 17 | rs6691526 | G/A | 1.4 (0.8, 2.4) | 0.262 | ACG | 0.245 | 1.4 (0.8, 2.4) | 0.262 | |
| 18 | rs3765965 | T/C | 1.1 (0.6, 2.0) | 0.744 | ACA | 0.072 | 1.8 (0.7, 4.6) | 0.249 | |
| 19 | rs2274334 | T/G | 2.4 (0.7, 8.8) | 0.172 | |||||
| 20 | rs10864376* | T/C | 1.9 (1.1, 3.2) | 0.015 | Block 4 | CCC* | 0.461 | 0.5 (0.3, 0.8) | 0.004 |
| 21 | rs3737665* | T/C | 1.4 (0.8, 2.4) | 0.316 | CCG | 0.222 | 1.4 (0.8, 2.5) | 0.499 | |
| 22 | rs12138897* | G/C | 2.0 (1.2, 3.1) | 0.004 | TTG* | 0.206 | 1.4 (0.8, 2.4) | 0.316 | |
| 23 | rs3765964 | A/G | 1.4 (0.9, 2.2) | 0.175 | TCG | 0.111 | 2.7 (1.1, 6.4) | 0.021 | |
| 24 | rs6577546 | G/A | 1.5 (0.8, 2.6) | 0.187 | Block 5 | AG | 0.460 | 0.5 (0.3, 0.8) | 0.004 |
| 25 | rs6680186 | A/G | 2.0 (1.3, 3.2) | 0.003 | AA | 0.306 | 1.6 (1.0, 2.7) | 0.068 | |
| 26 | rs7533137 | G/A | 1.6 (0.9, 2.8) | 0.099 | GA | 0.230 | 1.6 (0.9, 2.8) | 0.125 | |
| 27 | rs7513804 | T/C | 1.9 (1.2, 3.0) | 0.008 | |||||
Alleles in boldindicate significant positive associations and in italic negative associations at FDR < 0.25 (bold underline for significant differences at FDR ≤ 0.06). * indicates SNPs that also were associated with caries (see Table 4).
CA6 gene variation rs12138897 (G/C) and tooth biofilm microbiota
The association of the CCC-haploblock 4 and the TCG-haploblock 4 with S. mutans status (as well as caries status as described in the next paragraph) led us to evaluate the association between the C and G allele of rs12138897 (C representing the CCC- and G- the TTG haploblock) and overall tooth biofilm microbiota by multivariate Partial Least Squares (PLS) analysis of NGS determined taxa. PLS modelling (crossvalidated prediction Q2 = 24%) displayed separation of participants carrying the G versus C allele based on their tooth biofilm microbiota (Fig. 3a). The separation was explained by strong associations (Variable Importance in Projection (VIP) > 1.5) with the G allele for eight species, i.e., higher detection rate of Treponema sp. HOT268, Actinomyces sp. HOT896, Tannerella sp. HOT286, S. mutans, GN02[G-1] sp. HOT872, Scardovia wiggsiae, and species detected by Veillonella genus probe1 (Fig. 3b). The C allele associated with higher detection rate of Bacteroidales [G-2] sp. HOT274, Prevotella sp. HOT315, Tannerella forsythia, Fretibacterium fastidiosum, Filifactor alocis, and species detected by Treponema genus probe 2 (Fig. 3c). Five of the eight species associating with the G allele, i.e., Treponema sp. HOT268, Tannerella sp. HOT286, S. mutans, Veillonella gp.1 and S. wiggsiae were also significantly different between the G and C genotype (pchi2 < 0.05) in univariate analysis.
Figure 3.
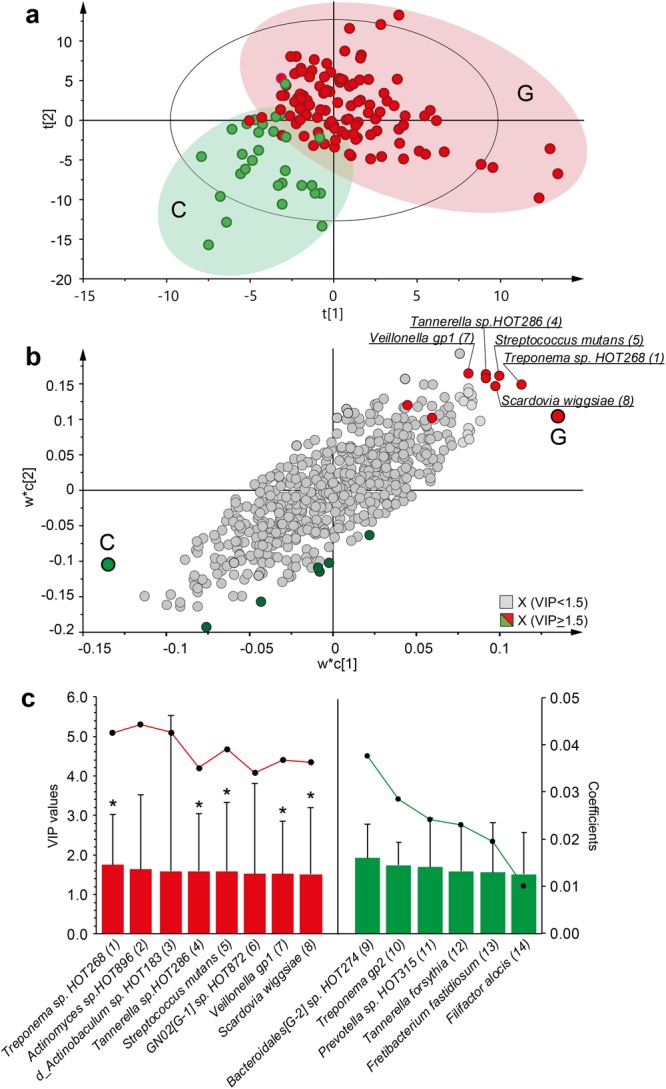
CA6 gene variation and tooth biofilm microbiota. (a) PLS scatter plot illustrating separation of participants carrying either the G or C of the rs12138897 SNP based on their tooth biofilm microbiota using the dominant SNP model (G/G + G/C vs. C/C); (b) PLS loading scatter plot of the X and Y weights (w* and c) illustrating the relation between the microbiota species and the G (red) and C (green) genotype of CA6. Red/green dots refer to taxa with a VIP-value > 1.5, and grey dots to taxa with a VIP value < 1.5. (c) Correlation coefficients (mean (95% CI; shown on the left x-axis) for taxa with a VIP value > 1.5 (shown on the right x-axis) in plot B. Red bars show association with G and green bars with C genotypes of CA6. The dots on the lines represent median prevalence for each taxon and the stars (*) statistically significant difference between the two genotypes using univariate Chi2 test and p-values < 0.05.
CA6 gene variation and caries status
Following that CA6 gene variation was associated with colonization of caries associated bacteria, such as S. mutans and S. wiggsiae, the association between the 27 SNPs and caries status (yes or no) was tested in an allelic association model. The rs12021597 (A), rs10864376 (T), rs3737665 (T), and rs12138897 (G) associated positively with having caries (Table 4). The strongest effects on having caries was observed for the rs10864376 (T) allele with an odds ratio (95% CI) of 2.1 (1.2, 3.7) (p = 0.009).
Table 4.
CA6 SNP and haploblock associations with being caries-affected or caries-free.
| SNP | Haploblock | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP number | Name | Allele | Caries-affected vs. Caries-free | Haplo-block number | Allele | Frequency | Caries-affected vs. Caries-free | ||
| OR (95%CI) | pchi2 | OR (95%CI) | pchi2 | ||||||
| 1 | rs6688840 | T/C | 1.1 (0.5, 2.3) | 0.895 | Block 1 | CT | 0.717 | 0.9 (0.5, 1.6) | 0.748 |
| 2 | rs2781087 | G/T | 1.1 (0.6, 1.9) | 0.749 | CG | 0.167 | 1.1 (0.6, 2.2) | 0.784 | |
| 3 | rs1475734 | G/T | 1.0 (0.5, 2.1) | 0.955 | TG | 0.116 | 1.1 (0.5, 2.3) | 0.894 | |
| 4 | rs6692694 | C/T | 1.1 (0.6, 2.1) | 0.783 | |||||
| 5 | rs17032875 | C/T | 1.1 (0.6, 1.8) | 0.744 | |||||
| 6 | rs12067941 | C/A | 1.2 (0.5, 2.6) | 0.663 | |||||
| 7 | rs12568902 | A/T | 1.0 (0.4, 2.5) | 0.955 | |||||
| 8 | rs6577541 | G/C | 1.1 (0.6, 1.8) | 0.821 | Block 2 | CCAC | 0.348 | 1.0 (0.6, 1.7) | 0.948 |
| 9 | rs2274327 | C/T | 1.2 (0.7, 2.0) | 0.514 | GTCC | 0.329 | 0.8 (0.5, 1.4) | 0.449 | |
| 10 | rs2274328 | C/A | 1.0 (0.5, 1.7) | 0.982 | GCAT | 0.220 | 0.9 (0.5, 1.7) | 0.828 | |
| 11 | rs17032907 | C/T | 1.1 (0.6, 1.9) | 0.815 | GCAC | 0.043 | 1.7 (0.5, 6.8) | 0.479 | |
| 12 | rs11121278 | G/C | 1.4 (0.6, 3.5) | 0.468 | GCCC | 0.036 | 5.4 (0.7, 43.5) | 0.065 | |
| 13 | rs12021597 | A/G | 1.9 (1.1, 3.4) | 0.023 | GCCT | 0.011 | 1.3 (0.1, 14.4) | 0.854 | |
| 14 | rs17032912 | G/C | 1.5 (0.8, 3.0) | 0.214 | |||||
| 15 | rs7545200 | G/A | 1.0 (0.6, 1.8) | 0.930 | Block 3 | GTA | 0.401 | 0.6 (0.4, 1.1) | 0.063 |
| 16 | rs1832262 | C/T | 1.7 (1.0, 2.8) | 0.057 | GCA | 0.296 | 1.9 (1.1, 3.3) | 0.037 | |
| 17 | rs6691526 | A/G | 1.2 (0.6, 2.1) | 0.637 | ACG | 0.240 | 0.9 (0.5, 1.6) | 0.636 | |
| 18 | rs3765965 | T/C | 1.2 (0.6, 2.3) | 0.538 | ACA | 0.062 | 1.4 (0.5, 4.3) | 0.508 | |
| 19 | rs2274334 | G/T | 1.1 (0.4, 3.6) | 0.850 | |||||
| 20 | rs10864376 | T/C | 2.1 (1.2, 3.7) | 0.009 | Block 4 | CCC | 0.461 | 0.6 (0.3, 0.9) | 0.028 |
| 21 | rs3737665 | T/C | 2.3 (1.2, 4.4) | 0.014 | CCG | 0.217 | 0.9 (0.5, 1.7) | 0.776 | |
| 22 | rs12138897 | G/C | 1.8 (1.1, 2.9) | 0.028 | TTG | 0.217 | 2.3 (1.2, 4.4) | 0.014 | |
| 23 | rs3765964 | A/G | 1.5 (0.9, 2.4) | 0.160 | TCG | 0.105 | 1.3 (0.6, 3.1) | 0.518 | |
| 24 | rs6577546 | A/G | 0.9 (0.5, 1.7) | 0.851 | Block 5 | AG | 0.475 | 0.8 (0.5, 1.3) | 0.314 |
| 25 | rs6680186 | A/G | 1.4 (0.8, 2.2) | 0.242 | AA | 0.301 | 1.3 (0.8, 2.4) | 0.319 | |
| 26 | rs7533137 | G/A | 1.1 (0.6, 2.1) | 0.697 | GA | 0.219 | 1.1 (0.6, 2.0) | 0.754 | |
| 27 | rs7513804 | T/C | 1.2 (0.7, 1.9) | 0.573 | |||||
Alleles in bold indicate significant positive associations and in italic negative associations at FDR of < 0.25 (bold underline for significant differences at FDR ≤ 0.06).
Of the 5 haploblocks, two (haploblock 3 and 4) were associated with caries status (Table 4). The GCA-haploblock 3 and TTG-haploblock 4 associated with increased odds to have caries, whereas CCC-haploblock 4 associated with decreased odds of having caries. Comparing the TTG-haploblock 4 with the CCC-haploblock 4 showed an odds ratio of 2.6 (1.3, 5.2) (p = 0.008) covering 66.7% of the participants.
CA6 gene variation and saliva CA6 protein levels
Saliva concentration of CA6 was measured by ELISA in the first 90 sampled participants (for further information see material and methods). The median (10%, 90% percentile) concentration of CA6 in saliva was 8.4 (0.3, 17.0) µg/mL and secreted CA6 11.0 (0.4, 25.4) µg/min (Table 1). High concentration tended to associate with rs10864376 (T) (p = 0.031) and rs3737665 (T) (p = 0.031), as well as the GCA-haploblock 3 (p = 0.030) and TTG-haploblock 4 (p = 0.051) but none was significant at FDR < 0.25 (Table S3).
CA6 in saliva (concentration or secreted amounts) was not associated with having S. mutans or not, but secreted amounts (µg/min) was significantly higher in caries-free than caries-affected (p = 0.012) (Table 1). In accordance, twice as many caries-free, compared to caries-affected, participants were identified in the highest tertile based on the distribution of secreted CA6 amounts (pbetween groups = 0.007) with an OR to be caries-free of 3.5 (1.4, 9.1).
Haploblock 4 summary
A summary of characteristics for the various allele variants in haploblock 4 is presented in Table 5 together with comparisons between the allele variants groups as well as trends among the allele variants. There were no significant differences between the alleles with respect to sex, BMI, smoking, frequency of sweet snacking, or number of tooth surfaces. The proportions with S. mutans by NGS and culturing, respectively, differed significantly between the allele variant groups (p < 0.001) as did the trend from the TCG to CCC variant (p < 0.001). The association with caries was less strong, but the proportions caries-affected participants tended to differ between the allele variant groups (p = 0.054) with a significant trend (p = 0.021), whereas caries prevalence (DeFS) did not differ between the groups though the trend did (p = 0.018).
Table 5.
Characteristics of the 17-year-old participants in Haploblock 4.
| Haploblock 4 | p value | |||||
|---|---|---|---|---|---|---|
| TCG | TTG | CCG | CCC | Group | Trend | |
| Frequency, % | 11.1 | 20.6 | 22.2 | 46.1 | — | — |
| Sex, % male | 77.8 | 58.9 | 55.6 | 54.6 | 0.166 | 0.067 |
| Smoker, % | 0.0 | 3.6 | 1.8 | 2.5 | 0.776 | 0.749 |
| BMI, kg/m2 | 20.8 (17.5, 26.2) | 21.1 (18.9, 25.8) | 21.0 (18.2, 24.3) | 21.8 (18.5, 25.2) | 0.240 | 0.149 |
| Sweet snack frequency/day | 0.9 (0.3, 2.1) | 0.7 (0.2, 1.3) | 0.9 (0.3, 2.1) | 0.7 (0.3, 2.2) | 0.126 | 0.176 |
| Number of tooth surfaces | 128 (108, 128) | 128 (108, 130) | 128 (108, 128) | 128 (113, 128) | 0.364 | 0.136 |
| S. mutans | ||||||
| Proportion with detection by NGS, % | 92.6 | 87.5 | 71.4 | 63.9 | <0.001 | <0.001 |
| Mutans streptococci | ||||||
| CFU/mL, log10 value | 3.0 (0, 5.2) | 2.8 (0, 4.6) | 2.9 (0, 5.1) | 0.7 (0, 4.4) | 0.021 | 0.014 |
| Proportion with detection by culture, % | 81.5 | 64.3 | 69.6 | 50.4 | 0.007 | 0.002 |
| Caries status | ||||||
| DeFS | 1.0 (0.0, 14.0) | 2.0 (0.0, 13.9) | 1.5 (0.0, 12.0) | 1.0 (0.0, 10.0) | 0.101 | 0.021 |
| Proportion caries-affected, % | 66.7 | 75.0 | 59.3 | 53.8 | 0.054 | 0.018 |
Continuous measures are presented as medians (10, 90 percentiles) and group differences were tested with Mann Whitney U test. Differences between group numbers (presented as %) were tested with Chi2 or Fisher’s exact test. P-values were considered significant at FDR < 0.25 but significant differences at FDR ≤0.06 are also indicated in bold. CFU for colony forming units and NGS for Next Generation Sequencing of DNA extracted from saliva (n = 152) or tooth biofilm (n = 139). All analysis included participants who reported brushing their teeth twice daily.
Discussion
This study tested the hypothesis that variation in the CA6 gene is relevant to dental health and found that CA6 gene polymorphisms and haploblocks of CA6 were associated with S. mutans colonization, overall microbiota composition and dental caries in Swedish adolescents. Secreted CA6 in saliva tended to be linked to CA6 gene variation. Thereby, it is the first study to demonstrate effects of CA6 gene polymorphisms on the oral bacterial communities in a population-based setting.
Collectively, the present findings support a role of CA6 polymorphisms in oral health, i.e., balancing less, against more, aciduric species, e.g., cariogenic S. mutans and S. wiggsiae, and de- and remineralization in the caries process. The results are in line with previous studies reporting a role of CA6 in caries development in children13–17. Though several of the physiological effects of CA6 are relevant for both oral microbiota ecology and the caries process, the detailed mechanisms for the effects remain to be elucidated. The underlying mechanisms for oral health of CA6 and CA6 gene variations may, alone or in concert, include tooth formation, local pH regulation, bacteria adhesion and metabolism and taste induced food preferences.
A tooth formation mechanism appears plausible because CA6 is expressed in the bud, cup, and bell stage of early tooth development, and by pre- and early differentiated odontoblasts and dental papilla mesenchyme cells, but most avidly in late tooth development with formation and maturation of the enamel2. Differences in enamel quality affects the initial tooth covering protein pellicle, and consequently the profile of attachment epitopes for bacteria19 and possibly oral microbiota composition. Differences in enamel quality also affects resistance of tooth tissue to destruction in an acidic environment.
Given the importance of pH homeostasis for the oral environment, local pH regulation by CA6 may be a contributory mechanism. Several of the most abundant bacteria in the mouth are glycolytic with avid acid formation, leading to pH fall and enrichment of aciduric, cariogenic species (dysbiosis)20. The CA6 protein is suggested to regulate pH in secreted fluids1, and saliva buffer capacity was reported to differ by CA6 gene variation at rs227432716, and CA6 protein activity in some12, but not other5, studies. This aspect could not be tested in the present study as neither saliva buffer capacity nor pH were analyzed, but some gene variants were associated with presence of aciduric and acidogenic species, such as S. mutans and S. wiggisae, which are promoted at low pH. Since reported intake of sweets did not differ by CA6 gene variation this may reflect a less well functioning pH regulation by some CA6 gene variants. Impaired pH regulation may also increase or decrease the risk of tooth de- and remineralisation.
Other important factors in oral biofilm ecology include access to attachment sites on host- and partner bacteria structures, and exposure to oxygen and nutrients. CA6 incorporated in pellicle and tooth biofilm may influence bacterial ecology and metabolism through effects unrelated to it’s enzymatic activity. Such effects may be two-fold; i.e., by proteolytic release of bioactive peptides or by providing a bacteria binding site. Here, the TTG and CCC associated SNPs were linked with mixed panels of oral bacterial such as S. mutans and S. wiggsiae, but also Treponema sp. HOT268, and Tannerella sp. HOT286 which thrive at a neutral to slightly basic pH. However, these suggested mechanisms are theoretical and need to be evaluated in future studies.
Finally, CA6 gene variation has been suggested to link to bitter taste and smell perception21–27. To date, reported associations with food selection are inconsistent and the present study did not find any effect of CA6 polymorphisms on sweet snack intake. This may reflect a “true” lack of association or the inherent error in measuring sugar intake in general and especially in the dental office. This aspect as well as the reported association between bitter taste receptor stimulation and innate immunity function28 should be addressed by a study design that is less prone to bias, such as using a biomarker or genetic marker reflecting sweet intake.
Compared to previous investigations, the present study included detailed genotyping of a relatively large number of SNPs within and near CA6, allowing fine-mapping of both single genetic variants and recombination blocks. The phenotypes were also detailed, including a range of salivary, microbiome, and host characteristics which may affect oral microbiota ecology and the caries process directly or indirectly. Additional strengths were that (i) all participants were of the same age (17 years old), (ii) participants were recruited in connection with their regular dental survey which reduces the risk for selection bias, (iii) participants permanent teeth have been exposed to the oral environment for years allowing ecology stabilization and potential development caries, and (iv) have minimal confounding by diseases or medications. Further, the study design, i.e., consecutive inclusion of participants as they attended their regular visit for a dental examination, and that virtually all accepted to participate, limited the risk of a selection bias.
There are some limitations in the study that should be acknowledged. The relative low number of participants, including that only a subset was available for assessing CA6 amounts, limits the possibility of comparisons in subgroups. Further, identification of microorganisms by NGS is associated with potential errors at various steps, such as the PCR amplification, sequencing per se, and bioinformatic accuracy. It should also be noted that in contrast to culturing, DNA based identification by NGS identifies dead and alive bacteria, and that saliva detection, in contrast to tooth biofilm samples, reflects bacteria from epithelial and tooth surfaces. These differences contributed to the variation in S. mutans detection frequency between the different identification strategies. To reduce the impact of the sampling source, being S. mutans positive was based on presence in either saliva or tooth biofilm by NGS. In addition, self-reported information on lifestyle habits, here tooth brushing, tobacco use and intake of sweet snacks, is prone to bias, such as the well-known under-reporting of diet intake29.
CA6 was originally found in the salivary glands, but has now been found to be expressed by the lacrimal, tracheobronchial, nasal, and mammary glands (with especially high content in colostrum30, and in epithelial linings of the oesophagus, stomach, and large intestine. This has led to a wider search of its functions. Thereby, CA6 has been linked to acinic cell carcinoma31, colorectal cancer27,32–34, and Sjögrens syndrome35–37. Based on the findings from the current study it may be hypothesized that CA6 and CA6 gene variations contribute to the microbial environment in both the nasopharynx and gastro-intestinal tracts.
In conclusion, the results from the present study support that CA6 gene polymorphisms rs10864376 (T), rs3737665 (T), rs12138897 (G) and haploblock TTG of CA6 are associated with S. mutans colonization, overall microbiota composition and dental caries in Swedish adolescents. Further studies need to elucidate the mechanisms.
Material and Methods
Participants
In this study 154 17-year-old adolescents were recruited from three public dental health care clinics in the city of Umeå, Sweden38. Adolescents who had other chronic medical conditions, used medication regularly, required antibiotic treatment during the latest six months, who did not consent, or were unable to communicate in Swedish or English were excluded. All participants had answered a questionnaire with information on general health status, medication use, oral hygiene, dietary habits, and tobacco use, and samples of whole simulated saliva and supragingival biofilm were collected and stored at −80 °C until use.
Ethical approval
The study was approved by the Regional Ethical Review Board in Umeå, Sweden (Dnr 2012-111-31 M, Dnr 2015-389-32 M and Dnr 2017-450-31) and abided by the Declaration of Helsinki and by the European General Data Protection Regulation (GDPR). Data collections and experiments were performed in accordance with relevant guidelines and regulations, including that all adolescents and their caregivers provided informed consent and data collection and handling followed the Helsinki declaration and the Swedish Law on personal data act (PuL).
CA6 genotyping
Single nucleotide polymorphisms in the region of the CA6 gene were selected and genotyped on DNA isolated from whole stimulated saliva. The criteria for SNP selection were a) minor allele frequency (MAF) ≥ 5% in the CEU population b) not in complete linkage disequilibrium (r2 ≤ 0.8) and c) in the 10,000 bp up-stream to 10,000 bp down-stream region of the CA6 gene. Using the NIH snptag resource (https://snpinfo.niehs.nih.gov/snpinfo/snptag.html) 30 SNPs met these criteria. Genotyping was performed using a multiplexed primer extension chemistry of the iPLEX assay with detection of the incorporated allele by mass spectrometry using a MassARRAY analyzer (Agena Bioscience, Hamburg, Germany). Raw data from the mass reader was converted to genotype data using the Typer software (Agena Bioscience)39–41. Two SNP markers had sample call rate 0%, rs2274333 and rs17032942, the reasons for the failure was low allele signals resulting in inaccurate genotype calls. An additional SNP (rs6697763) was excluded from analysis for deviation from Hardy-Weinberg equilibrium (p = 0.033), leaving 27 SNPs with valid genotype data, where the average call rate per sample was 99.4% and overall call rate was 99.8% (Table S1). Genotyping data are uploaded and available at figshare 10.6084/m9.figshare.6948020.
Sampling for microbiota analyses and CA6 determination
Whole simulated saliva was collected for 3 min into ice-chilled sterile test tubes while participants chewed on a 1 g piece of paraffin wax. Supragingival biofilm was collected from all accessible tooth surfaces using sterile wooden toothpicks and pooled by participant into 100 µL of TE buffer (10 mM Tris, 1 mM ethylenediaminetetraacetic acid [EDTA], pH 7.6). All study participants refrained from oral hygiene on the morning of their visit to the dental clinic. All samples, except those for culturing, were stored at −80 °C until used.
Oral microbiota genotyping
Genomic DNA was extracted from supragingival biofilm samples, saliva and from bacteria mock communities with the GenElute Bacterial Genomic DNA Kit (Sigma-Aldrich, Stockholm, Sweden) as previously described38. Briefly, samples were collected and lysed in buffer with lysozyme and mutanolysin (Sigma-Aldrich), treated with RNase and proteinase K, purified and washed. DNA quantity and quality were measured using NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific, Uppsala, Sweden). Multiplex 16S rDNA amplicon sequencing was performed with the Illumina MiSeq platform (http://www.illumina.com). 16S rDNA amplicon sequencing was conducted at the Forsyth Research Institute with the HOMINGS protocol42. Briefly, the V3-V4 hypervariable regions of the 16S rRNA gene were polymerase chain reaction (PCR) amplified with the 341 F (ACGGGAGGCAGCAG) forward primer and the 806 R (GGACTACHVGGGTWTCTAAT) reverse primer. Pair-end reads were fused using FLASH, and barcodes, primers, and ambiguous and chimeric sequences were removed within QIIME. Taxa were identified with the ProbeSeq customized BLAST program, which targeted recognition of 538 species (using 638 species-level probes) and 129 groups of closely-related species (using 129 genus-level probes)43. Taxa targeted by the genus probes (gp) are described in Table S2. Based on sequencing results for the mock communities, taxa that were represented by at least 100 sequences with >98.5% similarity were included in further analyses.
Caries scoring and lifestyle information
Visual and radiographic dental examinations were performed as described previously38. The numbers of tooth surfaces with caries in the enamel or into the dentine, or a filling, or were missing were recorded. The total numbers of decayed and filled tooth surfaces (DeFS) were calculated. Fissure sealants were not recorded. Missing surfaces were not considered to represent dental caries because tooth loss occurred for orthodontic reasons, tooth formation defects or aplasia in the study group.
Determination of CA6 concentration in saliva
For the first 90 participants’ saliva, samples were collected and stored at −80 °C until used for CA6 concentration measurements. CA6 protein concentration in saliva was measured by ELISA (CA6 ELISA Kit, OKCD01736, Nordic Biosite, Täby, Sweden) following the manufacturer’s instructions. Briefly, 100 µL saliva, diluted 1:5 was used for analysis, CA6 protein was captured by a target specific antibody, then detected by a biotinylated secondary antibody, followed by an avidin-horseradish peroxidase (HRP) conjugate and tetramethylbenzidine (TMB) for colour development. The colour development reaction was terminated by sulfuric acid addition and the optical density (OD) measured at 450 nm using a MultiscanGO spectrophotometry (Thermo Scientific, Abninolab, Upplands-Väsby, Sweden). A standard curve using purified CA6 protein ranging from 0–1,000 pg/mL was run.
Determination of total saliva protein concentration
Total saliva protein concentration was determined using Pierce Coomassie (Bradford) protein assay kit (ThermoFisher). Briefly, 5 µl diluted (1:1) saliva, was mixed with 250 µL protein dye solution and incubated for 30 min at room temperature. The optical density of each sample at 595 nm wavelength was assessed using a MultiscanGO spectrophotometry (Thermo Scientific) and compared to the optical density of a bovine serum albumin standard curve from known antigen concentrations to determine the sample concentration.
Data handling and statistical inference
NGS sequencing of saliva was performed in 139 tooth biofilm samples and 154 saliva samples where 2 samples failed. Caries information was missing for one participant who was excluded. All analyses with caries as outcome were restricted to the 129 subjects who reported brushing their teeth twice a day.
SPSS software version 24.0 was used for descriptive statistics, including medians (10%, 90% percentiles), frequencies (n), proportions (%) and odds ratios (OR) with 95% confidence intervals (CI). Group comparisons of continuous variables were done using Mann-Whitney U test and for categorical variables Chi2 or Fisher’s exact test. All tests were two-tailed. Correction for multiple comparison was done by the Benjamini Hochberg false discovery rate (FDR). P-values were considered significant at FDR < 0.25 and ≤0.06. The higher FDR was applied to avoid missing potential CA6 SNPs of interest as described earlier44,45.
Haploview software (version 4.2) was used to evaluate characteristics of SNPs (observed heterozygosity, predicted heterozygosity, Hardy-Weinberg equilibrium, minor allele frequency, pairwise linkage disequilibrium), as well as evaluation of potential haploblocks using the algorithm described by Gabriel et al.46. Haploblocks with frequencies <1% were not included in the analyses. Association between genetic variation (SNPs and haploblocks) and phenotypes was evaluated using Haploview and Chi2 tests. The tests were two-tailed tests and corrected for multiple comparison by FDR as described above. Phenotype penetrance was assessed in common allele, recessive and dominant models47.
Clustering of participants by presence (0/1) of bacterial taxa in saliva and tooth biofilm, respectively, was modelled using partial least square (PLS) regression. Saliva taxa did not yield a significant model and therefore PLS analyses employed tooth biofilm taxa. Variables with a Variable Importance in the Projection (VIP) > 1.5 were considered influential. SIMCA P+ version 14.1 (Umetrics AB) was used for these analyses.
Electronic supplementary material
Carbonic Anhydrase 6 Gene Variation influences Oral Microbiota Composition and Caries Risk in Swedish adolescents
Acknowledgements
The study was supported by the Swedish Patent Revenue Foundation, and TUA, the County Council of Västerbotten (Sweden). None of the funding bodies had any influence on the design, data collection, analysis, interpretation, or writing of the manuscript. Carina Öhman and Agnetha Rönnlund are acknowledged for skilful laboratory work and Linda Eriksson for clinical examinations.
Author Contributions
Esberg A. and Johansson I. contributed to conception, design and data analysis, drafted and critically revised the manuscript. Haworth S., Lif Holgerson P. and Brunius C. contributed to data analysis and critically revised the manuscript; All authors gave final approval and agree to be accountable for all aspects of the work.
Data Availability
NGS sequence data can be found at 10.6084/m9.figshare.5794989. Genotyping CA6 datasets generated and analysed during the current study are available from the corresponding author on reasonable request and with appropriate ethical approvals.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-36832-z.
References
- 1.Supuran CT. Carbonic anhydrases–an overview. Curr Pharm Des. 2008;14:603–614. doi: 10.2174/138161208783877884. [DOI] [PubMed] [Google Scholar]
- 2.Reibring CG, et al. Expression patterns and subcellular localization of carbonic anhydrases are developmentally regulated during tooth formation. PLoS One. 2014;9:e96007. doi: 10.1371/journal.pone.0096007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murakami H, Sly WS. Purification and characterization of human salivary carbonic anhydrase. J Biol Chem. 1987;262:1382–1388. [PubMed] [Google Scholar]
- 4.Kivela J, Parkkila S, Parkkila AK, Leinonen J, Rajaniemi H. Salivary carbonic anhydrase isoenzyme VI. J Physiol. 1999;520(Pt 2):315–320. doi: 10.1111/j.1469-7793.1999.t01-1-00315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kivela J, et al. Secretory carbonic anhydrase isoenzyme (CA VI) in human serum. Clin Chem. 1997;43:2318–2322. [PubMed] [Google Scholar]
- 6.Kimoto M, Kishino M, Yura Y, Ogawa Y. A role of salivary carbonic anhydrase VI in dental plaque. Arch Oral Biol. 2006;51:117–122. doi: 10.1016/j.archoralbio.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 7.Leinonen J, Kivela J, Parkkila S, Parkkila AK, Rajaniemi H. Salivary carbonic anhydrase isoenzyme VI is located in the human enamel pellicle. Caries Res. 1999;33:185–190. doi: 10.1159/000016515. [DOI] [PubMed] [Google Scholar]
- 8.Vitorino R, et al. In vitro hydroxyapatite adsorbed salivary proteins. Biochem Biophys Res Commun. 2004;320:342–346. doi: 10.1016/j.bbrc.2004.05.169. [DOI] [PubMed] [Google Scholar]
- 9.Bardow A, Moe D, Nyvad B, Nauntofte B. The buffer capacity and buffer systems of human whole saliva measured without loss of CO2. Arch Oral Biol. 2000;45:1–12. doi: 10.1016/S0003-9969(99)00119-3. [DOI] [PubMed] [Google Scholar]
- 10.Lips A, et al. Salivary protein polymorphisms and risk of dental caries: a systematic review. Braz Oral Res. 2017;31:e41. doi: 10.1590/1807-3107BOR-2017.vol31.0041. [DOI] [PubMed] [Google Scholar]
- 11.Picco DCR, et al. Children with a Higher Activity of Carbonic Anhydrase VI in Saliva Are More Likely to Develop Dental Caries. Caries Res. 2017;51:394–401. doi: 10.1159/000470849. [DOI] [PubMed] [Google Scholar]
- 12.Frasseto F, et al. Relationship among salivary carbonic anhydrase VI activity and flow rate, biofilm pH and caries in primary dentition. Caries Res. 2012;46:194–200. doi: 10.1159/000337275. [DOI] [PubMed] [Google Scholar]
- 13.Aidar M, et al. Effect of genetic polymorphisms in CA6 gene on the expression and catalytic activity of human salivary carbonic anhydrase VI. Caries Res. 2013;47:414–420. doi: 10.1159/000350414. [DOI] [PubMed] [Google Scholar]
- 14.Koc Ozturk L, et al. The investigation of genetic polymorphisms in the carbonic anhydrase VI gene exon 2 and salivary parameters in type 2 diabetic patients and healthy adults. Mol Biol Rep. 2012;39:5677–5682. doi: 10.1007/s11033-011-1374-1. [DOI] [PubMed] [Google Scholar]
- 15.Yildiz G, Ermis RB, Calapoglu NS, Celik EU, Turel GY. Gene-environment Interactions in the Etiology of Dental Caries. J Dent Res. 2016;95:74–79. doi: 10.1177/0022034515605281. [DOI] [PubMed] [Google Scholar]
- 16.Peres RC, et al. Association of polymorphisms in the carbonic anhydrase 6 gene with salivary buffer capacity, dental plaque pH, and caries index in children aged 7–9 years. Pharmacogenomics J. 2010;10:114–119. doi: 10.1038/tpj.2009.37. [DOI] [PubMed] [Google Scholar]
- 17.Li ZQ, Hu XP, Zhou JY, Xie XD, Zhang JM. Genetic polymorphisms in the carbonic anhydrase VI gene and dental caries susceptibility. Genet Mol Res. 2015;14:5986–5993. doi: 10.4238/2015.June.1.16. [DOI] [PubMed] [Google Scholar]
- 18.Yarat A, et al. Carbonic anhydrase VI exon 2 genetic polymorphism in Turkish subjects with low caries experience (preliminary study) In Vivo. 2011;25:941–944. [PubMed] [Google Scholar]
- 19.Sonju Clasen AB, Hannig M, Skjorland K, Sonju T. Analytical and ultrastructural studies of pellicle on primary teeth. Acta Odontol Scand. 1997;55:339–343. doi: 10.1080/00006357.1997.11978411. [DOI] [PubMed] [Google Scholar]
- 20.Marsh PD, Zaura E. Dental biofilm: ecological interactions in health and disease. J Clin Periodontol. 2017;44(Suppl 18):S12–S22. doi: 10.1111/jcpe.12679. [DOI] [PubMed] [Google Scholar]
- 21.Henkin RI, Martin BM, Agarwal RP. Decreased parotid saliva gustin/carbonic anhydrase VI secretion: an enzyme disorder manifested by gustatory and olfactory dysfunction. Am J Med Sci. 1999;318:380–391. doi: 10.1016/S0002-9629(15)40663-9. [DOI] [PubMed] [Google Scholar]
- 22.Melis M, et al. The gustin (CA6) gene polymorphism, rs2274333 (A/G), as a mechanistic link between PROP tasting and fungiform taste papilla density and maintenance. PLoS One. 2013;8:e74151. doi: 10.1371/journal.pone.0074151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patrikainen M, Pan P, Kulesskaya N, Voikar V, Parkkila S. The role of carbonic anhydrase VI in bitter taste perception: evidence from the Car6(−)/(−) mouse model. J Biomed Sci. 2014;21:82. doi: 10.1186/s12929-014-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barbarossa IT, et al. The gustin (CA6) gene polymorphism, rs2274333 (A/G), is associated with fungiform papilla density, whereas PROP bitterness is mostly due to TAS2R38 in an ethnically-mixed population. Physiol Behav. 2015;138:6–12. doi: 10.1016/j.physbeh.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 25.Feeney EL, Hayes JE. Exploring associations between taste perception, oral anatomy and polymorphisms in the carbonic anhydrase (gustin) gene CA6. Physiol Behav. 2014;128:148–154. doi: 10.1016/j.physbeh.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Genick UK, et al. Sensitivity of genome-wide-association signals to phenotyping strategy: the PROP-TAS2R38 taste association as a benchmark. PLoS One. 2011;6:e27745. doi: 10.1371/journal.pone.0027745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi JH, et al. Variations in the bitterness perception-related genes TAS2R38 and CA6 modify the risk for colorectal cancer in Koreans. Oncotarget. 2017;8:21253–21265. doi: 10.18632/oncotarget.15512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Workman AD, Palmer JN, Adappa ND, Cohen NA. The Role of Bitter and Sweet Taste Receptors in Upper Airway Immunity. Curr Allergy Asthma Rep. 2015;15:72. doi: 10.1007/s11882-015-0571-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johansson I, et al. Validation and calibration of food-frequency questionnaire measurements in the Northern Sweden Health and Disease cohort. Public Health Nutr. 2002;5:487–496. doi: 10.1079/PHNPHN2001315. [DOI] [PubMed] [Google Scholar]
- 30.Karhumaa P, et al. The identification of secreted carbonic anhydrase VI as a constitutive glycoprotein of human and rat milk. Proc Natl Acad Sci USA. 2001;98:11604–11608. doi: 10.1073/pnas.121172598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsieh MS, Jeng YM, Jhuang YL, Chou YH, Lin CY. Carbonic anhydrase VI: a novel marker for salivary serous acinar differentiation and its application to discriminate acinic cell carcinoma from mammary analogue secretory carcinoma of the salivary gland. Histopathology. 2016;68:641–647. doi: 10.1111/his.12792. [DOI] [PubMed] [Google Scholar]
- 32.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uhlen M, et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005;4:1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Pertovaara M, Bootorabi F, Kuuslahti M, Pasternack A, Parkkila S. Novel carbonic anhydrase autoantibodies and renal manifestations in patients with primary Sjogren’s syndrome. Rheumatology (Oxford) 2011;50:1453–1457. doi: 10.1093/rheumatology/ker118. [DOI] [PubMed] [Google Scholar]
- 36.Shen L, et al. Novel autoantibodies in Sjogren’s syndrome. Clin Immunol. 2012;145:251–255. doi: 10.1016/j.clim.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 37.Everett S, et al. Analysis of novel Sjogren’s syndrome autoantibodies in patients with dry eyes. BMC Ophthalmol. 2017;17:20. doi: 10.1186/s12886-017-0412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eriksson, L., Lif Holgerson, P., Esberg, A. & Johansson, I. Microbial Complexes and Caries in 17-Year-Olds with and without Streptococcus mutans. J Dent Res, 22034517731758, 10.1177/0022034517731758 (2017). [DOI] [PubMed]
- 39.Gabriel, S., Ziaugra, L. & Tabbaa, D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet Chapter 2(Unit2), 12, 10.1002/0471142905.hg0212s60 (2009). [DOI] [PubMed]
- 40.Ross P, Hall L, Smirnov I, Haff L. High level multiplex genotyping by MALDI-TOF mass spectrometry. Nat Biotechnol. 1998;16:1347–1351. doi: 10.1038/4328. [DOI] [PubMed] [Google Scholar]
- 41.Storm N, Darnhofer-Patel B, van den Boom D, Rodi CP. MALDI-TOF mass spectrometry-based SNP genotyping. Methods Mol Biol. 2003;212:241–262. doi: 10.1385/1-59259-327-5:241. [DOI] [PubMed] [Google Scholar]
- 42.Gomes BP, Berber VB, Kokaras AS, Chen T, Paster BJ. Microbiomes of Endodontic-Periodontal Lesions before and after Chemomechanical Preparation. J Endod. 2015;41:1975–1984. doi: 10.1016/j.joen.2015.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen T, et al. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010;2010:baq013. doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morgan XC, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu GD, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gabriel SB, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 47.Clarke GM, et al. Basic statistical analysis in genetic case-control studies. Nat Protoc. 2011;6:121–133. doi: 10.1038/nprot.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Carbonic Anhydrase 6 Gene Variation influences Oral Microbiota Composition and Caries Risk in Swedish adolescents
Data Availability Statement
NGS sequence data can be found at 10.6084/m9.figshare.5794989. Genotyping CA6 datasets generated and analysed during the current study are available from the corresponding author on reasonable request and with appropriate ethical approvals.