Abstract
Interpreting polygenic overlap between ADHD and both literacy-related and language-related impairments is challenging as genetic associations might be influenced by indirectly shared genetic factors. Here, we investigate genetic overlap between polygenic ADHD risk and multiple literacy-related and/or language-related abilities (LRAs), as assessed in UK children (N ≤ 5919), accounting for genetically predictable educational attainment (EA). Genome-wide summary statistics on clinical ADHD and years of schooling were obtained from large consortia (N ≤ 326,041). Our findings show that ADHD-polygenic scores (ADHD-PGS) were inversely associated with LRAs in ALSPAC, most consistently with reading-related abilities, and explained ≤1.6% phenotypic variation. These polygenic links were then dissected into both ADHD effects shared with and independent of EA, using multivariable regressions (MVR). Conditional on EA, polygenic ADHD risk remained associated with multiple reading and/or spelling abilities, phonemic awareness and verbal intelligence, but not listening comprehension and non-word repetition. Using conservative ADHD-instruments (P-threshold < 5 × 10−8), this corresponded, for example, to a 0.35 SD decrease in pooled reading performance per log-odds in ADHD-liability (P = 9.2 × 10−5). Using subthreshold ADHD-instruments (P-threshold < 0.0015), these effects became smaller, with a 0.03 SD decrease per log-odds in ADHD risk (P = 1.4 × 10−6), although the predictive accuracy increased. However, polygenic ADHD-effects shared with EA were of equal strength and at least equal magnitude compared to those independent of EA, for all LRAs studied, and detectable using subthreshold instruments. Thus, ADHD-related polygenic links with LRAs are to a large extent due to shared genetic effects with EA, although there is evidence for an ADHD-specific association profile, independent of EA, that primarily involves literacy-related impairments.
Introduction
Children with Attention-Deficit/Hyperactivity Disorder (ADHD) often experience difficulties mastering literacy-related and/or language-related abilities (LRAs)1–3. It has been estimated that up to 40% of children diagnosed with clinical ADHD also suffer from reading disability (RD, also known as developmental dyslexia) and vice versa4. The spectrum of affected LRAs in ADHD may, however, also include writing5,6, spelling7,8, syntactic9,10 and phonological9,10 abilities. Both clinical ADHD and RD are complex childhood-onset neurodevelopmental conditions that affect about 5% and 7% of the general population, respectively11,12. ADHD is characterised by hyperactive, inattentive and impulsive symptoms13, whereas decoding and/or reading comprehension deficits are prominent in individuals with RD14.
To interpret the comorbidity of ADHD and RD, a multiple-deficit model including shared underlying aetiologies has been proposed, involving both genetic and environmental influences15. This model is supported by twin studies suggesting that the co-occurrence of ADHD symptoms and reading deficits is, to a large extent, attributable to shared genetic influences16–18. Further twin research suggests that the genetic covariance between reading difficulties and ADHD is largely independent of genetic factors shared with IQ19, although it is not known whether these findings extend to a wider spectrum of LRAs, beyond reading abilities. Furthermore, the interpretation of polygenic ADHD-LRA overlap using markers on genotyping arrays is more challenging. There is strong evidence that genetically predicted educational attainment (EA)20 shares genetic variability with both ADHD21 and reading abilities22,23. Genetically predicted EA is a genetic proxy of cognitive abilities, but also socioeconomic status20 including, for example, associations with maternal smoking during pregnancy, parental smoking, household income or watching television24. Thus, observed genetic associations between ADHD and reading abilities may solely reflect shared genetic variation with EA, but not any other, more specific neuro-cognitive mechanisms. In other words, polygenic associations might be inflated or even induced25 by genetically predictable traits that are related to both, ADHD and reading abilities (or other LRAs).
Here, we (a) study polygenic links between clinical ADHD and a wide range of population-ascertained literacy-related and language-related measures as captured by common variation, (b) evaluate to what extent such links reflect a shared genetic basis with EA and (c) assess whether there is support for shared genetic factors between clinical ADHD and LRAs conditional on genetically predicted EA.
Studied ADHD polygenic scores (ADHD-PGS) are based on ADHD genome-wide association study (GWAS) summary statistics from two large independent ADHD samples, the Psychiatric Genomics Consortium (PGC) and the Danish Lundbeck Foundation Initiative for Integrative Psychiatric Research (iPSYCH), and a combination thereof. Associations between ADHD-PGS and a wide spectrum of population-based literacy-related and language-related measures related to reading, spelling, phonemic awareness, listening comprehension, non-word repetition and verbal intelligence skills, are examined in a sample of children from the UK Avon Longitudinal Study of Parents and Children (ALSPAC). Applying multivariable regression (MVR) techniques, analogous to Mendelian Randomisation (MR) approaches26, we report here disentangled associations between polygenic ADHD risk and LRA measures and estimate effects independent of and shared with genetically predicted years of schooling, using summary statistics from the Social Science Genetic Association Consortium (SSGAC).
Methods and materials
Literacy-related and language-related abilities in the general population
LRAs were assessed in children and adolescents from ALSPAC, a UK population-based longitudinal pregnancy-ascertained birth cohort (estimated birth date: 1991–1992, Supplementary Information)27,28. Ethical approval was obtained from the ALSPAC Law-and-Ethics Committee (IRB00003312) and the Local Research-Ethics Committees. Written informed consent was obtained from a parent or individual with parental responsibility and assent (and for older children consent) was obtained from the child participants.
Phenotype information
Thirteen measures capturing LRAs related to reading, spelling, phonemic awareness, listening comprehension, non-word repetition and verbal intelligence scores were assessed in 7 to 13 year-old ALSPAC participants (N ≤ 5919, Table 1) using both standardised and ALSPAC-specific instruments. Detailed descriptions of all LRA measures are available in Table 1 and the Supplementary Information.
Table 1.
Literacy-related and language-related abilities in the Avon Longitudinal Study of Parents and Children
| LRA (psychological instrument) | Mean Score (SE) | Mean Age (SE) | N (%males) | LRA combinations | |
|---|---|---|---|---|---|
| Reading accuracy and comprehension (WORD69), words | 28.44 (9.24) | 7.53 (0.31) | 5891 (50.6) | ---------------------Reading---------------- | --------------------------- --------Global LRAs------------- ----------------------------------- |
| Reading accuracy (ALSPAC specific: NBO70), words | 7.55 (2.44) | 9.87 (0.32) | 5738 (49.3) | ||
| Reading speeda (NARA II71), passages | 105.50 (12.47) | 9.88 (0.32) | 5189 (49.1) | ||
| Reading accuracya (NARA II71), passages | 104.11 (13.62) | 9.88 (0.32) | 5201 (49.1) | ||
| Reading speed (TOWRE72), words | 82.58 (10.28) | 13.83 (0.20) | 4247 (48.4) | ||
| Non-word reading accuracy (ALSPAC specific: NBO70) | 5.24 (2.48) | 9.87 (0.32) | 5731 (49.2) | ||
| Non-word reading speed (TOWRE72) | 50.82 (9.38) | 13.83 (0.20) | 4237 (48.3) | ||
| Spelling accuracy (ALSPAC specific: NB) | 7.89 (4.39) | 7.53 (0.31) | 5800 (50.2) | -Spelling- | |
| Spelling accuracy (ALSPAC specific: NB) | 10.27 (3.43) | 9.87 (0.32) | 5728 (49.2) | ||
| Phonemic awareness (AAT73) | 20.23 (9.51) | 7.53 (0.31) | 5919 (50.6) | ||
| Listening comprehension (WOLD74) | 7.50 (1.96) | 8.63 (0.30) | 5473 (49.9) | ||
| Non-word repetition (CNRep75) | 7.26 (2.51) | 8.63 (0.30) | 5464 (49.9) | ||
| Verbal intelligencea (WISC-III76) | 107.85 (16.74) | 8.64 (0.31) | 5456 (49.7) | ||
Note: Thirteen LRAs capturing aspects related to reading, spelling, phonemic awareness, listening comprehension, non-word repetition and verbal intelligence were assessed in 7 to 13 year-old ALSPAC participants using both standardised and ALSPAC-specific instruments (Supplementary Information)
LRAs literacy-related and language-related abilities, WORD Wechsler Objective Reading Dimension, ALSPAC Avon Longitudinal study of Parents and Children, NBO ALSPAC-specific assessment developed by Nunes, Bryant and Olson, NARA II The Neale Analysis of Reading Ability-Second Revised British Edition, TOWRE Test Of Word Reading Efficiency, NB ALSPAC-specific assessment developed by Nunes and Bryant, AAT Auditory Analysis Test, WOLD Wechsler Objective Language Dimensions, CNRep Children’s Test of Nonword Repetition, WISC-III Wechsler Intelligence Scale for Children III
aScores were derived using age norms and adjusted for sex and principal components only before transformation
All LRA scores were rank-transformed to allow for comparisons of genetic effects across different psychological instruments with different distributions (Supplementary Information). Phenotypic correlations, using Pearson-correlation coefficients, were comparable for untransformed and rank-transformed scores (Table S1). To account for multiple testing, we estimated the effective number of phenotypes studied using Matrix Spectral Decomposition29(MatSpD), revealing seven independent measures (experiment-wide error rate of 0.007).
For sensitivity analysis, we excluded 188 children with an ADHD diagnosis at age 7, based on the Development and Wellbeing Assessment (DAWBA)30 (Supplementary Information).
Genetic analyses
ALSPAC participants were genotyped using the Illumina HumanHap550 quad chip genotyping platforms, and genotypes were called using the Illumina GenomeStudio software. Genotyping, imputation and genome-wide association analysis details are described in the Supplementary Information and Table 2.
Table 2.
Sample description
| Phenotype | Sample | Source | Ethnicity | Imputation reference panel | N |
|---|---|---|---|---|---|
| LRAs | ALSPAC | General population | White European | HRC r1.1 | ≤5891 |
| ADHD | PGC | Clinical sample | Predominantly white European | HapMap phase 3 | 16,203 (Ncases = 4163) |
| ADHD | iPSYCH | Clinical sample | White European | 1000 Genomes phase 3 | 37,076 (Ncases = 14,584) |
| ADHD | PGC + iPSYCH (EUR) | Clinical sample | White European | 1000 Genomes phase 3 | 53,293 (Ncases = 19,099) |
| ADHD | PGC + iPSYCH | Clinical sample | Predominantly white European | 1000 Genomes phase 3 | 55,374 (Ncases = 20,183) |
| EA | SSGAC | Predominantly general population | White European | 1000 Genomes phase 3a | 326,041 |
aPredominantly 1000 Genomes phase 3 (20)
Abbreviations: LRAs literacy-related and language-related abilities, ADHD Attention-Deficit/Hyperactivity Disorder, EA educational attainment, ALSPAC Avon Longitudinal study of Parents and Children, PGC Psychiatric Genomics Consortium, iPSYCH The Lundbeck Foundation Initiative for Integrative Psychiatric Research, EUR European ancestry, SSGAC Social Science Genetic Consortium, HRC The Haplotype Reference Consortium
Note: There is no overlap between LRA, ADHD and EA samples
Clinical ADHD summary statistics
Psychiatric Genomics Consortium (PGC). GWAS summary statistics were obtained from a mega-analysis of clinical ADHD31, conducted by the PGC (4163 cases and 12,040 controls/pseudo-controls) (Table 2, Supplementary Information, www.med.unc.edu/pgc/).
The Lundbeck Foundation Initiative for Integrative Psychiatric Research (iPSYCH). An independent set of ADHD GWAS summary statistics were accessed through the Danish iPSYCH project32 (14,584 ADHD cases, 22,492 controls) (Table 2, Supplementary Information), using samples from the Danish Neonatal Screening Biobank hosted by Statens Serum Institute21,33.
Combined PGC and iPSYCH ADHD sample (PGC + iPSYCH). To maximise power, we also analysed meta-GWAS summary statistics from an ADHD sample containing both PGC and iPSYCH participants21 (20,183 cases, 35,191 controls/pseudo-controls) (Table 2, www.med.unc.edu/pgc/) and its European-only subset (PGC + iPSYCH(EUR), 19,099 cases, 34,194 controls/pseudo-controls) (Table 2, www.med.unc.edu/pgc/).
Detailed sample descriptions are available in Table 2 and the Supplementary Information.
Educational attainment summary statistics
GWAS summary statistics for EA20 (discovery and replication sample combined, excluding ALSPAC and 23andMe samples, N = 326,041) were obtained from the SSGAC consortium. EA was assessed as years of schooling20. A detailed sample description is available in Table 2 and the Supplementary Information.
Genome-wide complex trait analysis
SNP-h2 and genetic correlations (rg) between LRAs were estimated using Restricted Maximum Likelihood (REML) analyses34,35 as implemented in Genome-wide Complex Trait Analysis (GCTA) software36, including individuals with a genetic relationship < 0.0534. For this study, we selected only LRAs with evidence for SNP-h2 and sample size N > 4000 (Table S2).
Linkage disequilibrium score regression and correlation
Linkage Disequilibrium Score (LDSC) regression37 was used to distinguish confounding biases from polygenic influences by examining the LDSC regression intercept. Unconstrained LD-score correlation38 analysis was applied to estimate rg (Supplementary Information).
Polygenic scoring analyses
ADHD-PGS39,40 were created in ALSPAC using the independent PGC and iPSYCH GWAS summary statistics, and, to maximise power, also for GWAS summary statistics from the combined PGC + iPSYCH sample (Supplementary Information). ADHD-PGS have been previously linked to ADHD symptoms in ALSPAC participants41. Rank-transformed LRAs were regressed on Z-standardised ADHD-PGS (aligned to measure risk-increasing alleles) using ordinary least square (OLS) regression (R:stats library, Rv3.2.0). The proportion of phenotypic variance explained is reported as OLS-regression-R2. Beta-coefficients (β) for ADHD-PGS quantify here the change in standard deviation (SD) units of LRA performance per one SD increase in ADHD-PGS.
Multivariable regression analysis
To study the genetic association between ADHD and LRAs conditional on genetic influences shared with EA, we applied MVR. This technique is analogous to MR methodologies26 and controls for collider bias42 through the use of GWAS summary statistics. Technically, it involves the regression of regression estimates from independent samples on each other26 (Supplementary Information). Within this study we use MVR without inferring causality due to violations of classical MR assumptions26 (see below).
Genetic variant selection: To disentangle ADHD-LRA associations, we selected two sets of instruments from the most powerful ADHD GWAS summary statistics (PGC + iPSYCH). The first set contained genome-wide significant variants (P < 5 × 10−8, conservative). The second set included variants passing a more lenient P-value threshold (P < 0.0015, subthreshold) to increase power, consistent with current guidelines for the selection of genetic instruments in MR (F-statistic < 10)43. All sets included independent (PLINK44 clumping: LD-r2 < 0.25, ± 500 kb), well imputed (INFO45 > 0.8) and common (EAF > 0.01) variants. This resulted in 15 conservative and 2689 < NSNPs ≤ 2692 subthreshold ADHD-instruments (Table S8).
Estimation of ADHD effects: We extracted regression estimates for selected ADHD-instruments (conservative and subthreshold) from ADHD (PGC + iPSYCH), EA (SSGAC) and 13 LRA (ALSPAC) GWAS summary statistics. Analysing each set of variants independently, regression estimates for individual LRA measures (β) were regressed on both ADHD (β as lnOR) and EA regression estimates (β) using an OLS regression framework (R:stats library, Rv3.2.0). Outcomes were (1) a MVR regression estimate quantifying the change in SD units of LRA performance per log odds increase in ADHD risk conditional on years of schooling (ADHD effect independent of EA), and (2) a MVR regression estimate quantifying the change in SD units of LRA performance per year of schooling as captured by ADHD instruments (ADHD effect shared with EA). Latter MVR regression estimates capture here shared genetic effects between ADHD, EA and LRAs, including (1) genetic confounding (i.e., genetically predictable EA influences both ADHD and LRAs), (2) mediation (i.e., genetically predictable ADHD influences LRA indirectly through EA) and (3) biological pleiotropy (i.e., ADHD risk variants affect ADHD and EA through independent biological pathways). As ADHD risk and EA are inversely genetically related with each other21, they were reported to quantify change per missing year of schooling. To compare the magnitude of both MVR estimates, we also conducted analyses using fully standardised EA, ADHD and LRA regression estimates (Supplementary Information).
Finally, MVR regression estimates were meta-analysed and contrasted across reading-related, spelling-related and all LRA measures (excluding the composite measure verbal intelligence) (Table 1) using random-effects meta-regression, accounting for phenotypic correlations between LRAs (R:metafor library46, Rv3.2.0; Supplementary Information).
Sensitivity analyses
As the directionality of the relationship between ADHD, EA and LRAs cannot be inferred in this study, we also examined the genetic association between EA and LRAs, conditional on ADHD, using MVR. Two sets of EA instruments (conservative and subthreshold, Table S8) were selected from EA (SSGAC) GWAS summary statistics, analogous to the selection of ADHD instruments, and MVR was conducted as described above. Note that we did not create LRA instrument sets, as GWAS summary statistics of LRAs were underpowered.
Attrition analysis
We carried out an attrition analysis in ALSPAC studying the genetic association between LRA-missingness and polygenic ADHD risk, using both polygenic scoring analyses and MVR (Supplementary Information).
Results
Genetic architecture of literacy-related and language-related abilities and clinical ADHD
Phenotypic variation in literacy-related and language-related measures (Table 1), including reading abilities (comprehension, accuracy and speed) assessed in words/passages and non-words, spelling abilities (accuracy), phonemic awareness, listening comprehension, non-word repetition and verbal intelligence scores, can be tagged by common variants, with SNP-h2 estimates between 0.32 (SE = 0.07, non-word repetition age 8) and 0.54 (SE = 0.07, verbal intelligence age 8) (Table S2; GCTA-based and LDSC-based estimations). Importantly, all LRAs are phenotypically (Table S1) and genetically (Table S3) moderately to strongly interrelated. The observed LDSC-based evidence for genetic liability of clinical ADHD within the PGC (LDSC-h2 = 0.08(SE = 0.03)), iPSYCH (LDSC-h2 = 0.26(SE = 0.02)) and PGC + iPSYCH samples (Table S4) is consistent with previous reports21.
Association between ADHD polygenic risk scores and literacy-realted and language-related abilities
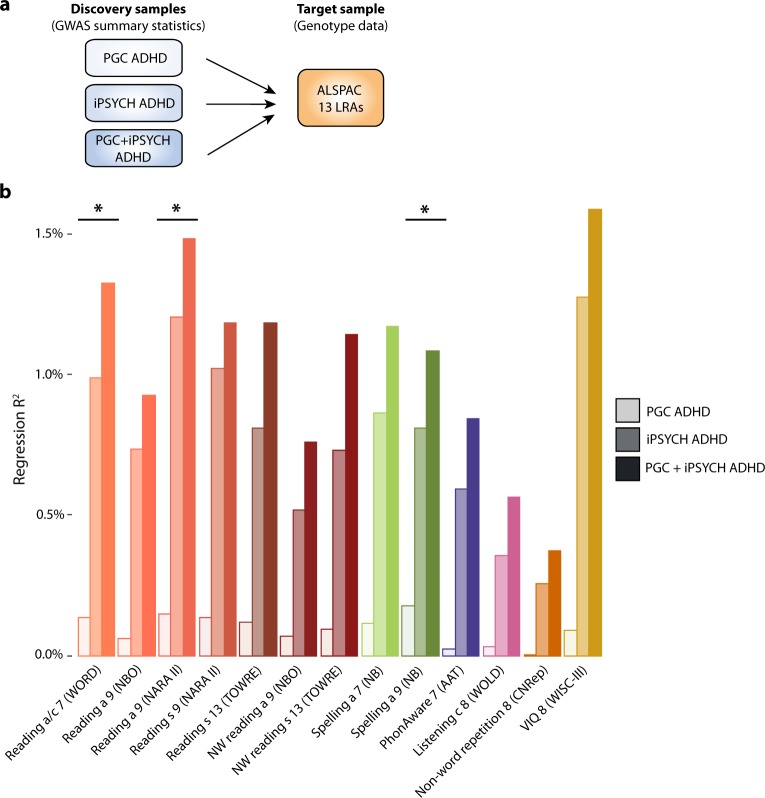
We observed robust evidence for an inverse genetic association between ADHD-PGS and reading accuracy/comprehension age 7 (PGC: OLS-R² = 0.1%, P = 4.6 × 10-3; iPSYCH: OLS-R² = 1.0%, P < 1 × 10−10), reading accuracy age 9 (PGC: OLS-R² = 0.1%, P = 5.7 × 10−3; iPSYCH: OLS-R² = 1.2%, P < 1 × 10−10), and spelling accuracy age 9 (PGC: OLS-R² = 0.2%, P = 1.5 × 10−3; iPSYCH: OLS-R² = 0.8%, P < 1 × 10−10) using independent ADHD discovery samples (Fig. 1, Table S5). The strongest evidence for association was observed when ADHD discovery samples were combined (PGC + iPSYCH; Fig. 1), including those of European ancestry only (PGC + iPSYCH(EUR)), with genetic trait-disorder overlap present for all LRAs studied (Table S5). For example, ADHD-PGS explain 1.49% phenotypic variation in reading accuracy age 9, translating into a genetic covariance of −0.11(95%-CI: −0.14; −0.09) (Supplementary Information). Polygenic scoring results are presented for a P-value threshold of 0.1, but other thresholds provided similar results (data not shown). Results were not affected by the exclusion of children with an ADHD diagnosis in ALSPAC (Table S6).
Fig. 1. Phenotypic variance in literacy-related and language-related abilities explained by polygenic ADHD risk.
a accuracy, c comprehension, s speed, WORD Wechsler Objective Reading Dimension, NBO Nunes, Bryant and Olson (ALSPAC specific instrument), NARA II The Neale Analysis of Reading Ability-Second Revised British Edition, TOWRE Test Of Word Reading Efficiency, NW non-word, NB Nunes and Bryant (ALSPAC specific instrument), PhonAware phonemic awareness, AAT Auditory Analysis Test, WOLD Wechsler Objective Language Dimensions, CNRep Children’s Test of Nonword Repetition, VIQ verbal intelligence quotient, WISC-III Wechsler Intelligence Scale for Children III, PGC Psychiatric Genomics Consortium, iPSYCH The Lundbeck Foundation Initiative for Integrative Psychiatric Research, ADHD Attention-Deficit/Hyperactivity Disorder a Schematic representation of polygenic scoring analyses. ADHD polygenic scores were created in ALSPAC using PGC, iPSYCH and PGC + iPSYCH GWAS summary statistics. Rank-transformed LRAs were regressed on Z-standardised ADHD-PGS using ordinary least square regression. b Phenotypic variance in literacy-related and language-related abilities explained by polygenic ADHD risk. *Evidence for association between LRAs and polygenic ADHD risk as observed in PGC ADHD, iPSYCH ADHD and PGC + iPSYCH ADHD samples. Note that all LRAs were associated with polygenic ADHD risk in iPSYCH ADHD and PGC + iPSYCH ADHD passing the experiment-wide error rate (P < 0.007)
Shared genetic liability between ADHD and LRA with EA
There was strong evidence for a moderate negative genetic correlation (rg = –0.53(SE = 0.03), P < 1 × 10−10) between genetically predicted ADHD, as captured by the largest ADHD discovery sample (PGC + iPSYCH), and EA (LDSC-h2 = 0.11(SE = 0.004)), consistent with previous findings21. Likewise, LRAs were moderately to highly positively correlated with EA (e.g., reading speed age 13 rg = 0.80(SE = 0.22), P = 3.0 × 10−4; Table S7), as previously reported22,23. Additionally, two independent variants reached genome-wide significance for both ADHD21 and EA20, consistent with biological pleiotropy (i.e., single genetic loci influencing multiple traits)47. These findings indicate complex, potentially reciprocal cross-trait relationships (Fig. 2a) and violate MR causal modelling assumptions26. Consequently, ADHD instruments are not valid MR instruments as they are not independent of EA.
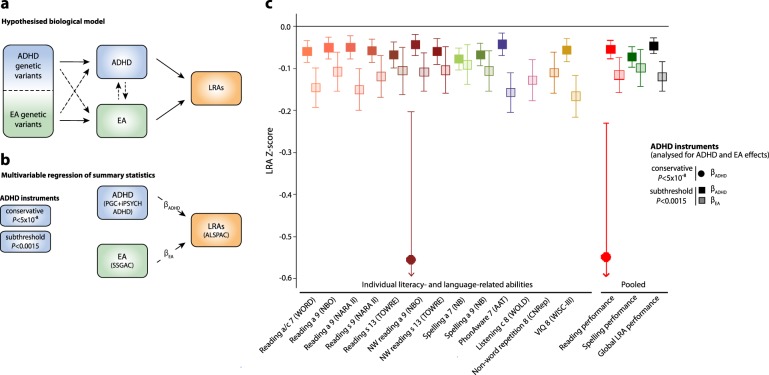
Fig. 2. Genetic relationships between ADHD, educational attainment and literacy-related and language-related abilities.
ADHD Attention-Deficit/Hyperactivity Disorder, EA educational attainment, LRAs literacy and language-related abilities, PGC Psychiatric Genomics Consortium, iPSYCH The Lundbeck Foundation Initiative for Integrative Psychiatric Research; SSGAC Science Genetic Association Consortium, ALSPAC Avon Longitudinal Study of Parents And Children, MVR multivariable regression. a Hypothesised biological model of genetic relationships between ADHD, EA, and LRAs reflecting complex, pleiotropic and reciprocal genetic links that prevent causal inferences. b Schematic MVR model assessing polygenic ADHD-LRA overlap independent of and shared with genetic effects for EA. c MVR estimates of ADHD-specific effects independent of EA and ADHD effects shared with EA on LRAs using standardised ADHD instruments: Sets of conservative (P < 5 × 10-8) and subthreshold (P < 0.0015) ADHD instruments were extracted from ADHD (PGC + iPSYCH), EA (SSGAC) and LRAs (ALSPAC) GWAS summary statistics. ADHD-specific effects independent of EA (βADHD) and ADHD effects shared with EA (βEA) on LRAs were estimated with MVRs. To compare the magnitude of βADHD and βEA, MVR analyses were conducted using standardised regression estimates (Supplementary Methods). βADHD estimates measure the change in LRA Z-score per Z-score in ADHD liability. βEA estimates measure the change in LRA Z-scores per Z-score in missing school years. MVR estimates based on raw genetic effect estimates are provided in Table 3. Pooled estimates for reading, spelling and global LRA measures (Table 1) were obtained through random-effects meta-regression. Only effects passing the experiment-wide significance threshold (P < 0.007) are shown with corresponding 95% confidence intervals. There is no causality inferred
Multivariable regression analyses
To disentangle the genetic overlap of polygenic ADHD risk with literacy-related and language-related measures into ADHD genetic effects independent of and shared with EA, we applied MVR26 using ADHD instruments based on the most powerful ADHD discovery sample (PGC + iPSYCH) (Fig. 2b).
Using conservative ADHD instruments (Table S8), non-word reading accuracy at age 9 and pooled reading-related abilities were associated with polygenic ADHD risk, conditional on EA (Table 3). The latter translates into, for example, a decrease of 0.35 SD in pooled reading performance per log-odds increase in ADHD risk (βADHD = -0.35(SE = 0.09), P = 9.2 × 10-5, Phet = 0.19), an effect that was considerably stronger than for other LRAs (Pmod = 0.011, Table S10).
Table 3.
Multivariable regression analysis of polygenic associations between ADHD and literacy-related and language-related abilities (raw estimates)
| LRAs | ADHD-specific effects independent of EA (βADHD) | EA genetic effects of ADHD-associated variantsa (βEA) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Conservative instruments (Pthr < 5 × 10-8) | Subthreshold instruments (Pthr < 0.0015) | Conservative instruments (Pthr < 5 × 10-8) | Subthreshold instruments (Pthr < 0.0015) | |||||||||
| β (SE) | P | P het | β (SE) | P | P het | β (SE) | P | P het | β (SE) | P | P het | |
| Reading a/c 7 (WORD) | –0.18 (0.13) | 0.20 | — | –0.03 (0.01) | 1.4 × 10−5 | — | −0.09 (1.19) | 0.94 | — | −0.63(0.10) | 1.2 × 10−9 | — |
| Reading a 9 (NBO) | −0.28 (0.09) | 0.01 | — | −0.03 (0.01) | 2.1 × 10−4 | — | 1.33 (0.84) | 0.14 | — | −0.46 (0.10) | 6.3 × 10−6 | — |
| Reading s 9 (NARA II) | −0.29 (0.13) | 0.04 | — | −0.03 (0.01) | 3.0 × 10−5 | — | 1.55 (1.16) | 0.21 | — | −0.51 (0.11) | 2.5 × 10−6 | — |
| Reading a 9 (NARA II) | −0.34 (0.12) | 0.01 | — | −0.03 (0.01) | 3.8 × 10−4 | — | 1.57 (1.11) | 0.18 | — | −0.64 (0.11) | 4.1 × 10−9 | — |
| Reading s 13 (TOWRE) | −0.36 (0.15) | 0.03 | — | −0.04 (0.01) | 2.7 × 10−5 | — | 2.06 (1.35) | 0.15 | — | −0.46 (0.12) | 1.8 × 10−4 | — |
| NW reading a 9 (NBO) | −0.37 (0.09) | 0.002 | — | −0.03 (0.01) | 2.4 × 10−4 | — | 1.54 (0.85) | 0.09 | — | −0.46 (0.10) | 6.1 × 10−6 | — |
| NW reading s 13 (TOWRE) | −0.21 (0.17) | 0.25 | — | −0.03 (0.01) | 2.4 × 10−4 | — | 1.43 (1.58) | 0.38 | — | −0.45 (0.12) | 1.9 × 10-4 | — |
| Spelling a 7 (NB) | −0.05 (0.12) | 0.72 | — | −0.05 (0.01) | 1.4 × 10−5 | — | -1.12 (1.13) | 0.34 | — | −0.63 (0.10) | 1.2 × 10−9 | — |
| Spelling a 9 (NB) | −0.23 (0.08) | 0.01 | — | −0.04 (0.01) | 8.7 × 10−7 | — | 1.43 (0.72) | 0.38 | — | −0.45 (0.10) | 1.3 × 10−5 | — |
| PhonAware 7 (AAT) | −0.22 (0.15) | 0.16 | — | −0.02 (0.01) | 0.002 | — | −0.31 (1.33) | 0.82 | — | −0.67 (0.10) | < 1 × 10−10 | — |
| Listening c 8 (WOLD) | −0.03 (0.08) | 0.75 | — | −0.02 (0.01) | 0.02 | — | −1.47 (0.70) | 0.06 | — | −0.55 (0.11) | 4.5 × 10−7 | — |
| Non-word repetition 8 (CNRep) | −0.14 (0.14) | 0.34 | — | −0.02 (0.01) | 0.01 | — | −0.16 (1.30) | 0.90 | — | −0.47 (0.11) | 1.5 × 10−5 | — |
| VIQ 8 (WISC-III) | −0.23 (0.15) | 0.16 | — | −0.03 (0.01) | 5.0 × 10−5 | — | −0.36 (1.37) | 0.80 | — | −0.71 (0.11) | < 1 × 10−10 | — |
| Pooled reading | −0.35 (0.09) | 9.2 × 10−5 | 0.19 | −0.03 (0.01) | 1.4 × 10−6 | 0.79 | 1.67 (0.78) | 0.03 | 0.31 | −0.50 (0.09) | 4.9 × 10−8 | 0.09 |
| Pooled spelling | −0.18 (0.11) | 0.10 | 0.03 | −0.04 (0.01) | 1.1 × 10−8 | 0.28 | 0.40 (1.25) | 0.75 | 0.001 | −0.42 (0.10) | 1.3 × 10−5 | 0.39 |
| Pooled LRAs | −0.18 (0.07) | 0.01 | 0.005 | −0.03 (0.01) | 1.9 × 10−6 | 0.05 | 0.29 (0.69) | 0.67 | 0.001 | −0.49 (0.08) | < 1 × 10−10 | 0.004 |
Note: Sets of conservative (P < 5 × 10−8) and subthreshold (P < 0.0015) ADHD instruments were extracted from ADHD (PGC + iPSYCH), EA (SSGAC) and LRAs (ALSPAC) GWAS summary statistics. ADHD-specific effects independent of EA (βADHD) and ADHD effects shared with EA (βEA) on LRAs were estimated with MVRs (Fig. 2b). ADHD effects shared with EA were assessed through EA genetic effect estimates of ADHD-associated variants and presented with respect to missing school years. βADHD quantifies the change in LRA Z-score per log odds increase in ADHD liability. βEA quantifies the change in LRA Z-score per missing year of schooling. Pooled estimates for reading, spelling and global LRAs (Table 1) were obtained through random-effects meta-regression. Evidence for effect heterogeneity (Phet) was monitored through Cochran’s Q-test. To compare effect sizes of βADHD and βEA, MVR was carried out using standardised genetic effect estimates for which results are provided in Fig. 2c.
LRAs literacy-related and language-related abilities, ADHD Attention-Deficit/Hyperactivity Disorder, EA educational attainment, PthrP-value threshold, Phet heterogeneity P-value, a accuracy, c comprehension, s speed, WORD Wechsler Objective Reading Dimension, NBO Nunes, Bryant and Olson (ALSPAC specific instrument), NARA II The Neale Analysis of Reading Ability-Second Revised British Edition, TOWRE Test Of Word Reading Efficiency, NW non-word, NB Nunes and Bryant (ALSPAC specific instrument), PhonAware phonemic awareness, AAT Auditory Analysis Test, WOLD Wechsler Objective Language Dimensions, CNRep Children’s Test of Nonword Repetition, VIQ verbal intelligence quotient, WISC-III Wechsler Intelligence Scale for Children III, MVR Multivariable regression
aADHD genetic effects shared with EA as assessed through EA genetic effect estimates of ADHD-associated variants
Using subthreshold ADHD instruments (Table S8), polygenic ADHD effects on LRA performance, conditional on EA, were detectable for all reading-related and spelling-related measures, phonemic awareness and verbal intelligence, but not other LRAs such as listening comprehension and non-word repetition (Table 3). Evidence was strongest for pooled reading and spelling abilities (Table 3, minimum P = 1.1 × 10−8). However, observable effects were smaller in magnitude compared to those captured by conservative ADHD instruments with, for example, a 0.03 SD decrease in pooled reading performance per log-odds increase in ADHD risk (βADHD = -0.03(SE = 0.01), P = 1.4 × 10−6, Table 3). Comparing ADHD-specific effects on both reading and spelling with ADHD-specific effects on all other LRAs provided evidence for effect differences (Pmod = 0.016), with stronger ADHD effects on literacy-related abilities, in particular spelling (Table S10).
Polygenic ADHD effects that are shared with EA were identified for all LRAs studied using subthreshold, but not conservative ADHD instruments (Table 3). This translates into, for example, a further 0.50 SD units decrease in pooled reading performance per missing school year (βEA = −0.50(SE = 0.09), P = 4.9 × 10−8, Table 3). Thus, the observed association between polygenic ADHD risk and listening comprehension and non-word repetition is fully attributable to genetic effects shared with EA (Table 3). Contrary to ADHD-specific effects, ADHD effects shared with EA showed no evidence for effect differences between literacy-related versus other LRAs (P = 0.31). Conducting MVR with fully standardised estimates showed that ADHD effects shared with EA were as large as or even larger compared to ADHD-specific effects (Fig. 2c, Table S9).
Using an analogous approach, we disentangled the genetic overlap between polygenic EA and LRAs into genetic EA effects independent of and shared with ADHD, based on EA instruments (Fig. S1). There was strong evidence for EA effects shared with ADHD using subthreshold, but not conservative EA instruments (Table S11). The magnitude of ADHD genetic effects shared with EA, captured by ADHD genetic instruments, compared to the magnitude of EA genetic effects shared with ADHD, captured by EA instruments, was largely consistent with each other in fully standardised analyses (Tables S9 and S11).
There was little evidence supporting the inclusion of regression intercepts in MVR that would imply additional genetic effect variation in LRAs estimates, not yet captured by either ADHD or EA effect estimates, based on the selected instruments. Therefore, all MVRs were performed using constrained intercepts26.
Attrition in ALSPAC
Analyses of sample drop-out in ALSPAC, exemplified by missing reading accuracy and comprehension scores at age 7 (WORD), revealed a positive genetic association between missingness and polygenic ADHD risk (min P = 1.4 × 10−8, Supplementary Information, Table S12, Table S13).
Discussion
This study identified strong and replicated evidence for an inverse association between polygenic ADHD risk and multiple population-based LRAs using a polygenic scoring approach. However, these associations involve shared genetic variation with genetically predictable EA. Accurate modelling of polygenic links using MVR techniques, conditional on EA, revealed an ADHD-specific association profile that primarily involves literacy-related impairments. Once shared genetic effects with EA were accounted for, polygenic ADHD risk was most strongly inversely associated with reading and/or spelling abilities, in addition to phonemic awareness and verbal intelligence, but not listening comprehension and non-word repetition abilities. Importantly, genetic overlap between polygenic ADHD risk and all of the LRAs studied was inflated by genetic effects shared with EA.
Using independent ADHD discovery samples, these findings show that genetic overlap between ADHD and literacy-related impairments observed in twin and family studies16–18 can be extended to genetic associations, as captured by common variation in general population samples. The identified association profile suggests that not only reading-related abilities (including both word and nonword reading skills), but also phonological and spelling-related abilities share genetic aetiology with ADHD. These interrelated LRAs may, as hypothesised for RD, arise from a phonological impairment48,49, which affects decoding and reading skills50, but also spelling abilities51. However, reading abilities can, once developed, also shape phonological skills52.
In addition, this study suggests that genetic associations between polygenic ADHD and LRAs reflect, at least partially, shared genetic influences with genetically predictable EA and that, equally likely, genetic associations between polygenic EA and LRAs share genetic influences with ADHD. The magnitude of these shared effects, modelled with different MVR approaches, was comparable with each other. This is consistent with reciprocal genetic influences between EA and ADHD (Fig. 2a) and supports an intergenerational multiple-deficit model proposed for reading disability15,53. Children growing up in disadvantaged environments, genetically predictable through polygenic EA scores54, might be more vulnerable to psychiatric illness including ADHD55 that affects, in turn, their LRAs. In addition, adolescents with ADHD might be more likely to leave school at an earlier age, with lower LRA performance and EA, and pass on an increased genetic load to their own children56.
Here, we demonstrate that disentangling multivariate genetic interrelationships between ADHD, EA and LRAs using MVR can aid the interpretation of genetic overlap, while controlling for collider bias42. However, using MVR, the detection of these polygenic associations was strongly governed by the choice of genetic variants. Conservative ADHD instruments identified large ADHD-specific effects on reading as a domain and little evidence for genetic effects that are shared with EA, although they had limited power57. They comprised 15 independent SNPs only, including variation within FOXP2, a gene that has been implicated in childhood apraxia of speech and expressive and receptive language impairments (http://omim.org/entry/602081)58. On the other hand, subthreshold instruments, including thousands of variants, tagged ADHD-specific polygenic links with LRAs (conditional on EA) with smaller effects, but with higher predictive accuracy. However, these instruments also captured shared genetic effects with EA, affecting polygenic links between ADHD and all of the LRAs studied. These shared genetic influences were of equal strength and at least equal magnitude compared to ADHD-LRA associations independent of EA. Contrary, a previous twin study showed that the genetic covariance between ADHD and reading difficulties was largely independent of genetic effects shared with IQ19, suggesting that our findings may also reflect socio-economic influences. Thus, in order to improve reading and, more generally, literacy-related deficits in children with ADHD, there is potentially a need for further intervention programmes targeting EA-independent underlying neurocognitive deficits, beyond general training programmes aiming at schooling outcomes59.
In general, our findings are consistent with an omnigenic60 model of complex trait architectures, compatible with a general factor model of psychopathology61, including ADHD62. The omnigenic model construes that only the largest-effect variants will reflect ADHD specificity, and may thus tag the most trait-specific associations between ADHD and reading, independent of EA. The majority of variants, however, will capture pleiotropic (omnigenic) influences pointing to highly interconnected neural networks60 that give rise to genetic confounding. Consequently, the majority of subthreshold variants, captured by both ADHD and EA subthreshold variants, are likely to represent highly powerful cross-trait genetic predictors that may enhance and induce genetic overlap.
Finally, the methodological framework within this work has not only relevance for studies investigating polygenic links between ADHD and LRAs, but for many studies examining multivariate trait interrelationships that involve shared genetic effects with a genetically predictable confounder. Specifically, our findings suggest that lower variant selection thresholds can introduce genetic variance sharing that is unspecific and needs to be accounted for before identified associations can be interpreted in terms of underlying mechanisms, including shared genetic aetiologies. This is especially important as current guidelines for studying polygenic links with allelic scores recommend aggregating genetic variants across less stringent significance thresholds to maximise genetic association between discovery and target samples63,64.
This study has several limitations. Firstly, ALSPAC, as other cohort studies, suffers from attrition65,66. Sensitivity analyses showed that this is unlikely to bias our findings based on conservative instruments. However, links identified using subthreshold ADHD variants, might have been underestimated given that individuals with a genetic predisposition to ADHD (but also smoking initiation, higher body mass index, neuroticism, schizophrenia and depression) are more likely to drop out66. Secondly, the strength of the genetic overlap between polygenic ADHD risk and LRAs may vary according to ADHD symptom domain levels, implicating especially inattentiveness67, as well as the nature of the literacy-related or language-related ability involved (as we observed evidence for effect heterogeneity when combining all LRAs). It is conceivable that also other verbal abilities, not investigated in this study, such as grammar, expressive vocabulary or pragmatic skills, may genetically overlap with ADHD. Furthermore, we only studied the extent to which shared genetic variance with EA affects the genetic association between ADHD and LRAs. However, we found little evidence for the presence of additional unaccounted for genetic influences using these instruments, i.e., effects that are not yet captured by either genetically predicted ADHD or EA. Finally, the power of available LRA GWAS summary statistics is still too low to generate genetic instruments supporting reverse models. Larger and more detailed clinical and population-based samples, as well as extensive multivariate variance analyses of the spectrum of LRAs (that are currently computationally expensive68) will help to further characterise the overlap between ADHD and literacy-related and language-related cognitive processes.
Conclusion
Polygenic associations of clinical ADHD and a range of LRAs are to a large extent attributable to genetic effects that are also shared with EA, especially when investigated with genetic variants typically selected for polygenic scoring approaches. Adjusting for these unspecific genetic effects reveals an ADHD-specific association profile that primarily involves literacy-related impairments.
Supplementary information
Acknowledgements
We are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. The UK Medical Research Council and the Wellcome Trust (Grant ref: 102215/2/13/2) and the University of Bristol provide core support for ALSPAC. The ALSPAC GWAS data was generated by Sample Logistics and Genotyping Facilities at the Wellcome Trust Sanger Institute and LabCorp (Laboratory Corporation of America) using support from 23andMe. This research was facilitated by the Social Science Genetic Association Consortium (SSGAC) and Psychiatric Genomics Consortium (PGC) by providing access to genome-wide summary statistics. This publication is the work of the authors and E.V. and B.S.T.P. will serve as guarantors for the contents of this paper. E.V., C.Y.S., B.S.T.P. and S.E.F. are supported by the Max Planck Society. The iPSYCH project is funded by the Lundbeck Foundation (grant no R102-A9118 and R155−014−1724) and the universities and university hospitals of Aarhus and Copenhagen. Genotyping of iPSYCH and PGC samples was supported by grants from the Lundbeck Foundation, the Stanley Foundation, the Simons Foundation (SFARI 311789 to M.J.D.), and NIMH (5U01MH094432−02 to M.J.D.). The Danish National Biobank resource was supported by the Novo Nordisk Foundation. Data handling and analysis was supported by NIMH (1U01MH109514−01 to Michael O’Donovan and Anders D Børglum). High-performance computer capacity for handling and statistical analysis of iPSYCH data on the GenomeDK HPC facility was provided by the Centre for Integrative Sequencing, iSEQ, Aarhus University, Denmark (grant to Anders D Børglum). An earlier version of this manuscript is available as a preprint on bioRxiv.
iPSYCH-Broad-PGC ADHD Consortium members:
Esben Agerbo3,19,20, Thomas Damm Als3,4,5,Marie Bækved-Hansen3,21, Rich Belliveau12, Anders D. Børglum3,4,5, Jonas Bybjerg-Grauholm3,21, Felecia Cerrato12, Kimberly Chambert12, Claire Churchhouse11,12,13, Søren Dalsgaard19, Mark J. Daly11,12,13, Ditte Demontis3,4,5, Ashley Dumont12, Jacqueline Goldstein11,12,13, Jakob Grove3,4,5,22, Christine S. Hansen3,21,23, Mads Engel Hauberg3,4,5, Mads V. Hollegaard3,21, David M. Hougaard3,21, Daniel P. Howrigan11,12, Hailiang Huang11,12, Julian Maller12,24, Alicia R. Martin11,12,13, Joanna Martin12,25,26, Manuel Mattheisen3,4,5,27,28, Jennifer Moran12, Ole Mors3,29, Preben Bo Mortensen3,4,19,20, Benjamin M. Neale11,12,13, Merete Nordentoft3,30, Jonatan Pallesen3,4,5, Duncan S. Palmer11,12, Carsten Bøcker Pedersen3,19,20, Marianne Giørtz Pedersen3,19,20, Timothy Poterba11,12,13, Jesper Buchhave Poulsen3,21, Stephan Ripke11,12,13,31, Elise B. Robinson11,32, F. Kyle Satterstrom11,12,13, Christine Stevens12, Patrick Turley11,12, Raymond K. Walters11,12, Thomas Werge3,23,33
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ellen Verhoef, Ditte Demontis
These authors jointly supervised this work: Anders D. Børglum, Beate St Pourcain
Contributor Information
Ellen Verhoef, Phone: +31 24 3521946, Email: Ellen.Verhoef@mpi.nl.
Beate St Pourcain, Phone: +31 24 3521964, Email: Beate.StPourcain@mpi.nl.
iPSYCH-Broad-PGC ADHD Consortium:
Esben Agerbo, Thomas Damm Als, Marie Bækved-Hansen, Rich Belliveau, Anders D. Børglum, Jonas Bybjerg-Grauholm, Felecia Cerrato, Kimberly Chambert, Claire Churchhouse, Søren Dalsgaard, Mark J. Daly, Ditte Demontis, Ashley Dumont, Jacqueline Goldstein, Jakob Grove, Christine S. Hansen, Mads Engel Hauberg, Mads V. Hollegaard, David M. Hougaard, Daniel P. Howrigan, Hailiang Huang, Julian Maller, Alicia R. Martin, Joanna Martin, Manuel Mattheisen, Jennifer Moran, Ole Mors, Preben Bo Mortensen, Benjamin M. Neale, Merete Nordentoft, Jonatan Pallesen, Duncan S. Palmer, Carsten Bøcker Pedersen, Marianne Giørtz Pedersen, Timothy Poterba, Jesper Buchhave Poulsen, Stephan Ripke, Elise B. Robinson, F. Kyle Satterstrom, Christine Stevens, Patrick Turley, Raymond K. Walters, and Thomas Werge
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41398-018-0324-2).
References
- 1.Geurts HM, Embrechts M. Language profiles in ASD, SLI, and ADHD. J. Autism Dev. Disord. 2008;38:1931–1943. doi: 10.1007/s10803-008-0587-1. [DOI] [PubMed] [Google Scholar]
- 2.Helland, W. A., Posserud, M.-B., Helland, T., Heimann, M., Lundervold, A. J. Language impairments in children with ADHD and in children with reading disorder. J. Atten. Disord. 1087054712461530 (2012). [DOI] [PubMed]
- 3.Germanò E, Gagliano A, Curatolo P. Comorbidity of ADHD and dyslexia. Dev. Neuropsychol. 2010;35:475–493. doi: 10.1080/87565641.2010.494748. [DOI] [PubMed] [Google Scholar]
- 4.Maughan B, Carroll J. Literacy and mental disorders. Curr. Opin. Psychiatry. 2006;19:350–354. doi: 10.1097/01.yco.0000228752.79990.41. [DOI] [PubMed] [Google Scholar]
- 5.Mayes SD, Calhoun SL. Learning, attention, writing, and processing speed in typical children and children with ADHD, autism, anxiety, depression, and oppositional-defiant disorder. Child Neuropsychol. J. Norm. Abnorm Dev. Child Adolesc. 2007;13:469–493. doi: 10.1080/09297040601112773. [DOI] [PubMed] [Google Scholar]
- 6.Mayes SD, Calhoun SL. Frequency of reading, math, and writing disabilities in children with clinical disorders. Learn Individ Differ. 2006;16:145–157. doi: 10.1016/j.lindif.2005.07.004. [DOI] [Google Scholar]
- 7.Czamara D, et al. Children with ADHD symptoms have a higher risk for reading, spelling and math difficulties in the GINIplus and LISAplus Cohort Studies. PLoS ONE. 2013;8:e63859. doi: 10.1371/journal.pone.0063859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Åsberg J, Kopp S, Berg-Kelly K, Gillberg C. Reading comprehension, word decoding and spelling in girls with autism spectrum disorders (ASD) or attention-deficit/hyperactivity disorder (AD/HD): performance and predictors. Int. J. Lang. Commun. Disord. 2010;45:61–71. doi: 10.3109/13682820902745438. [DOI] [PubMed] [Google Scholar]
- 9.Green BC, Johnson KA, Bretherton L. Pragmatic language difficulties in children with hyperactivity and attention problems: an integrated review. Int. J. Lang. Commun. Disord. 2014;49:15–29. doi: 10.1111/1460-6984.12056. [DOI] [PubMed] [Google Scholar]
- 10.Hawkins E, Gathercole S, Astle D, Holmes J. The Calm Team. Language problems and ADHD symptoms: how specific are the links? Brain Sci. 2016;6:50. doi: 10.3390/brainsci6040050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polanczyk G, de Lima MS, Horta BL, Biederman J, Rohde LA. The worldwide prevalence of ADHD: a systematic review and metaregression analysis. Am. J. Psychiatry. 2007;164:942–948. doi: 10.1176/ajp.2007.164.6.942. [DOI] [PubMed] [Google Scholar]
- 12.Peterson RL, Pennington BF. Developmental dyslexia. Lancet Lond. Engl. 2012;379:1997–2007. doi: 10.1016/S0140-6736(12)60198-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thapar A, Cooper M. Attention deficit hyperactivity disorder. Lancet. 2016;387:1240–1250. doi: 10.1016/S0140-6736(15)00238-X. [DOI] [PubMed] [Google Scholar]
- 14.Gough PB, Tunmer WE. Decoding, reading, and reading disability. Remedial Spec. Educ. 1986;7:6–10. doi: 10.1177/074193258600700104. [DOI] [Google Scholar]
- 15.Pennington BF. From single to multiple deficit models of developmental disorders. Cognition. 2006;101:385–413. doi: 10.1016/j.cognition.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 16.Martin NC, Levy F, Pieka J, Hay DA. A genetic study of attention deficit hyperactivity disorder, conduct disorder, oppositional defiant disorder and reading disability: aetiological overlaps and implications. Int J. Disabil. Dev. Educ. 2006;53:21–34. doi: 10.1080/10349120500509992. [DOI] [Google Scholar]
- 17.Willcutt EG, Pennington BF, DeFries JC. Twin study of the etiology of comorbidity between reading disability and attention-deficit/hyperactivity disorder. Am. J. Med. Genet. 2000;96:293–301. doi: 10.1002/1096-8628(20000612)96:3<293::AID-AJMG12>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 18.Willcutt EG, Pennington BF, Olson RK, DeFries JC. Understanding comorbidity: a twin study of reading disability and attention-deficit/hyperactivity disorder. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2007;144B:709–714. doi: 10.1002/ajmg.b.30310. [DOI] [PubMed] [Google Scholar]
- 19.Paloyelis Y, Rijsdijk F, Wood AC, Asherson P, Kuntsi J. The genetic association between ADHD symptoms and reading difficulties: the role of inattentiveness and IQ. J. Abnorm. Child Psychol. 2010;38:1083–1095. doi: 10.1007/s10802-010-9429-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okbay A, et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016;533:539–542. doi: 10.1038/nature17671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demontis D, et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet. 2019;51:63. doi: 10.1038/s41588-018-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selzam S, et al. Genome-wide polygenic scores predict reading performance throughout the school years. Sci. Stud. Read. 2017;21:334–349. doi: 10.1080/10888438.2017.1299152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luciano M, et al. Single nucleotide polymorphisms associated with reading ability show connection to socio-economic outcomes. Behav. Genet. 2017;47:469–479. doi: 10.1007/s10519-017-9859-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krapohl E, et al. Widespread covariation of early environmental exposures and trait-associated polygenic variation. Proc. Natl Acad. Sci. 2017;114:11727–11732. doi: 10.1073/pnas.1707178114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krapohl E, et al. Multi-polygenic score approach to trait prediction. Mol. Psychiatry. 2017;23:1368–1374. doi: 10.1038/mp.2017.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burgess S, et al. Dissecting causal pathways using mendelian randomization with summarized genetic data: application to age at menarche and risk of breast cancer. Genetics. 2017;207:481–487. doi: 10.1534/genetics.117.300191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fraser A, et al. Cohort profile: the avon longitudinal study of parents and children: ALSPAC mothers cohort. Int. J. Epidemiol. 2013;42:97–110. doi: 10.1093/ije/dys066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyd A, et al. Cohort Profile: the’children of the 90s’--the index offspring of the Avon Longitudinal Study of Parents and Children. Int. J. Epidemiol. 2013;42:111–127. doi: 10.1093/ije/dys064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 30.Goodman R, Ford T, Richards H, Gatward R, Meltzer H. The development and well-being assessment: description and initial validation of an integrated assessment of child and adolescent psychopathology. J. Child Psychol. Psychiatry. 2000;41:645–655. doi: 10.1111/j.1469-7610.2000.tb02345.x. [DOI] [PubMed] [Google Scholar]
- 31.Cross-Disorder Group of the Psychiatric Genomics Consortium. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat. Genet. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mors O, Perto GP, Mortensen PB. The Danish psychiatric central research register. Scand. J. Public Health. 2011;39(7 Suppl):54–57. doi: 10.1177/1403494810395825. [DOI] [PubMed] [Google Scholar]
- 33.Pedersen CB, et al. The iPSYCH2012 case-cohort sample: new directions for unravelling genetic and environmental architectures of severe mental disorders. Mol. Psychiatry. 2017;23:6–14. doi: 10.1038/mp.2017.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee SH, Yang J, Goddard ME, Visscher PM, Wray NR. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinforma. Oxf. Engl. 2012;28:2540–2542. doi: 10.1093/bioinformatics/bts474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J. Schizophrenia Working Group of the Psychiatric Genomics Consortium, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.International Schizophrenia Consortium. Purcell SM, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS. Genet. 2013;9:e1003348. doi: 10.1371/journal.pgen.1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stergiakouli E, et al. Shared genetic influences between dimensional ASD and ADHD symptoms during child and adolescent development. Mol. Autism. 2017;8:18. doi: 10.1186/s13229-017-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aschard H, Vilhjálmsson BJ, Joshi AD, Price AL, Kraft P. Adjusting for heritable covariates can bias effect estimates in genome-wide association studies. Am. J. Hum. Genet. 2015;96:329–339. doi: 10.1016/j.ajhg.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pierce BL, Ahsan H, Vanderweele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int. J. Epidemiol. 2011;40:740–752. doi: 10.1093/ije/dyq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS. Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Viechtbauer W. Conducting meta-analyses in R with the metafor package. J. Stat. Softw. 2010;36:1–48. doi: 10.18637/jss.v036.i03. [DOI] [Google Scholar]
- 47.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 2014;23(R1):R89–R98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carroll JM, Snowling MJ. Language and phonological skills in children at high risk of reading difficulties. J. Child Psychol. Psychiatry. 2004;45:631–640. doi: 10.1111/j.1469-7610.2004.00252.x. [DOI] [PubMed] [Google Scholar]
- 49.Hulme, C., Snowling, M. J. Developmental Disorders of Language Learning and Cognition. 1 edn. (Wiley-Blackwell, Chichester, Malden, 2009). 446 p.
- 50.Rack J, Snowling M, Olson R. The nonword reading deficit in developmental. Dyslexia a Rev. Read. Res Q. 1992;27:28–53. doi: 10.2307/747832. [DOI] [Google Scholar]
- 51.Treiman, R. Beginning to Spell: A Study of First-grade Children. (Oxford University Press, New York, 1992). 380 p.
- 52.Kamhi AG, Catts HW. Language and Reading Disabilities. 3 edn. Boston: (Pearson; 2011. [Google Scholar]
- 53.van Bergen E, van der Leij A, de Jong PF. The intergenerational multiple deficit model and the case of dyslexia. Front. Hum. Neurosci. 2014;8:346. doi: 10.3389/fnhum.2014.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ayorech Z, Krapohl E, Plomin R, Stumm Svon. Genetic influence on intergenerational educational attainment. Psychol. Sci. 2017;28:1302–1310. doi: 10.1177/0956797617707270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reiss F. Socioeconomic inequalities and mental health problems in children and adolescents: a systematic review. Soc. Sci. Med. 1982;90:24–31. doi: 10.1016/j.socscimed.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 56.Russell G, Ford T, Rosenberg R, Kelly S. The association of attention deficit hyperactivity disorder with socioeconomic disadvantage: alternative explanations and evidence. J. Child Psychol. Psychiatry. 2014;55:436–445. doi: 10.1111/jcpp.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schatzkin A, et al. Mendelian randomization: how it can—and cannot—help confirm causal relations between nutrition and cancer. Cancer Prev. Res. 2009;2:104–113. doi: 10.1158/1940-6207.CAPR-08-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–523. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- 59.Daley D, Birchwood J. ADHD and academic performance: why does ADHD impact on academic performance and what can be done to support ADHD children in the classroom? Child Care Health Dev. 2010;36:455–464. doi: 10.1111/j.1365-2214.2009.01046.x. [DOI] [PubMed] [Google Scholar]
- 60.Boyle EA, Li YI, Pritchard JK. An expanded view of complex traits: from polygenic to omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lahey BB, et al. Is there a general factor of prevalent psychopathology during adulthood? J. Abnorm. Psychol. 2012;121:971–977. doi: 10.1037/a0028355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Franke B. What’s in a name: the “omnigenic” model as a point of departure for polygenic psychiatric disorders. JPBS. 2017;2:S7. [Google Scholar]
- 63.Euesden J, Lewis CM, O’Reilly PF. PRSice: polygenic risk score software. Bioinformatics. 2015;31:1466–1468. doi: 10.1093/bioinformatics/btu848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wray NR, et al. Research review: polygenic methods and their application to psychiatric traits. J. Child Psychol. Psychiatry. 2014;55:1068–1087. doi: 10.1111/jcpp.12295. [DOI] [PubMed] [Google Scholar]
- 65.Martin J, et al. Association of genetic risk for schizophrenia with nonparticipation over time in a population-based cohort study. Am. J. Epidemiol. 2016;183:1149–1158. doi: 10.1093/aje/kww009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taylor A, et al. Exploring the association of genetic factors with participation in the Avon Longitudinal Study of Parents and Children. Int. J. Epidemiol. 2018;47:1207–1216. doi: 10.1093/ije/dyy060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Greven CU, Harlaar N, Dale PS, Plomin R. Genetic overlap between ADHD symptoms and reading is largely driven by inattentiveness rather than hyperactivity-impulsivity. J. Can. Acad. Child Adolesc. Psychiatry. 2011;20:6–14. [PMC free article] [PubMed] [Google Scholar]
- 68.St Pourcain B, et al. Developmental changes within the genetic architecture of social communication behaviour: a multivariate study of genetic variance in unrelated individuals. Biol. Psychiatry. 2017;83:598–606. doi: 10.1016/j.biopsych.2017.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.WORD. Wechsler Objective Reading Dimensions Manual. (Psychological Corporation, London, 1993) 146 p.
- 70.Nunes T, Bryant P, Olsson J. Learning morphological and phonological spelling rules: an intervention study. Sci. Stud. Read. 2003;7:289–307. doi: 10.1207/S1532799XSSR0703_6. [DOI] [Google Scholar]
- 71.Neale, M. D. Neale analysis of reading ability: second revised British Edition. (NFER-Nelson, London, 1997).
- 72.TOWRE. Test of word reading efficiency: examiner’s manual. PRO-ED (1999) 108 p.
- 73.Rosner J, Simon DP. The auditory analysis test: an initial report. J. Learn. Disabil. 1971;4:384–392. doi: 10.1177/002221947100400706. [DOI] [Google Scholar]
- 74.Rust, J., Wechsler, D. WOLD: Wechsler Objective Language Dimensions. (Psychological Corp., London, Artsberg Enterprises Ltd., Hong Kong, 1996).
- 75.Gathercole SE, Willis CS, Baddeley AD, Emslie H. The children’s test of nonword repetition: a test of phonological working memory. Mem. Hove Engl. 1994;2:103–127. doi: 10.1080/09658219408258940. [DOI] [PubMed] [Google Scholar]
- 76.Wechsler, D., Golombok, S., Rust, J. WISC-III U. K. Wechsler Intelligence Scale for Children–Third Edition UK Manual. (The Psychological Corporation, Sidcup, 1992).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.