Abstract
Current approaches to predicting a cardiovascular disease (CVD) event rely on conventional risk factors and cross-sectional data. In this study, we applied machine learning and deep learning models to 10-year CVD event prediction by using longitudinal electronic health record (EHR) and genetic data. Our study cohort included 109, 490 individuals. In the first experiment, we extracted aggregated and longitudinal features from EHR. We applied logistic regression, random forests, gradient boosting trees, convolutional neural networks (CNN) and recurrent neural networks with long short-term memory (LSTM) units. In the second experiment, we applied a late-fusion approach to incorporate genetic features. We compared the performance with approaches currently utilized in routine clinical practice – American College of Cardiology and the American Heart Association (ACC/AHA) Pooled Cohort Risk Equation. Our results indicated that incorporating longitudinal feature lead to better event prediction. Combining genetic features through a late-fusion approach can further improve CVD prediction, underscoring the importance of integrating relevant genetic data whenever available.
Introduction
Cardiovascular disease (CVD) is the leading cause of morbidity and mortality, accounting for one-third of all global deaths1,2. Several prediction models have been proposed, including the Framingham risk score3, American College of Cardiology/American Heart Association (ACC/AHA) Pooled Cohort Risk Equations4, and QRISK25. These models are typically built on a combination of cross-sectional risk factors such as hypertension, diabetes, cholesterol, and smoking status. Although the importance of conventional models cannot be ignored, well-known clinical risk factors for CVD explain only 50–75% of the variance in major adverse cardiovascular events6. About 15–20% of patients who experienced myocardial infarction (MI) had only one or two of these risk factors and were not identified as being at “risk” of CVD according to current prediction models7. Given the fact that CVD is preventable, and that its first manifestation may be fatal, a new strategy to enhance risk prediction beyond conventional factors is critical for public health.
Electronic health records (EHRs) contain a wealth of detailed clinical information and provide several distinct advantages for clinical research, including cost efficiency, big data scalability, and the ability to analyze data over time. With the wide implementation in the United States, accumulated EHR data has become an important resource for clinical studies8. Meanwhile, the recent convergence of two rapidly developing technologies—high-throughput genotyping and deep phenotyping within EHRs – presents an unprecedented opportunity to utilize routine healthcare data and genetic information to accelerate the healthcare. Many institutions and health care systems have been building EHR-linked DNA biobanks to enable such a vision. For example, the BioVU at Vanderbilt University Medical Center (VUMC), as of May 2018, has genotype data of over 50,000 individuals available for research.
Machine learning and deep learning approaches are particularly suited to exploiting such big data for individual outcome prediction, especially when EHRs can be linked to genetic data9,10. A recent study from the United Kingdom (UK) applied machine learning to conventional CVD risk factors on a large UK population and improved the prediction accuracy by 4.9%11. In this study, we examined: i) the performance of machine learning and deep learning on longitudinal EHR data for the prediction of 10-year CVD event, compared to a gold standard achieved by (ACC/AHA) Pooled Cohort Risk Equations, and ii) the benefits of incorporating extra genetic information.
Results
Machine learning and deep learning models with longitudinal EHR data (Experiment I)
This experiment involved EHR data of 109, 490 individuals (9,824 cases and 99, 666 controls, mean age [standard deviation, SD] 47.4 [14.7] years; 64.5% female and 86.3% European). We applied machine learning models using only ACC/AHA features, aggregate and longitudinal EHR features for CVD prediction, respectively. We compared the approaches with a baseline - ACC/AHA equation. We measured the area under a receiver operating characteristic curve (AUROC) and area under a precision recall curve (AUPRC).
The results were reported in Fig. 1 and Supplementary Table 1. Machine learning models all outperformed the baseline ACC/AHA equation. The ACC/AHA equation achieved an average AUROC of 0.732, while machine learning models using only ACC/AHA features obtained an AUROC of 0.738–0.751. By incorporating EHR features, the performance metrics were improved further. Machine learning models using aggregate EHR features achieved an AUROC of 0.765–0.782. By using temporal features, logistic regression (LR), gradient boosting trees (GBT) and deep learning models improved the AUROC to 0.781–0.790. Particularly, GBT and convolutional neural networks (CNN) achieved the highest AUROC of 0.790 (i.e. 7.9% improvement from baseline). For AUPRC, machine learning using temporal features remarkably improved the baseline (i.e., 0.246–0.285 vs. 0.186, a 32.8–44.1% improvement).
Figure 1.
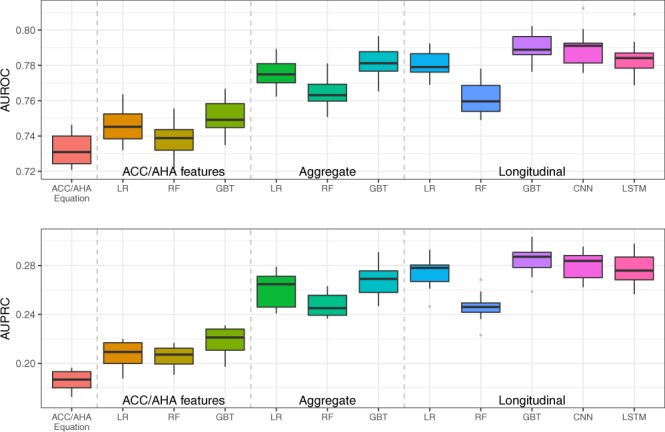
AUROC and AUPRC of gold standard and machine learning/deep learning models for predicting 10-year CVD risk on 10-fold cross validation in Experiment I. The mean values of the AUROC and AUPRC and the standard error are provided in Supplementary Table 1.
Feature analysis
We listed the top ten features for each optimized machine learning model (i.e. defined as the selected best model using grid-search on 10-fold cross-validation) in Table 1. For LR, the rank of features was determined by their coefficients (weights). For random forest trees (RF) and GBT, the features were ranked according to the impurity (information gain/entropy) decreasing from a feature. Because CNN and recurrent neural network with long short-term memory (LSTM) units are black box models, estimation of each feature’s contribution to predicting CVD risk remains difficult. We were not able to analyze the feature importance of the deep learning models in this study. We also used the recursive-backwards feature elimination on aggregate features with 5-fold cross-validation to select the top 10 features, shown in Supplementary Table 4.
Table 1.
Top 10 features for machine learning prediction in descending order of coefficients or feature importance returned by RF and GBT.
| LR with aggregate features | RF with aggregate features | GBT with aggregate features | LR with longitudinal features | RF with longitudinal features | GBT with longitudinal features |
|---|---|---|---|---|---|
| EHR length | EHR length | Age | EHR length | EHR length | Age |
| Max LDL-C | Age | EHR length | Age | Age | EHR length |
| Min Creatinine | Max BMI | SD Creatinine | SD Glucose in 2000 | Aspirin in 2006 | Smoking |
| Age | Min BMI | Smoking | SD Creatinine in 2000 | Max SBP in 2006 | Heart valve disorders in 2006 |
| Max HDL-C | Median BMI | Min BMI | Max HDL-C 2005 | Min BMI in 2006 | Hypertension in 2006 |
| Max BMI | Max SBP | Heart valve disorders (Phecode 395) | SD Glucose in 2006 | Median BMI in 2005 | Aspirin in 2006 |
| Max Total Cholesterol | Median SBP | Min Glucose | Median LDL-C in 2006 | Median SBP in 2006 | Disorders of lipoid metabolism in 2006 |
| Max DBP | SD BMI | Max SBP | Median BMI in 2006 | Max BMI in 2006 | Clopidogrel in 2006 |
| Median Triglycerides | MIN SBP | Max Triglycerides | Median Total Cholesterol in 2006 | Min BMI in 2001 | Max SBP in 2006 |
| Min Cholesterol | Max DBP | Aspirin | Heart valve disorders in 2006 | Min BMI in 2002 | SD Glucose in 2006 |
LDL-C (LDL Cholesterol); HDL-C (HDL Cholesterol); Systolic Blood Pressure (SBP); Diastolic Blood Pressure (DBP); Body mass index (BMI).
The top features in all machine learning models include some conventional risk factors such as age, blood pressure (BP), and total cholesterol, as well as several new features not included in ACC/AHA features such as body mass index (BMI), creatinine, glucose, and antiplatelet therapy (e.g. Aspirin, and Clopidogrel). Moreover, the maximum, minimum, and SD for laboratory values (e.g. fasting lipid values) and physical measurements (e.g. BMI and BP) contribute more than median values to the models. GBT preferred historically-entered diagnostic codes such as heart valve disorders, lipid disorders, and hypertension) over other features.
For machine learning models with longitudinal features, LR selected laboratory values in the years 2000 and 2006 (e.g. SD Glucose from the observation window). RF chose BMI in multiple years. GBT prioritized the medical conditions obtained from the most recent year before the prediction window (the year 2006).
Evaluation benefits of additional genetic features (Experiment II)
In this second experiment, we developed a two-stage late-fusion approach to combine genetic and longitudinal EHR features for machine learning models. This is possible at large academic centers where genome-wide data are rapidly moving into EHRs. We compared the approach with ACC/AHA equations, as well as with machine learning models using ACC/AHA features and longitudinal EHR features. The study was conducted using a genotyped cohort of 10,162 individuals (2,452 cases and 7,710 controls with both genotyped data and EHRs from 2000–2016, Supplementary Table 5).
The results are shown in Fig. 2 and Supplementary Table 2. GBT using ACC/AHA features generated similar results with ACC/AHA equations. Conversely, GBT using longitudinal EHR features outperformed ACC/AHA equations (AUROC of 0.71 vs. 0.698, AUPRC of 0.427 vs. 0.396). Our innovative late-fusion approach combining additional genetic features then further significantly improved the AUROC and AUPRC by 2.1% and 9.1%.
Figure 2.
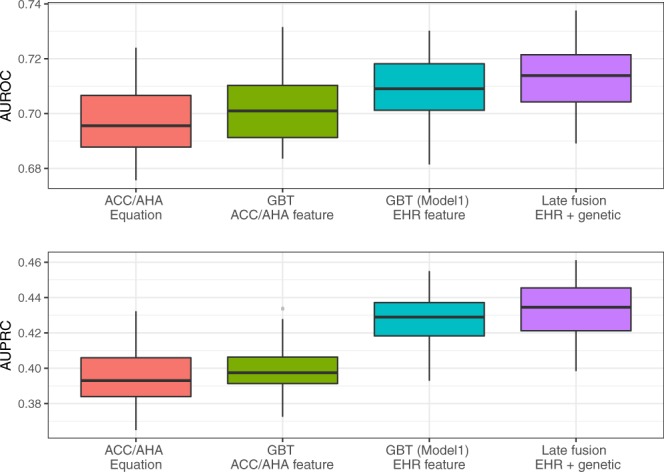
AUROC and AUPRC of gold standard, GBT model on EHR feature and late fusion on EHR and genetic feature for predicting 10-year CVD risk on 50 iterations in Experiment II. The mean values of the AUROC and AUPRC and the standard error are provided in Supplementary Table 2.
We listed the top ten features in the pre-trained model with genetic data in Table 2. A single nucleotide polymorphisms (SNP) (rs17465637) was ranked as the second most important feature after age. Many loci that emerged as informative during this approach (MIA3, CXCL12, and LPA) have previously been identified as predictors of early coronary artery disease12. We have previously shown that LPA is a strong independent predictor of CVD events on lipid lowering therapy13. Chemokines (CXCL12) and check point genes (CDKN2A) flagged by this approach may alter CVD through their role in angiogenesis. Other loci (GGCX) are known to impact response to anticoagulation14.
Table 2.
Top 10 features in pre-trained model 2 with genetic data. Features were ranked according to descending order of absolute value of coefficient effect size.
| Features | Reference gene | Coefficient |
|---|---|---|
| Age | — | 0.747 |
| rs17465637 | MIA3 | −0.334 |
| rs67180937 | MIA3 | 0.301 |
| Gender | — | −0.270 |
| EHR length | — | 0.180 |
| rs7568458 | GGCX | 0.103 |
| rs4977574 | CDKN2A | 0.095 |
| rs10455872 | LPA | 0.093 |
| rs1412444 | LPA | 0.092 |
| rs501120 | CXCL12 | −0.079 |
We chose the result from the iteration which generated the closest result from the average AUROC and AUPRC.
Discussion
This study leveraged a large EHR dataset and EHR-linked biobank in the U.S. to assess the added value of longitudinal data without and with extant genotype. The machine learning models outperformed a traditional clinically-used predictive model for CVD risk prediction (i.e. ACC/AHA equation). Consistent with previous reports11,15, we initially observed a benchmark predictability for the ACC/AHA equation of AUROC 0.732 (benchmark for AUPRC was 0.186). When we applied machine learning models using the same ACC/AHA features, they demonstrated better performance (improved AUROC of 0.738–0.751). Because we were interested in whether machine learning models incorporating longitudinal EHR features and genetic features could enhance the prediction, we designed two experiments: i) modeling EHR features with aggregated and temporal features for machine learning and deep learning models, and ii) developing a two-stage late-fusion model to incorporate these longitudinal EHR data with genetic features. The results showed that machine learning models incorporating longitudinal EHR features such as BMI, lab values, and medication history significantly outperformed the baseline approach (improved AUROC 0.761–0.790). Even using a strictly metric AUPRC, the improvement is still significant. We also observed that the late-fusion approach with incorporating genetic data can improve the prediction performance (AUROC of 0.713 vs. 0.698, AUPRC of 0.432 vs. 0.396).
The top features selected in our machine learning models include several conventional risk factors such as age, blood pressure, and total cholesterol. BMI, creatinine, and glucose values (not used in the ACC/AHA equations) were found to be important features, which confirmed previous reports that these features may be independent risk factors for CVD16–18. Moreover, the maximum, minimum, and SD of laboratory values (e.g. lipids) and physical measurements (e.g. BMI and BP) provide more discriminating abilities than median values19. This finding, that temporal instability in body weight and hemodynamics may be a stronger predictor of risk than cross-sectional estimates of the same parameters, is clinically important.
Longitudinal data more accurately reflect the fluctuation of physiological factors over time. Recently, the STABILITY trial suggested the higher visit-to-visit variation in both systolic and diastolic blood pressure is a strong predictor of CVD20. When we narrow our observation window to a one-year slice in time, we captured the longitudinal EHR features year by year. Although the most recent value of some features was often preferred, our machine learning models also have antecedent laboratory tests (e.g. glucose and creatinine in 2000) in their top features when applied to LR. Incorporating these features enhanced the overall performance.
Overall, the study evaluated LR, RF, and GBT using temporal features via two novel deep learning models– CNN and LSTM. CNN is particularly suited to learning local patterns in raw features from input like images, where the intensity of pixels can be combined into higher level features. By comparison, LSTM is designed to learn long/short term dependency of data in a sequence. Both CNN and LSTM outperformed LR and RF but had no measurable advantage over GBT. The reason is that GBT can balance bias and variance to yield better generalization by using a boosting strategy. Although we built the multivariate temporal matrix for each patient, like an 1D image, reduced sequential dependency in the data may not fulfill the advantage of CNN and LSTM. A future study is required to compare GBT, CNN and LSTM on a dataset of more detailed clinical events in a consequent manner.
Lastly, our study also underscores the importance of including genetic variants. CVD has a sizeable hereditary component3, and many contributing loci are now being validated through functional studies in vitro; the result is a deeper understanding of the biology underlying CVD21–24. To date, polygenic risk scoring, a method for summarizing genetic effects for diseases, are being incorporated into clinical practice25. However, how to combine genetic variants with other biological and lifestyle factors remain a challenge26. We performed a two-stage late-fusion approach and evaluated the predictive power of 204 SNPs with longitudinal EHR data in CVD prediction. Through access to BioVU at the Vanderbilt, the largest single-site biobank in the U.S., we have identified 10,162 individuals with both EHR data and selected SNPs. To enlarge the power of these data, we took advantage of another 34, 926 subjects genotyped cohort in BioVU with selected SNPs available. Our late-fusion approach trained multiple classifiers separately on longitudinal EHR and genetic data, fusing the prediction results across classifiers. Our results demonstrated that genetic features offer benefits to clinical features, resulting in an improved AUROC (i.e. 2.1%) and AUPRC (i.e. 9.1%).
For the model trained with only genetic and demographic features, age remains the strongest predictor for CVD (coefficient 0.747), followed two variants from the MIA3 gene, gender length of the EHR. This locus has previously been identified as a predictor of early coronary artery disease12. Our approach also underscored the importance of the LPA gene, a known predictor of CVD events on lipid lowering therapy13. Interestingly, in that prior work we reported that variability in the LPA gene predicted CVD events independent of circulating lipid levels. Our current observation highlights the importance of this locus in quantifying residual CVD risk even after models have considered lipids.
We acknowledge the limitations that, (1) this study was restricted to data obtained during routine clinical practice, (2) we only used 204 SNPs in our genetic experiment, and (3) that some of the effects of the SNPs may also be modeled directly by phenotypes. Yet, paradoxically, some SNPs for endophenotypes are more predictive of CVD events than the endophenotype itself13. We believe that, with even denser phenotypic and genetic information available in growing EHR cohorts, prediction would continue to improve. This study confirmed that combining phenotypic and genetic information with robust computational models can improve disease prediction.
Methods
Study setting
We conducted the study using data derived from Synthetic Derivative, a de-identified copy of whole EHRs at VUMC. Synthetic Derivative maintains rich and longitudinal EHR data from over 3 million unique individuals, including demographic details, physical measurements, history of diagnosis, prescription drugs, and laboratory test results. As of May 2018, over 50,000 of these individuals have genotype data available.
We focused our analysis on individuals with European or African ancestry. To ensure each individual to have some EHR data, we required an individual to meet the definitions of medical home27. We set the baseline date as 01/01/2007 to allow all individuals within the cohort to be followed-up for 10 years. For each individual, we split the EHR into: (i) the observation window (01/01/2000 to 12/31/2006; 7 years) and, (ii) the prediction window (01/01/2007 to 12/31/2016; 10 years). We extracted EHR data in the 7-year observation window (2000–2006) to train a classifier to classify whether the individual would have CVD event in the 10-year prediction window 2007–2016.
We define cases as individuals with ≥1 CVD diagnosis codes (the International Classification of Diseases, Ninth Revision, Clinical Modification [ICD-9-CM]: 411. * and 433. *) within the 10-year prediction window. Controls were individuals without any ICD-9-CM code 411. * or 433. * during the 10-year prediction window.
Study cohort
The study cohort included patients between the ages of 18 to 78 on 01/01/2000 (beginning of the observation window). Individuals with any CVD diagnosis (ICD-9-CM 411. * or 433. *) prior to the baseline date (i.e. 01/01/2007) were excluded. To reduce chart fragmentation and optimize the density of the longitudinal EHR data, we required each individual to have >=1 visit and >=1 blood pressure measurement during the observation window28,29. We excluded inpatient physical or laboratory measures for all individuals.
In total, we identified 109, 490 individuals, including 9,824 cases and 99, 666 controls (mean [SD] age 47.4 [14.7] years; 64.5% female and 86.3% European). The case/control ratio was consistent with a previous report from a large EHR cohort11. Among these 109, 490 individuals, 10,162 individuals (2,452 cases and 7,710 controls) had genotype data.
Data preprocessing and feature extraction
Phenotypic data: we extracted features including demographics, variables in the ACC/AHA equations (e.g. blood pressure measurements), physical measurements (e.g. BMI), and laboratory tests including glucose, triglyceride levels, and creatinine level (as a marker of renal function); such laboratory features have previously been reported relevant to CVD11. In addition, we applied chi-square (chi2)30, a common feature selection methods to select independent features on EHR data, and identified 40 relevant diagnostic codes and medication codes (Table 3).
Table 3.
Features included in the machine-learning models.
| Feature type | Features | Values |
|---|---|---|
| Demographic | Age* | Continuous |
| Gender* | Binary | |
| Race | Categorical | |
| Life styles | Body mass index (BMI) | Summarized data† |
| Smoking* | Binary | |
| Physical or lab measurements | Systolic blood pressure (SBP)* | Summarized data† |
| Diastolic blood pressure (DBP)* | Summarized data† | |
| Total Cholesterol (Cholesterol)* | Summarized data† | |
| HDL Cholesterol (HDL-C)* | Summarized data† | |
| LDL Cholesterol (LDL-C) | Summarized data† | |
| Creatinine | Summarized data† | |
| Glucose | Summarized data† | |
| Triglyceride | Summarized data† | |
| Diagnosis | Other tests (phecode33 1010) | Binary |
| Benign neoplasm of skin (216) | ||
| Diabetes mellitus* (250) | ||
| Disorders of lipoid metabolism (272) | ||
| Other mental disorder, random mental disorder (306) | ||
| Heart valve disorders (395) | ||
| Hypertension (401) | ||
| Cardiomyopathy (425) | ||
| Congestive heart failure; nonhypertensive (428) | ||
| Atherosclerosis (440) | ||
| Acute upper respiratory infections of multiple or unspecified sites (465) | ||
| Chronic airway obstruction (496) | ||
| Disorders of menstruation and other abnormal bleeding from female genital tract (626) | ||
| Medication | Warfarin (RXCUI 11289) | Binary |
| Aspirin (1191) | ||
| Atenolol (1202) | ||
| Amlodipine (17767) | ||
| Carvedilol (20352) | ||
| Lisinopril(29046) | ||
| Adenosine(296) | ||
| Clopidogrel (32968) | ||
| Digoxin (3407) | ||
| Diltiazem (3443) | ||
| Ramipril (35296) | ||
| Diuretics (3567) | ||
| Dobutamine (3616) | ||
| Simvastatin(36567) | ||
| Enalapril (3827) | ||
| Sestamibi (408081) | ||
| Ethinyl Estradiol (4124) | ||
| Furosemide (4603) | ||
| Nitroglycerin (4917) | ||
| Hydrochlorothiazide(5487) | ||
| Ibuprofen (5640) | ||
| Metoprolol (6918) | ||
| Acellular pertussis vaccine (798302) | ||
| Atorvastatin(83367) | ||
| ACE inhibitors (836) | ||
| Thallium(1311633) | ||
| Clonidine (2599) | ||
| Genetic | 204 SNPs# | Categorical |
| Others | EHR length | Continuous |
*Features in ACC/AHA Equations.
†Summarized data includes minimum, maximum, median and SD within a time window.
#204 SNPs are listed in the Supplementary Data.
We represented a physical measurement or laboratory feature with summarized data, e.g. minimum, maximum, median, and SD. We removed the outliers (>5 SD from the mean) to avoid unintended incorrect measurements (e.g. using lb. instead of kg. for body weight)31. If an individual had no such measure within the EHR, we imputed the missing value with the median value of the group with the same age and gender32. We also added a dummy variable for each measure to indicate whether the test value was imputed.
For disease phenotypes, we followed a standard approach and grouped relevant ICD-9-CM codes into distinct phecodes33. For medications, we collapsed brand names and generic names into groups by their composition (ingredients) and represented the groups using the RxNorm34 concepts (RxCUIs) for this variable. For example, ‘Tylenol Caplet, 325 mg oral tablet’ and ‘Tylenol Caplet, 500 mg oral tablet’ were both mapped to ‘Acetaminophen’ (RxCUI 161). We used a binary value to indicate whether or not an individual had each diagnosis or prescription.
For genetic data, we selected 248 SNPs reported to be associated with CVD in two large meta-analyses23,24. Among these SNPs, genotype data were available for 204 SNPs in our cohort and were included as features. Each SNP had a value 0, 1, or 2 to represent the count of minor alleles for an individual. Table 3 shows the features used in the machine learning models.
Experiment
Baseline
We chose the ACC/AHA equation for 10-year CVD risk as our baseline. For physical measurements or laboratory features (i.e. SBP/DBP and HDL-c level), we used the most recent values prior to the split date, 01/01/2007.
Machine learning and deep learning with longitudinal EHR data (Experiment I)
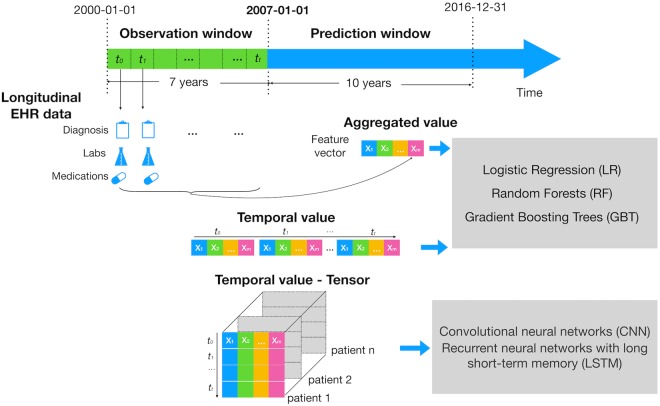
In this experiment, we used two different ways to model EHR data– extracting aggregate and longitudinal EHR features–for machine learning models (Fig. 3). We compared their performance of 10-year CVD prediction with baseline.
Figure 3.
Study design of experiment I. The figure illustrates how we defined the observation and prediction window. It also shows how we modeled longitudinal EHR features: i) We aggregated each feature across the 7-year observation window (e.g. median, max, min and SD of HDL); ii) we extracted each year value of each feature and concatenated the temporal values from all patients into a two-dimensional matrix for a classifier (e.g. LR, RF, GBT); we then constructed a tensor representation on temporal values from all patients for CNN and LSTM.
Aggregate features
We aggregated features across the 7-year observation window (e.g. median, max, min and SD of HDL from 01/01/2000 to 12/31/2006).
Longitudinal features (Multivariate temporal features)
We exploited the temporal information in the longitudinal EHR data by dividing the whole observation window into one-year slice window. Specifically, for physical or laboratory features, we extracted the median, max, min and SD values within one-year slice window. We replaced the missing physical or laboratory measures with the individual’s measurement on the closest date, e.g. using the HDL cholesterol result on 12/20/2005 instead if the individual had no HDL test in 2006. For diagnosis and medication features, we used a binary value to indicate whether or not an individual had each diagnosis or prescription in one-year slice window.
Machine learning and deep learning models
Three machine learning models, LR, RF and GBT were used in both aggregate and longitudinal features. Two deep learning models, CNN35 and LSTM36 were applied to the longitudinal features.
Implementation detail
We used CNN and LSTM on longitudinal features and concatenated an auxiliary input of demographic features to feed into a multilayer perceptron (MLP) with two hidden layers. More details can be found in Supplementary Table 3. LR, RF, and GBT were implemented with Python Scikit-Learn 0.19.1 (http://scikit-learn.org/stable/)37. The CNN and LSTM models were implemented with Keras 2.1.3 (https://keras.io/) using Tensorflow1.6.1 as the backend. The backward recursive feature elimination was implemented with mlxtend 0.13 (https://rasbt.github.io/mlxtend/).
Evaluation
We randomly divided the dataset into a training and a test set with a 90/10 split. We first trained models (LR, RF, GBT) using a grid search with 10-fold stratified cross-validation on the training set to select the best model with the maximum AUROC. Then we tested the selected model on test set. We repeated the above process ten times. For deep learning models, we randomly divided the data into training, validation, and testing sets with a ratio of 8:1:1 and iterated the process ten times. For each iteration, we calculated AUROC and AUPRC38 values after applying the model on the test set. We reported the average and SD of both values. To see whether there was a significance difference in the performance, we performed a paired t-test with the level of confidence 0.05.
Evaluation benefits of additional genetic features (Experiment II)
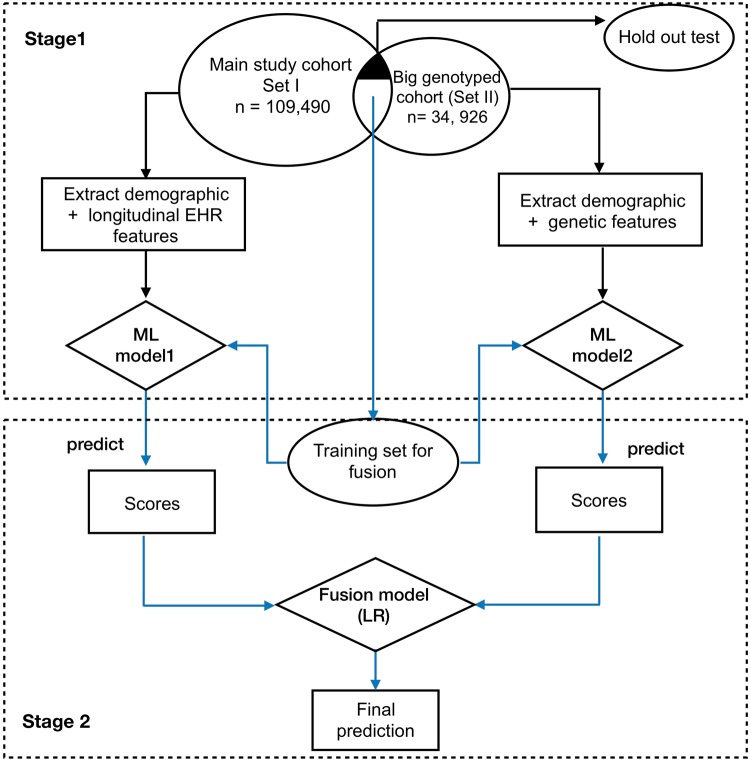
In this experiment, we examined combining genetic features with demographic and longitudinal EHR data for 10- year CVD prediction. We developed a two-stage late-fusion approach to incorporate EHR and genotyped features. Late-fusion is an effective approach to enhance prediction accuracy by combining the prediction results of multiple models trained separately by a group of features39. We trained two sperate models on EHR data and genotyped data, respectively. Then the approach fuses the prediction results by taking the prediction scores as input features and training a fusion model (e.g. LR) for final prediction.
To enlarge the data power, besides the main study cohort (set I, n = 109, 490) used in experiment I, the study utilized another big genotyped cohort (set II) including 34,926 individuals without restricting >1 record of SBP in the observation window (Supplementary Table 5). We used the same criteria (i.e. ≥1 CVD diagnosis codes in the prediction window) to label each individual in the genotyped cohort as a case or control. The set II had 204 SNPs features and basic demographic features (e.g. age, gender, and race). There is an overlap of 10,162 individuals between the two sets (denoted as intersect set), i.e. these individuals have both EHR and genotyped features (Fig. 4). We randomly split the intersect set into a training set (i.e. 8,129 individuals) used for training the fusion model and a holdout test set (i.e. 2, 033 individuals) with an 80/20 split.
Figure 4.
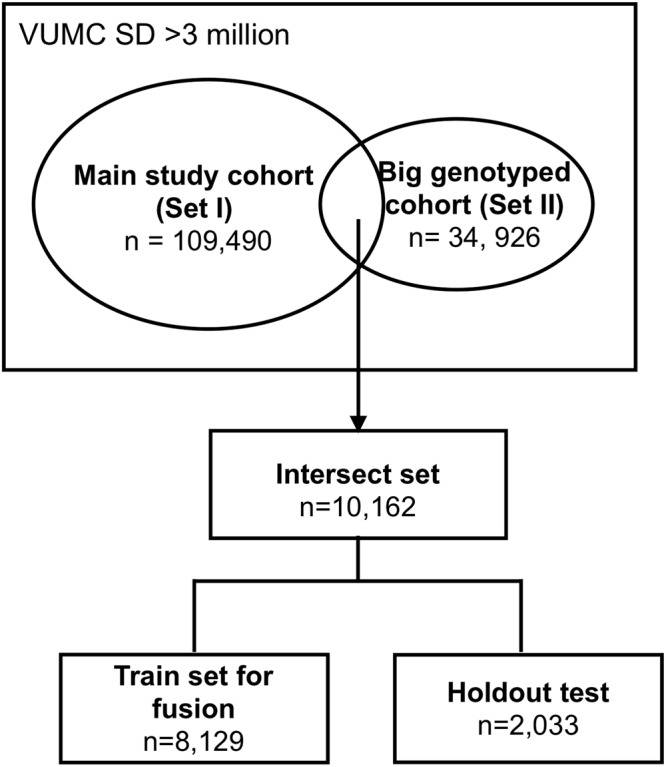
Flowchart of selecting cohort for late-fusion approach.
The workflow of the framework is presented in Fig. 5. In the first stage of the framework, we firstly trained a classifier (model1) with longitudinal EHR features on the set I (with holdout test set removed), and trained a another classifier (model 2) using 204 SNPs features on the set II (with holdout test set removed). In the second stage, we applied the model 1 and 2 to the training set from intersect cohort to get prediction scores, which then used as features in fusion model for final prediction.
Figure 5.
Framework for proposed late fusion approach to combine the genetic features with longitudinal EHR features.
We used GBT for the model 1 as GBT has good generalizability as an ensemble approach. We used the LR for model 2 and fusion model. To compare the performance, we tested model 1 and fusion model on the holdout test set (2,033 individuals). We performed 5-fold cross-validation and repeated the process ten times. We reported the mean and SD of AUROC and AUPRC.
Electronic supplementary material
Acknowledgements
This study was supported by GM120523, GM109145, HL133786, 5T32GM080178-09, T15 LM007450, T32 GM007347, K23AR064768, Rheumatology Research Foundation (K-supplement), American Heart Association (18AMTG34280063, 16SDG27490014 and 15MCPRP25620006), HG008672, R01 LM010685, GM115305 and Vanderbilt Faculty Research Scholar Fund. The dataset used for the analyses described were obtained from Vanderbilt University Medical Center’s resources, BioVU and the Synthetic Derivative, which are supported by institutional funding and by the Vanderbilt National Center for Advancing Translational Science grant 2UL1 TR000445-06 from NCATS/NIH. Existing genotypes in BioVU were funded by NIH grants RC2GM092618 from NIGMS/OD and U01HG004603 from NHGRI/NIGMS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
J.Z. and W.Q.W. initiated the idea and conducted the research. J.Z. performed the data processing, implemented the algorithms and results analyses. F.Q. initiated the genetic related idea and collected genetic data. P.W. helped investigate the research and analyze results. J.Z., W.Q.W. and F.Q. wrote the manuscript in consultation with R.A.W., Q.S.W. and R.A.L. W.Q.W. and J.C.D. supervised and supported the research. All authors edited and reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-36745-x.
References
- 1.WHO | The top 10 causes of death. WHO (2018). Available at: http://www.who.int/en/news-room/fact-sheets/detail/the-top-10-causes-of-death.
- 2.Benjamin EJ, et al. Heart Disease and Stroke Statistics—2017 Update: A Report From the American Heart Association. Circulation. 2017;135:e146–e603. doi: 10.1161/CIR.0000000000000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D’Agostino RB, et al. General Cardiovascular Risk Profile for Use in Primary Care: The Framingham Heart Study. Circulation. 2008;117:743–753. doi: 10.1161/CIRCULATIONAHA.107.699579. [DOI] [PubMed] [Google Scholar]
- 4.Goff DC, et al. 2013 ACC/AHA Guideline on the Assessment of Cardiovascular Risk: A Report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation. 2013;63:2935–2959. doi: 10.1016/j.jacc.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hippisley-Cox J, et al. Predicting cardiovascular risk in England and Wales: prospective derivation and validation of QRISK2. BMJ. 2008;336:1475–1482. doi: 10.1136/bmj.39609.449676.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kannel WB, Vasan RS. Adverse consequences of the 50% misconception. Am J Cardiol. 2009;103:426–7. doi: 10.1016/j.amjcard.2008.09.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khot UN. Prevalence of Conventional Risk Factors in Patients With Coronary Heart Disease. JAMA. 2003;290:898. doi: 10.1001/jama.290.7.898. [DOI] [PubMed] [Google Scholar]
- 8.Wei, W. -Q. & Denny, J. C. Extracting research-quality phenotypes from electronic health records to support precision medicine. Genome Med7 (2015). [DOI] [PMC free article] [PubMed]
- 9.Choi E, Schuetz A, Stewart WF, Sun J. Using recurrent neural network models for early detection of heart failure onset. Journal of the American Medical Informatics Association. 2017;24:361–370. doi: 10.1093/jamia/ocw112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh A, et al. Incorporating temporal EHR data in predictive models for risk stratification of renal function deterioration. Journal of Biomedical Informatics. 2015;53:220–228. doi: 10.1016/j.jbi.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weng SF, Reps J, Kai J, Garibaldi JM, Qureshi N. Can machine-learning improve cardiovascular risk prediction using routine clinical data? PLOS ONE. 2017;12:1–14. doi: 10.1371/journal.pone.0174944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ansari, W. M. et al. Effect of Coronary Artery Disease risk SNPs on serum cytokine levels and cytokine imbalance in Premature Coronary Artery Disease. Cytokine10.1016/j.cyto.2017.05.013 (2017). [DOI] [PubMed]
- 13.Wei, W. -Q. et al. LPA Variants are Associated with Residual Cardiovascular Risk in Patients Receiving Statins. Circulation, 10.1161/CIRCULATIONAHA.117.031356 (2018). [DOI] [PMC free article] [PubMed]
- 14.Tang X-Y, et al. The association between GGCX, miR-133 genetic polymorphisms and warfarin stable dosage in Han Chinese patients with mechanical heart valve replacement. J Clin Pharm Ther. 2017;42:438–445. doi: 10.1111/jcpt.12527. [DOI] [PubMed] [Google Scholar]
- 15.Tillin T, et al. Ethnicity and prediction of cardiovascular disease: performance of QRISK2 and Framingham scores in a U.K. tri-ethnic prospective cohort study (SABRE–Southall And Brent REvisited) Heart. 2014;100:60–67. doi: 10.1136/heartjnl-2013-304474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan SS, et al. Association of Body Mass Index With Lifetime Risk of Cardiovascular Disease and Compression of Morbidity. JAMA Cardiol. 2018;3:280–287. doi: 10.1001/jamacardio.2018.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wannamethee, S. G., Shaper, A. G. & Perry, I. J. Serum Creatinine Concentration and Risk of Cardiovascular Disease. Stroke (1997). [DOI] [PubMed]
- 18.Reusch JEB, Wang CCL. Cardiovascular Disease in Diabetes: Where Does Glucose Fit In? J Clin Endocrinol Metab. 2011;96:2367–2376. doi: 10.1210/jc.2010-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan UI, Rieder J, Cohen HW, Coupey SM, Wildman RP. Effect of modest changes in BMI on cardiovascular disease risk markers in severely obese, minority adolescents. Obes Res Clin Pract. 2010;4:e163–246. doi: 10.1016/j.orcp.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 20.Vidal-Petiot E, et al. Visit-to-visit variability of blood pressure and cardiovascular outcomes in patients with stable coronary heart disease. Insights from the STABILITY trial. Eur. Heart J. 2017;38:2813–2822. doi: 10.1093/eurheartj/ehx250. [DOI] [PubMed] [Google Scholar]
- 21.Gaulton KJ, et al. Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat. Genet. 2015;47:1415–1425. doi: 10.1038/ng.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCarthy MI. Genomics, Type 2 Diabetes, and Obesity. New England Journal of Medicine. 2010;363:2339–2350. doi: 10.1056/NEJMra0906948. [DOI] [PubMed] [Google Scholar]
- 23.Paquette M, et al. Polygenic risk score predicts prevalence of cardiovascular disease in patients with familial hypercholesterolemia. Journal of Clinical Lipidology. 2017;11:725–732.e5. doi: 10.1016/j.jacl.2017.03.019. [DOI] [PubMed] [Google Scholar]
- 24.Khera AV, et al. Genetic Risk, Adherence to a Healthy Lifestyle, and Coronary Disease. New England Journal of Medicine. 2016;375:2349–2358. doi: 10.1056/NEJMoa1605086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knowles JW, Ashley EA. Cardiovascular disease: The rise of the genetic risk score. PLOS Medicine. 2018;15:e1002546. doi: 10.1371/journal.pmed.1002546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Müller B, et al. Improved prediction of complex diseases by common genetic markers: state of the art and further perspectives. Hum Genet. 2016;135:259–272. doi: 10.1007/s00439-016-1636-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schildcrout JS, et al. Optimizing drug outcomes through pharmacogenetics: A case for preemptive genotyping. Clin Pharmacol Ther. 2012;92:235–242. doi: 10.1038/clpt.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei W-Q, et al. Impact of data fragmentation across healthcare centers on the accuracy of a high-throughput clinical phenotyping algorithm for specifying subjects with type 2 diabetes mellitus. Journal of the American Medical Informatics Association. 2012;19:219–224. doi: 10.1136/amiajnl-2011-000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei W-Q, Leibson CL, Ransom JE, Kho AN, Chute CG. The absence of longitudinal data limits the accuracy of high-throughput clinical phenotyping for identifying type 2 diabetes mellitus subjects. Int J Med Inform. 2013;82:239–247. doi: 10.1016/j.ijmedinf.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, H. & Setiono, R. Chi2: feature selection and discretization of numeric attributes. in Proceedings of 7th IEEE International Conference on Tools with Artificial Intelligence 388–391, 10.1109/TAI.1995.479783 (1995).
- 31.Yackel TR, Embi PJ. Unintended errors with EHR-based result management: a case series. J Am Med Inform Assoc. 2010;17:104–107. doi: 10.1197/jamia.M3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Batista GEAPA, Monard MC. An analysis of four missing data treatment methods for supervised learning. Applied Artificial Intelligence. 2003;17:519–533. doi: 10.1080/713827181. [DOI] [Google Scholar]
- 33.Wei W-Q, et al. Evaluating phecodes, clinical classification software, and ICD-9-CM codes for phenome-wide association studies in the electronic health record. PLOS ONE. 2017;12:1–16. doi: 10.1371/journal.pone.0175508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Normalized names for clinical drugs: RxNorm at 6 years. Available at, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3128404/ (Accessed: 18th May 2018) [DOI] [PMC free article] [PubMed]
- 35.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 36.Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput. 1997;9:1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 37.Pedregosa F, et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 38.Saito T, Rehmsmeier M. The Precision-Recall Plot Is More Informative than the ROC Plot When Evaluating Binary Classifiers on Imbalanced Datasets. PLOS ONE. 2015;10:e0118432. doi: 10.1371/journal.pone.0118432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lai KT, Liu D, Chang SF, Chen MS. Learning Sample Specific Weights for Late Fusion. IEEE Transactions on Image Processing. 2015;24:2772–2783. doi: 10.1109/TIP.2015.2423560. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.